Abstract
SUMOylation is an important posttranslational modification that regulates protein function in diverse biological processes. However, its role in early T cell development has not been genetically studied. UBC9 is the only E2 enzyme for all SUMOylation. In this study, by selectively deleting Ubc9 gene in T cells, we have investigated the functional roles of SUMOylation in T cell development. Loss of Ubc9 results in a significant reduction of CD4 and CD8 single-positive lymphocytes in both thymus and periphery. Ubc9-deficient cells exhibit defective late-stage maturation post the initial positive selection with increased apoptosis and impaired proliferation, among which attenuated IL-7 signaling was correlated with the decreased survival of Ubc9-deficent CD8 single-positive cells. Furthermore, NFAT nuclear retention induced by TCR signals was regulated by SUMOylation during thymocytes development. Our study thus reveals a novel posttranslational mechanism underlying T cell development.
Introduction
The CD4+ and CD8+ lineage αβ T cells are essential components of the adaptive immune system. They originate from a large cohort of double-positive (DP) precursor cells (CD4+ CD8+ DP) in the thymus through a highly ordered developmental process called positive selection, during which engagement of TCR on DP cells by self-peptide MHC complex (pMHC) from thymic epithelial cells with appropriate avidity will deliver signals to DP cells and enable these cells to proceed into CD4 or CD8 single-positive (SP) thymocytes (1–4). Most DP cells die either because of too strong (during negative selection) or too weak interaction (during death by neglect) between their TCR and pMHC (5).
Positive selection is a multistage process that requires persistent TCR signaling (6). It includes phases of initiation, phenotypic differentiation, and postselection maturation (7). Earlier stages of positive selection markers, such as upregulation of TCRβ, CD5, and CD69, are strictly dependent on TCR-proximal signaling (8–11). The engagement of TCR also induces coreceptor downregulation, pushing DP cells toward CD4+CD8int developmental intermediate stage, at which either CD4 or CD8 fate decisions are made (12). Positive selection also induces antiapoptotic BCL-2 protein, CCR7, and IL-7R expression. In the later stage of positive selection, expression of heat-stable Ag (HSA; mouse CD24) and CD69 on the cell surface are downregulated, and newly matured SP thymocytes emigrate from the thymus into peripheral lymphoid tissue (13, 14).
In addition to phosphorylation and ubiquitination, protein modification by small ubiquitin-like modifier (SUMO) proteins is another major type of posttranslational mechanism regulating protein function in eukaryotic organisms. It is characterized by covalent attachment of SUMO moiety to lysines within protein targets (15). Like ubiquitination, SUMO conjugation uses a cascade of factors that include E1–E3; only one E2 called UBC9 is used by the SUMO pathway as a conjugation enzyme that transfers SUMO moiety to SUMOylation substrates. Therefore, deletion of Ubc9 impairs the whole SUMOylation pathway. By influencing stability, intracellular localization, interaction with partners, and activity of target proteins, SUMOylation pathway affects many biological processes such as cell cycle, DNA repair, chromatin dynamics, and gene transcription. SUMOylation plays important roles in the immune system. In myeloid cells, SUMOylation restrains TLR-induced production of inflammatory cytokines and massive expression of type I IFN signature genes (16). In addition, SUMOylation is required for regulatory T cell expansion and function by regulating TCR signal transduction (17). However, the functional role of SUMOylation in the early T cell development is still not clear.
In this study, we conducted a genetic study on the role of SUMO in the adaptive immune system by specifically inactivating the Ubc9 gene in T cells in mice. We found that Ubc9 deficiency profoundly perturbed early T cell development, leading to a significant reduction of both CD4 and CD8 SP cells in the thymus and peripheral lymphoid tissues. When investigating positive selection of T cells, we observed that the late stage of T cell maturation in the thymus was defective in the absence of Ubc9 with increased apoptosis and impaired proliferation. IL-7 signaling was attenuated in CD8 SP cells. Moreover, NFAT nuclear retention was regulated by SUMOylation in thymocytes. Our study therefore has demonstrated that the SUMOylation pathway is essential for T cell development.
Materials and Methods
Mice and reagents
Mice with Ubc9fl/fl allele have been described previously (18). Primer 23 (5′-AAG CTG TAG CAG GGA TGT GCT CTG G-3′) and primer 24 (5′-TTG ACA AGG CCC TTA GGT GAA CAC CTC TC-3′) were used to distinguish wild-type (WT) Ubc9 (480 bp) from floxed Ubc9 allele (535 bp), whereas primer 22 (5′-CAG CAG ATG GGG ATG AGT AAG-3′) and primer 23 were used to confirm null allele (320 bp). Cd4-Cre mice were obtained from Dr. C. Wilson. The Ubc9fl/fl strain has been backcrossed with the C57BL/6 strain for 10 generations before crossing with the Cd4-Cre strain. Rag1−/−, OT-II, and HY transgenic strains were obtained from Taconic Farms or Jackson Laboratories. All animal experiments were performed by using sex- and age-matched littermates and following protocols approved by Institutional Animal Care and Use Committee.
Cell staining and flow cytometry analysis
Single-cell suspension was obtained from the thymus, spleen, and peripheral lymph nodes (inguinal plus axillary lymph nodes) of 4- to 10-wk-old mice. Cells were treated with Fc block (eBioscience) before surface staining with Abs to CD3, CD4, CD5, CD8α, TCRβ, HSA (CD24), CD69, IL-7Rα, Vα2, T3.70, CD45.2, and CCR7 that were obtained from BD Biosciences, eBioscience, or BioLegend. FOXP3 and BCL-2 Abs were ordered from eBioscience. Intracellular staining for these Ags was performed by using the FOXP3 intracellular staining kit from eBioscience. Annexin V was obtained from BD, and apoptosis was stained according to the kit protocol. For Phosphoflow staining, thymocytes were stimulated with IL-7 for 15 min and fixed by BD Phosphoflow staining buffer followed by methanol permeabilization. p-STAT5 (from BD) was stained with surface markers. Data were acquired on LSR II flow cytometry and analyzed by Tri-Star FlowJo software. Detailed information of flow Abs can be found in Supplemental Table I.
Thymocytes activation and Western blot analysis
Bulk thymocytes (20 million) were stimulated with 5.0 μg/ml anti-CD3 (2C11) and anti-CD28 (37.51) in RPMI 1640 medium for 20 min on ice, followed by washing once with RPMI 1640 medium. Bound Abs were then cross-linked by the addition of 40 μg/ml goat anti-hamster IgG in RPMI 1640 complete medium and incubated at 37°C for different times. In some experiments, stimulation of thymocytes was achieved by preincubating thymocytes with biotin-conjugated anti-CD4 and anti-CD3 (5 μg/ml) for 20 min on ice before one wash and cross-linking with streptavidin (40 μg/ml). Stimulation was terminated by the addition of cold PBS into reactions followed by placement of cells on ice. Cell pellets were obtained by centrifuging nonstimulated or stimulated thymocytes at 4°C and then subjecting to cell lysis with RIPA buffer. Protein assay was performed with BCA protein assay kit (Thermo Scientific). Cell lysates containing 5–20 μg of protein were used for Western blot analysis and were detected by the following Abs: anti-UBC9 (BD Biosciences), anti-IκBα (Santa Cruz), anti-ERK1 (Santa Cruz), anti–p-ERK1/2 (Cell Signaling), anti-JNK (Cell Signaling), anti–p-JNK (Cell Signaling), and anti-NFATc1 (Santa Cruz). Detailed information on the Abs used for Western blot analysis can be found in Supplemental Table I.
