Abstract
Tafazzin is a mitochondrial enzyme that transfers fatty acids from phospholipids to lysophospholipids. Mutations in tafazzin cause abnormal molecular species of cardiolipin and the clinical phenotype of Barth syndrome. However, the mechanism by which tafazzin creates acyl specificity has been controversial. We have shown that the lipid phase state can produce acyl specificity in the tafazzin reaction, but others have reported that tafazzin itself carries enzymatic specificity. To resolve this issue, we replicated and expanded the controversial experiments, i.e. the transfer of different acyl groups from phosphatidylcholine to monolysocardiolipin by yeast tafazzin. Our data show that this reaction requires the presence of detergent and does not take place in liposomes but in mixed micelles. To separate thermodynamic (lipid-dependent) from kinetic (enzyme-dependent) parameters, we followed the accumulation of cardiolipin during the reaction from the initial state to the equilibrium state. The transacylation rates of different acyl groups varied over 2 orders of magnitude and correlated tightly with the concentration of cardiolipin in the equilibrium state (lipid-dependent parameter). In contrast, the rates by which different transacylations approached the equilibrium state were very similar (enzyme-dependent parameter). Furthermore, we found that tafazzin catalyzes the remodeling of cardiolipin by combinations of forward and reverse transacylations, essentially creating an equilibrium distribution of acyl groups. These data strongly support the idea that the acyl specificity of the tafazzin reaction results from the physical properties of lipids.
Keywords: acyltransferase, cardiolipin, enzyme kinetics, lipid, mitochondria
Introduction
Tafazzin was first described as the protein mutated in Barth syndrome (1) and subsequently found to be homologous to acyltransferases (2). When Vreken et al. (3) demonstrated an abnormal fatty acid composition of cardiolipin (CL)2 in fibroblasts from patients with Barth syndrome, it appeared that tafazzin is a monolysocardiolipin (MLCL)-specific acyltransferase.
Acyltransferases catalyze the formation of phospholipids from lysophospholipids and acyl-CoA, an essentially irreversible reaction. The acyl specificity of this reaction, an important condition to maintain unique molecular species patterns in different phospholipids and organelles, arises largely from the distinct catalytic properties of individual members of the acyltransferase family (4–6). However, it turned out that tafazzin does not belong to this family; instead it is a transacylase that transfers fatty acids from phospholipids to lysophospholipids (7). In contrast to acyl-CoA-dependent acyltransferases, tafazzin catalyzes a reversible chemical reaction. In such reactions, the net flux rate depends not only on the enzyme but also on the thermodynamic driving force (8–11).
Interestingly, tafazzin reacts with all phospholipids and is able to transfer different types of fatty acids, including saturated and unsaturated fatty acids (12, 13). This was surprising given the specific effect of tafazzin in vivo, which selectively increases the abundance of unsaturated CL. We were able to demonstrate, however, that substrate specificity in the tafazzin reaction may arise from the physical properties of lipids even if the reaction itself is promiscuous. This specificity is caused by a better fit of certain molecular species into the lipid packing arrangement, which naturally leads to an accumulation of those species under the condition of free acyl exchange. For instance, we were able to induce the formation of physiologic CL species in vitro by providing a lipid-water interphase with strong negative curvature (13).
The idea that CL remodeling is based on thermodynamic forces has raised questions partly because it runs against the time-honored principle of enzymes conferring specificity to all metabolic pathways and partly because it is at odds with evidence showing mitochondrial lipids to be mostly in the bilayer state (14, 15). Recently, Abe et al. (16) reported that glutathione S-transferase (GST)-tagged yeast tafazzin can react with rigid bilayers and that it displays enzymatic acyl specificity. Because the conclusion by Abe et al. (16) directly contradicts ours (13), we reproduced their data and investigated the source of the discrepancy. The present study provides new insight into the mechanism of tafazzin and helps to explore the general concept of specificity for reversible enzymatic reactions.
Results and Discussion
The Physical State of the Substrates of Tafazzin
Abe et al. (16) have demonstrated transacylation activity of GST-tagged tafazzin from Saccharomyces cerevisiae with the substrates phosphatidylcholine (PC) and MLCL. Because these two lipids have a strong propensity to form bilayers, the data seem to be at odds with our conclusion that tafazzin can only react with lipids in the non-bilayer state (13). We thus prepared GST-tagged yeast tafazzin and let it react with liposomes formed from PC and MLCL (PC/MLCL = 9:1) as described by Abe et al. (16). In contrast to those authors, we did not observe any transacylation activity (Fig. 1A).
FIGURE 1.
