Abstract
The Obg protein family belongs to the TRAFAC (translation factor) class of P-loop GTPases and is conserved from bacteria to eukaryotes. Essential roles in many different cellular processes have been suggested for the Obg protein from Escherichia coli (ObgE), and we recently showed that it is a central regulator of bacterial persistence. Here, we report the first crystal structure of ObgE at 1.85-Å resolution in the GDP-bound state, showing the characteristic N-terminal domain and a central G domain that are common to all Obg proteins. ObgE also contains an intrinsically disordered C-terminal domain, and we show here that this domain specifically contributed to GTP binding, whereas it did not influence GDP binding or GTP hydrolysis. Biophysical analysis, using small angle X-ray scattering and multi-angle light scattering experiments, revealed that ObgE is a monomer in solution, regardless of the bound nucleotide. In contrast to recent suggestions, our biochemical analyses further indicate that ObgE is neither activated by K+ ions nor by homodimerization. However, the ObgE GTPase activity was stimulated upon binding to the ribosome, confirming the ribosome-dependent GTPase activity of the Obg family. Combined, our data represent an important step toward further unraveling the detailed molecular mechanism of ObgE, which might pave the way to further studies into how this GTPase regulates bacterial physiology, including persistence.
Keywords: biophysics, crystal structure, enzyme kinetics, enzyme structure, GTPase, X-ray crystallography, X-ray scattering
Introduction
Guanine nucleotide-binding proteins (GNBPs, G proteins, or GTPases) are ubiquitously found in all living organisms where they are known to play essential roles in a myriad of cellular processes, including protein synthesis and translocation, membrane trafficking, development, signal transduction, and cell cycle control (1–3). G proteins share a common structural module, the G domain that acts as a molecular switch by cycling between a GDP-bound “off” state and a GTP-bound “on” state. Both states mainly differ in the conformation of two regions (Switch I and II), allowing reversible interaction and activation of downstream effector proteins (4, 5).
In the intensively studied small GTPases of the Ras family, nucleotide exchange is very slow and exchange of GDP for GTP is stimulated by guanine nucleotide exchange factors (6, 7). Moreover, the intrinsically slow GTP hydrolysis is accelerated through the interaction with GTPase activating proteins (GAPs)3 (6, 8). The latter proteins stabilize and/or complement the active site of the GTPase by providing one or two catalytic residues in trans that counter negative charge development at the phosphate groups of GTP or orient the nucleophilic water molecule during hydrolysis (4, 6, 8). More recently a number of other activation mechanisms of G proteins have been described. One class of G proteins is activated through homo- or heterodimerization of their G domains, hence reciprocally activating each other (9–11). This class of G proteins is known as the G proteins activated by nucleotide-dependent dimerization (12). Another expanding family of G proteins recruits specific cations (such as K+) to stabilize the GTPase transition state (13–16). Some G proteins use a combination of the two above strategies, as is the case for MnmE, a G protein involved in tRNA modification (17).
The Obg family belongs to the TRAFAC (for translation factor) class of P-loop GTPases and consists of high molecular weight proteins like Obg/CgtA, YchF/YyaF, Drg/Rbg, Nog1, and Ygr210 (3). Among them, Obg is a highly versatile GTPase that is conserved in bacteria, as well as in many eukaryotes (18). Obg displays a three-domain arrangement with an N-terminal glycine-rich domain typical for Obg (Obg domain), a central G domain and a C-terminal domain that is highly variable in length and sequence among Obg proteins from different species (19, 20). Obg homologs are widely present in eukaryotic organelles, including chloroplasts and mitochondria (21–23). However, in contrast to bacterial Obg, the rice homolog OsYchF and the human homolog hOLA1 (human Obg-like ATPase 1) were found to bind and hydrolyze ATP more efficiently than GTP (24, 25). Obg displays a very low GTP hydrolysis rate in combination with a high guanine nucleotide exchange rate, seemingly abrogating the need for guanine nucleotide exchange factors (3, 26). No “classical” Obg-specific GAP proteins have so far been described, although the 50S ribosomal subunit has recently been found to increase the GTPase activity of Obg (27). Furthermore, it has been proposed, based on sequence analysis, that Obg might belong to the class of G proteins that is specifically activated by K+ ions (15).
Obg was originally discovered in Bacillus subtilis, where it was shown to have a critical role in sporulation (28). Moreover, Obg expression was found to be essential for viability in nearly all bacterial species (3, 26), and various functions including ribosome assembly and maturation, cell cycle control, DNA replication, stress response, sporulation and morphological development have been proposed (29–34). The role of Obg in the cellular response to environmental stress is particularly interesting (27, 33, 35–37). Recently, we showed that, in Escherichia coli as well as in Pseudomonas aeruginosa, Obg plays a central role in the regulation of bacterial persistence in response to nutrient starvation (38). Although the underlying mechanisms need further elucidation, Obg-mediated persistence depends on the presence of the alarmone (p)ppGpp and proceeds through induction of the expression of the HokB toxin.
Here, we report the first crystal structure of Obg from E. coli (ObgE) bound to a GDP molecule at 1.85-Å resolution. SAXS and MALS experiments reveal that ObgE behaves as a monomer in solution. A detailed biochemical analysis confirms that ObgE has a modest affinity for nucleotides and ppGpp and fast guanine nucleotide dissociation rates, combined with a low GTP hydrolysis rate. Interestingly, we show that the C-terminal intrinsically disordered domain plays a role in GTP binding, whereas deletion of this region has a negligible effect on GDP and ppGpp binding and GTP turnover. Finally, kinetic experiments show that ObgE is neither activated by K+ ions, nor by homodimerization, in contrast to what was previously suggested (15, 20). However, we do observe a weak but significant stimulation of the ObgE GTPase activity by the (70S) ribosome. Together, these data provide new insights into the structure and function of ObgE and will be instrumental to drive further studies aiming to unravel the role of Obg in bacterial persistence.
Results and discussion
Overall structure of GDP-bound ObgE
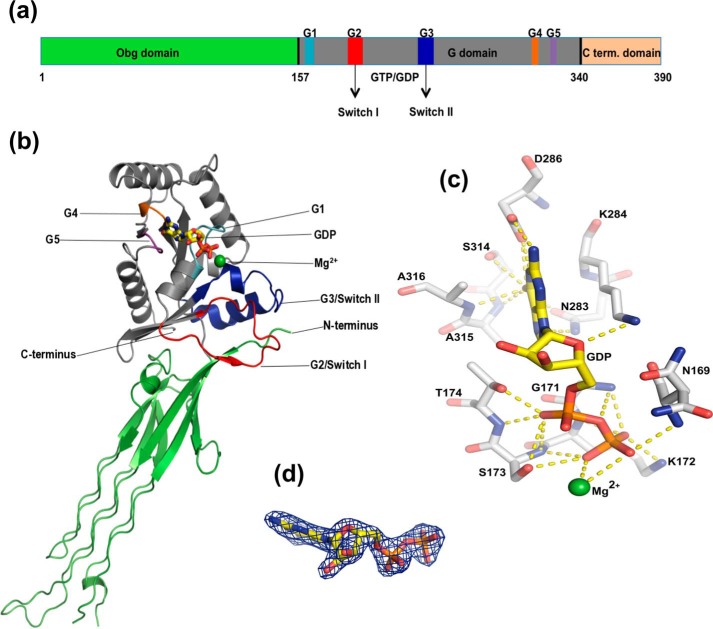
E. coli Obg (ObgE) is a 390-amino acid protein that is predicted to consist of a conserved N-terminal glycine-rich Obg domain (amino acids 1–157) followed by a central Ras-like G domain (amino acids 158–340), in analogy to the previously reported crystal structures of Obg from B. subtilis and Thermus thermophilus (19, 20). In most Obg proteins, these two domains are followed by a third domain, which is much less conserved. Unlike the proteins from B. subtilis and T. thermophilus, in ObgE this domain consists of a stretch of amino acids (341–390) that are composed for 60% of charged amino acids and are predicted to be intrinsically disordered (Fig. 1a) (39, 40). So far, the function of the latter domain remains largely enigmatic.
Figure 1.
