Abstract
Rotavirus causes significant morbidity and mortality among children worldwide. Stool samples from a previous hospital-based surveillance study to detect diarrhea etiology at the National Pediatric Hospital in Phnom Penh, Cambodia, by Meng and others in 2011 were tested for rotavirus by real-time reverse transcription polymerase chain reaction (PCR) targeting vp6 gene and characterized for G- and P-genotypes of positive samples based on vp7 and vp4 genes, respectively. Rotavirus was detected in 159/531 (30%) of children with diarrhea and none was detected in 287 nondiarrhea controls. All but three of the rotavirus-positive cases were children under the age of 2. The most common genotypes characterized by PCR and sequencing were G1P[8] (69%), G9P[8] (11%), and G2P[4] (11%). Genotype G9 was detected at a relatively high percentage that is consistent with the global trend and found to be associated with hospitalization. Data on disease burden and genotypic distribution are required information for the planning of rotavirus vaccine implementation in Cambodia.
Introduction
Infections with rotavirus result in significant morbidity and mortality in young children worldwide. Rotavirus is responsible for the majority of severe diarrhea comprising a large number of cases in children admitted to hospitals and hospital-based deaths due to diarrhea.1 A report in 2016 by the World Health Organization estimated that 215,000 deaths (3.4%) in children worldwide in 2013 were due to rotavirus.2 Spreading by person to person, rotavirus has continued to be the leading cause of diarrhea in children in both developing and developed countries despite improvements to water and sanitation systems.1
The most common rotavirus infecting humans belongs to genogroup A and is classified based on the outer capsid proteins VP7 and VP4.3 Sequences of vp7 and vp4 genes are used in genotypic characterization into G- and P-genotypes, respectively. Currently, there are 27 G-genotypes and 35 P-genotypes identified4 and at least 73 G/P-genotype combinations.5 Genotypes G1-G4 and G9 have been the most commonly detected G-genotypes among children worldwide.6,7
The distribution of different rotavirus genotype combinations is geographically variable. The common genotype combination, G1P[8], has been reported to be the predominant strain in North America, Europe, and Australia, whereas prevalent strains in other regions, for example, South America, Asia, or Africa vary including other common genotype combinations such as G2P[4] and G3P[8], and uncommon genotypes in 30–50% of reported cases.6,7 The currently available rotavirus vaccines, RotaTeq® (Merck and Co Inc., Kenilworth, NJ) and Rotarix® (GlaxoSmithKline Inc., Brentford, United Kingdom), were developed from rotavirus genotypes G1-G4 with P[5] and G1P[8], respectively.3 Country-specific data on the burden of diarrhea due to rotavirus are important as it provides information for country leaders and health policy makers to make informed decisions on whether to prioritize the introduction of these vaccines in their countries.8,9 It is also highly critical that surveillance for genotypic distribution continues after the implementation of the vaccines to determine the decrease in disease burden and a potential shift of genotypic distribution from selective pressure of vaccine to better understand the effectiveness and efficacy of the vaccine.
In this study, rotavirus was detected by real-time reverse transcriptase (RT) polymerase chain reaction (PCR) from stool samples collected in a previous diarrhea etiology surveillance study in young children at the National Pediatric Hospital (NPH) in Phnom Penh, Cambodia, from November 2004 to October 2006.10 Molecular epidemiology and genotypic distribution of rotavirus and the emergence of genotype G9 in Cambodia are reported here.
Materials and Methods
Source of specimens.
Stool samples used in this study were a subset of case and control samples from a diarrhea etiology surveillance study in children aged 3 months to 5 years conducted at the NPH in Phnom Penh, Cambodia, from November 2004 to October 2006.10 Cases were inpatient and outpatient with acute diarrhea of no more than 72 hours. Unmatched controls were inpatient and outpatient children who attended the hospital for other reasons and had not had diarrhea within the previous 2 weeks. Informed consent was obtained from one parent or a guardian for each participant. Rotavirus detection by enzyme-linked immunosorbent assay (ELISA) was previously performed on these samples.10 The study was approved by the National Ethics Committee for Health Research, Cambodia, and the Walter Reed Army Institute of Research Institutional Review Board, United States.
