Abstract
Junctional adhesion molecule C (JAM-C) is an immunoglobulin superfamily protein expressed in epithelial cells, endothelial cells, and leukocytes. JAM-C has been implicated in leukocyte transendothelial migration, angiogenesis, cell adhesion, cell polarity, spermatogenesis, and metastasis. Here, we show that JAM-C undergoes S-palmitoylation on two juxtamembrane cysteine residues, Cys-264 and Cys-265. We have identified DHHC7 as a JAM-C palmitoylating enzyme by screening all known palmitoyltransferases (DHHCs). Ectopic expression of DHHC7, but not a DHHC7 catalytic mutant, enhances JAM-C S-palmitoylation. Moreover, DHHC7 knockdown decreases the S-palmitoylation level of JAM-C. Palmitoylation of JAM-C promotes its localization to tight junctions and inhibits transwell migration of A549 lung cancer cells. These results suggest that S-palmitoylation of JAM-C can be potentially targeted to control cancer metastasis.
Keywords: cell adhesion, cell migration, chemical biology, post-translational modification (PTM), protein palmitoylation, protein-lipid interaction, tight junction
Introduction
Junctional adhesion molecule C (JAM-C)3 is a member of the JAM family and is localized at the cell-cell contact sites, in particular the tight junction (TJ) region of endothelial and epithelial cells (1–3). JAM-C is a type I transmembrane protein with two conserved immunoglobulin-like domains in the extracellular amino (N)-terminal region (Fig. 1A). At its intracellular carboxyl (C)-terminal region, JAM-C contains a PDZ-binding domain motif, which allows JAM-C to interact with other PDZ motif-containing proteins at the TJ, including ZO-1 and PAR-3 (1–3).
FIGURE 1.
JAM-C is an immunoglobulin superfamily protein containing two conserved cysteine residues (Cys-264 and Cys-265, human sequence). A, schematic structure of JAM-C transmembrane protein with two Ig-like domains in the extracellular region and one PDZ-binding motif in the cytoplasmic region. B, multiple sequence alignment by Clustal Omega showing the two cytoplasmic cysteine residues that are conserved across multiple species.
JAM-C has been found to regulate leukocyte adhesion and transmigration across endothelial cells through a heterophilic interaction between endothelial JAM-C and leukocyte integrin MAC-1 (αMβ2) (4). JAM-C is essential for polarized round spermatids, and JAM-C-deficient mice have defective spermatid differentiation (5). JAM-C deletion in mice leads to neuropathy, and some Jam-C mutations affect the integrity of the vascular system and brain (6, 7).
Recently, JAM-C has been shown to play a role in metastasis and development of certain cancer cells (8–11). In melanoma cells, the homophilic JAM-C/JAM-C trans-interaction between melanoma cells and endothelial cells promotes metastasis (12). Additionally, the JAM-C expression level in fibrosarcoma and lung cancer cells is reported to be positively correlated to metastasis. Knocking down JAM-C in highly metastatic lung cancer cells leads to a decrease in cell migration (8, 13). JAM-C, therefore, could be a therapeutic target for certain cancers.
Cys palmitoylation (S-palmitoylation), a reversible lipid post-translational modification, is the addition of a 16-carbon palmitoyl group onto cysteine residues of proteins via a labile thioester bond. S-Palmitoylation plays a crucial role in cell signaling, localization, and protein-protein interactions (14, 15). Palmitoyltransferases (DHHCs) catalyze S-palmitoylation. To date, 23 mammalian DHHCs have been identified (16, 17). Here we showed that JAM-C undergoes S-palmitoylation on two membrane-proximal cysteine residues (Cys-264 and Cys-265), and this modification can be catalyzed by DHHC7. We found that S-palmitoylation of JAM-C promotes its localization to the cell-cell contact region and regulates cell migration.
Results
JAM-C Is S-Palmitoylated on Two Conserved Juxtamembrane Cysteine Residues, Cys-264 and Cys-265
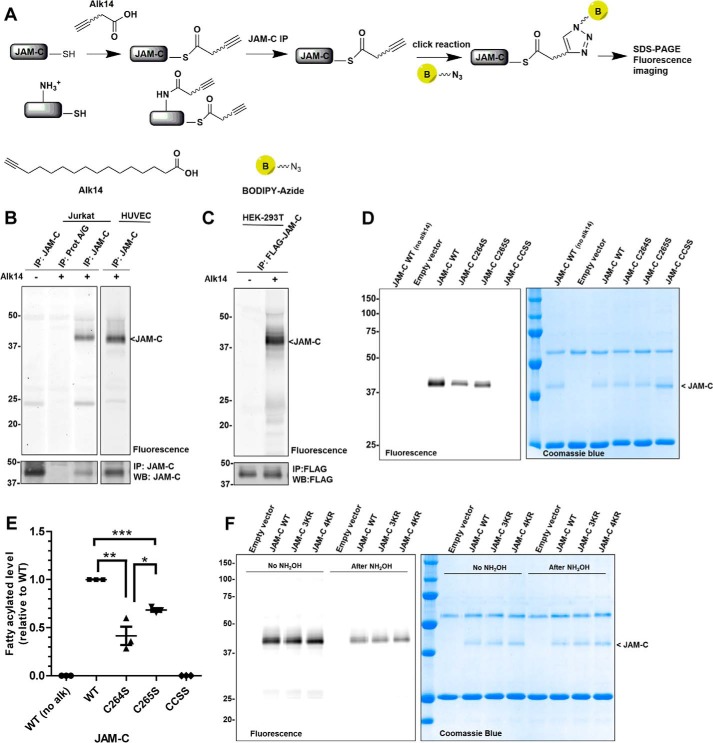
In addition to a PDZ-binding domain, the JAM-C intracellular C terminus contains two transmembrane proximal cysteine residues (Fig. 1A). These two cysteine residues are conserved across multiple mammalian species, suggesting these residues may have an important role (Fig. 1B). Moreover, JAM-C has been identified in palmitoyl-protein proteomics studies as a protein that potentially has Cys palmitoylation, but it was not validated (18). To test whether S-palmitoylation occurs on JAM-C, we used a metabolic labeling strategy using an alkyne-tagged palmitic acid analogue (Alk14) (19, 20). Jurkat cells and human umbilical vein endothelial cells were cultured in the presence of Alk14, and endogenous JAM-C was immunoprecipitated and conjugated to BODIPY-azide via click chemistry (Fig. 2A). JAM-C had fluorescent labeling, suggesting that it contains fatty acylation (Fig. 2B). Similar results were obtained from ectopically expressed FLAG-tagged JAM-C in human embryonic kidney 293T (HEK-293T) cells (Fig. 2C and supplemental Fig. S1).
