Abstract
Diverse protein import pathways into mitochondria use translocons on the outer membrane (TOM) and inner membrane (TIM). We adapted a genetic screen, based on Ura3 mistargeting from mitochondria to the cytosol, to identify small molecules that attenuated protein import. Small molecule mitochondrial import blockers of the Carla Koehler laboratory (MB)-10 inhibited import of substrates that require the TIM23 translocon. Mutational analysis coupled with molecular docking and molecular dynamics modeling revealed that MB-10 binds to a specific pocket in the C-terminal domain of Tim44 of the protein-associated motor (PAM) complex. This region was proposed to anchor Tim44 to the membrane, but biochemical studies with MB-10 show that this region is required for binding to the translocating precursor and binding to mtHsp70 in low ATP conditions. This study also supports a direct role for the PAM complex in the import of substrates that are laterally sorted to the inner membrane, as well as the mitochondrial matrix. Thus, MB-10 is the first small molecule modulator to attenuate PAM complex activity, likely through binding to the C-terminal region of Tim44.
Keywords: chemical biology, mitochondria, mitochondrial transport, protein translocation, small molecule
Introduction
Protein import into mitochondria is a highly regulated process that is coordinated by translocons on the outer membrane and the inner membrane (1). The translocase of the outer membrane (TOM)2 is the major gateway for proteins to enter mitochondria. Tom70, Tom20, Tom22, and Tom5 function as receptors to guide the precursor to the translocation pore that is formed by Tom40. The translocase of the inner membrane (TIM23) mediates import for proteins that typically have an N-terminal targeting sequence and reside in the matrix, whereas the TIM22 translocon directs the import of the carrier proteins. Together the translocation and assembly complexes coordinate assembly of the mitochondrion.
The TIM23 core complex consists of membrane-embedded subunits Tim50, Tim23, and Tim17 (see Fig. 7D for schematic) (2). The channel is formed by Tim17 and the C-terminal region of Tim23 (3), whereas the N-terminal regions of Tim23 and Tim50 have a receptor function to guide the precursor from the TOM complex to the TIM23 complex (4). The accessory subunit Tim21 assembles with the TIM23 core complex to serve as a conduit to promote the transfer of substrates with a hydrophobic stop transfer signal into the inner membrane in a membrane-potential dependent manner (5). Complete translocation across the inner membrane and into the matrix requires the membrane potential and the ATP-driven protein-associated motor (PAM) (2). Components of the PAM complex include Tim44, mitochondrial Hsp70, the nucleotide exchange factor mGrpE (Mge1), and J-proteins Pam16 and Pam18. mtHsp70 is the central component of the PAM complex because it pulls the precursor into the matrix. Co-chaperone Pam18 stimulates the ATPase activity of mtHsp70, whereas co-chaperone Pam16 regulates Pam18 (6). Tim44 is crucial for mtHsp70 and J-protein binding and association of the PAM complex with the membrane (2). Mge1 is the nucleotide exchange factor that is required for completion of the mtHsp70 reaction cycle. Thus, the TIM23 translocon is a dynamic and highly regulated machine.
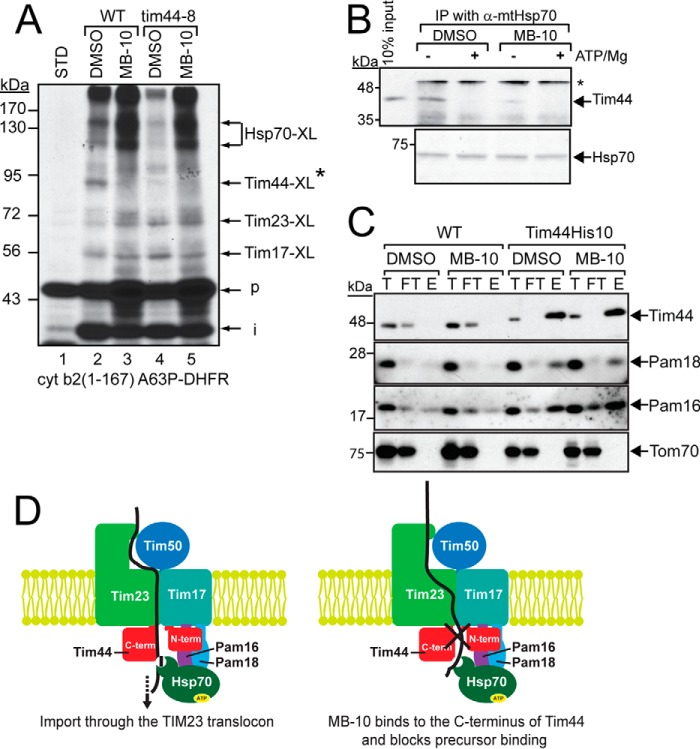
FIGURE 7.
MB-10 inhibits Tim44 binding to the precursor and to Hsp70, but not other components of the PAM complex. A, radiolabeled cyt b2 (167)A63P-DHFR was imported into mitochondria isolated from WT or the tim44-8 mutant in the presence of 1 μm MTX. Reactions were treated with cross-linker 200 μm DSS. An asterisk indicates the Tim44-cyt b2(167)A63P-DHFR cross-link that is absent in mitochondria from the tim44-8 mutant strain (22). p, precursor; i, intermediate. B, 20 μg of mitochondrial lysate was treated with 1% DMSO or 100 μm MB-10 and immunoprecipitated with anti-mitochondrial Hsp70 antibody in the absence of or presence of 5 mm MgCl2 and 1 mm ATP (+ATP/Mg). Tim44 was detected with anti-Tim44 antibody, and mtHsp70 was visualized by Ponceau S staining. An asterisk indicates the IgG heavy chain. C, mitochondria (400 μg) from a strain in which Tim44 contained a C-terminal His10 tag and a WT control were treated with 1% DMSO or 100 μm MB-10, followed by lysis in 1% digitonin. Tim44-His10 was purified with Ni2+ agarose, and immunoblot analysis was performed with antibodies against Tim44, Pam16, Pam18, and Tom70. 50 μg of input (T) and flow through (FT) and 300 μg of eluate (E) were loaded. D, schematic showing the TIM23 translocon and associated PAM complex in the absence of MB-10 (left panel). MB-10 addition blocks interactions between Tim44 and the precursor and increases interactions between Hsp70 and the precursor while blocking translocation into the matrix (right panel, marked with an X). STD, standard.
Mutations in components of the TIM23 protein import machinery result in a variety of diseases. Mutant DNAJC19, the homolog of Pam18, causes the dilated cardiomyopathy with ataxia syndrome (7). The human homolog of yeast Pam16, Magmas, is overexpressed in adenocarcinoma (8). TIMM44 is up-regulated in diabetes (9), whereas a point mutation (P308Q) in the C-terminal domain of TIMM44 is linked to an autosomal dominant familial oncocytic thyroid carcinoma (10). Gene delivery of TIM44 into diabetic models facilitates the import of antioxidative enzymes such as manganese superoxide dismutase (SOD2) and glutathione peroxidase, thereby reducing ROS production and protecting the cells (11).
Accordingly, the development of small molecule modulators for the translocation system would facilitate characterization of the role of protein translocation in complex diseases. To this end, we have completed two small molecule screens in which we have identified inhibitors for Tim10 and Erv1 (essential for respiration and vegetative growth) (12, 13). The small molecules worked well in both yeast and mammalian cells. The Erv1 inhibitor revealed an unexpected role for augmenter of liver regeneration as a survival factor in pluripotent stem cells (13). Here, we report the results of a screen in which we have adapted positive selection for growth that is caused by mislocalization of the Su9-Ura3 fusion protein from mitochondria to the cytosol (14). One hit compound, designated MB-10, was characterized in detail. MB-10 inhibited the import of substrates that use the TIM23 translocon but not those imported via other pathways. MB-10 likely targets Tim44 of the PAM complex and reduced the import of substrates that require the PAM complex activity. In mammalian cells, MB-10 treatment supports an important role for TIMM44 in the import of SOD2. Finally, TIMM44 was required for cardiac development in zebrafish. Therefore, MB-10 is a useful tool to investigate the role of TIMM44 in disease studies, as well as the mechanistic role of the PAM complex in protein translocation.
Results
An in Vivo Screen to Identify Inhibitors of Mitochondrial Protein Translocation
To identify modulators of the TOM/TIM23 translocation system, we adapted the genetic screen that resulted in the identification of import components Tim44, Tim23, and Tim17, based on mislocalization of the Su9-Ura3 fusion protein from mitochondria to the cytosol (14); the targeting sequence, abbreviated Su9, is derived from subunit 9 of Neurospora crassa F1Fo ATPase and confers robust import. If Su9-Ura3 is targeted to the mitochondrial matrix, yeast fails to grow in medium lacking uracil because Ura3 must be localized to the cytosol to participate in the uracil biosynthetic pathway. When protein translocation is attenuated, the Su9-Ura3 protein remains in the cytosol, and growth in medium lacking uracil is restored to the yeast cells (14). A plasmid encoding Su9-Ura3 with a C-terminal Myc tag was integrated into WT yeast and the tim23-2 temperature-sensitive mutant at the LEU2 locus (15) (supplemental Table S1). In addition, the multidrug resistance ABC transporters SNQ2 and PDR5 were deleted to increase the concentration of small molecules in the cells (12). The Su9-Ura3 protein was detected as the mature form in a whole cell lysate of the WT strain grown at 30 °C, whereas a small portion of unprocessed Su9-Ura3 was present in the whole cell lysate of the tim23-2 strain grown at 30 °C, verifying compromised import of Su9-Ura3 in the tim23-2 mutant (Fig. 1A).
