Abstract
DNA double strand breaks (DSBs) severely disrupt DNA integrity. 53BP1 plays critical roles in determining DSB repair. Whereas the recruitment of 53BP1 to the DSB site is key for its function, recent evidence suggests that 53BP1's abundance also plays an important role in DSB repair because recruitment to damage sites will be influenced by protein availability. Initial evidence has pointed to three proteins, the ubiquitin-conjugating enzyme UbcH7, the cysteine protease cathepsin L (CTSL), and the nuclear structure protein lamin A/C, that may impact 53BP1 levels, but the roles of each protein and any interplay between them were unclear. Here we report that UbcH7-dependent degradation plays a major role in controlling 53BP1 levels both under normal growth conditions and during DNA damage. CTSL influenced 53BP1 degradation during DNA damage while having little effect under normal growth conditions. Interestingly, both the protein and the mRNA levels of CTSL were reduced in UbcH7-depleted cells. Lamin A/C interacted with 53BP1 under normal conditions. DNA damage disrupted the lamin A/C-53BP1 interaction, which preceded the degradation of 53BP1 in soluble, but not chromatin-enriched, cellular fractions. Inhibition of 53BP1 degradation by a proteasome inhibitor or by UbcH7 depletion restored the 53BP1-lamin A/C interaction. Depletion of lamin A/C, but not CTSL, caused a similar enhancement in cell sensitivity to DNA damage as UbcH7 depletion. These data suggest that multiple pathways collectively fine-tune the cellular levels of 53BP1 protein to ensure proper DSB repair and cell survival.
Keywords: cell signaling, checkpoint control, DNA damage, DNA damage response, DNA repair, degradation
Introduction
DNA of eukaryotic cells is under continuous attack by a variety of stressors that exist either in the environment or within the cell. Among all forms of DNA damage, DNA double strand break (DSB)3 represents the most lethal stressor as it completely disrupts the integrity of the DNA double helix. Therefore, accurate repair of DSBs is essential for the maintenance of genome integrity and cell survival. DSBs are repaired by a combination of homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways. HR uses the sister chromatid as the template to repair DSBs, whereas NHEJ fuses two broken ends with little or no homology needed (1). In this regard, HR mainly occurs in S and G2 phases when sister chromatids are available and is generally considered error-free. On the other hand, NHEJ can take place from G1 through G2 phases and is regarded as error-prone. Abnormalities in the DSB repair pathway often lead to the development of cancers and/or resistance to anticancer therapies (1).
Recent research findings suggest that 53BP1 and BRCA1 play critical roles in the balance of DSB repair pathways by mutually affecting each other's recruitment to DSB sites (2). 53BP1, together with its partner proteins Rif1 and Mad2L2/Rev7, inhibit BRCA1 foci formation in G1 phase, thereby suppressing HR while promoting NHEJ (3–9). In contrast, BRCA1 and the associated E3 ligase UHRF1 suppress formation of Rif1 and Mad2L2/Rev7 foci in S and G2 phases (4, 7–10), favoring HR at these cell cycle stages.
Formation of 53BP1 foci is directly involved in the choice of DSB repair pathway. Increasing evidence also suggests that the cellular levels of 53BP1 influence its accumulation at DSB sites. Accordingly, manipulating the level of 53BP1 showed nearly as strong an impact as foci formation on DSB repair as well as in establishing cellular sensitivity to anticancer therapies. In the absence of BRCA1, cells exclusively use the error-prone NHEJ to repair DSBs due to the dominance of 53BP1, and this leads to cell death (11, 12). Consistently, BRCA1 null mice were early embryonic lethal (13, 14). However, co-depletion of 53BP1 rescued the lethality phenotype of BRCA1 loss as a result of restoring HR (13–15). UbcH7 degrades 53BP1, and this is necessary for cells to select HR to repair one-ended DSBs generated by replicative stress (16). Low levels of 53BP1 were observed in some triple negative breast cancers and correlated with poor prognosis and therapy resistance to poly ADP-ribose polymerase inhibitors (13, 17). Such resistance implicates a process of selecting cells with low/little 53BP1 or its downstream factors like Artemis. This would then allow the tumor cells to survive as a result of re-establishing HR conveyed by the loss or reduction of the 53BP1 pathway (13, 18).
On the other hand, increasing the cellular levels of 53BP1 enhanced the mutagenic NHEJ in the absence or presence of BRCA1, leading to increased sensitivity to DNA damage (16, 19). Surprisingly, increased expression levels of 53BP1 in certain breast or ovarian cancers also correlated with reduced survival rate and poor prognosis (20, 21), although the underlying mechanisms remain unclear. Notably, only a small portion of cancer patients had increased 53BP1 mRNA levels in The Cancer Genome Atlas. Nonetheless, these data suggest that maintaining a precise level of 53BP1 protein is critical for cell survival, and altering 53BP1 expression levels (increasing or decreasing) is able to shift the balance of DSB repair choice, which consequently affects tumor development and therapeutic outcome. Mechanisms controlling the protein level of 53BP1 are not well understood but are vital for controlling DSB repair choice and, more broadly, cellular sensitivity to anticancer therapies.
The protein level of 53BP1 at different stages of the cell division cycle is relatively stable with only minor oscillations (22–24), suggesting that 53BP1 is being constantly transcribed and translated during the cell cycle. On the other hand, a number of studies reported post-translational regulation of 53BP1. An endosomal/lysosomal protease cathepsin L (CTSL) cleaves 53BP1 in the absence of lamin A/C (25–27). In addition, we recently reported that UbcH7 regulates the proteasome-dependent degradation of 53BP1, especially when cells were treated with agents that cause replication stress (16). Furthermore, depletion of lamin A/C reduces the stability of 53BP1 (28). These three pathways were independently identified, and it was not known if they might collaboratively regulate the cellular levels of 53BP1. Here we report that UbcH7-dependent degradation, but not CTSL-dependent cleavage, regulates the basal turnover of 53BP1 under normal growth conditions. In contrast, interaction with lamin A/C stabilizes 53BP1. DNA damage reduces the interaction between 53BP1 and lamin A/C, which leads to UbcH7-dependent degradation of 53BP1. Paralleling the degradation by UbcH7, 53BP1 is also cleaved by CTSL in the presence of DNA damage.
Results
Proteasome-dependent degradation controls the basal level of 53BP1
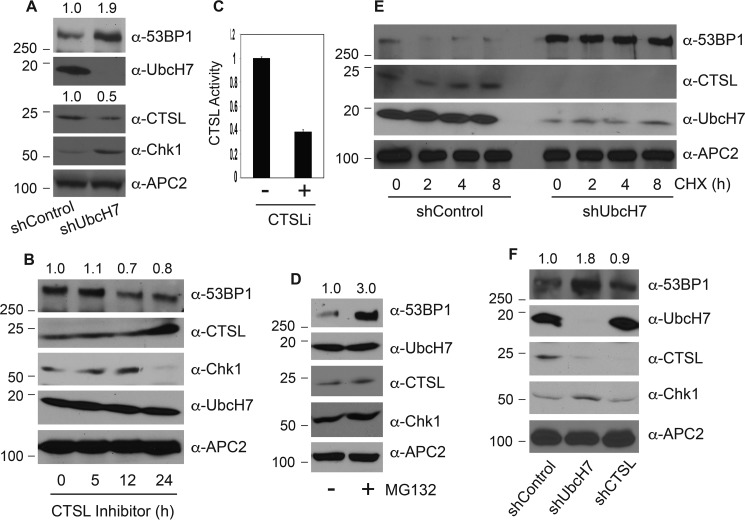
We recently reported that UbcH7 regulates proteasome-dependent degradation of 53BP1 (16). Previously, the cysteine proteinase CTSL was demonstrated to cleave 53BP1 in the absence of lamin A/C (25–27). However, it is unclear whether these two pathways cooperate to regulate 53BP1 protein levels or not. In addition, what determines which pathway(s) regulates the levels of 53BP1 under any specific condition is not well understood. To answer these questions, we first examined the expression profiles of 53BP1, UbcH7, and CTSL proteins in control or UbcH7 stably depleted cells. Depletion of UbcH7 significantly elevated the level of 53BP1 as well as another UbcH7 target, Chk1 (Fig. 1A), as we previously reported (16, 29). Interestingly, we observed a marked reduction in the levels of CTSL proteins in UbcH7-depleted cells (Fig. 1A), indicating that UbcH7 may also control the expression level of CTSL.