The crystal structure of E. coli ObgE_340 bound to GDP. a, domain organization of ObgE, showing the Obg domain (green), G domain (gray), and C-terminal domain (orange) and the conserved sequence motifs in the G domain (G1–G5, Switch I and II). The structure of a construct lacking the C-terminal domain (ObgE_340) was solved. b, schematic representation of the ObgE_340-GDP crystal structure. The Obg domain and G domain are shown in green and gray, respectively, with sequence motifs colored in the same way as in a. GDP is shown as sticks with carbon bonds colored yellow. Mg2+ is shown as a green sphere. c, close-up view of the GDP binding site of ObgE_340. GDP and Mg2+ are colored as in b. Hydrogen bonds between the protein and the nucleotide are indicated by yellow dotted lines. d, GDP bound to ObgE_340 with the clearly defined “omit” electron density map, contoured at 2σ, shown as a blue mesh.
To allow a straightforward interpretation of biochemical and biological data on E. coli ObgE we set out to solve its crystal structure. However, all attempts to crystallize the full-length protein (ObgE_FL) have failed so far, probably due to the presence of the intrinsically disordered C terminus. Therefore, we decided to generate a C-terminal deletion construct (ObgE_340) lacking the last 50 amino acids, by replacing the codon for amino acid Glu-341 by a stop codon. This protein was crystallized successfully in the presence of Mg2+ ions and GDP in space group C2221 with one molecule in the asymmetric unit, allowing to solve its crystal structure at a resolution of 1.85 Å by molecular replacement. We were able to confidently build 334 of the 340 amino acids in the electron density map. Residues 133–136, corresponding to a loop region in the Obg domain and C-terminal residues 339–340, were not built due to weak electron density. The crystal structure clearly shows density for a molecule of GDP and a Mg2+ ion in the G domain. The final model has a crystallographic R factor of 0.198 and a free R factor of 0.239 (Table 1).
TABLE 1.
Data collection and refinement statistics
| ObgE_340 - GDP | |
|---|---|
| Data collection and processing | |
| X-ray source | SOLEIL PROXIMA II |
| Wavelength (Å) | 1.07812 |
| Resolution range (Å)a | 49.00–1.85 (1.90–1.85) |
| Total/unique reflections | 280,027/41,090 (20,519/1,943) |
| Rmeas (%) | 9.7 (182.3) |
| I/σI | 9.67 (1.00) |
| CC1/2 | 99.8 (48.9) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 6.81 (6.97) |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 64.6, 83.0, 177.4 |
| a, b, g (°) | 90, 90, 90 |
| Model refinement | |
| Rwork/Rfree (%)b | 19.8/23.9 |
| R.m.s. deviation bond length (Å) | 0.0154 |
| R.m.s. deviation bond angle (°) | 1.7629 |
| Ramachandran | |
| Favored/allowed/disallowed regions (%) | 98.5/0.9/0.6 |
| PDB code | 5M04 |
a Values for the highest-resolution shell are given in between brackets. CC1/2 values were used as a guide for selecting the highest usable resolution shell (67).
b A subset of 5% of the reflections was used for calculating Rfree.
The crystal structure of ObgE_340 shows the typical arrangement of an N-terminal “Obg domain” followed by a Ras-like G domain (Fig. 1b). Similar to B. subtilis and T. thermophilus Obg, two regions can be discerned in the Obg domain: a region with six left-handed type II helices connected on one side by long loops, and an eight-stranded β-barrel containing an α-helix between the second and third strand (19, 20). This β-barrel makes extensive contacts with the G domain. The largest differences in this domain comparing ObgE to the proteins from B. subtilis and T. thermophilus are present in two of the three long loops connecting each pair of type II helices.
The G domain of ObgE displays a Ras-like fold, consisting of a six-stranded β-sheet and five α-helices. Superposition of the G domain of ObgE on the corresponding domains of T. thermophilus and B. subtilis Obg (supplemental Fig. S1) shows that the biggest differences in conformation are localized in the Switch I and II regions. In the GDP-bound structure of ObgE, both Switch loops could be fully traced, although they are in an “open” conformation where they do not interact with the nucleotide. However, interactions are made between both Switch regions and the N-terminal Obg domain. The Switch II region is folded in two helices, classically termed α1′ and α2′. The conformation of the Switch loops of GDP-bound ObgE most closely resembles the conformation of these loops in the nucleotide-free protomer of the B. subtilis Obg crystal structure, whereas especially Switch II adopted a different conformation in the T. thermophilus crystal structure, mainly due to partial unwinding of helix α1′. In the ppGpp-bound protomer of B. subtilis Obg the Switch regions could not be fully traced (19, 20).
Crystal structures of Obg available in the PDB are either bound to ppGpp (B. subtilis Obg) or show Obg in the apo state (B. subtilis and T. thermophilus Obg). Here we report the first high resolution structure of ObgE bound to GDP. GDP is anchored tightly to the G domain of ObgE_340 via interactions with residues from the P-loop and the G4 and G5 motif (Fig. 1, c and d). The α- and β-phosphates are bound by the P-loop, with the α-phosphate interacting with Thr-174 (side chain and main chain) and the β-phosphate with Asn-169 (main chain), Gly-171 (main chain), Lys-172 (side chain and main chain), and Ser-173 (side chain and main chain). Specificity for the guanine nucleotide is provided by the G4 motif via interactions with Asn-283 (with the N7 of guanine) and Asp-286 (interactions with the N1 and N6 of guanine), and by the G5 motif via interactions of the guanine O6 carbonyl with Ser-314, Ala-315, and Ala-316 (main chain).
ObgE is a monomer in solution
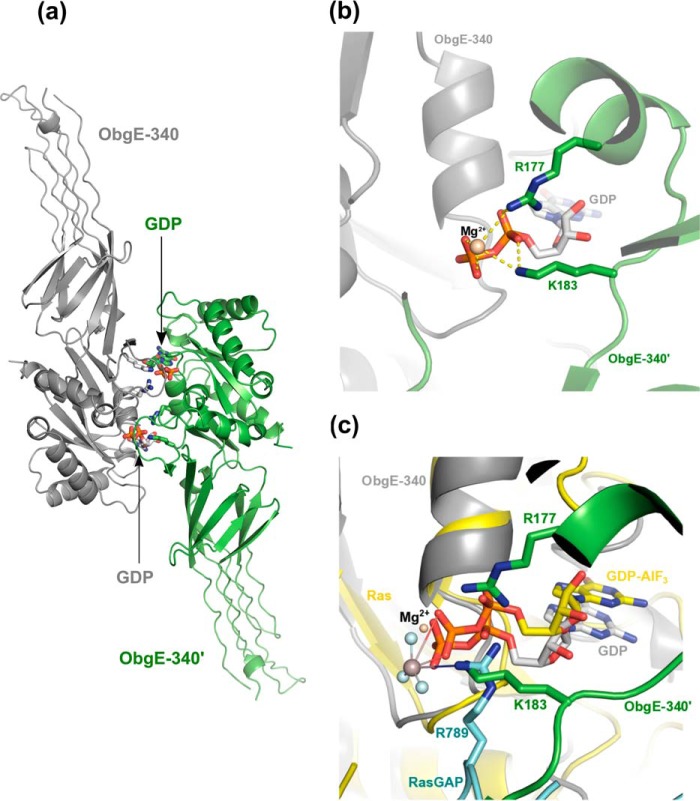
In the crystal structure, the G domain of ObgE_340 interacts with the G domain of the crystal symmetry neighbor. This interface buries a surface area of 836 Å2 and is mainly formed by interactions between residues of the P-loop and the first α-helix (6 residues), Switch I (10 residues), and Switch II (7 residues), where Switch I of one protomer interacts with Switch II of the adjacent protomer and vice versa (Fig. 2a and supplemental Fig. S2). This orientation places one arginine residue (Arg-177) of the first α-helix as well as one lysine residue (Lys-183) of Switch I into the GDP/GTP binding pocket of the neighboring protomer (Fig. 2b). The orientation of the amino group of Lys-183 is reminiscent of the position of the guanido group of the catalytic “arginine finger” (Arg-789) in the Ras-RasGAP complex (Fig. 2c, see below) (41). This observation raises the intriguing question whether ObgE could form a dimer in solution, where two adjacent G domains could reciprocally enhance the GTPase activity of each other. Interestingly, a functional dimerization was previously also suggested for the Obg protein from T. thermophilus, where the C-terminal domain of one protomer was found to interact with the G domain of the adjacent protomer (20).
Figure 2.