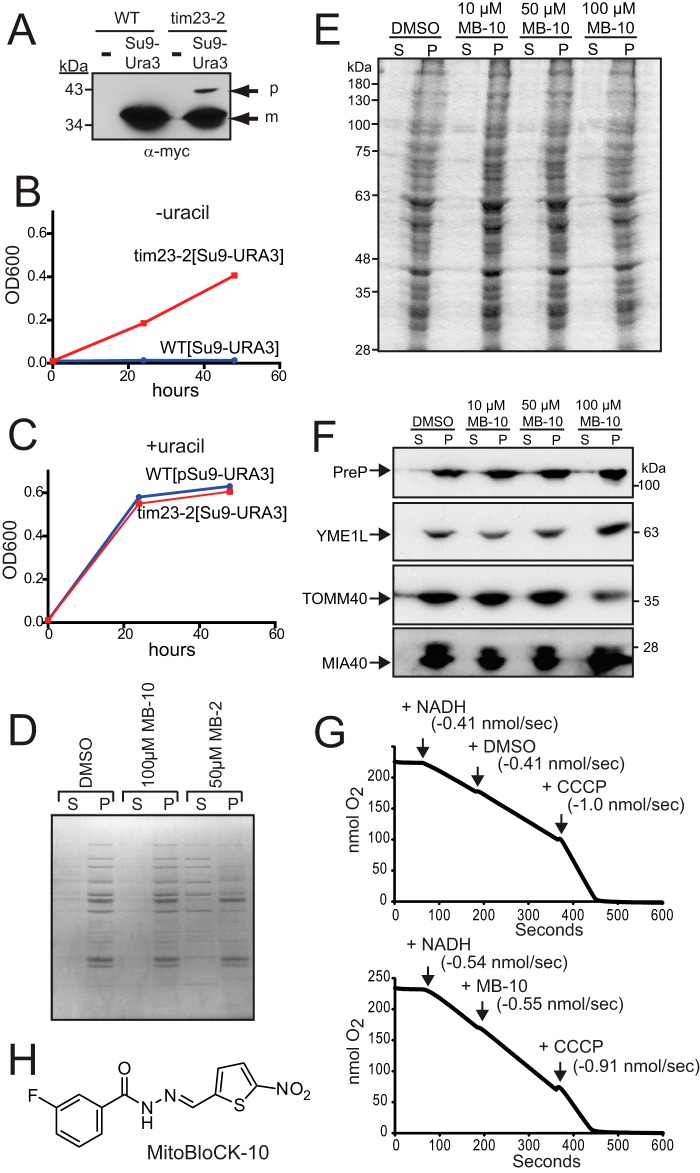
FIGURE 1.
MB-10 is a potential attenuator of protein import into mitochondria. A, a plasmid expressing Su9-Ura3-myc was integrated at the LEU2 locus in WT and tim23-2 strains, and the strains were grown at 30 °C. A whole cell lysate from WT and the tim23-2 mutant, either without (−) or with [Su9-Ura3], was separated by SDS-PAGE followed by immunoblotting with an anti-Myc antibody. p, precursor; m, mature. B, growth analysis of the WT and tim23-2 mutant with integrated [Su9-URA3] in synthetic dextrose medium in the absence of uracil. Growth was measured based on the density at A600 (OD600). C, as in B, in the presence of uracil. D, 1% DMSO, 100 μm MB-10, or 50 μm MB-2 was added to purified 100 μg/ml WT yeast mitochondria for 30 min at 25 °C in import buffer, and released proteins (S) were separated from mitochondria (P) by centrifugation. The proteins were visualized by Coomassie staining. E, as in B, 1% DMSO or the indicated concentration of MB-10 was added to purified mitochondria from HEK293T cells followed by Coomassie staining. F, as in E, the release of proteins was detected by immunoblotting; proteins included PreP (matrix), YME1L (inner membrane), TOMM40 (outer membrane), and MIA40 (intermembrane space). G, respiration measurements were performed with a Clark-type oxygen electrode using 100 μg/ml WT yeast mitochondria in the presence of 1% DMSO or MB-10. Respiration was initiated with NADH addition. 100 μm MB-10 (with DMSO at a 1% final concentration) or 1% DMSO was added once steady-state respiration was established. As a control, CCCP was added to uncouple the mitochondria. The respiration rate for each treatment has been included. H, the structure of MitoBloCK-10 (MB-10).
We tested the growth properties of the strains in minimal glucose medium (Fig. 1, B and C). In medium lacking uracil at 30 °C, the WT strain expressing Su9-Ura3 failed to grow, whereas the tim23-2 mutant expressing Su9-Ura3 grew much faster (Fig. 1B). The strains grew at a similar rate when the medium was supplemented with uracil (Fig. 1C). These data confirm that the import defect in the tim23-2 mutant conferred growth in medium lacking uracil. Thus, the WT[Su9-URA3] strain was amenable for screening with small molecules libraries.
A small molecule screen was conducted with an integrated robotic system. Briefly, a diversity-oriented commercial library (Chembridge) of drug-like compounds at a concentration of ∼10 μm was screened against the WT[Su9-URA3]. The screen encompassed a total of 30,000 compounds dissolved in DMSO (supplemental Table S2). The strain in minimal glucose medium lacking uracil was aliquoted into 384-well pates (consisting of 24 columns) followed by compound addition with robotic pinning into the assay wells (column 3–22). For a negative control of cell growth, column 2 contained WT[Su9-URA3] strain with 1% DMSO. For a positive control of cell growth, column 23 contained the tim23-2[Su9-URA3] strain with 1% DMSO. After incubation at 30 °C for 24 h, cultures in each well were measured for A600 to assess growth. Wells that had a growth increase of greater than 30% compared with the negative control were selected as potential candidates. Of 25 candidates that were reconfirmed by repeating the screening assay, 6 compounds reproducibly increased the growth of the WT[Su9-URA3] strain (supplemental Table S2).
Because the potential hits might nonspecifically damage mitochondria, we investigated mitochondrial membrane integrity and oxidative phosphorylation properties in the presence of the small molecules (Fig. 1, D–G). To assess membrane integrity, each compound (100 μm) was incubated with mitochondria followed by centrifugation. Released proteins were recovered in the supernatant fraction (S), whereas the pellet (P) contained intact mitochondria. The proteins were analyzed by Coomassie staining and immunoblotting. As a positive control, MB-2, a compound from a tim10-1 synthetic lethal screen that permeabilized mitochondrial membranes, was included (12). Treatment with the six compounds did not result in release of proteins from yeast mitochondria (one hit is shown as an example), in contrast to the release of proteins in the presence of MB-2 (Fig. 1D). In addition, MB-10 incubation with purified mitochondria from HEK293T cells did not cause release of proteins as detected by Coomassie staining (Fig. 1E) or immunoblot analysis (Fig. 1F). Respiration in the presence of the compounds with the Clark-type oxygen electrode was also tested (Fig. 1G). Respiration was initiated by the addition of NADH. The rate of oxygen consumption was representative of mitochondria that were well coupled. The addition of vehicle DMSO did not affect the respiration rate (−0.41 nmol/s in the presence of NADH and DMSO), but CCCP addition uncoupled the mitochondria, increasing the rate of oxygen consumption. The addition of four of the potential hits at 100 μm stopped oxygen consumption, and these compounds were dropped from the analysis. However, two compounds (designated MitoBloCK-10/MB-10 and MitoBloCK-11/MB-11) did not alter respiration. (Note that respiration for MB-10 is shown, and the rate of respiration is −0.55 nmol/s after MB-10 addition, which was similar to that in the presence of NADH (Fig. 1G).) The chemical name for MitoBloCK-10 is 3-fluoro-N′-[(5-nitro-2-thienyl)methylene]benzohydrazide, a hydrazide-hydrazone derivative (Fig. 1H), and it was selected for detailed characterization.
MB-10 Targets the TIM23 Translocon
We repeated the growth assay from the screen with varying concentrations of MB-10 to assess the growth properties of the WT[Su9-URA3] strain (Fig. 2A). At 2 μm MB-10, the growth of the WT[Su9-URA3] strain increased robustly (Fig. 2A). Because a complete block in protein translocation is predicted to be lethal, the minimum inhibitory concentration required to inhibit the growth of 50% of the yeast (MIC50) for MB-10 was determined for the WT, tim23-2, and tim10-1 strains. These strains were tested because they lacked the multidrug resistance pumps Pdr5 and Snq2, and the strains were used previously in a screen to identify modulators for the TIM22 pathway (12). We predicted that a yeast strain with a mutation in the translocon that MB-10 targets would display increased sensitivity to MB-10, based on a previous screening strategy to identify TIM22 modulators (12). The MIC50 of MB-10 was 1 μm for the tim23-2 strain, in contrast to MIC50 values of 7.1 and 4.7 μm for the WT and tim10-1 strains, respectively (Fig. 2B). The synthetic lethal analysis suggests that MB-10 likely targets the TIM23 translocon and not the TIM22 translocation system.
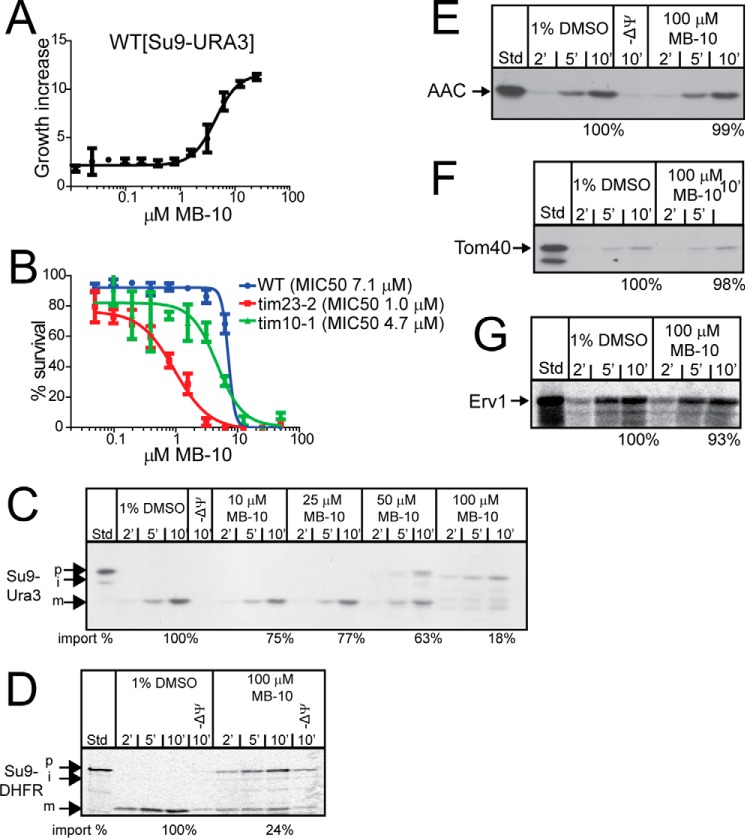
FIGURE 2.