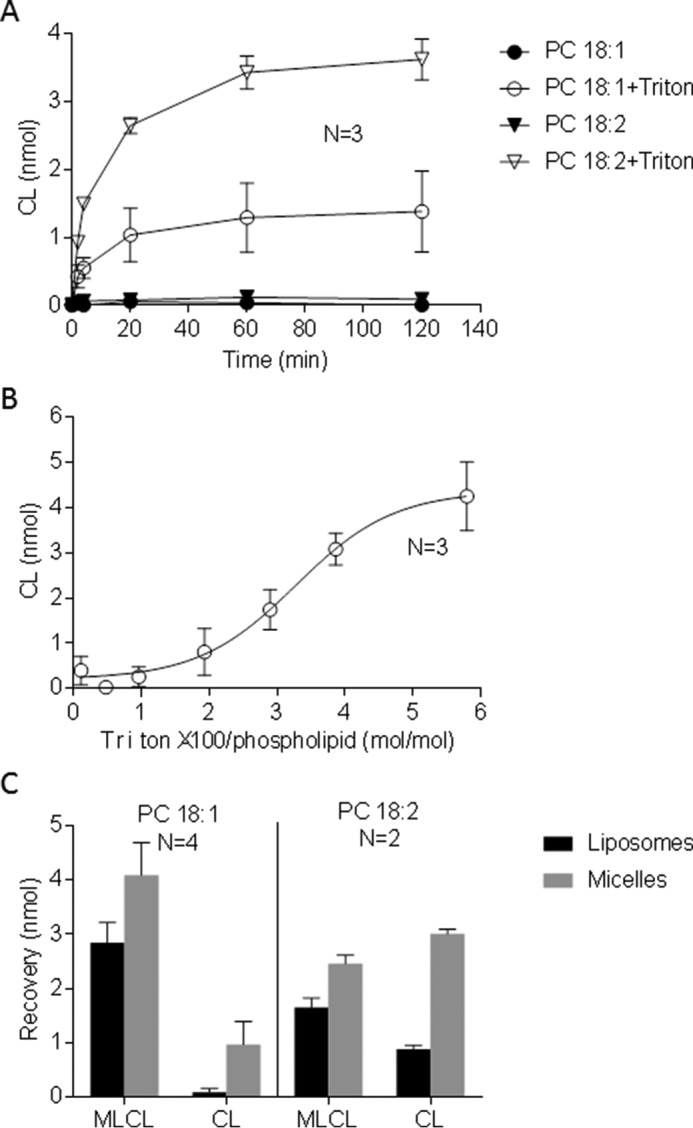
Tafazzin requires detergent to react with PC and MLCL in vitro. GST-tagged yeast tafazzin was incubated with liposomes containing 72 nmol of PC and 8 nmol of MLCL in a total volume of 0.1 ml. The formation of CL was measured by mass spectrometry. A, the acyl donors PC 18:1/18:1 and PC 18:2/18:2 were compared in the absence and presence of 0.2% Triton X-100. Transacylation activity with either substrate required detergent. B, CL was measured after 30-min incubation in the presence of PC 18:1/18:1 and different concentrations of Triton X-100. The detergent increased the transacylation activity. C, the reaction was performed for 30 min in the presence of 0.12% Triton X-100, liposomes were separated from micelles, and lipids were analyzed in the two fractions. The substrate (MLCL) was recovered in both fractions, but the product (CL) was recovered mostly in micelles. The data are mean values of the indicated number of experiments (n); the error bars represent S.E.
Abe et al. (16) have performed all steps of their enzyme purification in the presence of Triton X-100 and, in their own estimation, introduced as much as 0.08% (1.24 mm) of the detergent into the reaction mixture, which amounts to a Triton/phospholipid ratio of 2 mol/mol. In contrast, we have omitted any detergent from the buffers in the final two purification steps. As a result, only protein-bound Triton remained in the tafazzin preparation (265 ± 18 Triton molecules/enzyme molecule, n = 4), and the amount of Triton introduced by the enzyme into the reaction mixture was 0.0083 ± 0.0007% (0.13 mm), which corresponded to a Triton/phospholipid ratio of 0.16 mol/mol. Because the discrepancy in enzyme activity seemed to correlate with the amount of Triton introduced into the reaction, we wondered whether the detergent affects the transacylation. Indeed, when we added Triton X-100 to the PC-MLCL liposomes, we could replicate the data by Abe et al. (16) both with regard to transacylation activity and with regard to its acyl preference (18:2 > 18:1)3 (Fig. 1A). Further experiments established a sigmoidal response of the reaction to the concentration of Triton (Fig. 1B). Specifically, there was no activity below a Triton/phospholipid ratio of 1 mol/mol, whereas maximal activity was reached at a Triton/phospholipid ratio of about 5 mol/mol (4 mm = 0.26%).
When Triton X-100 is added to phospholipid bilayers, substantial structural alterations occur (17). Below a Triton/lipid ratio of 1, the bilayer state is largely preserved, although the incorporation of detergent molecules increases its fluidity. At Triton/lipid ratios between 1 and 2, bilayers coexist with micelles, but above Triton/lipid ratios of 2, mixed micelles become the dominant structure. To determine the phase preference of the reaction at a Triton/phospholipid ratio of 2 mol/mol, we separated micelles from liposomes and measured lipid recoveries in the two fractions. We recovered products primarily in micelles, whereas substrates were recovered equally in liposomes and micelles (Fig. 1C). The data demonstrate that tafazzin reacts with PC and MLCL only if the substrates are solubilized in mixed detergent micelles. These results identify the cause of the discrepancy between previous studies (13, 16) and support the idea that phospholipids have to assemble in a non-bilayer state to react with tafazzin.
Kinetic and Thermodynamic Aspects of the Tafazzin Reaction
Because tafazzin catalyzes a reversible reaction, its rate depends not only on the catalytic efficiency of the enzyme but also on the difference in free energy between substrates and products. In fact, it has been shown that the rate of such reactions can be calculated by multiplying separate kinetic and thermodynamic expressions (11). Acyl specificity in the tafazzin reaction, defined as preferential transfer of one acyl group over another, can also arise from kinetic and from thermodynamic causes. On the one hand, tafazzin may transfer acyl chains with different catalytic efficiency; on the other hand, product species may have different natural abundances in the chemical equilibrium state. Although either phenomenon produces acyl specificity, only the former constitutes enzymatic specificity; the latter is a consequence of the physical behavior of lipids, although one that requires the presence of tafazzin to manifest itself. Thus, when dealing with the issue of enzymatic specificity, it is crucial to rigorously separate kinetic from thermodynamic factors.