Nucleic acid isolation from stool specimens.
A 10% (wt/vol) stool suspension was prepared with sterile deionized water and clarified by centrifugation at 600 × g for 15 minutes. Total nucleic acid was isolated from 300 μL of stool suspension using the NucliSens® Magnetic Nucleic Acid Isolation Kit (BioMérieux, Inc., Durham, NC) according to manufacturer's instruction. Total nucleic acid was eluted from magnetic silica beads into 100 μL of elution buffer. Purified nucleic acid was used immediately in real-time RT PCR or stored at −70°C until use.
Real-time one-step RT PCR assays for rotavirus detection.
Probes and primers used for rotavirus detection were combinations of probes and primers designed for this study or modified from published literature (Table 1).11 Real-time RT PCR assays were conducted in a 15 μL reaction volume containing 1-μL nucleic acid template, TaqMan EZ buffer (Applied Biosystems, Foster City, CA), 0.3 mM of dATP, dCTP, dGTP each, 0.6 mM dUTP, 3.0 mM manganese acetate, 0.3 μM of forward and reverse primer each, 50 nM of TaqMan probe (Table 1), 1.5 U rTth DNA polymerase, and 0.15 U uracyl N-glycosylase (UNG). Reaction mixtures were incubated in an ABI7900 Sequence Detection System (Applied Biosystems). Amplification profile consisted of UNG treatment at 50°C for 2 minutes, RT at 60°C for 30 minutes, heat activation at 95°C for 5 minutes, followed by 40 cycles of 95°C for 20 seconds and 60°C for 60 seconds. The samples were considered positive when the number of cycles was less than 40.
Table 1.
Sequences of primers and probes used for the detection, amplification for genotyping, and sequencing of rotavirus in this study
| Primer | Gene/type | Sequence 5′ to 3′ | Position | Reference |
|---|---|---|---|---|
| VP6 screening primers/probes | ||||
| VP6-F1b | vp6 | GGATGTCCTGTACTCCTTRTCAAAA | 26-50 | Modified11 |
| VP6-R1 | vp6 | TCCAGTTTGGAACTCATTTCCA | 170-149 | 11 |
| VP6-P1 (Probe) | vp6 | ATAATGTGCCTTCGACAAT | 93-75 | 11 |
| VP6-F2b | vp6 | AGTCTTCGACATGGAGGTTCTGTA | 14-37 | This study |
| VP6-R2c | vp6 | CCAATTCCTCCAGTTTGAAAGTC | 178-156 | This study |
| VP6-P2 (Probe) | vp6 | AATATAATGTACCTTCAACAAT | 96-75 | 11 |
| G-genotyping (VP7) primers | ||||
| Beg9 | vp7 | GGCTTTAAAAGAGAGAATTTCCGTCTGG | 1-28 | 12 |
| End9 | vp7 | GGTCACATCATACAATTCTAATCTAAG | 1062-1036 | 12 |
| 9Con1 | vp7 | TAGCTCCTTTTAATGTATGG | 37-56 | 13 |
| 9Con2d | vp7 | GTATAAAAHACTTGCCACCA | 941-922 | Modified13 |
| 9T1-1d | G1 | TCTTGTCAARGCAAATAATG | 195-176 | Modified13 |
| 9T1-2 | G2 | GTTAGAAATGATTCTCCACT | 281-262 | 13 |
| 9T-3Pd | G3 | ATGTCYAGTTGCAGTGTAGC | 503-484 | Modified13 |
| 9T-4 | G4 | GGGTCGATGGAAAATTCT | 440-423 | 13 |
| 9T-9B | G9 | TATAAAGTCCATTGCAC | 147-131 | 13 |