SUMOylation analysis
In 293T cells, vectors containing FLAG-tagged targeted protein, HA-tagged UBC9, and His-tagged SUMO protein were transfected via phosphonate calcium. Cells were harvested 36 h after transfection and lysed by 1% SDS containing 20 mM NEM to preserve SUMOylation. Then cell lysates were denatured at 97°C for 10 min with 50 mM DTT followed by sonication to reduce viscosity. After dilution with RIPA buffer 10-fold, FLAG immunoprecipitation was performed using FLAG-M2 magnetic beads to enrich the targeted protein, and SUMOylated bands were detected by anti-FLAG (Sigma) and anti-His (Bioeasy) Abs.
In CD4+ primary T cells, CD4+ T cells were enriched from mouse lymph nodes and spleen by MACS technology (Miltenyi); then cells were activated by plate-coated anti-CD3 (5 μg/ml) and anti-CD28 (5 μg/ml) for 24 h and infected with retrovirus containing FLAG-tagged targeted protein. FLAG IP was performed as that in 293T cells; SUMOylated bands were detected using anti-FLAG (Sigma) and anti-SUMO2/3 (Invitrogen) Abs.
Real-time RT-PCR
RNA was extracted with TRIzol (Invitrogen) and reverse transcribed to cDNA with the SuperScript III First Strand kit (Invitrogen). The mRNA level of Thpok and Prf1 was assessed relative to Actin by real-time PCR with SYBR Green real-time PCR Master Mix (Bio-Rad). The data shown were relative values. Primers used for real-time PCR were as follows: Thpok (5′-TGCAGCTCCAGCGAACGGAC-3′, 5′-ACA GCC CTG TGG GTG CGG TA-3′) and Prf1 (5′-CAA TAA CGA CTG GCG TGT GG-3′, 5′-TGT TAA AGT TGC GGG GGA GG-3′).
Bone marrow chimera
Bone marrow cells, freshly harvested from femurs of WT and Ubc9 conditional knockout (KO) mice, were treated with anti-Thy1 plus complement to remove mature T cells, and ∼10 million purified bone marrow cells were injected into each irradiated recipient. Two months after bone marrow cell transfer, mice were sacrificed and analyzed.
Calcium influx
Thymocytes were loaded with 2 μM indo-1 AM (Invitrogen) for 30 min at 37°C in RPMI 1640 medium without serum, washed twice with RPMI 1640 containing 1% FBS, and then surface-stained with anti-CD4 (clone RM4-4; eBioscience) and anti-CD8 (clone 53-6.7; BD Biosciences) for 20 min on ice. Cells were washed twice and incubated for 30 min at room temperature with biotinylated anti-CD3 (10 μg/ml) and biotinylated anti-CD4 (clone GK1.5, 10 μg/ml; BioLegend). Cells were washed twice before being suspended in medium and warmed at 37°C for 10 min before analysis. A total of 200 μl of streptavidin (1 μg/ml; Roche) was added at the 1-min time point after baseline recording started. Fluorescence was collected over 9 min and analyzed using FlowJo.
Statistics
For two sets of data, we used Student t test, and for three or more sets of data, we used one-way ANOVA with a post hoc analysis. Asterisks denote statistical significance compared with the indicated controls: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Statistical analysis was performed in GraphPad PRISM 6.
Results
Disruption of the Ubc9 gene in T cells
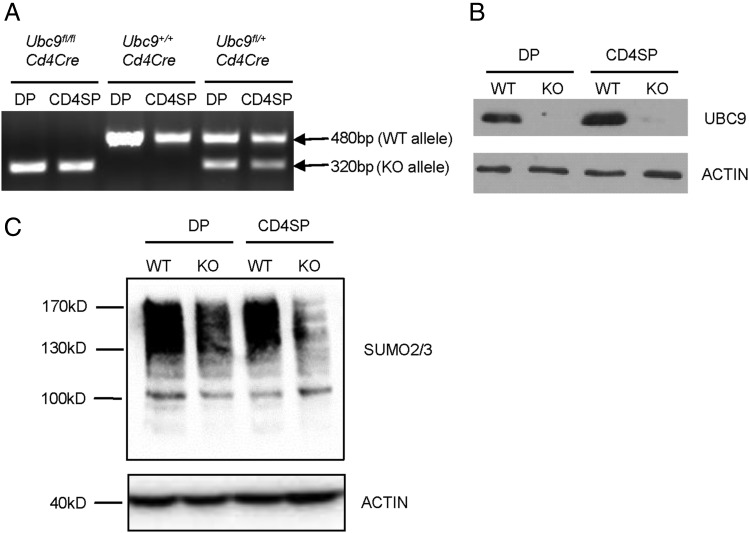
Deletion of Ubc9 results in embryonic lethality in mice (18, 19). To investigate the role of Ubc9-mediated SUMOylation in T cells, we crossed mice bearing an Ubc9 conditional allele (18) with the Cd4Cre strain, in which Cre expression is initiated at the DP stage of T cells (20). To investigate the deletion efficiency of Ubc9 gene, we sorted by FACS DP or SP thymocytes from Cd4Cre transgenic mice with WT or floxed (KO) Ubc9 allele. PCR analysis showed that floxed allele (535 bp) was completely cleaved and converted into null allele (320 bp) in both DP and SP cells sorted from KO mice (Fig. 1A). In addition, UBC9 protein was barely detected in these cells (Fig. 1B). As the only E2 in the SUMOylation cycle, loss of Ubc9 led to the substantial reduction of global SUMOylation level in DP and SP cells (Fig. 1C). These data demonstrated that UBC9-mediated SUMOylation was efficiently inactivated in DP and SP thymocytes from Ubc9fl/flCd4Cre mice.
FIGURE 1.
Specific deletion of Ubc9 in thymocytes. (A) Genomic DNA was extracted from CD4+ SP and CD4+ CD8+ DP thymocytes of Ubc9fl/fl Cd4Cre, Ubc9fl/+Cd4Cre, and Ubc9+/+ Cd4Cre mice (7 wk old). PCR analysis was performed to show WT Ubc9 (480 bp), floxed Ubc9 (535 bp), and Ubc9 null allele (320 bp). These data represent two separate experiments. (B and C) Whole-cell lysates were prepared from sorted CD4 SP and DP thymocytes and subjected to Western blot analysis of UBC9 (B) and SUMO2/3 (C). These data represent three separate experiments.
