Abstract
Helicases play a critical role in processes such as replication or recombination by unwinding double-stranded DNA; mutations of these genes can therefore have devastating biological consequences. In humans, mutations in genes of three members of the RecQ family helicases (blm, wrn, and recq4) give rise to three strikingly distinctive clinical phenotypes: Bloom syndrome, Werner syndrome, and Rothmund-Thomson syndrome, respectively. However, the molecular basis for these varying phenotypic outcomes is unclear, in part because a full mechanistic description of helicase activity is lacking. Because the helicase core domains are highly conserved, it has been postulated that functional differences among family members might be explained by significant differences in the N-terminal domains, but these domains are poorly characterized. To help fill this gap, we now describe bioinformatics, biochemical, and structural data for three vertebrate BLM proteins. We pair high resolution crystal structures with SAXS analysis to describe an internal, highly conserved sequence we term the dimerization helical bundle in N-terminal domain (DHBN). We show that, despite the N-terminal domain being loosely structured and potentially lacking a defined three-dimensional structure in general, the DHBN exists as a dimeric structure required for higher order oligomer assembly. Interestingly, the unwinding amplitude and rate decrease as BLM is assembled from dimer into hexamer, and also, the stable DHBN dimer can be dissociated upon ATP hydrolysis. Thus, the structural and biochemical characterizations of N-terminal domains will provide new insights into how the N-terminal domain affects the structural and functional organization of the full BLM molecule.
Keywords: DNA helicase, enzyme kinetics, genetic disease, protein self-assembly, X-ray crystallography, dimerization helical bundle, small angle X-ray scattering
Introduction
Helicases are the ubiquitous enzymes that unwind double-stranded DNA (dsDNA) into single-stranded DNA (ssDNA) during cellular processes, such as replication, transcription, or repair (1, 2). RecQ family helicases, which are named after the recQ gene of E. coli, play essential roles in preserving genome stability from bacteria to higher eukaryotes, especially in DNA damage response and repair processes (3–5). There are five dominant RecQ family members in humans: BLM, WRN, and RecQ4, along with RecQ1 and RecQ5. Mutations in the former three proteins lead to high genomic instabilities and cause Bloom syndrome (BS),3 Werner syndrome, and Rothmund-Thomson syndrome, respectively, each of which displays a unique clinical phenotype (6, 7).
The BLM helicase core is flanked by ancillary N- and C-terminal domains (Fig. 1A). Sequence alignment analysis reveals that three regions are highly conserved in the helicase core and can be identified in most bacterial and eukaryotic RecQ proteins: the helicase domain, the RecQ carboxyl-terminal (RQC) domain, and the Helicase and RNase D C-terminal domain (HRDC) (8, 9). The helicase domain is essential for ATP binding and hydrolysis and defines the RecQ family. The RQC domain is also restricted to RecQ family members and is considered important for both the structural integrity of the protein and dsDNA binding; it might also have a role in mediating interactions with other proteins (10, 11). The HRDC probably has an auxiliary role in nucleic acid binding and is found in other helicase families and in some RNA nucleases (12, 13).
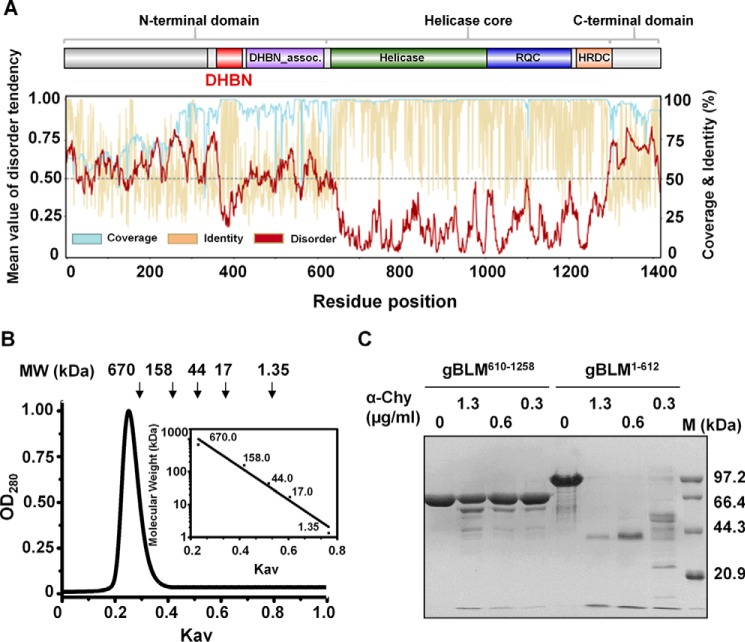
FIGURE 1.
DHBN is a highly conserved domain within the N-terminal domains of the vertebrate BLMs. A, sequences scanning and structure conservation of the vertebrate BLMs. The BLM helicase core is flanked by ancillary N-terminal domain and C-terminal domain. DHBN (red), DHBN_assoc. (violet), helicase (deep green), RQC (blue), and HRDC (orange) domains are indicated with different colors. Disorder tendency of each position is the average value of all available sites in the same column provided by IUPred prediction of intrinsically unstructured regions of proteins. B, size exclusion chromatography analysis of gBLM(1–612) by Superdex 200 10/300 GL. Inset, elution profiles of calibration kit proteins on the same column. The apparent molecular mass of gBLM(1–612) was 800 kDa, more or less that of a dodecamer. C, comparative limited proteolysis of gBLM(610–1258) and gBLM(1–612). A 6.5 μm concentration of each gBLM protein was cleaved with α-chymotrypsin at 0, 0.3, 0.6, and 1.3 μg/ml at 18 °C for 1 h, respectively. The corresponding molecular weights of protein marker M are shown on the right.
Because the helicase core of RecQ family helicases is highly conserved with similar DNA unwinding activities in vitro, a fundamental question that arises is how a deficiency or mutation of a particular RecQ helicase gives rise to strikingly different clinical syndromes. Furthermore, a large amount of literature has indicated that many differences, including characteristic biochemical, cellular, genetic, and organismal consequences, are observed among different RecQ helicase mutants (14, 15). Given that the ATPase and unwinding activities are common to all DNA helicases, some other physical properties of each helicase must govern its functional specificity.
Besides the well conserved helicase core domain, the N-terminal domains present in RecQ enzymes are different and may confer important and specific functions (10, 16). For example, the N-terminal domain of WRN and its orthologues are unique in containing an exonuclease function (17). In this regard, elucidation of the structure and function of N-terminal domains of other helicases may be one of the most important strategies to provide some new insights for understanding the functional specificity of a given RecQ helicase in cells.
BS is an autosomal recessive genetic disorder characterized by short stature and a skin rash that develops after sun exposure (18). BLM protein has been extensively characterized due to the fact that patients with BS are predisposed to the development of many cancer types (19–21). Investigations have shown that the N-terminal domain of human BLM interacts with a broad range of accessory proteins, such as ATM, ATR (22), 53BP1 (23), Exo1 (24), BRCA1 complex (25), DNA2-RPA-MRN (MRE11, RAD50, NBS1) complex (26), and TOP3α (27), and these interactions are mainly controlled by a phosphorylation process to eliminate illegitimate events (28). Despite its biological importance, the N-terminal structural characterization remains to be elucidated. To facilitate the study, we have expressed and purified fragments in the N-terminal domain of human BLM protein and orthologous BLM proteins from Gallus gallus (gBLM) and Pelecanus crispus (pBLM). In this work, by a combination of bioinformatics, biochemical, and structural approaches, we reveal that the N-terminal domain of BLM protein is highly divergent both in sequence and structure. In general, the N-terminal domain is loosely structured and potentially lacks conserved global three-dimensional structures. However, a highly conserved DHBN forms a dimer and is implicated in oligomerization of BLM protein. Moreover, the unwinding activity of gBLM is regulated by the different oligomers, and the dimer is the basic unit to form a hexamer. These findings shed new light on the enzymatic properties and structure of BLM protein and deepen our understanding of the molecular basis of the functional specificity of BLM in cells.