Figure 1.
Mechanisms controlling the basal levels of 53BP1 proteins. A, protein expression in A549 control or UbcH7 stably depleted cells by immunoblotting with the indicated antibodies. The molecular weight marker is shown on the left. B, A549 control cells were treated with 100 nm CTSLi for the indicated times, and protein expression was examined. C, CTSL activity in A549 control cells treated or not with 100 nm CTSL inhibitor for 24 h was monitored as described under “Experimental Procedures.” Data represent the means and S.D. from two independent experiments in triplicate. D, A549 parent cells were treated with 10 μm MG132 or vehicle for 6 h, and protein expression was examined. E, A549 stable cells infected with shRNA targeting control or UbcH7 were treated with 320 μm CHX for 0, 2, 4, and 8 h. Protein expression was monitored. F, A549 cells were stably infected with a control shRNA or those targeting UbcH7 or CTSL, and protein expression was examined. For protein quantitation, the 53BP1 or CTSL band intensity was quantified using NIH Image J software, and relative levels of 53BP1 or CTSL were normalized to that in the control lane and shown above the blot.
Because CTSL was reported to cleave 53BP1, we asked whether the increased levels of 53BP1 in UbcH7-depleted cells are the result of reduced expression of CTSL or not. Despite numerous attempts, we were not able to stably overexpress CTSL in UbcH7-depleted cells. Instead, we determined if inhibiting the enzyme of CTSL would also increase 53BP1 protein levels as observed by UbcH7 depletion. Treatment with a selective CTSL inhibitor (CTSLi) Z-FY(t-Bu)-DMK (Z-Phe-Tyr(tBu)-diazomethyl ketone) stabilized the CTSL protein (Fig. 1B), a common effect of competitive inhibitors on their cognate receptors. Although the inhibitor significantly suppressed the enzymatic activity of CTSL (Fig. 1C), it did not increase the protein level of 53BP1 (Fig. 1B). The inhibitor also failed to alter the expression level of UbcH7 (Fig. 1B), suggesting that CTSL does not regulate UbcH7 or 53BP1 levels. On the other hand, treatment of cells with the proteasome inhibitor MG132 significantly increased 53BP1 protein levels (Fig. 1D), mimicking the effect of UbcH7 depletion (Fig. 1A).
We subsequently examined the protein stability of 53BP1 in the presence of the protein synthesis inhibitor, cycloheximide (CHX). 53BP1 underwent time-dependent degradation in the presence of CHX in control cells, confirming that it is an unstable protein (Fig. 1E). However, depletion of UbcH7 almost completely blocked its degradation (Fig. 1E), reinforcing the idea that UbcH7 is the major factor that controls the basal turnover of 53BP1 in the absence of DNA damage (16). In contrast, UbcH7 and CTSL were relatively stable under this particular experimental setting (Fig. 1E).
To further confirm these results, we used a pool of two shRNA lentiviral vectors to stably inhibit the expression of CTSL and measured the levels of 53BP1. We repeatedly observed that depletion of UbcH7, but not CTSL, increased the basal level of 53BP1 (Fig. 1F). Again, inhibition of CTSL expression had no effect on the expression of UbcH7 (Fig. 1F). Together, these data suggest that UbcH7-dependent proteasomal degradation plays a major role in controlling the basal level of 53BP1 under normal growth conditions.
UbcH7 regulates 53BP1 and CTSL by distinct mechanisms
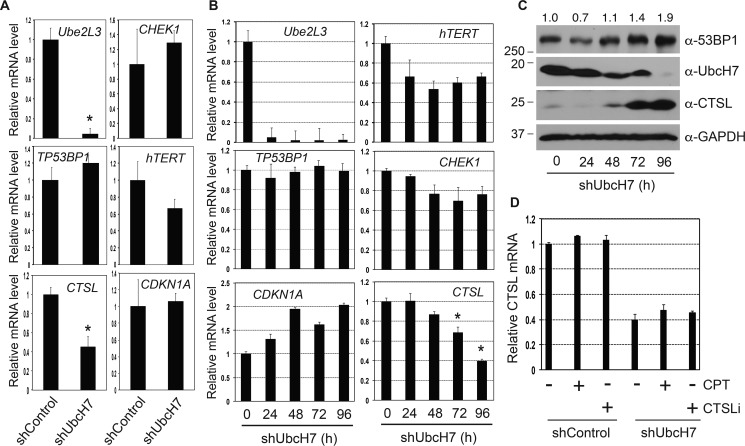
Depletion of UbcH7 resulted in a significant increase and decrease in the levels of 53BP1 and CTSL proteins, respectively (Fig. 1). Although increased protein stability led to the rise in the levels of 53BP1 proteins, the underlying mechanism that causes the decrease in CTSL is completely unknown. Because CTSL is a relatively stable protein (Fig. 1E), we asked if UbcH7 regulates expression of the CTSL gene. We first analyzed the mRNA level of CTSL in stable UbcH7-depleted cells by quantitative polymerase chain reaction (qPCR). The mRNA level of UBE2L3, the gene that encodes UbcH7, was nearly undetectable in UbcH7-depleted cells (Fig. 2A), confirming efficient knockdown of UbcH7 in these cells. We found that the mRNA level of CTSL was also significantly reduced in UbcH7-depleted cells compared with control cells (Fig. 2A). The human telomerase reverse transcriptase (TERT) mRNA level was also reduced (Fig. 2A), albeit to a much lesser extent than that of CTSL. However, a number of other cell cycle and DNA damage genes including TP53BP1, CHEK1, and CDKN1A (encoding human 53BP1, Chk1 and p21 proteins, respectively) were only moderately affected by UbcH7 depletion (Fig. 2A).
Figure 2.
UbcH7-dependent transcriptional regulation. A, relative mRNA levels of indicated genes in A549 cells stably depleted of control shRNA or shUbcH7. Data were acquired as described under “Experimental Procedures” from two independent experiments in triplicate. Student's t test was used to determine the statistical significance (*, p < 0.001). B, A549 cells were infected with lentiviral particles targeting UbcH7 for 24, 48, 72, and 96 h. Relative mRNA levels of indicated genes were obtained using methods described under “Experimental Procedures.” Data were collected from two independent experiments in triplicate. *, p < 0.001. C, parallel samples from B were analyzed for protein expression. Protein quantitation was carried out as in Fig. 1A. D, A549 cells stably infected with control shRNA or shUbcH7 were treated with 500 nm CPT, 100 nm CTSLi, or vehicle for 8 and 12 h, respectively. The CTSL mRNA level was measured as described in A. Data represent mean and S.D. from two experiments in triplicate.
The reduction in CTSL mRNA in response to UbcH7 silencing could be the result of three distinct mechanisms. One is that UbcH7 controls the transcription of the CTSL gene, another is that it controls CTSL mRNA stability, and the last is that the reduction in CTSL mRNA is a consequence of cell adaptation to stable lentiviral infection. To answer these questions, we first analyzed the mRNA levels of these genes after acute UbcH7 silencing. The levels of UBE2L3 were dramatically reduced by shRNA-mediated knockdown in these cells (Fig. 2B). Importantly, transient depletion of UbcH7 reduced the mRNA levels of CTSL and TERT (Fig. 2B), although the reduction kinetics is different. These data indicate that UbcH7 is likely involved in the transcriptional regulation of these two genes. Parallel samples showed that transient depletion of UbcH7 resulted in a time-dependent accumulation of 53BP1 protein, correlating inversely with that of UbcH7 (Fig. 2C). Notably, although the level of CTSL protein was reduced significantly after 24 h of UbcH7 depletion, it rebounded to much higher levels at later time points despite the fact that its mRNA levels were significantly reduced (Fig. 2C, 72–96 h of UbcH7 depletion). An increase in CTSL protein expression was also frequently observed when cells were grown over confluent regardless of the status of UbcH7 expression. Therefore, we propose that the increase in the CTSL protein at later time points of UbcH7 shRNA transduction occurs as a result of over-confluence of the cultures that may trigger an unknown growth stress-dependent mechanism that increases CTSL protein. Stable UbcH7-depleted cells were passaged normally; hence, this stress-induced CTSL up-regulation pathway is absent. Nonetheless, these data affirm the postulate that increased CTSL protein levels do not necessarily lead to decreased expression of 53BP1 in the absence of DNA damage.