FIGURE 2.
S-Palmitoylation of JAM-C depends on Cys-264 and Cys-265. A, method for the detection of S-palmitoylation in JAM-C with Alk14, the palmitic acid analogue. Cells were cultured with Alk14 for metabolic labeling. JAM-C was immunoprecipitated from total lysate, and BODIPY-azide was then conjugated to the alkyne group using click chemistry. The fluorescence signal was imaged after SDS-PAGE. B, endogenous JAM-C in both Jurkat and human umbilical vein endothelial cells (HUVEC) was labeled by Alk14. C, overexpressed FLAG-tagged JAM-C in HEK-293T cells also contained fatty acylation. D, the C264S and C265S mutations of JAM-C decreased the Alk14 labeling signal. Compared with the wild type JAM-C, palmitoylation in the single cysteine mutants (C264S and C265S) was reduced, whereas it was abolished in the double cysteine mutant (CCSS). E, quantified fatty acylation level of the JAM-C mutants relative to WT (n = 3; error bars represent S.D.). The palmitoylation signal was quantified and normalized by the protein level on the Coomassie Blue gel using Quantity One software. *, p < 0.05; **, p < 0.01; ***, p < 0.001. F, the cytosolic lysine to arginine mutants 3KR and 4KR of JAM-C did not significantly reduce the Alk14 labeling signal. Representative results from two independent experiments are shown. IP, immunoprecipitation; WB, Western blotting; alk, Alk14.
To further confirm that this modification occurs on cysteine residues, we mutated Cys-264 and Cys-265 to serine. We observed a significant decrease in the fluorescence signal of the single cysteine mutants C264S and C265S. However, the C264S mutation had a more profound effect on the fluorescence signal. Furthermore, we observed no fluorescence signal for the double cysteine mutant C264S/C265S (CCSS for short). These results indicated that S-palmitoylation on JAM-C depends on both Cys-264 and Cys-265 (Fig. 2, D and E). After the samples were treated with 0.5 m hydroxylamine (pH 10.0), most of the fluorescence signal on FLAG-JAM-C was removed (Fig. 2F), further confirming that JAM-C has cysteine palmitoylation. However, there was some hydroxylamine-resistant signal, similar to what was observed with TNF-α, a protein with lysine myristoylation (21).
To ensure that the hydroxylamine treatment was sufficient, we compared the labeling of FLAG-JAM-C before and after hydroxylamine treatment with the labeling of the FLAG-TNFα mutant (K19R/K20R), a protein that has only cysteine palmitoylation (21), as well as other membrane proteins reported to have S-palmitoylation such as transferrin receptor 1 (22) and Syntaxin6 (18). After hydroxylamine treatment, the majority, if not all, of the labeling on the other proteins was removed. However, some FLAG-JAM-C fluorescence signal remained (supplemental Fig. S2, A and B). However, there was a noticeable decrease in fluorescence signal after hydroxylamine treatment, supporting that JAM-C has cysteine palmitoylation.
To determine whether JAM-C has lysine fatty acylation, we mutated all the cytosolic lysine residues to arginine (K276R/K283R/K287R (3KR) and K276R/K283R/K287R/K305R (4KR)) and examined the fatty acylation signal. There was no significant change in the fluorescence signal with the 3KR and 4KR mutants compared with WT JAM-C, suggesting that the fatty acylation we detected for JAM-C does not come from lysine fatty acylation (Fig. 2F).
JAM-C Is a DHHC7 Target
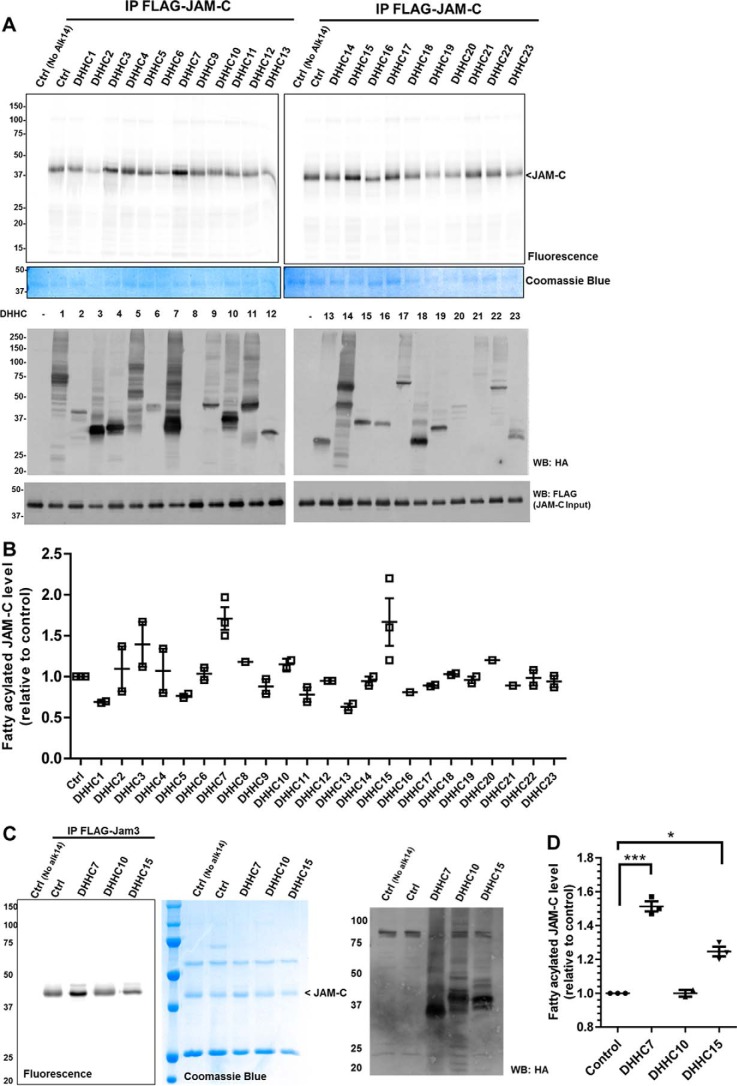
We next wanted to identify palmitoyltransferases that can control JAM-C S-palmitoylation. In mammals, there are 23 DHHC enzymes known to act as palmitoyltransferases. We co-overexpressed HA-tagged DHHC1–23 with FLAG-tagged JAM-C in HEK-293T cells and examined the fatty acylation level using the Alk14 metabolic labeling approach. From the initial DHHC screening and subsequent comparison of JAM-C palmitoylation levels with DHHC7 and DHHC15 overexpressed (Fig. 3, A and B, and supplemental Fig. S3), we decided to further validate DHHC7 as a palmitoyltransferase of JAM-C. However, because the screening result might be complicated by DHHC and JAM-C expression levels, we could not rule out that other DHHCs may also palmitoylate JAM-C.
