Abstract
Ion mobility-mass spectrometry (IM-MS) has proven to be a highly informative technique for the characterization of lipids from cells and tissues. We report the combination of hydrophilic-interaction liquid chromatography (HILIC) with traveling-wave IM-MS (TWIM-MS) for comprehensive lipidomics analysis. Main lipid categories such as glycerolipids, sphingolipids, and glycerophospholipids are separated on the basis of their lipid backbones in the IM dimension, whereas subclasses of each category are mostly separated on the basis of their headgroups in the HILIC dimension, demonstrating the orthogonality of HILIC and IM separations. Using our previously established lipid calibrants for collision cross-section (CCS) measurements in TWIM, we measured over 250 CCS values covering 12 lipid classes in positive and negative modes. The coverage of the HILIC-IM-MS method is demonstrated in the analysis of Neuro2a neuroblastoma cells exposed to benzalkonium chlorides (BACs) with C10 or C16 alkyl chains, which we have previously shown to affect gene expression related to cholesterol and lipid homeostasis. We found that BAC exposure resulted in significant changes to several lipid classes, including glycerides, sphingomyelins, phosphatidylcholines, and phosphatidylethanolamines. Our results indicate that BAC exposure modifies lipid homeostasis in a manner that is dependent upon the length of the BAC alkyl chain.
Keywords: liquid chromatography, cholesterol/biosynthesis, phospholipids, collision cross-section, benzalkonium chloride, AY9944, neuro2a cells, hydrophilic interaction liquid chromatography
Lipids play important roles in maintaining membrane structures and mediating signaling pathways (1, 2). Dysregulated lipid biosynthesis and metabolism have been implicated in a number of human diseases, such as atherosclerosis (3), nonalcoholic fatty liver (4), diabetes (5), Alzheimer’s disease (6), and cancer (7, 8). Advances in lipidomic techniques and strategies, led by the LIPID MAPS consortium, have greatly enhanced our understanding of the distribution and biological roles of lipids (9–13). Modern mass spectrometry (MS), coupled with electrospray ionization (ESI), is the key to qualitative and quantitative lipidomic analysis. Typical MS-based lipidomic strategies are shotgun (i.e., direct infusion) lipidomics (9, 14) and liquid chromatography (LC)-MS lipidomics (11, 15, 16). Shotgun lipidomics relies on partial intrasource separation of lipid classes through varying the pH of the lipid solution and identification of lipid species by their characteristic fragmentation in tandem MS analysis (9, 17). This approach has the advantage of being high-throughput, but it also has several disadvantages: a) suppression of low-abundant species by major polar lipids such as phosphatidylcholines; b) difficulty in analysis of lipid species that are poorly ionized by ESI; and c) inability to provide structural information on isobaric and isomeric species. The LC-MS-based strategy utilizes targeted analysis of each lipid class under conditions that are optimized for that particular class (11, 15, 16, 18). This strategy has the advantages of being specific, sensitive, and comprehensive, but it also has limitations, such as being time consuming and less cost effective. To increase the throughput of the LC-MS lipidomics while maintaining the specificity and sensitivity, we desire improved chromatographic techniques and additional dimensions of separation that are orthogonal to LC and MS.
Ion mobility spectrometry (IMS) provides such orthogonal separation (19–24). IMS separates ions on the basis of the mobility of the ions as they travel through a neutral background gas (most commonly helium and nitrogen), which is governed by the collision frequency between the ions and the neutral gas, i.e., the ion-neutral collision cross-section (CCS; Ω). The CCS is determined by the size and shape of the ion in the gas phase and the specific neutral gas. When ion mobility is coupled with mass spectrometry, a two-dimensional separation is achieved on the basis of the CCS-to-charge (Ω/z) and the mass-to-charge (m/z), respectively. Several ion mobility (IM)-MS techniques have been applied to lipidomic analysis, including drift tube ion mobility (DTIM) (25–27), traveling wave ion mobility (TWIM) (28, 29), and differential mobility spectrometry (DMS) (30–32). In DTIM-MS, the ions travel through the neutral gas-filled drift tube under a low and static electric field, which leads to separation of ions on the scale of micro- to milliseconds (21). DTIM offers high-IM resolving power and allows direct measurement of CCS. Using DTIM-MS, the Woods and McLean laboratories have independently reported separation of different polar lipid classes on the basis of their headgroups and the acyl chain composition (25–27). DMS, also called field asymmetric ion mobility spectrometry, separates ions on the basis of their different mobilities in the high and low electric fields of an asymmetric voltage waveform (33). Recently, the groups of Ekroos (31) and Dennis (32) successfully applied DMS to both shotgun and LC-based lipidomics, achieving a more specific and sensitive analysis. DMS offers the highest resolving power (30, 33), but in comparison with the time-dispersive DTIM and TWIM, it has the drawbacks of the following: a) the separation is mobility selective, i.e., only ions with certain mobilities can pass through the device, and b) the separation in DMS does not correlate with the CCS of the ions, i.e., no structural information can be obtained directly and the behavior of certain ions cannot be predicted by CCS values. TWIM also separates ions on the basis of their CCSs, similar to DTIM, but it uses a migrating low-voltage wave to push the ions through the inert gas (34, 35) instead of a static electric field as in DTIM. TWIM offers higher sensitivity than does traditional DTIM and a faster duty cycle, but has slightly lower IM resolution and does not allow direct measurement of CCS values. However, CCS of unknown ions can be indirectly calculated by calibrating against appropriate ions with known CCS values (29). Calibrants that are of similar physical properties to the analytes are desired to achieve the highest accuracy of CCS measurements in TWIM. Recently, we established a series of phospholipid calibrants for measurement of lipid CCSs using TWIM, which gave CCS values that are within 2% of the values measured on DTIM (close to the errors of DTIM measurements) (36). The commercial TWIM platform is well integrated with different chromatographic separation techniques and MS analysis and is used in this study.