Results
Bioinformatics analysis reveals that DHBN is the only highly conserved domain in N-terminal domains of the vertebrate BLM homologues
To probe whether the N-terminal domains are evolutionarily conserved in sequence and structure, we performed multiple-sequence alignment of most homologous proteins of BLM (BLMs) in the reference proteome database (see “Experimental Procedures”) and constructed a phylogenetic tree (supplemental Fig. S1). These data reveal that the N-terminal domain of BLM displays three striking features. (i) It lacks sequence conservation among orthologs. By scanning over 78 BLM sequences from different species, we found that the sequences of the helicase core region were highly conserved with an average 81.7% identity and 89.9% similarity, whereas those of the N-terminal domain varied greatly within and across species. Moreover, the divergence of the N-terminal sequences was particularly evident in invertebrates. For example, the sequence identity and similarity in flies were as low as 9.5 and 17.0%, respectively. (ii) According to the calculated coverage, identity of each residue position based on the alignment and disordered tendency (Fig. 1A), we found that the N-terminal domain contained a large proportion of random coils but a low proportion of α-helices or/and β-strands. Furthermore, the distribution of α-helices and β-strands was highly random among the N-terminal sequences. Therefore, the N-terminal domain is poorly structured or intrinsically unstructured. Indeed, the purified N-terminal domain of gBLM (gBLM(1–612)) was eluted as a single peak in the dead volume, indicating that it is a high order oligomer (≥800 kDa) (Fig. 1B). Furthermore, limited proteolysis of the N-terminal domain and helicase core showed that the helicase core fragment was more resistant to protease digestion than the N-terminal domain (Fig. 1C), indicating loose structural organization of the N-terminal domain. (iii) Despite the N-terminal domain lacking the general sequence and structural conservation, a protein fragment harboring about 50–60 amino acids with a potentially 2–3 α-helix conformation is highly conserved (Fig. 2A), which contains the previously named BDHCT domain (Bloom's syndrome DEAD helicase C-terminal domain) (29). In this study, we propose to refer it as the dimerization helical bundle in N-terminal domain (DHBN) to emphasize that this domain with helical bundle structure in the N-terminal domain but not in the C-terminal domain regulates the protein dimerization. Phylogenetic analysis reveals that many DHBNs locate in the middle of the N-terminal domain, whereas those of P. crispus and Calypte anna exist just at the beginning of the N terminus.
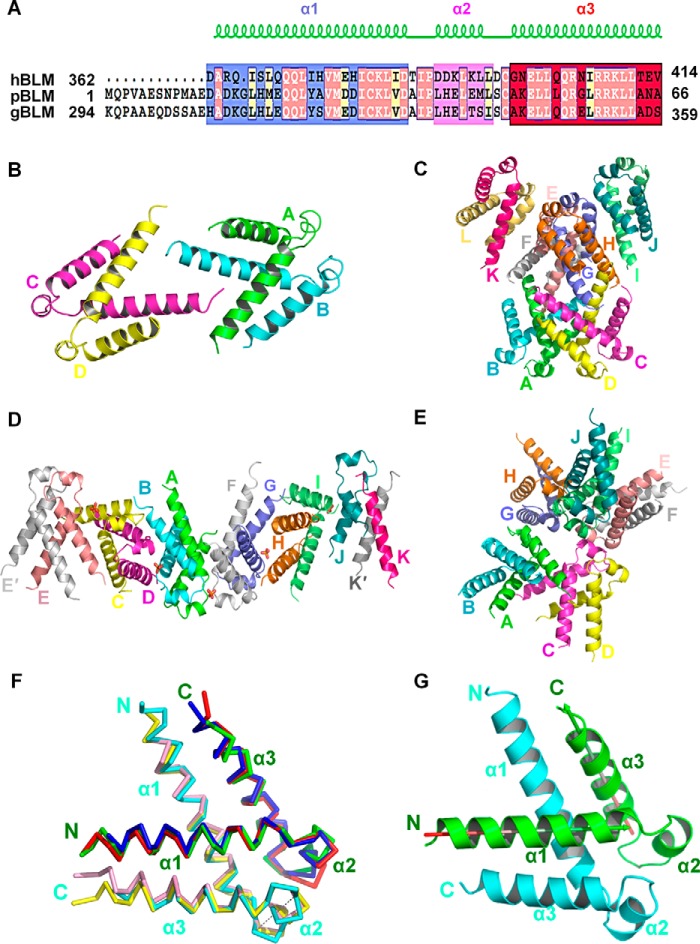
FIGURE 2.
Structure of the DHBN dimer. A, sequence alignment of DHBNs of hBLM, pBLM, and gBLM proteins. The detailed amino acid sites and the resolved secondary structures are displayed. B, asymmetric unit of hDHBN in space group C2221. C, asymmetric unit of hDHBN in space group P62. D, asymmetric unit of gDHBN in space group P21212. E, asymmetric unit of pDHBN in space group P21. F, orthologous DHBN dimers have a conserved structure. hDHBN (green/cyan), gDHBN (red/yellow), and pDHBN (blue/pink) are superimposed. The average r.m.s.d. over 96 superimposed Cα is 1.8 Å. G, structure of hDHBN dimer (cyan and green). The direction of helix α1 and helix α3 is shown as arrows on the green monomer. The angle between helix α1 and helix α3 is 120°.
Altogether, according to the above bioinformatics and biochemical analysis, along with the general rule that proteins with similar secondary elements (α-helices and β-strands) adopt similar three-dimensional structures (30, 31), we conclude that the N-terminal domain of BLM is loosely structured, and the overall three-dimensional structures should not be conserved within the N-terminal domains, except for the highly conserved DHBN.
Structure determination of DHBNs
hDHBN (human BLM (362–414)) crystals were resolved in two space groups, C2221 and P62 (Fig. 2A and Table 1). The asymmetric unit of hDHBN in space groups C2221 and P62 contains 4 and 12 molecules, respectively (Fig. 2, B and C). The common unit between the two space groups is a dimer. Two dimers and six dimers are present in the asymmetric unit in C2221 and P62, respectively. The two dimers in C2221 interact with each other by means of loose contacts between the two centrally located N termini. Although this kind of association is also present in P62 crystal form, the dimer of dimers cannot be exactly superimposed, and different symmetry is found for the six dimers. Dimers association is thus dependent on crystal packing.
TABLE 1.
Data collection and refinement statistics of the three BLM DHBN proteins
| Parameter | hDHBN |
gDHBN SeMet | pDHBN Native | |
|---|---|---|---|---|
| Native | Native | |||
| PDB code | 5LUP | 5MK5 | 5LUT | 5LUS |
| Phasing method | SIRAS | Molecular replacement | SAD | Molecular replacement |
| Data collection | ||||
| Space group | P 62 | C 2 2 21 | P 21 21 2 | P 21 |
| Cell dimensions a, b, c (Å) | 132.8, 132.8, 65.0 | 34.1, 144.7, 96.8 | 76.7, 230.8, 50.9 | 62.2, 72.4, 79.4 |
| Wavelength (Å) | 0.9754 | 1.7 | 0.9754 | 0.9754 |
| Resolution (Å) | 43.47–2.032 (2.105–2.032)a | 27.84–2.16 (2.237–2.16) | 54.30–2.72 (2.817–2.718) | 36.20–1.43 (1.484–1.433) |
| Total reflections | 338,112 (11,778) | 68,723 (7278) | 292,931 (30,951) | 34,0631 (16,186) |
| Unique reflections | 41,409 (3643) | 12,345 (1292) | 25,190 (2297) | 105,436 (6793) |
| Completeness (%) | 0.98 (0.87) | 0.93 (0.99) | 0.98 (0.93) | 0.83 (0.54) |
| Mean I/σ(I) | 18.34 (1.84) | 19.63 (3.75) | 18.81 (2.32) | 18.56 (2.26) |
| Rmerge | 0.07402 (0.5345) | 0.08213 (0.1837) | 0.1069 (1.896) | 0.03409 (0.3467) |
| CC1/2 | 0.999 (0.767) | 0.996 (0.988) | 0.999 (0.826) | 0.998 (0.893) |
| Phasing | ||||
| No. of sites | 22 | 12 | ||
| FOM | 0.17 | 0.21 | ||
| FOM after solvent flattening | 0.62 | 0.73 | ||
| No. of molecules in the asymmetric unit | 12 | 4 | 11 | 10 |
| Data refinement | ||||
| Reflections (Rwork) | 41,411 (3643) | 12,342 (1293) | 24,903 (2295) | 105,372 (6786) |
| Reflections (Rfree) | 1987 (167) | 1064 (112) | 1234 (136) | 5243 (343) |
| Rwork | 0.1818 (0.2628) | 0.2238 (0.2540) | 0.2334 (0.3184) | 0.1574 (0.2256) |
| Rfree | 0.2246 (0.3253) | 0.3012 (0.3294) | 0.2828 (0.3740) | 0.1712 (0.2563) |
| Solvent | 42.92 | 47.26 | 64.86 | 40.99 |
| Protein residues | 617 | 205 | 530 | 494 |
| B-factor | 40.23 | 43.0 | 76.21 | 33.02 |
| r.m.s.d., bonds (Å) | 0.007 | 0.002 | 0.009 | 0.009 |
| r.m.s.d., angles (degrees) | 0.96 | 0.40 | 1.12 | 1.23 |
| Ramachandran favored (%) | 98 | 99 | 99 | 1e+02 |
| Ramachandran allowed (%) | 1.20 | 1.00 | 1.00 | 0.21 |
| Ramachandran disallowed (%) | 0.34 | 0 | 0 | 0 |
a Statistics for the highest resolution shell are shown in parentheses.