| G12Fd | G12 | TYGTCATGCTGCCATTTA | 173-190 | Modified14 |
| G12Rd | G12 | GTCCARTCGGGRTCAGTT | 344-327 | Modified14 |
| P-genotyping (VP4) primers | ||||
| Con3d | vp4 | TGGCTTCRCTCATTTATAGACA | 11-32 | 15 |
| Con2d | vp4 | ATTTCDGACCATTTATAACC | 887-868 | 15 |
| VP4F | vp4 | TATGCTCCAGTNAATTGG | 132-149 | 16 |
| VP4R | vp4 | ATTGCATTTCTTTCCATAATG | 775-795 | 16 |
| P[4]d | P[4] | CTATTRTTAGAGGTTARAGTC | 494-474 | Modified15 |
| P[6]d | P[6] | TGTTGATYAGTTGGATTCAA | 278-259 | Modified15 |
| P[8] | P[8] | TCTACTGGRTTRACNTGC | 356-339 | 17 |
| P[8]m | P[8] | TCTACTGGATYGACGTGC | 356-339 | Modified15 |
| P[8]G1 | P[8] | TATATTGTCTATCTACTGGAT | 357-339 | This study |
| P[9] | P[9] | TGAGACATGCAATTGGAC | 402-385 | 15 |
| P[10] | P[10] | ATCATAGTTAGTAGTCGG | 594-575 | 15 |
| P[11] | P[11] | GTAAACATCCAGAATGTG | 323-306 | 17 |
| Sequencing primers | ||||
| Beg9–End 9 | vp7 | |||
| 9con1–9con2d | vp7 | |||
| Con3d–Con2d | vp4 | |||
| VP4F–VP4R | vp4 | |||
D = A/G/T; H = A/C/T; M = A/C; N = A/C/G/T; R = A/G; Y = C/T.
Genotyping of rotavirus by conventional RT PCR.
Two microliters of nucleic acid template was mixed with 0.5 μL of 10× DNase buffer, 1 μL of 10−4 diluted DNaseI (stock 1 U/μL; Invitrogen, Sao Paulo, Brazil) and 1.5 μL of deionized water and incubated at room temperature for 15 minutes. Then, 0.5 μL of 25 mM ethylenediaminetetraacetic acid was added and the reaction was incubated at 65°C for 10 minutes to inactivate the DNase.
RNA was reverse transcribed with the following primers: Con-3d for vp4 and 9Con1 for vp7 (Table 1). RT reactions were conducted in a total volume of 20 μL containing 1× of RT-PCR buffer, 1.0 mM of each dNTP, 2.5 mM of MgCl2, 10 mM of DTT, 15 U of Multiscribe reverse transcriptase (Applied Biosystems), 10 U of RNase inhibitor, 300 nM of primer, 5 μL of DNase-treated RNA. The reactions were then incubated at 42°C for 20 minutes.
A 20 μL reaction contained 1× PCR buffer, 0.8 mM dNTP mixture, 2.5 mM MgCl2, 0.5 U Taq DNA polymerase (Applied Biosystems), 3 μL of RT mixture, and 200 nM of each genotyping primer; P-typing: Con-3d and P[4]d, P[6]d, P[8], P[8]m, P[8]G1, P[9], P[10], or P[11]; for G-typing: 9Con1 and 9T1-1d, 9T1-2, 9T1-3Pd, 9T-4, or 9T-9B; for G12-typing: G12Fd and G12Rd (Table 1) was mixed and subjected to 40 cycles of amplification consisting of denaturation for 30 seconds at 95°C, annealing for 2 minutes at 50°C and extension for 1 minute at 72°C.
The resulting PCR products were analyzed on a 1.5% agarose gel along with 100-base pair DNA ladder (Invitrogen), and visualized by ethidium bromide staining. The size of each band indicated its specific genotype.
Nucleotide sequencing and phylogenetic analysis of nontypeable rotavirus strains.