MB-10 inhibits the import of substrates that use the TIM23 import pathway. A, the WT yeast strain expressing Su9-Ura3 was cultured in synthetic dextrose medium lacking uracil in the indicated concentrations of MB-10 for 24 h. The cell density is indicated as fold increase compared with growth in the presence of DMSO. The data represent the averages ± S.D. of n = 3 trials. B, MIC50 analysis of WT, tim23-2, and tim10-1 strains with MB-10. Each strain was cultured in rich ethanol-glycerol medium in the presence of various concentrations of MB-10 for 24 h. 100% was set as A600 of each strain grown in the presence of the vehicle control (1% DMSO). Average percentage of survival ± S.D. of n = 3 trials is shown. C–G, import assays were performed with radiolabeled precursors into mitochondria from the WT strain in the presence of MB-10 or the vehicle (1% DMSO). Non-imported precursor was removed by trypsin treatment. Precursors include Su9-Ura3 (C), Su9-DHFR (D), AAC (E), Tom40 (F), and Erv1 (G). 10% standard (Std) from the translation reaction is included. A representative gel is shown for each assay (n = 3). The last time point in the import reactions was quantified with ImageJ software; 100% was set as the amount of precursor imported in the presence of DMSO at end point in the time course. p, precursor; m, mature; i, intermediate.
Because MB-10 likely impairs the translocation of a subset of precursors that use a particular translocon, the ability of MB-10 to inhibit protein import was tested using the in vitro import assay with radiolabeled substrates. Mitochondria were preincubated with MB-10, and then substrates Su9-Ura3 and Su9-DHFR (TIM23 substrates), AAC (TIM22 substrate), Tom40 (SAM substrate), and Erv1 (Erv1/Mia40 substrate) were imported. A titration assay with MB-10 indicated that 100 μm inhibited Su9-Ura3 import by ∼80% (Fig. 2C). Similarly, 100 μm MB-10 reduced import of Su9-DHFR by 75% (Fig. 2D). In contrast, MB-10 did not interfere with the import of AAC, Tom40, and Erv1, substrates that do not require the TIM23 translocon (Fig. 2, E–G). Therefore, MB-10 specifically inhibits the import of TIM23 substrates.
The mitochondrial targeting sequence of Su9 from N. crassa has an interesting feature of two processing sites at amino acids 35 and 66 that are cleaved by the matrix processing peptidase (16). The precursor and intermediate forms of Su9-DHFR, and Su9-Ura3 to a lesser extent, were resistant to added protease in the presence of 50–100 μm MB-10 (Fig. 2, C and D). These unprocessed forms may have arrested during translocation as an intermembrane space intermediate, similar to that generated during import under low ATP conditions (17). Alternatively, the precursor form with a tightly folded DHFR domain may be held tightly against the TOM complex and therefore resistant to cleavage by the added protease (18).
We followed the import assay of Su9-Ura3 with osmotic shock to disrupt the mitochondrial outer membrane (generating mitoplasts) and subsequently added protease (19). The precursor and intermediate forms accumulated in the presence of MB-10, but not the vehicle control DMSO (Fig. 3, A and B). The intermediates were mostly degraded, indicating that the precursor and intermediate forms arrest as translocation intermediates in the intermembrane space (Fig. 3B). As a control, samples treated with Triton X-100 were generally degraded, except for a small amount of a lower molecular weight species that may represent a tightly folded domain of Ura3 (indicated by asterisks in Fig. 3, A and B). Thus, MB-10 addition caused the translocating precursor to arrest in transit, potentially in the TIM23 translocon.
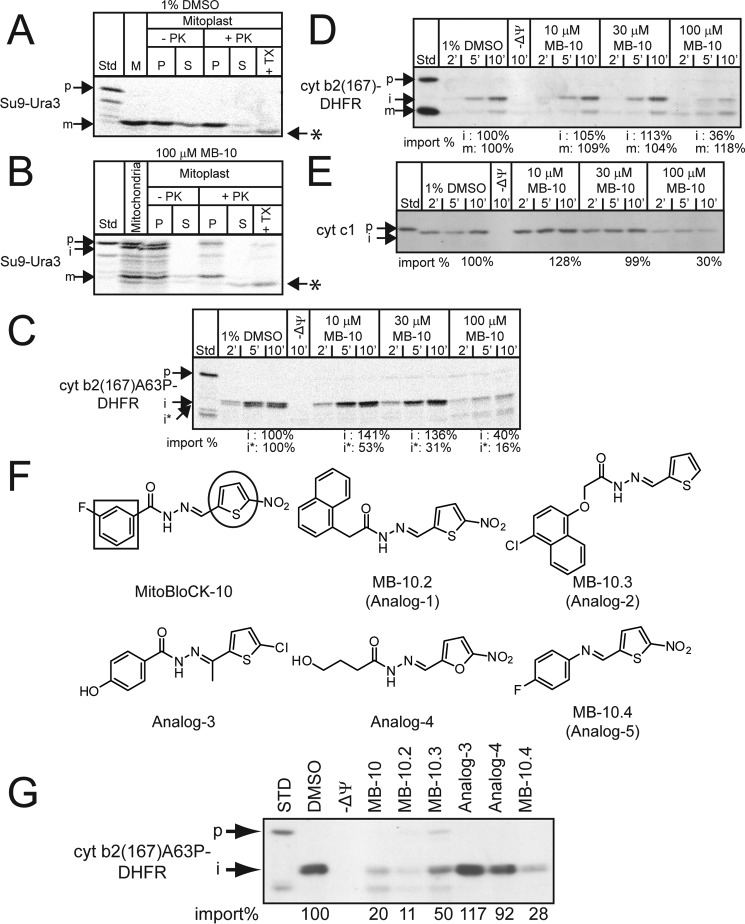
FIGURE 3.
MB-10 inhibition of import depends on specific chemical characteristics. A and B, Su9-Ura3 was imported as in Fig. 2C. After import in the presence of 1% DMSO (A), or 100 μm MB-10 (B), samples were incubated in hypotonic buffer to swell the outer membrane (generating mitoplasts) in the presence and absence of proteinase K (PK) followed by inactivation with PMSF. Mitoplasts were recovered by centrifugation (P) and separated from supernatant (S), which contains the soluble intermembrane space contents. As a control, samples were treated with Triton X-100 (TX). Asterisks mark lower molecular weight products that were resistant to protease in the presence of Triton X-100. C–E, import assays of precursors cyt b2(167)A63P-DHFR (mis-sorting mutant to matrix) (C), cyt b2(167)-DHFR (intermembrane space) (D), and cyt c1 (intermembrane space) (E) were performed. p, precursor; m, mature; i, intermediate; i*, intermediate* that is generated from secondary processing in the matrix (21). F, structures of MB-10 and analogs for SAR studies. The square and circle indicate the benzene ring and the thiophene ring, respectively. G, import of cyt b2(167)A63P-DHFR was performed as in C for 10 min in the presence of 1% DMSO, 100 μm MB-10, or 100 μm each of the analogs. The analogs that markedly inhibited import have been designated MB-10.2, MB-10.3, and MB-10.4. p, precursor; i, intermediate. Import reactions were quantified by ImageJ software; 100% was set as the amount of precursor imported in the presence of DMSO at the end point in the time course.
MB-10 Impairs the Import of Precursors Sorted to the Matrix and the Intermembrane Space
Next, we checked the effect of MB-10 on sorting of precursors to the intermembrane space and matrix, using cytochrome (cyt) b2 variants and cyt c1. The precursor, cyt b2 (167)A63P-DHFR, is targeted to the mitochondrial matrix, because of the mutation at amino acid 63 (a proline is substituted for alanine) in the stop transfer sequence (20) and processed in two stages. Immediately after translocation across the inner membrane, the targeting sequence is cleaved by matrix processing peptidase and the processing intermediate (designated i) is formed. In a subsequent step, eight additional amino acids are removed, yielding the intermediate i* form (21). In the presence of 10 and 30 μm MB-10, formation of the i* intermediate seemed to be reduced, resulting in accumulation of the i form (Fig. 3C). In the presence of 100 μm MB-10, import of cyt b2(167)A63P-DHFR was inhibited (Fig. 3C). The inhibition of i* formation has been observed with mutants of the components of PAM complex, such as Tim44 and Mge1 (22–24), suggesting that MB-10 may target the PAM complex. In contrast, for precursors cyt b2(167)-DHFR and cyt c1, which are laterally sorted to intermembrane space by the TIM23SORT complex in an ATP-independent manner (5, 19, 25), generation of the import intermediate was not inhibited by treatment with low concentrations of MB-10 (Fig. 3, D and E). Only in the presence of 100 μm MB-10 was the import of cyt b2(167)-DHFR and cyt c1 decreased (Fig. 3, D and E). Note that formation of the mature form of cyt c1 is typically not detected because processing by the inner membrane protease is slow (5). The PAM complex also associates with the laterally inserted precursors even when ATP is not required (26), indicating that the PAM complex plays a role in the import of precursors into the matrix and inner membrane. Taken together, we interpret these results to show that MB-10, in low concentration, inhibits the essential function of the PAM complex to import matrix proteins and that MB-10, in high concentration, further inhibits the PAM complex in facilitating the import of precursors laterally sorted to intermembrane space.
