Abstract
The type III secretion system is a highly conserved virulence mechanism that is widely distributed in Gram-negative bacteria. It has a syringe-like structure composed of a multi-ring basal body that spans the bacterial envelope and a projecting needle that delivers virulence effectors into host cells. Here, we showed that the Yersinia inner rod protein YscI directly interacts with the needle protein YscF inside the bacterial cells and that this interaction depends on amino acid residues 83–102 in the carboxyl terminus of YscI. Alanine substitution of Trp-85 or Ser-86 abrogated the binding of YscI to YscF as well as needle assembly and the secretion of effectors (Yops) and the needle tip protein LcrV. However, yscI null mutants that were trans-complemented with YscI mutants that bind YscF still assembled the needle and secreted Yops, demonstrating that a direct interaction between YscF and YscI is critical for these processes. Consistently, YscI mutants that did not bind YscF resulted in greatly decreased HeLa cell cytotoxicity. Together, these results show that YscI participates in needle assembly by directly interacting with YscF.
Keywords: bacterial pathogenesis, protein assembly, protein-protein interaction, type III secretion system (T3SS), Western blotting, Yersinia pestis, inner rod protein, needle assembly, needle protein
Introduction
Bacteria have evolved various secretion mechanisms to interact with their external environment to benefit their survival. Type III secretion systems (T3SSs)3 are widely distributed in Gram-negative bacteria, and they are highly conserved among plant and mammalian pathogens as well as some symbiotic bacteria (1). Human pathogens, such as Salmonella enterica, Shigella flexneri, pathogenic Escherichia coli, and the opportunistic pathogen Pseudomonas aeruginosa, use T3SS machines to deliver various virulence effectors into the host cell cytosol to alter the host cell physiology and cause infectious diseases. The sequences and biological activities of T3SS effectors are diversified, but the T3SS secretion apparatus called injectisome is highly conserved, and it is thought to be evolutionarily related to the bacterial flagellum (2–4).
Three human pathogenic Yersinia species, Yersinia pestis, Yersinia enterocolitica, and Yersinia pseudotuberculosis, share a common T3SS encoded in a 70-kb plasmid (called pCD1 or pYV) that is indispensable for pathogenesis (5). Electron microscopic analyses of injectisome purified from S. enterica serovar Typhimurium or injectisomes that are embedded in bacterial membranes of Y. enterocolitica or S. flexneri revealed a syringe-like structure protruding from the bacterial surface (1, 6). The Yersinia T3SS is composed of >20 proteins, and it is responsible for the delivery of virulence effectors (called Yops) into the host cell cytosol, where they hijack critical cellular signaling pathways (1, 7–9). The structure of the Yersinia T3SS apparatus includes an YscC secretin ring embedded in the outer membrane (10), an MS (membrane-spanning) ring composed of YscD and YscJ in the inner membrane (11, 12), and a needle that is ∼60 nm long that protrudes from the bacterial envelope (13). The export apparatus within the MS ring contains five transmembrane proteins: YscR, -S, -T, -U, and -V (14). The YscN ATPase, together with two ancillary proteins, YscK and YscL, provides the energy for substrate translocation (15). The multimeric protein YscQ forms the cytosolic part of the basal body that resembles the flagellar cytoplasmic (C)-ring (16). Diepold et al. (16) used a fluorescent fusion protein to monitor the dynamic process of Yersinia T3SS assembly. They demonstrated that the assembly process starts with the formation of the YscC secretin in the outer membrane and progresses inward to the MS ring in the inner membrane and then to the cytosolic components, ATPase, and C-ring.
After the secretion apparatus is assembled, the needle subunit protein YscF is exported by the nascent injectisome, and it polymerizes into a hollow needle via a poorly characterized process. The T3SS needle has a helical structure, and the needle of Yersinia T3SS consists of a few hundred copies of YscF (17–19). Before needle assembly, YscF binds two chaperone proteins, YscG and YscE, to avoid premature polymerization in the bacterial cytosol (20). The molecular ruler protein YscP, which switches the substrate specificity of the T3SS after the needle reaches its proper length, is responsible for controlling the length of the injectisome needle (13, 21, 22). A yscP mutant produces needles with undefined lengths that are unable to secrete Yop effectors (13), and YscU and YscP work coordinately to regulate the substrate specificity of the Yersinia T3SS by controlling the secretion of the inner rod protein YscI (23, 24). YopR is required for YscF secretion, but it does not directly participate in YscF polymerization (25). The inner rod protein YscI and its homologues, such as PscI in Pseudomonas, MxiI in Shigella, and PrgJ in Salmonella, constitute an inner rod protein family (26). The inner rod protein forms a rodlike structure that connects the needle and the center of the basal body (27). The PrgJ inner rod protein is required for injectisome assembly in S. enterica serovar Typhimurium, and a prgJ mutant can only produce the basal body structure without the extended needle (12). Zilkenat et al. (28) showed that not more than six PrgJ form the inner rod of the needle complex Salmonella. In Shigella and Salmonella, the assembly of the MxiI/PrgJ inner rod controls substrate switching and needle length (27, 29, 30). PscI in Pseudomonas aeruginosa anchors the PscF needle within the secretion apparatus but is not involved in substrate specificity switching (26). Taken together, previous studies have shown that the inner rod protein is required for needle assembly in different bacteria; however, the underlying mechanism is poorly understood. In this study, we demonstrate that the Yersinia inner rod protein YscI directly interacts with YscF and that this interaction depends on the conserved C-terminal domain of YscI. Point mutants of YscI amino acid residues Trp-85 and Ile-86 completely disrupt the binding of YscI to YscF, leading to defective YscF polymerization and substrate export as well as greatly decreased cytotoxicity to HeLa cells.
Results
Amino Acids 83–92 at the C Terminus of YscI Are Essential for YscF Binding
In a yeast two-hybrid screening that aimed to identify interactions among potential components of the T3SS, we found that YscI interacted with YscF. A GST pull-down assay using GST-tagged YscI truncants found that the C-terminal domain of YscI, which is highly conserved among different inner rod protein family members, was essential for binding YscF, whereas GST alone or the N-terminal domain of YscI could not bind to YscF (31). In the present study, we first wanted to clarify which regions of the YscI C terminus are essential for binding YscF. Four plasmids expressing various glutathione S-transferase (GST)-tagged YscI mutants with deletions of amino acids 73–82, 83–92, 93–102, or 103–115 were constructed, and the truncated YscI proteins were expressed in E. coli. Purified, GST-tagged YscI and its various mutants were immobilized onto Sepharose 4B beads and incubated overnight with purified, His-tagged YscF. The beads were washed intensively to remove unbound proteins, and bound proteins were detected by immunoblotting. Deletion of amino acid residues 83–92 or 93–102 almost abrogated the binding of YscI to YscF, whereas deletions of residues 73–82 or 103–115 dramatically decreased but did not eliminate binding (Fig. 1). These results demonstrate that amino acid residues 83–102 and 93–102 in the C-terminal domain of YscI are critical for the in vitro interaction between YscF and YscI, whereas the other tested regions had limited effects on binding.
FIGURE 1.
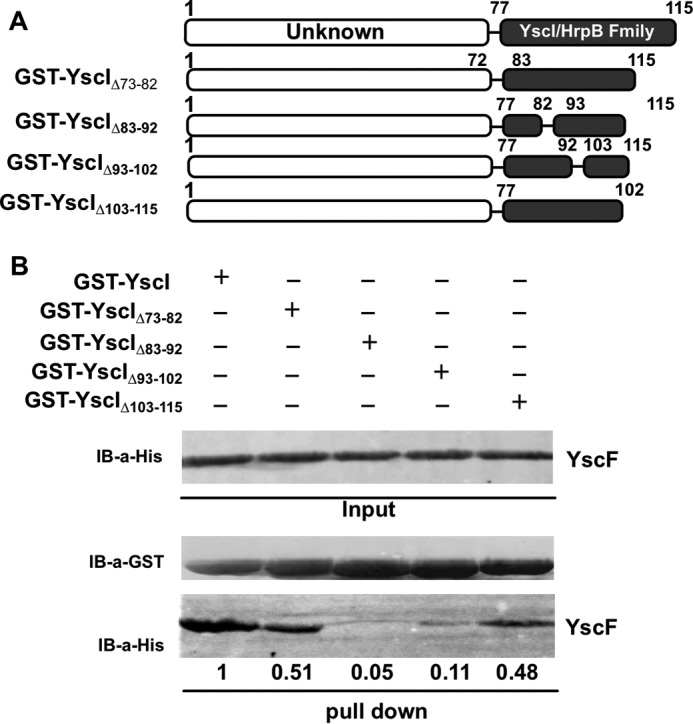
C-terminal amino acids residues 83–92 and 93–103 of YscI are essential for YscF binding. A, schematic diagrams of various GST-tagged YscI truncants. B, a GST pull-down assay was used to identify the amino acids that are critical for the binding of YscI to YscF. GST-tagged YscI proteins were bound to glutathione-Sepharose 4B beads, and the purified His-tagged YscF protein was added, followed by incubation at 4 °C overnight. The beads were intensively washed, 2× SDS loading buffer was added, and the proteins bound to the beads were analyzed by SDS-PAGE and immunoblotting. At least triplicate experiments were performed, and similar results were obtained; a representative result is shown here. Intensity of the YscF bands was quantitated by using ImageJ software.
