Abstract
DRB sensitivity-inducing factor (DSIF or Spt4/5) is a conserved transcription elongation factor that both inhibits and stimulates transcription elongation in metazoans. In Drosophila and vertebrates, DSIF together with negative elongation factor (NELF) associates with RNA polymerase II during early elongation and causes RNA polymerase II to pause in the promoter-proximal region of genes. The mechanism of how DSIF establishes pausing is not known. We constructed Spt5 mutant forms of DSIF and tested their capacity to restore promoter-proximal pausing to DSIF-depleted Drosophila nuclear extracts. The C-terminal repeat region of Spt5, which has been implicated in both inhibition and stimulation of elongation, is dispensable for promoter-proximal pausing. A region encompassing KOW4 and KOW5 of Spt5 is essential for pausing, and mutations in KOW5 specifically shift the location of the pause. RNA cross-linking analysis reveals that KOW5 directly contacts the nascent transcript, and deletion of KOW5 disrupts this interaction. Our results suggest that KOW5 is involved in promoter-proximal pausing through contact with the nascent RNA.
Keywords: mutagenesis, RNA polymerase II, RNA-protein interaction, transcription elongation factor, transcription regulation, DSIF, KOW domain, Spt4/5, promoter-proximal pausing
Introduction
The transcription cycle consists of initiation, elongation, and termination. In addition to these well defined stages of the transcription cycle, analyses of the distribution of Pol II2 genome-wide in vertebrates and Drosophila reveal that an additional stage in the transcription cycle occurs within the first 60 nucleotides of a gene after Pol II has initiated transcription (1–7). Pol II pauses in this region, and the duration of this pause can regulate the level of gene expression (8). The molecular mechanisms that initiate the pause and reactivate the paused Pol II are not understood. Three proteins appear to constitute the heart of this mechanism: DSIF, NELF, and P-TEFb. DSIF and NELF associate with the elongation complex sometime after it has transcribed at least 20 nucleotides and cause Pol II to pause (9, 10). Association of NELF depends on DSIF, and whereas DSIF can form a stable complex with the elongation complex in the absence of NELF, the two bind in a cooperative fashion (9). P-TEFb is a kinase that can phosphorylate DSIF (11, 12), NELF (13, 14), and the C-terminal domain (CTD) of Pol II (15). Although P-TEFb is considered essential for reactivating Pol II, it is not clear how each phosphorylation event contributes to pause release (16, 17). Phosphorylation of DSIF has been shown to change DSIF from an inhibitor to an activator of transcription elongation (11, 18), and it may be essential for the release of NELF (14). Both NELF-E and NELF-A can be phosphorylated by P-TEFb and are implicated in pause release at the HIV-LTR (13, 14). Phosphorylation of the Pol II CTD modulates the interactions of many factors with Pol II, but how these interactions regulate pause release is not known (19, 20). Moreover, a recent study indicated that P-TEFb-mediated Ser-2 phosphorylation of Pol II CTD is not essential for pause release on the majority of paused genes in mammalian cells (14).
Biochemical evidence indicates that DSIF plays a central role in promoter-proximal pausing (21, 22), but how it does so is not known. DSIF is composed of two subunits: Spt4 and Spt5. Spt5 and its homolog NusG are conserved across the three domains of life (23, 24). In eukaryotes and archaea, the NusG N-terminal (NGN) domain of Spt5 interacts with Spt4 to form a heterodimer (25–27). The crystal structure of archaeal RNAP clamp in complex with Spt4/5 suggests that the Spt5 NGN domain closes the RNAP active center cleft and renders the elongation complex stable and processive (27, 28). In support of this model, a recent structural study of mammalian Pol II-DSIF complex also places Spt4 and Spt5 NGN domain over the active center cleft (29). Biochemical studies showed that yeast Spt5 contributes to elongation by preventing Pol II arrest, and this involves contact between the non-transcribed strand of the transcription bubble and a conserved basic surface in the NGN domain of Spt5 (30). In eukaryotes, the NGN domain of Spt5 is followed by five KOW domains. Cross-linking analysis in Saccharomyces cerevisiae reveals extensive interactions between KOW4-5 domains of Spt5 and Rpb4/7 stalk of Pol II (31). Moreover, deletion of KOW4-5 impairs transcription elongation and derepresses transcription-coupled repair (31). Toward the C terminus of Spt5 is a serine-, threonine-, and proline-rich repeating region called the CTR, which is further divided into CTR1 and CTR2 based on repeating amino acid sequences (11, 32). CTR1 can be phosphorylated by P-TEFb at Thr-4 of the consensus repeat GS(R/Q)TP, and this phosphorylation is critical for DSIF-mediated stimulation of elongation (18). The Spt5 CTR also serves as a platform for the recruitment of factors involved in elongation and RNA processing, such as the Paf1 complex, mRNA capping enzyme, and pre-mRNA cleavage factor (33–36).
Efforts to identify regions of Spt5 involved in inhibiting elongation have yielded conflicting results. Two biochemical studies found that the Spt4-interacting NGN domain and the Pol II-interacting region downstream of KOW1 were important (11, 32). However, whereas one of these studies concluded that the CTR was dispensable for the repressive function (32), the other suggested that it was critical (11). Genetic screens have identified mutations in Spt5 that impair the repressive activity of DSIF. Single amino acid mutations close to the C terminus of Spt5 in zebrafish and Drosophila were found to cause developmental defects and impair the inhibitory activity of DSIF in in vitro transcription reactions (25, 37). None of these studies, however, directly analyzed the impact of these mutations on promoter-proximal pausing.
We set out to identify regions of Spt5 that are involved in promoter-proximal pausing. Given that Spt5 makes multiple contacts with Pol II and regulates the expression of many genes, we have taken a biochemical approach in an effort to focus on its function in pausing Pol II in the promoter-proximal region. Most biochemical assays measuring the inhibitory activity of DSIF have monitored the effects of DSIF on inhibiting transcription over distances of several hundred nucleotides. However, the relevance of this to promoter-proximal pausing is uncertain because promoter-proximal pausing typically occurs after Pol II has transcribed 20–60 nucleotides. We have succeeded in reconstituting promoter-proximal pausing on the hsp70 heat shock gene of Drosophila using nuclear extracts from Drosophila embryos (7). In this system, the location of the pause matches well with the location of the pause observed in vivo and recapitulates the DSIF, NELF, and GAGA factor dependence that has been observed in vivo (7). By measuring the pausing activity of mutant versions of Spt5 that have been co-expressed with Spt4 in bacteria and subsequently purified, we have determined that a region encompassing KOW4 and KOW5 of Spt5 contributes to the pausing activity, whereas the C-terminal region containing the CTR is dispensable for promoter-proximal pausing in vitro. In addition, we find that mutations in KOW5 shift Pol II downstream from the location where it normally pauses.
Results
In Vitro Transcription of hsp70 Reveals That the KOW4-KOW5 Region of Spt5 Participates in Promoter-proximal Pausing
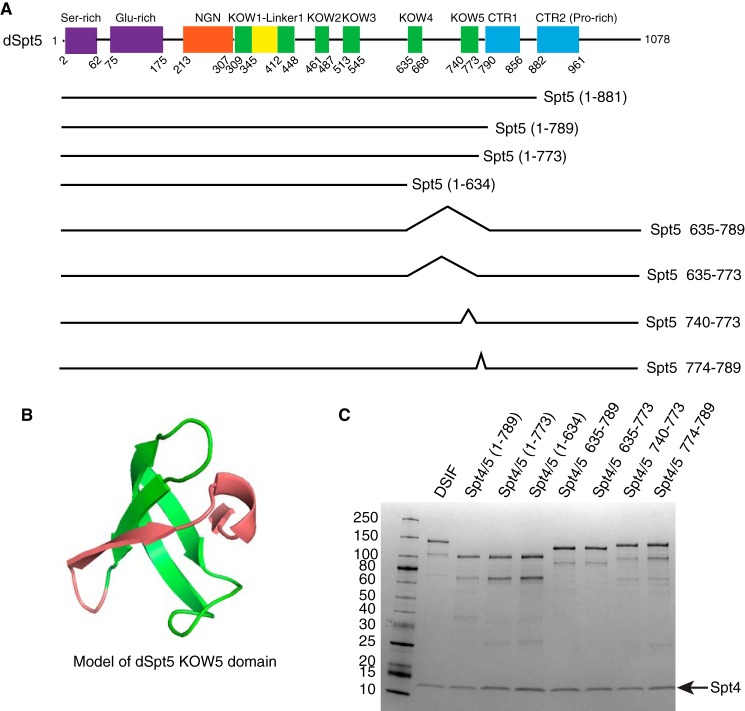
Sequence homology allowed us to identify domains in Drosophila Spt5 (dSpt5) that match domains previously noted in human Spt5 (hSpt5) (11, 32) (Fig. 1A). Modification of the domain structure was made to reflect the KOW1-Linker1 domain structure recently determined by crystallography of S. cerevisiae Spt5 (ySpt5) (38). In the schematic, KOW2–5 represent the conserved KOW motif that was initially identified in Nus and RL proteins (39). NMR structures of hSpt5 KOW2, KOW3, and KOW5 (PDB entries 2E6Z, 2DO3, and 2E70) as well as the crystal structure of ySpt5 KOW2–3 (38) indicate that the KOW motif forms a Tudor-like domain with two additional β strands. Modeling of the KOW5 domain of dSpt5 predicts a structure similar to KOW5 of hSpt5 (Fig. 1B). The three β strands shown in green represent the conserved KOW motif, which forms a broader KOW domain with two additional β strands shown in red. In this study, the two additional β strands are referred to as the “edge” of the KOW5 domain.
FIGURE 1.
Design of Spt5 mutants and purification of DSIF derivatives. A, diagram showing domains of Drosophila Spt5 and some of the Spt5 mutants. The region encompassing KOW4, KOW5, and the CTR were subjected to mutagenesis. B, the structure of dSpt5 KOW5 domain modeled by Phyre2 (59) using the NMR structure of hSpt5 KOW5 domain (PDB code 2E70) as a template, displayed in PyMOL (version 1.7.4, Schrödinger, LLC). The green part corresponds to the KOW motif spanning from amino acid 740 to 773. The red part corresponds to the “edge” of KOW5 spanning from amino acid 774 to 789. C, DSIF with different Spt5 derivatives was expressed in E. coli cells and purified through two affinity columns, binding Spt4-His and Spt5-FLAG subunits, respectively. The largest polypeptide corresponds to full-length Spt5, and the next largest polypeptide is an Spt5 breakdown product caused by loss of part of the N terminus. Purified proteins were analyzed on a 4–20% SDS-polyacrylamide gel and stained with Coomassie Blue.