Potential dimer interface of ObgE_340 generated though crystal symmetry operations. a, potential dimer organization of ObgE_340 via a face-to-face interaction of its G domain with the G domain of a neighboring protein molecule in the crystal lattice. The two symmetry variants are colored gray and green, and are labeled ObgE_340 and ObgE_340′, respectively. The C-atoms of the bound GDP molecules are colored according to the protomer to which they belong. b, close up view of the interaction surface shown in a. Lys-183 and Arg-177, located on the switch I and the first α-helix of the G domain of one protomer, interact with the phosphate groups of GDP from the adjacent promoter. c, superposition of the structure of the Ras-RasGAP complex (in presence of GDP-AlF3, PDB code 1WQ1 (62)) with the ObgE_340-GDP structure. The two symmetry related protomers of ObgE_340 and GDP are colored and labeled according to b. Ras and the bound GDP-AlF3 are colored yellow. The arginine finger of RasGAP (R789) is shown in cyan. This superposition shows a spatially similar position of the amino group of Lys-183 in the ObgE_340-ObgE_340′ homodimer and the guanido group of Arg-789 of RasGAP in the Ras-RasGAP complex.
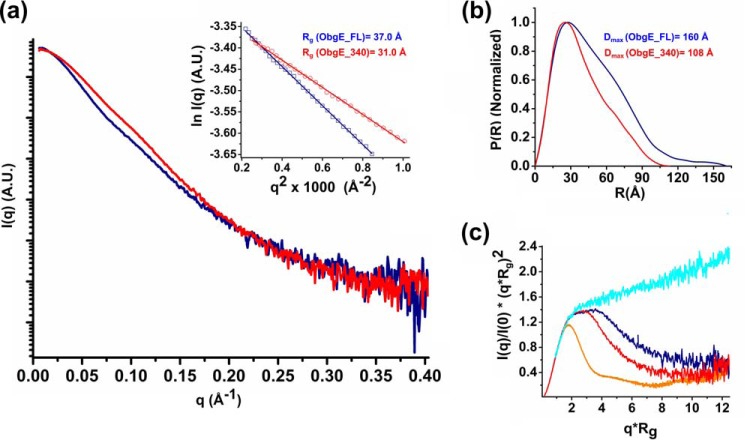
To determine the oligomeric state of ObgE_340 and ObgE_FL in solution, first small angle X-ray scattering experiments coupled to size exclusion chromatography (SEC-SAXS) were performed using the proteins in a nucleotide-free form (Fig. 3). The SEC-SAXS profile of ObgE_340 shows a symmetric peak with a constant radius of gyration (Rg) across the elution peak (supplemental Fig. S3a). An average Rg (from Guinier analysis) and maximal intramolecular distance (Dmax) of 30.9 and 108 Å, respectively, is obtained that is very close to the Rg and Dmax values calculated from the crystal structure of an ObgE_340 monomer (Rg = 29 Å, Dmax = 110 Å). Furthermore, the molecular mass obtained from the Porod volume is 41 kDa, whereas the molecular mass derived from the Porod invariant (QR) is 36 kDa (42) (supplemental Table S1). These molecular mass estimations match very well with the theoretical monomer molecular mass of 39.1 kDa calculated from the protein sequence, indicating that ObgE_340 mainly adopts a monomeric form in solution. Subsequently, SAXS was also used to determine the molecular mass of ObgE_FL (Fig. 3 and supplemental Fig. S4a). The SAXS data of ObgE_FL in a nucleotide-free state yield Rg (from Guinier analysis) and Dmax values of 37.0 and 160 Å, respectively, whereas the molecular mass estimation from the Porod volume gives a value of 60 kDa (supplemental Table S1). The latter value is higher than the expected value for an ObgE_FL monomer (45.4 kDa). The overestimation of the molecular mass using the Porod volume and the large values of Rg and Dmax are, however, in agreement with the presence of an intrinsically disordered C-terminal domain that occupies a large volume in solution (see further). Correspondingly, calculation of the molecular mass using the Porod invariant (QR) gives a molecular mass of 44.8 kDa, which is much closer to the theoretical molecular mass of an ObgE_FL monomer. Together these data show that ObgE in its nucleotide-free state behaves as a monomer in solution.
Figure 3.
SAXS analysis of ObgE_FL and ObgE_340 in their nucleotide-free state, providing information regarding the Rg, maximum particle dimension (Dmax), and molecular mass. a, averaged scattering profile of ObgE_340 (red) and ObgE_FL (blue). The inset shows the linear Guinier region for both constructs, indicative of a non-aggregated protein sample. The deduced Rg values are given. b, normalized P(R) profile of ObgE_340 (red) and ObgE_FL (blue). The broader distribution of intramolecular distances and larger value of Dmax of ObgE_FL reflect the disordered nature of its C-terminal domain. c, dimensionless Kratky plot for ObgE_340 (red) and ObgE_FL (blue) in comparison with an intrinsically disordered protein (hTau40wt, cyan trace) and a globular protein (BSA, orange trace) (68). Although the shapes of both curves are typical for an elongated protein such as ObgE, comparison of the curves indicates a higher degree of flexibility in ObgE_FL. Raw data underlying the averaged scattering profiles are shown in supplemental Figs. S3 and S4.
To further test whether dimerization could potentially occur upon nucleotide binding, we also collected SAXS data of ObgE_340 and ObgE_FL in the presence of an excess of GDP or the nucleotide triphosphate analogue Gpp(NH)p (supplemental Figs. S3–S6). Calculation of the molecular mass using the Porod invariant (QR) yields values of 47.3 and 34 kDa for ObgE_FL and ObgE_340 in the presence of GDP and 45.1 and 37.7 kDa for ObgE_FL and ObgE_340 in the presence of Gpp(NH)p, again very close to the expected monomer molecular masses of 45.4 and 39.1 kDa (supplemental Table S1). Finally, the molecular mass of ObgE_FL (apo), ObgE_FL in the presence of Gpp(NH)p, and ObgE_FL in the presence of the transition state analogue GDP-AlFx was further validated using SEC-MALS. SEC-MALS gave a monodisperse peak with apparent molecular mass in the range of 49–51 kDa (supplemental Fig. S7), again close to the value expected for a monomer (45.4 kDa). Together, we can conclude from these experiments that ObgE behaves mainly as a monomer in solution, regardless of concentration and nucleotide state. This is a strong indication that the dimeric form observed in the crystal structure, generated from crystallographic symmetry operations, is a crystallographic artifact. However, at this point we cannot completely exclude that very transient dimer interactions do occur (see below).
Interdomain orientation and influence of nucleotides on the conformation of ObgE
Although the individual Obg and G domains of the Obg crystal structures from E. coli (this study), B. subtilis (19), and T. thermophilus (20) superimpose very well (see above), the relative orientation of these domains in the crystal structures differs. Indeed, it had been noted before that upon superimposing the G domains of B. subtilis (nucleotide-free or ppGpp bound) and T. thermophilus (nucleotide-free) Obg, the respective Obg domains are rotated by about 180° around the G domain axis (20). Moreover, in a low resolution (5.5 Å) EM structure of E. coli Obg, bound to the 50S ribosomal subunit, an orientation of the Obg domain intermediate to that observed in B. subtilis Obg and T. thermophilus Obg was observed (27). A recent molecular dynamics study suggested that the relative orientation of the Obg domain, vis à vis the G domain changes depending on the nucleotide state of the protein, with the largest difference occurring in the GDP-bound state compared with the apo- and GTP-bound states (43). However, in the current high resolution structure of ObgE_340 bound to GDP, the orientation of the Obg domain with respect to the G domain is very similar to the B. subtilis Obg structure, either in the apo state or bound to ppGpp (supplemental Fig. S1).
To further investigate the influence of nucleotides on the conformation of E. coli ObgE_340 and ObgE_FL in solution we again turned to SAXS. To test whether our crystal structure corresponds to a main conformation in solution, we first compared the theoretical scatter curve of the ObgE_340 crystal structure, after flexible modeling of the N-terminal purification tag, with the experimental scatter curve (supplemental Fig. S8). Because both curves overlay very well, we can conclude that the crystal structure indeed represents a relevant conformation in solution. Next, we compared the scattering curves of both ObgE_340 and ObgE_FL bound to GDP or Gpp(NH)p to the corresponding curve of the nucleotide-free protein (supplemental Figs. S5 and S6). This superposition does not reveal any significant nucleotide-dependent changes in the scattering curves, although small differences are observable when comparing the pair-distance distribution functions and dimensionless Kratky plots of ObgE_FL in different nucleotide states (supplemental Figs. S5 and S6). These data seem to indicate that no large-scale nucleotide-induced conformational changes are taking place in E. coli ObgE. However, alternatively, it remains possible that such changes only take place in the presence of certain partner proteins, such as the ribosome (27), or that the suggested rotation of the Obg domain vis à vis the G-domain does not lead to an observable difference in the low-resolution spherically averaged SAXS profiles. High-resolution crystal structures of ObgE in different nucleotide-bound states could shed further light on this issue.