YscI Interacts with YscF inside Bacterial Cells
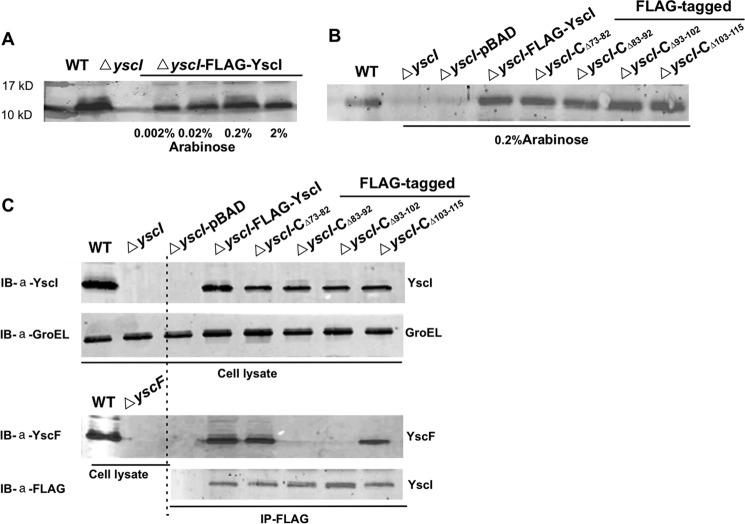
To determine whether YscI can bind YscF inside bacterial cells, a co-immunoprecipitation (co-IP) assay was performed using Y. pestis strains expressing different versions of YscI mutants that were fused to a FLAG tag. We first constructed a yscI null mutant, named ΔyscI, of Y. pestis strain 201. A series of plasmids, in which expression of the YscI deletion mutants was driven by the arabinose-inducible promoter in the pBAD plasmid, was constructed and electrotransformed into the ΔyscI strain to generate strains expressing YscIΔ73–82, YscIΔ83–92, YscIΔ93–102, or YscIΔ103–115 (Table 1). Different concentrations of arabinose were tested for their ability to induce YscI expression, and the level of FLAG-YscI that was induced by 0.2% arabinose was similar to that of the wild-type strain (Fig. 2A); therefore, 0.2% arabinose was used in the following experiments. Immunoblotting detection using an anti-YscI antibody confirmed that the yscI mutant strains that were trans-complemented with the pBAD-YscI plasmids could successfully express native YscI and the various YscI mutants (Fig. 2B). For the co-IP assay, the bacterial strains were grown to the exponential phase in TMH medium at 26 °C and then transferred to 37 °C to fully induce the expression of T3SS. The bacterial cells were lysed, and aliquots of an anti-FLAG M2 affinity gel were added to capture FLAG-tagged YscI and the YscI mutants; the presence of YscF in the immunoprecipitates was detected using a rabbit anti-YscF antibody. We found that YscF was successfully co-immunoprecipitated by FLAG-YscI as well as by the YscIΔ73–82 and YscIΔ103–115 mutants (Fig. 2C). However, no YscF band was detected when using Y. pestis strains expressing the YscIΔ83–92 or YscIΔ93–102 proteins to perform the co-IP assay (Fig. 2C). These results indicate that YscI can bind to YscF and that amino acid residues 83–92 and 93–103 of YscI are essential for binding YscF inside bacteria under physiologically relevant conditions. However, we cannot exclude the possibility that a non-identified third party inside the bacterial cell is involved in this interaction. Although YscIΔ73–82 and YscIΔ103–115 exhibited decreased binding to YscF in the GST pull-down assay, they bound almost as well as wild-type YscI inside bacteria.
TABLE 1.
Strains and plasmids used in this study
| Strains or plasmids | Descriptions | Sources |
|---|---|---|
| Plasmids | ||
| pKD46 | Temperature-sensitive plasmid expressing λRed recombinase under the control of arabinose; Apr | Ref. 36 |
| pGEX-4T-2 | Expression vectors GST fusion protein | Amersham Biosciences |
| pGEX-4T-YscI | yscI gene was inserted into pGEX-4T-2; Apr | This study |
| pGEX-4T-YscIΔ73–82 | yscI gene lacking amino acid 73–82 coding sequence was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscIΔ83–92 | yscI gene lacking amino acid 83–92 coding sequence was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscIΔ93–102 | yscI gene lacking amino acid 93–102 coding sequence was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscIΔ103–115 | yscI gene lacking amino acid 103–115 coding sequence was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscIW85A | yscI gene with W85 to A mutation was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscIS86A | yscI gene with Ser-86 to Ala mutation was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscII88A | yscI gene with Ile-88 to Ala mutation was inserted into pGEX-4T-2 | This study |
| pGEX-4T-YscII92A | yscI gene with Ile-92 to Ala mutation was inserted into pGEX-4T-2 | This study |
| pET28a-YscF | yscF gene was inserted into pET28a; Kmr | This study |
| pET28a-YscI | yscI gene was inserted into pET28a; Kmr | This study |
| pBAD24 | Plasmids for expression of the cloned gene under the control of arabinose; Apr | Laboratory collection |
| pBAD-YscI | yscI gene fused with a FLAG coding sequence at the C terminus was inserted into NheI-HindIII sites of pBAD24 | This study |
| pBAD-YscIΔ73–82 | yscI gene lacking 73–82 amino acids coding sequence was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscIΔ83–92 | yscI gene lacking amino acid 83–92 coding sequence was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscIΔ93–102 | yscI gene lacking amino acid 93–102 coding sequence was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscIΔ103–115 | yscI gene lacking amino acid 103–115 coding sequence was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscIW85A | yscI gene with Trp-85 to Ala mutation was inserted into NheI-HindIII of pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscIS86A | yscI gene with Ser-86 to Ala mutation was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscII88A | yscI gene with Ile-88 to Ala mutation was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| pBAD-YscII92A | yscI gene with Ile-92 to Ala mutation was inserted into pBAD24, fused with a FLAG tag at the C terminus | This study |
| Strains | ||
| Y. pestis | ||
| 201 strain | Wild-type Y. pestis strain | Ref. 38 |
| ΔyscI | yscI gene was replaced by Kanamycin cassette, Kmr | This study |
| ΔyscF | yscF gene was replaced by Kanamycin cassette, Kmr | This study |
| ΔyscI-CΔ73–82 | ΔyscI containing plasmid pBAD-YscIΔ73–82 | This study |
| ΔyscI-CΔ83–92 | ΔyscI containing plasmid pBAD-YscIΔ83–92 | This study |
| ΔyscI-CΔ93–102 | ΔyscI containing plasmid pBAD-YscIΔ93–102 | This study |
| ΔyscI-CΔ103–115 | ΔyscI containing plasmid pBAD-YscIΔ103–115 | This study |
| ΔyscI-CW85A | ΔyscI containing plasmid pBADW85A | This study |
| ΔyscI-CS86A | ΔyscI containing plasmid pBADS86A | This study |
| ΔyscI-CI88A | ΔyscI containing plasmid pBADI88A | This study |
| ΔyscI-CI92A | ΔyscI containing plasmid pBADI92A | This study |
| ΔyscI-pBAD | ΔyscI containing plasmid pBAD24 | This study |
| ΔyscI-FLAG-YscI | ΔyscI containing plasmid pBAD-YscI | This study |
FIGURE 2.