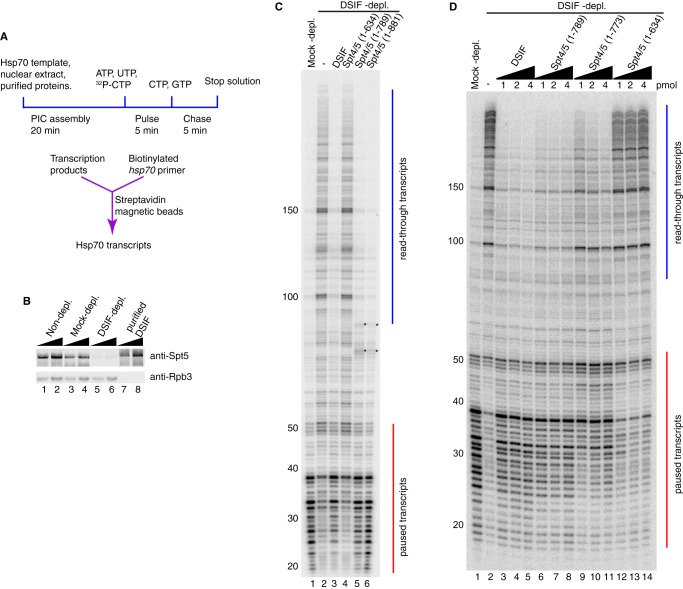
We utilized a polycistronic vector system (40) to co-express His6-tagged dSpt4 and various derivatives of FLAG-tagged dSpt5 in E. coli cells and purified DSIF through successive affinity columns (Fig. 1C). We then tested various DSIF derivatives for their capacity to restore promoter-proximal pausing at the hsp70 promoter in the DSIF-depleted nuclear extract (Fig. 2A). Drosophila embryo nuclear extracts that contain Pol II and general transcription factors (41) were either mock-depleted or DSIF-depleted by pre-immune antibody or Spt5 antibody. Western blotting analysis showed that the level of the Spt5 subunit was reduced >10-fold, whereas the level of the Rpb3 subunit of Pol II was unchanged (Fig. 2B, compare lanes 3 and 4 with lanes 5 and 6). Next, we determined whether the recombinant DSIF had pausing activity in the in vitro transcription reactions. Transcription from the hsp70 promoter in the mock-depleted nuclear extract produced transcripts corresponding primarily to pausing in the promoter-proximal region, with the strongest signals between +25 and +38 (Fig. 2C, lane 1). This is consistent with pausing previously observed at the hsp70 promoter in vitro and in vivo (7). For the DSIF-depleted nuclear extract, the paused transcripts proximal to the promoter were greatly diminished, whereas the read-through transcripts at +100, +150, and beyond were prevalent (Fig. 2C, lane 2). When an amount of recombinant DSIF comparable with what was depleted from the extract was added back to the DSIF-depleted nuclear extract, the paused transcripts were restored, and read-through transcripts diminished to the level of the mock-depleted nuclear extract (Fig. 2C, lane 3). Thus, the recombinant DSIF purified from E. coli has promoter-proximal pausing activity.
FIGURE 2.
In vitro transcription with different versions of DSIF reveals a region in Spt5 important for promoter-proximal pausing. A, diagram of the in vitro transcription assay. Normal and mutant DSIF proteins were added to nuclear extracts depleted of endogenous DSIF and then incubated with a plasmid containing the hsp70 promoter. Transcripts were radiolabeled by a pulse-chase procedure and purified by hybridizing to biotinylated hsp70 oligonucleotides that were then captured by streptavidin magnetic beads. Hsp70 transcripts were analyzed on a 10% polyacrylamide gel containing 8 m urea. B, Western blotting analysis of nuclear extracts and recombinant DSIF probed with antiserum against Spt5. Lanes 1–6 show two amounts each of non-depleted extract, mock-depleted extract, and DSIF-depleted extract, corresponding to one-eighth or one-fourth of nuclear extract used in each in vitro transcription reaction. Lanes 7 and 8 correspond to 0.125 and 0.25 pmol of recombinant DSIF. Rpb3 is a subunit of RNA polymerase II. C and D, DSIF with different Spt5 mutations was compared with normal DSIF for pausing activity. Red vertical lines from +20 to +50 indicate paused transcripts. Blue vertical lines from +100 and beyond indicate read-through transcripts. C, truncation mutants Spt4/5(1–881), Spt4/5(1–789), and Spt4/5(1–635) were first tested. Spt4/5(1–881) and Spt4/5(1–789) restored pausing similarly as normal DSIF. Spt4/5(1–635) failed to restore pausing. The asterisks in lanes 5 and 6 indicate contaminating radiolabeled nucleic acids generated by the extract independently of Pol II (7). The results shown in C are representative of three independent experiments. D, 1, 2, and 4 pmol of Spt4/5(1–789), Spt4/5(1–773), and Spt4/5(1–635) were further tested. The amount of protein used in each reaction is shown above each lane. Spt4/5(1–789) restored pausing. Spt4/5(1–635) failed to restore pausing. Spt4/5(1–773) partially restored pausing and shifted the pause sites downstream. The results shown in D are representative of two independent experiments.
To identify regions of Spt5 that are important for pausing, we initially generated C-terminally truncated versions of Spt5 and tested whether they were able to restore pausing in the DSIF-depleted nuclear extract. Previous mutations in CTR1 or CTR2 showed defects in transcription elongation, but the role of the CTR in promoter-proximal pausing was unclear (11, 32). Here we found that both the CTR2 deletion (residues 1–881) and the entire CTR deletion (residues 1–789) were able to restore pausing (Fig. 2C, lanes 5 and 6). This indicates that the CTR is not essential for promoter-proximal pausing. The bands marked by asterisks are contaminating RNAs from the nuclear extract that become radiolabeled independently from Pol II (7). We then removed an additional upstream region encompassing the KOW4 and KOW5 domains and tested whether this DSIF derivative Spt4/5(1–635) restored pausing. The paused transcripts were diminished, whereas the read-through transcripts were enhanced to the same level as in the DSIF-depleted extract (Fig. 2C, compare lane 4 with lane 2). Therefore, this additional deletion caused a nearly complete loss of pausing activity, suggesting that the region encompassing KOW4 and KOW5 is important for pausing.
We added increasing amounts of DSIF derivatives to determine whether any pausing activity could be detected. Normal DSIF was able to restore pausing at as low as 1 pmol, and adding 4-fold this amount did not alter the level of paused transcripts (Fig. 2D, lanes 3–5). The CTR deletion (residues 1–789) also restored pausing at 1 pmol, although there were slightly more read-through transcripts and slightly fewer paused transcripts (Fig. 2D, lanes 6–8). In contrast, the 1–634 mutant lacked pausing activity, even when 4-fold the amount sufficient for normal DSIF to restore pausing was added (Fig. 2D, lanes 12–14). To further define the region important for pausing, we deleted the edge of KOW5 in addition to the CTR. This deletion, Spt4/5(1–773), diminished the pausing activity (Fig. 2D, lanes 9–11). Because amino acids 774–789 are predicted to contribute to the folding of the KOW5 domain (Fig. 1B), the loss of pausing activity caused by the deletion of these amino acids suggests that KOW5 is involved in pausing.
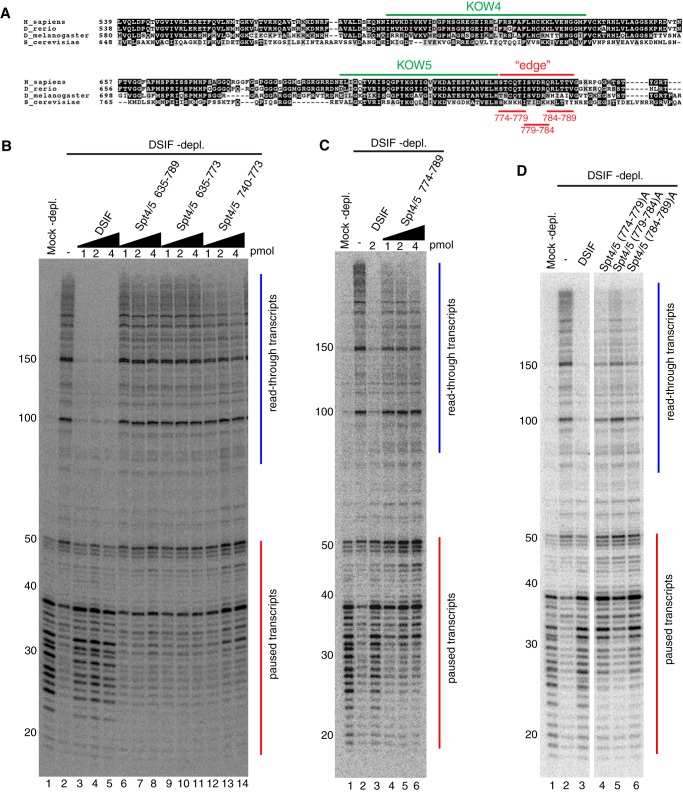
The KOW4 and KOW5 domains are conserved among eukaryotes. Alignment of the KOW4-5 region reveals striking similarity between Drosophila and vertebrates but some divergence between Drosophila and S. cerevisiae (Fig. 3A), suggesting that KOW4 and KOW5 of metazoan Spt5 may play a unique role in promoter-proximal pausing that is not seen in S. cerevisiae. Because some previous reports indicated that the CTR contributes to the inhibitory activity of Spt5 (11, 42), we tested the effects of internal deletions of KOW4 and KOW5 in the presence of the CTR to ascertain whether mutations in the KOW4-5 region alone would impair pausing. Deletions Δ635–789 and Δ635–773, both of which remove KOW4 and KOW5, caused complete loss of pausing activity (Fig. 3B, lanes 6–8 and lanes 9–11). In contrast, deletion of the KOW5 motif alone (Δ740–773) caused a partial loss of pausing activity (Fig. 3B, lanes 12–14). We attempted to construct a deletion of the KOW4 motif alone, but this was unsuccessful because the protein was poorly expressed. Our results from the internal deletions are in good agreement with the C-terminal truncations and identify the KOW4-5 region as essential for promoter-proximal pausing and KOW5 as contributing to promoter-proximal pausing.
FIGURE 3.