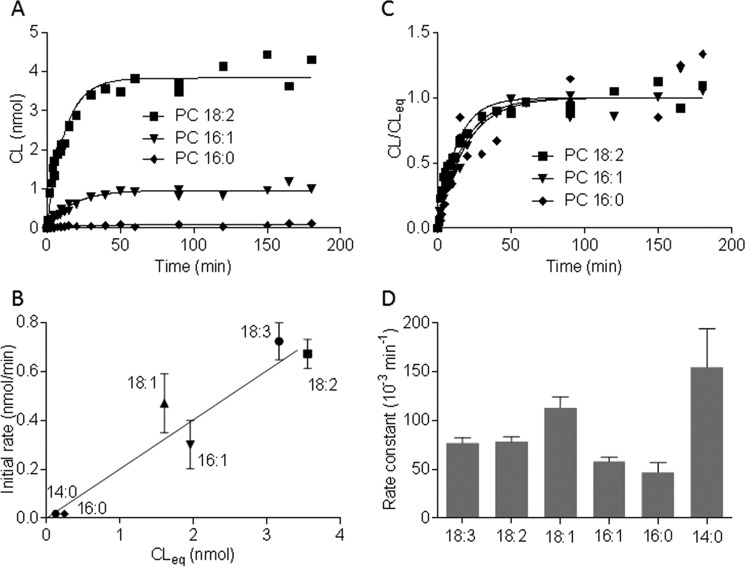
For instance, Abe et al. (16) have shown and we have confirmed that tafazzin transfers 18:2 groups faster than 16:1 groups and 16:1 groups faster than 16:0 groups from PC to MLCL. However, this does not necessarily imply catalytic specificity because the observation can just as easily be explained by different abundances of CL in the chemical equilibrium state of the reactions (Fig. 2A). In fact, we found a strong correlation between the initial rates of transacylation and the chemical composition at equilibrium when we compared six different acyl groups (Fig. 2B). This clearly indicates that thermodynamic factors are important for the acyl specificity of the tafazzin reaction.
FIGURE 2.
The acyl specificity of the tafazzin reaction is caused by thermodynamic properties of lipids. GST-tagged yeast tafazzin was incubated with 72 nmol of PC and 8 nmol of MLCL in 0.1 ml of buffer containing 0.18% Triton X-100. The concentration of CL was measured at different time points in the presence of different PC species. A, the initial rate of CL formation varied in the order 18:2 > 16:1 > 16:0. B, comparison of different acyl groups showed a linear correlation (r2 = 0.917) between the initial rate of CL formation (0–4 min) and the concentration of CL at equilibrium (CLeq). The data are mean values; error bars represent S.D. (n = 3). C, the CL/CLeq ratio was plotted over time. Reactions with 18:2, 16:1, and 16:0 approached the chemical equilibrium at similar rates. D, the rate constants by which reactions approached the chemical equilibrium were determined by non-linear regression. Similar rate constants were estimated for six different acyl groups. Each constant is the best fit value of 24 data points obtained from three independent experiments; error bars represent S.E.
Catalytic Efficiency of Tafazzin for Different Acyl Groups
Thermodynamic factors aside, it is still possible, however, that enzyme kinetics contributed to the acyl specificity. To examine this idea, we have to define the term “enzymatic specificity” (18) for reversible enzyme-catalyzed reactions, the kinetics of which are given by the Haldane relation (8).
| (Eq. 1) |
In this equation, V+ and V− are the limiting forward and reverse velocities, KS and KP are the Michaelis constants of substrate and product, and K′ is the equilibrium constant representing the ratio of substrate to product concentration at equilibrium. As has been recognized by Haldane (8) and further elaborated on in recent publications (9, 10, 19), the equation forms an important connection between biology and chemistry because the four kinetic parameters (V+, V−, KS, and KP) are properties of the enzyme, but K′ is a property of the reactants. Hence, the enzyme kinetics is constrained by its inherent dependence on reaction thermodynamics.
Let us presume there is true acyl specificity of the enzyme such that acyl group 1 is preferred over acyl group 2 in the forward reaction (MLCL + PC → CL + LPC).
| (Eq. 2) |
In accordance with the Haldane relation (8) we obtain Equation 3.
| (Eq. 3) |
If there is indeed no thermodynamic difference between the two reactions (K′1 = K′2), Equation 4 follows.
| (Eq. 4) |
Thus, kinetic preference for any acyl group in the forward reaction inevitably requires preference for the same acyl group in the reverse reaction (CL + LPC → MLCL + PC). If forward and reverse reactions of one acyl group proceed faster than the corresponding reactions of another, the reaction of the preferred acyl group will approach its chemical equilibrium faster. This is consistent with the general concept that catalytic specificity manifests itself in the rate by which the catalyst establishes chemical equilibrium for various reactants. We can therefore isolate kinetic specificity from thermodynamic specificity by measuring the time it takes to reach the chemical equilibrium state.
When we measured the transfer of different acyl groups from PC to MLCL, we found that they reached the chemical equilibrium within a similar period of time (Fig. 2C). Rate constants estimated by fitting the time dependence of the CL/CLeq ratio, where CLeq is the CL concentration at equilibrium, confirmed that the reactions approached the equilibrium state with similar velocities. Importantly, the existing variations in the rate constants did not match the observed acyl specificities and therefore did not provide a rational explanation for those specificities (Fig. 2D). Collectively, the data of Fig. 2 demonstrate that the acyl specificity in the CL formation from MLCL and PC is due to the thermodynamic properties of lipids but not the kinetic properties of tafazzin.