Conformation and role of the C-terminal domain
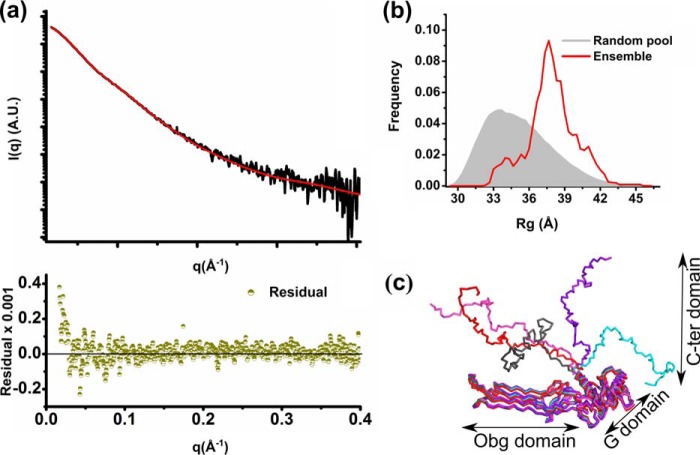
The last 50 C-terminal amino acids of ObgE_FL are mainly composed of charged amino acids (about 60%) and are predicted to be intrinsically disordered (39, 40). The flexible disordered nature of this peptide region is also in agreement with our SAXS data for ObgE_FL (see above), which show that (i) the calculated Porod volume is at least 15% higher than would be expected for a compact monomeric protein, and (ii) the normalized Kratky plot has a broader peak and converges at higher angle than the corresponding plot for ObgE_340 (Fig. 3c). Correspondingly, we did not succeed in crystallizing the full-length ObgE protein. Consequently, so far no information regarding the structure and function of this C-terminal domain is available. To get an idea of the conformational space adopted by the C-terminal domain in solution, we used the experimental SAXS data of the full-length ObgE, in combination with our partial crystal structure and the amino acid sequence, to perform ensemble modeling. The conformation of the last 50 residues was probed using all-atom modeling, followed by model validation using the Molprobity score and further refinement using iterative normal mode analysis (NMA). Model selection based on the experimental SAXS data indicated that the latter could be accounted for by an ensemble of 5 models as depicted in Fig. 4. However, we would like to stress that the presented ensemble only gives a representative subset of possible orientation of the C-terminal domain in solution that is consistent with the experimental scattering data.
Figure 4.
SAXS-based ensemble modeling of ObgE_FL taking into account the flexibility of the C-terminal domain. a, ensemble fit of ObgE_FL (red line) to the experimental ObgE_FL SAXS profile (black trace). The residuals are shown at the bottom of the fit (dark green). b, the Rg distribution of the ensemble (red curve) compared with the Rg distribution of a random pool of models (gray filled area) generated using an ensemble optimization method coupled to all-atom modeling, model validation, and NMA refinement. c, ribbon representation of 5 ensemble models, selected by the genetic algorithm, which give an average theoretical curve that fits the experimental ObgE_FL SAXS profile as shown in a.
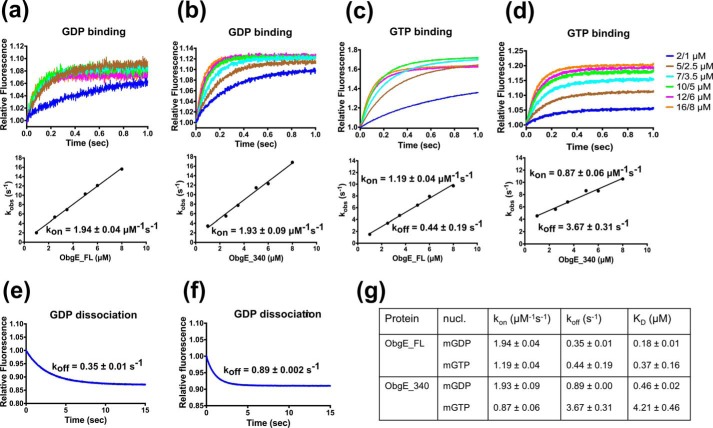
To investigate the function of this intrinsically disordered C-terminal domain we determined its contribution to nucleotide binding and hydrolysis by comparing the full-length protein with the C-terminal deletion construct ObgE_340. First, we determined its contribution to nucleotide binding affinity and kinetics using stopped flow fluorescence experiments (using mant-labeled nucleotides). In agreement with previous reports (35), we find that ObgE_FL displays a relatively low affinity for GDP (KD = 0.18 ± 0.01 μm) and GTP (KD = 0.37 ± 0.16 μm) due to fast nucleotide binding and dissociation (see Fig. 5 for all data). Deletion of the C-terminal domain (ObgE_340) leads to a small decrease in affinity for GDP (KD = 0.46 ± 0.02 μm), whereas the affinity for GTP is decreased more than 10-fold (KD = 4.21 ± 0.46 μm). This decrease in affinity is nearly entirely due to an increased GTP dissociation rate (koff(GTP, ObgE_FL) = 0.44 ± 0.19; koff(GTP, ObgE_340) = 3.67 ± 0.31; Fig. 5).
Figure 5.
GDP and GTP binding and dissociation kinetics of ObgE_FL (a, c, and e) and ObgE_340 (b, d, and f) determined via stopped flow fluorescence analysis. a and b, transients obtained by following mGDP (0.2/0.1 μm, before/after mixing) fluorescence upon rapid mixing with different concentrations of ObgE_FL (a) and ObgE_340 (b). Concentration values given on the graph represent ObgE concentrations before and after mixing. The lower panels show the concentration dependence of the observed rate constant (kobs). The slope of the linear fit yields kon. c and d, transients obtained by following mGTP (0.2/0.1 μm, before/after mixing) fluorescence upon rapid mixing with different concentrations of ObgE_FL (c) and ObgE_340 (d). The same concentrations as in a and b are used. The lower panels show the concentration dependence of the observed rate constant (kobs). The slope and intercept of the linear fit yield kon and koff, respectively. e and f, direct determination of koff by following the release of mGDP from ObgE_FL (e) and ObgE_340 (f) upon mixing 200 μm unlabeled GDP with a mixture of 0.4 μm ObgE and 1.5 μm mGDP. g, summary of nucleotide binding/dissociation kinetics and the deduced KD values of ObgE_FL and ObgE_340.
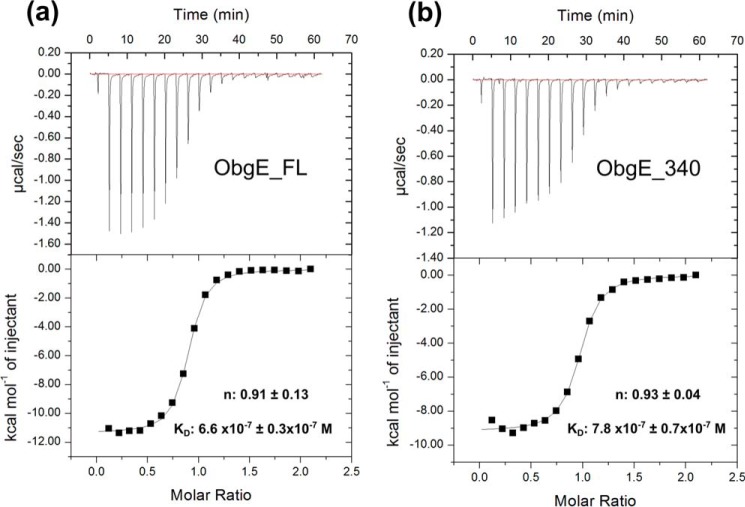
Subsequently, considering that ppGpp is required for ObgE-mediated persistence (38), we also determined the affinity of ObgE_FL and ObgE_340 for ppGpp using ITC experiments (Fig. 6 and supplemental Fig. S9). ObgE_FL binds ppGpp with a KD = 0.66 ± 0.03 μm. Deletion of the C terminus leads to a very small decrease in affinity for ppGpp (KD = 0.78 ± 0.07 μm), in agreement with the observed small effect of the C-terminal domain on GDP binding.