Mutations in KOW5 shift the pause site downstream. A, alignment of the amino acid sequences of Spt5 from four species was performed with Clustal Omega (60). The region encompassing KOW4 and KOW5 is shown. The similarity is displayed by BoxShade (version 3.21), with black or gray background indicating identical or similar residues, respectively. KOW4 and KOW5 represent the initially defined KOW motif. Additional amino acids that form a Tudor-like domain with the KOW5 motif are noted as the “edge” of the KOW5 domain. The alanine substitutions in the edge region are shown in red. B–D, in vitro transcription of hsp70 was done as described in the legend to Fig. 2. B, 1, 2, or 4 pmol of each protein was used as indicated above each lane. Δ635–789 and Δ635–773 failed to restore pausing with all three amounts. Δ740–773 partially restored pausing and shifted the pause location downstream. The addition of more protein increased the intensity of paused transcripts but did not shift them back to their normal location. C, 1, 2, or 4 pmol of Δ774–789 was used as indicated above each lane. 2 pmol of normal DSIF was used as a positive control. Δ774–789 partially restored pausing and shifted the pause sites downstream. The addition of more protein increased the intensity of paused transcripts but did not shift them back to the normal pause location. The results shown in B and C are representative of two independent experiments. D, three alanine substitution mutants in the edge of KOW5 (A) were tested along with normal DSIF for pausing activity. 2 pmol of each protein was used. All three mutants partially restored pausing and shifted the pause sites downstream. Mutant (779–784)A shows a more severe defect in pausing compared with the other two mutants. The results shown in D are representative of three independent experiments.
Mutations in KOW5 Cause the Location of Pausing to Shift Downstream
Close inspection of the pausing activity associated with the KOW5 motif deletion Spt4/5Δ740–773 (Fig. 3B, lanes 12–14) and KOW5 edge truncation Spt4/5(1–773) (Fig. 2D, lanes 9–11) reveals that the location where Pol II pauses was shifted downstream from where Pol II pauses when intact DSIF or the Spt4/5(1–789) mutant was added. This suggests that KOW5 influences the location of the pause. To investigate this further, we targeted mutations to the edge of KOW5. Deletion of the edge Δ774–789 reduced pausing activity and shifted the location of the pause downstream from the normal location, with the strongest pause signals between +32 and +50 (Fig. 3C, lanes 4–6). The addition of 4-fold the amount of Δ774–789 relative to the amount of normal DSIF sufficient for normal promoter-proximal pausing did not shift the pause back to its normal location. This suggests that the KOW5 edge deletion is not simply affecting pausing kinetically via delayed binding to the elongation complex, but via another mechanism, perhaps by altering the structure of the DISF-bound elongation complex.
Because of the multivalent nature of DSIF-Pol II elongation complex interactions (9, 28–31), we were concerned that this edge deletion might have affected adjacent Spt5 regions by changing the spacing between them. To address this, we constructed three alanine substitution mutations, (774–779)A, (779–784)A, and (784–789)A that each changed six consecutive amino acids in the edge region (Fig. 3A). All of the alanine mutants partially restored pausing but shifted the strongest pause sites downstream (Fig. 3D, lanes 4–6). Of the three mutants, (779–784)A exhibited the most severe defect in pausing activity. Therefore, it was later selected for analyses in vivo. Together, these results establish that the edge of KOW5 impacts the location of the pause.
Mutants Defective in Pausing Also Have Reduced Affinity for Elongation Complexes
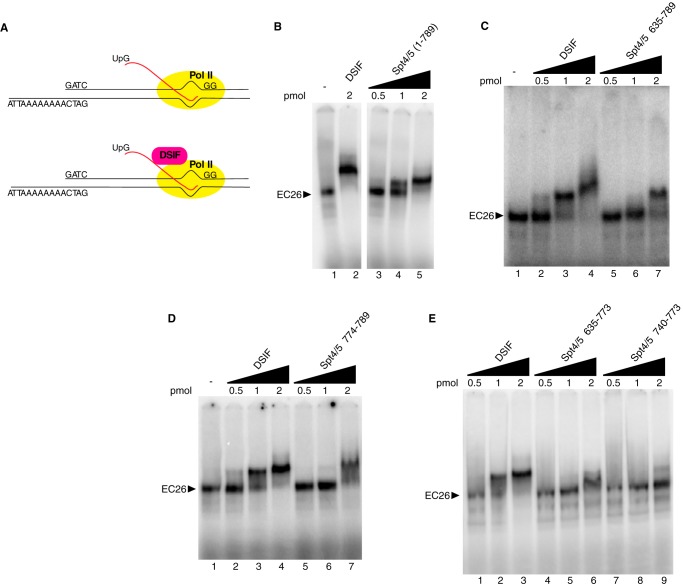
One way the mutations in Spt5 could be impacting the pausing activity of DSIF is by preventing DSIF from binding the Pol II elongation complex (EC). To test this, we used an electrophoretic mobility shift assay (EMSA) to measure binding of DSIF to reconstituted Pol II elongation complexes. We generated elongation complexes with 26-nucleotide-long nascent transcripts (EC26) by initiating transcription on a tailed template with purified Drosophila Pol II, UpG, and nucleotides. Pol II was stalled at the end of a G-less cassette by O-methyl-GTP (Fig. 4A). A previous study using Drosophila Pol II and the same type of tailed template showed that the RNA-DNA hybrid formed by the nascent transcript and the template strand was restricted to the transcription bubble (43). Normal or mutant DSIF was then added to reconstituted EC26, and the complexes were analyzed on 4% native polyacrylamide gels.
FIGURE 4.
DSIF mutants defective in pausing have lower binding affinity for elongation complexes. A, elongation complexes were formed by initiating transcription with purified Pol II, UpG, ATP, UTP, radioactive CTP, and O-methyl-GTP on a tailed template with a 26-nucleotide G-less cassette. After the complexes were stalled at the end of the G-less cassette, they were incubated with increasing amounts of purified normal or mutant DSIF proteins and then analyzed on 4% native gels. B–E, normal or mutant DSIF was added to the stalled EC at 0.5, 1, and 2 pmol to compare their affinity with EC26. DSIF shifted more than 50% of EC26 at 1 pmol and 100% of EC26 at 2 pmol. Spt4/5(1–789) showed similar affinity as DSIF. Spt4/5Δ635–789 did not shift EC26 at 1 pmol and partially shifted it at 2 pmol. Spt5Δ774–789 shifted most of EC26 at 2 pmol, forming an indistinct band. Spt5Δ635–773 and Spt5Δ740–773 partially shifted EC26 at 2 pmol. The results shown are representative of two independent experiments.
Normal DSIF started to shift EC26 at 0.5 pmol, with more than 50% shifted at 1 pmol and a complete shift at 2 pmol (Fig. 4, B (lane 2), C (lanes 2–4), D (lanes 2–4), and E (lanes 1–3)). The addition of CTR deletion mutant Spt4/5(1–789) caused a shift similar to normal DSIF, with a 50% shift at 1 pmol and a complete shift at 2 pmol (Fig. 4B, lanes 3–5). In accordance with its lack of an effect on pausing, the Spt5 CTR is not required for the stable association with the Pol II elongation complex.
Next we tested whether the DSIF mutants that are defective in pausing still associate with the Pol II elongation complex. The KOW4-5 deletion Spt4/5Δ635–789 was unable to shift EC26 at 0.5 or 1 pmol and resulted in a partial shift at 2 pmol (Fig. 4C, lanes 5–7), suggesting a decrease in affinity for EC26 compared with normal DSIF. The KOW5 edge deletion Spt4/5Δ774–789 did not shift EC26 at 0.5 pmol, started to shift at 1 pmol, and caused a nearly complete shift at 2 pmol (Fig. 4D, lanes 5–7). The shifted band is not as distinct as that resulting from the addition of normal DSIF, suggesting that its association with EC26 is less stable compared with normal DSIF. Spt4/5Δ635–773 and Spt4/5Δ740–773 showed no shift at 0.5 or 1 pmol and a partial shift at 2 pmol (Fig. 4E, lanes 4–9). These results indicate that mutations in the KOW4-5 region that cause defects in pausing also result in decreased affinity for the elongation complex.
UV-cross-linking Analysis Reveals That KOW5 Contacts the Nascent Transcript in the Elongation Complex
The results from EMSA indicate that the deletion of the region encompassing KOW4-5 diminished the binding of DSIF to the Pol II elongation complex but did not disrupt the association completely. This is consistent with previous findings that Spt5 makes multiple contacts with Pol II through its NGN and various KOW domains (11, 28–31), and these contacts could persist even when the KOW4-5 region was removed. Although the KOW5 edge deletion reduced the affinity of the mutant for the EC, sufficient binding remained that we were prompted to use protein-RNA cross-linking to see if we could identify contacts that this mutation might be disrupting. UV cross-linking previously showed that Spt5 cross-links to the nascent transcript as it first emerges from the elongation complex (9). Here we investigated whether the KOW5 edge deletion mutant is competent to bind the nascent RNA. EC26 was assembled as described above except that UTP was replaced with photoreactive bromo-UTP. After the addition of Spt4/5, samples were subjected to UV irradiation to induce cross-links between protein and radiolabeled RNA. The reaction mixes were then digested with nucleases and subjected to SDS-PAGE.
When no Spt4/5 protein was added, the most prominent cross-linked bands were Rpb1 at 245 kDa and Rpb2 at 135 kDa (Fig. 5A, lane 1). These two bands were seen in all lanes. When DSIF was added, the appearance of a band slightly above Rpb2 indicated cross-linking of RNA to full-length Spt5 (Fig. 5A, lane 2, red dot). A second band corresponding to a breakdown product of Spt5 was also observed (Fig. 5A, lane 2, orange dot). As expected, CTR-deleted Spt5(1–789) also cross-linked to nascent RNA, with the full-length band at around 100 kDa and the breakdown product slightly below 80 kDa (Fig. 5A, lane 5, green and blue dots, respectively). When KOW5 edge deletion Δ774–789 was added, only Rpb1 and Rpb2 cross-links were detected. No cross-linking of RNA to Spt5 was observed (Fig. 5A, lane 3). Similarly, no cross-linking was detected for the other KOW5 mutant, Δ740–773 (Fig. 5A, lane 4). These results suggest that KOW5 is important for the contact between Spt5 and nascent RNA, and deletion of the KOW5 edge is sufficient to abrogate cross-linking.
FIGURE 5.