Forward and Reverse Transacylation of CL
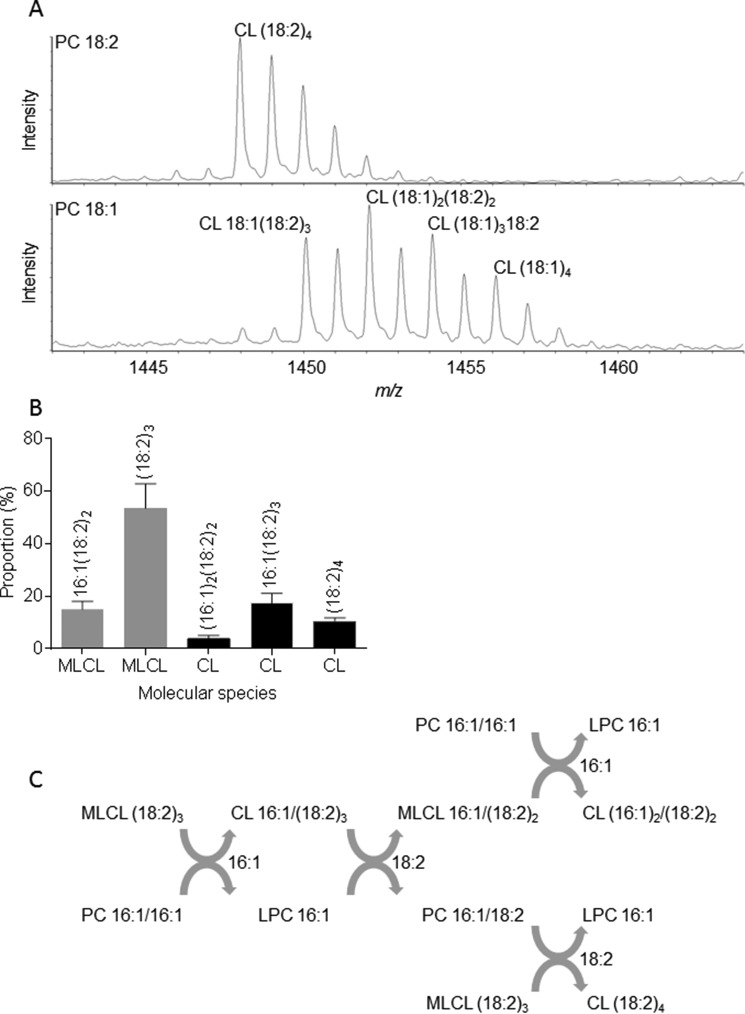
Incubation of tafazzin with PC (18:2)2 and MLCL (18:2)3 produced CL (18:2)4 as expected, but incubation of tafazzin with PC (18:1)2 and MLCL (18:2)3 produced four different species, including CL 18:1(18:2)3, CL (18:1)2(18:2)2, CL (18:1)318:2, and CL (18:1)4 (Fig. 3A). The incorporation of multiple 18:1 residues into CL is only possible if tafazzin transfers not only 18:1 groups from PC to MLCL (PC + MLCL → LPC + CL) but also 18:2 groups from CL to LPC (CL + LPC→ MLCL + PC). Likewise, incubation of tafazzin with PC (16:1)2 and MLCL (18:2)3 produced multiple molecular species (Fig. 3B), indicating the transfer of acyl groups from PC to MLCL and back from CL to LPC (Fig. 3C). The data demonstrate that tafazzin catalyzes the bidirectional exchange of acyl groups between PC and CL species rather than the linear incorporation of a specific acyl group into CL. By its very nature, such acyl exchange cannot be under kinetic but only under thermodynamic control because different fatty acids have to be transferred concurrently, which renders any kinetic advantage of one over the other ineffective for the overall process. As fatty acids shuttle in and out of CL, the reaction will inevitably seek the species distribution with the lowest free energy. The kinetic properties of tafazzin will determine how fast that occurs but will not influence the outcome of the reaction.
FIGURE 3.
Tafazzin changes the CL composition by combinations of forward and reverse transacylations. GST-tagged yeast tafazzin was incubated for 3 h with 0.72 mm PC, 0.08 mm MLCL (18:2)3, and 0.12% Triton X-100 in a total volume of 0.1 ml. The transacylation products were analyzed by mass spectrometry. A, CL (18:2)4 was the only transacylation product in the presence of PC 18:2/18:2, but multiple CL species were formed in the presence of PC 18:1/18:1. B, multiple CL and MLCL species were formed in the presence of PC 16:1/16:1. Data are mean values; error bars represent S.E. (n = 4). C, the scheme shows a sequence of transacylation reactions that convert PC 16:1/16:1 and MLCL (18:2)3 into the transacylation products identified above.
Remodeling of Mitochondrial CL by Multiple Transacylations
We wanted to determine whether multiple forward and reverse transacylations are also detectable in mitochondrial membranes, the native environment of tafazzin. To this end, we expressed Drosophila tafazzin in Sf9 insect cells with a baculovirus expression system, which yields mitochondria with a high tafazzin density (13). Upon lipid addition to isolated mitochondria, we found acyl transfer from endogenous phospholipids to exogenous lysophospholipids and from exogenous phospholipids to endogenous lysophospholipids and between exogenous substrates. For example, 16:1 and 18:1 groups were transferred from endogenous phospholipids to exogenous LPC (Fig. 4A), and 18:3 groups were transferred from exogenous PC to endogenous lipids (Fig. 4B) but also from exogenous PC to exogenous LPC (Fig. 4C). Even in exogenous-to-exogenous transacylations, 93–98% of the reaction products remained associated with mitochondria, suggesting that the reactions required fusion of the exogenous lipids with mitochondrial membranes. Consistent with previous findings (13), only lipids with the potential to form non-bilayer structures, such as phosphatidylethanolamine (PE) and CL + Ca2+, were effective acyl donors, whereas the obligatory bilayer lipid PC was not (Fig. 5). Conversely, PC became an acyl donor when exogenous lysophospholipids were added, enabling the formation of micelles (Fig. 4, B and C).
FIGURE 4.