FIGURE 3.
DHHC7 overexpression enhances the palmitoylation of JAM-C. A, the palmitoylation levels of FLAG-tagged JAM-C co-overexpressed with different HA-tagged DHHC1–23 in HEK-293T cells. B, quantification of the results shown in A (mean ± S.D., n = 2). The palmitoylation signal was quantified and normalized with the protein levels on the Coomassie Blue gel using Quantity One software. The signal in control cells without DHHC overexpression was set to 1.00 and served as the reference point for all other samples. C, DHHC7 overexpression most significantly increased JAM-C palmitoylation. A representative result from two independent experiments is shown. D, the quantified fatty acylation level of JAM-C co-overexpressed with DHHC7, DHHC10, or DHHC15 relative to control (n = 2; error bars represent S.D.). *, p < 0.05; ***, p < 0.001. IP, immunoprecipitation; WB, Western blotting; Ctrl, control.
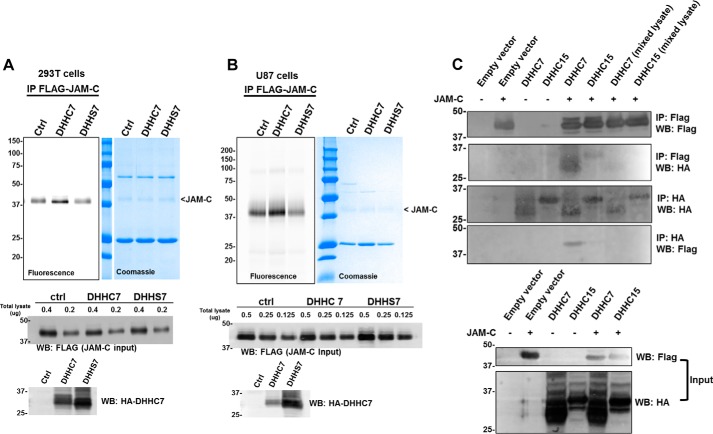
To confirm that DHHC7 can directly catalyze the palmitoylation of JAM-C, we examined the S-palmitoylation of JAM-C with co-expression of wild type DHHC7 and a catalytic dead mutant, DHHS7, in which the conserved catalytic cysteine residue is mutated to serine. As expected, only overexpression of DHHC7, but not DHHS7, augmented the JAM-C fatty acylation level in HEK-293T cells (Fig. 4A). Similar results were also obtained in U87 cells (Fig. 4B). These results suggest that the catalytic activity of DHHC7 is required for the S-palmitoylation of JAM-C.
FIGURE 4.
DHHC7 interacts with JAM-C, and its catalytic activity is required for JAM-C S-palmitoylation. A, overexpression of DHHC7, but not DHHS7, increased JAM-C S-palmitoylation in HEK-293T cells. HA-tagged DHHC7 or HA-tagged DHHS7 was co-overexpressed with FLAG-tagged JAM-C. FLAG-tagged JAM-C was pulled down for palmitoylation detection. B, the same experiment as in A was performed in U87 cells. The full FLAG JAM-C input Western blot can be found in supplemental Fig. S5. C, DHHC7 interacts with JAM-C. HA-tagged DHHC7 was co-overexpressed with FLAG-tagged JAM-C in HEK-293T cells. HA-tagged DHHC7 was pulled down, and FLAG-tagged JAM-C was detected by Western blotting and vice versa. In the two lanes labeled with mixed lysate, JAM-C and DHHC7 or DHHC15 were expressed in different cells, and then the cell lysate was mixed before IP. A representative result from two independent experiments is shown. IP, immunoprecipitation; WB, Western blotting; Ctrl, control.
Additionally, we investigated whether DHHC7 could interact with JAM-C. HA-tagged DHHC7 was co-overexpressed with FLAG-tagged JAM-C in HEK-293T cells. We carried out HA and FLAG pulldown experiments. Indeed, HA-tagged DHHC7 was able to pull down FLAG-tagged JAM-C and vice versa, suggesting that JAM-C physically interacts with DHHC7 (Fig. 4C). In contrast, FLAG-JAM-C and HA-DHHC15 did not exhibit a strong interaction as the signal is only slightly above the background on the Western blots (Fig. 4C).
Knockdown of DHHC7 Decreases JAM-C S-Palmitoylation
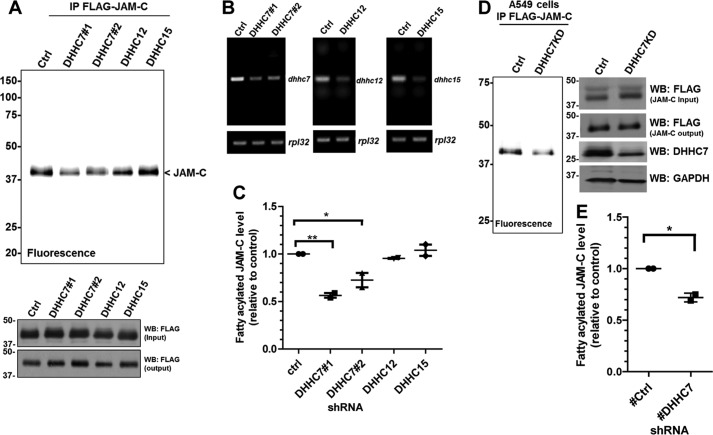
Ectopic expression of the DHHCs might affect the selectivity of the enzyme and possibly lead to an increase in S-palmitoylation of nonspecific targets (23). To verify that JAM-C is a palmitoylation target of endogenous DHHC7, we generated DHHC7, DHHC12, and DHHC15 stable knockdown HEK-293T cells using shRNAs targeting DHHC7, DHHC12, and DHHC15 mRNA, respectively. We then examined the JAM-C palmitoylation level (Fig. 5, A–C). Knockdown of DHHC7 led to a decrease in the JAM-C palmitoylation level compared with the control with scrambled shRNA, whereas no significant difference in JAM-C palmitoylation was observed in DHHC12 and DHHC15 knockdown cells. These results demonstrate that JAM-C is a palmitoylation target of endogenous DHHC7. Based on the BioGPS gene database, the DHHC7 gene expression level in the lung is relatively high compared with other tissues. Thus, we also looked at JAM-C palmitoylation in A549 lung cancer cells. DHHC7 stable knockdown in A549 cells decreased JAM-C palmitoylation (Fig. 5, D and E).