Figure 6.
ITC experiments for binding of ppGpp to ObgE_FL (a) and ObgE_340 (b), respectively. Experiments were performed at 25 °C by titrating ppGpp from a stock solution of 750 μm into a solution of 75 μm nucleotide-free ObgE. Equilibrium dissociation constants (KD) and binding stoichiometries (n) are given as the mean ± S.D. of 3 independent measurements (all isotherms are given in supplemental Fig. S9).
Finally, we also assessed the influence of the C-terminal domain on GTP turnover. To this end we measured Michaelis-Menten kinetics. ObgE_FL has a kcat = 0.064 ± 0.001 min−1 and a Km = 10.40 ± 0.70 μm, compared with a kcat = 0.046 ± 0.001 min−1 and a Km = 11.07 ± 0.92 μm for ObgE_340 (Table 2 and supplemental Fig. S10). This indicates that although the C terminus is involved in GTP binding, it is not substantially involved in GTP turnover.
TABLE 2.
Steady state kinetic parameters of GTP hydrolysis by wild-type ObgE_FL, ObgE_340, and mutant ObgE_FL
Assays were performed in buffer containing 150 mm NaCl or KCl.
| Protein | Salt | kcat | Km |
|---|---|---|---|
| min−1 | μm | ||
| ObgE_FL | NaCl | 0.064 ± 0.001 | 10.40 ± 0.70 |
| ObgE_FL | KCl | 0.045 ± 0.001 | 5.64 ± 0.56 |
| ObgE_FL_K183A | NaCl | 0.020 ± 0.000 | 10.27 ± 0.75 |
| ObgE_FL_R177A | NaCl | 0.060 ± 0.002 | 6.63 ± 0.69 |
| ObgE_340 | NaCl | 0.046 ± 0.001 | 11.07 ± 0.92 |
In conclusion, we show here that the intrinsically disordered C-terminal domain of E. coli ObgE specifically contributes to GTP binding with very little effect on GDP or ppGpp binding. Such a discriminatory effect of the C-terminal domain on GTP versus GDP binding seems to indicate that this domain transiently folds back on the G domain in the GTP-bound state. Such a conformational change of the C-terminal domain is, however, not reflected in large changes in our SAXS profiles (see above), and further research is required to reveal the underlying mechanisms.
Testing potential GTPase-activating mechanisms of ObgE
ObgE, and Obg proteins in general, show a very low intrinsic GTPase activity (kcat (ObgE) = 0.064 min−1, Table 2). Classically, the rate of GTP hydrolysis of small Ras-like GTPases is enhanced by dedicated GAPs (6, 8). However, so far no such GAPs for Obg have been reported, although it was recently described that the GTPase activity of Obg is increased upon binding to the ribosome (27, 43). Moreover, recently a number of alternative mechanisms to increase the GTPase activity of G proteins have been reported, including binding of specific ions and reciprocal complementation of active sites by homodimerization (9–16).
With respect to a potential mechanism of GTPase activation though homodimerization, we were particularly triggered by the observation of a potential homodimer in the ObgE_340 crystal structure, formed through crystal symmetry (see above). This dimeric organization would place both an arginine (Arg-177) and a lysine residue (Lys-183) coming from, respectively, the first α-helix and the Switch I region of one subunit into the GTP binding pocket of an adjacent subunit (Fig. 2b). Despite our observation that ObgE_340 is mainly monomeric in solution (see above), it can a priori not be excluded that a transient dimeric complex could be formed, where Lys-183 and Arg-177 would function as a catalytic lysine or arginine “finger” by electrostatically stabilizing the GTPase transition state in the adjacent subunit. Such a catalytic role of Lys-183 and/or Arg-177 would be reminiscent of the catalytic arginine finger (Arg-789) used by the RasGAP protein for activation of the Ras GTPase activity (41). Even more intriguingly in this respect we find that superposition of the G domain of ObgE_340 onto Ras in the Ras-RasGAP complex (41) places the amino group of Lys-183 from the adjacent ObgE subunit nearly perfectly on the guanidino group of the catalytic arginine finger of the RasGAP protein (Fig. 2c). To test whether Lys-183 or Arg-177 contribute to GTP hydrolysis, we mutated both residues to alanine in ObgE_FL, and compared the steady state kinetic parameters to the wild type protein (Table 2 and supplemental Fig. S10). These data show that the K183A mutation lowers kcat by a factor of 3, whereas the Km value is nearly unaffected (kcat = 0.020 ± 0.000 min−1; Km = 10.27 ± 0.75 μm). On the other hand, the R177A mutant has unaffected kcat and Km values (kcat = 0.060 ± 0.002 min−1; Km = 6.63 ± 0.69 μm). We thus conclude that, although the K183A mutation has a somewhat lowered kcat value, the observed effects are too small to account for a genuine catalytic finger.
Although many small GTPases use an arginine finger from a GAP protein to stabilize the transition state of GTP hydrolysis, other G proteins including MnmE, YqeH, FeoB, RbgA, and EngA bind a K+ ion in the active site to neutralize the negative charge of the transition state (13, 14, 44–46). Based on sequence analysis and the presence of an asparagine residue in the so-called K-loop of the Switch I region, Obg proteins were recently proposed to be potential members of these so-called potassium-selective cation-dependent GTPases (15). However, so far this hypothesis has not been experimentally validated. To investigate the effect of K+ ions on the kinetic constants of ObgE, the Michaelis-Menten kinetic parameters of ObgE_FL were determined in the presence of 150 mm KCl and compared with those in the presence of 150 mm NaCl (Table 2 and supplemental Fig. S10). A small (less than 2-fold) decrease in kcat was observed in the presence of KCl compared with NaCl. Therefore, we can rule out a mechanism whereby K+ acts as a GTPase activating factor.
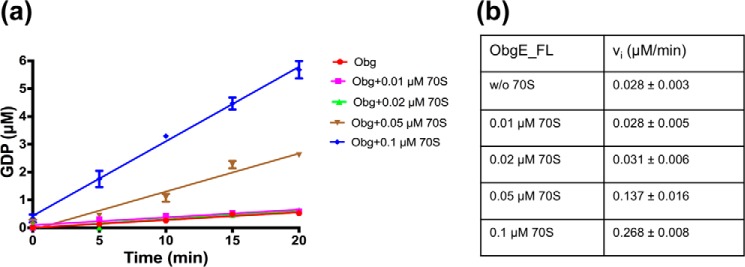
Because we did not find any activation via dimerization or K+ binding, we also reinvestigated the reported effect of the ribosome. Indeed, it was recently shown that a 1:1 complex of ObgE and the 50S ribosomal particle stimulates the ObgE GTPase activity about 120-fold (27). In another study it was shown that the GTPase activity of Vibrio cholerae Obg (or CgtA) was also stimulated up to 5-fold by catalytic amounts of 50S or 70S ribosomes (43). Hereto we measured the ObgE_FL activity upon addition of increasing amounts of ribosome. However, at the highest ribosome concentrations we found a significant GTPase activity in the ribosome preparation, most likely due to GTPases that bind and co-purify with the ribosome. After subtracting this background activity we found a gradual increase in the GTPase activity of ObgE with increasing (catalytic) amounts of ribosome (Fig. 7). Addition of 0.1 μm 70S ribosome to 0.5 μm ObgE increases the initial rate (at 100 μm GTP) about 10-fold. We can thus conclude that although ObgE does not seem to be activated by homodimerization or K+ ions as previously suggested, it is activated upon binding to the ribosome.
Figure 7.
Stimulation of ObgE GTPase activity by the 70S ribosome. Initial rate kinetic traces (a) and deduced initial rates (b) for GTP (100 μm) hydrolysis by ObgE_FL (0.5 μm) without and with increasing concentrations of 70S ribosome. Time traces and initial rates are shown after subtraction of a background GTPase activity in the ribosomal preparation, probably due to a contaminating GTPase that copurified with the ribosome. Each data point represents the average ± S.D. of 3 independent measurements.