RNA samples that were positive for rotavirus but their genotypes could not be determined by conventional RT PCR were reverse transcribed and amplified using sequencing primers as shown in Table 1 and sequenced by Macrogen Inc., Korea. DNA sequencing data were verified for consensus sequences by using Sequencher software, version 4.1.2 (Gene Codes Corporation, Ann Arbor, MI). Multiple sequence alignments were generated using consensus sequences and sequences of rotavirus P-genotype prototype with Clustal W18 (EMBL-EBI, Hinxton, United Kingdom) and a phylogenetic tree was constructed using the neighbor-joining method with the Kimura 2-parameter model with 1,000 bootstrap replicates in MEGA, version 6.19
Statistical analysis.
The differences among proportions were analyzed by χ2 test in IBM SPSS® Statistics, version 22 (IBM Corporation, Armonk, NY).
Results
A total of 818 stools samples (531 cases and 287 controls) were available for real-time RT PCR detection of rotavirus. In this study, rotavirus was detected in 159/531 (30%) of diarrheal cases and none was detected from control samples by real-time PCR. A comparison of the proportion of stool samples positive by real-time RT PCR (159/531, 30%) to the same samples tested by ELISA (142/531, 26.7%) showed no statistically significant difference (P = 0.25).
All rotavirus-positive samples were from children less than 2 years of age except for three cases (Table 2). Rotavirus was detected significantly higher among inpatient cases at 54% (22/41) comparing to 28% (137/490) of outpatient cases (P < 0.001). Clinical presentations of cases with rotavirus diarrhea revealed that approximately 78% of cases had diarrhea five or more times per day, 99% had watery diarrhea, 94% had history of fever, 94% had fatigue, and 82% had vomiting.
Table 2.
Detection of rotavirus in diarrhea case and nondiarrhea control samples by age group, at the National Pediatric Hospital, Phnom Penh, Cambodia, from November 2004 to October 2006
| Age (months) | Case | Control |
|---|---|---|
| No. of rotavirus positive/no. of tested (%) | No. of rotavirus positive/no. of tested (%) | |
| N = 531 | N = 287 | |
| 0–5 | 21/89 (23.6) | 0/6 (0) |
| 6–11 | 81/239 (33.9) | 0/29 (0) |
| 12–23 | 54/173 (31.2) | 0/51 (0) |
| 24–35 | 3/16 (18.8) | 0/60 (0) |
| > 36 | 0/14 (0) | 0/141 (0) |
| Total | 159/531 (30) | 0/287 (0) |
Genotyping of rotavirus-positive samples showed that G1 was the predominant genotype representing 70% of all rotavirus samples, followed by G2 at 14%, G9 at 12%, G3 at 2%, and G4 at 2%. All samples were typeable for G-genotype. For P-genotype, P[8] was the most common genotype with 83% followed by P[4] at 12%. Two samples could not be characterized for P-genotype by conventional RT PCR. The most common G- and P-genotypes combination was G1P[8] (69%) followed by G2P[4] and G9P[8] each contributing at 11.3% of rotavirus-positive samples. Genotype G1P[8] (75.2%) predominated in outpatient cases, whereas G9P[8] (54.6%) predominated in hospitalized cases. The genotypic distribution of outpatient cases was significantly different from hospitalized cases, P < 0.001 (Table 3).
Table 3.
Genotypic distribution of rotavirus among inpatient and outpatient cases at the National Pediatric Hospital in Phnom Penh, Cambodia, from November 2004 to October 2006
| Genotype | No. of Inpatient (%) | No. of Outpatient (%) |
|---|---|---|
| G1P[8] | 6 (27.3)* | 103 (75.2)* |
| G1PNT | – | 2 (1.5) |
| G2P[4] | – | 18 (13.1) |
| G2P[8] | 1 (4.6) | 3 (2.2) |
| G3P[6] | – | 1 (0.7) |
| G3P[8] | 1 (4.6) | – |
| G3P[19] | – | 1 (0.7) |
| G4P[4] | – | 1 (0.7) |
| G4P[6] | – | 2 (1.5) |
| G4P[8] | 1 (4.6) | – |
| G9P[8] | 12 (54.6)* | 6 (4.4)* |
| G9P[19] | 1 (4.6) | – |
| Total | 22 (100) | 137 (100) |
PNT = nontypeable P-genotype.