Ubc9 is required for T cell development in the thymus and periphery
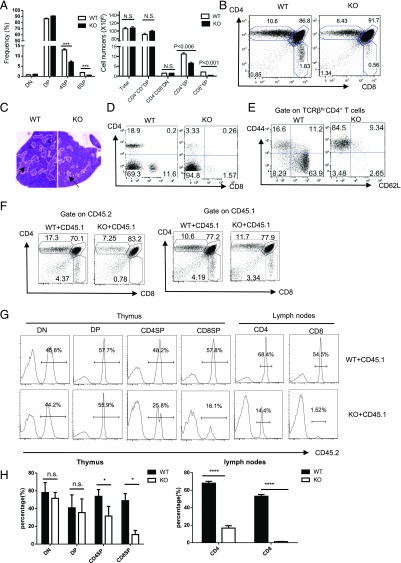
No gross abnormality was observed in Ubc9fl/fl Cd4Cre mice for at least 1 y of maintenance (data not shown). We focused on the T cell development in these mice. Because the phenotypes of Ubc9fl/+ Cd4Cre and Ubc9fl/fl (no Cd4Cre) mice were the same (data not shown), we used either of them as WT control throughout our study. In comparison with WT littermate control, cellularity of total thymocytes and double-negative (DN) cells were unaltered, whereas the proportion and cell number of DP cells were slightly increased in Ubc9fl/fl Cd4Cre (KO) mice (Fig. 2A, 2B). Importantly, KO mice displayed a 2-fold reduction both in frequencies and cell numbers of CD4 SP cells, with a more severe reduction of CD8 SP cells (Fig. 2A, 2B). Mature SP or DP thymocytes undergoing positive selection normally have higher expression levels of TCRβ than their more immature precursors. Cellularity of TCRβhigh thymocytes was also significantly lower in KO mice (Supplemental Fig. 1A). Consistent with this, as an area populated by SP thymocytes after positive selection, the region of thymic medulla was considerably reduced in KO mice (Fig. 2C).
FIGURE 2.
Ubc9 deficiency impairs thymic and peripheral T cell development. (A and B) Percentages and absolute cell numbers (A) and representative dot plots (B) of CD4+ SP, CD8+ SP, CD4+ CD8+ DP, and CD4− CD8− DN subset in 4- to 6-wk-old mice (WT, Ubc9fl/+Cd4Cre, n = 5; KO, Ubc9fl/fl Cd4Cre, n = 4). (C) H&E staining of thymus section from Ubc9fl/fl (WT) and Ubc9fl/fl Cd4Cre (KO) mice. The cortex (dark zone) and medulla region (light zone, indicated by arrow) can be distinguished by the intensity of staining (original magnification ×10). (D and E) Phenotype of peripheral T cells in Ubc9fl/fl (WT) and Ubc9fl/fl Cd4Cre (KO) mice. Representative CD4 and CD8 staining on TCRβhi splenocytes (D), and CD62L and CD44 staining on TCRβhi CD4+ (E) are shown (WT, Ubc9fl/fl, n = 3; KO, Ubc9fl/fl Cd4Cre, n = 3). (F) Flow cytometry analysis of CD4 and CD8 thymocytes by gating on tester (CD45.2+, left) or competitor compartments (CD45.1+, right) in Rag1−/− recipient mice. Numbers adjacent to dot plots indicate the percentage of each subset among total thymocytes (WT, Ubc9fl/fl, n = 3; KO, Ubc9fl/fl Cd4Cre, n = 3). (G and H) Contribution of tester bone marrow–derived progeny (CD45.2+) to T cell compartments in mixed bone marrow chimeric animals. Thymocytes and lymph node cells from bone marrow chimera were stained with anti-CD4, -CD8, and -CD45.2 (G). Numbers in histograms indicate percentages of CD45.2+ cells in each T cell subset shown on top of histograms; statistic results are shown in (H) (WT, Ubc9fl/fl, n = 3; KO, Ubc9fl/fl Cd4Cre, n = 3). Asterisks denote statistical significance compared with the indicated controls: *p < 0.05, ***p < 0.001, ****p < 0.0001.
γδ T cells diverge from conventional T cells at the DN3 stage (21), when Ubc9 deletion is not initiated. Not surprisingly, no difference was found in this population between WT and KO mice (Supplemental Fig. 1B). In contrast, NKT cells (22) and FOXP3+ CD25+ regulatory T cells (23) that generated associated with positive selection were significantly reduced in the thymus of KO mice (Supplemental Fig. 1C, 1D). In peripheral lymph nodes and spleens, Ubc9 inactivation resulted in fewer T cells, whereas B cells were unaffected (Fig. 2D, Supplemental Fig. 1E, 1F). In addition, floxed Ubc9 allele was only partially cleaved in the residual peripheral T cells (Supplemental Fig. 1G), indicating that there was a selection pressure against cells that had deleted Ubc9. Residual T cells were predominant with the CD62Llo CD44hi memory-like population, which likely resulted from homeostatic expansion in the lymphopenic environment (Fig. 2E). Collectively, these data suggest that Ubc9 inactivation impaired both thymic generation of mature SP cells and T cell lymphopenia in the peripheral lymphoid tissue of these mice.
Thymocytes receive other signals from the environment in addition to TCR-pMHC ligation (5). To determine whether T cell developmental block in Ubc9fl/fl Cd4Cre mice was cell intrinsic, we generated mixed bone marrow chimera by injecting irradiated Rag1−/− mice with 1:1 ratio of tester bone marrow cells either from Ubc9fl/fl Cd4Cre mice or Ubc9fl/fl mice (CD45.2+) along with congenic B6-SJL bone marrow cells (CD45.1+) as competitors. Ubc9-deficient CD4 SP thymocytes showed >2-fold reduction in percentage than that of WT counterparts, with CD8 SP thymocytes exhibiting more severe phenotype (Fig. 2F). Competitor thymocytes (CD45.1+) in bone marrow chimera animals, however, exhibited a similar CD4 versus CD8 profile (Fig. 2F). Furthermore, in DN and DP thymocytes, contribution of Ubc9 KO CD45.2+ cells was around 50%, indicating that Ubc9 is dispensable for maintenance of the DP population (Fig. 2G), whereas in SP thymocytes, Ubc9-deficient cells were not comparable with their CD45.1+ counterparts. The biased reduction of KO T cells was even more aggravated in peripheral lymph nodes (Fig. 2G). These data demonstrated that T cell developmental block in Ubc9fl/fl Cd4Cre mice occurring from DP to SP transition was due to a cell-intrinsic mechanism.