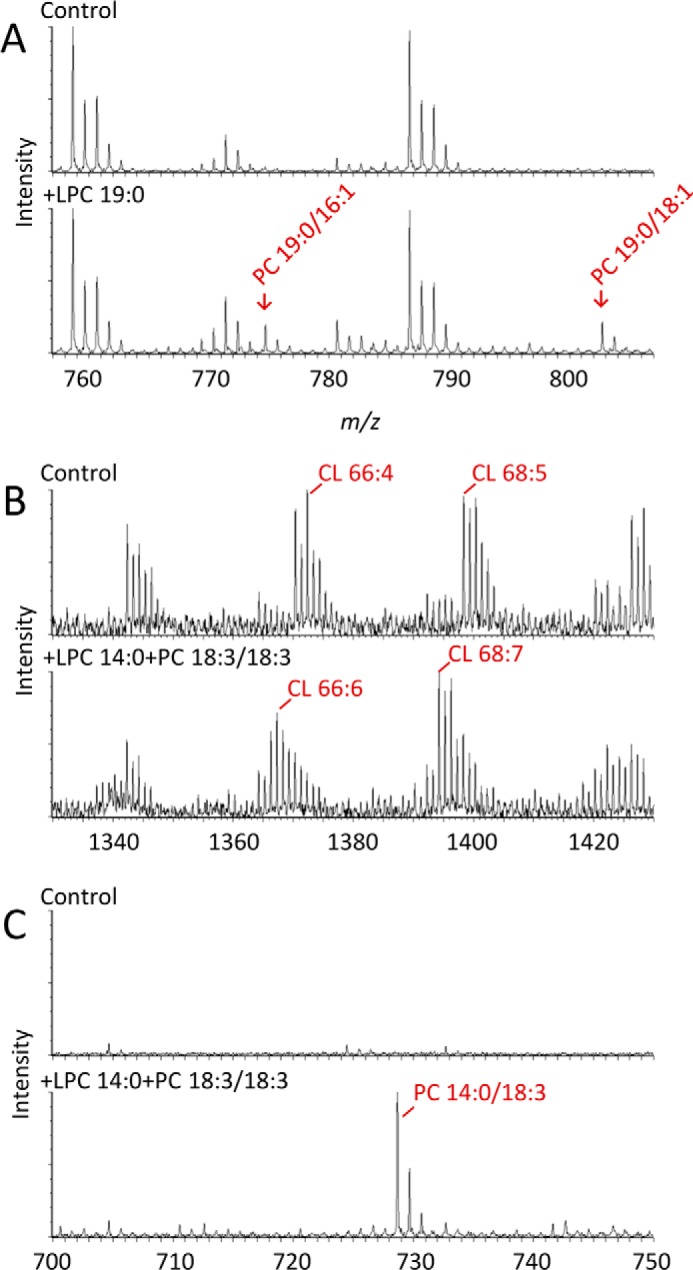
Mitochondrial tafazzin catalyzes transacylations between exogenous and endogenous lipids. Mitochondria were isolated from Sf9 cells expressing isoform A of Drosophila tafazzin. Isolated mitochondria (0.25 mg of protein) were incubated with exogenous lipids for 90 min and analyzed by mass spectrometry. A, new molecular species of PC were formed upon addition of 8 nmol of LPC 19:0, including PC 19:0/16:1 (m/z = 774.6) and PC 19:0/18:1 (m/z = 802.6). B, the molecular species pattern of endogenous CL was altered in the presence of 8 nmol of LPC 14:0 and 80 nmol of PC (18:3)2. C, PC 14:0/18:3 (m/z = 728.5) was formed in the presence of 300 nmol of LPC 14:0 and 80 nmol of PC (18:3)2.
FIGURE 5.
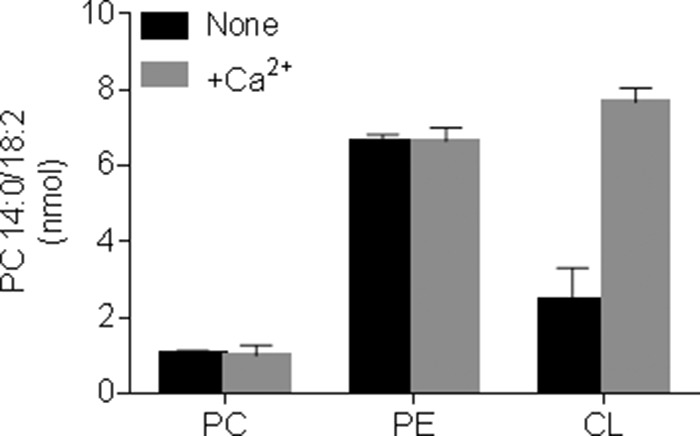
Mitochondrial tafazzin requires substrates that can form non-bilayer structures. Mitochondria were isolated from Sf9 cells expressing isoform A of Drosophila tafazzin. Isolated mitochondria (0.05 mg of protein) were incubated with 90 nmol of PC (18:2)2, PE (18:2)2, or CL (18:2)4 and 10 nmol of LPC 14:0 in the absence or presence of 15 mm CaCl2 for 90 min. The amount of newly formed PC 14:0/18:2 was measured by mass spectrometry. Data are mean values; error bars represent ranges (n = 2).
When we added exogenous bovine heart CL, we observed remodeling of endogenous mitochondrial lipids, including PE, PC, and CL, which resulted in the formation of new molecular species. Those species became more abundant in the presence of Ca2+, which increased the propensity of CL to form non-bilayer assemblies (Fig. 6). Importantly, the amount of newly formed species far exceeded the amount of endogenous lysophospholipids, including LPC, lyso-PE, and MLCL. This is only possible if fatty acid exchange took place between exogenous CL and endogenous phospholipids by combinations of forward and reverse transacylations. CL remodeling also occurred in response to exogenous PC + LPC, which also required bidirectional transacylation via the forward (CL + LPC → MLCL + PC) and the reverse (MLCL + PC → CL + LPC) reactions (Fig. 4B). These data provide direct evidence for the remodeling of mitochondrial CL by combinations of forward and reverse transacylations, showing that tafazzin alone can catalyze CL remodeling.
FIGURE 6.