P < 0.001, χ2 test.
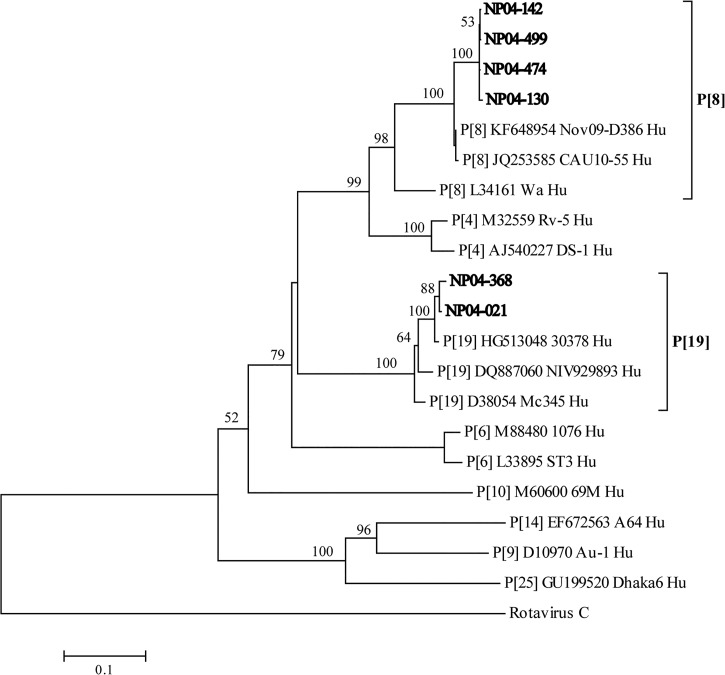
Phylogenetic analysis of sequenced samples clustered four samples (NP04-130, NP04-142, NP04-474, and NP04-499) into P[8] genotype and two samples (NP04-021 and NP04-368) into P[19] genotype (Figure 1 ). Nucleotide sequences of vp4 gene of P-genotype have been submitted to GenBank (Bethesda, MD) under these accession numbers with sample numbers indicated in parentheses: JQ360846 (NP04-130), JQ360847 (NP04-142), JQ360848 (NP04-474), JQ360849 (NP04-499), JQ360850 (NP04-021), and JQ360851 (NP04-368).
Figure 1.
Phylogenetic tree of vp4 gene fragment amplified from six nontypeable P-genotype samples with sequencing primers (Table 1). Fourteen reference rotavirus strains representing P[4]: RV-5 (M32559), DS-1 (AJ540227); P[6]: 1076 (M88480), ST3 (L33895); P[8]: Wa (L34161), Nov09-D386 (KF648954), CAU10-55 (JQ253585); P[9]: AU-1 (D10970); P[10]: 69M (M60600); P[14]: A64 (EF672563); P[19]: Mc345 (D38054), 30378 (HG513048), NIV929893 (DQ887060); P[25]: Dhaka6 (GU199520); and rotavirus serotype C (AB648917) from GenBank were used in this analysis. Bootstrap values (1,000 replicates) above 50 are shown. Sequenced nontypeable P-genotype samples are indicated in bold by their samples names. The human rotavirus serotype C sequence was used as an outgroup.
Discussion
In this study, rotavirus was detected by real-time RT PCR in 30% of diarrhea cases. The rotavirus prevalence among hospitalized cases of 54% is similar to the reported 56% by Nyambat and others, whose 2-year surveillance study was also conducted at overlapping period at the NPH in Phnom Penh, Cambodia, but using ELISA techniques for detection.20 Molecular technique has been widely accepted as a more sensitive method for rotavirus detection; however, ELISA is a more practical and cost-effective method to detect rotavirus as recommended by the U.S. Center for Disease Control especially in resource-limited areas.21
Unlike many of the studies on rotavirus surveillance that focus on hospitalized cases, this study included outpatient diarrhea cases and nondiarrhea controls for comparison. Our findings support rotavirus as an important causative agent of diarrhea and its association with a severe disease requiring hospitalization. Moreover, rotavirus was not detected by a sensitive method as real-time PCR in any of the 287 stool samples of children without diarrhea in this study. However, this finding may be biased by the age distribution of cases and nondiarrhea controls as more than half of controls were children older than 3 years old while over 90% of cases in this study were children less than 2 years of age.