Positive selection of T cells was impaired in the absence of Ubc9
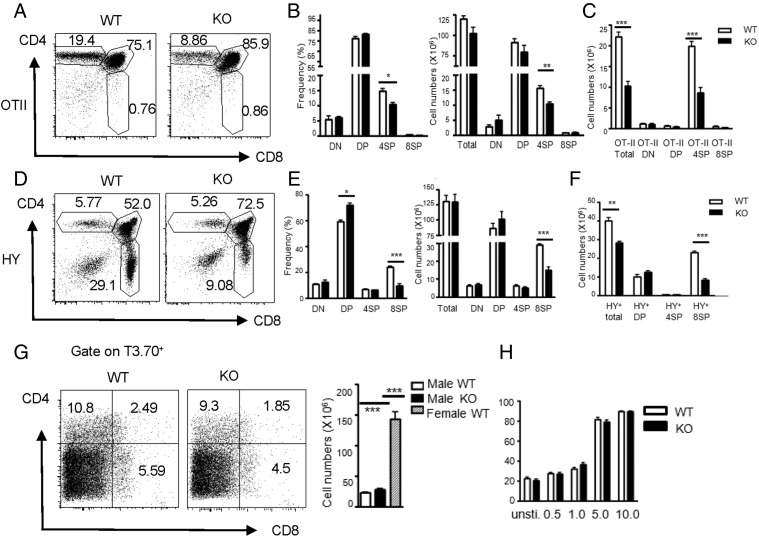
T cell fate decision by positive versus negative selection is dictated by the affinity/avidity of the interaction between TCR on DP cells and pMHC delivered by APCs (5). To investigate whether the effect of Ubc9 deficiency on T cell development is associated with positive selection, we crossed Ubc9fl/fl Cd4Cre mice with transgenic OT-II strain, which carries MHC class II (MHC II)–restricted TCR (24). Compared with Ubc9fl/fl OT-II mice (WT), Ubc9fl/fl Cd4Cre OT-II mice (KO) showed a significant reduction in frequencies and absolute numbers of CD4 SP thymocytes. However, Ubc9 deficiency failed to affect DN, DP, and CD8 SP populations or even total thymic cellularity (Fig. 3A, 3B). Furthermore, the cell number of Vα2+ thymocytes in total and SP cell populations was substantially reduced (Fig. 3C). We also crossed Ubc9fl/fl Cd4Cre mice with the transgenic HY strain, which bears MHC I–restricted TCR (25). HY is a self-antigen expressed only in male mice. Therefore, female or male mice on HY transgenic background are usually used as a positive or negative selection model, respectively. In female mice on HY background, the CD8 SP population was dramatically reduced both in percentages and in cell numbers by Ubc9 inactivation, whereas the CD4 SP population was unaltered (Fig. 3D, 3E). In addition, HY-specific TCR staining (clone T3.70) showed >2-fold reduction occurring in the CD8 SP compartment imposed by Ubc9 deficiency (Fig. 3F). These data with single MHC II– or MHC I–restricted TCR suggest that positive selection was severely impaired by Ubc9 inactivation.
FIGURE 3.
Positive selection was disturbed in Ubc9-deficient thymocytes. (A–C) Surface staining of CD4 and CD8 on total thymocytes was shown as dot plots. Frequencies and total cell numbers of DN, DP, CD4 SP, and CD8 SP subset (B) or total cell numbers of Va2+ subset (C) were calculated and statistically presented as bar graphs (WT, Ubc9fl/fl OT-II+, n = 8; KO, Ubc9fl/fl Cd4Cre OT-II+, n = 5). (D–F) Surface staining of CD4 and CD8 on total thymocytes from female mice is shown as dot plots (D). Frequencies and total cell numbers of DN, DP, CD4 SP, and CD8 SP subsets (E) or total cell numbers of various T3.70+ subsets (F) were calculated and statistically presented as bar graphs (WT, Ubc9fl/fl HY+, n = 5; KO, Ubc9fl/fl Cd4Cre HY+, n = 4). (G) Surface staining of CD4 and CD8 on T3.70hi population from male Ubc9fl/fl HY+ (WT) or Ubc9fl/fl Cd4-Cre+ HY+ mice (KO) were shown as dot plots (left) (WT, n = 4; KO, n = 3). Total thymic cellularity of these male HY transgenic mice was calculated and statistically presented as a bar graph. Thymic cellularity from female Ubc9fl/fl HY+ (WT) mice included as a control (right). (H) Apoptosis of thymocytes from female Ubc9fl/fl (WT) or Ubc9fl/fl Cd4Cre mice (KO) mice in response to anti-CD3 (1×)/anti-CD28 (5×) stimulation for 24 h in vitro was assessed by Annexin V staining. Concentration of anti-CD3 (μg/ml) is shown on x-axis. The data are representative of at least two (A–G) or three (H) experiments. Asterisks denote statistical significance compared with the indicated controls: *p < 0.05, **p < 0.01, ***p < 0.001.
It is possible that Ubc9 inactivation somehow reduces the threshold for negative selection like Schnurri 2, rendering DP cells to undergo negative selection in response to positive selecting signals (26). To assess this possibility, we investigated the effect of Ubc9 deletion on negative selection by using male mice on HY background. Ubc9-deficent mice eliminated T3.70+ TCR-specific DP thymocytes as efficiently as their WT littermate control. Both male WT and KO HY transgenic mice showed comparable numbers of thymocytes, although their total thymic cell numbers were significantly lower than their female counterparts (Fig. 3G). In addition, Ubc9-deficient DP cells exhibited the same levels of apoptosis in vitro as WT DP cells when stimulated by a wide range of concentrations of anti-CD3/CD28 (Fig. 3H). Therefore, negative selection was intact in the absence of Ubc9.
It was possible that diminished thymic SP generation induced by Ubc9 deficiency was due to the redirection of pMHC I– or pMHC II–signaled DP cells into the opposite lineage. However, in Vα2+ cells from OT-II transgenic mice, the diminished CD4 SP population caused by Ubc9 deletion was not replaced by the expansion of CD8 SP thymocytes (Supplemental Fig. 2A). Likewise, in the T3.70+ cell compartment from HY-transgenic female mice, the diminished CD8 SP population induced by Ubc9 deficiency was not replaced by CD4 SP thymocytes either (Supplemental Fig. 2B). Concordantly, CD4 SP from WT and KO mice on TCR polyclonal background showed comparable levels of mRNA of Th-POK, a master transcriptional factor for CD4 SP lineage commitment (27), whereas CD8 SP from these mice exhibited no difference in the expression of Perforin, an effector molecule specifically produced by mature CD8 cells (28) (Supplemental Fig. 2C). These data demonstrated that CD4 versus CD8 lineage commitment was intact in the absence of Ubc9. Collectively, these data indicate that only positive selection was impaired in Ubc9-deficient mice.