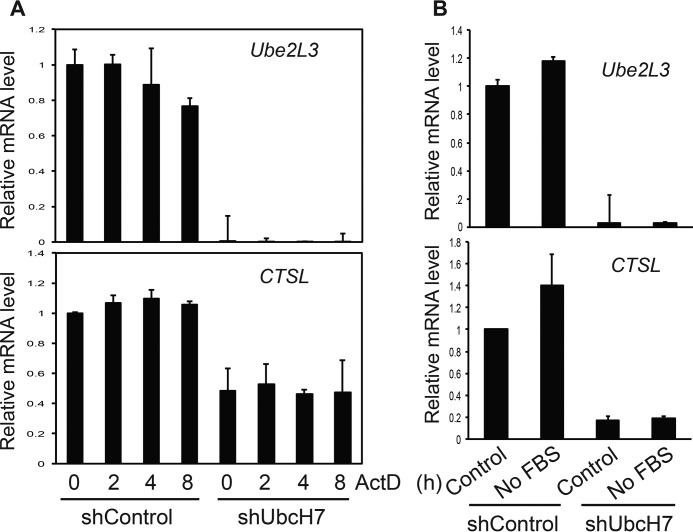
We then asked if exogenous stressors would alter the mRNA levels of CTSL similar to that observed with UbcH7 silencing. To this end, we treated control or stable UbcH7-depleted cells with the CTSLi or a DNA-damaging agent, camptothecin (CPT), a topoisomerase inhibitor that causes replicative stress and stimulates UbcH7-dependent degradation of 53BP1 (16). Neither CTSLi nor CPT affected the mRNA level of CTSL in the presence or absence of UbcH7 (Fig. 2D), indicating that CTSL mRNA levels are regulated by UbcH7 through an independent mechanism. To further understand the mRNA regulation of CTSL by UbcH7, we asked if UbcH7 stabilizes the CTSL mRNA. To address this question, we treated control or UbcH7-depleted cells with actinomycin D (ActD), an agent that blocks transcription, and measured the decay of CTSL over time by qPCR. The mRNA level of UBE2L3 was slightly reduced by ActD in control cells; however, ActD did not change CTSL mRNA levels (Fig. 3A). Furthermore, depletion of UbcH7 showed no effect on the mRNA level of CTSL in the presence of ActD (Fig. 3A). These data suggest that the CTSL transcript is highly stable and that UbcH7 does not seem to regulate its stability. We then asked if any stresses would induce transcription of CTSL. We found that serum deprivation induced a consistent, albeit not statistically significant, increase in the levels of both CTSL and UBE2L3 in control cells (Fig. 3B). However, this increase was absent in UbcH7 depleted cells (Fig. 3B). Together, these data suggest that UbcH7 regulates the protein levels of 53BP1 and CTSL through posttranslational and transcriptional mechanisms, respectively.
Figure 3.
UbcH7 regulated CTSL transcription. A, A549 control or UbcH7-depleted cells were treated with 1 μg/ml ActD for 0, 2, 4, and 8 h, RNAs were collected, and mRNA levels of Ube2l3 (the gene encoding UbcH7) and CTSL were measured by qPCR. Data represent the mean and S.D. from two independent experiments in duplicate. B, A549 control or UbcH7-depleted cells were cultured in regular media or without serum for 24 h, and gene expression levels were assessed as in A. Data represent the mean and S.D. from two independent experiments in duplicate.
CTSL regulates expression of 53BP1 during DNA damage
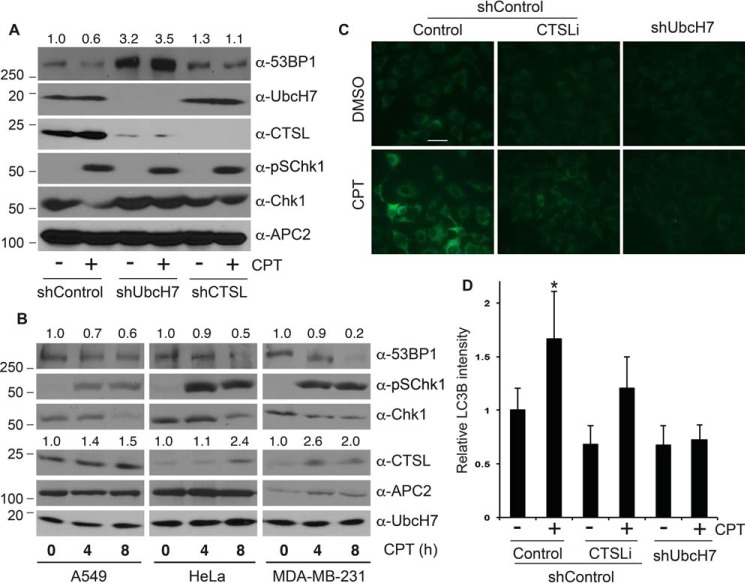
We recently showed that replicative stress (e.g. CPT treatment) induced UbcH7-dependent degradation of 53BP1 (16). On the other hand, CTSL was reported to cleave 53BP1 in the absence of lamin A/C (26, 27). Thus, we asked whether CTSL is involved in 53BP1 degradation in response to DNA damage. To answer this question, we treated control and UbcH7- or CTSL-depleted cells with CPT and examined the levels of 53BP1 proteins. CPT treatment reduced the level of 53BP1 in control cells (Fig. 4A), as we previously reported (16). Depletion of UbcH7 nearly completely prevented the CPT-induced degradation of 53BP1 (Fig. 4A) (16). We consistently noticed that depletion of CTSL partially inhibited the CPT-induced degradation of 53BP1 (Fig. 4A).
Figure 4.
CPT caused 53BP1 degradation, CTSL up-regulation, and activation of autophagy. A, A549 cells stably transduced with control shRNA or those targeting UbcH7 or CTSL were treated with 500 nm CPT or vehicle for 8 h, and protein expression was examined. Protein quantitation was performed as in Fig. 1A. B, parental A549, HeLa, or MDA-MB-231 cells were treated with 500 nm CPT for 0, 4, and 8 h, and protein expression was examined. Protein quantitation was carried out as in Fig. 1A. C, A549 shControl cells were pretreated or not with 100 nm CTSLi for 2 h, then together with UbcH7 depleted cells they were treated with or without 500 nm CPT for 8 h, fixed, and stained with anti-LC3B antibodies. Representative images are shown. The scale bar is 10 μm. D, quantitation of results from C by the NIH Image J software. Data represent the mean relative LC3B intensities normalized to the 0-h control sample and S.D. from >100 cells from two independent experiments. *, p < 0.001.
Interestingly, CPT treatment increased the protein level of CTSL (Fig. 4A), although it had little effect on its mRNA expression (Fig. 2D). The increase in CTSL was not due to over-confluence of cell culture as we carefully controlled the culture conditions for these experiments. Instead, it may represent a common mechanism by which CPT or similar drugs increase the protein levels of the lysosomal cathepsin protease family members. Consistent with this idea, CPT has been reported to increase the protein level of another cathepsin member, CTSS (30). To further test the role of CTSL in CPT-induced 53BP1 degradation, we treated a number of cancer cell lines that originated from different tissues with CPT and examined the protein levels of CTSL and 53BP1. The results showed that CPT treatment induced a time-dependent increase and decrease in the levels of CTSL and 53BP1, respectively, in all cell lines tested (Fig. 4B), indicating potential involvement of CTSL in the CPT-induced degradation of 53BP1.
CPT treatment induced activation of autophagy as revealed by anti-LC3B antibody based immunostaining of autophagosomes (31, 32) (Fig. 4, C and D). Interestingly, activation of autophagy was significantly reduced in UbcH7-depleted cells (Fig. 4, C and D). CTSLi also reduced the CPT-activated autophagy; however, the inhibitory effect was much less than UbcH7 depletion (Fig. 4, C and D). These results suggest that CPT-induced autophagy activation largely depends on UbcH7 and, to a lesser extent, CTSL.