In a previous study reporting the genotypic distribution of rotavirus in Cambodia among 10% of randomly selected samples showed G1P[8] as the predominating genotype (53%), followed by G2P[4] (10%) and 14% and 29% of nontypeable G- and P-genotypes, respectively.20 In our study, in addition to G1P[8] and G2P[4] that were the first and second most common genotypes detected, G9P[8] was also identified in 11.3%, at the same proportion as G2P[4]. Moreover, G9P[8] was detected at a significantly higher percentage (54.6%) in inpatient cases where as only 4.4% of outpatient cases were infected with G9P[8] (P < 0.001) indicating that G9 might play an important role in more severe diarrhea cases requiring hospitalization. Other studies also reported an increased predominance of G9 and its association with severe diarrhea in Latin America22 and in the United Kingdom where 71% of patients infected with G9P2A[6] required hospitalization and 33% of those who were admitted were severely dehydrated.23 However, G9 association with increased severity is debatable as others reported that G1 had greater association with severe dehydration when compared with G9,24,25 whereas other studies showed no difference of severity with any specific G-genotypes.26–28 Differences in clinical severity associated G-genotypes may rely on the geographical origin of the strain or associated P-genotypes.23 In addition, a significant difference of genotype distribution of rotavirus between outpatient and hospitalized cases may suggest that surveillance of rotavirus in only hospitalized severe diarrhea cases may not accurately represent distribution of rotavirus strains circulating in the area.
The currently available vaccines that include G1-G4 proteins and a live-attenuated G1P[8] strains will have a potential impact on the reduction of morbidity of rotavirus infections in Cambodia as supported by our data that approximately of 88% of rotavirus in this study belonged to genotypes that are covered by the vaccines. G9 has not been included in any commercially available rotavirus vaccines, except for the Rotavac® Bharat Biotech, Hyderabad, India which is licensed for use only in India.29 This incomplete vaccine coverage of rotavirus genotypes could raise concerns on vaccine efficacy though some cross protection between genotypes has been reported.30–32 The worldwide emergence of G9 rotavirus and its potential association with more severe disease should be considered in the future development of rotavirus vaccines to improve vaccine efficacy especially in areas where genotype G9 has been shown to be prevalent.
ACKNOWLEDGMENTS
We would like to thank Bryan Smith and the staff members of the National Pediatric Hospital, Phnom Penh, Cambodia, and the Department of Enteric Diseases, AFRIMS for their support on the conduct of the study and laboratory testing.
Disclaimer: The view expressed in this manuscript are those of the author(s) and do not reflect the official policy of the Department of the Army, Department of Defense, or the U.S. Government. Trade names are used for identification purposes only and do not imply endorsement.
Footnotes
Financial support: This project was supported by Armed Forces Health Surveillance Center-Global Emerging Infections Surveillance (AFHSC-GEIS).
Authors' addresses: Sasikorn Silapong, Pimmada Sakpaisal, Ladaporn Bodhidatta, Paphavee Lertsethtakarn, Orntipa Sethabutr, and Brett E. Swierczewski, Department of Enteric Diseases, Armed Forces Research Institute of Medical Sciences (AFRIMS) Bangkok, Thailand, E-mails: sasikorns@afrims.org, pimmadaJ@afrims.org, ladapornb@afrims.org, paphaveeL@afrims.org, aey.orntipa@gmail.com, and brett.swierczewski.mil@afrims.org. Ket Vansith and Chhour Y. Meng, National Pediatric Hospital, Phnom Penh, Cambodia, E-mails: vansith52@gmail.com and cymeng.nph@online.com. Carl J. Mason, Seattle, WA, E-mail: carlmason@icloud.com.