Defective late-stage maturation in Ubc9-deficient thymocytes
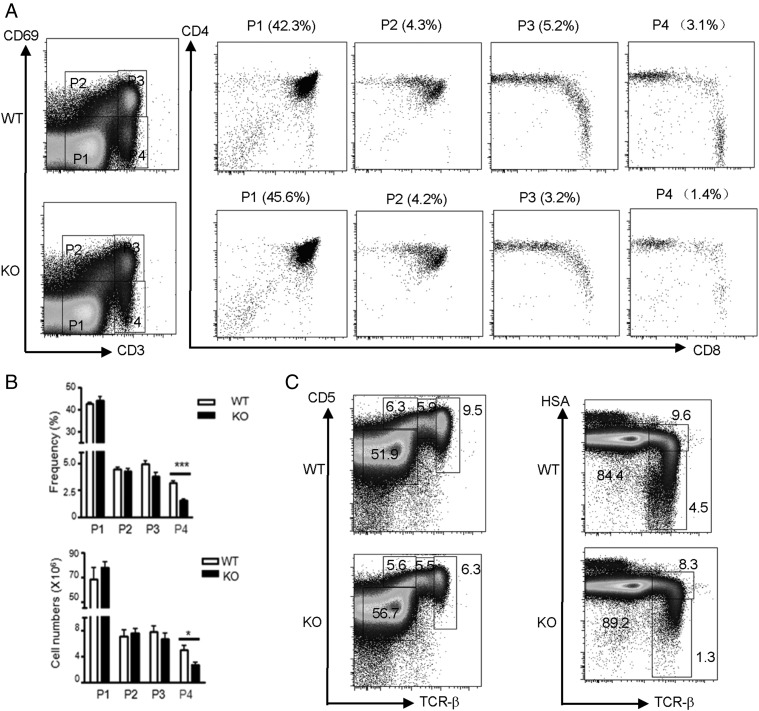
CD69 and CD5 are markers for positive selection. However, total thymocytes or DP cells from KO mice and their littermate control mice showed similar levels of CD69 and CD5. In addition, the transitional population CD4hi CD8int was unaltered between these mice (data not shown). To identify when thymocytes development was first affected in Ubc9-deficient mice, we performed two-parameter analysis, CD3 and CD69 staining, to delineate stages of this process. In this analysis, the numbers of CD3lo CD69− (P1) and CD3lo CD69int (P2) cells, which represented preselection DP thymocytes and DP thymocytes at the initial stage of positive selection, respectively, were normal in Ubc9-deficient mice (Fig. 4A). CD3hi CD69hi cells (P3), which were immature thymocytes that are post positive selection, are slightly reduced in KO mice. However, this difference failed to reach statistical significance. Strikingly, CD3hi CD69− cells (P4), the most mature thymocytes post positive selection consisting mainly of SP thymocytes, were significantly reduced in Ubc9-deficient mice (Fig. 4B). These results were confirmed with additional positive selection markers, TCRβ, CD5, and HSA. The most mature thymocytes, defined as TCRβhi CD5hi or TCRβhi HSAlo (Fig. 4C), were present in Ubc9-deficient mice at significantly reduced frequencies than those in littermate control mice. These data suggest that Ubc9-deficient DP thymocytes successfully initiated positive selection but failed to complete the maturation step.
FIGURE 4.
Impaired late-stage development of thymocytes in the absence of Ubc9. (A) Flow cytometry analysis of CD3 and CD69 on total live thymocytes from Ubc9fl/fl (WT) or Ubc9fl/fl Cd4Cre (KO) mice was used to define sequential developmental stages (P1–P4) of T cells in dot plots. Numbers in parentheses above plots indicate percentages of each developmentally defined subpopulation among total thymocytes. (B) Bar graph shows statistical analysis of frequencies and absolute numbers of these developmentally defined subpopulations in (A) (WT, n = 4; KO, n = 3). (C) Flow cytometry analysis of TCRβ versus CD5 (left dot plots) and TCRβ versus HSA (right dot plots) on total live thymocytes from Ubc9fl/fl (WT) or Ubc9fl/fl Cd4Cre (KO) mice. Numbers adjacent to dot plots indicate percentage of each developmentally defined subpopulation in total thymocytes (WT, n = 5; KO, n = 7). The data shown are representative of at least five experiments. Asterisks denote statistical significance compared with the indicated controls: *p < 0.05, ***p < 0.001.
Attenuated IL-7 signaling in Ubc9-deficient CD8 SP thymocytes
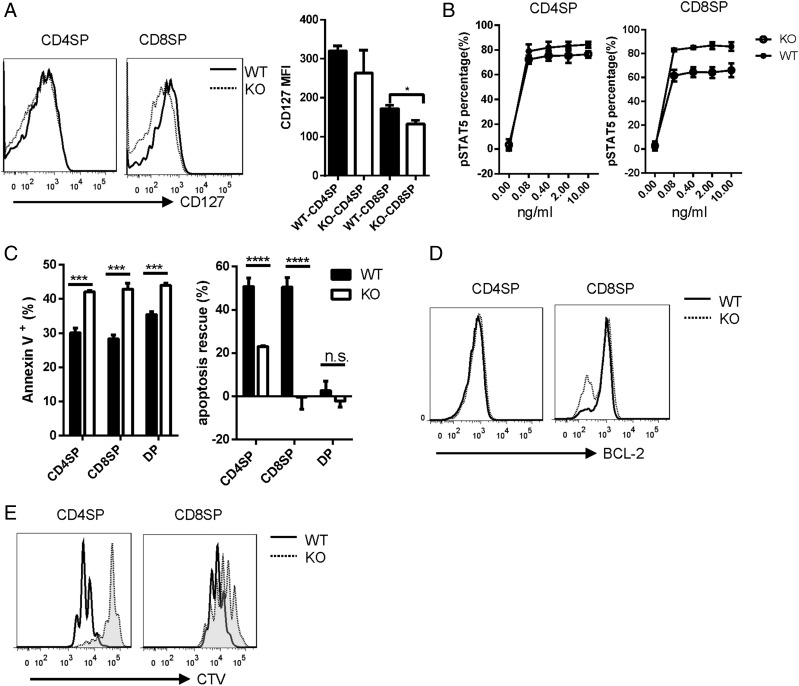
It was documented that IL-7Rα (CD127), a cytokine receptor upregulated during positive selection, controls the development and maturation of late-stage thymocytes, especially the CD8 SP thymocytes (29, 30). We asked whether defective maturation of thymocytes during a late stage of T cell development in Ubc9-deficienct mice is due to disrupted IL-7Rα expression during positive selection. We found that IL-7Rα expression was well maintained in Ubc9-deficient CD4 SP cells, but significantly reduced in Ubc9-deficient CD8 SP cells (Fig. 5A). Consistent with this, when treated with exogenous IL-7, the downstream STAT5 phosphorylation had little alteration in CD4 SP cells but was greatly reduced in CD8 SP cells in the absence of Ubc9 (Fig. 5B).
FIGURE 5.
Compromised IL-7 signaling in Ubc9-deficient CD8 SP cells. (A) Flow cytometry analysis of IL-7Rα (CD127) expression on CD4 SP or CD8 SP cells. MFI, mean fluorescence intensity (WT, n = 3; KO, n = 3). (B) CD4 SP or CD8 SP cells were stimulated with indicated concentration of IL-7 for 15 min, and STAT5 phosphorylation was analyzed by Phosphoflow. The percentage of p-STAT5 is shown (WT, n = 3; KO, n = 3). (C) CD4 SP, CD8 SP, or DP cells were sorted by FACS and cultured in medium for 18 h. Annexin V was stained to detect the apoptotic cells (left). A total of 10 ng/ml IL-7 was added to rescue the apoptotic cells, and the rescue percentage was shown (right) (WT, n = 3; KO, n = 3). (D) Flow cytometry analysis of BCL-2 expression in CD4 SP or CD8 SP cells (WT, n = 3; KO, n = 3). (E) FACS sorted CD4 SP or CD8 SP cells were labeled with CellTrace Violet (CTV) and cultured with plate-coated anti-CD3 and anti-CD28 for 3 d; dye dilution was detected by flow cytometry. (A–E) WT, Ubc9fl/fl; KO, Ubc9fl/fl Cd4-Cre. These are the representative data of at least three experiments. Asterisks denote statistical significance compared with the indicated controls: *p < 0.05, ***p < 0.001, ****p < 0.0001.