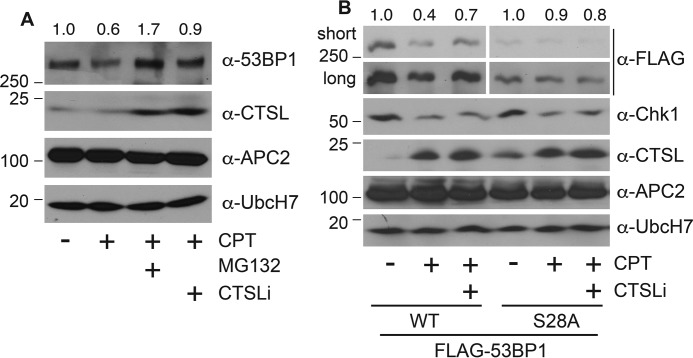
To discern how CTSL modulates the expression of 53BP1 in the presence of DNA damage, we asked whether the CTSL enzymatic activity was necessary for CPT-induced degradation of 53BP1. We observed that co-treatment with CTSLi partially suppressed 53BP1 degradation after CPT treatment, albeit to a much lesser extent than with the proteasome inhibitor MG132 (Fig. 5A). This inhibitory effect of CTSLi on CPT-induced 53BP1 down-regulation is similar to that caused by RNAi (Fig. 4A), further confirming the role of CTSL in CPT-induced 53BP1 degradation.
Figure 5.
CTSL participated in the degradation of 53BP1 that occurs in response to CPT. A, A549 parental cells were treated with 500 nm CPT or vehicle in the presence or absence of 10 μm MG132 or 100 nm CTSLi for 8 h, and expression of the indicated proteins was examined. Protein quantitation was performed as in Fig. 1A. B, HEK293T cells were transfected with FLAG-53BP1 WT or S28A for 48 h and treated with 500 nm CPT in the presence of absence of 100 nm CTSLi for 8 h, and expression of the indicated proteins was examined. Protein quantitation was performed as in Fig. 1A.
We previously reported that ATM (ataxia telangiectasia-mutated)-dependent phosphorylation at the amino terminus of 53BP1 is involved in its degradation (16). Thus, we also determined if phosphorylation functions as a trigger to initiate the CTSL-dependent cleavage of 53BP1. This involved assessing the impact of CPT in the presence or absence of CTSLi on the expression of wild type 53BP1 (WT) or a mutant form in which the 28 potential phosphorylation sites at the amino terminus of 53BP1 were mutated to Ala (S28A). CPT treatment reduced the level of 53BP1 WT, whereas it failed to alter the levels of the S28A mutant, consistent with our previous publication (16). The CTSLi partially blocked CPT-induced degradation of 53BP1 WT; however, it showed no effect on the level of the S28A mutant (Fig. 5B). These results suggest that DNA damage-induced phosphorylation of 53BP1 may be required for the CTSL-dependent cleavage of 53BP1 and further indicate that CTSL is at least partially responsible for CPT-induced degradation of 53BP1.
53BP1 is unstable in soluble, but not insoluble nuclear compartments
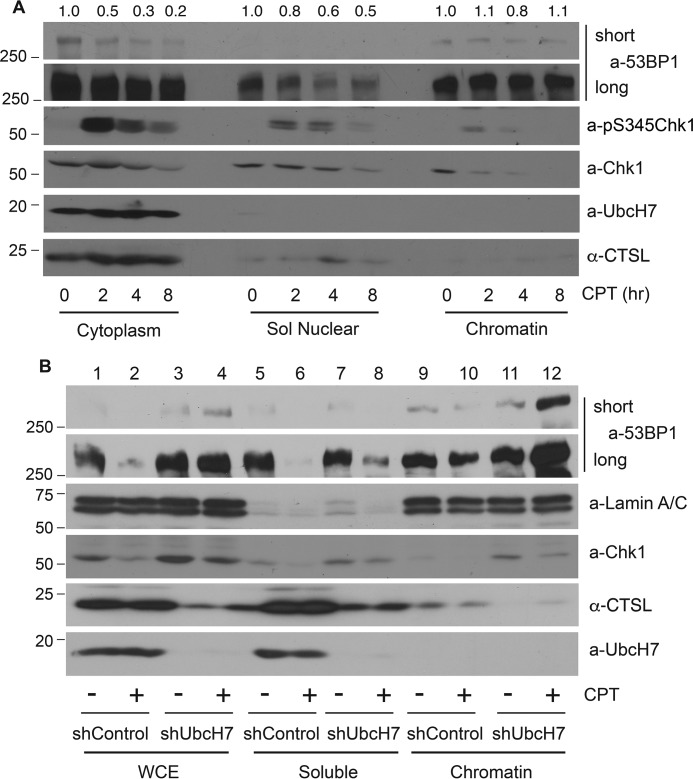
To further define the mechanisms underlying 53BP1 degradation, we treated cells with CPT over time and fractionated cells into cytoplasmic, soluble nuclear, and insoluble nuclear chromatin-enriched cellular fractions (33, 34). We found that 53BP1 was degraded in the cytoplasmic and soluble nuclear fractions in a time-dependent manner in the presence of CPT, although the degradation rate is greater in the cytoplasm than in the soluble nucleus (Fig. 6A); on the other hand, 53BP1 in the chromatin-enriched insoluble nuclear fraction remained fairly stable (Fig. 6A). In contrast, Chk1, the known UbcH7 target, was degraded in all cellular fractions. As a control, the phosphorylated form of Chk1 mainly resided in the cytoplasmic and soluble nuclear fractions, as we previously reported (34, 35). Although the majority of UbcH7 and CTSL proteins were located in the cytoplasmic fraction, there were detectable levels in the soluble nucleus (Fig. 6A), correlating with the degradation of 53BP1 by CPT in these fractions.
Figure 6.
Non-chromatin-associated 53BP1 was degraded in response to CPT. A, parental A549 cells were treated with 500 nm CPT for 0, 2, 4, and 8 h, fractionated into cytoplasmic, soluble nuclear, and insoluble chromatin-enriched nuclear compartments, and protein expression was examined. Protein quantitation was performed as in Fig. 1A. B, A549 control or UbcH7-depleted cells were treated with 500 nm CPT or vehicle for 8 h, fractionated into soluble (a combination of cytoplasm and soluble nucleus) and insoluble (chromatin-enriched) fractions, and protein expression was examined. Whole cell extracts (WCE) were also analyzed.
We then treated A549 control or UbcH7 stably depleted cells with CPT, fractionated cells into cytoplasmic, soluble nuclear, and insoluble nuclear fractions. Because 53BP1 was degraded in both cytoplasmic and soluble nuclear fractions (Fig. 6A), we combined these two fractions as the soluble fraction. We found that 53BP1 was extensively degraded in whole cell extracts as well as in soluble fractions in control cells (Fig. 6B, lanes 1 and 2 and lanes 5 and 6) with only moderate degradation occurring in the insoluble fraction (Fig. 6B, lanes 9 and 10). Interestingly, the level of 53BP1 in soluble fractions of UbcH7-depleted cells was also reduced by CPT treatment (Fig. 6B, lanes 7 and 8). However, this reduction was not due to protein degradation based on two reasons. First, unlike in control cells, the total level of 53BP1 remained unchanged in UbcH7-depleted cells (Fig. 6B, lanes 1 and 2 and lanes 3 and 4). Second, the level of 53BP1 in the insoluble fraction was dramatically increased by CPT in UbcH7-deleted cells (Fig. 6B, lanes 11 and 12). This type of protein redistribution appears to be specific to 53BP1, as we did not observe the same changes for Chk1 (Fig. 6B). Together, these data suggest that CPT treatment caused a relocalization of 53BP1 from soluble fractions into insoluble fraction in UbcH7-depleted cells, which may promote its stabilization.
Interaction of 53BP1 with lamin A/C is reduced by DNA damage
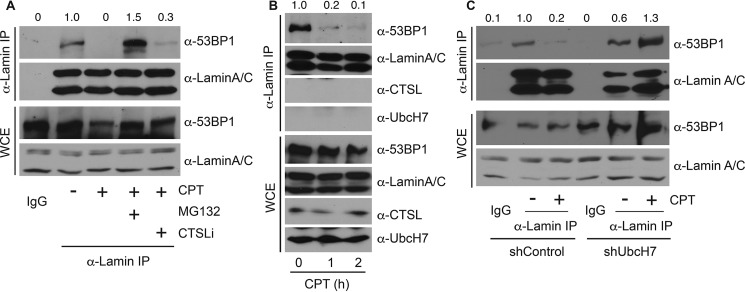
We then focused on understanding the mechanisms that contribute to the CPT-induced relocalization of 53BP1 in UbcH7-depleted cells. The nuclear architecture protein lamin A/C has previously been reported to be involved in stabilizing 53BP1 (27, 28). However, the mechanism by which lamin A/C regulates 53BP1 stability was unclear. We determined if the interaction between 53BP1 and lamin A/C is altered in the presence or absence of DNA damage. As reported (28), 53BP1 associated with lamin A/C under normal growth conditions, and DNA damage (i.e. CPT) significantly reduced this interaction (Fig. 7A). Co-treatment with the proteasome inhibitor MG132 not only restored, but also increased the interaction between 53BP1 and lamin A/C (Fig. 7A), indicating that stabilizing 53BP1 rescued its interaction with lamin A/C. On the other hand, the CTSLi only weakly restored the interaction between 53BP1 and lamin A/C (Fig. 7A). These data suggest that inhibiting 53BP1 degradation by the proteasome inhibitor and, to a much lesser extent the CTSLi, prevents CPT-induced dissociation between 53BP1 and lamin A/C.