FIGURE 5.
JAM-C palmitoylation level decreases in DHHC7 knockdown cells. A, HEK-293T cells were infected with lentiviruses containing scrambled shRNA (control) or DHHC7, DHHC12, or DHHC15 shRNAs. Puromycin-resistant cells were selected for stable DHHC knockdown cells and used for FLAG-tagged JAM-C overexpression. The FLAG-tagged JAM-C in the DHHC knockdown cells was then immunoprecipitated and detected for palmitoylation levels by fluorescence labeling. B, semiquantitative RT-PCR showing the mRNA levels of Dhhc7, Dhhc12, and Dhhc15 in the stable knockdown HEK-293T cells. C, quantified fatty acylation levels of FLAG-tagged JAM-C expressed in DHHC7, DHHC12, and DHHC15 knockdown HEK-293T cells relative to control (mean ± S.D., n = 2). The palmitoylation level from each group was quantified and normalized with the corresponding protein level on the Coomassie Blue gel using Quantity One software. The signal from FLAG-tagged JAM-C in the control knockdown cells was set to 1.00 and served as the reference point for the other samples. D, DHHC7 knockdown in A549 cells also decreased the palmitoylation of JAM-C. The experiments were carried out similar to that described in A. E, quantified fatty acylation levels of JAM-C in A549 cells (n = 2; error bars represent S.D.). *, p < 0.05; **, p < 0.01. IP, immunoprecipitation; WB, Western blotting; Ctrl, control.
S-Palmitoylation Promotes JAM-C Localization to the TJ
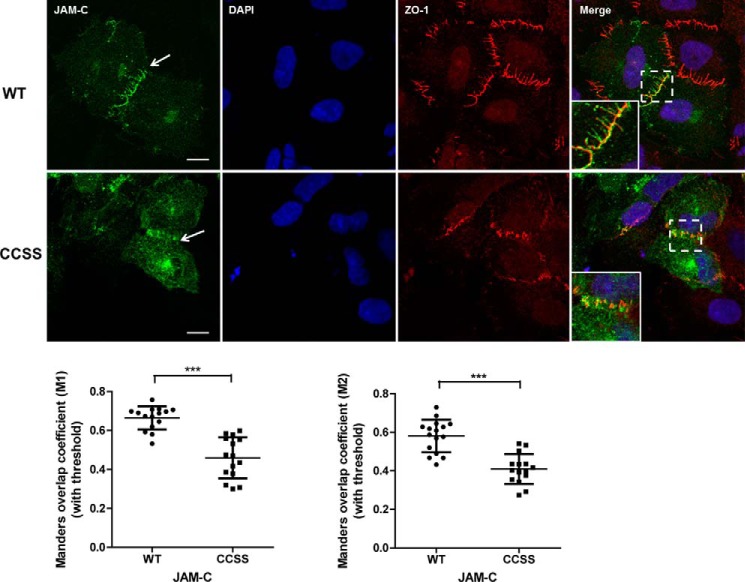
To further understand the physiological function of JAM-C S-palmitoylation, we investigated whether this modification is required for the TJ localization of JAM-C. FLAG-tagged JAM-C WT and the palmitoylation-deficient CCSS mutant were ectopically expressed in A549 lung cancer cells. The cells were stained with anti-FLAG and anti-ZO-1 antibodies to visualize JAM-C and the TJ, respectively. Images were viewed and analyzed using ZEN 2012 imaging software (Zeiss). Both WT and the CCSS mutant were localized on plasma membrane or intracellular organelles. However, JAM-C WT was more concentrated at the TJ (white arrow) as indicated by the co-localization with ZO-1, whereas the CCSS mutant had very little co-localization with ZO-1 (Fig. 6). The co-localization between JAM-C and ZO-1 was quantified using the Coloc2 plug-in in Fiji software and is presented as the mean of Manders' coefficient (M1 and M2) ±S.D. Thus, S-palmitoylation of JAM-C facilitates its localization to the TJ.
FIGURE 6.
S-Palmitoylation promotes JAM-C localization to the tight junction. Top, FLAG-tagged JAM-C WT and the CCSS mutant were ectopically expressed in A549 cells. The cells were immunofluorescently stained with anti-FLAG and anti-ZO1 antibodies after fixation. JAM-C WT was more co-localized with ZO-1 at the tight junction (white arrows), whereas the CCSS mutant had much less co-localization with ZO-1. The cells were visualized at room temperature with a Zeiss LSM 710 confocal microscope with a 63×/1.4 oil immersion objective. Images were viewed and analyzed using ZEN 2012 imaging software. These representative images were from three independent experiments with at least 15 FLAG-JAM-C-containing cells analyzed in each experiment. Scale bars, 10 μm. Bottom, the co-localization between JAM-C and ZO-1 was analyzed by the Coloc2 plug-in in Fiji software and is presented as the mean of Manders' coefficient (n = 15; error bars represent S.D.). ***, p ≤ 0.0001, Student's t test.
JAM-C S-Palmitoylation Affects Cell Migration
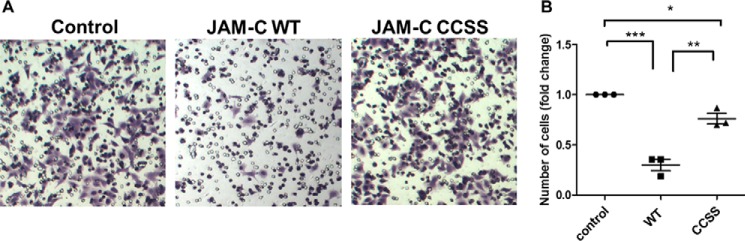
Because JAM-C S-palmitoylation affects its localization to the cell-cell contact TJ regions, we wondered whether it would also affect cell-cell adhesion. We therefore decided to check whether JAM-C S-palmitoylation could affect cell migration, which is influenced by cell-cell adhesion. A549 cells were transfected with FLAG-tagged JAM-C WT or the CCSS mutant for 24 h. We used a transwell migration assay, which was performed in a 24-well Transwell plate with 8-μm polycarbonate sterile membranes. Overexpression of JAM-C WT dramatically decreased cell migration compared with control cells that were transfected with an empty vector. In contrast, overexpression of the non-palmitoylatable CCSS mutant only slightly decreased the cell migration compared with control cells that were transfected with an empty vector (Fig. 7, A and B). The difference in cell migration was not due to the variation of cell proliferation as there was no significant difference in cell proliferation when JAM-C WT and the CCSS mutant were overexpressed (data not shown). Thus, S-palmitoylation of JAM-C affects cell migration.