References
- 1.Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) Estimated Rotavirus Deaths for Children Under 5 Years of Age: 2013, 215 000. 2016. http://www.who.int/immunization/monitoring_surveillance/burden/estimates/rotavirus/en/ Available at. Accessed July 13, 2016.
- 3.Dennehy PH. Rotavirus infection: an update on management and prevention. Adv Pediatr. 2012;59:47–74. doi: 10.1016/j.yapd.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthijnssens J, Bilcke J, Ciarlet M, Martella V, Banyai K, Rahman M, Zeller M, Beutels P, Van Damme P, Van Ranst M. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 2009;4:1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- 6.Patton JT. Rotavirus diversity and evolution in the post-vaccine world. Discov Med. 2012;13:85–97. [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization (WHO) Global Rotavirus Information and Surveillance Bulletin. Reporting Period: January to December. 2011. http://www.who.int/immunization/diseases/rotavirus/RV_bulletin_Jan_Dec_2011_FINAL.pdf Available at. Accessed September 29, 2016.
- 8.Widdowson MA, Steele D, Vojdani J, Wecker J, Parashar U. Global rotavirus surveillance: determining the need and measuring the impact of rotavirus vaccines. J Infect Dis. 2009;200:S1–S8. doi: 10.1086/605061. [DOI] [PubMed] [Google Scholar]
- 9.Patel MM, Parashar UD. Assessing the effectiveness and public health impact of rotavirus vaccines after introduction in immunization programs. J Infect Dis. 2009;200:S291–S299. doi: 10.1086/605059. [DOI] [PubMed] [Google Scholar]
- 10.Meng CY, Smith BL, Bodhidatta L, Richard SA, Vansith K, Thy B, Srijan A, Serichantalergs O, Mason CJ. Etiology of diarrhea in young children and patterns of antibiotic resistance in Cambodia. Pediatr Infect Dis J. 2011;30:331–335. doi: 10.1097/INF.0b013e3181fb6f82. [DOI] [PubMed] [Google Scholar]
- 11.Logan C, O'Leary JJ, O'Sullivan N. Real-time reverse transcription-PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children. J Clin Microbiol. 2006;44:3189–3195. doi: 10.1128/JCM.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer TK, Steinsland H, Molbak K, Ca R, Gentsch JR, Valentiner-Branth P, Aaby P, Sommerfelt H. Genotype profiles of rotavirus strains from children in a suburban community in Guinea-Bissau, Western Africa. J Clin Microbiol. 2000;38:264–267. doi: 10.1128/jcm.38.1.264-267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castello AA, Arguelles MH, Rota RP, Olthoff A, Jiang B, Glass RI, Gentsch JR, Glikmann G. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J Clin Microbiol. 2006;44:2046–2050. doi: 10.1128/JCM.02436-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, Gentsch JR, Gray JJ, Kirkwood C, Page N, Iturriza-Gomara M. New oligonucleotide primers for P-typing of rotavirus strains: strategies for typing previously untypeable strains. J Clin Virol. 2008;42:368–373. doi: 10.1016/j.jcv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Iturriza-Gomara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31:259–265. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyambat B, Meng CY, Vansith K, Vuthy U, Rin E, Kirkwood C, Bogdanovic-Sakran N, Kilgore PE. Hospital-based surveillance for rotavirus diarrhoea in Phnom Penh, Cambodia, March 2005 through February 2007. Vaccine. 2009;27:F81–F84. doi: 10.1016/j.vaccine.2009.08.085. [DOI] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention (CDC) Rotavirus: Clinical Information. 2014. http://www.cdc.gov/rotavirus/clinical.html Available at. Accessed July 13, 2016.