Figure 7.
DNA damage reduced the interaction between 53BP1 and lamin A/C. A, A549 parental cells were treated with 500 nm CPT or vehicle in the presence or absence of 10 μm MG132 or 100 nm CTSLi for 8 h, and cell lysates were collected and immunoprecipitated (IP) with anti-lamin A/C antibodies, run on SDS-PAGE, and immunoblotted with anti-53BP1 or anti-lamin A/C antibodies. WCE were examined for protein expression. B, A549 parental cells were treated with 500 nm CPT for 0, 1, and 2 h, immunoprecipitated with anti-lamin A/C antibodies, and blotted with anti-53BP1, anti-lamin A/C, and anti-CTSL or anti-UbcH7 antibodies. Protein expression in WCE was also monitored. C, A549 cells stably depleted with control shRNA or shUbcH7 were treated with 500 nm CPT or vehicle for 8 h and processed as in B. The band intensities of the 53BP1 blots from both immunoprecipitate and WCE were quantified using Image J software, and the relative intensity of immunoprecipitated 53BP1 was normalized to that of 53BP1 proteins in WCE.
We then examined the time course of dissociation of 53BP1 from lamin A/C after CPT treatment. Dissociation occurred relatively rapidly as the majority of lamin A/C-bound 53BP1 was already released from lamin A/C within 1 h of CPT treatment (Fig. 7B). However, we did not observe UbcH7 or CTSL association with lamin A/C in the presence or absence of CPT (Fig. 7B), suggesting that UbcH7 or CTSL does not compete with lamin A/C for 53BP1 binding. We previously reported that CPT treatment increased the interaction between 53BP1 and UbcH7 (16); hence, we propose that CPT treatment shifts the association of 53BP1 from lamin A/C to UbcH7, resulting in its subsequent degradation. Consistent with this idea, blocking 53BP1 degradation by the proteasome inhibitor MG132 restored the interaction between 53BP1 and lamin A/C (Fig. 7A).
If the interaction of 53BP1 with lamin A/C prevents its degradation by UbcH7, then depletion of UbcH7 should also stabilize the interaction between 53BP1 and lamin A/C, as observed by treatment with MG132. For this experiment, we treated the cells with CPT for only 1 h, a time period that is insufficient to cause notable degradation of 53BP1 in control cells (16)(Fig. 7C, WCE). Under this condition, any reduction in the level of lamin A/C-bound 53BP1 should not be attributed to the loss of cellular levels of 53BP1 proteins. Again, CPT treatment significantly reduced the interaction between 53BP1 and lamin A/C in control cells (Fig. 7C). However, CPT failed to dissociate 53BP1 from lamin A/C in UbcH7-depleted cells (Fig. 7C). In fact, the 53BP1-lamin A/C interaction was increased when UbcH7 was depleted (Fig. 7C), similar to MG132 treatment (Fig. 7A). A possible explanation for the increase in 53BP1-lamin A/C interaction is that blocking 53BP1 degradation by MG132 or UbcH7 depletion increased total abundance of 53BP1, consequently facilitating its association with lamin A/C. These data suggest that DNA damage reduces the interaction between 53BP1 and lamin A/C, whereas stabilization of 53BP1 by either proteasome inhibition or UbcH7 depletion restores the 53BP1-lamin A/C interaction.
Lamin A/C contributes to 53BP1 protein stability and cellular sensitivity to DNA damage
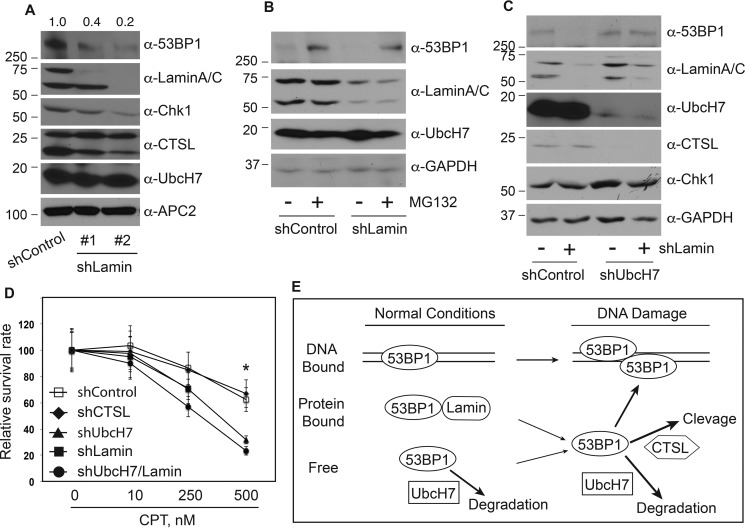
The dissociation of 53BP1 from lamin A/C precedes its degradation induced by CPT (e.g. Fig. 7B). This led us to ask if lamin A/C stabilizes 53BP1. To address this issue, we first asked if depletion of lamin A/C would affect the expression level of 53BP1. Using two shRNA vectors targeting different regions of the LAMN gene, we found that the protein level of 53BP1 was greatly reduced when lamin A/C was silenced (Fig. 8A), indicating that lamin A/C is involved in stabilizing 53BP1 under normal growth conditions. Then we asked if the reduction of 53BP1 in lamin A/C-depleted cells is through proteasome-dependent degradation. We treated control or lamin A/C-depleted cells with MG132 and observed that MG132 indeed stabilized 53BP1 in the absence of lamin A/C (Fig. 8B), confirming that loss of lamin A/C leads to proteasome-dependent degradation of 53BP1. Furthermore, to determine if UbcH7-dependent degradation is responsible for the reduced levels of 53BP1 that occurs in the absence of lamin A/C, we co-depleted UbcH7 and lamin A/C. Depletion of UbcH7 greatly suppressed the reduction in 53BP1 in the absence of lamin A/C (Fig. 8C), indicating that UbcH7 is responsible for degradation of 53BP1 following release from lamin A/C.
Figure 8.
Lamin A/C stabilized 53BP1. A, A549 cells were stably infected with a control shRNA or two independent shRNAs targeting the LMNA (the lamin A/C gene) that encodes lamin A/C. Protein expression was then determined using the indicated antibodies. Protein quantitation was performed as in Fig. 1A. B, A549 stable cells infected with control shRNA or shLamin were treated with 10 μm MG132 or vehicle for 6 h, and protein expression was examined. C, protein expression in A549 cells stably transduced with control shRNA, shLamin, shUbcH7, or ShUbcH7 plus shLamin. D, clonogenic survival assay in A549 stable cell lines transduced with control shRNA, shCTSL, shUbcH7, shLamin, or shUbcH7 plus shLamin were performed as described under “Experimental Procedures.” Data represent the means and S.D. from two experiments in duplicate. *, p < 0.001. E, model of 53BP1 protein level regulation. Under normal growth conditions, 53BP1 exists in at least three forms including chromatin-bound and lamin A/C-bound and free forms. UbcH7 regulates the proteasomal degradation of free forms of 53BP1 proteins. When cells are treated with DNA-damaging agents, 53BP1 in the lamin A/C-bound form will be released into the free form, which not only facilitates its recruitment to DSB sites but is also available for UbcH7-dependent degradation and to a much lesser extent, CTSL-dependent cleavage.