Meanwhile, to rule out the possibility that the above dimeric structure is an artifact of crystal packing, we characterized two additional DHBNs from G. gallus (gDHBN) and P. crispus (pDHBN), the latter of which is located at the N terminus rather than between the N terminus and helicase core (Fig. 2A). gDHBN crystals belong to space group P21212 with unit cell a = 76.7 Å, b = 230.8 Å, c = 50.9 Å, and α = β = γ = 90°. Native Patterson analysis showed the presence of a translational non-crystallographic symmetry (tNCS) vector, corresponding to 22% of the origin peak with coordinates (0.112, 0.5, 0). The structure was solved by SAD on a SeMet derivative. The tNCS may explain the high Rfree value (28.2%) at the end of refinement. The asymmetric unit contains 11 chains named A–K having slightly different lengths (Table 1 and Fig. 2D). Most chains encompass residues 310–359, but chains J and K, which are less well defined and built without side chains, are shorter, comprising residues 320–359 and residues 311–332, respectively. The chains are packed in a continuous arrangement in the asymmetric unit. Dimers are formed between molecules A and B, C and D, F and G, and H and I. Chain E forms a dimer with its crystallographic 2-fold symmetry mate E′. Chains J and K form a more complicated dimer; chain K consists of a single α-helix in interaction with chain J, but its crystallographic 2-fold symmetry mate K′ is also in contact with chain J. Therefore, this unusual asymmetric unit contains 10.5 molecules: dimers of A and B, C and D, F and G, and H and I contribute 8 molecules; dimer E-E′ contributes 1 molecule by crystallographic 2-fold axis; dimer K-J contributes 1.5 molecules; and molecule K and K′ mimic an entire monomer.
Additionally, pDHBN was crystallized in space group P21 with unit cell a = 62.2 Å, b = 72.4 Å, c = 79.2 Å, α = 90°, β = 99.4°, γ = 90°. The structure was solved by molecular replacement with hDHBN structure. The asymmetric unit contains 10 molecules, which form five dimers between chains A and B, C and D, E and F, G and H, and I and J (Table 1 and Fig. 2E). Contacts between molecules in the asymmetric unit and crystallographic symmetry mates are different from hDHBN and gDHBN.
Thus, we have crystallized DHBN orthologous domains in four crystal forms, which have distinct crystal packing, and the common unit among all the crystals of DHBN orthologs is the dimer structure, which is extremely conserved. Superimposition of the different hDHBN, gDHBN, and pDHBN dimers gives an average r.m.s.d. of 1.8 Å over 96 Cα (Fig. 2F). Altogether, these results indicate that the dimer is the basic unit of biological structure. Statistics processing and refinement data of the above three high resolution DHBN structures are summarized in Table 1.
DHBN dimer is mainly stabilized by hydrophobic interactions
As shown in Fig. 2F, each DHBN dimer is composed of two “head-to-tail” V-shaped monomers formed of three α-helices. The helix α1 (362–384 for hDHBN, 306–329 for gDHBN, and 20–36 for pDHBN) and helix α3 (397–414 for hDHBN, 342–359 for gDHBN, and 49–66 for pDHBN) are separated by a junction formed by the short α2 helix (388–393 for hDHBN, 333–338 for gDHBN, and 40–45 for pDHBN). The angle between α1 and α3 is 120° (Fig. 2G). DHBN monomer has an internal pseudosymmetry; when the monomers are superimposed in the N to C orientation on the reverse C to N orientation, the r.m.s.d. is 3 Å for the Cα. Therefore, it seems at a first glance that the two monomers are related only by a 10-Å translation. However, the two monomers are arranged in an anti-parallel manner and are related by a 2-fold axis rotation. Thus, the interface is symmetric (isologous), with each monomer engaging the same interface in the dimer.
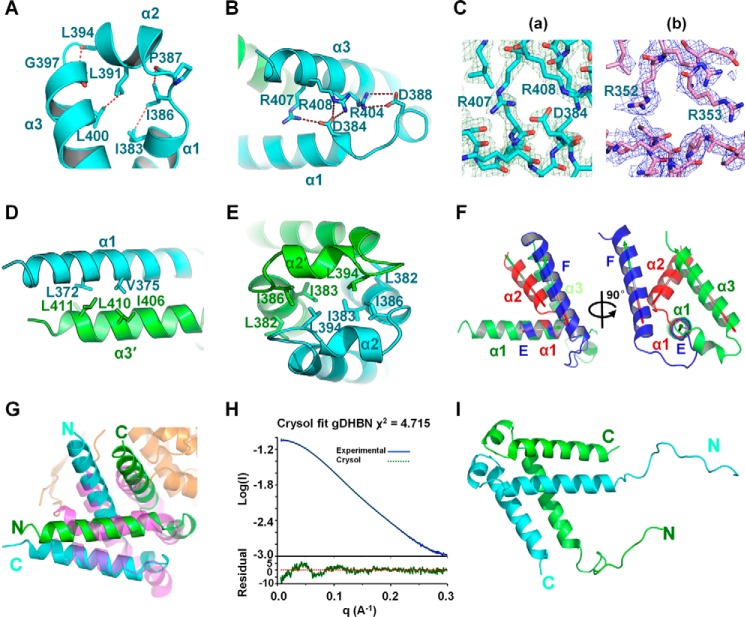
One helix in one monomer is packed against another one of the other monomer; α1 of molecule A is packed against α3′ of molecule B, and α2 helices are packed against each other. This dimer has, therefore, an extensive buried surface of an average of 1300 Å2 for all DHBN structures (Fig. 2F). The comparison of DHBN orthologous structures reveals conserved features. The V-shaped conformation of DHBN monomer is stabilized by several interactions. A set of hydrophobic amino acids residues, including Ile-386, Pro-387, Leu-391, Leu-394, Gly-397, and Leu-400 (hDHBN numbering), constitute a hydrophobic core at the junction of α1, α2, and α3 helices (Fig. 3A). The relative position of α1 and α3 is further stabilized by the conserved acidic residues Asp-384 and Asp-388, which are in electrostatic interaction with conserved basic residues Arg-404, Arg-407, and Arg-408 in α3 (Fig. 3B), and the electrostatic interactions between Asp-384 and Arg-407 and between Asp-384 and Arg-408 are shown with the experimental electron density map obtained by SIRAS KI phasing in the final refined hDHBN model (contour level is at 2σ) (Fig. 3C (a)), and the same region of gDHBN is also representatively displayed (Fig. 3C (b)). DHBN dimer formation is mediated by hydrophobic interactions between the α-helices. α1 interacts with α3′ of the other monomer by hydrophobic interactions involving Leu-372 and Val-375 in α1 and Ile-406, Leu-410, and Leu-411 in α3′ (Fig. 3D). The structure does not have the characteristics of a canonical coiled-coil, because the specific spacing of hydrophobic residues is not present. Interactions between α2 and α2′ involve residues Leu-382, Ile-383, Ile-386, and Leu-394 of both monomers (Fig. 3E).
FIGURE 3.
DHBN dimer stabilization and characterization by SAXS. A, a set of hydrophobic interactions at the junction of α1, α2, and α3 helices. The distances (Å) between each two hydrophobic residues are indicated. B, the DHBN monomer is stabilized by electrostatic interactions between α1 and α3. C, a, representative experimental electron density map of electrostatic interactions in B obtained by SIRAS KI phasing with the final refined hDHBN model. Contour level is at 2σ. b, representative experimental SAD electron density map of gDHBN in the same zone, which is contoured at 2σ. D, hydrophobic interactions between α1 and α3′. E, interactions between α2 and α2′. The isologous interface in DHBN dimer involves D and E. F, superimposition of hDHBN monomer (green) with EF-hand (blue) and helix-turn-helix (red). The view (right) rotated by 90° shows that the helices are not located at the same position. G, superimposition of hDHBN dimer (cyan and green) on Syngap GAPex domain (purple). H, DHBN dimer was characterized in solution by HPLC SAXS. The crystal structure was fitted into the SAXS curve (χ2 = 4.72) by introducing flexibility in the N-terminal part that is not seen in the crystallographic structure. I, SAXS model of gDHBN without ab initio envelope. All of the amino acids are shown in one-letter code.