YscF can be immunoprecipitated by YscI inside bacterial cells. A, expression levels of FLAG-tagged YscI in the presence of 0.002–2% arabinose were detected by immunoblotting using an anti-YscI antibody. B, successful construction of the ΔyscI strain and various mutants expressing different YscI deletion mutants was confirmed by immunoblotting using an anti-YscI antibody. C, co-IP of YscF by different YscI mutants. Strains were cultured in TMH without calcium at 37 °C, and arabinose was added to induce the expression of the FLAG-tagged YscI mutants. Bacterial cells were collected and lysed, and anti-FLAG M2 affinity gel was added to immunoprecipitate FLAG-tagged YscI and the various YscI mutants. Wild-type YscI was co-immunoprecipitated with the endogenous YscF protein, whereas the YscI mutants with deletions of amino acid residues 83–92 or 93–102 no longer bound YscF. Immunoblotting (IB) detection of GroEL showed that equal amounts of the bacterial lysates from the different strains were used in the co-IP assay. Triplicate experiments were performed, and similar results were obtained; a representative result is shown here. Images of the immunoblotting results were taken by an Odyssey SA imaging system.
YscI Mutations That Disrupt YscF Binding Abrogate Needle Assembly
The needle structure of the Yersinia T3SS is a long, hollow tube that is made up of a single polymerized protein, YscF. In the presence of the cross-linker bis-sulfosuccinimidyl suberate (BS3), bacterial surface-exposed YscF can be cross-linked, and it forms ladders of multimers after SDS-PAGE separation (24). After BS3 cross-linking, samples of bacterial cells were separated by SDS-PAGE, and the polymerization of YscF was detected by immunoblotting using a rabbit anti-YscF antibody. To observe the influence of YscI mutations on needle assembly, we used BS3 treatment to detect the polymerization of YscF in ΔyscI strains that were trans-complemented with plasmids expressing different YscI truncated mutants. It should be noted that the Y. pestis strains used in this study contain the pPCP1 plasmid that encodes the Pla protease, and because surface-exported YscF can be degraded by Pla (32), we did not observe bands of YscF monomers or multimers with the expected molecular weights. Nevertheless, we unambiguously observed a ladder of bands of polymerized YscF in the wild-type Y. pestis strain (Fig. 3A). YscF polymerization was basically normal in the ΔyscI-CΔ73–82 and ΔyscI-CΔ103–115 strains. In contrast, no ladders of polymerized YscF could be observed in BS3-treated ΔyscI-CΔ83–92 and ΔyscI-CΔ93–102 strains (Fig. 3A). These results imply that the interaction between YscI and YscF might be important for the assembly of the YscF needle, because the ΔyscI-CΔ83–92 and ΔyscI-CΔ93–102 strains, which did not co-immunoprecipitate YscF (Fig. 3), did not produce polymerized YscF (Fig. 3A).
FIGURE 3.
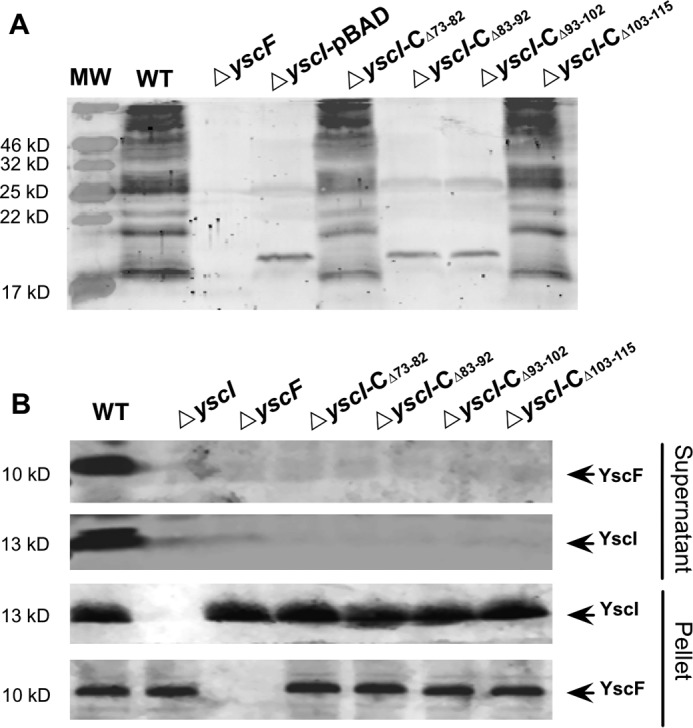
Deletion of amino acid residues 83–92 and 93–102 of YscI leads to defects in YscF polymerization. A, detection of polymerized YscF in Y. pestis strains containing various yscI mutations. Bacterial strains were cultured under T3SS-inducing conditions, and the bacterial cells were treated with the cross-linker BS3. YscF multimers that were produced in different strains were separated by SDS-PAGE, followed by immunoblotting detection using an anti-YscF antibody. The ΔyscI-FLAG-YscI strain has not been included in this experiment; see Fig. 5 for the result of this strain. B, YscF and YscI that were secreted into the culture medium of different Y. pestis strains were detected by immunoblotting using anti-YscF and anti-YscI antibodies. At least triplicate experiments were performed, and similar results were obtained; a representative result is shown here.
We hypothesized that the defect in needle assembly of the ΔyscI-CΔ83–92 and ΔyscI-CΔ93–102 strains were caused by the failure of YscF secretion. To test this hypothesis, proteins that were present in the culture medium of Y. pestis strains harboring various YscI mutations were analyzed by immunoblotting. YscI and YscF are the early substrates of the Yersinia T3SS (23, 33). As expected, large amounts of YscI and YscF were detected in the culture medium of the wild-type strain (Fig. 3B). In contrast, deleting any region of YscI, not only amino acids 83–92 and 93–102, but also residues 73–82 and 103–115, prevented YscF secretion (Fig. 3B). Surprisingly, although the ΔyscI-CΔ73–82 and ΔyscI-CΔ103–115 strains seemed to assemble the YscF needle normally, no secreted YscF was detected in their culture supernatants. These results imply that needle assembly requires the participation of residues 83–102 but not 73–82 and 103–115 of YscI; however, export of YscF requires an intact YscI C-terminal domain that is highly conserved in the rod protein family to regulate the substrate specificity of the Yersinia T3SS (22, 23). YscI secretion is totally blocked in the ΔyscF strain, and YscF secretion is also blocked in the ΔyscI strain, which is consistent with previous results showing that these proteins are essential for secretion by the T3SS (24, 32). Together, these results suggest that YscI might assist YscF polymerization and that deleting any region of the C terminus of YscI completely blocks YscF secretion.