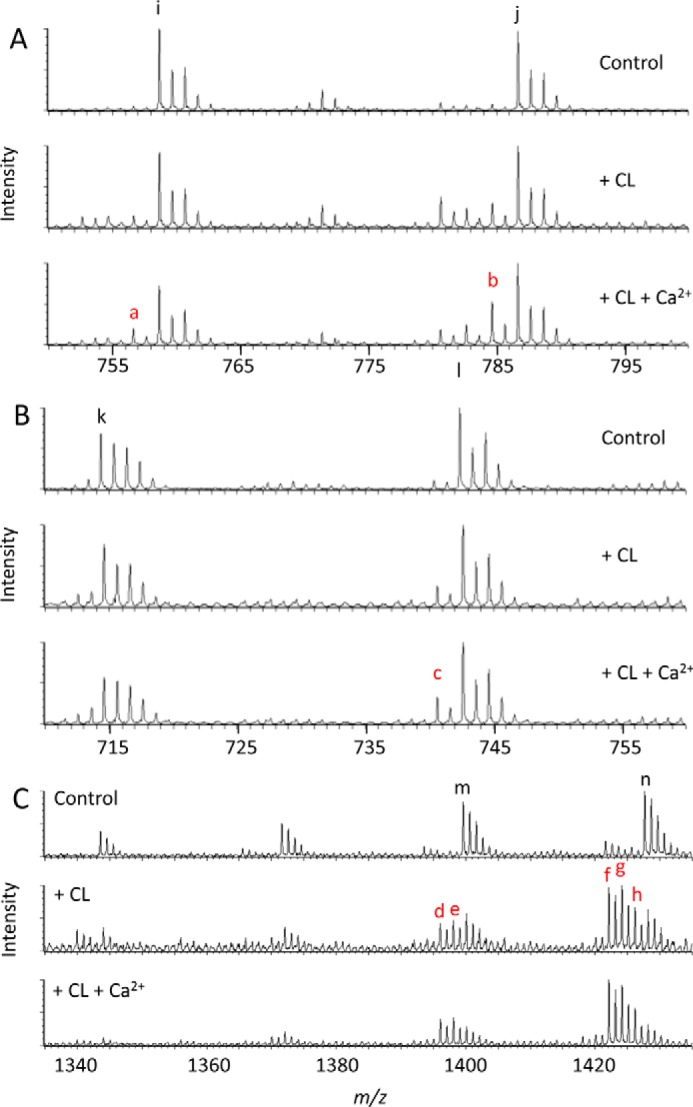
Mitochondrial tafazzin remodels cardiolipin by forward and reverse transacylations. Mitochondria were isolated from Sf9 cells expressing isoform A of Drosophila tafazzin. Isolated mitochondria (0.25 mg of protein) were incubated with 10 nmol of exogenous bovine heart CL with or without 15 mm CaCl2 for 90 min and analyzed by mass spectrometry. Exogenous CL triggered the formation of novel molecular species, including PC 16:1/18:2 (a; m/z = 756.6), PC 18:1/18:2 (b; m/z = 784.6), PE 18:1/18:2 (c; m/z = 740.5), CL (16:1)2-(18:2)2 (d; m/z = 1395.9), CL (16:1)2-18:1-18:2 (e; m/z = 1397.9), CL 16:1-(18:2)3 (f; m/z = 1421.9), CL 16:1-18:1-(18:2)2 (g; m/z = 1424.0), and CL 16:1-(18:1)2-18:2 (h; m/z = 1426.0). Native species of Sf9 mitochondria included PC 16:1/18:1 (i; m/z = 758.6), PC 18:1/18:1 (j; m/z = 786.6), PE 16:1/18:1 (k; m/z = 714.5), PE 18:1/18:1 (l; m/z = 742.5), CL (16:1)2-(18:1)2 (m; m/z = 1400.0), and CL 16:1-(18:1)3 (n; m/z = 1428.0). Addition of CaCl2 increased the concentration of several transacylation products. A, PC region (positive ion mode). B, PE region (negative ion mode). C, CL region (negative ion mode). Colored letters indicate novel molecular species.
We asked whether the outcome of CL remodeling changed if the availability of fatty acids was altered. This was indeed suggested by a recent study in which the treatment of yeast cells with different fatty acids led to large changes in the CL composition (20). Consistent with those data, we found that the apparent specificity of CL remodeling, assessed by comparing the wild type with the tafazzin deletion mutant, varied drastically when different exogenous fatty acids were added to the medium (supplemental Table S1). These data show that the tafazzin reaction alters its apparent acyl specificity in vivo when presented with different substrates, which is consistent with our thermodynamic model. However, we have not been able to identify the mechanism that drives the acyl selection in mitochondrial membranes. Candidates include for instance the assembly of protein complexes, the formation of membrane folds and crista junctions, or fission-fusion events.
Conclusions
In summary, we studied the formation of CL species by tafazzin-catalyzed transacylations and found (i) that the reaction cannot take place in rigid lipid bilayers, (ii) that acyl specificity arises from the thermodynamic properties of lipids, and (iii) that CL species can be remodeled by combinations of forward and reverse transacylations. The data strongly support the idea that the molecular composition of mitochondrial CL is born out of a transacylation equilibrium in which two components manifest themselves, the pattern of available fatty acids and the specific packing conditions of lipids. In a mathematical model of this equilibrium, we presented the two components as matrices M0 (fatty acid composition) and Φ (lipid packing constraints), which together determine the molecular composition of CL (21). Tafazzin reshuffles fatty acids between lipids until the molecular species composition with the lowest free energy emerges; this composition is the one that confers optimal packing conditions. Although tafazzin provides the acyl exchange mechanism, it is the process of lipid assembly that determines the acyl specificity. This conclusion is consistent with transgenic experiments showing that human tafazzin reproduces native yeast CL when expressed in S. cerevisiae (22) but native fly CL when expressed in Drosophila melanogaster (23).
Finally, the present study emphasizes the significance of thermodynamics for reversible biochemical reactions (9–11). This notion has received little attention in the current literature because most studies have focused on signal transductions and other irreversible steps in which a large drop of free energy occurs. In those situations, thermodynamic considerations of the chemical equilibrium can be ignored for the sake of simplicity (11). However, in reversible reactions it is necessary to consider the thermodynamic properties and to acknowledge that rate and specificity are not solely controlled by the catalyzing proteins. Here it is necessary to separate thermodynamic from kinetic factors, to apply rigorously the concept of enzymatic specificity (defined as differential catalytic efficiency), and to consider its biological implications. For tafazzin in biological membranes, where many competing transacylations take place, thermodynamics is more important than kinetics simply because the chemical equilibrium determines which CL composition is formed, whereas the enzyme kinetics merely determines how fast that occurs.