The V-shape with the interhelical angle of 120° between the helices α1 and α3 is reminiscent of the helix-turn-helix (HTH) (32) and EF-hand motifs (33), which are 110° (Trp repressor, PDB code 1TRO) and 117° (S100A10, PDB code 1BT6), respectively. However, when the structures are superimposed on the first helix, the second helix has a different spatial location. The DHBN has, therefore, a different conformation from those of the HTH and EF-hands (Fig. 3F). A search of structural homologs of the dimeric DHBN with DALI gave a significant hit (Z-score 7.4) with Syngap, a Ras GTPase-activating protein (PDB code 3BXJ) (Fig. 3G). The homolog structural elements found in Syngap are in the GAPex domain between GTPase domain and C2 domain (34). When superimposed, the r.m.s.d. is 3.7 Å calculated on 94 Cα. This result suggests that the DHBN may possess an intrinsic structural property that is involved in protein-protein interaction either intermolecularly or intramolecularly.
Characterization of DHBN dimer in solution
To distinguish whether the observed molecular association in the asymmetric unit reflects real quaternary structure or a crystal packing artifact, gel filtration chromatography, dynamic light scattering, and small angle X-ray scattering (SAXS) were performed to gain insight into the intrinsic oligomeric property of gDHBN in solution. Surprisingly, on gel filtration chromatography, the gDHBN was eluted at the fraction corresponding to a tetramer (27.6 kDa) (the crystal structures are compatible with a dimer of dimer); similarly, the hydrodynamic radius of gDHBN determined by dynamic light scattering (DLS) also displays an apparent molecular mass of 26.1 kDa. To clarify the situation, the solution conformation of gDHBN was further determined by SAXS with SEC-HPLC coupled with SAXS data collection (Fig. 3H). A single peak was eluted, corresponding exactly to the molecular mass of the dimer. The modeling results demonstrate that gDHBN has a conformation in a triangle-like conformation and fits well with our crystal model with a final χ2 of 4.72 (Fig. 3, H and I). All of the SAXS parameters are summarized in Table 2. Therefore, gDHBN has a conformation in solution close to the one observed in the crystal structure of dimeric form gDHBN. The higher apparent molecular weight deduced from gel filtration and DLS might be caused by the unstructured parts of gDHBN in solution, which create the non-spherical dimeric structure.
TABLE 2.
SAXS data collection and processing of gDHBN
| Parameters | Values |
|---|---|
| Data collection parameters | |
| Instrument | SWING |
| Beam geometry (mm) | 0.4 × 0.1 |
| Wavelength (Å) | 1.03 |
| q range (Å−1) | 0.007–0.5 |
| Exposure time (s)/no. of frames | 1/100 |
| Concentration range (mg/ml) | 10 |
| Temperature (K) | 288 |
| Structural parameters | |
| I(0) (cm−1) (from P(r)) | 0.091 |
| Rg (Å) (from P(r)) | 20.9 |
| I(0) (cm−1) (from Guinier) | 0.091 ± 2.7.10−5 |
| Rg (Å) (from Guinier) | 20.4 ± 0.028 |
| Dmax (Å) | 71.53 |
| Porod estimate (Å3) | 28,041 |
| Molecular mass determination | |
| Partial specific volume (cm3·g−1) | 0.745 |
| Contrast (Δρ × 1010 cm−2) | 2.736 |
| Mr (from I(0)) | 15,500 |
| Calculated monomeric Mr from sequence | 7787.88 |
| Data processing | |
| Primary data reduction | FOXTROT |
| Data processing | PRIMUS |
| Ab initio analysis | DAMMIF |
| Number of models | 50 |
| Model χ2 | 2.196 ± 0.020 |
| Validation and averaging | DAMAVER |
| Normalized spatial discrepancy | 0.467 ± 0.208 |
| Rigid body modeling | DADIMODO |
| Computation of model intensities | CRYSOL |
| Model χ2 | 4.715 |
DHBN plays an essential role in oligomerization and regulates unwinding activity of BLM
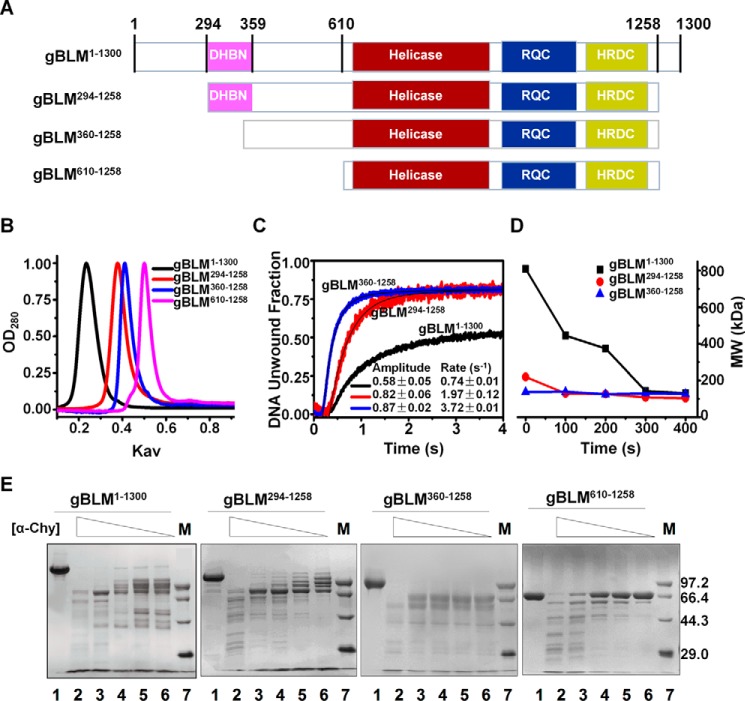
To facilitate our studies in understanding the function of the DHBN, we expressed and purified G. gallus BLM protein (gBLM(1–1300)), which shares 80.3% sequences identity with human BLM in the helicase core and 25.2% sequence identity in the N-terminal domain, respectively. The purified gBLM(1–1300) exhibits biochemical properties and catalytic activities (DNA binding, unwinding, and ATPase activities) comparable with those of human BLM protein (supplemental Table S1). To evaluate the functional effects of the DHBN, the N-terminal amino acids were successively truncated, which resulted in three mutants containing the DHBN and DHBN-associated domain (gBLM(294–1258)), without the DHBN (gBLM(360–1258)), and with the entire N terminus truncated (gBLM(610–1258)), respectively (Fig. 4A). We first determined the oligomeric states of those proteins by gel filtration chromatography (Fig. 4B). The apparent molecular weights of gBLM(294–1258) and gBLM(610–1258) were determined with reasonable accuracy and corresponded to dimer and monomer, respectively, whereas gBLM(1–1300) was eluted in the dead volume, indicating that this protein is a higher order oligomer with a molecular mass of >680 kDa (the dead volume). Furthermore, the molecular weight of gBLM(360–1258) appears to be higher than that of a monomer but lower than that of a dimer. Considering the fact that the deletion of DHBN from the N-terminal domain of BLM might cause the N-terminal structure to be more loose (Fig 4E) and give out an unusual elution profile, gBLM(360–1258) was considered as a monomer.
FIGURE 4.
gDHBN plays an essential role in oligomerization of gBLM helicase and DNA unwinding regulation. A, schematic representation of gBLM(1–1300) and three N-terminal truncated gBLM proteins. B, size exclusion chromatography analysis of the above four gBLM truncates. The experiment was performed and analyzed as described under “Experimental Procedures.” C, multiple-turnover unwinding kinetics of gBLM(1–1300), gBLM(294–1258), and gBLM(360–1258). A 100 nm concentration of each gBLM truncate protein and 4 nm DNA, 1.0 mm ATP were simultaneously preincubated at 25 °C for 5 min, respectively. Data were collected and fitted as described under “Experimental Procedures.” D, kinetics of conformational transition of the full-length and truncated gBLM proteins determined by DLS. The assays were performed as described under “Experimental Procedures.” E, comparative limited proteolysis of gBLM(1–1300), gBLM(294–1258), gBLM(360–1258), and gBLM(610–1258). A 6.5 μm concentration of each gBLM truncate protein was cleaved with α-chymotrypsin at 0, 5.0, 2.5, 1.3, 0.6, and 0.3 μg/ml (lanes 1–6) at 18 °C for 1 h, respectively. The corresponding protein marker M is shown in lane 7.