Secretion of T3SS Substrates Is Impaired When YscI Cannot Bind YscF
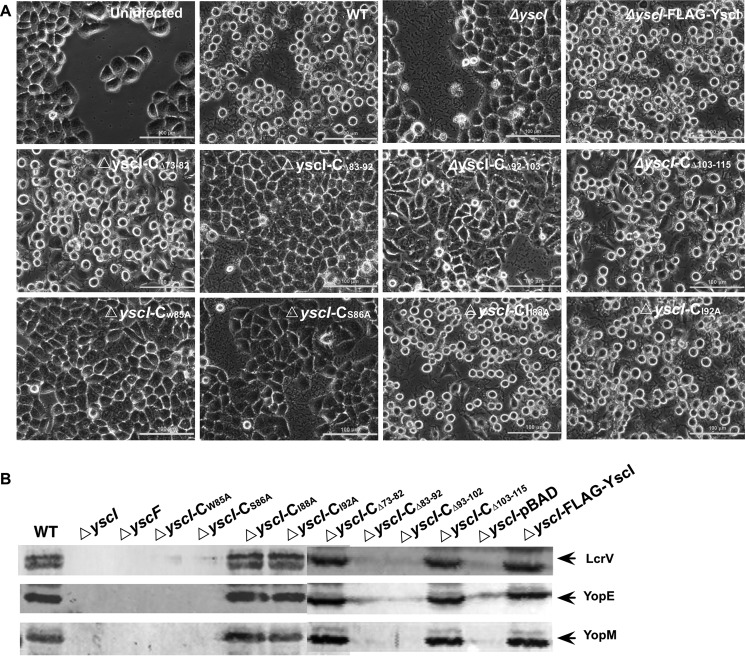
Knowing that YscF needle assembly requires binding between YscI and YscF, we wondered how substrate secretion would be affected if the binding of YscI to YscF was disrupted. We detected the secretion of several T3SS substrates, including the needle tip protein LcrV that binds to the YopB/D translocon at the tip of the needle, the gatekeeper protein YopN that plugs the secretion channel under T3SS secretion non-permissive conditions, and the effectors YopE, YopK, and YopM, in the culture supernatants of yscI null mutants that were trans-complemented with plasmids expressing different YscI mutants. As expected, when grown under the T3SS secretion permissive condition, the yscI null mutant that cannot form the inner rod structure failed to export any of the substrates (Fig. 4). The secretion of YopE, YopK, YopM, and LcrV was only marginally affected or was unaffected in the ΔyscI-CΔ73–82 and ΔyscI-CΔ103–115 strains, but it was completely abolished in the ΔyscI-CΔ83–92 strain. The secretion of tip protein LcrV was dramatically decreased in the ΔyscI-CΔ93–102 strain; however, it was almost unaffected for the secretion of the effectors YopE, YopM, and YopK. Repeated experiments showed that secretion of the gatekeeper protein YopN was greatly decreased in the ΔyscI-CΔ83–92 strain, but not in the other mutants. These results indicate that the impact of YscI deletion mutations on YopN secretion is limited even when Yop secretion is blocked (Fig. 4), suggesting that YopN has a higher secretion priority than Yops, which is consistent with previous findings (34). Surprisingly, although both the amino acids 83–92 and 93–102 are essential for the YscF binding and the needle assembly, Yop secretion is only abolished in the ΔyscI-CΔ83–92 strain and not in the ΔyscI-CΔ93–102 strain, suggesting that Yops can be secreted even when there are no polymerized needles. Notably, the ability of ΔyscI-CΔ93–102 to secret Yops indicates that YscIΔ93–102 probably forms an assembled inner rod structure and is able to switch substrate, whereas YscIΔ83–92 could not assemble rod structure or is unable to switch substrate. We speculate that the inner rod structure made of YscIΔ93–102 is in a conformation impairing a proper anchoring of YscF, due to the inability of YscIΔ93–102 to bind YscF.
FIGURE 4.
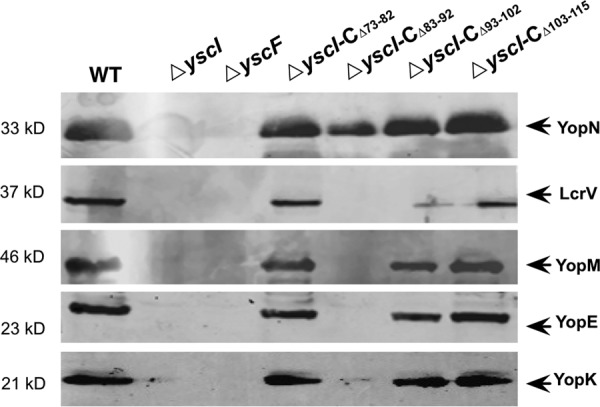
Deletion of amino acids residues 83–92 or 93–102 of YscI affects substrate secretion by the T3SS. Y. pestis strains were cultured under T3SS-inducing conditions, and the secreted proteins in the culture supernatants were separated by SDS-PAGE, followed by immunoblotting detection using anti-YopN, anti-LcrV, anti-YopE, anti-YopM, and anti-YopK antibodies. Strains ΔyscI-CΔ73–82 and ΔyscI-CΔ103–115 secreted substrates as well as the wild-type strain. However, deletion of amino acids 83–92 completely abrogated the secretion of Yops and LcrV, and deleting amino acids 93–102 significantly decreased LcrV secretion and slightly affected the secretion of YopM, YopE, and YopK. At least triplicate experiments were performed, and similar results were obtained; a representative result is shown here.
Single Amino Acid Mutations in YscI Disrupt YscF Binding
The above results showed that deleting the amino acids 83–102 of YscI abolishes YscF binding and needle assembly and impairs or blocks Yop secretion as well. However, deleting tens of amino acids might have unexpected effects on the secretion apparatus, other than influencing the binding of YscI to YscF. Thus, we decided to identify the amino acid residues in YscI that are critical for YscF binding without affecting the structure of the secretion apparatus. A previous study reported that the inner protein and needle protein are homologous to each other (24). A sequence alignment of YscI and YscF showed that they shared high amino acid identity at residues 85–92, and amino acid residues 85, 86, 88, and 92 of YscI are identical to those of YscF (Fig. 5A). Given that mutant strains expressing YscI lacking residues 83–92 are defective for YscF polymerization and Yop secretion, we wanted to determine whether these residues are important for YscF binding. First, we cloned various sequences encoding YscI W85A, S86A, I88A, or I92A mutants with a FLAG-coding sequence fused at the C terminus into the pBAD24 vector; thus, the expression of FLAG-tagged YscI mutants was under the control of the arabinose-inducible promoter. Then the plasmids were introduced into the ΔyscI mutant, and the expression of the FLAG-tagged YscI mutants was confirmed by immunoblotting (Fig. 5B). The binding of the YscI mutants to YscF was analyzed by co-IP. Bacterial strains were grown in TMH medium without calcium at 37 °C, and expression of the FLAG-tagged YscI mutants was induced by the addition of 0.2% arabinose. After a 4-h incubation, bacterial cells were lysed and subjected to a co-IP analysis. GroEL was detected by immunoblotting to show that equal amounts of the bacterial lysates were used in co-IP experiments. As shown in Fig. 5C, a strong band corresponding to the YscF protein demonstrated that it was successfully co-immunoprecipitated by FLAG-tagged YscI. However, the ΔyscI-CW85A and ΔyscI-CS86A mutants did not co-immunoprecipitate YscF (Fig. 5C), whereas the ΔyscI-CI88A and ΔyscI-CI92A mutants did not exhibit any obvious defects in binding.
FIGURE 5.
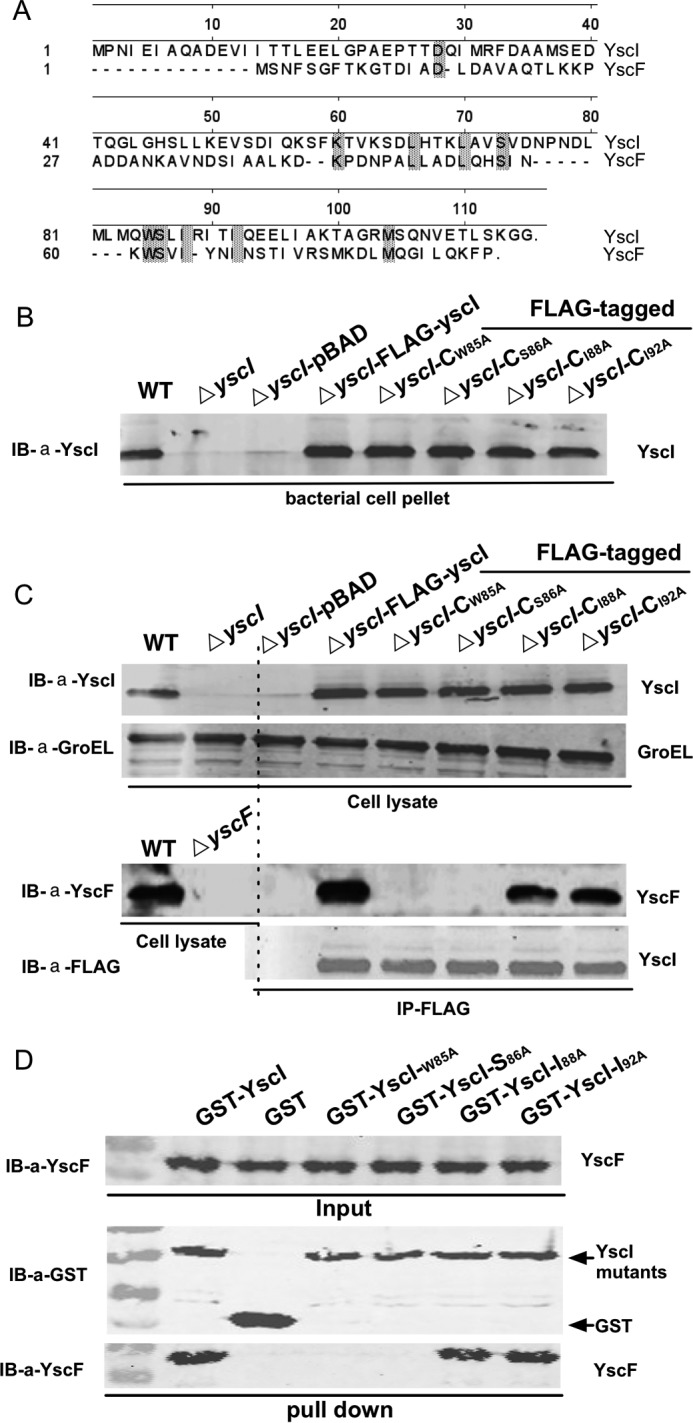
Single amino acid mutations of YscI abolish YscF binding. A, sequence alignment analysis of YscI and YscF. B, expression of various YscI point mutants in bacterial strains was analyzed by immunoblotting (IB) detection. C, analysis of binding of various YscI point mutants to YscF inside bacterial cells by co-IP. Y. pestis strains were cultured in TMH without calcium at 37 °C, and arabinose was added to induce the expression of FLAG-tagged YscI mutants. Co-IP experiments were performed as described in Fig. 2 except using the strains expressing different YscI point mutants as indicated. Mutations of Trp-85 or Ser-86 abrogated YscF binding. Immunoblotting detection of GroEL showed that equal amounts of the bacterial lysates from the different strains were used in the co-IP assay. D, a GST pull-down assay was used for analyzing the binding of YscI point mutants to YscF. Experiments were performed as described in the legend to Fig. 1. Triplicate experiments were performed, and a representative result is shown here.