We previously reported that depletion of UbcH7 sensitizes cancer cells to DNA-damaging agents, especially replicative stresses like CPT (16). To assess the impact of the pathways that regulate 53BP1 protein levels on cell sensitivity to anticancer drugs, we measured cell survival after CPT treatment using the clonogenic survival assay. Depletion of CTSL had a minimal effect on cell survival in the presence of CPT compared with control cells (Fig. 8D). This is consistent with CTSL only playing a minor role in regulating the level of 53BP1. On the other hand, depletion of UbcH7 or lamin A/C significantly increased cell sensitivity to CPT especially at higher doses of the drug (Fig. 8D). Co-depletion of lamin A/C and UbcH7 induced a stronger effect than depletion of either one alone, although the effect was not statistically different (Fig. 8D), suggesting that UbcH7 and lamin A/C overlap in promoting cell survival.
Discussion
Given its key roles in the choice of DSB repair pathways, 53BP1 is tightly associated with therapeutic response to anticancer regimens that induce DSBs (14). Therefore, regulation of 53BP1 influences the outcomes of anticancer treatment of cells in vitro. Mechanistically, accumulation of 53BP1 proteins at the DSB site directly determines the DSB repair choice. However, for any given cell line, the pre-established cellular level of 53BP1 likely defines the extent of 53BP1 recruitment to DSB sites, which are a result of the equilibrium between production and degradation of 53BP1 proteins. Any abnormalities in the level of 53BP1 will likely lead to either inadequate or excessive recruitment of 53BP1 to DSB sites, which then alters the DSB repair choice and consequently affects the anticancer therapy outcome. For example, we recently reported that increasing the levels of 53BP1 by inhibiting its degradation or simply overexpressing the WT protein forced cells to primarily use NHEJ to repair DSBs (16), consistent with enhanced recruitment of 53BP1 to damage sites (16, 36). Accordingly, these cells were much more sensitive to DSB-inducing agents than parental cells due to the error-prone nature of NHEJ (16). Similarly, blocking the nuclear import of 53BP1 by depleting the nuclear pore complex protein NUP153 led to impaired foci formation of 53BP1 followed by defective DSB repair and increased sensitivity to DNA damaging agents (37). In this regard, pathways that control the cellular level and localization of 53BP1 probably play as important a role as its foci formation in determining the DSB repair choice as well as cell sensitivity to anticancer treatments.
Here we provide novel insights into the regulation of 53BP1 protein levels both under normal growth conditions and during DNA damage. We showed that UbcH7-dependent proteasomal degradation is the major pathway that controls the level of 53BP1 in the absence or presence of DNA damage in a wide range of cultured human cell lines. Lamin A/C associates with 53BP1 to stabilize 53BP1 under normal growth conditions. This likely involves shielding 53BP1 from the UbcH7-dependent degradation machinery. This seems to be a general mechanism by which lamin A/C protects 53BP1, as similar results were recently reported in human dermal fibroblasts (28). Such stabilizing effect of lamin A/C was also observed for other proteins, including the retinoblastoma Rb (38), suggesting that lamin A/C proteins are involved in the stabilization of many nuclear proteins. These findings lead us to present a model in which the protein levels of 53BP1 play important roles in regulating DSB repair choice (Fig. 8E). Under normal growth conditions, 53BP1 exists in at least three forms: chromatin-bound through the recognition of di-methylated H4K20 (39–41), lamin A/C-bound (Ref. 28 and data herein), and free forms. It is anticipated that 53BP1 proteins achieve a balance among these three locales under normal growth conditions. The free form undergoes UbcH7-dependent proteasomal degradation, which then controls the equilibrium of 53BP1 production and degradation. In the presence of DNA damage, the chromatin-bound 53BP1 may relocate to damage sites to form foci. In parallel, the lamin A/C-associated 53BP1 proteins will be released, which can then either be recruited to damage sites to form foci or serve as the target for UbcH7 and, to a lesser extent, CTSL, for proteasome-dependent degradation or protease-dependent cleavage. Ultimately, these regulatory mechanisms fine-tune the cellular level of 53BP1 to mount an effective DSB response and repair.
Our data suggest that when cells encounter DNA damage (herein, CPT treatment), CSTL is involved in regulating the levels of 53BP1 by cleaving the protein, although it only played a minor role. A similar role of CTSL in cleaving 53BP1 was observed when lamin A/C was depleted (26, 27). We noticed that when cells were overly confluent, the protein level of CTSL increased dramatically. However, this increased CTSL does not seem to regulate the protein level of 53BP1 in the absence of DNA damage.
An intriguing finding is that mechanisms regulating the mRNA level of CTSL appear to be uncoupled from those that regulate its protein levels. Treatment of cells with the DNA-damaging agent CPT or a CTSL-specific inhibitor did not affect the mRNA level of CTSL; however, they both stabilized the CTSL protein. On the other hand, serum deprivation induced a slight increase in the mRNA level of CTSL. These data suggest that regulation of CTSL depends on both the cellular context and the stimulating stress. Nonetheless, these hypotheses are currently under investigation in independent projects.
Our data also revealed surprising yet intriguing results that need further investigation. The levels of CTSL proteins were dramatically decreased upon silencing of UbcH7 (transient or stable knockdown). We further showed that these changes in CTSL are most likely through transcriptional regulation, as the mRNA level of CTSL was reduced in UbcH7-depleted cells. The other DNA damage response gene whose mRNA level also depended on UbcH7 was hTERT. These observations are unlikely due to off-target effects of stable lentiviral transduction, as transient inhibition of UbcH7 by shRNA also resulted in a significant reduction in mRNA levels of CTSL and hTERT. We further showed that UbcH7 seems to modulate CTSL transcription but not mRNA stabilization. The current paradigm suggests that UbcH7 almost exclusively functions in posttranslational modifications, e.g. protein ubiquitination and proteasomal degradation (16, 29). To the best of our knowledge this is the first observation that UbcH7 regulates gene transcription. Such a transcriptional effect of UbcH7 might be direct or indirect, and most likely more genes will be identified as UbcH7 targets in the future. Nonetheless, they represent novel research directions that await further investigation.
In conclusion, here we present evidence that cells have evolved multiple layers of regulation to control the protein level of 53BP1 both under normal growth conditions and during DNA damage. These pathways cooperatively control the reservoir of 53BP1 proteins so that suitable responses will be mounted to counteract genotoxic stress. This is likely critical for activating the best combination of DSB repair choices to promote cell survival. Accordingly, manipulating the protein level of 53BP1 offers a unique strategy in altering the DSB repair choice and consequently the outcome of anticancer therapies.
Experimental procedures
Cell cultures, chemicals, and transfection
HEK293T, A549, HeLa, MDA-MB-231, and U2-OS cells were cultured in DMEM with 10% fetal bovine serum (FBS). CHX and CPT were purchased from Arcos Organics. CHX was freshly dissolved in phosphate-buffered saline before use, whereas CPT was dissolved in dimethyl sulfoxide at a stock concentration of 10 mm. MG132 was purchased from Selleck Chemical LLC (Houston, TX) and dissolved in dimethyl sulfoxide at the stock concentration of 10 mm. Cell transfection was performed with the X-tremeGENE HP transfection reagent (Sigma) according to the manufacture's protocols.
RNAi
For RNAi, lentiviral shRNA vectors targeting UbcH7, CTSL, and lamin A/C were obtained from Sigma. The following are targeting sequences for: UbcH7: CCAGCAGAGTACCCATTCAAA (TRCN0000007209) and CCACCGAAGATCACATTTAAA (TRCN0000007211); CTSL: CCAAAGACCGGAGAAACCATT (TRCN0000349635) and AGGCGATGCACAACAGATTAT (TRCN0000318682); lamin A/C: GATGATCCCTTGCTGACTTAC (TRCN0000262697) and AGAAGGAGGGTGACCTGATAG (TRCN0000262764).
For production of lentivirus, HEK293T cells were seeded onto 10-cm dishes at ∼40–50% confluency in growth medium. For each dish, the transfection complex was prepared as follows: 12 μg of DNA (3 μg of shRNA plasmid, 3 μg of pMD2.G (gag/pol vector), 3 μg of pMDLg (VSV-G vector), and 3 μg of pRSV-Rev (rev vector)) in 250 μl of sterile ultra pure H20 containing 62 μl of 2 m CaCl2. After 5 min, 250 μl of 2× Hanks' balanced salt solution was added and incubated for 20 min at room temperature. The transfection mix was directly added to HEK293T cells. 72 h after transfection the medium was collected into a 15-ml sterile centrifuge tube. Viral supernatant was centrifuged at 2000 rpm for 10 min at room temperature and transferred to a new tube. For transduction, cells were seeded in 35-mm dishes at 30% confluency and incubated at 37 °C. Twelve hours before infection the culture medium was removed and replaced by fresh complete medium containing 10 μg/ml Polybrene. Then the appropriate amounts of virus particles were added to the cells and incubated for 3 days. Stable shRNA-expressing cells were selected with 3 μg/ml puromycin for 2 weeks.