In accordance with the above results, DLS analysis further revealed that those proteins were monodisperse in solution and were characterized by hydrodynamic radii of 10.42, 5.96, and 4.81 nm, which corresponded to the molecular masses of a hexamer gBLM(1–1300) (810 kDa), dimer gBLM(294–1258) (220 kDa), and monomer gBLM(360–1258) (138 kDa), respectively (Table 3). The above results suggest that the gDHBN plays an important role in oligomerization, and essentially in the dimerization process of BLM protein.
TABLE 3.
DLS analysis of the gBLM truncate proteins treated with ATP and its analogues
The DLS determination was performed at 25 °C by a DynaPro Nano Star instrument, and the buffer used was 25 mm Tris-HCl, pH 7.5, 80 mm NaCl, 3 mm MgCl2, 1 mm DTT, and 23-nucleotide ssDNA whose concentration was equal to that of each protein assayed. Each gBLM protein was treated with or without 2 mm ATP or its analogues (AMPNP and ATPrS), and treatment data were collected from 10 repeats.
| gBLM | Radius | Mass | Molecular mass |
|---|---|---|---|
| nm | % | kDa | |
| gBLM(1–1300) | 10.42 | 99.5 | 810 |
| gBLM(1–1300) + 2 mm ATP | 4.62 | 62.2 | 121 |
| 11.89 | 36.9 | 1104 | |
| gBLM(1–1300) + 2 mm AMPNP | 10.32 | 99.5 | 793 |
| gBLM(1–1300) + 2 mm ATPrS | 9.31 | 99.6 | 623 |
| gBLM(294–1258) | 5.96 | 99.9 | 220 |
| gBLM(294–1258) + 2 mm ATP | 4.4 | 99.4 | 108 |
| gBLM(294–1258) + 2 mm AMPNP | 5.87 | 99.8 | 212 |
| gBLM(294–1258) + 2 mm ATPrS | 5.92 | 99.7 | 216 |
| gBLM(360–1258) | 4.81 | 96.8 | 138 |
| gBLM(360–1258) + 2 mm ATP | 4.72 | 100 | 130 |
| gBLM(360–1258) + 2 mm AMPNP | 4.90 | 99.9 | 139 |
| gBLM(360–1258) + 2 mm ATPrS | 4.84 | 99.9 | 135 |
To further confirm whether the full-length gBLM could be dissociated upon the addition of ATP or non-hydrolyzable ATP analogues as observed previously with human BLM, the oligomeric states of the above three proteins were investigated in the presence of ATP or non-hydrolyzable ATP analogues. As shown in Table 3, both the hexameric and dimeric gBLM proteins are dissociated into monomer upon the addition of 2 mm ATP, as judged from the determined hydrodynamic radii, but not from the ATP analogues (AMPNP and ATPrS), and the phenomenon is similar to the previous observations with human BLM by Gyimesi et al. (35) and Xu et al. (36).
Because oligomeric state may influence helicase activity or substrate specificity (37), we next measured the helicase activities of different oligomeric gBLM proteins. By assaying the helicase activity under multiple-turnover conditions with stopped flow, we found that whereas the dimer (gBLM(294–1258)) rapidly arrived at the same unwinding amplitude of the monomer (gBLM(360–1258)), it appeared that the putative hexamer (gBLM(1–1300)) took more time to achieve a comparable level (Fig. 4C), suggesting that the observed differences at a given time in unwinding amplitudes may reflect the extent of dissociation from oligomer to monomer. To confirm this, the kinetics of transition was measured with DLS in the presence of ATP (Fig. 4D). There is a global kinetic correlation between the increasing in unwinding amplitude and the extent of dissociation (Fig. 4, compare C and D). We noticed that the DLS-determined kinetics of dissociation lagged behind the unwinding rate. This is, at least partially, because the unwinding activity is much more sensitive than the DLS assay in which the protein quantity must be accumulated to a certain level to be detected.
To rule out the possibility that the observed variations in DNA unwinding activities of different constructions come from alterations in DNA binding and/or ATPase activities, both activities were determined in parallel with DNA unwinding assays under the same experimental conditions. Our results showed that no significant differences in either activity were observed among the full-length BLM and three truncated forms (supplemental Fig. S2 and Table S2). The above observations indicate that the DHBN may associate with the other N-terminal motif to regulate different oligomeric states of BLM and, consequently, influence the corresponding unwinding amplitude and rate.
To ascertain whether DHBN could stabilize the global structure, the above three proteins were submitted to proteolytic digestions. The results showed that both gBLM(1–1300) and gBLM(294–1258) were more resistant to the protease digestion than gBLM(360–1258), which was rapidly digested into the helicase core (Fig. 4D). Because both gBLM(1–1300) and gBLM(294–1258) contain the DHBN, which is absent in gBLM(360–1258), we conclude that DHBN may stabilize and/or enhance the structural stability of the N-terminal domains.
gBLM subunits do not display cooperativity in DNA unwinding
The pioneering works of Gyimesi et al. (35) have revealed that the oligomeric states of human BLM can be regulated with different structured DNA substrates. Although they show that there is no cooperativity among the active sites for ATP hydrolysis, it is not clear whether gBLM displays cooperativity in DNA unwinding and whether DHBN could influence the cooperativity.
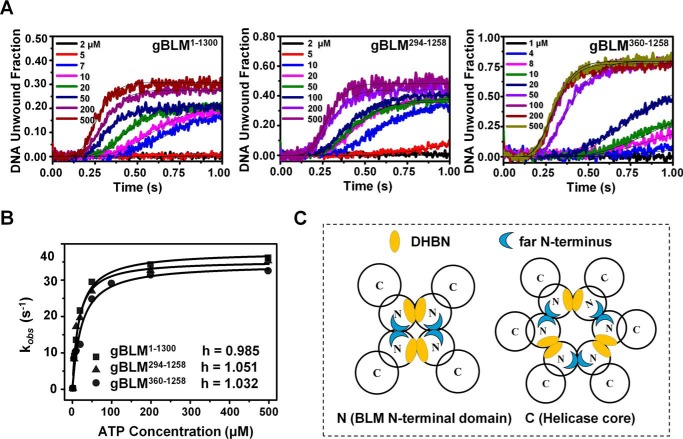
The experiments were performed under single-turnover kinetic experimental conditions with increasing ATP concentrations and the three types of gBLM: hexameric gBLM (gBLM(1–1300)), dimeric gBLM (gBLM(294–1258)), and monomeric gBLM (gBLM(360–1258)). The single-turnover unwinding kinetic data are presented in Fig. 5A. By fitting the data curves to the Hill equation, we obtained the unwinding rate constants at different ATP concentration. Interestingly, all data were best fitted by the Hill equation, yielding h = 0.99, 1.05, and 1.03 and Km = 17.11 ± 0.51, 14.96 ± 0.52, and 22.73 ± 0.66 for gBLM(1–1300), gBLM(294–1258), and gBLM(360–1258), respectively (Fig. 5B). These results are in accordance with the previous human BLM study that found that oligomeric gBLM helicase unwinds DNA substrate independently without cooperativity between the subunits (36).
FIGURE 5.
gDHBN can be dissociated upon ATP hydrolysis. A, single-turnover unwinding kinetics of the three gBLM truncate proteins at different ATP concentrations. 4 nm DNA substrate of D16S10 overhang duplex with 60 nm gBLM truncate and various concentrations of ATP with 2.5 mm dT56 were simultaneously preincubated, respectively. The unwinding data of the three gBLM truncate proteins were fitted by the sequential n steps equation with n = 10 as described under “Experimental Procedures.” The different ATP concentrations and curves are shown with distinct colors. B, fit of the observed unwinding rate constants of the three gBLM truncate proteins described above. The lines fit well to the Hill equation, kobs = kmax[ATP]h/([ATP]h + Kmh); all data were best fitted by the Hill equation, yielding h = 0.985, 1.051, and 1.032 and Km = 17.11 ± 0.51, 14.96 ± 0.52, and 22.73 ± 0.66 for gBLM(1–1300), gBLM(294–1258), and gBLM(360–1258), respectively, suggesting their monomer state in the unwinding processes. C, proposed oligomeric ring structures. The model shows that DHBN as the basic unit of a dimer collaborates with an unidentified far N terminus to constitute a tetramer or hexamer.