Positive co-IP results cannot exclude the possible presence of a third party in bacterial cells to mediate the indirect binding between YscI and YscF. Thus, we sought to determine the YscF binding affinity of purified YscI point mutants in an in vitro assay. GST-tagged YscI point mutants were expressed in E. coli and purified by using Sepharose 4B beads, and GST pull-down assays were performed as described above. GST-YscI successfully pulled down His-tagged YscF (Fig. 5D), and I88A or I92A YscI mutants exhibit no defect in YscF binding. In contrast, mutation of 85 or 86 amino acids of YscI resulted in the disruption of YscF binding (Fig. 5D). Taken together, these results indicate that YscI can bind directly to YscF and that mutation of Trp-85 or Ser-86 of YscI totally abolishes YscF binding.
The W85A and S86A YscI Mutants Impair Needle Assembly and Yop Secretion
To investigate whether the YscI mutations that disrupt YscF binding also affect needle assembly, YscF polymerization was analyzed by a cross-linking assay using Y. pestis strains expressing various YscI mutants. As expected, the yscI mutant that was trans-complemented with a plasmid expressing wild-type YscI assembled needles as well as the wild-type strain; however, the ΔyscI-CW85A and ΔyscI-CS86A strains no longer polymerized YscF (Fig. 6A). Normal YscF polymerization was found in the ΔyscI-CI88A and ΔyscI-CI92A strain. These results clearly demonstrate that mutation of Trp-85 or Ser-86 not only disrupts the binding of YscI to YscF but also results in the failure of needle assembly. This suggests that binding to YscI is required for YscF polymerization and/or the anchoring of the polymerized needle and supports our assumption that YscI is involved in needle assembly by directly interacting with YscF.
FIGURE 6.
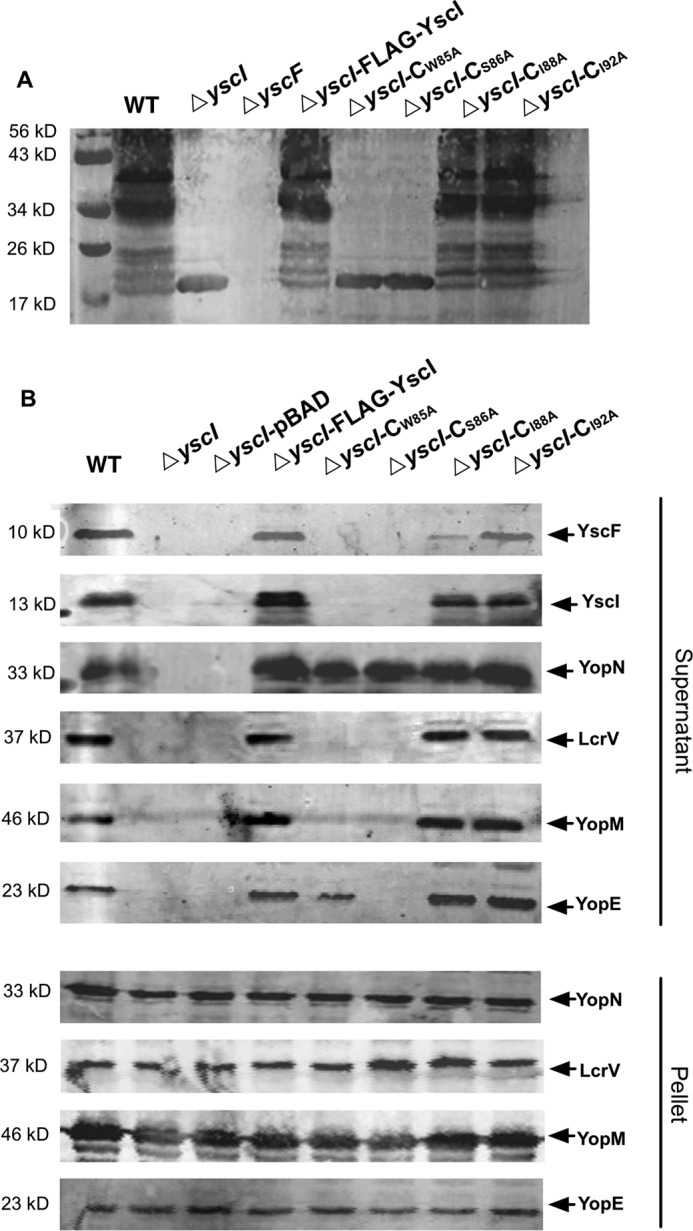
Mutation of YscI at Ser-86 or Trp-85 affects needle assembly and substrate secretion. Detection of polymerized YscF in various YscI point mutants of Y. pestis (A). Bacterial strains were cultured under T3SS-inducing conditions, and the bacterial cells were treated with the cross-linker BS3. Y. pestis strains expressing YscIS86A and YscIW85A cannot produce polymerized YscF, whereas those expressing YscII88A and YscII92A can still polymerize YscF, although the mutation of Ile-88 led to a slight decrease in the multimer bands. B, T3SS substrate secretion analysis of Y. pestis strains expressing single point mutants of YscI. Strains were grown under T3SS-inducing conditions, and the secreted proteins in the culture medium and the cell pellets were detected by immunoblotting using specific polyclonal antibodies. Alanine substitutions of Ser-86 or Trp-85 of YscI completely abolished the secretion of Yops and LcrV, whereas mutations of Ile-88 or Ile-92 did not have obvious effects, except that YscF secretion was significantly decreased in the ΔyscI-CI88A strain. Triplicate experiments were performed, and a representative result is shown here.
Next, we analyzed substrate secretion by Y. pestis strains expressing YscI single amino acid mutants. The yscI mutant that was trans-complemented with pBAD-YscI exhibited wild-type levels of Yops, LcrV, YscF, and YscI secretion; however, secretion was completely abrogated in the ΔyscI-CW85A and ΔyscI-CS86A strains, except that the ΔyscI-CW85A strain still secreted a small amount of YopE (Fig. 6B). Immunoblotting analysis of Yops and LcrV in the bacterial cell pellets demonstrated that all of the tested strains exhibited wild-type level expression of those proteins. Substrate secretion is almost blocked in ΔyscI-CW85A and ΔyscI-CS86A strains, except that they still secret YopN, suggesting that Trp-85 or Ser-86 mutations might result in an inner rod structure with conformation losing specificity for the secretion substrates. The small amount of secretion of YopE in ΔyscI-CW85A and the decreased secretion of YscF in ΔyscI-CI88A mutant imply that YscI might play critical roles in secretion substrate specificity and hierarchy, and the single mutations of the key region can lead to the deregulated function of the inner rod. In contrast, mutations of residues Ile-88 and Ile-92 basically did not affect substrate secretion, which is consistent with the above results showing that YscII88A and YscII92A bind YscF and that strains expressing YscII88A or YscII92A can assemble the YscF needle.