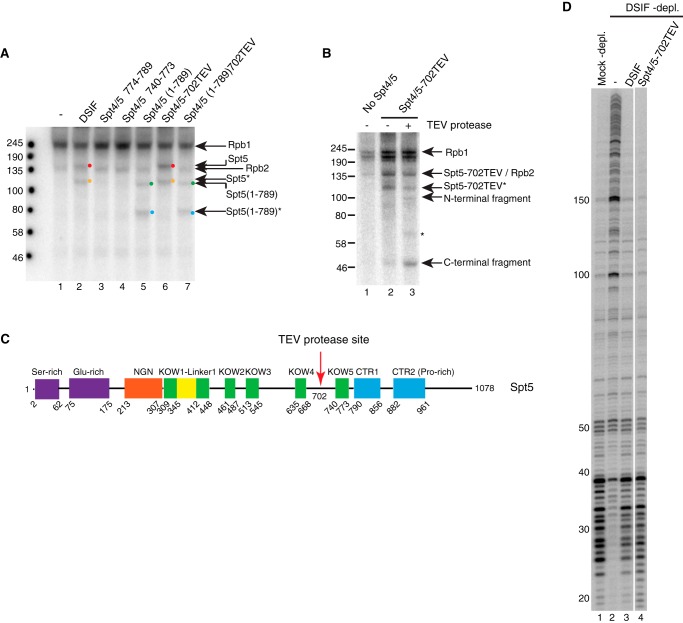
UV cross-linking suggests that KOW5 is important for the contact with the nascent RNA. Elongation complexes were generated as described in Fig. 4 except that UTP was replaced with photoreactive bromo-UTP. After UV treatment, the samples were digested with DNase I and RNase A. Proteins were analyzed by 7% Tris acetate PAGE. A, UV cross-linking in the absence (lane 1) or presence of various Spt4/5 derivatives (lanes 2–7). Cross-linked Rpb1 (245 kDa) and Rpb2 (135 kDa) are detected in every lane. The addition of DSIF results in a cross-linked band just above Rpb2, corresponding to full-length Spt5 (lane 2, red dot). The addition of Spt4/5(1–789) results in a cross-linked band at 100 kDa (lane 5, green dot). The addition of mutant Δ774–789 or Δ740–773 did not result in cross-linked Spt5 (lanes 3 and 4). The orange dot in lane 2 and blue dot in lane 5 identify the breakdown products of Spt5 derivatives that are evident in Fig. 1C. Spt5–702TEV and Spt5(1–789)702TEV cross-linked in the same way as the corresponding proteins lacking the TEV site (compare lanes 6 and 7 with lanes 2 and 5). B, TEV digestion of cross-linked Spt5–702TEV reveals cross-linking of RNA on both sides of the 702TEV site. Proteins were digested with TEV protease after UV cross-linking and before nuclease treatment. Lanes 2 and 3 show the cross-linked products before and after TEV digestion, respectively. After digestion, two bands at 100 kDa and around 46 kDa corresponding to predicted N-terminal and C-terminal fragments of Spt5–702TEV were detected (lane 3). The band between 58 and 80 kDa in lane 3 (asterisk) is likely to be the N-terminal fragment from the breakdown product marked as Spt5–702TEV*. C, schematic showing the location of a TEV protease site inserted between KOW4 and KOW5 domains in full-length Spt5. D, in vitro transcription assay indicates that Spt4/5–702TEV has pausing activity similar to that of normal DSIF.
The loss of contact to RNA resulting from mutations in KOW5 could be caused by either a direct loss of the contact point or an allosteric change to a different region of Spt5 that contacts RNA. To determine whether KOW5 directly contacts nascent RNA, we introduced a TEV protease site into Spt5 to map the regions of Spt5 that cross-link to the nascent RNA. A single TEV site was placed between KOW4 and KOW5, after amino acid 702 (Fig. 5C). The insertion of a TEV site at this position did not affect the function of Spt4/5 in pausing because Spt4/5–702TEV fully restored pausing to the DSIF-depleted nuclear extract (Fig. 5D, lane 4). After UV cross-linking, TEV protease was added to cut Spt5 into two parts (Fig. 5C). Both the N-terminal and C-terminal fragments cross-linked to RNA (Fig. 5B, lane 3), suggesting that both the KOW4 and KOW5 sides contact the nascent transcripts. The band between 58 and 80 kDa (marked by an asterisk) in lane 3 of Fig. 5B is likely to be the N-terminal part of the breakdown component marked as Spt5–702TEV* in Fig. 5B.
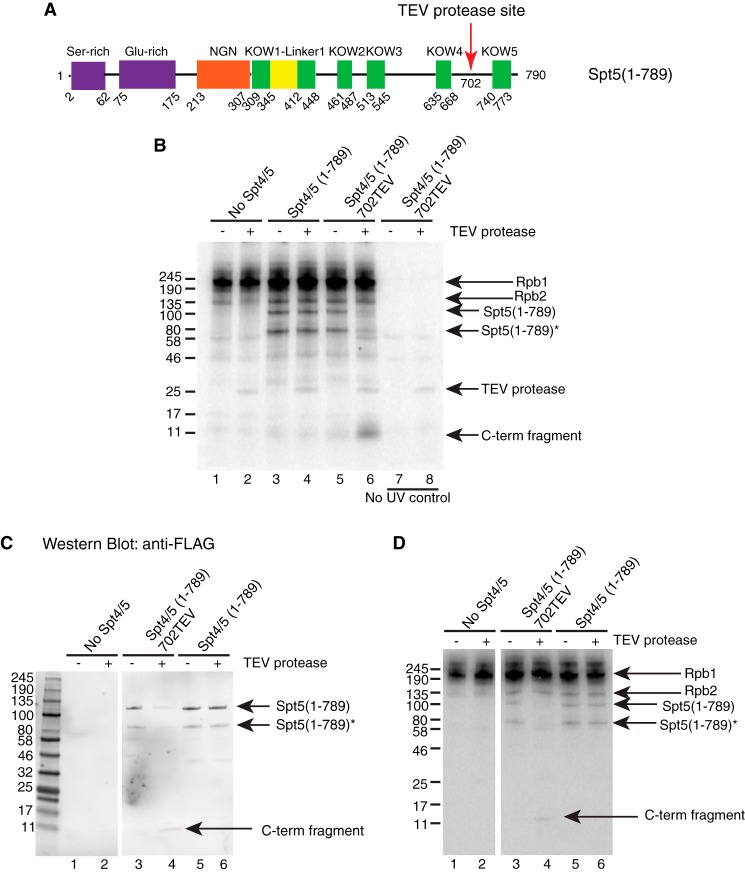
The KOW4 side of the TEV site contains several KOW domains as well as the NGN domain, whereas the KOW5 side contains only KOW5 and the CTR. To narrow down where contact was being made on the KOW5 side of the TEV site, we performed cross-linking with an Spt5 derivative that had a TEV site between KOW4 and KOW5 but lacked the CTR (Fig. 6A). After TEV digestion, the predicted C-terminal fragment of 10 kDa from Spt5(1–789)702TEV was evident (Fig. 6B, lanes 5 and 6). The 10-kDa fragment was absent in the sample without UV treatment (Fig. 6B, lane 8), indicating that this signal was indeed from cross-linked RNA. TEV digestion of Spt5(1–789) that lacked the TEV site did not generate the same fragment (Fig. 6B, compare lanes 4 and 6), suggesting that it was a specific product from TEV digestion. Western blotting analysis for the FLAG tag on the C terminus of Spt5 derivatives confirmed that the radioactively tagged 10-kDa fragment was FLAG-tagged (Fig. 6, C (lane 4) and D (lane 4)). These results indicate that the 10-kDa fragment cross-linked to nascent RNA was the C-terminal product from Spt5(1–789)702TEV. Therefore, KOW5 directly contacts nascent RNA in the elongation complex.
FIGURE 6.
UV cross-linking indicates that KOW5 directly contacts with the nascent RNA. A UV-cross-linking assay was performed as described in the legend to Fig. 5. For TEV-digested samples, TEV protease was added 1 h before nuclease treatment. Proteins were analyzed by 4–20% SDS-PAGE. A, diagram showing the location of a TEV protease site inserted between the KOW4 and KOW5 domains in CTR-deleted Spt5. B, TEV digestion following UV cross-linking of the CTR-deleted Spt5 with the 702TEV site. Before TEV digestion, cross-linked Spt5(1–789) and its breakdown product were detected in lanes 3 and 5. After digestion, a ∼10 kDa band corresponding to the predicted C-terminal fragment of Spt5(1–789)702TEV was detected in lane 6 but not in lane 4. The 10 kDa band was not detected in samples without UV treatment (lanes 7 and 8). It is not known why the TEV protease is labeled with radioactivity even in the absence of UV treatment. C and D, after UV cross-linking and TEV digestion, the products were separated on a 4–20% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane for Western blotting. The blot was probed with FLAG antibody and later exposed to a phosphor screen to detect radiolabeled proteins. C, Western blotting image showing that the 10-kDa digestion fragment, as well as the full-length and breakdown versions of Spt5(1–789), is FLAG-tagged. D, phosphor image showing proteins that are cross-linked to radiolabeled RNA.
Mutations in KOW4-5 Region Impair Spt5 Function in Vivo
To determine whether mutations that affected pausing in vitro also impaired Spt5 function in vivo, we generated transgenic flies that express normal or mutant forms of FLAG-tagged Spt5 under the control of a Gal4-driven promoter. Three mutants, the KOW4-5 deletion Δ635–789, the KOW5 edge deletion Δ774–789, and the KOW5 edge mutation (779–784)A, were selected. The transgenes encoding these ectopically expressed versions of Spt5 were modified with synonymous mutations so that they would be resistant to an Spt5 RNAi that is also under the control of a Gal4-driven promoter. Expression of transgenes was driven by mating the transgenic fly lines to an actin-Gal4 driver line, BDSC 4414 (Fig. 7A), or a salivary gland Gal4 driver line, BDSC 1824 (Fig. 7B), and analyzed by Western blotting. BDSC 4414 or BDSC 1824 alone was used as a negative control. The levels of ectopically expressed Spt5 were determined by a FLAG antibody (Fig. 7, A and B). Comparable FLAG signals were detected for each Spt5 derivative line but not the control lines (Fig. 7 (A and B), compare lanes 2–5 with lane 1). The levels of total Spt5 were determined by an Spt5 antibody (Fig. 7, A and B). The total Spt5 signal of Spt5 derivative lines were comparable with each other and much stronger than that of the control lines (Fig. 7 (A and B), compare lanes 2–5 with lane 1). These results indicate that the levels of ectopically expressed Spt5 derivatives were similar to each other but higher than that of endogenous Spt5. Despite being highly expressed, none of the mutants exhibited dominant negative effects when expressed ubiquitously with an actin-Gal4 driver (BDSC 4414) or in salivary glands with a salivary gland Gal4 driver (BDSC 1824).
FIGURE 7.