Discussion
Although biochemical and genetic evidence supports roles for all five human RecQ helicases in DNA replication, DNA recombination, and the biological responses to DNA damage, the question of how a particular helicase deficiency gives rise to a syndrome with a unique phenotype remains unanswered (38, 39). A simple sequence inspection reveals that those proteins differ greatly from one another in their N-terminal domains in sequence composition and length. Whereas the N-terminal domain of WRN has been shown to possess exonuclease activity, functional analysis of the N-terminal domain of BLM has been hindered in part by the lack of sequence conservation between orthologs. In agreement with previous observations (28, 40), our bioinformatics analysis reveals that sequences of the N-terminal domain of BLM orthologs are highly divergent and highly unstructured. In addition to the high disorder tendency, the quantities and the distributions of the predicted secondary structures (α-helices and β-strands) vary greatly among the BLM orthologs. It is unlikely that the N-terminal domains of BLM helicases possess a highly conserved three-dimensional structure. Furthermore, the previously identified residues implicated in phosphorylation (Thr-99, Thr-122, and Ser-144) (41) and sumoylation (Lys-317 and Lys-331) (42) of human BLM are localized in the random coil area and not conserved between hBLM and gBLM, suggesting that the structures and functions of the BLM N-terminal domain may be species-specific. More studies are required to understand this issue.
However, a highly conserved DHBN was identified in all analyzed vertebrate BLMs. The structure of the dimeric DHBN is unique. The space group of gDHBN is unusual, and this raises the question of how to distinguish biologically relevant contacts from crystal packing. To this end, we further crystallized Homo sapiens and P. crispus DHBNs. Although the space groups of the three proteins are different, the spatial conformation of dimer is the basic unit and highly structurally conserved, indicating that the dimer is a biologically relevant structure. This conclusion is consistent with the observations of Gyimesi et al. (35), in which most of the observed oligomers are dimers.
The dimer is formed by isologous (symmetric) association of two identical chains in an antiparallel manner. Each α-helix is packed by hydrophobic interactions with another opposite one. The hydrophobic nature of the residues involved in contacts is conserved in the DHBN orthologs and throughout sequences analyzed.
The above results may provide new insights into potential functions of the conserved DHBN. First, the main function of the DHBN should be to induce dimerization of BLM monomer. This is confirmed by the three different N-terminal truncation proteins of gBLM (Fig. 4A). Only in the presence of the DHBN does gBLM form a stable dimeric BLM (residues 294–1258). By combination with the far N terminus (within 1–293) and the DHBN, gBLM becomes a stable hexamer, as confirmed by gel filtration and DLS. Previous electron microscopy experiments have shown that the full-length hBLM protein can form hexameric ring-like structures in the absence of ATP and DNA (43). Interestingly, the same study also described the presence of a 4-fold symmetric structure that might represent an oligomeric form distinct from the hexameric ring (such as a tetramer or an octamer). Gel filtration studies showed that BLM has a native molecular mass of ∼700 kDa, consistent with an oligomeric state comprising tetramers or hexamers (43). Furthermore, by analyzing a purified N-terminal fragment of hBLM (residues 1–431), Beresten et al. (40) concluded that the isolated N terminus was a hexamer and was implicated in oligomerization of hBLM. Based on the previous studies and our structural and biochemical results presented here, we suggest that the DHBN is a key element for inducing BLM dimerization/oligomerization.
Second, the function of gDHBN is to mediate the DNA unwinding amplitude and rate through regulating dimer and hexamer formation (Fig. 4C). Assembly states ranging from monomers to hexamers have been described previously for various RecQ enzymes (10). The relationship between the different multimeric states and enzymatic activities of RecQ enzymes in the cell remains to be established. The results obtained for human RECQ1 helicase indicate that RecQ1 tetramer possesses HJ branch migration and DNA strand annealing activities that are not shared with the homodimer, which has only 3′-5′ fork-unwinding activity (44, 45). In this study, we clearly show that gDHBN controls the DNA unwinding amplitude and rate of monomer, dimer, and hexamer by regulating their oligomeric states. Gyimesi et al. (35) have combined electron microscopic and atomic force microscopic measurements and solution biophysical techniques to study the assembly state of BLM. Whereas they found that human BLM existed mainly as a monomer under extremely diluted conditions (1–100 nm), we both found that apo-BLMs existed as a high order oligomer in solution and that the oligomeric BLM was dissociated into monomer upon ATP hydrolysis, whereas the non-hydrolyzable ATP analogue (AMPNP) was unable to do so (Table 3) (35).
Third, DHBN should be an essential factor in regulating BLM oligomerization. This conclusion is based on the following results: (i) the structure of the isolated DHBN is a dimer; (ii) the BLM fragment containing DHBN (gBLM(294–1258)), but not that without DHBN (gBLM(360–1258)), behaves as a dimer (as shown by gel filtration and DLS); and (iii) by different biochemical and biophysical techniques, we and others have shown that the full length of BLM is a hexamer (36, 43). We therefore propose a tentative model in which DHBN as a basic dimeric unit collaborates with the far N terminus to form tetramer or hexamer (Fig. 5C). This model can perfectly explain why hBLM can be observed as tetramer or hexamer but not trimer by electron microscopy (43). The proteolytic profiles of hexamer, dimer, and monomer gBLM show that in the presence of the DHBN, the proteins are resistant to proteolytic digestion. This indicates that the highly conserved DHBN is a central structural domain, which makes the N-terminal domain fold into a more compact structure that is resistant to proteolysis digestion. However, we must assess that the above tetramer and hexamer models are not constructed based on experimental structural information; they are just referenced from our DHBN dimerization structure. Therefore, the models are speculative.
Finally, we reveal that both hBLM and gBLM share the same property, that the hexamer is dissociated into monomer mainly upon ATP hydrolysis. Interestingly, it was reported that the hexameric WRN was also dissociated into monomer to catalyze DNA unwinding (46). In addition to the biological meaning of the BLM dissociation phenomenon, the underlying molecular mechanisms remain largely elusive. For example, how does ATP hydrolysis weaken gDHBN dimer interaction and lead to the dissociation? How was this ATP hydrolysis signal transmitted across a large distance within the BLM molecule from the ATP binding sites to the dimerization positions?
Experimental Procedures
Bioinformatics analysis of the conservation of the DHBN
All BLM homologous protein sequences have been downloaded from the Reference Proteomes database by hmmsearch. First, the sequences were aligned with the online MAFFT program after the truncated sequences were removed manually. Then the maximum likelihood phylogenetic tree was constructed by PhyML (JTT model, default parameters) (47), and the final tree with domain organization information was manipulated and rendered with evolview (48, 49). The disorder tendency of each residue of each sequence was calculated by IUPred, a web-based tool for the prediction of intrinsically unstructured regions of a protein. The coverage, identity, and average values of disorder tendency of each residue site of the multiple-sequence alignment were integrated with python scripts.
Plasmid construction, protein expression, and purification
According to the NCBI amino acid sequences of G. gallus bloom syndrome protein (accession number NP_001007088.2, gBLM for short), gBLM(1–1300) without the nuclear localization signal was amplified and constructed into pET21a-sumo vector. Then by taking the positive pET21a-sumo gBLM (1–1300) as the template, gBLM(1–612), gBLM(294–1258), gBLM(360–1258), and gDHBN(294–359) were amplified by the corresponding forward and reverse primers and constructed into pET15b-sumo plasmids and transformed into the expression strain of 2566, respectively. Similarly, gBLM(610–1258) consisting of the gBLM helicase core was amplified, constructed into pTWIN1 (New England Biolabs), and transformed into BL21 (DE3). Likewise, pET15b-sumo hDHBN (residues 362–414) and pET15b-sumo pDHBN (residues 1–66) were constructed according to the NCBI amino acid sequences of H. sapiens BLM helicase (accession number P54132, hBLM for short) and P. crispus bloom syndrome protein (accession number XP_009483195.1, pBLM for short). In the experiment, expression of all BLM truncated proteins were induced by 0.3 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 16 h. Because gBLM(1–612), gBLM(294–1258), gBLM(360–1258), and gBLM(1–1300) showed lower stabilities than gBLM(610–1258) and the DHBN proteins, they were purified to homogeneity sequentially by affinity chromatography with cOmplete His tag purification resin (CHTPR, Roche Applied Science) column and ion exchange chromatography on SP Sepharose Fast Flow (GE Healthcare) and Q Sepharose Fast Flow (GE Healthcare), respectively. Similarly, purification of gBLM(610–1258) was carried out through chitin affinity chromatography and SP ion exchange chromatography, substituting KCl for NaCl in the buffers. As to the DHBN proteins, they were purified to high purity and high yield by following two turns of CHTPR purification and then by gel filtration chromatography with a Superdex 200 column (Amersham Biosciences, AKTA FPLC system).