YscI Mutants That Cannot Bind YscF Exhibit Reduced Cytotoxicity to HeLa Cells
The most important function of the T3SS is to attack host biological processes by delivering virulence effectors into the host cell cytosol. Because the W85A and S86A mutations abrogated needle assembly and impaired substrate secretion, they should influence the translocation of Yops into host cells as well, thereby reducing the cytotoxicity to infected cells. To test this hypothesis, HeLa cells were infected with Y. pestis strains expressing various YscI mutants, and images of the infected HeLa cells were taken after 2 h of infection. As expected, HeLa cells that were infected with the wild-type strain rounded up significantly, which is consistent with previous reports showing that YscI is essential for the virulence of Y. pestis (30, 32). The ΔyscI-CI88A and ΔyscI-CI92A strains caused cytotoxic effects to HeLa cells similar to the wild-type strain, whereas the morphologies of cells that were infected with the ΔyscI strain were similar to that of the mock-infected control (Fig. 7A) The ΔyscI-CΔ83–92, ΔyscI-CΔ93–102, ΔyscI-CW85A, and ΔyscI-CS86A mutants exhibited greatly reduced cytotoxicity to HeLa cells, which is in line with the above results showing that YscIΔ82–93, YscI-CΔ93–102, YscIS86A, and YscIW85A do not bind YscF and that those strains do not form the polymerized YscF needle either. Although ΔyscI-CΔ93–102 is able to secrete LcrV and several tested Yops, it cannot form the polymerized YscF needle (Fig. 6). In line with those results, a HeLa cell cytotoxicity assay indicated that ΔyscI-CΔ93–102 failed to induce cell cytotoxicity, suggesting that it cannot deliver Yop effectors into the host cells.
FIGURE 7.
Disruption of binding between YscI and YscF reduces cytotoxicity to HeLa cells. HeLa cells were infected with different Y. pestis strains (MOI = 10) as indicated (A). After infection for 2 h, phase-contrast images of the infected cells were taken; scale bars, 100 μm. B, translocation assays of Yersinia T3SS substrates. HeLa cells were infected with Y. pestis strains and then lysed in 0.1% Triton X-100. The obtained cell lysates were centrifuged to separate the supernatants and precipitates containing insoluble debris and the bacterial cells. Proteins in the supernatants were separated by SDS-PAGE and then detected by immunoblotting analysis. At least triplicate experiments were performed, and similar results were obtained; a representative result is shown here.
HeLa cells were infected with Y. pestis strains expressing various YscI mutants. After 2 h of infection, cells were washed in PBS and lysed in 0.1% Triton X-100, and the cell lysates were centrifuged to separate the soluble fractions and precipitates containing insoluble cell debris and the bacterial cells. Proteins from the soluble fractions of the infected HeLa cells were analyzed for the presence of YopE, YopM, and LcrV. The wild-type strain and the yscI mutant that was trans-complemented with pBAD-YscI could successfully translocate YopE, YopM, and LcrV into the cytoplasm of HeLa cells (Fig. 7B); however, translocation was completely blocked in the ΔyscI-CW85A and ΔyscI-CS86A strains. ΔyscI-CΔ83–92 and ΔyscI-CΔ93–102 strains that are unable to assemble the YscF needle and exhibit reduced cytotoxicity to HeLa cells translocate neither YopE, YopM, nor LcrV into HeLa cells whereas ΔyscI-CΔ73–92, ΔyscI-CΔ103–115, ΔyscI-CI88A, and ΔyscI-CI92A strains can translocate those substrates as well as the wild-type strain.
Discussion
In this study, we demonstrated that YscI directly interacts with YscF within the bacterial cells and that the region spanning amino acids 82–103 in the conserved YscI C terminus is critical for this interaction. Trans-complementation of the yscI null strain with plasmids expressing YscI point mutants, as well as a co-IP analysis, showed that YscIW85A or YscIS86A could not bind YscF and that the ΔyscI-CW85A and ΔyscI-CS86A strains were defective in YscF needle assembly and LcrV and Yop secretion. Notably, these two strains did not secrete YscI and YscF, although they did secrete the gatekeeper protein YopN (Fig. 6). Similarly, the ΔyscI-CΔ83–92 strain was defective for needle assembly and LcrV and Yop secretion, but it still secreted YopN, albeit at a decreased level. This implies that the inner rod of the T3SS injectisome could be assembled to some extent, although the assembled inner rod is somehow distorted, in the different YscI mutants (except for the yscI null mutant) and that the injectisome secretion channel was still competent for YopN secretion. An additional possibility is that YopN secretion is independent of the inner rod; however, this conjecture can be easily excluded because the ΔyscI mutant did secret YopN (Fig. 4). It could be possible that the C-terminal region of YscI is required for coordinating protein secretion with needle assembly, besides influencing YscF polymerization, which can be supported by the previous report (29). In that report, Lefebre et al. (29) found that alanine substitutions of PrgJ at the C-terminal region resulted in the deregulated needle length and an altered hierarchy in protein secretion.
It was surprising that YscI mutants with deletions of amino acids 73–82 or 103–115 did not secrete YscF, yet still produced the polymerized YscF needle. It is likely that YscF was polymerized once it was exported onto the bacterial surface; thus, YscF polymerization occurred before the secretion of free YscF monomers into the culture media of the ΔyscI-CΔ73–82 and ΔyscI-CΔ103–115 strains; otherwise, it is not clear how these strains could produce polymerized YscF in the absence of YscF monomer secretion. If this assumption is correct, then it is easy to understand how the ΔyscI-CI88A and ΔyscI-CI92A strains could secrete YscF into the culture medium, because they are competent for needle assembly (Fig. 6). In addition, these strains translocated Yops into the host cytosol, as evidenced by the fact that the cytotoxic effects of the ΔyscI-CI88A and ΔyscI-CI92A strains were similar to that of the wild-type strain (Fig. 6). This is in accordance with the results showing that the polymerization and secretion of YscF in the ΔyscI-CI88A strain was slightly decreased, yet the strain still secreted substantial amounts of Yops.
We found that the ΔyscI-CΔ73–82 and ΔyscI-CΔ103–115 strains did not secrete YscI, whereas needle assembly and Yop secretion were not affected. This demonstrated that YscI secretion is not essential for YscF polymerization and Yop export in these strains. The ΔyscI-CW85A and ΔyscI-CS86A strains did not secrete YscI either, and both needle assembly and Yop secretion were disrupted, indicating that Trp-85 and Ser-86 are critical amino acids for needle assembly and Yop translocation by the Yersinia T3SS injectisome. Surprisingly, the ΔyscI-CΔ93–102 strain secreted Yops at a level comparable with that of the wild-type strain, but it was unable to assemble the YscF needle, implying that Yops could be secreted out of the bacteria, even when the needle was not assembled. Nevertheless, YscF is still essential for Yop secretion, because the ΔyscF strain did not secret any substrates (Fig. 5), which is in line with previous results showing that a Y. pestis yscF mutant strain expressing YscFD77A can neither export YscFD77A to the bacterial surface nor secrete Yops (32). There are several critical steps in the assembly of the YscF needle. First, YscF monomers need to be exported to the bacterial surface or to the site of YscF polymerization. Second, YscF monomers polymerize into a hollow tube that keeps increasing in length. Finally, after the needle attains its proper length, the YscF polymerization process is stopped via a YscP-YscU-YscI-mediated mechanism, which is accompanied by the secretion of Yop effectors (23). The ΔyscI-CW85A and ΔyscI-CS86A strains, which express YscI mutants that do not bind YscF, exhibited defects in needle assembly; thus, we conclude that YscI participates in YscF assembly by directly binding to YscF and that the needle assembly defect caused by the YscI point mutations could be due to the failure of YscF translocation or YscF polymerization. As has been reported, before assembly, YscF binds to two chaperone proteins, YscG and YscE, to prevent premature polymerization inside the bacterial cells (20). YscI binding to YscF increases the complexity of interactions between the needle protein and the other T3SS components, which implies that the precise needle assembly requires highly coordinated interactions among multiple participants.
Experimental Procedures
Bacterial Strains and Growth Conditions
The bacterial strains used in this study are listed in Table 1. Y. pestis 201 strain belongs to the biovar Microtus, and it is highly virulent to mice but avirulent to humans (35). Y. pestis strains were grown in Luria-Bertani (LB) broth at 26 °C or chemically defined TMH medium, with or without 2.5 mm calcium at 37 °C, as indicated for specific experiments. E. coli DH5α or BL21(DE3) were cultured in LB broth. Antibiotics were added to culture media when needed at the following concentrations: 100 mg ml−1 ampicillin, 20 mg ml−1 streptomycin, 10 mg ml−1 tetracycline, and 20 mg ml−1 kanamycin.