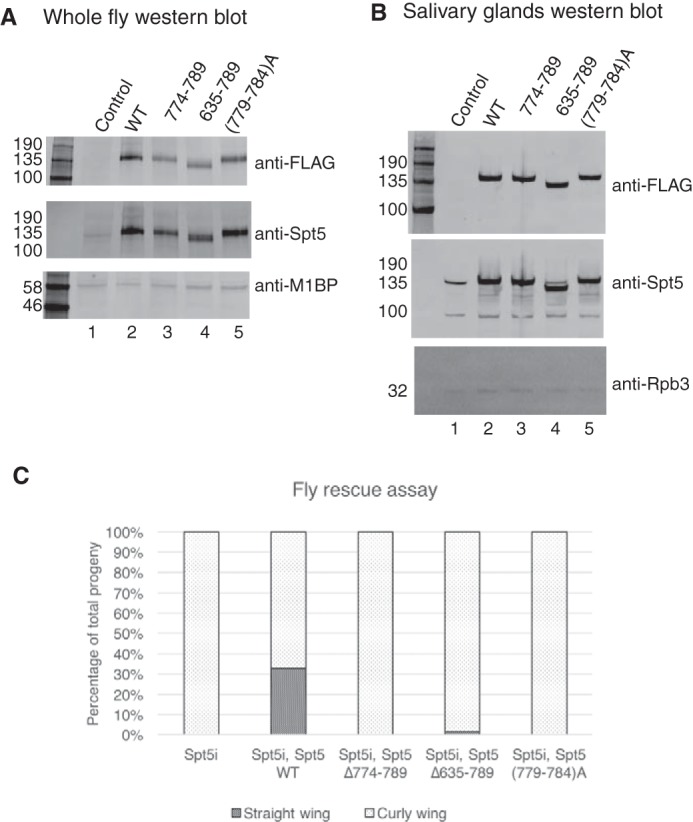
Mutations in KOW4-5 region of Spt5 fail to rescue the lethality caused by Spt5 RNAi in flies. A, Western blotting analysis for ubiquitous expression of Spt5 derivatives in flies. Whole fly samples were prepared from pupae of the matings between Spt5 derivative lines and an actin-Gal4 driver line (BDSC 4414). BDSC 4414 alone was used as a negative control that does not ectopically express Spt5 derivatives. FLAG antibody detects ectopically expressed Spt5 derivatives. Spt5 antibody detects both endogenous and ectopically expressed Spt5. M1BP antibody was used to detect M1BP as a loading control. B, Western blotting analysis for expression of Spt5 derivatives in salivary glands. Glands were isolated from third instar larvae of the matings between Spt5 derivative lines and a salivary gland Gal4 driver line (BDSC 1824). BDSC 1824 alone was used as a negative control. Rpb3 antibody was used to detect Rpb3 as a loading control. C, fly rescue assay for Spt5 derivatives. Spt5 lines with a Gal4-regulated Spt5 RNAi transgene or the Spt5 RNAi line alone were mated to actin-Gal4/CyO (BDSC 4414). In total, 158, 258, 157, 136, and 136 adult flies were generated from Spt5 RNAi, Spt5 WT, Spt5 Δ774–789, Spt5 Δ635–789, and Spt5 (779–784)A, respectively. The percentage of straight wing flies and curly wing flies in the progeny were calculated. Spt5 WT generated more than 30% of straight wing flies. Spt5 Δ635–789 generated less than 1.5% straight wing flies. Spt5 RNAi, Spt5 Δ774–789, and Spt5 (779–784)A generated no straight wing flies.
We recombined our various Gal4-regulated Spt5 transgenes with the Gal4-regulated Spt5 RNAi and then tested whether the expression of wild type or mutant Spt5 could rescue the lethality caused by Spt5 RNAi. The recombinants were mated to actin-Gal4/CyO flies, which ubiquitously express Gal4. In the progeny, flies carrying actin-Gal4 and thus expressing the Spt5 RNAi and Spt5 transgene have straight wings, whereas those carrying CyO and thus not expressing the Spt5 RNAi and the Spt5 transgene have curly wings. Gal4-driven expression of Spt5 RNAi alone caused lethality (Fig. 7C, Spt5i). Gal4-driven expression of wild type Spt5 was able to rescue the lethality caused by RNAi, generating both straight wing flies and curly wing flies (Fig. 7C, Spt5i and Spt5 WT). In contrast, the mutant forms of Spt5 were unable to rescue, generating no or only a few straight wing flies (Fig. 7C). Therefore, mutations in the KOW4-5 region not only impair pausing in vitro but also disrupt normal development of flies.
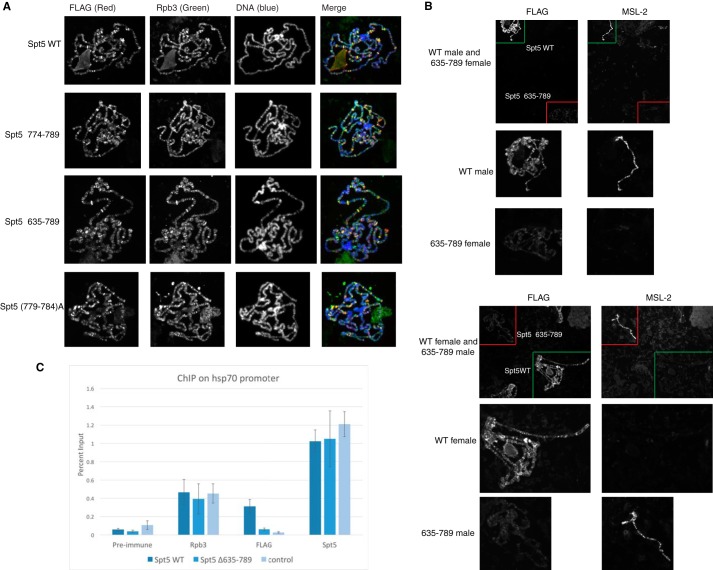
Spt5 has been shown to associate with polytene chromosomes and co-localize with Pol II on many genes (44, 45). Our results showed that mutations in the KOW4-5 region caused reduced binding of DSIF to Pol II elongation complexes in vitro. We then tested whether the mutations also affected the association with chromosomes in vivo. Fly lines expressing normal or mutant Spt5 were mated with a line expressing Gal4 in salivary glands. Polytene chromosomes from salivary glands were prepared and stained with FLAG antibody that detects ectopically expressed Spt5 as well as Rpb3 antibody that detects Pol II. We first stained chromosomes from each line individually. Both normal and mutant forms of Spt5 showed FLAG signal that co-localized with Rpb3 signal, suggesting that ectopically expressed Spt5 derivatives associate with chromosomes at sites occupied by Pol II (Fig. 8A). However, to achieve the similar image intensities shown in Fig. 8A, the KOW4-5 deletion Δ635–789 requires a longer exposure time than wild type, whereas the two KOW5 mutants require an exposure time similar to wild type. This suggests that the KOW4-5 deletion Δ635–789 has a weaker association with the chromosome compared with wild type Spt5. To make a direct comparison, we prepared polytene chromosomes from wild type and Δ635–789 on the same slide. To distinguish the two sets of chromosomes, the male larvae of one line and the female larvae of the other line were used and stained with an antibody against male-specific lethal-2 (MSL-2), which preferentially stains the X chromosomes of the male (Fig. 8B). The direct comparison showed a clear difference; the FLAG signal of Δ635–789 was significantly weaker than that of WT, regardless of whether male or female larvae of each line were analyzed (Fig. 8B). These results indicate that Spt5 lacking the KOW4-5 region associates with chromosomes more weakly than normal Spt5 despite being expressed at comparable levels (Fig. 7B). In contrast, the mutations that targeted KOW5 alone appeared to associate with chromosomes at levels similar to normal Spt5.
FIGURE 8.
KOW4-5 deletion causes reduced affinity of Spt5 for chromosomes. A, immunofluorescence micrographs of polytene chromosomes of larvae from normal and mutant Spt5 lines mated to the salivary gland driver line BDSC 1824. Signal from FLAG staining is shown in the red channel. Signal from Rpb3 staining is shown in the green channel. DNA staining is shown in the blue channel. All four Spt5 derivatives associated with chromosomes and co-localized with Pol II at many loci. The exposure time was optimized for each channel in each image. B, direct comparison between Spt5 WT and Spt5Δ635–789 on the same slide. MSL-2 staining for the X chromosome of male larvae was used to distinguish males containing one Spt5 transgene and females containing the other Spt5 transgene. A field with chromosomes from both lines was selected for the pictures in the first row. The Spt5 WT and Spt5Δ635–789 chromosomes are outlined with green and red boxes, respectively. Magnified views of the chromosomes in the first row are displayed in the second and third rows. Spt5Δ635–789 showed significantly weaker FLAG signal than Spt5 WT regardless of sex. C, ChIP analysis on the hsp70 promoter. Spt5 WT and Spt5Δ635–789 were mated to salivary gland Gal4 driver line BDSC1824, and glands were prepared for ChIP with FLAG, Rpb3, and Spt5 antibodies. Glands from the BDSC1824 line, which does not express FLAG-tagged protein, was used as a negative control. All three lines showed similar enrichment for Rpb3 and total Spt5. The ChIP signal for FLAG-Spt5 WT was well above background or pre-immune control, whereas the ChIP signal for FLAG-Spt5Δ635–789 was near background. Three or more biological replicates were used for each ChIP experiment. Error bars, S.E.
The diminished association of Spt5Δ635–789 with polytene chromosomes appears to be genome-wide. To determine whether this deletion affected the association of Spt5 with the hsp70 promoter used in our in vitro pausing assay, we monitored this association in salivary glands using chromatin immunoprecipitation (ChIP). The FLAG ChIP signal for FLAG-Spt5 WT was well above the pre-immune control or the background signal detected in glands that were not expressing FLAG-tagged protein (Fig. 8C). In contrast, the FLAG ChIP signal for FLAG-Spt5 Δ635–789 was near background. This loss of signal for FLAG-Spt5Δ635–789 was not due to poor sample preparation because the ChIP signals for an Rpb3 antibody or an antibody that recognizes both the endogenous and ectopically expressed forms of Spt5 were comparable among the progeny from the three matings. Note that these salivary glands were not expressing Spt5 RNAi because Spt5Δ635–789 failed to rescue the halted larvae development and the loss of glands caused by the Spt5 RNAi. Hence, although Spt5Δ635–789 is expressed at levels greater than the endogenous Spt5, it does not affect the level of endogenous Spt5 associating with the hsp70 promoter.
To summarize, the deletion of KOW4-5 not only disrupted pausing and interaction with the Pol II elongation complex in vitro; it also impaired the association of Spt5 with the hsp70 promoter and with many other sites on the chromosomes. Although there are multiple contacts between Spt5 and Pol II, the KOW4-5 region is essential for keeping Spt5 stably associated with the Pol II elongation complex.
Discussion
Promoter-proximal pausing has emerged as a requisite intermediate in the transcription cycle for most and possibly all protein-coding genes in Drosophila and vertebrates, but the underlying mechanisms are not known. An important step toward elucidating these mechanisms is identifying regions of proteins involved in causing promoter-proximal pausing and assessing how these regions might contribute to pausing. Here we have determined that a 155-amino acid region of Spt5 that encompasses KOW4 and 5 is essential for promoter-proximal pausing in Drosophila nuclear extracts. Within this 155-amino acid region is a 16-amino acid region at the edge of KOW5 that when mutated diminishes the pausing activity of Spt5 and shifts the location where Pol II pauses slightly downstream from where it normally pauses. Additionally, deletion of the 155-amino acid region or mutations within the 16-amino acid region impair the function of Spt5 in flies. Using UV cross-linking, we have determined that KOW5 is in intimate contact with the nascent transcript in reconstituted elongation complexes.