Immunoblotting, immunoprecipitation, immunofluorescence, and antibodies
Immunoblotting was performed as previously described (16, 42, 43). In brief, cells were collected in 1.5-ml centrifuge tube, and cell lysates were prepared using lysis buffer (50 mm Tris-HCl, pH 7.6, 300 mm NaCl, 10 mm NaF, 1% Nonidet P-40) with protease inhibitors for at least 30 min on ice. The lysate was sonicated for 10 s four times. The resulting cell lysate was centrifuged at 14,000 for 10 min at 4 °C. After determining the protein concentrations, samples were directly diluted by 4× SDS-PAGE loading buffer and separated in SDS-PAGE. Anti-Chk1 (DCS-310, #SC-56291), anti-CTSL (33/2, #SC-32320), anti-lamin A/C (636, #SC-7292), and anti-APC2 (H-295, #SC-20984) antibodies were from Santa Cruz (Dallas, TX). Anti-phospho-S345-Chk1 (133D3, #2348) and anti-LC3B (D11, XP) antibodies were from Cell Signaling (Danvers, MA). Anti-53BP1 (#NB100-304) and anti-UbcH7 (#NB100-2265) were from Novus Biologicals (Littleton, CO). Anti-UbcH7 (#A300-737A) was from Bethyl Laboratories (Montgomery, TX).
For immunoprecipitation, two 10-cm plates for each sample at ∼90% confluency were collected in 15-ml tubes. The cells were washed two times in phosphate-buffered saline. Cell extracts were then prepared by lysing the cells in Nonidet P-40 buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1 mm EDTA, 5% glycerol, 0.5% Nonidet P-40) with protease inhibitors (1 mm DTT, 1 mm PMSF, 1 mm β-glycerophosphate, 10 μg/ml aprotinin, and 1 μg/ml leupeptin). Cells in the lysis buffer were incubated on ice for 60 min. The cell lysate was then sonicated 5 times and centrifuged at 14,000 rpm for 10 min at 4 °C. Equal amounts of proteins (∼2 mg) were immunoprecipitated with 1 μg of lamin A/C antibodies overnight. Protein (A + G) beads were added into the lysates and incubated for additional 4 h. Beads were washed with Nonidet P-40 buffer 4 times, resuspended in 2× SDS sample buffer, and loaded onto an SDS-PAGE gel.
For immunofluorescence of LC3B, A549 control or UbcH7-depleted cells were grown on glass coverslips, treated with 500 nm CPT for 0, 2, 4, and 8 h, fixed in 3.7% formaldehyde for 15 min, blocked in 5% BSA with 0.2% Triton X-100 in PBS for 1 h, washed with PBS extensively, and incubated in rabbit anti-LC3B monoclonal antibody (1:200 dilution) in 1% BSA with 0.2% Triton X-100 in PBS at 4 °C overnight. The slides were washed in PBS extensively, incubated in goat anti-rabbit Alexa 488 (∼1:750) in 1% BSA with 0.2% Triton X-100 in PBS for 1 h, washed, mounted in anti-fade solutions with DAPI (Life Technologies), and visualized under fluorescence microscopy.
CTSL activity analysis
The activity of CTSL was measured by the Fluorogenic InnoZyme™ Cathepsin L Activity kit (CBA023) from EMD Millipore (Billerica, MA). In brief, cells treated with 100 nm CTSL inhibitor Z-FY(t-Bu)-DMK (Z-Phe-Tyr(tBu)-diazomethyl ketone) or vehicle for 12–24 h were lysed in lysis buffer. Two hundred nanograms of protein in 50 μl of lysis buffer from each sample was used for the reaction based on the protocol provided by the manufacturer. Fluorescence was measured in a plate reader with excitation and emission at 360–380 nm and 460–480 nm, respectively. Data were collected from at least three independent experiments with a replicate.
RNA isolation and quantitative real-time PCR
RNA was isolated using TRIzol Reagent (Life Technologies) and purified with the Qiagen RNAeasy Mini kit (Valencia, CA). Polyadenylated mRNA was reverse-transcribed using Thermo Scientific Maxima H Minus Reverse Transcriptase (Waltham, MA) according to the manufacturer's recommendations. Each reverse transcription reaction mixture contained 1 μg of total RNA. The cDNA samples were diluted (1:10), and gene expression analysis was carried out by qPCR using the Luminaris Color HiGreen High ROX qPCR Master Mix (2x) (Thermo Scientific). The expression ratio of mRNAs was calculated with the 2−ΔΔCt method using ACTB mRNA expression level for normalization.
The following primer sequences were used: CTSL forward 5′-TCCTGTGAAGAATCAGGGTCAG-3′; CTSL reverse 5′-CCATTAGGCCACCATTGCAG-3′; Ube2l3 forward 5′-GAGCAGCACCAAATCCAAGATG-3′; Ube2l3 reverse, 5′-GGGTTGTCAGGAACAATAAGCC-3′; ACTB forward, 5′-CTGGAACGGTGAAGGTGAGA-3′; ACTB reverse 5′-AAGGGACTTCCTGTAACAATGCA-3′; CDKN1A forward, 5′ATGTGGACCTGTCACTGTCTTG-3′; CDKN1A reverse 5′-TGGTAGAAATCTGTCATGCTGGT-3′; TERT forward: 5′-CGGAAGAGTGTCTGGAGCAA-3′; TERT reverse: 5′-GGATGAAGCGGAGTCTGGA-3′; CHEK1 forward 5′-GCCAAGCTTATGGCAGTGCCCTTTGTGGAAGACTG-3′; CHEK1 reverse 5′-GCGTCTAGATCAACAGTACTCCAGAAATAAATATTG-3′.
Cell colony formation assay
Cytotoxicity testing was determined using CellTiter 96® Non-Radioactive Cell Proliferation Assay according the manufacturer's protocol (Promega, Madison, WI). Cells were seeded at 5 × 104 cells/well in 6-well plates and allowed to grow in the growth medium containing escalating doses of CPT (0, 5, 100, 250 nm) for 3 days. Twenty μl of Dye Solution was added into each well and incubated in the plate for 1 h at 37 °C. Colored soluble product was aliquoted onto 96-well plates and measured at the 490-nm absorbance using a plate reader.
Author contributions
F. M. P. and J. T. performed the experiments and analyzed the data. K. W. B., R. A. K., and X. Y. assisted the experiments and provided insights into the study. F. M. P. and Y. Z. designed the experiments and analyzed the data. Y. Z. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA163214 (NCI; to Y. Z.), the Clinical and Translational Science Collaborative of Cleveland Grant UL1TR000439 (National Center for National Center for Translational Sciences (NCATS) (to Y. Z.), Case Cancer Center P30 Grant CA043703 (to Y. Z.), and Grant CA126710 (NCI; to R. A. K.). This work was also supported by American Cancer Society Grant ACS RSG-15-042 DMC (to Y. Z.) and United States Department of Defense Grant W81XWH-14-1-0354) (to R. A. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DSB
- DNA double strand break
- 53BP1
- p53-binding protein 1
- CTSL
- cathepsin L
- CTSLi
- CTSL inhibitor
- NHEJ
- non-Homologous end-joining
- HR
- homologous recombination
- UbcH7
- ubiquitin-conjugating enzyme H7
- CHX
- cycloheximide
- CPT
- camptothecin
- shRNA
- short hairpin RNA
- (h)TERT
- (human) telomerase reverse transcriptase
- qPCR
- quantitative PCR
- ActD
- actinomycin D
- WCE
- whole cell extracts.