Reduced cell numbers of mature SP cells in the thymus from Ubc9fl/fl Cd4Cre mice might be attributed to dysregulation of thymocytes migration, decreased cell survival, or defect in the acquisition of proliferation competence during maturation. CCR7 and CD62L expression are required for cortex-medulla intrathymic migration or thymic emigration, respectively (31, 32). However, both CCR7 and CD62L expression on CD4 SP cells were intact in the absence of Ubc9 (Supplemental Fig. 3A). To test whether Ubc9 deficiency might result in increased apoptosis, we cultured sorted SP and DP cells in medium for 18 h. Ubc9-deficient SP cells showed significantly increased apoptosis compared with normal cells (Fig. 5C). Furthermore, addition of exogenous IL-7 could rescue about half of the WT apoptosis cells; however, this rescue ability was significantly reduced in Ubc9-deficient CD4 SP cells and was completely abolished in Ubc9-deficient CD8 SP cells (Fig. 5C), consistent with the reduced IL-7Rα expression on these cells. It is reported that IL-7 endows the survival signal for CD8 SP thymocytes mainly through the upregulation of BCL-2 (33); indeed, we found that BCL-2 expression was selectively reduced in CD8 SP thymocytes (Fig. 5D). These results indicate that as an important survival signal, attenuated IL-7 signaling contributes to the decreased survival of Ubc9-deficent SP cells, especially CD8 SP cells.
UBC9-mediated SUMOylation is intrinsically involved in cell cycle progression in proliferating cells (34). To investigate whether Ubc9 deficiency affects the acquisition of proliferation competence when T cells immigrate to the periphery, we labeled sorted CD4 SP and CD8 SP cells with CellTrace Violet and stimulated them with anti-CD3 and anti-CD28 for 3 d. Compared with WT cells, Ubc9-deficient SP cells exhibited significantly impaired proliferation driven by TCR signals (Fig. 5E). Addition of exogenous IL-7 could not rescue this defect (data not shown), suggesting that other proliferation signals might be affected because of Ubc9 deletion in mature SP cells. Thus, these data indicate that Ubc9 is required for SP thymocytes to receive survival signals and differentiate to mature thymocytes with proliferation competence.
TCR-induced NFAT nuclear localization is regulated by SUMOylation in thymocytes
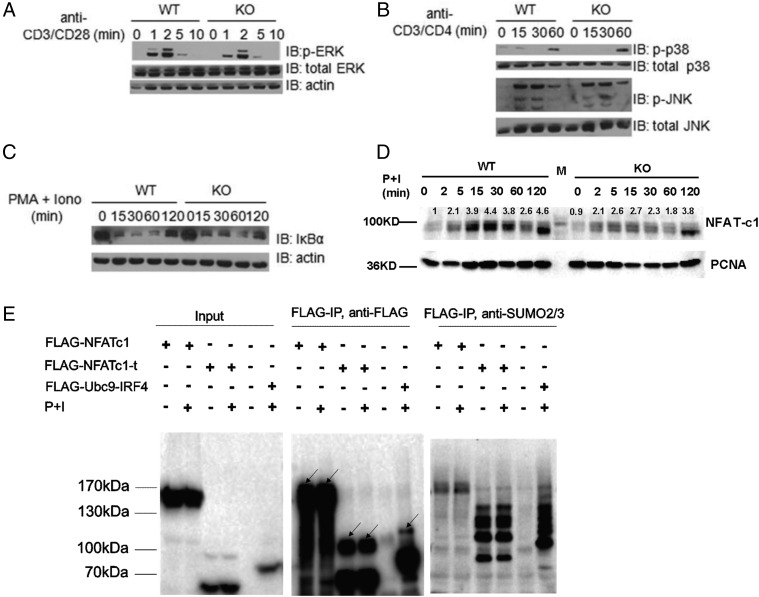
TCR signaling is the major driver for the progression of thymic positive selection (5). Because T cell development in Ubc9-deficient mice was blocked at a late stage of positive selection, and TCR-driven cell proliferation was impaired in the absence of Ubc9, we wanted to detect whether Ubc9 deficiency may affect TCR signal transduction. First, we measured the expression of CD69 on DP cells. CD69 is an early marker for positive selection, and its upregulation during positive selection is strictly dependent on TCR-proximate signaling (35). We found that presignaled DP thymocytes (CD69− CD4+ CD8+) from Ubc9-deficient mice and WT mice showed the same levels of CD69 upregulation upon stimulation with anti-CD3/CD28 in vitro (Supplemental Fig. 3B). It was highly possible that the MEK1-ERK1/2 pathway, a cascade downstream of TCR and essential for positive selection (36), would be affected by Ubc9 deficiency because MEK1 activity was modulated by Ubc9-mediated SUMOylation modification in cancer cells (37). However, this was not the case in thymocytes. Thymocytes from Ubc9-deficient mice showed the same levels of ERK activation as that from WT control mice upon stimulation by anti-CD3/CD28 cross-linking (Fig. 6A).
FIGURE 6.
NFAT nuclear retention was affected by Ubc9 deletion. (A–C) Bulk thymocytes from Ubc9fl/fl (WT) and Ubc9fl/fl Cd4Cre (KO) mice were stimulated as indicated for various times. Whole-cell lysates were obtained for Western blot analysis of indicated protein. (D) Bulk thymocytes from Ubc9fl/fl (WT) and Ubc9fl/fl Cd4Cre (KO) mice were stimulated with PMA and Ionomycin for indicated times, nuclear component was extracted, and nuclear NFATc1 was detected by Western blot. Relative expression of NFATc1 was normalized to the basal level of that in WT thymocytes using ImageJ. (E) Primary CD4+ T cells isolated from mouse lymph nodes and spleen were infected with retrovirus containing FLAG-tagged full-length NFATc1 or truncated NFATc1 (with 1–300 aa deletion at N terminus). Cells were stimulated by PMA and ionomycin for 2 h or left unstimulated before harvest. FLAG IP was performed in denaturing condition, and anti-FLAG and anti-SUMO2/3 Abs were used to detect the SUMOylated bands. In the middle panel, SUMOylated bands were indicated by arrows. FLAG-NFATc1, full-length NFATc1; FLAG-NFATc1-t, truncated NFATc1; P+I, PMA and Ionomycin; FLAG-Ubc9-IRF4 was used as a positive control. These data are representative of two times. (A–D) These data are representative of at least three experiments.
Several TCR signal regulators such as Themis and TAK1 have definitive roles during T cell development (38, 39). Mice with these genes deleted in T cells displayed similar phenotypes to Ubc9-deficient mice reported in this article. These mice showed impaired production of mature SP thymocytes and failure of downregulation of CD69/HSA at the SP stage (38, 39). Moreover, recently it was reported that NF-κB is involved in the late stages of T cell maturation in the thymus (40).We thus asked whether posttranslational modification or downstream target activity of these proteins was regulated by Ubc9-mediated SUMOylation pathway during positive selection. Themis, however, could not be modified by SUMO even if overexpressed together with SUMO1/2 and UBC9 in 293T cells (Supplemental Fig. 3C). Activation of NF-κB and JNK, two pathways modulated by TAK1 in thymocytes (39), were also intact in the absence of Ubc9 (Fig. 6B, 6C).