Specific Properties of MB-10 Inhibit Protein Import
We purchased an additional five compounds with similarity to MB-10 (Fig. 3F) for structural-activity relationship (SAR) studies. The import of cyt b2(167)A63P-DHFR was tested in the presence of the analogs at 100 μm. Analogs 1, 2, and 5 decreased the import of cyt b2(167)A63P-DHFR similarly to MB-10. We renamed these compounds MB-10.2, MB-10.3, and MB-10.4, respectively (Fig. 3G). In contrast, analogs 3 and 4 did not abrogate protein import (Fig. 3G). Analog 3 has a methyl group attached to the carbon atom of the hydrazone moiety, and analog 4 has an alkylalcohol group that replaces the fluorobenzene moiety and the thiophene exchanged for a furan group. In contrast, MB-10.2, MB-10.3, and MB-10.4 contain side groups similar to those of MB-10. Based on structural comparison, we assumed that the six-membered ring and thiophene linked by hydrazide-hydrazone bond would be critical to MB-10 activity. Methylation of the hydrazone interferes with MB-10 activity. To support this notion, we revisited the screening results. The Chembridge chemical library includes MB-10-like compounds with substitution of the thiophene or with methylation of the hydrazone group; these compounds did not confer the growth of WT[Su9-URA3] strain in the screening assay (supplemental Fig. S1), suggesting that these chemical alterations in MB-10-like compounds abolished their ability to inhibit protein import. Taken together, the substituted rings on MB-10 are important for inhibitory activity toward the PAM complex.
MB-10 Targets Tim44 of the PAM Complex
The protein target may display reduced sensitivity to an exogenous protease because the small molecule binds tightly to the protein and stabilizes the protein structure, thereby reducing the ability of the protease to cleave protein (27). We used this assay to determine whether Tim44 was a potential target, because our import studies and the published literature support that Tim44 is a probable target. A battery of proteases at varying concentrations were tested to identify conditions in which the protein was degraded in a stepwise manner that could be captured in a time course assay by immunoblotting; Pronase gave the best results (27). Isolated yeast mitochondria were lysed with buffer containing 0.2% Triton X-100. Then the lysates were incubated in the presence of 1% DMSO, 100 μm MB-10, or analog 4, followed by Pronase treatment and immunoblot analysis. In the presence of DMSO or analog 4, Tim44 was readily degraded to a 30-kDa truncation product after 10 min (marked by an asterisk), whereas MB-10 stabilized Tim44, slowing the rate of protease digestion (Fig. 4A, top panel). In contrast, Tim23 and matrix protein, α-ketoglutarate dehydrogenase, were digested by Pronase at the same rate, even in the presence of MB-10 (Fig. 4A, middle and bottom panels). MB-10 did not alter the protease degradation of another component of PAM complex, mitochondrial Hsp70 (Fig. 4B; Note that the antibody also identified a lower molecular weight species that was marked by an asterisk). Thus, these data support that MB-10 likely targets Tim44.
FIGURE 4.
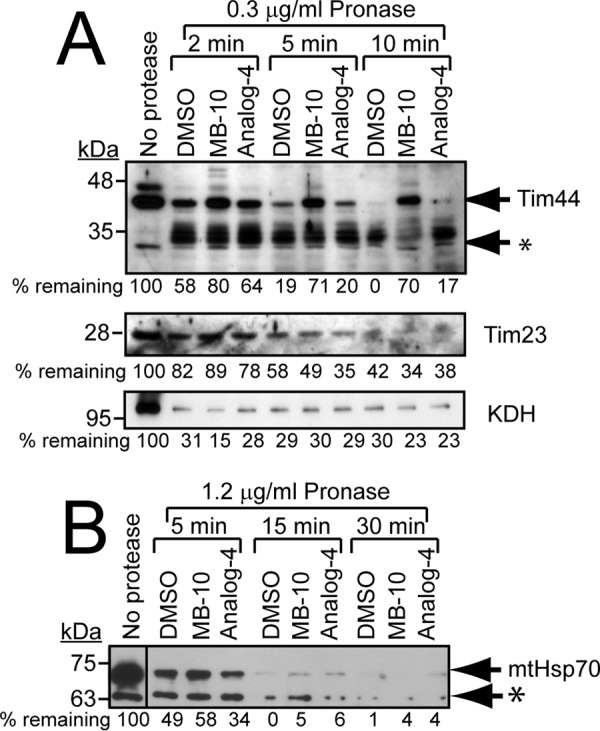
MB-10 targets Tim44. A, mitochondria were lysed in buffer containing 0.2% Triton X-100. The lysates were incubated with 1% DMSO, 100 μm MB-10, or 100 μm analog 4 for 15 min followed by treatment with 0.3 μg/ml Pronase at 25 °C. At the indicated time points, proteolysis was stopped by the addition of 0.2% SDS and incubation at 100 °C. Samples were analyzed by immunoblotting with antibodies against Tim44, Tim23, and α-ketoglutarate dehydrogenase (KDH). The asterisk indicates cleaved Tim44 products. A representative gel is shown; quantification of bands was performed with ImageJ software, and the percentage was calculated relative to the treatment with no protease. B, mitochondria were lysed in buffer containing 0.2% Triton X-100. The lysates were incubated with 1% DMSO, 100 μm MB-10, or 100 μm analog 4 for 15 min followed by treatment with 1.2 μg/ml Pronase at 25 °C. Samples were analyzed by immunoblotting with anti-mtHsp70 antibody. An asterisk mars a nonspecific band of lower molecular weight that was detected by the antibody.
Given that the crystal structure of the C-terminal domain of Tim44 has been solved (28), we initiated a collaboration with the Sha laboratory to determine whether MB-10 could be incorporated into crystallization trials. However, MB-10 was not soluble under the experimental conditions, and this line of investigation could not be developed.3 Instead, we used a genetic approach, based on random PCR mutagenesis, to isolate tim44 mutants that were resistant to MB-10 (29), presumably because the mutations altered interactions with MB-10. Mutants were selected that were resistant to 15 μm MB-10, which is higher than the MIC50 of 7.1 μm in WT. A concentration greater than 16 μm MB-10 failed to generate resistant yeast strains. The mutant plasmids were isolated and sequenced. The mutant tim44 plasmids were then transformed into a strain in which the genomic copy of TIM44 was deleted and viability was maintained with TIM44 expressed from a centromeric URA3 plasmid. Using a plasmid shuffling strategy (30), the URA3 plasmid with TIM44 was removed by selection on 5-fluororotic acid, and the ability of the strain with the mutagenized tim44 plasmid to grow was tested on selective medium that included uracil; this strategy was used to confirm that resistance to MB-10 was based on the mutagenized plasmid. Although mild conditions were used for mutagenesis, plasmids typically contained two or three mutations in TIM44. A region that was enriched in point mutations among several plasmids from the strains that were resistant to MB-10 centered on the helix designated α4 in the C-terminal domain that functions in membrane tethering (Fig. 5A). Specific mutations in this region were F284L, T290S, I297V, and V298A (Fig. 5B). This area is highly conserved across species, and residues 284 and 297 are invariable (31). The tim44 mutants with specific mutations, T290S and I297V, were reconstructed with the single mutation, respectively, and resistance to MB-10 was confirmed. The MIC50 of tim44 mutants T290S and I297V shifted from ∼8 μm to ∼12 and ∼16 μm, respectively (Fig. 5C). Protease sensitivity studies indicated that MB-10 had a decreased affinity for Tim44 I297V, because the Tim44 I297V mutant was degraded in the presence of a lower MB-10 concentration (60 μm) than the WT protein (greater than 80 μm; Fig. 5D). The genetic approach in yeast indicates that MB-10 inhibits Tim44 function through a specific interaction with the C-terminal domain of Tim44.
FIGURE 5.
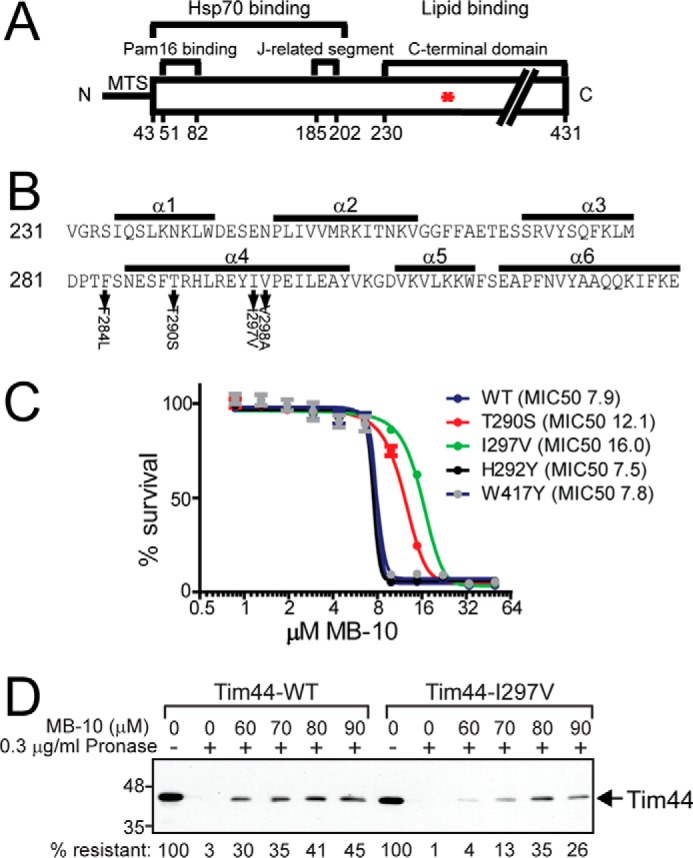
Mutations in the α4 helix of the C-terminal domain of Tim44 confer resistance to MB-10. A, schematic of Tim44 organization including the Hsp70 and Pam16 binding regions, the J-related segment, and the C-terminal domain that is implicated in lipid binding. The asterisk denotes the C-terminal region where the mutations that conferred MB-10 clustered. B, specific mutations in the α 4 helix in the C-terminal region of Tim44 were identified that conferred growth in the presence of 15 μm MB-10. C, MIC50 analysis of the WT Tim44 and Tim44 mutants T290S, I297V, H292Y, and W417Y. Each strain was cultured in rich ethanol-glycerol medium in the presence of various concentrations of MB-10 for 24 h. 100% was set as A600 of each strain grown in the presence of the vehicle control (1% DMSO). Average percentage of survival ± S.D. of n = 3 trials. D, as in Fig. 4A, protease sensitivity assays with lysates from WT and Tim44-I297V mitochondria. MB-10 was added from 60–90 μm, and Tim44 was detected by immunoblotting. The first lane is a control with DMSO and mitochondrial lysate that lacks protease and MB-10. Tim44 abundance was quantified with ImageJ software; 100% was set as the amount of Tim44 that lacked protease.