Experimental Procedures
Expression and Purification of GST-tagged Yeast Tafazzin
Tafazzin from S. cerevisiae was expressed and purified as described by Abe et al. (16) with some modifications. The open reading frame of the S. cerevisiae gene encoding tafazzin (TAZ1) was amplified by the polymerase chain reaction and inserted into pGEX-EF, a GST fusion protein expression vector. BL21 Escherichia coli cells were transformed with pGEX-EF-TAZ1 plasmid DNA. The cells were grown to the early logarithmic phase (A600 = 0.6) in 1.25 liters of LB broth, and protein expression was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were allowed to express the recombinant protein at 32 °C for 3.5 h after which they were harvested by centrifugation and stored frozen at −80 °C. Frozen cell pellets were resuspended in a total volume of 50 ml of ice-cold phosphate-buffered saline (PBS) containing 1 mm EDTA, 100 μg/ml lysozyme, 10 mm β-mercaptoethanol, 0.1% Triton X-100, and protease inhibitor mixture (Sigma). An aliquot of 0.25 ml of DNase I (3434 units/ml, protease-free; Thermo Fisher) was added, and the suspension was incubated on ice for 30 min. The cells were broken up with the Cell Disruption System (Taylor Scientific). Debris and unbroken cells were removed by centrifugation at 10,000 × g for 30 min at 4 °C. The supernatant was incubated for 2 h with 2-ml bed volumes of glutathione-Sepharose 4B resin (GE Healthcare) by gentle rotation at 4 °C. The resin was removed by centrifugation at 500 × g for 5 min and washed three times with cold PBS containing 1 mm EDTA and protease inhibitor mixture. The recombinant protein was eluted from the washed resin in 3 ml of elution buffer containing 50 mm Tris (pH 8.0), 1 mm EDTA, and 10 mm glutathione. The purity of the protein was assessed by SDS-PAGE. The protein concentration of the final enzyme preparations ranged from 0.4 to 1.0 g/liter.
Preparation of Mitochondria Expressing Drosophila Tafazzin
Sf9 insect cells were cultured at 27 °C in BaculoGoldTM TNM-FH insect medium from BD Biosciences. The cDNA encoding the full-length isoform of tafazzin from D. melanogaster was inserted into the NcoI-KpnI sites of the baculovirus shuttle vector pAc-XL, which we modified by eliminating the GST tag. Using calcium precipitation, 5 μg of vector DNA and 0.5 μg of BD BaculoGold linearized baculovirus DNA (BD Biosciences) were co-transfected in 2 million Sf9 cells by incubation for 4 h at 27 °C in TNM-FH medium. Recombinant baculovirus released into the medium was harvested and stored at 4 °C. To express tafazzin, Sf9 cells were infected with the recombinant baculovirus stock and then cultured at 27 °C for 3–5 days. The cells were harvested and homogenized with a tight fitting Teflon-glass homogenizer in a buffer containing 210 mm mannitol, 70 mm sucrose, 1 mm EGTA, and 5 mm Hepes (pH 7.2) at 4 °C. Cell debris and nuclei were removed by centrifugation at 750 × g for 5 min. Mitochondria were collected from the supernatant by centrifugation at 17,000 × g for 20 min. Mitochondria were washed in isolation buffer, recollected by centrifugation, and stored at −80 °C.
Transacylation Reactions with Purified Tafazzin
Transacylation reactions were performed in 20 mm Tris (pH 7.4) at 25 °C in a total volume of 100 μl. The final concentration of lipids was 0.8 mm (PC/MLCL = 9:1). Synthetic lipid species and MLCL were obtained from Avanti Polar Lipids. MLCL consisted of 96% MLCL (18:2)3 as shown by mass spectrometry. Liposomes were prepared by lipid extrusion using the Mini-Extruder from Avanti Polar Lipids. Triton X-100 was added from stock solutions as specified in the legends. The reactions were initiated by adding 3 μg of GST-tagged yeast tafazzin and stopped by adding 2 ml of 2-propanol/acetonitrile (3:2, v/v). The concentrations of transacylation products were determined by MALDI-TOF mass spectrometry using the method of Sun et al. (24). A small aliquot of the quenched sample was mixed 1:1 with matrix solution containing 20 g/liter 9-aminoacridine in 2-propanol/acetonitrile (3:2, v/v). For each sample, 1 μl of this mixture was spotted on a target plate. Measurements were performed with a MALDI Micro MX mass spectrometer (Waters) operated in reflectron mode. The pulse voltage was set to 2000 V, the detector voltage was set to 2200 V, and the time lag focusing delay was set to 700 ns. The nitrogen laser (337 nm) was fired at a rate of 5 Hz, and 10 laser shots were acquired per subspectrum. The instrument was operated in negative ion mode with a flight tube voltage of 12 kV, a reflectron voltage of 5.2 kV, and a negative anode voltage of 3.5 kV and calibrated daily. We typically acquired 100 subspectra (representing 1000 laser shots) per sample in a mass range from 400 to 2000 Da. Spectra were only acquired if their base peak intensity was within 10–95% of the saturation level. Data were analyzed with MassLynx 4.1 software.
Transacylation Reactions with Mitochondrial Membranes
Isolated Sf9 mitochondria that expressed Drosophila tafazzin were incubated with exogenous lipids in 50 mm Tris and 0.5 mm EDTA (pH 7.4) for 90 min at 37 °C. Lipids were dispersed by sonication before adding them to the mitochondria. The total volume was 0.1 ml; the amounts of lipids and protein are specified in the legends. The incubations were stopped by adding 2 ml of methanol and 1 ml of chloroform. Lipids were extracted according to Bligh and Dyer (25) and analyzed by MALDI-TOF mass spectrometry as described above. PC was analyzed in positive ion mode, and PE and CL were analyzed in negative ion mode.
Miscellaneous Methods
The concentration of Triton X-100 was measured by absorbance at 274 nm. A series of standard solutions with different Triton concentrations was used to calibrate the method. Protein concentrations were measured with the Bio-Rad Protein Assay kit, which is based on the method of Bradford (26). To separate liposomes from micelles, samples were diluted with water to a total volume of 0.6 ml and spun at 100,000 × g for 1 h in a TLA 100.3 rotor using the Optima Max XP table top ultracentrifuge (Beckman Coulter).
Author Contributions
M. S. performed experiments, analyzed data, wrote the paper, and coordinated the study. Y. X. performed experiments, analyzed data, and revised the manuscript. M. R. analyzed data and revised the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grant GM115593 (to M. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
Acyl groups are abbreviated by an x:y code in which x indicates the number of carbon atoms and y indicates the number of double bonds.