Conclusion
ObgE is an essential G protein implicated in ribosome maturation, cell cycle control, DNA replication, and bacterial persistence. The present study provides new insights into the structure and function of ObgE. We determined the first crystal structure of ObgE bound to GDP at 1.85-Å resolution and we unequivocally show, using SAXS and MALS, that ObgE behaves as monomer in solution. Biochemical and kinetic analysis unravels a role of the intrinsically disordered C-terminal domain in binding of GTP. Because this domain is less conserved among Obg family members, it remains to be determined whether such a role for this domain is general. Finally, it has been suggested lately that the GTPase activity of Obg proteins might be accelerated by an alternative mechanism, such as homodimerization or binding of K+ ions. We show that neither of these hypotheses are valid, but rather confirm that the GTPase activity is stimulated in the presence of ribosomes. These findings thus contribute to our understanding of the detailed molecular mechanism of Obg proteins, which in turn might lead to novel insights into the role of ObgE in cellular physiology, including bacterial persistence.
Experimental procedures
Protein expression and purification
The open reading frame (ORF) coding for full-length ObgE (ObgE_FL) was amplified by PCR from genomic E. coli DNA as described before (38), and subsequently cloned within the NdeI and HindIII restriction sites of a pET28 vector containing an N-terminal His6 tag. In addition, another ObgE_FL construct was cloned with a C-terminal strep tag in a pET22b vector within the NdeI and XhoI restriction sites. An ObgE construct, lacking the last 50 amino acids (ObgE_340), was generated by replacing the codon for Glu-341 by a stop codon in pET28-ObgE_FL using the QuikChange site-directed mutagenesis method (Stratagene).
All protein constructs were expressed in either E. coli BL21(DE3) or Rosetta (DE3) pLysS cells. Cells were grown in TB medium at 37 °C and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside when an A600 of 0.7 was reached. After induction for 6 h at 25 °C, cells were harvested by centrifugation and resuspended in buffer A (20 mm Hepes/NaOH, pH 7.5, 300 mm NaCl, 5 mm MgCl2, 5 mm β-mercaptoethanol, 5% glycerol) containing protease inhibitors (4-(2-aminoethyl)benzenesulfonyl fluoride and leupeptin, 0.1 mg/ml and 1 μg/ml final concentrations, respectively). After cell disruption using a cell disruptor system (Constant Systems) and clearance of the lysate by centrifugation at 30,000 × g, the supernatant with His6-tagged proteins was applied to a Ni2+-Sepharose HP column (GE Healthcare), equilibrated with buffer A. After extensive washing with buffer A containing increasing amounts of imidazole (20 and 30 mm), the protein was eluted with buffer A containing 250 mm imidazole. In the case of the C-terminal strep-tagged protein, the supernatant of the cell lysate was loaded on a Strep-tactin column (IBA Lifesciences), washed extensively with buffer A, and eluted with buffer A containing 2.5 mm desthiobiotin. Preparation of nucleotide-free ObgE was achieved by incubation with calf intestine alkaline phosphatase (Roche Diagnostics) followed by dialysis against buffer A. The nucleotide load of the protein was monitored via reversed phase chromatography on a C18 column (Jupiter, 25 cm × 4.6 mm) coupled to an Alliance HPLC (Waters) system as described previously (47). The residual alkaline phosphatase was removed by Ni2+-Sepharose HP (for His6-tagged ObgE) or a HiTrap Q HP anion exchange (for strep-tagged ObgE) column. Size-exclusion chromatography (Superdex 75, 16/60) in buffer containing 20 mm Hepes/NaOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, and 2 mm DTT was used as the final purification step. Fractions containing purified ObgE were concentrated, flash frozen in liquid nitrogen, and stored at −80 °C. All subsequent experiments were started from these nucleotide-free Obg protein batches.
Purification of 70S ribosome
15 g (wet weight) of E. coli MRE600 cells grown in LB medium were resuspended in a buffer solution containing 20 mm Tris/HCl, pH 7.5, 10 mm MgCl2, 100 mm NH4Cl, and 6 mm β-mercaptoethanol (buffer B). Cells were lysed by two passes through a cell disruptor system (Constant Systems). The lysate was centrifuged twice in a Beckman JA-20 rotor at 17,000 rpm for 30 min. Subsequently the supernatant was loaded onto two 35-ml sucrose cushions (1.1 m sucrose, 20 mm Tris/HCl, pH 7.5, 500 mm NH4Cl, 10 mm MgCl2, and 0.5 mm EDTA) in 75-ml ultracentrifuge polycarbonate tubes, and centrifuged for 20 h at 35,000 rpm at 4 °C in a Ti-45 Beckman rotor. The translucent ribosome pellet was gently washed with buffer B. The pellet was resuspended in 5 ml of the same buffer and the volume was adjusted to 100 ml with buffer B containing 0.5 m NH4Cl. An ultracentrifugation step for 6 h at 25,000 rpm was repeated and the pellet was washed with buffer B again. The purified ribosomes were resuspended in 1.6 ml of buffer B containing 50 mm Tris/HCl, pH 7.5. The ribosome concentration was calculated using a ϵ260 nm of 4.35 × 107 m−1 cm−1. Finally, the ribosomes were flash frozen in liquid nitrogen and stored at −80 °C.
Crystallization, data collection, phasing, and refinement
ObgE_340 was loaded with 1 mm GDP and crystals were obtained by mixing protein (at a concentration of 8 mg/ml) with an equal volume of crystallization buffer in a hanging drop vapor diffusion set-up. The crystallization solution contained 16% (w/v) polyethylene glycol (PEG) 6000, 15% (v/v) 2-propanol, and 100 mm sodium citrate/HCl, pH 5.6. Crystals were flash frozen in liquid nitrogen using crystallization buffer supplemented with 15% glycerol as cryoprotectant, and data were collected at 100 K at the PROXIMA II beamline of the Soleil Synchrotron (Paris, France). Data indexing, integration, and scaling were done using the XDS suite (48). Data quality was assessed in phenix.xtriage (49). Crystals belong to the space group C2221 with unit cell dimensions a = 64.6 Å, b = 83.0 Å, c = 177.4 Å and α = β = γ = 90° (Table 1).
Phases were obtained by molecular replacement using PHASER (50, 51) from the CCP4 software package (52) and using the structure of B. subtilis Obg (PDB code 1LNZ) as search model. ARP/wARP was used for automated model building (53). Model building was finalized by manual building cycles in COOT (54), alternated with refinement in Refmac (55). TLS refinement was implemented in the refinement protocol, using six individual TLS groups determined by the TLSMD server (56, 57). The obtained model was validated with the Molprobity server (58). All figures were prepared in PyMOL. Data collection and refinement statistics are summarized in Table 1.
SAXS measurement and modeling
SAXS data for ObgE_FL and ObgE_340 (C-terminal strep-tag and N terminally His6-tagged constructs, respectively) were collected either at the SWING beamline of the Soleil Synchrotron (Paris, France) or using an in house Rigaku BioSAXS-2000 instrument. Measurements of ObgE_FL and ObgE_340 in a nucleotide-free state and in the presence of Gpp(NH)p and ObgE_FL in the presence of GDP were performed using an in-line HPLC-SEC setup at the SWING beamline (59). The scattering intensities were recorded after injection of 70 μl of 8–15 mg/ml of protein on an Agilent Bio-SEC 3 column pre-equilibrated with 20 mm Hepes/NaOH, pH 7.5, 300 mm NaCl, 250 mm imidazole, 5 mm MgCl2, and 2 mm DTT (SEC buffer) using a flow rate of 0.2 ml/min. For measurements of ObgE_340 in the presence of GDP, 1 ml of concentrated protein sample (10 mg/ml) was injected on a S75 10/300 column pre-equilibrated with SEC buffer and connected to an AKTA purifier (GE Healthcare) using a flow rate of 1.0 ml/min, and subsequently the elution fractions of the protein peak were collected and concentrated to 2.0, 4.0, and 8.0 mg/ml. The SAXS data were immediately afterward collected in batch mode on a Rigaku BioSAXS-2000 instrument. For data collection of nucleotide-bound proteins, the protein was preincubated with 1 mm of the nucleotide, and 400 μm of the corresponding nucleotide was added to the running buffer of the size-exclusion chromatography.
The radial averaging and buffer subtraction of the resulting data frames were performed using FOXTROT for the data collected at the synchrotron and using Rigaku SAXSLab for the data collected on the Rigaku BioSAXS-2000 instrument. A final scattering curve used for processing of the data collected in house, was obtained by merging the lower q value data of the 4.0 mg/ml curve with the higher q value data of the 8.0 mg/ml curve. In case of data collection at the synchrotron, DATASW was used for calculation of the invariants (60). The averaged data, corresponding to a peak of interest, was further processed using ATSAS (61) and SCATTER (42) software packages. The molecular mass of the scattering particle was derived using the QR method (42) and Porod volume (62). Further interpretation of the SAXS data involved representation using the dimensionless Kratky plot and calculation of the pair-distance distribution function using the ATSAS program GNOM (63), whereas calculation of the theoretical scattering profile of the X-ray crystal structure was done using CRYSOL (64).