FIGURE 7.
JAM-C S-palmitoylation affects cell migration. A549 cells were transfected with FLAG-tagged JAM-C WT or the CCSS mutant for 24 h and then cultured in RPMI 1640 serum-free medium for 14 h. The cell migration assay was then performed in a 24-well Transwell plate with 8-μm polycarbonate sterile membranes. A total of 3.5 × 104 cells in 200 μl of RPMI 1640 serum-free medium were plated into each upper chamber and placed in wells containing 600 μl of RPMI 1640 medium supplemented with 10% FBS. After 24 h, cells on the upper surface were detached with a cotton swab. The chambers were fixed, and cells in the lower filter were stained with 0.1% crystal violet for 15 min and counted. The quantified results were calculated by counting three random fields of migrated cells. The control cells were transfected with an empty pCMV4a vector. A, representative images of migrated cells from three independent experiments are shown. B, the number of migrated cells per field was quantified and normalized by the value of the control (n = 3; error bars represent S.D.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
Protein lipidation has become a more widely identified class of protein post-translational modifications and has been found to be involved in numerous biological pathways (15, 24). Here, using a bio-orthogonal palmitic acid probe (19, 20), we demonstrated that both endogenous and ectopically expressed JAM-C contain S-palmitoylation. S-Palmitoylation occurs on Cys-264 and Cys-265 of JAM-C. The S-palmitoylation on JAM-C is relatively resistant to hydroxylamine treatment for which the reason remains unclear. Because the hydroxylamine cleavage depends on the environment (25), one plausible explanation is the S-palmitoylation in the membrane-proximal region of JAM-C may be shielded from hydroxylamine attack. However, the decrease in labeling after hydroxylamine treatment supports that JAM-C has S-palmitoylation, which is further supported by mutagenesis studies.
The palmitoylacyltransferase family members share a conserved Asp-His-His-Cys (DHHC)-cysteine-rich domain as a catalytic domain of the enzyme (26, 27). DHHCs have broad substrate specificity, and how substrate specificity is determined remains unclear. By screening the 23 DHHCs, we have found that overexpression of DHHC7 can substantially increase the JAM-C palmitoylation level. Moreover, we showed that knockdown of DHHC7 decreased the S-palmitoyl level of JAM-C, supporting that JAM-C is a direct palmitoylation target of endogenous DHHC7. Nevertheless, other DHHCs may also be able to regulate JAM-C palmitoylation as some DHHCs have redundant functions and act on the same targets (28).
JAM-C has previously been shown to be phosphorylated at Ser-281, and JAM-C localization at the cell-cell contact region is negatively regulated by Ser-281 phosphorylation (29, 30). The JAM-C S281A mutant led to mislocalization and diffusion from the cell-cell contact region. In our case, we observed that JAM-C WT was concentrated at the cell-cell contact region, whereas the CCSS mutant was distributed more evenly on the cell membrane and in the cytosol, which is similar to the effect of S281A. It is therefore possible that there is cross-talk between the phosphorylation and S-palmitoylation of JAM-C. However, we found that the non-phosphorylable JAM-C mutant (S281A) and the phosphomimetic JAM-C mutants (S281D and S281E) had no significant effect on the S-palmitoylation of JAM-C (supplemental Fig. S4). We could not determine whether the CCSS JAM-C mutant can affect the phosphorylation on JAM-C due to the lack of proper antibodies to detect Ser-281 phosphorylation.
JAM-C has been reported to have homophilic interactions with JAM-C and heterophilic interactions with other PDZ domain-containing proteins such as ZO-1 and PAR3 at the cell tight junction. A possible explanation for the effect of palmitoylation on JAM-C localization is that S-palmitoylation facilitates the interaction of JAM-C with its interacting partners, leading to the enrichment of JAM-C at the cell-cell contact regions.
We demonstrated for the first time that JAM-C palmitoylation can affect cell migration. Overexpression of WT JAM-C decreased migration of A549 cells, but overexpression of the non-palmitoylatable JAM-C mutant did not affect the migration much. S-Palmitoylation of CD44, an adhesion protein, has previously been found to decrease the migration of breast cancer cells, and the invasiveness of cancer is negatively correlated with palmitoylation status of CD44 (31). This is similar to our finding that WT JAM-C decreases the cell migration of A549 cells, but the non-palmitoylatable mutant did not. The effect of JAM-C palmitoylation on cell migration is likely connected to its effect on the cell-cell contact localization of JAM-C and cell tight junction integrity. However, exactly how JAM-C palmitoylation regulates cell migration remains elusive.
Currently, only a few substrate proteins for DHHC7 are known, including Fas (32), sex steroid receptors (33), phosphatidylinositol 4-kinase IIα (34), Gα (35), stress-regulated exon (28) and Scribble (36). Our findings expand the substrate scope of DHHC7, which eventually will help to understand the substrate specificity and function of different DHHCs.
Interestingly, a decrease in DHHC7 levels has been related to tumorigenesis (37), suggesting that DHHC7 may play a role in preventing tumorigenesis. Our findings that JAM-C palmitoylation affects cancer cell migration and that DHHC7 directly controls the palmitoylation level of JAM-C suggest that pharmacologically controlling DHHC7 could be a useful strategy to control cancer cell migration and possibly cancer metastasis. However, to fully take advantage of this, a more detailed understanding of the function of DHHC7 is needed.