- CL
- cardiolipin
- LPC
- lysophosphatidylcholine
- MLCL
- monolysocardiolipin
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine.
References
- 1. Bione S., D'Adamo P., Maestrini E., Gedeon A. K., Bolhuis P. A., and Toniolo D. (1996) A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12, 385–389 [DOI] [PubMed] [Google Scholar]
- 2. Neuwald A. F. (1997) Barth syndrome may be due to an acyltransferase deficiency. Curr. Biol. 7, R465–R466 [DOI] [PubMed] [Google Scholar]
- 3. Vreken P., Valianpour F., Nijtmans L. G., Grivell L. A., Plecko B., Wanders R. J., and Barth P. G. (2000) Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279, 378–382 [DOI] [PubMed] [Google Scholar]
- 4. Gijón M. A., Riekhof W. R., Zarini S., Murphy R. C., and Voelker D. R. (2008) Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J. Biol. Chem. 283, 30235–30245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shindou H., Hishikawa D., Harayama T., Eto M., and Shimizu T. (2013) Generation of membrane diversity by lysophospholipid acyltransferases. J. Biochem. 154, 21–28 [DOI] [PubMed] [Google Scholar]
- 6. Yamashita A., Hayashi Y., Nemoto-Sasaki Y., Ito M., Oka S., Tanikawa T., Waku K., and Sugiura T. (2014) Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 53, 18–81 [DOI] [PubMed] [Google Scholar]
- 7. Xu Y., Malhotra A., Ren M., and Schlame M. (2006) The enzymatic function of tafazzin. J. Biol. Chem. 281, 39217–39224 [DOI] [PubMed] [Google Scholar]
- 8. Haldane J. B. S. (1930) The course of enzymatic reactions and its mathematical theory, in Enzymes, pp. 74–92, Longmans, Green and Co. [Google Scholar]
- 9. Alberty R. A. (2006) Relations between biochemical thermodynamics and biochemical kinetics. Biophys. Chem. 124, 11–17 [DOI] [PubMed] [Google Scholar]
- 10. Alberty R. A., Cornish-Bowden A., Goldberg R. N., Hammes G. G., Tipton K., and Westerhoff H. V. (2011) Recommendations for terminology and databases for biochemical thermodynamics. Biophys. Chem. 155, 89–103 [DOI] [PubMed] [Google Scholar]
- 11. Noor E., Flamholz A., Liebermeister W., Bar-Even A., and Milo R. (2013) A note on the kinetics of enzyme action: a decomposition that highlights thermodynamic effects. FEBS Lett. 587, 2772–2777 [DOI] [PubMed] [Google Scholar]
- 12. Malhotra A., Xu Y., Ren M., and Schlame M. (2009) Formation of molecular species of mitochondrial cardiolipin. 1. A novel transacylation mechanism to shuttle fatty acids between sn-1 and sn-2 positions of multiple phospholipid species. Biochim. Biophys. Acta 1791, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schlame M., Acehan D., Berno B., Xu Y., Valvo S., Ren M., Stokes D. L., and Epand R. M. (2012) The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat. Chem. Biol. 8, 862–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cullis P. R., de Kruijff B., Hope M. J., Nayar R., Rietveld A., and Verkleij A. J. (1980) Structural properties of phospholipids in the rat liver inner mitochondrial membrane. Biochim. Biophys. Acta 600, 625–635 [DOI] [PubMed] [Google Scholar]
- 15. De Kruijff B., Nayar R., and Cullis P. R. (1982) 31P-NMR studies on phospholipid structure in membranes of intact, functionally-active, rat liver mitochondria. Biochim. Biophys. Acta 684, 47–52 [DOI] [PubMed] [Google Scholar]
- 16. Abe M., Hasegawa Y., Oku M., Sawada Y., Tanaka E., Sakai Y., and Miyoshi H. (2016) Mechanism for remodeling of the acyl chain composition of cardiolipin catalyzed by Saccharomyces cerevisiae tafazzin. J. Biol. Chem. 291, 15491–15502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dennis E. A. (1974) Formation and characterization of mixed micelles of the nonionic surfactant Triton X-100 with egg, dipalmitoyl, and dimyristoyl phosphatidylcholines. Arch. Biochem. Biophys. 165, 764–773 [DOI] [PubMed] [Google Scholar]
- 18. Cornish-Bowden A. (1984) Enzyme specificity: its meaning in the general case. J. Theor. Biol. 108, 451–457 [DOI] [PubMed] [Google Scholar]
- 19. Cornish-Bowden A. (2004) Fundamentals of Enzyme Kinetics, 3rd Ed., Portland Press, London [Google Scholar]
- 20. Tyurina Y. Y., Lou W., Qu F., Tyurin V. A., Mohammadyani D., Liu J., Hüttemann M., Frasso M. A., Wipf P., Bayir H., Greenberg M. L., and Kagan V. E. (2017) Lipidomics characterization of biosynthetic and remodeling pathways of cardiolipins in genetically and nutritionally manipulated yeast cells. ACS Chem. Biol. 12, 265–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schlame M. (2009) Formation of molecular species of mitochondrial cardiolipin 2. A mathematical model of pattern formation by phospholipid transacylation. Biochim. Biophys. Acta 1791, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaz F. M., Houtkooper R. H., Valianpour F., Barth P. G., and Wanders R. J. (2003) Only one splice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. J. Biol. Chem. 278, 43089–43094 [DOI] [PubMed] [Google Scholar]
- 23. Xu Y., Zhang S., Malhotra A., Edelman-Novemsky I., Ma J., Kruppa A., Cernicica C., Blais S., Neubert T. A., Ren M., and Schlame M. (2009) Characterization of tafazzin splice variants from humans and fruit flies. J. Biol. Chem. 284, 29230–29239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun G., Yang K., Zhao Z., Guan S., Han X., and Gross R. W. (2008) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80, 7576–7585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 26. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.