To determine the binding pocket for MB-10 in the C-terminal domain of Tim44, we used computational approaches including Brownian dynamics (BD) simulations and docking and molecular dynamics (MD) simulations (Fig. 6, A and B) (32–35). The GeomBD program was used to search for all potential MB-10 binding sites in Tim44, and a total six possible binding sites were identified (Fig. 6C). Because most binding sites suggested by BD diffusion may be nonspecific binding surfaces for a compound, we additionally used docking simulations to estimate binding affinities and to reveal the most possible binding pocket for MB-10. Fig. 6D illustrates the computed binding affinities for MB-10 and Tim44. Only one pocket between α4 helix, α5 helix, and a loop that connects the β3 and β4 strands showed a predicted binding affinity more negative than −9 kcal/mol, indicating a tight and specific binding site (Fig. 6, A and D). Notably, this is a novel binding site that is different from the large hydrophobic binding site for ligand pentaethylene glycol on the opposite face of Tim44, which has been suggested to interact with the membrane (Fig. 6A) (31). Because proteins are flexible and the crystal structure only illustrates one stable conformation, MD simulations were carried out to further investigate detailed MB-10 interactions with wild-type and mutant Tim44 (Fig. 6, E and F). The MD runs revealed that residues Ile-297, Phe-284, and Thr-290 have direct contact with MB-10, and Val-298 supports the β-sheets on the back of the major binding pocket to provide a stable three-dimensional structure for MB-10 binding (Fig. 6B). MD simulations and energy calculations (supplemental Table S3) for MB-10 and Tim44 with mutations at I297V, F284L, T290S, or V298A showed that binding of MB-10 and the Tim44 mutants was weakened, compared with MB-10 binding to the wild-type Tim44. The weaker interactions between MB-10 and mutant Tim44 explain the experimentally observed MB-10 resistance to the mutants. The agreement between the modeling work and experiments further confirmed the novel binding pocket identified in this study.
FIGURE 6.
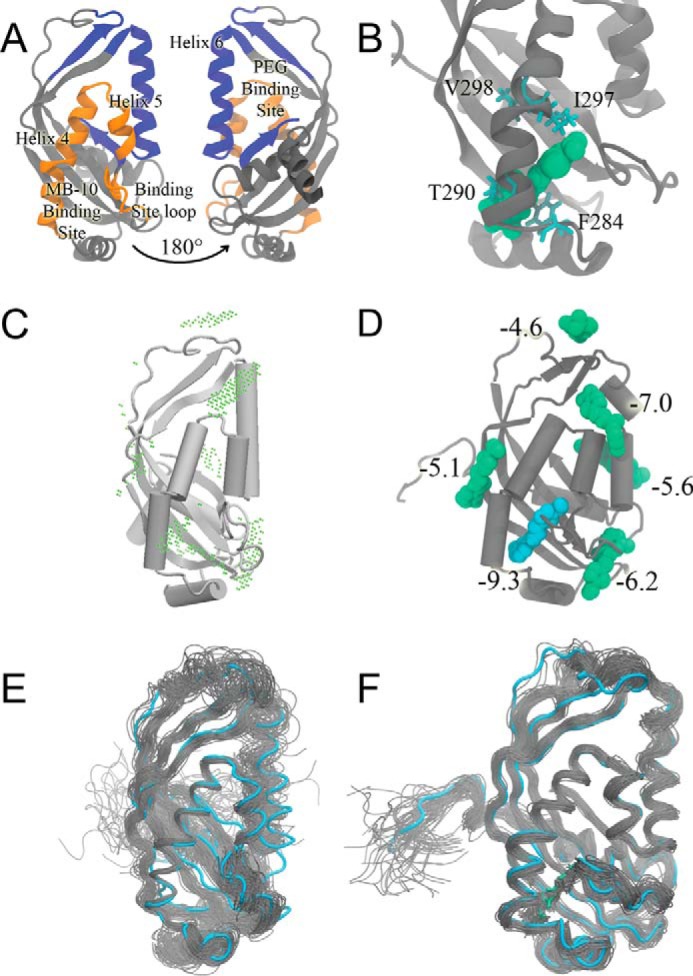
MB-10 fits in a binding pocket in the C-terminal domain of Tim44. A, a labeled diagram of Tim44. The MB-10 binding pocket composed of important regions is colored orange. On the opposite face is a large groove that binds to pentaethylene glycol and is postulated to bind to the membrane (rotated 180°). B, one snapshot of MD simulations for MB-10 found to wild-type Tim44. Gray, Tim44; cyan, residues Phe-284, Thr-290, Ile-297, and Val-298; green, MB-10. C and D, results from BD simulation and molecular docking. C, potential MB-10 binding sites on Tim44 by diffusing MB-10 to the protein using the GeomBD package. The association probability is shown by the dots, where the center of MB-10 has high possibility to encounter with Tim44. Tim44 structure was from PDB (code 2FXT). D, docking results to each potential binding sites found by BD simulations. Only one conformation with the best predicted binding affinities (kcal/mol) is shown for each potential binding site. E and F, conformations were sampled from MD simulations for the wild-type Tim44 in the absence (E) and presence (F) of MB-10. The initial conformation is presented in cyan (protein) and green (ligand), and the other conformations are presented in gray. Although the crystal structures only reveal one certain conformation, proteins are flexible and can adopt more than one conformation. Both figures have a total of 50 frames from a 100-ns MD simulations. The frames were superimposed to the initial conformation for clearer visualization.
Based on the docking analysis, we inserted conservative point mutations in conserved residues of Tim44 that were predicted to interfere with MB-10 binding. Specific mutations included H292A and H292Y of helix α4, W316A and W316Y of helix α 5, and W417A and W417Y of the loop that connects the β3 and β4 strands. The plasmids were transformed into the shuffling strain in which viability was maintained with TIM44 on a centromeric URA3 plasmid. When the cells were initially transformed, viable colonies expressing Tim44 H292A, W316A, W316Y, and W417A were not recovered despite numerous attempts (supplemental Fig. S2), signifying that these mutations likely yielded a dominant negative phenotype. In contrast, the Tim44 H292Y and W417Y strains grew similar to the WT strain (supplemental Fig. S2 and data not shown) and were amenable to plasmid shuffling to remove the plasmid for WT TIM44. The MIC50 of Tim44 H292Y and W417Y strains was ∼8 μm, similar to the WT strain (Fig. 5C). Thus, the H292Y and W417Y mutations were not sufficient to cause inviability or decrease MB-10 affinity for Tim44, whereas mutations H292A, W316A, W316Y, and W417A rendered Tim44 nonfunctional.
To determine whether MB-10 treatment altered interactions of a substrate during translocation, we performed cross-linking of methotrexate-arrested cyt b2(167)A63P-DHFR using 200 μm disuccinimidyl suberate (DSS), because the cross-linking pattern to TIM23 components has been well defined (36, 37). In WT mitochondria, methotrexate-arrested cyt b2(167)A63P-DHFR was cross-linked to several proteins by DSS (Fig. 7A, lane 2), indicating that the precursor was present in the translocation channel. In the presence of 100 μm MB-10, a cross-linked product at ∼90 kDa was strongly decreased (Fig. 7A, lane 3). According to published experiments, the missing cross-link is the Tim44 adduct (36–38). We confirmed this by comparison with cross-linking in mitochondria purified from the tim44-8 mutant, which also lacks a cross-link to Tim44 (22). In tim44-8 mitochondria incubated at 37 °C, the cross-linked product was not observed in both the presence and the absence of MB-10 (Fig. 7A, lanes 4 and 5), supporting that the ∼90-kDa cross-linked product is Tim44. In addition, the cross-linked products, representing precursor bound to mtHsp70, at ∼115–130 kDa were increased in abundance, denoting that MB-10 addition may cause an arrest or shift of the interaction of the precursor from Tim44 to mtHsp70.
We also tested the binding between Tim44 and Hsp70 in the presence of MB-10. In WT mitochondria, Tim44 and Hsp70 bound tightly under low ATP conditions, but the addition of ATP and Mg2+ subsequently stimulated release of Hsp70 from Tim44 (Fig. 7B), as has been reported previously (39). In contrast, MB-10 addition specifically inhibited Tim44 binding to Hsp70 in low ATP conditions (Fig. 7B). Tim44-Pam16 and Tim44-Pam18 interactions were not disrupted by MB-10 (Fig. 7C). A strain was generated in which a histidine tag was integrated at the 3′ end of TIM44, generating Tim44-His protein for pulldown experiments with Ni2+-agarose (40). Mitochondria were incubated in 1% DMSO or 100 μm MB-10 followed by solubilization in 1% digitonin. When Tim44-His was isolated, Pam16 and Pam18 co-purified to the same extent in the presence of DMSO or MB-10 (Fig. 7C), supporting the possibility that MB-10 did not disrupt interactions between Tim44 and Pam16/Pam18. Tom70 was included as a negative control. A schematic in Fig. 7D highlights the interactions at the TIM23 channel (see “Discussion” for details). Thus, MB-10 specifically inhibits Tim44 binding with Hsp70 and the precursor.