Prior to flexibility assessment, two short internal regions within the ObgE_340 structure (residues 133–136 and residues 339–340), missing from the crystal structure, were modeled using the “model missing loop” tool of MODELLER within CHIMERA (65). A similar procedure was performed for ObgE_FL before modeling the flexible C-terminal domain including the C-terminal purification tag. For flexibility assessment of ObgE_340, the crystal structure with modeled loops and the amino acid sequence of the 340 residues of the protein along with the 20 residues of N-terminal purification tag were used. On the other hand, for ObgE_FL, the amino acid sequence of all 390 residues together with the C-terminal purification tag was used. In both cases, a random pool of structures was generated using the ensemble optimization method suite (66), which gives the combination of an all-atom model, corresponding to the crystal structure, and the C-α traces of the missing flexible regions. Next, a computational pipeline FULCHER4 was used to convert the random pool of combined all-atom/C-α trace models to all-atom models, with subsequent model validation using the Molprobity clash score and iterative NMA refinement of the top 50% scoring models to probe the conformational dynamics of the molecule. Finally, the genetic algorithm GAJOE was used to obtain an ensemble of models that best describes the experimental SAXS data. Selected ensembles included refined models that had r.m.s. deviation <0.4 Å compared with the initial crystal structure.
MALS analysis
Multi-angle light scattering experiments coupled to SEC (SEC-MALS) were performed using a Dawn Heleos (Wyatt Technology) detector (using 9 angles) connected to an Agilent Bio-SEC 3 or Shodex KW-800 column attached to an HPLC system (Waters). 10–20 μl of ObgE_FL (with N-terminal His6 tag) at 4–8 mg/ml was injected on the column at a flow rate of 0.2 ml/min in 20 mm Hepes/NaOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, and 2 mm DTT. In case of the nucleotide-bound sample, the buffer was supplemented with 400 μm Gpp(NH)p or 1 mm GDP-AlFx. The molar mass was calculated with the ASTRA 5.3.4.20 software.
Steady state kinetic measurements
GTP hydrolysis rates of ObgE were measured by following the production of GDP in function of time on HPLC (Waters). All measurements were performed at 25 °C in 20 mm Hepes/NaOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, and 2 mm DTT. For multi-turnover analysis, 0.5 μm ObgE was incubated with different GTP concentrations ranging from 2.5 until 100 μm. At different time points 50-μl aliquots were taken and the reaction was stopped by heating for 5 min at 100 °C. Hydrolysis of GTP was monitored by separation of the nucleotides (GTP and GDP) using a C18 column (Jupiter, 25 cm × 4.6 mm) attached to an HPLC Alliance system (Waters). The nucleotides were eluted using a buffer solution containing 100 mm KH2PO4, pH 6.4, 10 mm tetrabutyl ammonium bromide, and 7.5% acetonitrile, as mobile phase. Nucleotides were detected at 254 nm and peak areas were converted to concentration using a standard curve derived from known GDP concentrations. Initial rates were obtained as the slope of the [GDP] versus time plot and fitted to the Michaelis-Menten equation. The influence of the 70S ribosome on ObgE GTPase activity was determined by preincubating increasing amounts of the 70S ribosome (0.01, 0.02, 0.05, and 0.1 μm) with 0.5 μm ObgE at 25 °C for 30 min. GTPase activity was determined at 100 μm GTP. Because a low GTPase activity co-purified with the ribosome, identical time traces were recorded in the absence of ObgE at each ribosome concentration. These time traces were subtracted from the measurements in the presence of ObgE before calculating initial rates.
Fluorescence stopped flow kinetics
Nucleotide binding kinetics were determined via stopped flow fluorescence (SX18.MV; Applied Photophysics). ObgE in a concentration range of 2–16 μm was rapidly mixed with 0.2 μm 2′,3′ N-methylanthraniloyl-labeled nucleotides (mant-GDP/mant-GTP; Jena Bioscience), providing conditions for pseudo-first order binding kinetics. Mant nucleotides were excited at 360 nm and the change in fluorescence was monitored through a 405-nm cut-off filter. For each protein concentration, the data of at least 5 time traces were averaged and fitted to a single exponential function, yielding the observed rate constant kobs. The association and dissociation rate constants (kon and koff) were obtained from the slope and intercept of the plot of kobs versus the protein concentration. Alternatively, koff was obtained by mixing 200 μm of unlabeled nucleotide with a mixture of 0.4 μm protein and 1.5 μm mant-nucleotide. The resulting time traces were fitted on a single exponential, yielding koff. The KD values were calculated from the ratio of koff and kon. The experiments were performed at 25 °C in 20 mm Hepes/NaOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, and 2 mm DTT.
Isothermal titration calorimetry
Binding affinity of ObgE for guanosine-3′,5′-bisdiphosphate (ppGpp; TriLink Biotechnologies) was determined by isothermal titration calorimetry (ITC), using the MicroCal iTC200 system (GE Healthcare) with a reference power of 10 microcal/s. The thermodynamic parameters upon titration of ppGpp (750 μm) to nucleotide-free ObgE (75 μm) were measured at 25 °C in a buffer consisting of 20 mm Hepes/NaOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, and 2 mm DTT. For each experiment a preliminary 0.4-μl injection (not included in data analysis) was followed by 20 injections of each 2 μl with duration of 4 s at a stirring speed of 400 rpm and intervals of 180 s. The initial delay was 120 s. All experiments were performed in triplicate. Origin version 7.0 software was used for data integration and fitting to a single binding site model using the standard Marquardt non-linear regression method as provided in the Microcal Origin routines. The equilibrium binding dissociation constant (KD) as well as the binding stoichiometry (n) reported in the main text are given as the mean ± S.D. of the three independent measurements.
Author contributions
S. G., R. K. S., M. F., N. V., J. M., and W. V. conceived and designed the experiments. S. G. solved the crystal structure. S. G., J. M., and W. V. performed and analyzed kinetic data. R. K. S. and A. V. S. performed and analyzed SAXS measurements. S. G., R. K. S., and W. V. wrote the paper. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank the staff of beamlines PROXIMA II and SWING of the Soleil Synchrotron (Paris, France) and Dr. Rodrigo Gallardo for helping with MALS experiments.
This work was supported in part by Fonds voor Wetenschappelijk Onderzoek Grants G0471.12N and G0B2515N, the Hercules Foundation, BioStruct-X by the European Community's Seventh Framework Programme, a Strategic Research Program (SRP34) from the VUB, and the Interuniversity Attraction Poles Program initiated by the Belgian Science Policy Office. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S10 and Table S1.
The atomic coordinates and structure factors (code 5M04) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Small angle X-ray scattering data were deposited at the Small Angle Scattering Biological Data Bank (SASDB) under accession codes SASDBS6, SASDBB9, SASDBT6, SASDBU6, SASDBC9, and SASDBV6.
A. Shkumatov, unpublished data.
- GAPs
- GTPase activating proteins
- TRAFAC
- translation factor
- SAXS
- small angle X-ray scattering
- MALS
- multi-angle light scattering
- pppGpp
- guanosine pentaphosphate
- ppGpp
- guanosine tetraphosphate
- TLS
- translation/libration/screw
- NMA
- normal mode analysis
- r.m.s.
- root mean square
- SEC
- size exclusion chromatography
- ITC
- isothermal titration calorimetry
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphate.