We also found that the entire C-terminal region, which has been implicated in both inhibitory and stimulatory elongation activities of Spt5 (11, 18, 32, 42), is not essential for promoter-proximal pausing. Our finding is at odds with two reports showing that CTR deletions result in loss of the inhibitory activity of vertebrate Spt4/5 (11, 42). However, the in vitro transcription assays in these previous studies monitored transcription over several hundred nucleotides, whereas we focused on the promoter-proximal region 20–60 nucleotides from the transcription start site. Besides, these previous studies placed the deletion end point immediately adjacent to the edge of KOW5 corresponding to our 789 breakpoint but did not include a C-terminal tag as we did. It is possible that the deletions that were intended to only remove the CTR of Spt5 also disrupted KOW5. In support of this possibility is the finding that an internal CTR deletion with the breakpoint located 5 amino acids further in the C-terminal direction from the KOW5 edge does not impair the inhibitory activity of Spt4/5 (46). Two closely positioned point mutations in the CTR have been found to impair the capacity of Spt4/5 to inhibit elongation over several hundred nucleotides, but the effect of these mutations on promoter-proximal pausing was not tested (25, 37). Nevertheless, these mutants raise the possibility that factors interacting with this region in vivo might regulate the inhibitory activity of Spt4/5.
Our finding that mutations in the KOW4-5 region reduce the binding affinity of Spt4/5 to the elongation complex is in excellent agreement with what has been observed for yeast Spt4/5 and the yeast Pol II elongation complex (30, 31). This dependence on KOW4 and -5 for EC binding provides a simple explanation for why this deletion completely eliminates the pausing activity of Spt4/5 in our assay. In contrast, several different mutations in KOW5 partially lose pausing activity but also have the intriguing effect of shifting the location of the pause downstream by 8–10 nucleotides. Our cross-linking analysis shows that KOW5 contacts the nascent transcript and that a small deletion at the edge of KOW5 results in a loss of this cross-linking. We propose that contact between KOW5 and the nascent transcript plays an important role in dictating both the efficiency and location of the pause. The decrease in efficiency caused by mutating KOW5 could be due to a decrease in affinity of Spt4/5 for the EC, as evidenced by our gel shift assay. Because pausing is affected by the kinetic competition between elongation and the association of pausing factors (7), it is possible that the downstream shift in the location of the pause is due to a decrease in the rate at which Spt4/5 associates with elongation complex. Alternatively, contact between KOW5 and the nascent transcript could impact the path of the RNA or the architecture of the elongation complex, and this in turn could affect where the Pol II pauses.
Protein-protein cross-linking studies show that KOW4 and -5 are located near the base of the Rpb4/7 stalk that projects from the body of Pol II (31), and RNA cross-linking shows that the nascent transcript contacts Rpb7 when the nascent transcript reaches a length of 24 nucleotides (47). One possibility could be that a network of RNA interactions with Rpb7, KOW5, and possibly KOW4 helps to restrain the extrusion of the RNA from the elongation complex and thus inhibits the forward translocation of the Pol II. Structural studies of RNA Pol II suggest that Rpb4/7 forms a wedge on the Pol II core and restricts the Pol II clamp to a closed conformation (48–50). A recent FRET analysis shows that the association of Spt4/5 with the elongation complex also affects the conformation of the RNAP clamp (51). Because the KOW4 and -5 domains of Spt5 are located near where Rpb4/7 interacts with the Pol II core (31), an alternative possibility could be that the contact with nascent RNA by KOW4-5 triggers conformational changes of the Pol II clamp in a way that promotes pausing.
The conformational changes induced by Spt4/5 might affect how NELF interacts with the elongation complex. Previously, we showed that immunodepletion of NELF from our nuclear extract impairs pausing, and this can be corrected by adding back purified NELF (7). Early studies have implicated an RNA recognition motif in the NELF-E subunit in repressing elongation (46), but we were unable to detect cross-linking of NELF subunits to the nascent transcript in EC22, EC27, or EC31 (9). Hence, the role of NELF in pausing could involve contacts elsewhere on Pol II or Spt4/5. Perhaps the basic elements of promoter-proximal pausing are provided by Spt4/5, because it was recently discovered that Spt4-dependent promoter-proximal pausing occurs in Schizosaccharomyces pombe, and S. pombe lacks NELF (52). NELF may have evolved in metazoans to provide regulatory mechanisms needed for the complex patterns of gene regulation that contribute to development.
In this study, our attention was directed at KOW4 and -5 as a consequence of systematically deleting regions from the C terminus of Spt5. Analysis of additional mutations in Spt5 and extension of this approach to the four subunits of NELF should provide important insights into the pausing mechanism. A biochemical approach is essential because of the pleiotropic effects that are likely to accompany mutagenesis studies in vivo. Ultimately, the results of the functional analysis of mutants will mesh with advances toward determining the structure of the elongation complex in association with pausing factors (29, 53) to yield a fuller understanding of the mechanistic basis for promoter-proximal pausing.
Experimental Procedures
Purification of Recombinant DSIF
The coding sequences of DSIF subunits, Spt4 and Spt5, were subcloned into the polycistronic vector pST44 (40) with a His6 tag at the C terminus of Spt4 and a FLAG tag at the C terminus of Spt5. Spt5 mutants were generated by Phusion site-directed mutagenesis (ThermoFisher). Sequence-confirmed plasmids were transformed into E. coli strain Rosetta (DE3) pLysS for expression of both subunits. For each protein, 2 liters of cell culture was grown at 37 °C until A600 reached 0.6–0.8 and then induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C overnight. Harvested cells were resuspended in 40 ml of TBS150 (50 mm Tris-HCl, 150 mm NaCl, 10% glycerol, pH 7.5). After freeze-thaw, the lysate was sonicated and cleared by centrifugation at 20,000 × g for 20 min (Sorvall RC-5B). The supernatant was incubated with Talon metal affinity resin (Clontech) and washed with TBS300 (50 mm Tris-HCl, 300 mm NaCl, 10% glycerol, pH 7.5) containing up to 5 mm imidazole. The proteins were eluted with 150 mm imidazole in TBS300 followed by 200 mm imidazole in TBS300. The eluates were combined and further purified with anti-FLAG M2 affinity resin (Sigma-Aldrich) or anti-DYKDDDDK G1 affinity resin (Genscript). Proteins were incubated with the resin, washed first with TBS300 and then with TBS150, and finally eluted with TBS150 containing 100 μg/μl 3×FLAG peptide (Sigma-Aldrich). Purified DSIF with various forms of Spt5 was analyzed on 4–20% SDS-polyacrylamide gels (Bio-Rad). The concentration of DSIF was determined by comparing Coomassie Blue staining intensity of the full-length Spt5 with that of BSA standards.
Preparation of Drosophila Embryo Nuclear Extracts
Drosophila embryos were collected every 12–14 h and stored at 4 °C for up to 72 h. Nuclear extracts were prepared as described (41) with modifications. The extracts were dialyzed against 40 mm KCl-HEMG (25 mm HEPES, 12.5 mm MgCl2, 0.1 mm EDTA, 10% glycerol, pH 7.6), supplemented with 1 mm DTT, 0.5 mm sodium bisulfite, and 0.1 mm PMSF, until the conductivity was equivalent to that of 150 mm KCl-HEMG (7). The nuclear extracts were then immunodepleted by pre-immune serum or anti-Spt5 serum-conjugated protein A-Sepharose beads as described previously (22). 500-μl portions of nuclear extracts were depleted two times for 2 h each time at 4 °C with 100 μl of antibody beads.
Promoter-proximal Pausing in Drosophila Nuclear Extracts
In vitro transcription was done using a pulse-chase labeling strategy as described previously (7) with modifications. Briefly, 14 μl of reaction pre-mix containing 20 mm HEPES (pH 7.5), 1 mm DTT, 100 ng of DNA template with hsp70 promoter, 2 μg of HaeIII-cut E. coli DNA, and 0.8 units/μl RNasin (Promega) was incubated with 16 μl of nuclear extract containing 0.75 μm flavopiridol and 4 μl of purified protein or TBS150 at room temperature (21–22 °C) for 20 min. Then a 2-μl solution containing 2 mm ATP, 2 mm UTP, and 2 μl of [α-32P]CTP (6000 Ci/mmol, 10 μCi/μl) was added to each sample for the pulse. After 5 min, 2 μl of solution containing 2 mm CTP and 2 mm GTP was added for the chase. The reaction was stopped 5 min later by 200 μl of stop solution (20 mm EDTA, pH 8.0, 500 mm NaCl, 1% SDS, 0.25 mg/ml Torula yeast RNA, and 0.1 mg/ml Proteinase K). After incubating at room temperature for 5 min, the reaction mix was extracted with phenol/chloroform/isoamyl alcohol (25:24:1). 100 μl of solution containing 30 mm Tris-HCl (pH 7.5), 500 mm NaCl, and 1.6 pmol of biotinylated oligonucleotide complementary to the hsp70 transcript from +1 to +44 was added to each sample for overnight incubation at room temperature. Then 150 μg of washed Dynabeads M-280 Streptavidin (Invitrogen) beads were added to each sample to isolate the hsp70 transcripts. After washing the beads twice with 300 μl of wash buffer (10 mm NaCl, 10 mm Tris-HCl (pH 7.5), 5 mm EDTA, 0.5 mg/ml yeast tRNA), the transcripts were eluted in 15 μl of sequencing gel loading buffer (98% deionized formamide, 10 mm EDTA (pH 8.0), 0.025% xylene cyanol FF, and 0.025% bromphenol blue) at 95 °C for 5 min and then analyzed on a 10% polyacrylamide gel containing 8 m urea.
Generation of Stalled Elongation Complexes and Binding of Proteins
The DNA template was assembled by annealing two chemically synthesized oligonucleotides containing a 25-nucleotide G-less cassette followed by a 24-nucleotide sequence containing all four nucleotides. The bottom strand contains an 11-nucleotide 3′ overhang serving as an initiation site for Pol II in the presence of dinucleotide UpG (Sigma-Aldrich or Tri-link). The double-stranded DNA template was purified from a 10% native polyacrylamide gel. The sequences of the oligonucleotides are as follows: EC26 top strand, 5′-GATCTTCATTTCTCATTCCACTCCCGGATCCTCTAGAGTCGACCTGCAG-3′; EC26 bottom strand, 3′-ATTAAAAAAAACTAGAAGTAAAGAGTAAGGTGAGGGCCTAGGAGATCTCAGCTGGACGTC-5′.