MB-10 Inhibits Protein Import into Mammalian Mitochondria
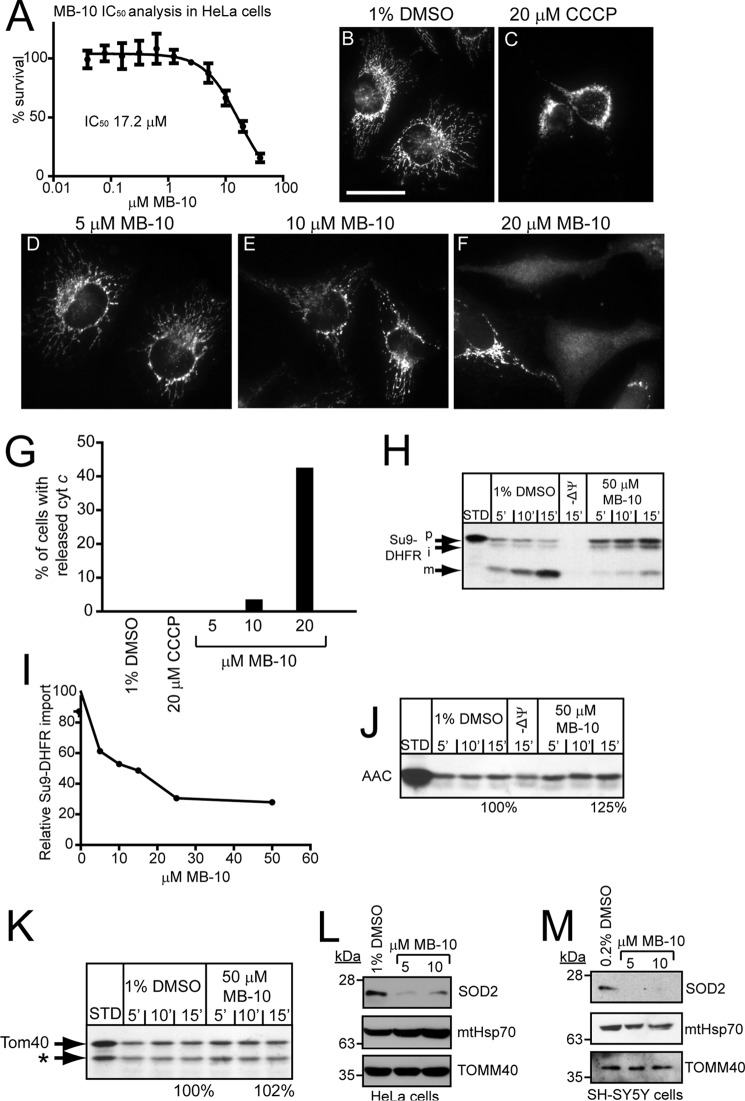
The long term goal of our chemical screens is to develop small molecule modulators that also work in other systems such as cultured cells and zebrafish. Therefore, we switched experimental systems to cultured mammalian cells. HeLa cells were treated with increasing concentrations of MB-10 for 24 h, and cell viability was evaluated with a 1-(4,5-dimethylthiazol-2-yl)-3,5-diphenylformazan (MTT) assay. MB-10 treatment reduced viability of HeLa cells in a dose-dependent manner with an MIC50 of 17.2 μm (Fig. 8A). Upon microscopic observation using an antibody against cyt c to mark mitochondria, mitochondrial morphology was normal in the presence of 5–10 μm MB-10 (below the MIC50) (Fig. 8, D and E). However, the addition of 20 μm MB-10 (just above the MIC50) caused the mitochondrial network to fragment with cyt c release to the cytosol in 40% of the cells (Fig. 8, F and G). As a control, 20 μm CCCP induced mitochondrial fragmentation but did not induce cyt c release in HeLa cells, and treatment with 1% DMSO did not perturb the mitochondrial network under our experimental conditions (Fig. 8, B and C).
FIGURE 8.
MB-10 inhibits protein import into mammalian mitochondria. A, cell viability of HeLa cells in the presence of MB-10 was determined using an MTT cell viability assay. HeLa cells were treated for 24 h with MB-10 at the indicated concentrations, and then viability was measured. 100% was defined as the signal from cells treated with 1% DMSO. Average percentage of survival ± S.D. of n = 3 trials. B–F, HeLa cells were treated with 1% DMSO (B), 20 μm CCCP (C), or MB-10 (D–F) for 24 h followed by immunostaining with anti-cyt c antibody. Note that 20 μm MB-10 is just above the MIC50 in mammalian cells. F, diffuse cyt c staining is indicative of release from mitochondria by apoptosis. Scale bar, 20 μm. G, quantitation of cyt c release in cells from experiments B–F. 200 cells were quantitated. H, an import assay was performed with Su9-DHFR into mitochondria isolated from HeLa cells as described in Fig. 3 in the presence of 50 μm MB-10 or the vehicle control (1% DMSO). I, as in H, import of Su9-DHFR was quantitated in the presence of the indicated concentrations of MB-10. J and K, precursors yeast AAC and Tom40 were also imported. The asterisk indicates a truncation product of Tom40 translation that is imported. L and M, HeLa cells (L) and SH-SY5Y cells (M) were treated with 0.2% DMSO or MB-10 for 24 h. A total cell lysate was analyzed by immunoblotting with antibodies against SOD2, mitochondrial Hsp70, and TOMM40 antibodies.
We then tested whether MB-10 inhibited protein import into mitochondria isolated from HeLa cells. In the presence of MB-10, import of Su9-DHFR was inhibited (Fig. 8H). Notably, the precursor and intermediate forms of Su9-DHFR were also detected as in yeast mitochondria (Fig. 2D), supporting that MB-10 behaves similarly in yeast and mammalian mitochondria. Furthermore, MB-10 inhibited protein import more effectively in mammalian mitochondria than in yeast mitochondria. An approximate 70% reduction of Su9-DHFR import was achieved with 25 μm MB-10 (Fig. 8I). Potential reasons for the increased sensitivity of mammalian cells to MB-10 may be that protein import is more dependent on TIMM44 in mammalian cells or that MB-10 has higher affinity to mammalian TIMM44 than yeast Tim44; the crystallization studies support that the structures of yeast and mammalian Tim44 are highly conserved (31), so MB-10 may bind more tightly to mammalian TIM44. Consistent with the results in yeast mitochondria, MB-10 did not inhibit the import of AAC and Tom40 that are imported independently of the TIM23 complex (Fig. 8, J and K). Therefore, we conclude that MB-10 inhibited Tim44 in both yeast and mammalian mitochondria. In addition, loss of TIMM44 activity caused cell death by apoptosis.
MB-10 Inhibits Biogenesis of SOD2 in Mammalian Culture Cells
Previous studies show that TIMM44 is up-regulated in diabetic mouse kidneys, and siRNA-mediated knockdown of TIMM44 in a variety of cells decreases the abundance of SOD2 and glutathione peroxidase (11, 41). We therefore measured SOD2 levels in the presence of MB-10 in HeLa cells and SH-SY5Y neuroblastoma cells (42). Sensitivity of SH-SY5Y cells to MB-10 was evaluated by the MTT assay and was similar to that of HeLa cells (data not shown). Upon treatment of 5 and 10 μm MB-10 to cells for 24 h, the abundance of SOD2 was markedly decreased, whereas the abundance of matrix protein mtHsp70 and outer membrane protein TOMM40 was not decreased (Fig. 8, L and M). Thus, MB-10 treatment recapitulates decreased SOD2 levels when TIMM44 levels are decreased by RNAi treatment.
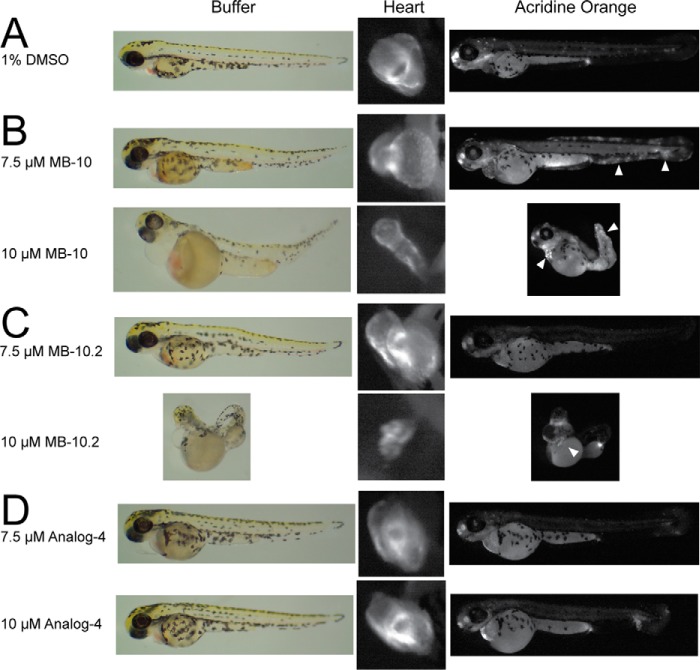
MB-10 Treatment Impairs Cardiac Development and Induces Apoptosis in Zebrafish Embryos
Mutations in DNAJC19, a component of the PAM complex, lead to dilated cardiomyopathy with ataxia syndrome, an autosomal recessive Barth syndrome-like condition in patients (43). We applied MB-10 to zebrafish embryos to determine how impaired TIMM44 altered development, particularly focusing on cardiac tissue. A transgenic zebrafish line in which DsRed is targeted to mitochondria under control of the heart specific cardiac myosin light chain promoter cmlc2 was used (13). Embryos were placed in either 1% DMSO, MB-10, MB-10.2, or analog 4 as indicated (Fig. 9) at 3 hpf and allowed to develop until 72 hpf. Embryos incubated with 10 μm MB-10 displayed dorsal body curvature and impaired heart development with excessive pericardiac effusion (Fig. 9) that is characteristic of zebrafish with dilated cardiomyopathy (44, 45). MB-10-treated embryos also showed an increase in apoptotic cells as visualized by acridine orange staining (Fig. 9, highlighted by arrowheads), consistent with the increased apoptosis observed in MB-10-treated HeLa cells (Fig. 8, D–G). In addition, embryos incubated with MB-10.2 had developmental defects similar to MB-10 (Fig. 9C), whereas embryos incubated with analog 4 looked similar to the zebrafish treated with DMSO (Fig. 9D). Thus, MB-10 is an effective tool for characterizing TIMM44 function in cultured cells and zebrafish as well as yeast.
FIGURE 9.
MB-10 treatment impairs cardiac development and induces apoptosis in zebrafish. Embryos (3 hpf) were treated with DMSO (A), MB-10 (B), MB-10.2 (C), or analog 4 (D) at the indicated concentrations. Development was observed by microscopy at 72 hpf. Zebrafish hearts were marked with mitochondrial-targeted DsRed expressed from the cmcl2 promoter. Apoptotic cells were visualized by acridine orange staining. Arrowheads indicate increased apoptotic cells in MB-10- and MB-10.2-treated embryos.