Limited proteolysis characterization
Digestion of the gBLM truncated proteins was compared with α-chymotrypsin (Invitrogen) at a series of concentrations in each 20-μl reaction buffer (20 mm Tris-HCl, pH 8.0, 250 mm NaCl, 10 mm CaCl2) at 18 °C for 1 h; then 1 mm PMSF was added into each treatment to stop the reactions; and finally the samples were analyzed by 10% SDS-PAGE.
Crystal screening and structure determination
After cycles of screening and optimization, SeMet-substituted gDHBN finally obtained high quality crystals from a condition comprising 0.1 m magnesium formate, 0.1 m trisodium citrate, pH 4.4, 12% PEG 1500, and 15% glycerol. As expected, the homologous pDHBN gave out high resolution crystals from the optimal condition of 0.2 m ammonium acetate, 0.1 m trisodium citrate, pH 3.6, and 16% PEG 3350. Crystals were loop-mounted, cryo-protected, and diffracted on BL18U1 and BL19U1 beamlines at SSRF (Shanghai Synchrotron Radiation Facility). Once data were collected and processed, the structures extending to 2.7 Å resolution of gDHBN and 1.4 Å resolution of pDHBN were ascertained by selenium and by the molecular replacement phasing method, respectively (50).
Moreover, by following the similar sitting drop crystallization and optimization methods of gDHBN and pDHBN, optimized crystals of hDHBN exhibited higher diffraction resolution in well conditions of 0.1 m trisodium citrate, pH 4.6, 0.9 m magnesium sulfate and 0.1 m Tris, pH 8.5, 26% PEG 1500, 16% glycerol, respectively. Although the optimized SeMet-substituted crystals of hDHBN also yielded good diffraction data, there was no signal of selenium in the collected data, and the structure was not solved until the data were collected from crystals soaked in the optimized well solution supplemented with 0.3 m KI. Different from the P21212 space group of gDHBN, the space groups of hDHBN were P62 and C2221. All of the structure refinements were carried out by using Coot (51) and Phenix (50).
SAXS
SAXS determination of gDHBN was performed at 1.0, 3.0, 5.0, 7.0, and 9.0 mg/ml at the BL19U2 beamline of the National Center for Protein Sciences Shanghai and in HPLC mode at beamline SWING (SOLEIL Synchrotron, Saint-Aubin, France). The buffer used was 25 mm Hepes, pH 7.5, 500 mm NaCl, and 5% glycerol. The corresponding data were collected and processed with buffer subtraction with programs available on the beamlines (BL19U2 pipeline and FOXTROT, respectively). The programs of ATSAS suite PRIMUS and GNOM were first used to extract the experimental Rg (radius of gyration) and Dmax (maximum particle dimension) values (52), and then the ab initio envelope was determined by using DAMMIF. Ab initio model quality was evaluated further using averaging with DAMAVER, and the atomic model of gDHBN was proposed by flexible modeling with DADIMODO (53). Finally, the atomic model profile was calculated, aligned, and fitted to the experimental data using CRYSOL and SUPCOMB.
DLS determination and gel filtration analysis
DLS measurements were performed at 25 °C using a DynaPro Nano Star instrument (Wyatt Technology Europe GmbH, Dernbach, Germany) with a microcuvette. The DLS buffer used was 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 1 mm DTT for the protein only. To further identify what impacts ATP would have on the different gBLM truncates, 2 mm ATP or its analogues were chosen to fulfill the different treatments. The assays were performed at the optimized protein concentration (1.5–4 μm) in the DLS buffer substituted with 25 mm Tris-HCl, pH 7.5, 80 mm NaCl, 3 mm MgCl2, 1 mm DTT, and 23-nucleotide ssDNA whose concentration is equal to that of each protein assayed. All of the above measurement data were stored every 10 s and analyzed with the Dynamics version 7.0 software by using regularization arithmetic (Wyatt Technology) as described previously (54).
Meanwhile, the Superdex 200 10/300 GL (24 ml) column was first calibrated by the Bio-Rad gel filtration standard of thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) according to the manufacturer's protocol, and then 100 μg of each gBLM truncate was loaded onto the column and eluted. Assuming similar spherical shape and charge factors, the corresponding molecular weights of the BLM truncates were calculated with the fitted standard curve of ln(Mr) = −11.5657 Kav + 16.4812 (R2 = 0.97).
Helicase unwinding activity assay
To identify whether the crystallized gDHBN has any biological function, helicase unwinding activity measurements were carried out. The assay was performed at 25 °C in a buffer of 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 3 mm MgCl2 and 1 mm DTT. Two complementary strands of each substrate were labeled with fluorescein (F) at the 3′-tail and hexachlorofluorescein (HF) at the 5′-tail, respectively. The D16S10 overhang duplex substrate was annealed by equal moles of S1 (CTCTGCTCGACGGATT-F) and S2 (HF-AATCCGTCGAGCAGAGTTTTTTTTTT); similarly, the fD20S12 substrate was formed by equal moles of S3 (CTCTGGCCGTCTTACGGTCGCTCTGCTCGACG-F) and S4 (HF-CGTCGAGCAGAGCGACCGTATTATTTTTTTTT). The parameters used in multiturnover unwinding were as follows: gBLM truncate was 100 nm, DNA substrate was 4 nm, and ATP was 1 mm. To find out what unwinding kinetics the gBLM truncates would have, single-turnover unwinding determinations were also performed with 4 nm D16S10 overhang duplex, 60 nm gBLM truncate, various concentrations of ATP, and 2.5 mm dT56. The DNA unwinding kinetic parameters were determined by a Bio-Logic SFM-400 mixer and the Bio-Logic MOS450/AF-CD optical system (FC-15, Bio-Logic, Seyssinet-Pariset, France) and analyzed as described previously (36).
For converting the output voltage to a percentage of unwinding, a calibration experiment was performed in a four-syringe mode, with helicase in syringe 1, hexachlorofluorescein-labeled and fluorescein labeled single-stranded oligonucleotides in syringe 2 and 3, respectively, and ATP ± protein trap in syringe 4. All samples were incubated in the unwinding buffer. The fluorescence signal of the mixed solution from the four syringes, after equilibration, corresponds to 100% unwinding. The fraction of unwound (η) at time t was calculated by Equation 1,
| (Eq. 1) |
where F(t) is the measured fluorescence signal at time t, Fmin is the minimum fluorescence signal after unwinding initiation, and F100% is the signal obtained from the calibration measurement.
Author contributions
J. S., S. R., and X.-G. X. designed the work, summarized the results, and wrote the paper; J. S. purified proteins and cultivated crystals; J. S., B. Z., X. A., and N. N. L. performed the biochemical experiments; S.-H. F. performed the bioinformatics analysis; and W.-F. C. and S. R. determined the X-ray structures and SAXS model.
Supplementary Material
Acknowledgments
We thank the staff of the BL17U1 beamline at Shanghai Synchrotron Radiation Facility and the BL18U1, BL19U1, and BL19U2 beamlines at the National Center of Protein Sciences Shanghai for assistance with structure data collection. We are also grateful to Dr. Aurelien Thureau and his team for SAXS data collection at the beamline SWING (SOLEIL Synchrotron). This research was conducted within the context of the “LIA helicase-mediated G-quadruplex DNA unwinding and genome stability.”
This work was supported by National Natural Science Foundation of China Grants 11574252 and 31370798 and Northwest A&F University Startup Funding for Xu-Guang Xi (Grants Z101021102 and Z111021103). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 5LUP, 5MK5, 5LUT, and 5LUS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- BS
- Bloom syndrome
- RQC
- RecQ C-terminal domain
- HRDC
- helicase and RNase D C-terminal domain
- DHBN
- dimerization helical bundle in N-terminal domain
- hBLM
- gBLM, and pBLM, H. sapiens, G. gallus, and P. crispus BLM, respectively
- DLS
- dynamic light scattering
- SAXS
- small angle X-ray scattering
- r.m.s.d.
- root mean square deviation
- tNCS
- translational non-crystallographic symmetry
- SAD
- single-wavelength anomalous dispersion
- SeMet
- selenomethionine
- F
- fluorescein
- HF
- hexachlorofluorescein
- PDB
- Protein Data Bank
- AMPNP
- β,γ-imidoadenosine 5′-triphosphate
- ATPrS
- adenosine 5′-(3-thiotriphosphate).