Elongation complexes were generated as described previously (9, 43). Briefly, 15 μl of transcription premix containing 50 mm HEPES (pH 7.5), 200 mm KCl, 1 mm MnCl2, 12% glycerol, 0.5 mm DTT, 0.5 mm UpG, 20 units of RNasin (Promega), and 100 ng of template was incubated with ∼100 ng of Drosophila Pol II (purification procedure described in Ref. 9) for 5 min at room temperature. Transcription was initiated by adding 5 μl of NTP mix containing 0.4 mm ATP, 0.4 mm UTP, 0.02 mm CTP, 0.02 mm 3′O-methyl-GTP, and 0.2 μl of [α-32P]CTP (6000 Ci/mmol, 10 μCi/μl). Each reaction was incubated at room temperature for 20 min.
Purified Spt4/5 proteins in TBS150 were added to transcription reactions. They were incubated for 15 min at room temperature before 5 μg of Torula yeast RNA was added to reduce the nonspecific binding between the proteins and nascent RNA. After 5 min, the samples were analyzed by native gel electrophoresis or processed for UV cross-linking.
Analysis of Elongation Complexes by Native Gel Electrophoresis
Samples were loaded onto a 4% native polyacrylamide (40:1 acrylamide/bisacrylamide) gel containing 50 mm Tris-HCl (pH 8.0), 380 mm glycine, 2 mm EDTA, 5 mm MgCl2, 2.5% glycerol, and 0.5 mm DTT (9). The gel was pre-run at 100 V for 90 min at 4 °C before loading and was run at 100 V for 20 min followed by 200 V for 4 h. After electrophoresis, the gel was transferred to filter paper, dried, and exposed in a PhosphorImager.
Protein-RNA UV Cross-linking and TEV Protease Digestion
In vitro transcription was conducted as described above except that UTP was replaced with photoreactive 5-bromo-UTP. After the binding of Spt4/5 proteins, samples were exposed to 300-nm wavelength UV for 10 min at 4 °C. After UV treatment, 1.5 μl of 10% Nonidet P-40, 2.5 μl of 2% Sarkosyl, 1 unit of DNase I (New England Biolabs), 1 mg of RNase A, and 3 μl of 10× DNase I buffer (New England Biolabs) were added to each sample to digest nucleic acids. Digestion occurred for 1 h at room temperature. For samples subjected to TEV protease digestion, 170 units of TEV protease (a gift from Dr. Song Tan) and 1.5 μl of 10% Nonidet P-40 were added and incubated for 2 h before the addition of Sarkosyl and nucleases. After digestion, protein-RNA complexes were precipitated with trichloroacetic acid, dissolved in SDS sample buffer, and analyzed by 4–20% SDS-PAGE or 7% Tris acetate PAGE (54).
Generation of Transgenic Fly Lines
The following fly lines used in this study were obtained from public sources. Two Gal4 driver lines were from the Bloomington Drosophila Stock Center. BDSC 4414 ubiquitously expresses Gal4 under the control of the Act5C promoter. BDSC 1824 expresses Gal4 in larval salivary glands. Spt5 RNAi line HMS 00153 was from the Transgenic RNAi Project (TRiP, Harvard Medical School). It expresses shRNA targeting a 21-nucleotide region in the coding sequence of Spt5 under the control of a Gal4-driven promoter upstream activating sequence. This line was outcrossed to a yw line to eliminate the recessive lethal phenotype associated with the original stock of Spt5 RNAi.
Transgenic fly lines expressing various forms of Spt5 under the control a Gal4-driven upstream activating sequence promoter were generated using the phiC31 site-specific integration system as described previously (55, 56). First, the coding sequence of Spt5 was subcloned into pUAST-attB (a gift from Dr. Graham Thomas). Two tandem FLAG tags were added to the C terminus of Spt5. The 21-nucleotide region targeted by Spt5 RNAi was modified with synonymous mutations (from GACAGAAGCTACAGTCCATTCAAT to GATAGGTCCTATTCCCCCTTCAAT) so that it was resistant to Spt5 RNAi. The plasmids carrying Spt5, Spt5Δ635–789, Spt5Δ774–789, or Spt5 (779–789)A were injected into fly line BDSC 24749, which contains an attP site and expresses phiC31 integrase (Rainbow Transgenic Flies). Each Spt5 transgene was inserted at 86Fb in the genome.
To recombine the Spt5 transgene and Spt5 RNAi gene into the same line, the various Spt5 transgenic lines were mated to the Spt5 RNAi line to allow homologous recombination. The Spt5 transgenes are on chromosome 3R; the Spt5 RNAi transgene is on chromosome 3L. The progeny carrying both transgenes were selected according to their eye color (Spt5 carries w+ marker) and body color (Spt5 RNAi carries y+ marker).
Western Blotting for Fly Samples
To monitor ubiquitous expression of Spt5 derivatives, matings were set up between Spt5 lines and actin-Gal4/CyO driver line BDSC 4414. Individuals harboring red-eyed progeny that carry both Spt5 transgene and actin-Gal4 were dissected from the pupal cases, homogenized in LDS sample buffer, and heated at 95 °C for 5 min.
To monitor expression of Spt5 derivatives in salivary glands, Spt5 lines were mated to salivary gland Gal4 driver BDSC1824. Salivary glands were isolated from late third instar larvae, treated with benzonase (New England Biolabs) in NEBuffer3 supplemented with 1% Triton X-100, and boiled in LDS sample buffer at 95 °C for 5 min.
After electrophoresis, proteins were transferred to nitrocellulose membranes. The blots were probed with FLAG M2 antibody (mouse; Sigma-Aldrich), Spt5 antibody (rabbit), Rpb-3 antibody (rabbit), or M1BP antibody (rabbit).
Fly Rescue Assay
Matings were set up with five male flies from the act-Gal4/CyO (BDSC 4414) line and five virgin female flies from each of Spt5 transgenic line containing Spt5 RNAi. The flies were raised at 24 °C. Between 14 and 21 days after the matings were set up, the numbers of adult flies in the progeny with curly wings or straight wings were scored. The total numbers are from two independent experiments.
Immunofluorescence Analyses of Polytene Chromosomes
Transgenic lines containing Spt5, Spt5Δ635–789, Spt5Δ774–789, or Spt5(779–789)A were mated to the salivary gland Gal4 driver line BDSC 1824. The BDSC 1824 line alone was used as negative control. Flies were grown at 18 °C to obtain larger salivary glands that gave better staining patterns. Polytene chromosomes were prepared as described previously (57, 58). Briefly, two pairs of salivary glands from each mating were dissected and then incubated in solution A (15 mm Tris-Cl (pH 7.4), 60 mm KCl, 15 mm spermine, 1.5 mm spermidine, 1% Triton X-100) for 5 s, solution B (15 mm Tris-Cl (pH 7.4), 60 mm KCl, 15 mm spermine, 1.5 mm spermidine, 1% Triton X-100, 3.7% formaldehyde) for 30 s, and solution G (50% glacial acetic acid) for 3 min, during which time the glands were transferred to a droplet of 10 μl of solution G on a siliconized coverslip. A glass slide was placed onto the coverslip and then flipped. Chromosomes were squashed until desired spreading was achieved. The slides were flash-frozen, and the coverslip was removed. Slides were used immediately or stored in 95% ethanol at 4 °C for up to 5 days.
The slides were rehydrated in TBS (10 mm Tris-HCl, 150 mm NaCl, pH 7.5) twice for 5 min each time and then incubated in blocking solution (10% fetal bovine serum in TBS) for 1 h at room temperature. The slides were washed with TBS for 5 min before incubation with primary antibodies. Rpb3 antibody (rabbit) and FLAG antibody (mouse; Sigma-Aldrich) were diluted 1:100 in blocking solution. The slides were incubated with primary antibody for 2 h and then washed twice with TBS for 10 min each time. Secondary antibodies anti-mouse Alexa Fluor 647 and anti-rabbit Alexa Fluor 488 (ThermoFisher) were diluted 1:200 in blocking solution. After incubation with secondary antibody solution for 1.5 h at room temperature in the dark, the slides were first washed with TBS containing 10 ng/ml Hoechst for 15 min to stain DNA and then washed with TBS for 15 min. The slides were mounted with a coverslip and 20 μl of mounting solution (2% n-propyl gallate, 80% glycerol, 100 mm Tris-HCl, pH 8.5). Chromosomes were viewed with a fluorescence microscope (Carl Zeiss Axioskop 40).
Chromatin Immunoprecipitation of Salivary Glands
Matings were set up as described above for the expression of Spt5 variants in salivary glands. Preparation of samples for immunoprecipitation was done as previously described (58). 10 pairs of salivary glands from third instar larvae were dissected and cross-linked by incubating with 1% formaldehyde in dissection buffer for 5 min on ice, followed by 7 min at room temperature. 2.5 m glycine was added to a final concentration of 125 mm to quench the cross-linking reaction, and the glands were incubated on ice for 2 min. The glands were then collected by centrifugation at 900 × g and washed with PBS. 100 μl of sonication buffer (20 mm Tris (pH 8.0), 0.5% SDS, 2 mm EDTA, 0.5 mm EGTA, 0.5 mm PMSF) and 2 μl of protease inhibitor mixture (1.6 mg/ml benzamidine-HCl, 1 mg/ml aprotinin, 1 mg/ml pepstatin A, and 1 mg/ml leupeptin) were added to each sample. The glands were incubated at room temperature for 10 min and then on ice for 10 min. The glands were shaken vigorously for 10 min and then homogenized manually with a small pestle. Lysates were sonicated at 4 °C in a Bioruptor (Diagenode) at maximum power twice for 7.5 min each time, with a cycle of 30 s on and 30 s off. The lysates were cleared by centrifugation at 14,000 × g for 7 min and stored at −80 °C.
For each immunoprecipitation (IP), 20 μl of lysate was diluted into 400 μl of IP dilution buffer (20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 2 mm EDTA (pH 8.0), 0.5% Triton X-100, 10% glycerol), precleared with a 15-μl slurry of 50% protein A-Sepharose beads suspended in TE plus 1 mg/ml acetylated BSA (Promega) at 4 °C for 2 h. The precleared lysates were separated from the beads by centrifugation at 900 × g for 4 min and then incubated with primary antibody at 4 °C overnight. 4 μl of pre-immune serum, 4 μl of Rpb3 antibody (rabbit), 2 μl of FLAG antibody (rabbit, Genscript), or 2 μl of Spt5 antibody (rabbit) was used, respectively. A 30-μl slurry of 50% protein A-Sepharose in TE was added to each sample and incubated for another 2 h at 4 °C. The beads were collected by centrifugation at 900 × g and washed once with low salt wash buffer (20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 2 mm EDTA (pH 8.0), 1% Triton X-100, 0.1% SDS), three times with high salt wash buffer (20 mm Tris-HCl (pH 8.0), 500 mm NaCl, 2 mm EDTA (pH 8.0), 1% Triton X-100, 0.1% SDS) and twice with lithium chloride wash buffer (10 mm Tris-HCl (pH 8.0), 250 mm LiCl, 2 mm EDTA (pH 8.0), 1% Nonidet P-40, 1% sodium deoxycholate). The beads were washed with TE once, transferred to fresh tubes, and then washed with TE once more. Each wash was carried out at 4 °C for 5 min with 400 μl of wash buffer, except that the first lithium chloride wash was carried out at 4 °C overnight. 100 μl of elution buffer (1% SDS and 0.1 m NaHCO3) was added to the beads and incubated at room temperature for 15 min. After centrifugation, the supernatant was collected. The process was repeated once more, and the eluates were combined. The eluates from IP as well as input lysate diluted in elution buffer were incubated at 65 °C for 4 h to reverse cross-linking. The samples were treated with Proteinase K for 30 min, extracted with phenol/chloroform/isoamyl alcohol (25:24:1), and ethanol-precipitated to isolate DNA.