References
- 1. Goodarzi A. A., and Jeggo P. A. (2013) The repair and signaling responses to DNA double-strand breaks. Adv. Genet. 82, 1–45 [DOI] [PubMed] [Google Scholar]
- 2. Daley J. M., and Sung P. (2014) 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 34, 1380–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zimmermann M., Lottersberger F., Buonomo S. B., Sfeir A., and de Lange T. (2013) 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339, 700–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chapman J. R., Barral P., Vannier J. B., Borel V., Steger M., Tomas-Loba A., Sartori A. A., Adams I. R., Batista F. D., and Boulton S. J. (2013) RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A. D., Feldhahn N., Resch W., Oliveira T. Y., Chait B. T., Nussenzweig A., Casellas R., Robbiani D. F., and Nussenzweig M. C. (2013) Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng L., Fong K. W., Wang J., Wang W., and Chen J. (2013) RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288, 11135–11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J. T., Tkáč J., Cook M. A., Rosebrock A. P., Munro M., Canny M. D., Xu D., and Durocher D. (2013) A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 [DOI] [PubMed] [Google Scholar]
- 8. Boersma V., Moatti N., Segura-Bayona S., Peuscher M. H., van der Torre J., Wevers B. A., Orthwein A., Durocher D., and Jacobs J. J. (2015) MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 521, 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu G., Chapman J. R., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M., Jaspers J. E., Watanabe K., Pieterse M., Kersbergen A., Sol W., et al. (2015) REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H., Liu H., Chen Y., Yang X., Wang P., Liu T., Deng M., Qin B., Correia C., Lee S., Kim J., Sparks M., Nair A. A., Evans D. L., Kalari K. R., et al. (2016) A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat. Commun 7, 10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saberi A., Hochegger H., Szuts D., Lan L., Yasui A., Sale J. E., Taniguchi Y., Murakawa Y., Zeng W., Yokomori K., Helleday T., Teraoka H., Arakawa H., Buerstedde J. M., and Takeda S. (2007) RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell. Biol. 27, 2562–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGlynn P., and Lloyd R. G. (2002) Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3, 859–870 [DOI] [PubMed] [Google Scholar]
- 13. Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., Haffty B. G., Tommiska J., Blomqvist C., Drapkin R., Adams D. J., et al. (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., and Nussenzweig A. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao L., Xu X., Bunting S. F., Liu J., Wang R. H., Cao L. L., Wu J. J., Peng T. N., Chen J., Nussenzweig A., Deng C. X., and Finkel T. (2009) A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han X., Zhang L., Chung J., Mayca Pozo F., Tran A., Seachrist D. D., Jacobberger J. W., Keri R. A., Gilmore H., and Zhang Y. (2014) UbcH7 regulates 53BP1 stability and DSB repair. Proc. Natl. Acad. Sci. U.S.A. 111, 17456–17461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neboori H. J., Haffty B. G., Wu H., Yang Q., Aly A., Goyal S., Schiff D., Moran M. S., Golhar R., Chen C., Moore D., and Ganesan S. (2012) Low p53 binding protein 1 (53BP1) expression is associated with increased local recurrence in breast cancer patients treated with breast-conserving surgery and radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 83, e677–e683 [DOI] [PubMed] [Google Scholar]
- 18. Wang J., Aroumougame A., Lobrich M., Li Y., Chen D., Chen J., and Gong Z. (2014) PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 28, 2693–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zong D., Callén E., Pegoraro G., Lukas C., Lukas J., and Nussenzweig A. (2015) Ectopic expression of RNF168 and 53BP1 increases mutagenic but not physiological non-homologous end joining. Nucleic Acids Res. 43, 4950–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pennington K. P., Wickramanayake A., Norquist B. M., Pennil C. C., Garcia R. L., Agnew K. J., Taniguchi T., Welcsh P., and Swisher E. M. (2013) 53BP1 expression in sporadic and inherited ovarian carcinoma: relationship to genetic status and clinical outcomes. Gynecol. Oncol. 128, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacot W., Thezenas S., Senal R., Viglianti C., Laberenne A. C., Lopez-Crapez E., Bibeau F., Bleuse J. P., Romieu G., and Lamy P. J. (2013) BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 13, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Vugt M. A., Gardino A. K., Linding R., Ostheimer G. J., Reinhardt H. C., Ong S. E., Tan C. S., Miao H., Keezer S. M., Li J., Pawson T., Lewis T. A., Carr S. A., Smerdon S. J., Brummelkamp T. R., and Yaffe M. B. (2010) A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G2/M DNA damage checkpoint. PLoS Biol. 8, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giunta S., Belotserkovskaya R., and Jackson S. P. (2010) DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jullien D., Vagnarelli P., Earnshaw W. C., and Adachi Y. (2002) Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci. 115, 71–79 [DOI] [PubMed] [Google Scholar]
- 25. Grotsky D. A., Gonzalez-Suarez I., Novell A., Neumann M. A., Yaddanapudi S. C., Croke M., Martinez-Alonso M., Redwood A. B., Ortega-Martinez S., Feng Z., Lerma E., Ramon y Cajal T., Zhang J., Matias-Guiu X., Dusso A., and Gonzalo S. (2013) BRCA1 loss activates cathepsin L-mediated degradation of 53BP1 in breast cancer cells. J. Cell Biol. 200, 187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez-Suarez I., Redwood A. B., Grotsky D. A., Neumann M. A., Cheng E. H., Stewart C. L., Dusso A., and Gonzalo S. (2011) A new pathway that regulates 53BP1 stability implicates cathepsin L and vitamin D in DNA repair. EMBO J. 30, 3383–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonzalez-Suarez I., Redwood A. B., Perkins S. M., Vermolen B., Lichtensztejin D., Grotsky D. A., Morgado-Palacin L., Gapud E. J., Sleckman B. P., Sullivan T., Sage J., Stewart C. L., Mai S., and Gonzalo S. (2009) Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 28, 2414–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibbs-Seymour I., Markiewicz E., Bekker-Jensen S., Mailand N., and Hutchison C. J. (2015) Lamin A/C-dependent interaction with 53BP1 promotes cellular responses to DNA damage. Aging Cell 14, 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitcomb E. A., Dudek E. J., Liu Q., and Taylor A. (2009) Novel control of S phase of the cell cycle by ubiquitin-conjugating enzyme H7. Mol. Biol. Cell 20, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burden R. E., Gormley J. A., Kuehn D., Ward C., Kwok H. F., Gazdoiu M., McClurg A., Jaquin T. J., Johnston J. A., Scott C. J., and Olwill S. A. (2012) Inhibition Of Cathepsin S by Fsn0503 enhances the efficacy of chemotherapy in colorectal carcinomas. Biochimie 94, 487–493 [DOI] [PubMed] [Google Scholar]
- 31. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He H., Dang Y., Dai F., Guo Z., Wu J., She X., Pei Y., Chen Y., Ling W., Wu C., Zhao S., Liu J. O., and Yu L. (2003) Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 278, 29278–29287 [DOI] [PubMed] [Google Scholar]
- 33. Méndez J., and Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y. W., Otterness D. M., Chiang G. G., Xie W., Liu Y. C., Mercurio F., and Abraham R. T. (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell 19, 607–618 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y. W., Brognard J., Coughlin C., You Z., Dolled-Filhart M., Aslanian A., Manning G., Abraham R. T., and Hunter T. (2009) The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol. Cell 35, 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grøfte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S., Mailand N., Neumann B., Heriche J. K., Shearer R., Saunders D., Bartek J., Lukas J., and Lukas C. (2012) TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 [DOI] [PubMed] [Google Scholar]
- 37. Moudry P., Lukas C., Macurek L., Neumann B., Heriche J. K., Pepperkok R., Ellenberg J., Hodny Z., Lukas J., and Bartek J. (2012) Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 19, 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson B. R., Nitta R. T., Frock R. L., Mounkes L., Barbie D. A., Stewart C. L., Harlow E., and Kennedy B. K. (2004) A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9677–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanders S. L., Portoso M., Mata J., Bähler J., Allshire R. C., and Kouzarides T. (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 [DOI] [PubMed] [Google Scholar]
- 40. Bothmer A., Robbiani D. F., Di Virgilio M., Bunting S. F., Klein I. A., Feldhahn N., Barlow J., Chen H. T., Bosque D., Callen E., Nussenzweig A., and Nussenzweig M. C. (2011) Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fradet-Turcotte A., Canny M. D., Escribano-Díaz C., Orthwein A., Leung C. C., Huang H., Landry M. C., Kitevski-LeBlanc J., Noordermeer S. M., Sicheri F., and Durocher D. (2013) 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J., Han X., Feng X., Wang Z., and Zhang Y. (2012) Coupling cellular localization and function of checkpoint kinase 1 (chk1) in checkpoints and cell viability. J. Biol. Chem. 287, 25501–25509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J., Han X., and Zhang Y. (2012) Autoregulatory mechanisms of phosphorylation of checkpoint kinase 1. Cancer Res. 72, 3786–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]