Experimental Procedures
JAM-C Cloning and Expression
Human JAM-C cDNA was purchased from Transomic (clone ID BC012147). The full-length cDNA was PCR-amplified by Platinum® Pfx DNA polymerase (ThermoFisher) and subcloned into the pCMV-tag 4a vector using the BamHI and EcoRV restriction sites using the following primers: sense, 5′-AGTCAGGGATCCATGGCGCTGAGGCGGCCA-3′; antisense, 5′-AGTCAGGATATCGATCACAAACGATGACTTGTGTCT-3′. JAM-C mutants (C264S, C265S, and CCSS) were generated by QuikChange mutagenesis. The JAM-C in pCMV-tag 4a vector was PCR-amplified using Phusion® high fidelity DNA polymerase and the following mutagenic primers (the underlined nucleotide sequences code for the mutated amino acid): C264S: sense, 5′-CCCTGATCACGTTGGGCATCAGCTGTGCATACAGACGTGGCTA-3′; antisense, 5′-GATGCCCAACGTGATCAGGG-3′; C265S: sense, 5′-TGATCACGTTGGGCATCTGCAGTGCATACAGACGTGGCTACTT-3′; antisense, 5′-GCAGATGCCCAACGTGATCA-3′; and CCSS: sense, 5′-CCCTGATCACGTTGGGCATCAGCAGTGCATACAGACGTGGCTACTT-3′; antisense, 5′-GATGCCCAACGTGATCAGGG-3′. The plasmids of DHHC1–23 in pEF-BOS-HA vector for screening were generously provided by Prof. Maurine Linder and Prof. Masaki Fukata.
Cell Culture and Transfection
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen), and Jurkat and A549 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). All cells were incubated in a humidified incubator at 37 °C with 5% CO2. FuGENE® 6 transfection reagent (Promega) was used for cell transfection according to the manufacturer's instruction.
Antibodies
Anti-FLAG® M2-peroxidase (HRP) antibody (mouse monoclonal IgG) for Western blotting and anti-FLAG M2 affinity gel for immunoprecipitation were purchased from Sigma (catalogue numbers A8592 and A2220, respectively). Anti-FLAG antibody (mouse monoclonal IgG1) for immunofluorescence was purchased from Cell Signaling Technology (catalogue number 8146). Secondary antibody Alexa Fluor® 488 conjugate (mouse polyclonal IgG) was purchased from ThermoFisher (catalogue number A-11001). Anti-HA-peroxidase (rat IgG1) was purchased from Roche Applied Science (catalogue number 12013819001). The anti-human JAM-C antibody (mouse monoclonal IgG) was purchased from Enzo Life Sciences (catalogue number ALX-803-306), and anti-DHHC7 (rabbit IgG) antibody was purchased from AssayBiotech (catalogue number R12-3691). All other peroxidase-conjugated secondary antibodies were from Santa Cruz Biotechnology.
Western Blotting
Cells were collected and lysed with 1% Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 1% (v/v) Nonidet P-40 (Igepal) containing protease inhibitor mixtures (Sigma)). Protein concentration was determined by Bradford assay (PierceTM Coomassie Protein Assay kit). Protein samples were separated by 12% SDS-PAGE and transferred to PVDF membrane (Bio-Rad) for 90 min. The membrane was blocked with 5% bovine serum albumin (BSA; Santa Cruz Biotechnology) in TBST (25 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 0.1% Tween 20) and incubated with the primary antibody for 3 h at room temperature or overnight at 4 °C. After washing five times with TBST, the membrane was incubated with the secondary antibody for 1 h at room temperature. The membrane was washed five more times with TBST before it was developed in ECL-Plus Western blotting detection reagent (GE Healthcare). The signal was visualized using a Typhoon 9400 variable mode imager (GE Healthcare) with 457-nm excitation and 526-nm detection filters using a photomultiplier tube setting of 600 V (normal sensitivity). The signal was analyzed by ImageQuant TL v2005 and Quantity One (Bio-Rad).
Metabolic Labeling of Palmitoylation on FLAG-tagged JAM-C
Cells were transfected with FLAG-JAM-C in pCMV4a vector. After 24 h, the cells were incubated with 50 μm Alk14 in medium supplemented with 10% FBS for 6 h. Cells were collected and washed with 1× phosphate-buffered saline (PBS) three times. The cell pellets were lysed with 1% Nonidet P-40 lysis buffer. The protein concentration was determined by Bradford assay (Pierce Coomassie Protein Assay kit). The expression level of FLAG-JAM-C was confirmed by Western blotting. For FLAG-JAM-C immunoprecipitation, 25 μl of anti-FLAG M2 affinity gel was added into 600 μg of total cell lysate with a final volume of 1 ml in an Eppendorf tube. The samples were gently agitated at 4 °C for 2 h. The samples were then centrifuged at 1,000 × g for 2 min at 4 °C to remove the supernatant. The beads were washed three times with 0.1% Nonidet P-40 lysis buffer. After the last wash, all the buffer was removed. The beads were resuspended in 15 μl of 0.1% Nonidet P-40 buffer. To perform the click reaction, 5.6 μl of the reaction master mixture was added to each sample. The click reaction master mixture contains the following: 3 μl of 1 mm BODIPY-azide in N,N-dimethylformamide, 1 μl of 50 mm tris(2-carboxyethyl)phosphine hydrochloride (Calbiochem) in water, 0.6 μl of 10 mm tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine (Sigma) in N,N-dimethylformamide, and 1 μl of 50 mm CuSO4 in water. After 1 h at room temperature, 20 μl of 3× protein loading buffer (187 mm Tris-HCl (pH 6.8), 6% SDS, 150 mm DTT, 30% (v/v) glycerol, and 0.006% bromphenol blue) was added to each sample, and the samples were boiled at 95 °C for 5 min and centrifuged at 17,000 × g for 2 min. The supernatants were transferred into new Eppendorf tubes and loaded for 12% SDS-PAGE. The fluorescence signal was visualized with a Typhoon 9400 variable mode imager using 488-nm excitation and 520-nm detection filters and a photomultiplier tube setting of 550 V (normal sensitivity). The signal was analyzed by ImageQuant TL v2005 and quantified by Quantity One within a linear range of exposure.