The calcineurin-NFAT pathway plays crucial roles in the thymocyte proliferation and maturation (41). Deficiency of calcineurin in T cell resulted in a reduced cell numbers of SP thymocytes and a defect of peripheral T cell activation (33). It was reported that NFAT1 and NFAT2 can be directly SUMOylated, and SUMOylation modification regulates their transcriptional activities by promoting their nuclear anchorage within nuclear bodies (42, 43). We observed that there was no difference in calcium flux between DP or SP cells from WT mice and Ubc9-deficient mice upon anti-CD3/CD4 cross-linking (Supplemental Fig. 3D), indicating the calcium signals for NFAT activation were intact. However, when stimulated with PMA and ionomycin, NFAT2 (NFATc1) failed to enter the nucleus efficiently in Ubc9-deficient thymocytes (Fig. 6D). We further confirmed that NFATc1 can be SUMOylated in primary T cells (Fig. 6E). Collectively, these results suggest that among numerous proximal signaling pathways driven by TCR stimulation, NFAT nuclear retention is affected in the absence of Ubc9 in thymocytes.
Discussion
SUMOylation is an evolutionarily conserved protein modification that is critical for a wide range of physiological processes in eukaryotic organism. In this study, we genetically investigated the role of SUMOylation in T cell development and demonstrated a critical role of this regulation in positive selection of thymic T cells, especially for the maturation step.
Previous studies in yeast, drosophila, zebrafish, and mouse showed that global inhibition of SUMOylation at the organismal level selectively affects mitotic cells, resulting in cell cycle arrest, chromosome defect, nuclear structure disintegration, or even apoptosis in highly proliferative cells (18, 19, 44–47). In this study, nondividing status of most DP thymocytes (∼90%) (48) allowed us to interrogate the definitive role of SUMOylation pathway during T cell development, making it unnecessary to consider the interference from general defects incurred by Ubc9 deletion as in dividing cells. Our genetic study revealed that T cell development was blocked from DP-to-SP transition when Ubc9 was deleted specifically in DP thymocytes because of defective positive selection. The reduction of CD8 SP in Ubc9-deficent mice was more severe than CD4 SP, indicating the positive selection process might be different in these two populations. In terms of peripheral T cells, homeostatic expansion will enforce the thymic recent emigrants into active cell cycle, which probably induces proliferation block and apoptosis in the absence of Ubc9. Therefore, it is likely that T cell lymphopenia in the periphery was secondary to defective T cell development in this study.
T cell maturation is important in generating a functional peripheral T cell pool. In addition to conventional T cells, development of other mature thymocyte populations including NKT and FOXP3+ regulatory T cells was also impaired in the absence of Ubc9, indicating Ubc9 is essential for the late-stage maturation stage after positive selection. The molecular mechanism underlying the maturation stage is not entirely understood so far. In our study, we found increased apoptosis and defect in proliferation were associated with the immature phenotype. IL-7Rα was selectively reduced in CD8 SP thymocytes, accompanied with reduced expression of antiapoptotic BCL-2, which might account for the inferior survival and severe cell number reduction of these cells. However, IL-7 signaling and BCL-2 expression were quite normal in CD4 SP thymocytes; other signals that regulate cell survival may be regulated by Ubc9 in CD4 SP cells.
The NF-κB pathway plays important roles in positive selection and postselection maturation (40, 49). Moreover, many components of the NF-κB pathway have been reported to be SUMO targets (50). However, we did not observe any defect in this pathway in Ubc9-deficient thymocytes or Ubc9-deficient peripheral T cells (17), suggesting that SUMOylation regulation may play distinctive roles in different cell types or at different developmental stages. In our study, we found that most of the proximate signaling downstream of TCR was intact in the absence of Ubc9. However, TCR-induced NFAT nuclear localization was significantly affected because of Ubc9 deletion, indicating the dysfunctional role of this signaling pathway in Ubc9-deficent thymocytes. The direct targets altered by a defective NFAT pathway during thymocytes development remain to be determined.
In summary, our study has revealed an obligatory role of SUMOylation in T cell development. Further investigations on Ubc9 deletion in other immune cells or at different stages of T cell development would enrich our knowledge on the posttranslational regulation of immune cell function and help to devise new strategies to treat immune diseases.
Supplementary Material
Acknowledgments
We thank the flow cytometry facility of the Center for Immunology, Tsinghua University, for help in cell sorting, Dionne Prescod for help in maintaining the mouse colony, and members of Dr. Chen Dong's laboratory for technical support and assistance.
This work was supported by National Natural Science Foundation of China Grant 041321082.
The online version of this article contains supplemental material.
- DN
- double-negative
- DP
- double-positive
- HSA
- heat-stable Ag
- KO
- knockout
- MHC II
- MHC class II
- pMHC
- self-peptide MHC complex
- SP
- single-positive
- SUMO
- small ubiquitin-like modifier
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Zinkernagel R. M., Callahan G. N., Klein J., Dennert G. 1978. Cytotoxic T cells learn specificity for self H-2 during differentiation in the thymus. Nature 271: 251–253. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald H. R., Lees R. K., Schneider R., Zinkernagel R. M., Hengartner H. 1988. Positive selection of CD4+ thymocytes controlled by MHC class II gene products. Nature 336: 471–473. [DOI] [PubMed] [Google Scholar]
- 3.Kisielow P., Teh H. S., Blüthmann H., von Boehmer H. 1988. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature 335: 730–733. [DOI] [PubMed] [Google Scholar]
- 4.Teh H. S., Kisielow P., Scott B., Kishi H., Uematsu Y., Blüthmann H., von Boehmer H. 1988. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature 335: 229–233. [DOI] [PubMed] [Google Scholar]
- 5.Starr T. K., Jameson S. C., Hogquist K. A. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21: 139–176. [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Adams A., Wildt K. F., Aronow B., Feigenbaum L., Bosselut R. 2003. Restricting Zap70 expression to CD4+CD8+ thymocytes reveals a T cell receptor-dependent proofreading mechanism controlling the completion of positive selection. J. Exp. Med. 197: 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fink P. J., Hendricks D. W. 2011. Post-thymic maturation: young T cells assert their individuality. Nat. Rev. Immunol. 11: 544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mick V. E., Starr T. K., McCaughtry T. M., McNeil L. K., Hogquist K. A. 2004. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J. Immunol. 173: 5434–5444. [DOI] [PubMed] [Google Scholar]
- 9.Azzam H. S., Grinberg A., Lui K., Shen H., Shores E. W., Love P. E. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarakhovsky A., Kanner S. B., Hombach J., Ledbetter J. A., Müller W., Killeen N., Rajewsky K. 1995. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science 269: 535–537. [DOI] [PubMed] [Google Scholar]