Discussion
We completed a screen to identify inhibitors of the general import pathway by adapting a genetic screen in yeast (14). Because this assay selects for a gain in growth, false-positive compounds that are generally lethal are omitted. Metrics for evaluating and validating a robust high throughput include a low hit rate typically less than 0.14% and a screening window coefficient (referred to as the Z′ factor) greater than 0.5 (46). Our screening approach was strong with a high Z′ factor (>0.8) and low hit rate (0.02%). Another benefit of this screening approach is that a WT yeast strain, instead of mutants (12), was used. In addition, as shown with previous inhibitors MB-1 and MB-6 (12, 13), MB-10 inhibits import in both yeast and mammalian mitochondria. The SAR studies in the protein import assays (Fig. 3) and zebrafish (Fig. 9) support that MB-10 seems specific for targeting Tim44. Indeed, MB-10 and MB-10.2 specifically induced cardiac deficiency in developing zebrafish embryos and inhibited import. In contrast, analog 4 did not alter zebrafish development, although the compound shared a similar structure to MB-10 and MB-10.2 (Fig. 9). Thus, these probes are valuable for study in a broad range of model systems.
A multifaceted strategy illustrates that MB-10 likely targets Tim44, but interactions at the interface with other components of the PAM and TIM23 complex cannot be ruled out completely. With a genetic approach, the tim23-2 mutant was more sensitive to MB-10 than the tim10-1 mutant. A battery of import assays using substrates of each translocon (TOM, TIM23, and MIA (mitochondrial intermembrane space assembly)) narrowed the candidate to five or six factors of the PAM complex. Subsequent detailed studies including cross-linking to the cyt b2(167)-DHFR and protease protection corroborated that MB-10 likely targeted Tim44. Whereas the direct approach of incorporating MB-10 into crystallography studies was not successful, a genetic approach identified Tim44 mutants that were not sensitive to MB-10. In conjunction with docking and MD simulations, MB-10 binds in a pocket in the C-terminal region that is on the opposite face from a large groove that may interact with the membrane (31). The N-terminal region of Tim44 interacts with motor partners Hsp70, Mge1, Pam18, and Pam16 (47), but our study shows that the C-terminal region is also essential for Tim44 function. Indeed, the C-terminal region is highly conserved, mutations at invariable residues resulted in a dominant-negative phenotype, and MB-10 addition blocked interactions of Tim44 with the imported precursor and Hsp70. As shown in Fig. 7D, we propose that MB-10 blocks binding of the C-terminal domain of Tim44 and the precursor, thereby blocking protein translocation. The cross-linking studies indicate that binding of precursor to Hsp70 increases, but this intermediate is not productive for protein import. Based on our pulldown studies, MB-10 does not disrupt interactions among components of the TIM23 translocon and PAM complex. Our studies thus agree with those of Mokranjac and co-workers (47) in which they reported that the C-terminal domain interacted with the translocating protein. Thus, MB-10 is a specific probe to block interactions of the translocating precursor with the PAM complex.
From the import studies with substrates that have a stop transfer motif, MB-10 at 100 μm inhibited the import of precursors sorted to the intermembrane space (Fig. 3). This import inhibition is similar to the phenotype of tim44 temperature-sensitive mutants reported previously (22, 48). Recently, the role of the import motor in the translocation of precursors that are laterally sorted (i.e. cyt b2(167)-DHFR and cyt c1) has been under debate (49). The “modular” model proposes that the TIM23 complex exists in two forms: one with Tim21 that lacks the import motor and mediates lateral insertion and the second one with the import motor that lacks Tim21 and mediates transport into the matrix (50, 51). The “single-entity” model proposes that all essential subunits of the translocase function as one complex that may be actively remodeled based on targeting sequences of the translocating substrate (49, 52). Given that MB-10 inhibits the import of cyt b2(167)-DHFR and cyt c1, our studies are tilted to support the “single-entity” model, because Tim44 of the motor is required for the import of laterally sorted precursors. MB-10 will be a useful tool to continue mechanistic studies about how the dynamic TIM23 translocon mediates protein import.
Experimental Procedures
High Throughput Screening
Screening was performed using the WT[Su9-URA3] strain that grown in SD medium supplemented with uracil and without leucine. Yeast were thoroughly washed with sterile H2O twice to remove uracil and diluted in SD medium lacking uracil to an A600 of 0.05. The cell suspension was kept on ice throughout the screening run. A Titertek multidrop instrument was used to dispense 50 μl of cell suspension into column 2–22 of each clear 384-well plate (Greiner Bio-One). The Biomek FX (Beckman Coulter) was used to pin transfer 0.5 μl of compounds from the 1 mm stock or DMSO to respective wells. The approximate screening concentration of the small molecules was 10 μm. As controls, column 2 and 23 consisted of the WT[Su9-URA3] and tim23-2[Su9-URA3] strains, respectively, supplemented with the vehicle 1% DMSO. After completing compound transfer, all plates were incubated at 30 °C in a humidified incubator for 24 h. Each plate was shaken in a Beckman orbital shaker to resuspend the settled cells, and the A600 in each well was read by a Wallac Victor plate reader (PerkinElmer Life Sciences). The compounds that augmented growth of the WT[Su9-URA3] strain by more than 30% of that of the tim23-2[Su9-URA3] strain were identified and cherry-picked into one plate for rescreening. Hit compounds that conferred growth in the repeated assay were reordered from Chembridge for follow-up analysis. Purchased compounds that conferred growth to the WT[Su9-URA3] strain were chosen for additional analysis (Fig. 3F).
Molecular Dynamics and Docking Simulations
Starting from the crystal structure of yeast Tim44 (PDB code 2FXT) for the MD run, we constructed the wild-type protein and four mutants (T290S, I297V, V298A, and F284L). The structure was aligned with PDB code 3QK9 for better visualization. The structure of MB-10 was generated using Vega ZZ. The AMBER 99SB force field was used for the protein (53, 54). The AMBER12 package was used for standard setup, and the GPU acceleration was used to run the MD simulations. After the energy minimization, the systems were solvated using a box of TIP3P water of at least 12 Å around the protein by the leap module in AMBER12. The systems were composed of ∼43,400 atoms. All production runs were performed for 100 ns at 298 K, and the temperature was maintained at 298 K by Langevin dynamics (55). The particle mesh Ewald was used for the long range electrostatic interactions, whereas the van der Waals interactions were cut off using amber default 8.0 Å. A time step of 2.0 fs was used, and MD frames were saved every 1 ps. Molecular docking of the compound was performed using AutoDock (56). Briefly, multiple frames selected from the MD simulation for wild-type Tim44 were used to dock the ligand into the binding site. Residues 315, 316, 404, 415, and 417 were kept flexible during docking. The Lamarckian genetic algorithm was used to search for minimum energy ligand conformations and orientations. A total of 20 unique docking results were generated and analyzed, and the lowest energy conformation that placed MB-10 in the binding site was used for Fig. 6B. All three-dimensional molecular structures were generated using VMD (57). Computational details for the simulations are provided in the supplemental “Experimental Procedures.”
General Assays with Mitochondria
Detailed methods are provided in the supplemental “Experimental Procedures.”
Author Contributions
N. M. conceived and designed the study; acquired biological, biochemical, and biophysical data; analyzed and interpreted the data; and drafted or revised the article. Z. T. conceived and performed molecular modeling and docking, acquired biochemical and biophysical data, and analyzed and interpreted the data. M. A. C. acquired biological, biochemical, and biophysical data; analyzed and interpreted the data; and drafted or revised the article. M. E. J. acquired biological data, analyzed and interpreted the data, and drafted or revised the article. C. J. D. analyzed and interpreted the data and drafted or revised the article. S. A. H. conceived and designed the biological screen, acquired data, and analyzed and interpreted the data. R. D. assisted with screen design and implementation, acquired data, and analyzed and interpreted the data. C. A. C. conceived and performed molecular modeling and docking, analyzed and interpreted the data, and drafted or revised the article. C. M. K. supervised the project, conceived and designed the study, analyzed and interpreted the data, and drafted or revised the article.
Supplementary Material
Acknowledgments
We thank Juwina Wijaya, Jong Sang Lee, and Winnie Hwong for technical assistance and N. Pfanner for the tim23-2 and tim44-8 mutants.
This work was supported by National Institutes of Health Grants GM073981 and GM61721 (to C. M. K.), California Institute of Regenerative Medicine Grants RS1-00313 and RB1-01397 (to C. M. K.), National Science Foundation Grant MCB-0919586 (to C. A. C.), and U.S. Public Health Service NRSA Grants GM07185 (to M. E. J., M. A. C., and C. J. D.) and GM08496 (to S. A. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental text, Tables S1–S3, and Figs. S1 and S2.
B. Sha, unpublished results.
- TOM
- translocase of outer membrane
- BD
- Brownian dynamics
- DHFR
- dihydrofolate reductase
- DSS
- disuccinimidyl suberate
- MD
- molecular dynamics
- MitoBloCK or MB
- mitochondrial protein import blocker from the Carla Koehler laboratory
- MTT
- 1-(4,5-dimethylthiazol-2-yl)-3,5-diphenylformazan
- PAM
- protein associated motor
- SAR
- structure activity relationship
- TIM
- translocase of inner membrane
- TIMM
- translocase of inner mitochondrial membrane
- TOMM
- translocase of outer mitochondrial membrane
- AAC
- ADP/ATP carrier
- cyt
- cytochrome
- hpf
- h post-fertilization
- PDB
- Protein Data Bank.