Cell Cultures, Reagents, and Antibodies
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 2 mm l-glutamine at 37 °C in a 5% CO2 incubator. Anti-FLAG M2 affinity gel, anti-FLAG M2 antibody, anti-GST antibody, anti-His antibody, anti-GroEL antibody, and BS3 were purchased from Sigma-Aldrich. IRDye 800CW-conjugated goat anti-rabbit and donkey anti-mouse antibodies were purchased from LI-COR Biosciences (Lincoln, NE). Rabbit antibodies specific for YscF, YscI, LcrV, YopN, YopE, YopM, and YopK were prepared in our laboratory as described previously (31). The Pierce BCA protein assay kit was purchased from Thermo Fisher Scientific (Waltham, MA). Nickel-nitrilotriacetic acid-agarose was purchased from Qiagen (Valencia, CA). Glutathione-Sepharose 4B was purchased from GE Healthcare (Little Chalfont, UK). An Amicon Ultra-15 regenerated cellulose centrifugal filter device (3,000-Da molecular mass cutoff) and Immobilon-P transfer membrane were purchased from Millipore (Bedford, MA).
Construction of the yscI and yscF Mutant Strains
The λ Red-mediated recombinant system was used to construct the yscI and yscF mutants using a method described previously (36). Briefly, a kanamycin resistance cassette that was used for homologous recombination was amplified using primers containing sequences that were complementary to each target gene (supplemental Table 1). pKD46 was introduced into the wild-type Y. pestis strain 201 by electroporation. Then the PCR-amplified kanamycin resistance cassettes were electroporated into the Y. pestis strain containing pKD46, and arabinose was added to induce the expression of the recombinase. Clones containing the desired mutations were identified by PCR, and pKD46 was eliminated by cultivation at 42 °C overnight.
Construction of Plasmids Expressing Various YscI Mutants
An overlapping PCR strategy was used to amplify deletion-containing yscI genes (supplemental Table 1). The PCR products were inserted into the pGEX4T-2 plasmid, and GST-tagged YscI proteins were expressed and purified by GST affinity chromatography as described previously (37).
Site-directed mutagenesis of yscI was performed by overlapping PCR using the primers listed in supplemental Table 1. A FLAG-encoding sequence was contained in the PCR primers (to obtain a FLAG tag at the C terminus of YscI or its truncants). The PCR products containing the point mutations were inserted into pBAD24 so that the expression of the YscI mutants was under the control of the arabinose-inducible promoter. After verification of the inserted sequences by DNA sequencing analysis, the recombinant plasmids were introduced into the ΔyscI strain by electroporation, generating yscI strains that were trans-complemented with the plasmids expressing different yscI mutants (Table 1).
GST Pull-down Assay
E. coli BL2(DE3) strains expressing His-tagged YscF or GST-tagged YscI deletion mutants were grown in LB broth at 37 °C, and protein expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside. His-tagged YscF was purified by affinity chromatography on nickel-nitrilotriacetic acid-agarose, and the GST-tagged proteins were purified with glutathione-Sepharose beads 4B as described previously (31). For the pull-down experiments, the GST-tagged recombinant proteins were incubated with glutathione-Sepharose 4B beads for 4 h at 4 °C. The beads were washed thoroughly with PBS and added to the His-tagged proteins (0.1 mg in PBS), followed by incubation at 4 °C overnight. After washing, bound proteins were analyzed by SDS-PAGE and immunoblotting using anti-GST and anti-His monoclonal antibodies. Intensities of the YscF bands were quantitated by using ImageJ 1.50i software (National Institutes of Health).
Co-IP
Y. pestis strains were grown at 26 °C in TMH medium without calcium to A600 of 1.0. Arabinose at a concentration of 0.2% was added to induce the expression of YscI or its mutants, and the bacteria were allowed to grow for an additional 3 h at 37 °C. Bacterial cells were harvested by centrifugation and resuspended in PBS and then lysed by ultrasonication on ice. The bacteria lysates were centrifuged at 12,000 rpm for 20 min, and the cell pellets were discarded. A small amount of lysate was used for Western blotting analysis. Aliquots (40 μl) of anti-FLAG M2 affinity gel were washed thoroughly in Tris-buffered saline (50 mm Tris HCl, 150 mm NaCl, pH 7.4) and then added to the bacterial lysates. The samples were shaken slowly for 2–4 h at 4 °C and then centrifuged, and the supernatants were carefully removed. The affinity gel was washed 3–4 times with 1 ml of PBS. Finally, 15 μl of 2× SDS loading buffer was added, and the samples were boiled for 5 min before running SDS-PAGE gels. Proteins were visualized using rabbit antibodies specific for recombinant YscF or YscI or a mouse monoclonal FLAG M2 antibody, followed by incubation with IRDye 800CW-conjugated goat anti-rabbit or donkey anti-mouse secondary antibodies. Images of the immunoblotting results were taken by an Odyssey SA imaging system (LI-COR Biosciences).
T3SS Secretion and Translocation Assays
Y. pestis strains were grown at 26 °C in TMH medium without calcium to an A600 of 1.0. Arabinose was added at a concentration of 0.2% to induce the expression of YscI and its mutants, and the strains were cultured for an additional 3 h at 37 °C to induce T3SS expression and secretion. Cultures were centrifuged at 12,000 rpm for 10 min, and the supernatants containing the secreted proteins were concentrated by centrifugation through an Ultra-15 centrifugal filter device. The concentrations of the protein samples were determined by the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). The bacterial pellets were resuspended in PBS. Based on the optical densities of the bacterial cultures, protein sample volumes were adjusted to ensure equal loading of the different samples. Equal amounts of culture supernatants and cell pellets were separated by SDS-PAGE and transferred onto an Immobilon-P transfer membrane. Proteins were visualized using rabbit antibodies specific for recombinant YscF, YscI, LcrV, YopN, YopE, YopM, and YopK, followed by incubation with an IRDye 800CW-conjugated goat anti-rabbit secondary antibody. All of the rabbit antibodies were prepared in our laboratory as described previously (37). GroEL was detected with a mouse monoclonal antibody and an IRDye 800CW-conjugated donkey anti-mouse secondary antibody.
For the translocation assay, HeLa cells were seeded into 24-well plates and grown to 80–90% confluence. HeLa cells were then infected with Y. pestis strains at an MOI of 10. After 2 h of infection, HeLa cells were washed once in PBS and lysed in 0.1% Triton X-100 in sterile H2O for 15 min, and the bacterial cells were intact under these conditions. The obtained cell lysates were centrifuged at 12,000 rpm × 30 min at 4 °C to separate the supernatant and precipitates containing cell debris and the bacterial cells. Proteins in the supernatants were precipitated overnight by 10% trichloroacetic acid (TCA). The precipitated proteins were separated by SDS-PAGE and then analyzed as described above.
Cross-linking of YscF
Y. pestis strains were grown in TMH medium at 26 °C overnight. Cultures were inoculated into fresh TMH medium without calcium, and arabinose was added at a final concentration of 0.2%, followed by culturing for an additional 3 h at 37 °C. Aliquots of 1-ml cultures were washed three times in PBS and resuspended in 0.5 ml of PBS. An equal amount of PBS containing 5 mm BS3 was added and mixed thoroughly, and the bacterial suspensions were incubated for 30 min at 37 °C. Excess cross-linker was quenched by the addition of 50 μl of 1 m Tris-Cl (pH 8.0) for 15 min. Then the samples were centrifuged at 12,000 rpm for 5 min, and the bacterial pellets were resuspended in 20 μl of deionized water. An equal volume of 2× SDS loading buffer was added, and the samples were boiled for 10 min. Then the samples were separated on SDS-polyacrylamide gels, and polymerized YscF was detected using a rabbit anti-YscF antibody and an IRDye 800CW-conjugated goat anti-rabbit secondary antibody.