- 11.Swat W., Dessing M., von Boehmer H., Kisielow P. 1993. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur. J. Immunol. 23: 739–746. [DOI] [PubMed] [Google Scholar]
- 12.Singer A., Adoro S., Park J. H. 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8: 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita I., Nagata T., Tada T., Nakayama T. 1993. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int. Immunol. 5: 1139–1150. [DOI] [PubMed] [Google Scholar]
- 14.Ramsdell F., Jenkins M., Dinh Q., Fowlkes B. J. 1991. The majority of CD4+8- thymocytes are functionally immature. J. Immunol. 147: 1779–1785. [PubMed] [Google Scholar]
- 15.Geiss-Friedlander R., Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956. [DOI] [PubMed] [Google Scholar]
- 16.Decque A., Joffre O., Magalhaes J. G., Cossec J. C., Blecher-Gonen R., Lapaquette P., Silvin A., Manel N., Joubert P. E., Seeler J. S., et al. 2016. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat. Immunol. 17: 140–149. [DOI] [PubMed] [Google Scholar]
- 17.Ding X., Wang A., Ma X., Demarque M., Jin W., Xin H., Dejean A., Dong C. 2016. Protein SUMOylation is required for regulatory T cell expansion and function. Cell Rep. 16: 1055–1066. [DOI] [PubMed] [Google Scholar]
- 18.Demarque M. D., Nacerddine K., Neyret-Kahn H., Andrieux A., Danenberg E., Jouvion G., Bomme P., Hamard G., Romagnolo B., Terris B., et al. 2011. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology 140: 286–296. [DOI] [PubMed] [Google Scholar]
- 19.Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9: 769–779. [DOI] [PubMed] [Google Scholar]
- 20.Lee P. P., Fitzpatrick D. R., Beard C., Jessup H. K., Lehar S., Makar K. W., Pérez-Melgosa M., Sweetser M. T., Schlissel M. S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774. [DOI] [PubMed] [Google Scholar]
- 21.Prinz I., Sansoni A., Kissenpfennig A., Ardouin L., Malissen M., Malissen B. 2006. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat. Immunol. 7: 995–1003. [DOI] [PubMed] [Google Scholar]
- 22.Chun T., Page M. J., Gapin L., Matsuda J. L., Xu H., Nguyen H., Kang H. S., Stanic A. K., Joyce S., Koltun W. A., et al. 2003. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J. Exp. Med. 197: 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan M. S., Boesteanu A., Reed A. J., Petrone A. L., Holenbeck A. E., Lerman M. A., Naji A., Caton A. J. 2001. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2: 301–306. [DOI] [PubMed] [Google Scholar]
- 24.Barnden M. J., Allison J., Heath W. R., Carbone F. R. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76: 34–40. [DOI] [PubMed] [Google Scholar]
- 25.Kisielow P., Blüthmann H., Staerz U. D., Steinmetz M., von Boehmer H. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333: 742–746. [DOI] [PubMed] [Google Scholar]
- 26.Staton T. L., Lazarevic V., Jones D. C., Lanser A. J., Takagi T., Ishii S., Glimcher L. H. 2011. Dampening of death pathways by schnurri-2 is essential for T-cell development. Nature 472: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He X., He X., Dave V. P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B. A., Kappes D. J. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433: 826–833. [DOI] [PubMed] [Google Scholar]
- 28.Held W., MacDonald H. R., Mueller C. 1990. Expression of genes encoding cytotoxic cell-associated serine proteases in thymocytes. Int. Immunol. 2: 57–62. [DOI] [PubMed] [Google Scholar]
- 29.Yu Q., Park J. H., Doan L. L., Erman B., Feigenbaum L., Singer A. 2006. Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J. Exp. Med. 203: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tani-ichi S., Shimba A., Wagatsuma K., Miyachi H., Kitano S., Imai K., Hara T., Ikuta K. 2013. Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc. Natl. Acad. Sci. USA 110: 612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurobe H., Liu C., Ueno T., Saito F., Ohigashi I., Seach N., Arakaki R., Hayashi Y., Kitagawa T., Lipp M., et al. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity 24: 165–177. [DOI] [PubMed] [Google Scholar]
- 32.Spertini O., Kansas G. S., Munro J. M., Griffin J. D., Tedder T. F. 1991. Regulation of leukocyte migration by activation of the leukocyte adhesion molecule-1 (LAM-1) selectin. Nature 349: 691–694. [DOI] [PubMed] [Google Scholar]
- 33.Bueno O. F., Brandt E. B., Rothenberg M. E., Molkentin J. D. 2002. Defective T cell development and function in calcineurin A beta-deficient mice. Proc. Natl. Acad. Sci. USA 99: 9398–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flotho A., Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82: 357–385. [DOI] [PubMed] [Google Scholar]
- 35.Bendelac A., Matzinger P., Seder R. A., Paul W. E., Schwartz R. H. 1992. Activation events during thymic selection. J. Exp. Med. 175: 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer A. M., Katayama C. D., Pagès G., Pouysségur J., Hedrick S. M. 2005. The role of erk1 and erk2 in multiple stages of T cell development. Immunity 23: 431–443. [DOI] [PubMed] [Google Scholar]
- 37.Kubota Y., O’Grady P., Saito H., Takekawa M. 2011. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat. Cell Biol. 13: 282–291. [DOI] [PubMed] [Google Scholar]
- 38.Lesourne R., Uehara S., Lee J., Song K. D., Li L., Pinkhasov J., Zhang Y., Weng N. P., Wildt K. F., Wang L., et al. 2009. Themis, a T cell-specific protein important for late thymocyte development. Nat. Immunol. 10: 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H.-H., Xie M., Schneider M. D., Chen Z. J. 2006. Essential role of TAK1 in thymocyte development and activation. Proc. Natl. Acad. Sci. USA 103: 11677–11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing Y., Wang X., Jameson S. C., Hogquist K. A. 2016. Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macian F. 2005. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 5: 472–484. [DOI] [PubMed] [Google Scholar]
- 42.Nayak A., Glöckner-Pagel J., Vaeth M., Schumann J. E., Buttmann M., Bopp T., Schmitt E., Serfling E., Berberich-Siebelt F. 2009. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J. Biol. Chem. 284: 10935–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terui Y., Saad N., Jia S., McKeon F., Yuan J. 2004. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 279: 28257–28265. [DOI] [PubMed] [Google Scholar]
- 44.Kaikkonen S., Makkonen H., Rytinki M., Palvimo J. J. 2010. SUMOylation can regulate the activity of ETS-like transcription factor 4. Biochim. Biophys. Acta 1799: 555–560. [DOI] [PubMed] [Google Scholar]
- 45.Nowak M., Hammerschmidt M. 2006. Ubc9 regulates mitosis and cell survival during zebrafish development. Mol. Biol. Cell 17: 5324–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasso M. 2008. Emerging roles of the SUMO pathway in mitosis. Cell Div. 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanakousaki K., Gibson M. C. 2012. A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development 139: 2751–2762. [DOI] [PubMed] [Google Scholar]
- 48.Seitan V. C., Hao B., Tachibana-Konwalski K., Lavagnolli T., Mira-Bontenbal H., Brown K. E., Teng G., Carroll T., Terry A., Horan K., et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimi E., Strickland I., Voll R. E., Long M., Ghosh S. 2008. Differential role of the transcription factor NF-kappaB in selection and survival of CD4+ and CD8+ thymocytes. Immunity 29: 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mabb A. M., Miyamoto S. 2007. SUMO and NF-kappaB ties. Cell. Mol. Life Sci. 64: 1979–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.