Quantitative PCR analysis was performed using SYBR Green PCR master mix from SensiMix (Bioline) as described previously (58). The reactions were performed on an Applied Biosystems 7300 real-time PCR system. Data were analyzed with Applied Biosystems 7300 software. Primers targeting −72 to +29 of hsp70Bc were used for hsp70 promoter ChIP. All data were obtained from the linear range of the amplification. Standard curves were generated from a serial dilution of input samples. The percentage input for each IP was calculated from the standard curves and presented in the chart.
Author Contributions
Y. Q. and D. S. G. designed the study and wrote the manuscript. Y. Q. performed the experiments and analyzed the data.
Acknowledgments
We thank Song Tan for providing the polycistronic expression vector, Mitzi Kuroda for providing the MSL-2 antibody, and the TRiP at Harvard Medical School (National Institutes of Health (NIH)/NIGMS Grant R01-GM084947) for providing the transgenic RNAi stock. Stocks obtained from the Bloomington Drosophila Stock Center (NIH Grant P40OD018537) were used in this study. We thank Joseph Reese for critiquing the manuscript.
This work was supported by National Institutes of Health Grant GM047477 (to D. S. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Pol II
- RNA polymerase II
- DSIF
- DRB sensitivity-inducing factor
- NELF
- negative elongation factor
- CTR
- C-terminal repeats of Spt5
- CTD
- C-terminal domain of Pol II
- P-TEFb
- positive elongation factor b
- NGN
- NusG N-terminal
- KOW
- Kyrpides-Ouzounis-Woese
- EC
- elongation complex
- MSL
- male-specific lethal
- hSpt5 and ySpt5
- human and yeast Spt5, respectively
- IP
- immunoprecipitation
- TEV
- tobacco etch virus
- RNAP
- RNA polymerase.
References
- 1. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., and Adelman K. (2007) RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeitlinger J., Stark A., Kellis M., Hong J.-W., Nechaev S., Adelman K., Levine M., and Young R. A. (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Core L. J., Waterfall J. J., and Lis J. T. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee C., Li X., Hechmer A., Eisen M., Biggin M. D., Venters B. J., Jiang C., Li J., Pugh B. F., and Gilmour D. S. (2008) NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 28, 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., and Adelman K. (2010) Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kwak H., Fuda N. J., Core L. J., and Lis J. T. (2013) Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J., Liu Y., Rhee H. S., Ghosh S. K. B., Bai L., Pugh B. F., and Gilmour D. S. (2013) Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol. Cell 50, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fuda N. J., Ardehali M. B., and Lis J. T. (2009) Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Missra A., and Gilmour D. (2010) Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc. Natl. Acad. Sci. U.S.A. 107, 11301–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng B., and Price D. H. (2008) Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 36, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivanov D., Kwak Y. T., Guo J., and Gaynor R. B. (2000) Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20, 2970–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J. B., and Sharp P. A. (2001) Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276, 12317–12323 [DOI] [PubMed] [Google Scholar]
- 13. Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., and Peterlin B. M. (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu X., Zhu X., Li Y., Liu M., Yu B., Wang Y., Rao M., Yang H., Zhou K., Wang Y., Chen Y., Chen M., Zhuang S., Chen L.-F., Liu R., and Chen R. (2016) Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 44, 6853–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marshall N. F., Peng J., Xie Z., and Price D. H. (1996) Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271, 27176–27183 [DOI] [PubMed] [Google Scholar]
- 16. Peterlin B. M., and Price D. H. (2006) Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 [DOI] [PubMed] [Google Scholar]
- 17. Jonkers I., and Lis J. T. (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., and Handa H. (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 [DOI] [PubMed] [Google Scholar]
- 19. Eick D., and Geyer M. (2013) The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490 [DOI] [PubMed] [Google Scholar]
- 20. Adelman K., and Lis J. T. (2012) Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J., and Gilmour D. S. (2011) Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 21, 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu C.-H., Lee C., Fan R., Smith M. J., Yamaguchi Y., Handa H., and Gilmour D. S. (2005) Molecular characterization of Drosophila NELF. Nucleic Acids Res. 33, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Werner F. (2012) A nexus for gene expression: molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 417, 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartzog G. A., and Fu J. (2013) The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim. Biophys. Acta 1829, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo S., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., and Rosenthal A. (2000) A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408, 366–369 [DOI] [PubMed] [Google Scholar]
- 26. Hirtreiter A., Damsma G. E., Cheung A. C. M., Klose D., Grohmann D., Vojnic E., Martin A. C. R., Cramer P., and Werner F. (2010) Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 38, 4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klein B. J., Bose D., Baker K. J., Yusoff Z. M., Zhang X., and Murakami K. S. (2011) RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc. Natl. Acad. Sci. U.S.A. 108, 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez-Rucobo F. W., Sainsbury S., Cheung A. C. M., and Cramer P. (2011) Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 30, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bernecky C., Herzog F., Baumeister W., Plitzko J. M., and Cramer P. (2016) Structure of transcribing mammalian RNA polymerase II. Nature 529, 551–554 [DOI] [PubMed] [Google Scholar]
- 30. Crickard J. B., Fu J., and Reese J. C. (2016) Biochemical analysis of yeast suppressor of Ty 4/5 (Spt4/5) reveals the importance of nucleic acid interactions in the prevention of RNA polymerase II arrest. J. Biol. Chem. 291, 9853–9870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W., Giles C., and Li S. (2014) Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 42, 7069–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamaguchi Y., Wada T., Watanabe D., Takagi T., Hasegawa J., and Handa H. (1999) Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274, 8085–8092 [DOI] [PubMed] [Google Scholar]
- 33. Lindstrom D. L., Squazzo S. L., Muster N., Burckin T. A., Wachter K. C., Emigh C. A., McCleery J. A., Yates J. R. 3rd, and Hartzog G. A. (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23, 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W. H., Ranish J., Shokat K. M., and Hahn S. (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29, 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schneider S., Pei Y., Shuman S., and Schwer B. (2010) Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol. Cell. Biol. 30, 2353–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayer A., Schreieck A., Lidschreiber M., Leike K., Martin D. E., and Cramer P. (2012) The Spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol. Cell. Biol. 32, 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jennings B. H., Shah S., Yamaguchi Y., Seki M., Phillips R. G., Handa H., and Ish-Horowicz D. (2004) Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr. Biol. 14, 1680–1684 [DOI] [PubMed] [Google Scholar]
- 38. Meyer P. A., Li S., Zhang M., Yamada K., Takagi Y., Hartzog G. A., and Fu J. (2015) Structures and functions of the multiple KOW domains of transcription elongation factor Spt5. Mol. Cell. Biol. 35, 3354–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kyrpides N. C., Woese C. R., and Ouzounis C. A. (1996) KOW: a novel motif linding a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426 [DOI] [PubMed] [Google Scholar]
- 40. Tan S., Kern R. C., and Selleck W. (2005) The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395 [DOI] [PubMed] [Google Scholar]
- 41. Biggin M. D., and Tjian R. (1988) Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53, 699–711 [DOI] [PubMed] [Google Scholar]
- 42. Chen H., Contreras X., Yamaguchi Y., Handa H., Peterlin B. M., and Guo S. (2009) Repression of RNA polymerase II elongation in vivo is critically dependent on the C-terminus of Spt5. PLoS One 4, e6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Z., Wu C.-H., and Gilmour D. S. (2004) Analysis of polymerase II elongation complexes by native gel electrophoresis. Evidence for a novel carboxyl-terminal domain-mediated termination mechanism. J. Biol. Chem. 279, 23223–23228 [DOI] [PubMed] [Google Scholar]
- 44. Wu C.-H., Yamaguchi Y., Benjamin L. R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., and Gilmour D. (2003) NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17, 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Andrulis E. D., Guzmán E., Döring P., Werner J., and Lis J. T. (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14, 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamaguchi Y., Inukai N., Narita T., Wada T., and Handa H. (2002) Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22, 2918–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ujvári A, and Luse D. S. (2006) RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 13, 49–54 [DOI] [PubMed] [Google Scholar]
- 48. Bushnell D. A., and Kornberg R. D. (2003) Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. U.S.A. 100, 6969–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Armache K.-J., Kettenberger H., and Cramer P. (2003) Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 100, 6964–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choder M. (2004) Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 29, 674–681 [DOI] [PubMed] [Google Scholar]
- 51. Schulz S., Gietl A., Smollett K., Tinnefeld P., Werner F., and Grohmann D. (2016) TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. Proc. Natl. Acad. Sci. U.S.A. 113, E1816–E1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Booth G. T., Wang I. X., Cheung V. G., and Lis J. T. (2016) Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 26, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vos S. M., Pöllmann D., Caizzi L., Hofmann K. B., Rombaut P., Zimniak T., Herzog F., and Cramer P. (2016) Architecture and RNA binding of the human negative elongation factor. Elife 10.7554/eLife.14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cubillos-Rojas M., Amair-Pinedo F., Tato I., Bartrons R., Ventura F., and Rosa J. L. (2010) Simultaneous electrophoretic analysis of proteins of very high and low molecular mass using Tris-acetate polyacrylamide gels. Electrophoresis 31, 1318–1321 [DOI] [PubMed] [Google Scholar]
- 55. Groth A. C., Fish M., Nusse R., and Calos M. P. (2004) Construction of transgenic drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bischof J., Maeda R. K., Hediger M., Karch F., and Basler K. (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Champlin D. T., Frasch M., Saumweber H., and Lis J. T. (1991) Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev. 5, 1611–1621 [DOI] [PubMed] [Google Scholar]
- 58. Ghosh S. K. B., Missra A., and Gilmour D. S. (2011) Negative elongation factor accelerates the rate at which heat shock genes are shut off by facilitating dissociation of heat shock factor. Mol. Cell. Biol. 31, 4232–4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–539 [DOI] [PMC free article] [PubMed] [Google Scholar]