Generation of DHHC7, DHHC12, and DHHC15 Stable Knockdown in HEK-293T Cell Line
Different DHHC shRNA lentiviral plasmids in pLKO.1-puro vector were purchased from Sigma (catalogue numbers DHHC7-SHCLNG-NM_017740, shRNA1-TRCN0000143647, shRNA2-TRCN0000275633; DHHC12-SHCLNG-NM_025428; and DHHC15-SHCLNG-NM_144969). To generate the lentiviruses, low passage number HEK-293T cells in a 10-cm3 plates were transfected with 6 μg of each DHHC shRNA, 4 μg of pCMV-dR8.2, and 2 μg of pM2D.G mixed with 36 μl of FuGENE 6 (Promega). The cell media containing the lentiviruses were collected after 48 h by centrifugation and filtered through 0.45-μm syringe filter to remove cell debris. The day before infection, 3 × 105 cells of low passage number HEK-293T cells were split into each well of a 6-well plate and grown overnight in 2 ml of DMEM with 10% FBS. The media were then removed and replaced with a mixture of 1.5 ml of the lentivirus medium collected above, 0.5 ml of fresh DMEM with 10% FBS, and 6 μg/μl Polybrene (Sigma). After 6 h, 3 ml of DMEM with 10% FBS was added to each well, and after 48 h, 1.5 μg/μl puromycin dihydrochloride (Santa Cruz Biotechnology) was added to the cells to select for stable DHHC knockdown cells.
Cell Migration Assay
A549 cells were transfected with FLAG-JAM-C WT in pCMV4a, the FLAG-tagged CCSS mutant in pCMV4a, or the empty pCMV4a vector (control) for 24 h and then cultured in RPMI 1640 serum-free medium for an additional 14 h. The cell migration assay was performed in a 24-well Transwell plate with 8-μm polycarbonate sterile membrane (Corning Inc.). A total of 3.5 × 104 cells in 200 μl of RPMI 1640 serum-free medium were plated in each upper chamber and then placed in wells containing 600 μl of RPMI 1640 medium supplemented with 10% FBS. After 24 h, the cells on the upper surface were detached with a cotton swab, and the upper chambers were fixed. The cells in the lower filter were stained with 0.1% crystal violet for 15 min and then counted. The quantified results represent three random fields of migrated cells.
Immunofluorescence Microscopy
A total of 2 × 105 A549 cells were split into each glass bottom culture dish (MatTek) and cultured overnight. The cells were then transfected with either the FLAG-JAM-C WT in pCMV4a vector or the CCSS mutant. After 24 h, cells were washed twice with 2 ml of 1× PBS (ThermoFisher), fixed with 4% paraformaldehyde in PBS on ice for 15 min, and then kept at room temperature for 5 min. Then cells were subsequently washed three times with 2 ml of 1× PBS and incubated with 0.1% saponin (TCI America) with 3% BSA in 1× PBS at room temperature. After 1 h, the blocking solution was removed, and the anti-FLAG antibody (rabbit monoclonal IgG1; 1:1,000; Cell Signaling Technology) and the anti-ZO1 antibody (mouse monoclonal IgG1; 1:800; BD Biosciences) in a saponin-BSA solution were added to the cells and incubated at 4 °C overnight. The cells were washed with 1 ml of saponin-BSA solution five times and then incubated with the secondary antibodies conjugated with Alexa Fluor 488 (goat anti-rabbit IgG polyclonal antibody; ThermoFisher; 1:1,000) or Texas Red® (goat anti-mouse IgG polyclonal antibody; ThermoFisher; 1:1,000) in the saponin-BSA solution at room temperature for 1 h. The cells were washed with 1 ml of saponin-BSA solution five times, then mounted with 200 μl of DAPI Fluoromount-G (Southern Biotech), and covered with a cover glass. After 24 h, the cells were visualized with a Zeiss LSM 710 confocal microscope with a 63×/1.4 oil immersion objective. Images were viewed and analyzed using ZEN 2012 imaging software. For JAM-C and ZO-1 co-localization analysis, the Manders' overlap coefficient with automatic thresholds (M1 and M2) was calculated using the Coloc2 plug-in in Fiji (background subtraction; rolling ball radius, 50 pixels) (38).
Statistical Analysis
Data are shown as mean ± S.D. Differences were analyzed by two-tailed Student's t test between two groups: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author Contributions
P. A. and H. L. designed the experiments and wrote the paper. N. A. S. carried out the experiment shown in Figs. 3, A and B, and 4C, and supplemental Figs. S1–S3 and synthesized Alk14. J. C. performed and analyzed the experiment shown in Fig. 7. All authors reviewed and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
The imaging data were enabled by the Cornell University Biotechnology Resource Center with National Institutes of Health Grant 1S10RR025502 funding for the Zeiss LSM 710 confocal microscope.
This work was supported in part by National Institutes of Health Grant GM098596. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S5.
- JAM
- junctional adhesion molecule
- TJ
- tight junction
- DHHC
- palmitoyltransferase
- Alk14
- alkyne14 (palmitic acid analogue)
- CCSS
- C264S/C265S
- 3KR
- K276R/K283R/K287R
- 4KR
- K276R/K283R/K287R/K305R.