Cytotoxicity Assay
HeLa cells were seeded into 24-well plates and grown to 70–80% confluence before infection. Y. pestis strains were grown in BHI at 26 °C overnight, and the bacterial cells were harvested by centrifugation and resuspended in DMEM. HeLa cells were infected with Y. pestis strains at an MOI of 10, and 0.2% arabinose was added to induce the expression of various YscI mutants. After 2 h of infection, phase-contrast images of cells were photographed under an Eclipse Ti-U inverted microscope system (Nikon Instruments Inc., Tokyo, Japan).
Author Contributions
Z.-M. D. and R.-F. Y. designed the study. S.-Y. C. and W.-B. L. performed the experiments shown in Figs. 2–7. T.-T. Z. and H.-Y. Y. performed the experiment shown in Fig. 1. Y.-F. T., T. W., X.-Y. W., and Y.-J. S. provided technical assistance. Z.-M. D. and S.-Y. C. analyzed the data. Z.-M. D. and S.-Y. C. wrote the manuscript.
Supplementary Material
This work was supported by National Basic Research Program of China (973 program) Grants 2013CB910804 and 2012CB518704 and National Natural Science Foundation of China Grants 31470242 and 81501716. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table 1.
- T3SS
- type III secretion system
- C-ring
- cytoplasmic ring
- MOI
- multiplicity of infection
- BS3
- bis-sulfosuccinimidyl suberate
- co-IP
- co-immunoprecipitation
- MS
- membrane-spanning.
References
- 1. Galán J. E., and Wolf-Watz H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 2. Blocker A., Komoriya K., and Aizawa S. (2003) Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. U.S.A. 100, 3027–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yip C. K., and Strynadka N. C. (2006) New structural insights into the bacterial type III secretion system. Trends Biochem. Sci. 31, 223–230 [DOI] [PubMed] [Google Scholar]
- 4. Tampakaki A. P., Fadouloglou V. E., Gazi A. D., Panopoulos N. J., and Kokkinidis M. (2004) Conserved features of type III secretion. Cell. Microbiol. 6, 805–816 [DOI] [PubMed] [Google Scholar]
- 5. Cornelis G. R., Boland A., Boyd A. P., Geuijen C., Iriarte M., Neyt C., Sory M. P., and Stainier I. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kudryashev M., Stenta M., Schmelz S., Amstutz M., Wiesand U., Castaño-Díez D., Degiacomi M. T., Münnich S., Bleck C. K., Kowal J., Diepold A., Heinz D. W., Dal Peraro M., Cornelis G. R., and Stahlberg H. (2013) In situ structural analysis of the Yersinia enterocolitica injectisome. Elife 2, e00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh P. (2004) Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cornelis G. R. (2010) The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol. Chem. 391, 745–751 [DOI] [PubMed] [Google Scholar]
- 9. Viboud G. I., and Bliska J. B. (2005) Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59, 69–89 [DOI] [PubMed] [Google Scholar]
- 10. Burghout P., van Boxtel R., Van Gelder P., Ringler P., Müller S. A., Tommassen J., and Koster M. (2004) Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 186, 4645–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diepold A., Amstutz M., Abel S., Sorg I., Jenal U., and Cornelis G. R. (2010) Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29, 1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimbrough T. G., and Miller S. I. (2000) Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U.S.A. 97, 11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Journet L., Agrain C., Broz P., and Cornelis G. R. (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760 [DOI] [PubMed] [Google Scholar]
- 14. Diepold A., Wiesand U., and Cornelis G. R. (2011) The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82, 502–514 [DOI] [PubMed] [Google Scholar]
- 15. Blaylock B., Riordan K. E., Missiakas D. M., and Schneewind O. (2006) Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188, 3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diepold A., Kudryashev M., Delalez N. J., Berry R. M., and Armitage J. P. (2015) Composition, formation, and regulation of the cytosolic C-ring, a dynamic component of the type III secretion injectisome. PLoS Biol. 13, e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loquet A., Sgourakis N. G., Gupta R., Giller K., Riedel D., Goosmann C., Griesinger C., Kolbe M., Baker D., Becker S., and Lange A. (2012) Atomic model of the type III secretion system needle. Nature 486, 276–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujii T., Cheung M., Blanco A., Kato T., Blocker A. J., and Namba K. (2012) Structure of a type III secretion needle at 7-Å resolution provides insights into its assembly and signaling mechanisms. Proc. Natl. Acad. Sci. U.S.A. 109, 4461–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoiczyk E., and Blobel G. (2001) Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 98, 4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun P., Tropea J. E., Austin B. P., Cherry S., and Waugh D. S. (2008) Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 377, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner S., Sorg I., Degiacomi M., Journet L., Dal Peraro M., and Cornelis G. R. (2009) The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome. Mol. Microbiol. 71, 692–701 [DOI] [PubMed] [Google Scholar]
- 22. Agrain C., Sorg I., Paroz C., and Cornelis G. R. (2005) Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 23. Edqvist P. J., Olsson J., Lavander M., Sundberg L., Forsberg A., Wolf-Watz H., and Lloyd S. A. (2003) YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185, 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wood S. E., Jin J., and Lloyd S. A. (2008) YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190, 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blaylock B., Berube B. J., and Schneewind O. (2010) YopR impacts type III needle polymerization in Yersinia species. Mol. Microbiol. 75, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Monlezun L., Liebl D., Fenel D., Grandjean T., Berry A., Schoehn G., Dessein R., Faudry E., and Attree I. (2015) PscI is a type III secretion needle anchoring protein with in vitro polymerization capacities. Mol. Microbiol. 96, 419–436 [DOI] [PubMed] [Google Scholar]
- 27. Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., and Galán J. E. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 [DOI] [PubMed] [Google Scholar]
- 28. Zilkenat S., Franz-Wachtel M., Stierhof Y. D., Galán J. E., Macek B., and Wagner S. (2016) Determination of the stoichiometry of the complete bacterial type III secretion needle complex using a combined quantitative proteomic approach. Mol. Cell. Proteomics 15, 1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lefebre M. D., and Galán J. E. (2014) The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 111, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cherradi Y., Schiavolin L., Moussa S., Meghraoui A., Meksem A., Biskri L., Azarkan M., Allaoui A., and Botteaux A. (2013) Interplay between predicted inner-rod and gatekeeper in controlling substrate specificity of the type III secretion system. Mol. Microbiol. 87, 1183–1199 [DOI] [PubMed] [Google Scholar]
- 31. Yang H., Tan Y., Zhang T., Tang L., Wang J., Ke Y., Guo Z., Yang X., Yang R., and Du Z. (2013) Identification of novel protein-protein interactions of Yersinia pestis type III secretion system by yeast two hybrid system. PLoS One 8, e54121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torruellas J., Jackson M. W., Pennock J. W., and Plano G. V. (2005) The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol. Microbiol. 57, 1719–1733 [DOI] [PubMed] [Google Scholar]
- 33. Björnfot A. C., Lavander M., Forsberg A., and Wolf-Watz H. (2009) Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J. Bacteriol. 191, 4259–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamad M. A., and Nilles M. L. (2007) Roles of YopN, LcrG and LcrV in controlling Yops secretion by Yersinia pestis. Adv. Exp. Med. Biol. 603, 225–234 [DOI] [PubMed] [Google Scholar]
- 35. Zhou D., Han Y., Dai E., Song Y., Pei D., Zhai J., Du Z., Wang J., Guo Z., and Yang R. (2004) Defining the genome content of live plague vaccines by use of whole-genome DNA microarray. Vaccine 22, 3367–3374 [DOI] [PubMed] [Google Scholar]
- 36. Du Z., Tan Y., Yang H., Qiu J., Qin L., Wang T., Liu H., Bi Y., Song Y., Guo Z., Han Y., Zhou D., Wang X., and Yang R. (2009) Gene expression profiling of Yersinia pestis with deletion of lcrG, a known negative regulator for Yop secretion of type III secretion system. Int. J. Med. Microbiol. 299, 355–366 [DOI] [PubMed] [Google Scholar]
- 37. Yang F., Ke Y., Tan Y., Bi Y., Shi Q., Yang H., Qiu J., Wang X., Guo Z., Ling H., Yang R., and Du Z. (2010) Cell membrane is impaired, accompanied by enhanced type III secretion system expression in Yersinia pestis deficient in RovA regulator. PLoS One 5, e12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Song Y., Tong Z., Wang J., Wang L., Guo Z., Han Y., Zhang J., Pei D., Zhou D., Qin H., Pang X., Zhai J., Li M., Cui B., Qi Z., et al. (2004) Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11, 179–197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.