References
- 1. Singleton M. R., Dillingham M. S., and Wigley D. B. (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 2. Abdelhaleem M. (2010) Helicases: an overview. Methods Mol. Biol. 587, 1–12 [DOI] [PubMed] [Google Scholar]
- 3. Khakhar R. R., Cobb J. A., Bjergbaek L., Hickson I. D., and Gasser S. M. (2003) RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 13, 493–501 [DOI] [PubMed] [Google Scholar]
- 4. Hickson I. D. (2003) RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3, 169–178 [DOI] [PubMed] [Google Scholar]
- 5. Larsen N. B., and Hickson I. D. (2013) RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 767, 161–184 [DOI] [PubMed] [Google Scholar]
- 6. Bachrati C. Z., and Hickson I. D. (2003) RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 374, 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harrigan J. A., and Bohr V. A. (2003) Human diseases deficient in RecQ helicases. Biochimie 85, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 8. Killoran M. P., and Keck J. L. (2006) Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 34, 4098–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rezazadeh S. (2012) RecQ helicases; at the crossroad of genome replication, repair, and recombination. Mol. Biol. Rep. 39, 4527–4543 [DOI] [PubMed] [Google Scholar]
- 10. Vindigni A., Marino F., and Gileadi O. (2010) Probing the structural basis of RecQ helicase function. Biophys. Chem. 149, 67–77 [DOI] [PubMed] [Google Scholar]
- 11. Bernstein D. A., Zittel M. C., and Keck J. L. (2003) High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 22, 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernstein D. A., and Keck J. L. (2005) Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure 13, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 13. Kitano K. (2014) Structural mechanisms of human RecQ helicases WRN and BLM. Front. Genet. 5, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suhasini A. N., and Brosh R. M. Jr. (2013) Disease-causing missense mutations in human DNA helicase disorders. Mutat. Res. 752, 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ben Salah G., Hadj Salem I., Masmoudi A., Kallabi F., Turki H., Fakhfakh F., Ayadi H., and Kamoun H. (2014) A novel frameshift mutation in BLM gene associated with high sister chromatid exchanges (SCE) in heterozygous family members. Mol. Biol. Rep. 41, 7373–7380 [DOI] [PubMed] [Google Scholar]
- 16. Vindigni A., and Hickson I. D. (2009) RecQ helicases: multiple structures for multiple functions? HFSP J. 3, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue Y., Ratcliff G. C., Wang H., Davis-Searles P. R., Gray M. D., Erie D. A., and Redinbo M. R. (2002) A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′–5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry 41, 2901–2912 [DOI] [PubMed] [Google Scholar]
- 18. Rodermund O. E., and Hausmann D. (1978) Bloom syndrome: review and definition. Fortschr. Med. 96, 1852–1858 [PubMed] [Google Scholar]
- 19. Nicotera T. M. (1991) Molecular and biochemical aspects of Bloom's syndrome. Cancer Genet. Cytogenet. 53, 1–13 [DOI] [PubMed] [Google Scholar]
- 20. Arora H., Chacon A. H., Choudhary S., McLeod M. P., Meshkov L., Nouri K., and Izakovic J. (2014) Bloom syndrome. Int. J. Dermatol. 53, 798–802 [DOI] [PubMed] [Google Scholar]
- 21. Swan M. K., Legris V., Tanner A., Reaper P. M., Vial S., Bordas R., Pollard J. R., Charlton P. A., Golec J. M., and Bertrand J. A. (2014) Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA. Acta Crystallogr. D Biol. Crystallogr. 70, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 22. Rao V. A., Conti C., Guirouilh-Barbat J., Nakamura A., Miao Z. H., Davies S. L., Saccá B., Hickson I. D., Bensimon A., and Pommier Y. (2007) Endogenous γ-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 5, 713–724 [DOI] [PubMed] [Google Scholar]
- 23. Sengupta S., Robles A. I., Linke S. P., Sinogeeva N. I., Zhang R., Pedeux R., Ward I. M., Celeste A., Nussenzweig A., Chen J., Halazonetis T. D., and Harris C. C. (2004) Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 166, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nimonkar A. V., Ozsoy A. Z., Genschel J., Modrich P., and Kowalczykowski S. C. (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U.S.A. 105, 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., and Qin J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14, 927–939 [PMC free article] [PubMed] [Google Scholar]
- 26. Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., and Kowalczykowski S. C. (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu L., Davies S. L., North P. S., Goulaouic H., Riou J. F., Turley H., Gatter K. C., and Hickson I. D. (2000) The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275, 9636–9644 [DOI] [PubMed] [Google Scholar]
- 28. Böhm S., and Bernstein K. A. (2014) The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair 22, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Staub E., Fiziev P., Rosenthal A., and Hinzmann B. (2004) Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays 26, 567–581 [DOI] [PubMed] [Google Scholar]
- 30. Chothia C., Lesk A. M. (1986) The relation between the divergence of sequence and structure in proteins. EMBO J. 5, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doolittle R. F. (1981) Similar amino acid sequences: chance or common ancestry? Science 214, 149–159 [DOI] [PubMed] [Google Scholar]
- 32. Wintjens R., and Rooman M. (1996) Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262, 294–313 [DOI] [PubMed] [Google Scholar]
- 33. Lewit-Bentley A., and Réty S. (2000) EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 10, 637–643 [DOI] [PubMed] [Google Scholar]
- 34. Pena V., Hothorn M., Eberth A., Kaschau N., Parret A., Gremer L., Bonneau F., Ahmadian M. R., and Scheffzek K. (2008) The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep. 9, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gyimesi M., Pires R. H., Billington N., Sarlós K., Kocsis Z. S., Módos K., Kellermayer M. S., and Kovács M. (2013) Visualization of human Bloom's syndrome helicase molecules bound to homologous recombination intermediates. FASEB J. 27, 4954–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu Y. N., Bazeille N., Ding X. Y., Lu X. M., Wang P. Y., Bugnard E., Grondin V., Dou S. X., and Xi X. G. (2012) Multimeric BLM is dissociated upon ATP hydrolysis and functions as monomers in resolving DNA structures. Nucleic Acids Res. 40, 9802–9814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cui S., Arosio D., Doherty K. M., Brosh R. M. Jr., Falaschi A., and Vindigni A. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 32, 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma S., Doherty K. M., and Brosh R. M. Jr. (2006) Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398, 319–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Croteau D. L., Popuri V., Opresko P. L., and Bohr V. A. (2014) Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83, 519–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beresten S. F., Stan R., van Brabant A. J., Ye T., Naureckiene S., and Ellis N. A. (1999) Purification of overexpressed hexahistidine-tagged BLM N431 as oligomeric complexes. Protein Expr. Purif. 17, 239–248 [DOI] [PubMed] [Google Scholar]
- 41. Rao V. A., Fan A. M., Meng L., Doe C. F., North P. S., Hickson I. D., and Pommier Y. (2005) Phosphorylation of BLM, dissociation from topoisomerase IIIα, and colocalization with γ-H2AX after topoisomerase I-induced replication damage. Mol. Cell. Biol. 25, 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eladad S., Ye T. Z., Hu P., Leversha M., Beresten S., Matunis M. J., and Ellis N. A. (2005) Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14, 1351–1365 [DOI] [PubMed] [Google Scholar]
- 43. Karow J. K., Newman R. H., Freemont P. S., and Hickson I. D. (1999) Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 9, 597–600 [DOI] [PubMed] [Google Scholar]
- 44. Muzzolini L., Beuron F., Patwardhan A., Popuri V., Cui S., Niccolini B., Rappas M., Freemont P. S., and Vindigni A. (2007) Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 5, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pike A. C., Gomathinayagam S., Swuec P., Berti M., Zhang Y., Schnecke C., Marino F., von Delft F., Renault L., Costa A., Gileadi O., and Vindigni A. (2015) Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: insights from DNA complex structures. Proc. Natl. Acad. Sci. U.S.A. 112, 4286–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Choudhary S., Sommers J. A., and Brosh R. M. Jr. (2004) Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J. Biol. Chem. 279, 34603–34613 [DOI] [PubMed] [Google Scholar]
- 47. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., and Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 48. Zhang H., Gao S., Lercher M. J., Hu S., and Chen W. H. (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. He Z., Zhang H., Gao S., Lercher M. J., Chen W. H., and Hu S. (2016) Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 52. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Evrard G., Mareuil F., Bontems F., Sizun C., and Perez J. (2011) DADIMODO: a program for refining the structure of multidomain proteins and complexes against small-angle scattering data and NMR-derived restraints. J. Appl. Crystallogr. 44, 1264–1271 [Google Scholar]
- 54. Duan X. L., Liu N. N., Yang Y. T., Li H. H., Li M., Dou S. X., and Xi X. G. (2015) G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J. Biol. Chem. 290, 7722–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.