References
- 1. Ebnet K., Suzuki A., Ohno S., and Vestweber D. (2004) Junctional adhesion molecules (JAMs): more molecules with dual functions? J. Cell Sci. 117, 19–29 [DOI] [PubMed] [Google Scholar]
- 2. Keiper T., Santoso S., Nawroth P. P., Orlova V., and Chavakis T. (2005) The role of junctional adhesion molecules in cell-cell interactions. Histol. Histopathol. 20, 197–203 [DOI] [PubMed] [Google Scholar]
- 3. Bazzoni G. (2003) The JAM family of junctional adhesion molecules. Curr. Opin. Cell Biol. 15, 525–530 [DOI] [PubMed] [Google Scholar]
- 4. Weber C., Fraemohs L., and Dejana E. (2007) The role of junctional adhesion molecules in vascular inflammation. Nat. Rev. Immunol. 7, 467–477 [DOI] [PubMed] [Google Scholar]
- 5. Gliki G., Ebnet K., Aurrand-Lions M., Imhof B. A., and Adams R. H. (2004) Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature 431, 320–324 [DOI] [PubMed] [Google Scholar]
- 6. Guo J., Wang L., Zhang Y., Wu J., Arpag S., Hu B., Imhof B. A., Tian X., Carter B. D., Suter U., and Li J. (2014) Abnormal junctions and permeability of myelin in PMP22-deficient nerves. Ann. Neurol. 75, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akawi N. A., Canpolat F. E., White S. M., Quilis-Esquerra J., Morales Sanchez M., Gamundi M. J., Mochida G. H., Walsh C. A., Ali B. R., and Al-Gazali L. (2013) Delineation of the clinical, molecular and cellular aspects of novel JAM3 mutations underlying the autosomal recessive hemorrhagic destruction of the brain, subependymal calcification, and congenital cataracts. Hum. Mutat. 34, 498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hao S., Yang Y., Liu Y., Yang S., Wang G., Xiao J., and Liu H. (2014) JAM-C promotes lymphangiogenesis and nodal metastasis in non-small cell lung cancer. Tumour Biol. 35, 5675–5687 [DOI] [PubMed] [Google Scholar]
- 9. Leinster D. A., Colom B., Whiteford J. R., Ennis D. P., Lockley M., McNeish I. A., Aurrand-Lions M., Chavakis T., Imhof B. A., Balkwill F. R., and Nourshargh S. (2013) Endothelial cell junctional adhesion molecule C plays a key role in the development of tumors in a murine model of ovarian cancer. FASEB J. 27, 4244–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arcangeli M. L., Frontera V., Bardin F., Thomassin J., Chetaille B., Adams S., Adams R. H., and Aurrand-Lions M. (2012) The junctional adhesion molecule-B regulates JAM-C-dependent melanoma cell metastasis. FEBS Lett. 586, 4046–4051 [DOI] [PubMed] [Google Scholar]
- 11. Tenan M., Aurrand-Lions M., Widmer V., Alimenti A., Burkhardt K., Lazeyras F., Belkouch M. C., Hammel P., Walker P. R., Duchosal M. A., Imhof B. A., and Dietrich P. Y. (2010) Cooperative expression of junctional adhesion molecule-C and -B supports growth and invasion of glioma. Glia 58, 524–537 [DOI] [PubMed] [Google Scholar]
- 12. Langer H. F., Orlova V. V., Xie C., Kaul S., Schneider D., Lonsdorf A. S., Fahrleitner M., Choi E. Y., Dutoit V., Pellegrini M., Grossklaus S., Nawroth P. P., Baretton G., Santoso S., Hwang S. T., et al. (2011) A novel function of junctional adhesion molecule-C in mediating melanoma cell metastasis. Cancer Res. 71, 4096–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuse C., Ishida Y., Hikita T., Asai T., and Oku N. (2007) Junctional adhesion molecule-C promotes metastatic potential of HT1080 human fibrosarcoma. J. Biol. Chem. 282, 8276–8283 [DOI] [PubMed] [Google Scholar]
- 14. Smotrys J. E., and Linder M. E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 15. Resh M. D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 16. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., and Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 17. Linder M. E., and Jennings B. C. (2013) Mechanism and function of DHHC S-acyltransferases. Biochem. Soc. Trans. 41, 29–34 [DOI] [PubMed] [Google Scholar]
- 18. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R. 3rd, Davis N. G., and El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yount J. S., Zhang M. M., and Hang H. C. (2011) Visualization and identification of fatty acylated proteins using chemical reporters. Curr. Protoc. Chem. Biol. 3, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson J. P., Raghavan A. S., Yang Y. Y., Charron G., and Hang H. C. (2011) Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell. Proteomics 10, M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R., Hang H. C., Hao Q., and Lin H. (2013) SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jing S. Q., and Trowbridge I. S. (1987) Identification of the intermolecular disulfide bonds of the human transferrin receptor and its lipid-attachment site. EMBO J. 6, 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dietrich L. E., and Ungermann C. (2004) On the mechanism of protein palmitoylation. EMBO Rep. 5, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Resh M. D. (2012) Targeting protein lipidation in disease. Trends Mol. Med. 18, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iwamoto M., Sudo Y., Shimono K., and Kamo N. (2001) Selective reaction of hydroxylamine with chromophore during the photocycle of pharaonis phoborhodopsin. Biochim. Biophys. Acta 1514, 152–158 [DOI] [PubMed] [Google Scholar]
- 26. Roth A. F., Feng Y., Chen L., and Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lobo S., Greentree W. K., Linder M. E., and Deschenes R. J. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 28. Tian L., McClafferty H., Jeffries O., and Shipston M. J. (2010) Multiple palmitoyltransferases are required for palmitoylation-dependent regulation of large conductance calcium- and voltage-activated potassium channels. J. Biol. Chem. 285, 23954–23962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mandicourt G., Iden S., Ebnet K., Aurrand-Lions M., and Imhof B. A. (2007) JAM-C regulates tight junctions and integrin-mediated cell adhesion and migration. J. Biol. Chem. 282, 1830–1837 [DOI] [PubMed] [Google Scholar]
- 30. Ebnet K., Aurrand-Lions M., Kuhn A., Kiefer F., Butz S., Zander K., Meyer zu Brickwedde M. K., Suzuki A., Imhof B. A., and Vestweber D. (2003) The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity. J. Cell Sci. 116, 3879–3891 [DOI] [PubMed] [Google Scholar]
- 31. Babina I. S., McSherry E. A., Donatello S., Hill A. D., and Hopkins A. M. (2014) A novel mechanism of regulating breast cancer cell migration via palmitoylation-dependent alterations in the lipid raft affiliation of CD44. Breast Cancer Res. 16, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rossin A., Durivault J., Chakhtoura-Feghali T., Lounnas N., Gagnoux-Palacios L., and Hueber A. O. (2015) Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability. Cell Death Differ. 22, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pedram A., Razandi M., Deschenes R. J., and Levin E. R. (2012) DHHC-7 and -21 are palmitoylacyltransferases for sex steroid receptors. Mol. Biol. Cell 23, 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu D., Sun H. Q., Wang H., Barylko B., Fukata Y., Fukata M., Albanesi J. P., and Yin H. L. (2012) Phosphatidylinositol 4-kinase IIα is palmitoylated by Golgi-localized palmitoyltransferases in cholesterol-dependent manner. J. Biol. Chem. 287, 21856–21865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., and Fukata M. (2009) Identification of G protein α subunit-palmitoylating enzyme. Mol. Cell. Biol. 29, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen B., Zheng B., DeRan M., Jarugumilli G. K., Fu J., Brooks Y. S., and Wu X. (2016) ZDHHC7-mediated S-palmitoylation of Scribble regulates cell polarity. Nat. Chem. Biol. 12, 686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaspar C., Cardoso J., Franken P., Molenaar L., Morreau H., Möslein G., Sampson J., Boer J. M., de Menezes R. X., and Fodde R. (2008) Cross-species comparison of human and mouse intestinal polyps reveals conserved mechanisms in adenomatous polyposis coli (APC)-driven tumorigenesis. Am. J. Pathol. 172, 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.