References
- 1. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., and Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neupert W., and Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 3. Alder N. N., Jensen R. E., and Johnson A. E. (2008) Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 134, 439–450 [DOI] [PubMed] [Google Scholar]
- 4. Meinecke M., Wagner R., Kovermann P., Guiard B., Mick D. U., Hutu D. P., Voos W., Truscott K. N., Chacinska A., Pfanner N., and Rehling P. (2006) Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 5. van der Laan M., Meinecke M., Dudek J., Hutu D. P., Lind M., Perschil I., Guiard B., Wagner R., Pfanner N., and Rehling P. (2007) Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9, 1152–1159 [DOI] [PubMed] [Google Scholar]
- 6. D'Silva P. R., Schilke B., Hayashi M., and Craig E. A. (2008) Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol. Biol. Cell 19, 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ojala T., Polinati P., Manninen T., Hiippala A., Rajantie J., Karikoski R., Suomalainen A., and Tyni T. (2012) New mutation of mitochondrial DNAJC19 causing dilated and noncompaction cardiomyopathy, anemia, ataxia, and male genital anomalies. Pediatr. Res. 72, 432–437 [DOI] [PubMed] [Google Scholar]
- 8. Tagliati F., Gentilin E., Buratto M., Molè D., degli Uberti E. C., and Zatelli M. C. (2010) Magmas, a gene newly identified as overexpressed in human and mouse ACTH-secreting pituitary adenomas, protects pituitary cells from apoptotic stimuli. Endocrinology 151, 4635–4642 [DOI] [PubMed] [Google Scholar]
- 9. Wada J., and Kanwar Y. S. (1998) Characterization of mammalian translocase of inner mitochondrial membrane (Tim44) isolated from diabetic newborn mouse kidney. Proc. Natl. Acad. Sci. U.S.A. 95, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonora E., Evangelisti C., Bonichon F., Tallini G., and Romeo G. (2006) Novel germline variants identified in the inner mitochondrial membrane transporter TIMM44 and their role in predisposition to oncocytic thyroid carcinomas. Br. J. Cancer 95, 1529–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Wada J., Hashimoto I., Eguchi J., Yasuhara A., Kanwar Y. S., Shikata K., and Makino H. (2006) Therapeutic approach for diabetic nephropathy using gene delivery of translocase of inner mitochondrial membrane 44 by reducing mitochondrial superoxide production. J. Am. Soc. Nephrol. 17, 1090–1101 [DOI] [PubMed] [Google Scholar]
- 12. Hasson S. A., Damoiseaux R., Glavin J. D., Dabir D. V., Walker S. S., and Koehler C. M. (2010) Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc. Natl. Acad. Sci. U.S.A. 107, 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dabir D. V., Hasson S. A., Setoguchi K., Johnson M. E., Wongkongkathep P., Douglas C. J., Zimmerman J., Damoiseaux R., Teitell M. A., and Koehler C. M. (2013) A small molecule inhibitor of redox-regulated protein translocation into mitochondria. Dev. Cell 25, 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maarse A. C., Blom J., Grivell L. A., and Meijer M. (1992) MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO J. 11, 3619–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dekker P. J., Martin F., Maarse A. C., Bömer U., Müller H., Guiard B., Meijer M., Rassow J., and Pfanner N. (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16, 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ungermann C., Neupert W., and Cyr D. M. (1994) The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science 266, 1250–1253 [DOI] [PubMed] [Google Scholar]
- 17. Hwang S. T., Wachter C., and Schatz G. (1991) Protein import into the yeast mitochondrial matrix. A new translocation intermediate between the two mitochondrial membranes. J. Biol. Chem. 266, 21083–21089 [PubMed] [Google Scholar]
- 18. Matouschek A., Pfanner N., and Voos W. (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep. 1, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glick B. S., Brandt A., Cunningham K., Müller S., Hallberg R. L., and Schatz G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69, 809–822 [DOI] [PubMed] [Google Scholar]
- 20. Beasley E. M., Müller S., and Schatz G. (1993) The signal that sorts yeast cytochrome b2 to the mitochondrial intermembrane space contains three distinct functional regions. EMBO J. 12, 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Voos W., Gambill B. D., Laloraya S., Ang D., Craig E. A., and Pfanner N. (1994) Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol. Cell Biol. 14, 6627–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bömer U., Maarse A. C., Martin F., Geissler A., Merlin A., Schönfisch B., Meijer M., Pfanner N., and Rassow J. (1998) Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J. 17, 4226–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laloraya S., Dekker P. J., Voos W., Craig E. A., and Pfanner N. (1995) Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol. Cell Biol. 15, 7098–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Milisav I., Moro F., Neupert W., and Brunner M. (2001) Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem. 276, 25856–25861 [DOI] [PubMed] [Google Scholar]
- 25. Voos W., Gambill B. D., Guiard B., Pfanner N., and Craig E. A. (1993) Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popov-Celeketić D., Mapa K., Neupert W., and Mokranjac D. (2008) Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27, 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lomenick B., Hao R., Jonai N., Chin R. M., Aghajan M., Warburton S., Wang J., Wu R. P., Gomez F., Loo J. A., Wohlschlegel J. A., Vondriska T. M., Pelletier J., Herschman H. R., Clardy J., et al. (2009) Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. U.S.A. 106, 21984–21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Josyula R., Jin Z., Fu Z., and Sha B. (2006) Crystal structure of yeast mitochondrial peripheral membrane protein Tim44p C-terminal domain. J. Mol. Biol. 359, 798–804 [DOI] [PubMed] [Google Scholar]
- 29. Koehler C. M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R. J., and Schatz G. (1998) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279, 369–373 [DOI] [PubMed] [Google Scholar]
- 30. Tong A. H., and Boone C. (2006) Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313, 171–192 [DOI] [PubMed] [Google Scholar]
- 31. Handa N., Kishishita S., Morita S., Akasaka R., Jin Z., Chrzas J., Chen L., Liu Z. J., Wang B. C., Sugano S., Tanaka A., Terada T., Shirouzu M., and Yokoyama S. (2007) Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form. Acta Crystallogr. D Biol. Crystallogr. 63, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 32. Ai R., Qaiser Fatmi M., and Chang C. E. (2010) T-Analyst: a program for efficient analysis of protein conformational changes by torsion angles. J. Comput. Aided Mol. Des. 24, 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang C. E., and Gilson M. K. (2003) Tork: Conformational analysis method for molecules and complexes. J. Comput. Chem. 24, 1987–1998 [DOI] [PubMed] [Google Scholar]
- 34. Roberts C. C., and Chang C. E. (2015) Modeling of enhanced catalysis in multienzyme nanostructures: effect of molecular scaffolds, spatial organization, and concentration. J. Chem. Theory Comput. 11, 286–292 [DOI] [PubMed] [Google Scholar]
- 35. Roberts C. C., and Chang C. E. (2016) Analysis of ligand-receptor association and intermediate transfer rates in multienzyme nanostructures with all-atom Brownian dynamics simulations. J. Phys. Chem. B 120, 8518–8531 [DOI] [PubMed] [Google Scholar]
- 36. Schneider H. C., Berthold J., Bauer M. F., Dietmeier K., Guiard B., Brunner M., and Neupert W. (1994) Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371, 768–774 [DOI] [PubMed] [Google Scholar]
- 37. Rassow J., Maarse A. C., Krainer E., Kübrich M., Müller H., Meijer M., Craig E. A., and Pfanner N. (1994) Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ungermann C., Guiard B., Neupert W., and Cyr D. M. (1996) The delta psi- and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO J. 15, 735–744 [PMC free article] [PubMed] [Google Scholar]
- 39. D'Silva P., Liu Q., Walter W., and Craig E. A. (2004) Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 11, 1084–1091 [DOI] [PubMed] [Google Scholar]
- 40. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 41. Matsuoka T., Wada J., Hashimoto I., Zhang Y., Eguchi J., Ogawa N., Shikata K., Kanwar Y. S., and Makino H. (2005) Gene delivery of Tim44 reduces mitochondrial superoxide production and ameliorates neointimal proliferation of injured carotid artery in diabetic rats. Diabetes 54, 2882–2890 [DOI] [PubMed] [Google Scholar]
- 42. Biedler J. L., Roffler-Tarlov S., Schachner M., and Freedman L. S. (1978) Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 38, 3751–3757 [PubMed] [Google Scholar]
- 43. Davey K. M., Parboosingh J. S., McLeod D. R., Chan A., Casey R., Ferreira P., Snyder F. F., Bridge P. J., and Bernier F. P. (2006) Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J. Med. Genet. 43, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vogel B., Meder B., Just S., Laufer C., Berger I., Weber S., Katus H. A., and Rottbauer W. (2009) In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem. Biophys. Res. Commun. 390, 516–522 [DOI] [PubMed] [Google Scholar]
- 45. Haas J., Frese K. S., Park Y. J., Keller A., Vogel B., Lindroth A. M., Weichenhan D., Franke J., Fischer S., Bauer A., Marquart S., Sedaghat-Hamedani F., Kayvanpour E., Köhler D., Wolf N. M., et al. (2013) Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 5, 413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang J. H., Chung T. D., and Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 47. Banerjee R., Gladkova C., Mapa K., Witte G., and Mokranjac D. (2015) Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein. Elife 4, e11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hutu D. P., Guiard B., Chacinska A., Becker D., Pfanner N., Rehling P., and van der Laan M. (2008) Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol. Biol. Cell 19, 2642–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Popov-Čeleketić D., Waegemann K., Mapa K., Neupert W., and Mokranjac D. (2011) Role of the import motor in insertion of transmembrane segments by the mitochondrial TIM23 complex. EMBO Rep. 12, 542–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chacinska A., Lind M., Frazier A. E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H. E., Truscott K. N., Guiard B., Pfanner N., and Rehling P. (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829 [DOI] [PubMed] [Google Scholar]
- 51. Chacinska A., van der Laan M., Mehnert C. S., Guiard B., Mick D. U., Hutu D. P., Truscott K. N., Wiedemann N., Meisinger C., Pfanner N., and Rehling P. (2010) Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell Biol. 30, 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., Yamamoto H., Sesaki H., and Endo T. (2009) Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. DePaul A. J., Thompson E. J., Patel S. S., Haldeman K., and Sorin E. J. (2010) Equilibrium conformational dynamics in an RNA tetraloop from massively parallel molecular dynamics. Nucleic Acids Res. 38, 4856–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loncharich R. J., Brooks B. R., and Pastor R. W. (1992) Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 32, 523–535 [DOI] [PubMed] [Google Scholar]
- 56. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.