References
- 1. Bourne H. R., Sanders D. A., and McCormick F. (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348, 125–132 [DOI] [PubMed] [Google Scholar]
- 2. Caldon C. E., and March P. E. (2003) Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6, 135–139 [DOI] [PubMed] [Google Scholar]
- 3. Verstraeten N., Fauvart M., Versées W., and Michiels J. (2011) The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 75, 507–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vetter I. R., and Wittinghofer A. (2001) The guanine nucleotide: binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 5. Wittinghofer A., and Vetter I. R. (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 [DOI] [PubMed] [Google Scholar]
- 6. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 7. Maracci C., and Rodnina M. V. (2016) Translational GTPases. Biopolymers 105, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mishra A. K., and Lambright D. G. (2016) Small GTPases and their GAPs. Biopolymers 105, 431–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghosh A., Praefcke G. J., Renault L., Wittinghofer A., and Herrmann C. (2006) How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440, 101–104 [DOI] [PubMed] [Google Scholar]
- 10. Meyer S., Wittinghofer A., and Versées W. (2009) G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J. Mol. Biol. 392, 910–922 [DOI] [PubMed] [Google Scholar]
- 11. Byrnes L. J., and Sondermann H. (2011) Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. U.S.A. 108, 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gasper R., Meyer S., Gotthardt K., Sirajuddin M., and Wittinghofer A. (2009) It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell Biol. 10, 423–429 [DOI] [PubMed] [Google Scholar]
- 13. Scrima A., and Wittinghofer A. (2006) Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 25, 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anand B., Surana P., and Prakash B. (2010) Deciphering the catalytic machinery in 30S ribosome assembly GTPase YqeH. PLoS ONE 5, e9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ash M.-R., Maher M. J., Mitchell Guss J., and Jormakka M. (2012) The cation-dependent G-proteins: in a class of their own. FEBS Lett. 586, 2218–2224 [DOI] [PubMed] [Google Scholar]
- 16. Rafay A., Majumdar S., and Prakash B. (2012) Exploring potassium-dependent GTP hydrolysis in TEES family GTPases. FEBS Open Bio. 2, 173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fislage M., Wauters L., and Versées W. (2016) MnmE, a GTPase that drives a complex tRNA modification reaction. Biopolymers 105, 568–579 [DOI] [PubMed] [Google Scholar]
- 18. Leipe D. D., Wolf Y. I., Koonin E. V., and Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 19. Buglino J., Shen V., Hakimian P., and Lima C. D. (2002) Structural and biochemical analysis of the Obg GTP binding protein. Structure 10, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 20. Kukimoto-Niino M., Murayama K., Inoue M., Terada T., Tame J. R., Kuramitsu S., Shirouzu M., and Yokoyama S. (2004) Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8. J. Mol. Biol. 337, 761–770 [DOI] [PubMed] [Google Scholar]
- 21. Hirano Y., Ohniwa R. L., Wada C., Yoshimura S. H., and Takeyasu K. (2006) Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes to Cells. 11, 1295–1304 [DOI] [PubMed] [Google Scholar]
- 22. Bang W. Y., Hata A., Jeong I. S., Umeda T., Masuda T., Chen J., Yoko I., Suwastika I. N., Kim D. W., Im C. H., Lee B. H., Lee Y., Lee K. W., Shiina T., and Bahk J. D. (2009) AtObgC, a plant ortholog of bacterial Obg, is a chloroplast-targeting GTPase essential for early embryogenesis. Plant Mol. Biol. 71, 379–390 [DOI] [PubMed] [Google Scholar]
- 23. Kotani T., Akabane S., Takeyasu K., Ueda T., and Takeuchi N. (2013) Human G-proteins, ObgH1 and Mtg1, associate with the large mitochondrial ribosome subunit and are involved in translation and assembly of respiratory complexes. Nucleic Acids Res. 41, 3713–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koller-Eichhorn R., Marquardt T., Gail R., Wittinghofer A., Kostrewa D., Kutay U., and Kambach C. (2007) Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 282, 19928–19937 [DOI] [PubMed] [Google Scholar]
- 25. Cheung M.-Y., Li X., Miao R., Fong Y.-H., Li K.-P., Yung Y.-L., Yu M.-H., Wong K.-B., Chen Z., and Lam H.-M. (2016) ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses. Proc. Natl. Acad. Sci. U.S.A. 113, 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kint C., Verstraeten N., Hofkens J., Fauvart M., and Michiels J. (2014) Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit. Rev. Microbiol. 40, 207–224 [DOI] [PubMed] [Google Scholar]
- 27. Feng B., Mandava C. S., Guo Q., Wang J., Cao W., Li N., Zhang Y., Zhang Y., Wang Z., Wu J., Sanyal S., Lei J., and Gao N. (2014) Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLos Biol. 12, e1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trach K., and Hoch J. A. (1989) The Bacillus subtilis spo0B Stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171, 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kok J., Trach K. A., and Hoch J. A. (1994) Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J. Bacteriol. 176, 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okamoto S., and Ochi K. (1998) An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol. Microbiol. 30, 107–119 [DOI] [PubMed] [Google Scholar]
- 31. Datta K., Skidmore J. M., Pu K., and Maddock J. R. (2004) The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol. Microbiol. 54, 1379–1392 [DOI] [PubMed] [Google Scholar]
- 32. Foti J. J., Schienda J., and Sutera V. A Jr., and Lovett S. T. (2005) A bacterial G protein-mediated response to replication arrest. Mol. Cell. 17, 549–560 [DOI] [PubMed] [Google Scholar]
- 33. Raskin D. M., Judson N., and Mekalanos J. J. (2007) Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 104, 4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kint C. I., Verstraeten N., Wens I., Liebens V. R., Hofkens J., Versées W., Fauvart M., and Michiels J. (2012) The Escherichia coli GTPase ObgE modulates hydroxyl radical levels in response to DNA replication fork arrest. FEBS J. 279, 3692–3704 [DOI] [PubMed] [Google Scholar]
- 35. Wout P., Pu K., Sullivan S. M., Reese V., Zhou S., Lin B., and Maddock J. R. (2004) The Escherichia coli GTPase CgtA cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthase/hydrolase. J. Bacteriol. 186, 5249–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang M., Sullivan S. M., Wout P. K., and Maddock J. R. (2007) G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 189, 6140–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Persky N. S., Ferullo D. J., Cooper D. L., Moore H. R., and Lovett S. T. (2009) The Obg/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 73, 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verstraeten N., Knapen W. J., Kint C. I., Liebens V., Van den Bergh B., Dewachter L., Michiels J. E., Fu Q., David C. C., Fierro A. C., Marchal K., Beirlant J., Versées W., Hofkens J., Jansen M., Fauvart M., and Michiels J. (2015) Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell. 59, 9–21 [DOI] [PubMed] [Google Scholar]
- 39. Dosztányi Z., Csizmok V., Tompa P., and Simon I. (2005) IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 40. Dosztányi Z., Csizmók V., Tompa P., and Simon I. (2005) The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347, 827–839 [DOI] [PubMed] [Google Scholar]
- 41. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., and Wittinghofer A. (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 42. Rambo R. P., and Tainer J. A. (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chatterjee A., and Datta P. P. (2015) Two conserved amino acids of juxtaposed domains of a ribosomal maturation protein CgtA sustain its optimal GTPase activity. Biochem. Biophys. Res. Commun. 461, 636–641 [DOI] [PubMed] [Google Scholar]
- 44. Ash M. R., Guilfoyle A., Clarke R. J., Guss J. M., Maher M. J., and Jormakka M. (2010) Potassium-activated GTPase reaction in the G protein-coupled ferrous iron transporter B. J. Biol. Chem. 285, 14594–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Achila D., Gulati M., Jain N., and Britton R. A. (2012) Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis. J. Biol. Chem. 287, 8417–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Foucher A. E., Reiser J. B., Ebel C., Housset D., and Jault J. M. (2012) Potassium acts as a GTPase-activating element on each nucleotide-binding domain of the essential Bacillus subtilis EngA. PLoS ONE 7, e46795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fislage M., Brosens E., Deyaert E., Spilotros A., Pardon E., Loris R., Steyaert J., Garcia-Pino A., and Versées W. (2014) SAXS analysis of the tRNA-modifying enzyme complex MnmE/MnmG reveals a novel interaction mode and GTP-induced oligomerization. Nucleic Acids Res. 42, 5978–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 55. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 56. Painter J., and Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 57. Painter J., and Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 58. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. David G., and Perez J. (2009) Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 42, 892–900 [Google Scholar]
- 60. Shkumatov A. V., and Strelkov S. V. (2015) DATASW, a tool for HPLC-SAXS data analysis. Acta Crystallogr. D Biol. Crystallogr. 71, 1347–1350 [DOI] [PubMed] [Google Scholar]
- 61. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rambo R. P., and Tainer J. A. (2011) Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Svergun D. I. (1992) Determination of the regularization parameter in indirect: transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 64. Barberato C., Koch M. H. J., and Svergun D. I. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 65. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., and Šali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 66. Tria G., Mertens H. D., Kachala M., and Svergun D. I. (2015) Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shkumatov A. V., Chinnathambi S., Mandelkow E., and Svergun D. I. (2011) Structural memory of natively unfolded tau protein detected by small-angle X-ray scattering. Proteins 79, 2122–2131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.