- 22.Linhares AC, Verstraeten T, Wolleswinkel-van den Bosch J, Clemens R, Breuer T. Rotavirus serotype G9 is associated with more-severe disease in Latin America. Clin Infect Dis. 2006;43:312–314. doi: 10.1086/505493. [DOI] [PubMed] [Google Scholar]
- 23.Cubitt WD, Steele AD, Iturriza M. Characterisation of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6] J Med Virol. 2000;61:150–154. doi: 10.1002/(sici)1096-9071(200005)61:1<150::aid-jmv24>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 24.Bahl R, Ray P, Subodh S, Shambharkar P, Saxena M, Parashar U, Gentsch J, Glass R, Bhan MK, Grp DRS. Incidence of severe rotavirus diarrhea in New Delhi, India, and G and P types of the infecting rotavirus strains. J Infect Dis. 2005;192:S114–S119. doi: 10.1086/431497. [DOI] [PubMed] [Google Scholar]
- 25.Sudarmo SM, Shigemura K, Athiyyah AF, Osawa K, Wardana OP, Darma A, Ranuh R, Raharjo D, Arakawa S, Fujisawa M, Shirakawa T. Genotyping and clinical factors in pediatric diarrhea caused by rotaviruses: one-year surveillance in Surabaya, Indonesia. Gut Pathog. 2015;7:3. doi: 10.1186/s13099-015-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aupiais C, de Rougemont A, Menager C, Vallet C, Brasme JF, Kaplon J, Pothier P, Gendrel D. Severity of acute gastroenteritis in infants infected by G1 or G9 rotaviruses. J Clin Virol. 2009;46:282–285. doi: 10.1016/j.jcv.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Rivera R, Forney K, Castro MR, Rebolledo PA, Mamani N, Patzi M, Halkyer P, Leon JS, Iniguez V. Rotavirus genotype distribution during the pre-vaccine period in Bolivia: 2007–2008. Int J Infect Dis. 2013;17:e762–e767. doi: 10.1016/j.ijid.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Rougemont A, Kaplon J, Pillet S, Mory O, Gagneur A, Minoui-Tran A, Meritet JF, Mollat C, Lorrot M, Foulongne V, Gillet Y, Nguyen-Bourgain C, Alain S, Agius G, Lazrek M, Colimon R, Fontana C, Gendrel D, Pothier P, French Rotavirus Network. Molecular and clinical characterization of rotavirus from diarrheal infants admitted to pediatric emergency units in France. Pediatr Infect Dis J. 2011;30:118–124. doi: 10.1097/INF.0b013e3181ef034e. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organization (WHO) Global Advisory Committee on Vaccine Safety, 11–12 June 2014. 2014. http://www.who.int/vaccine_safety/committee/reports/jul_2014/en/ Available at. Accessed December 13, 2016.
- 30.Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, Shinefield HR, Christie CD, Ylitalo S, Itzler RF, Coia ML, Onorato MT, Adeyi BA, Marshall GS, Gothefors L, Campens D, Karvonen A, Watt JP, O'Brien KL, DiNubile MJ, Clark HF, Boslego JW, Offit PA, Heaton PM. Rotavirus Efficacy Safety Trial Study Team Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, Cervantes Y, Linhares AC, Lopez P, Macias-Parra M, Ortega-Barria E, Richardson V, Rivera-Medina DM, Rivera L, Salinas B, Pavia-Ruz N, Salmeron J, Ruttimann R, Tinoco JC, Rubio P, Nunez E, Guerrero ML, Yarzabal JP, Damaso S, Tornieporth N, Saez-Llorens X, Vergara RF, Vesikari T, Bouckenooghe A, Clemens R, De Vos B, O'Ryan M. Human Rotavirus Vaccine Study Group Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Haq N, Amjad M, McGrath E, Chearskul P, Amer A, Salimnia H, Asmar BI. Emergence of human rotavirus genotype G9 in metropolitan Detroit between 2007 and 2009. J Med Microbiol. 2011;60:761–767. doi: 10.1099/jmm.0.026807-0. [DOI] [PubMed] [Google Scholar]