The chromatographic resolution of lipid classes was previously achieved by normal-phase separations using a silica or amino stationary phase and a mobile phase comprised of complex organic solvents (11, 15, 16, 37, 38). Lipid separations based on reverse-phase liquid chromatography have also been developed and typically use C8 or C18 columns with a highly organic gradient separation (11, 15, 37). The order of separation of lipids in normal- and reverse-phase chromatography are distinct but complimentary in that lipids separate by headgroup in normal-phase chromatography and by acyl chain length and unsaturation in reverse-phase chromatography. However, gradients for these separations tend to be long (30 min or longer), and sometimes two separate separations need to be performed to have complete coverage. LC separations by hydrophilic-interaction liquid chromatography (HILIC), which combines a hydrophilic stationary phase and reverse-phase mobile phase, were initially widely adopted for the analysis of small polar metabolites because of their increased retention on HILIC columns in relation to reverse-phase C18 columns (39–41). Recently, HILIC has been applied to the separation of phospholipid species as an alternative to lipid separations by normal- and reverse-phase chromatography (32, 42–45). HILIC separations offer the benefits of both reverse-phase and normal-phase lipid separations in that lipid species are first resolved on the basis of polar headgroup, similar to a normal-phase separation, and then by acyl chain length and unsaturation within a lipid class, as seen in reverse-phase separations (32, 43).
In this work, we report the establishment of a three-dimensional HILIC-IM-MS method for comprehensive lipidomics analysis, which achieves the resolution of isobaric lipid species by structure and hydrophilicity. Using this lipidomics methodology in combination with lipid CCS calibrants, we have determined over 250 lipid CCS values in positive and negative ionization modes. Application of the lipidomics method is demonstrated in assessing the changes to lipid homeostasis in a neuroblastoma cell line that was exposed to different small molecules, including the environmental chemicals, benzalkonium chlorides (BACs), and a known inhibitor of cholesterol biosynthesis, AY9944.
MATERIALS AND METHODS
Reagents
HPLC grade solvents (water, acetonitrile, methylene chloride, chloroform, and methanol), ammonium acetate (Optima LC/MS), and sodium chloride were purchased from Thermo Fisher Scientific. BAC with alkyl chain lengths of 10 and 16 (BAC-C10 and BAC-C16, respectively), butylated hydroxytoluene (BHT), and triphenylphoshine (PPh3) were purchased from Sigma-Aldrich. AY9944 [trans-1,4-bis(2-chlorobenzylaminomethyl) cyclohexane dihydrochloride] (purity of >99%) was prepared as described previously (46). AY9944, BAC-C10, and BAC-C16 were each dissolved in dimethyl sulfoxide (DMSO) to make a 1-mM stock solution and stored at −80°C. The following extracts were purchased from Avanti Polar Lipids: ceramide (Cer, 860052P), cerebroside (hexosyl Cer, HexCer, 131303P), sphingomyelin (SM, 860062C), L-α-phosphatidic acid (PA, 840101C), L-α-phosphatidylcholine (PC, 840051C), L-α-lysophosphatidylcholine (LysoPC, 830071P), L-α-phosphatidylethanolamine (PE, 840022C), L-α-lysophosphatidylethanolamine (LysoPE, 850095P), L-α-phosphatidylglycerol (PG, 841138C), L-α-phosphatidylinositol (PI, 840042C), and L-α-phosphatidylserine (PS, 840032C). Diacylglycerides standards were purchased from Nu-Chek Prep: 1,3-dimyristoyldiacylglyceride (1,3-DG 14:0, D-142), 1,3-dipalmitoyldiacylglycerol (1,3-DG 16:0, D-152), 1,3-distearoyldiacylglyceride (1,3-DG 18:0, D-162), 1,3-dioleoyldiacylglyceride (1,3-DG 18:1, D-237), cis-1,3-dilinoleoyldiacylglyceride (cis-1,3-DG 18:2, D-252), 1,3-diarachidoyldiacylglyceride (1,3-DG 20:0, D-172), and 1,3-diarachidonoyldiacylglyceride (1,3-DG 20:4, D-297). Stock solutions of lipid standards and standard extracts were prepared at 1 mM in chloroform. For the analysis, a 5-µM mixture of lipid standards in buffer B was prepared by dilution from the stock solutions.
Cell culture, treatment, and lipid extraction
Neuro2a cells were purchased from the American Type Culture Collection (Rockville, MD). Cultures were maintained in DMEM high-glucose media (ThermoFisher Scientific Gibco), supplemented with L-glutamine (GE Healthcare Hyclone), 10% FBS (GE Healthcare Hyclone), and penicillin/streptomycin (ThermoFisher Scientific Gibco) at 37°C and 5% CO2. For the experiment, cells were seeded at a density of 1.0 × 106 cells per 100-mm plate and left to adhere overnight. The next day, the media were replaced with DMEM high-glucose media containing N2-supplement (ThermoFisher Scientific Gibco), L-glutamine, penicillin/streptomycin, and one of the following treatments: 0.1% DMSO, as vehicle control; 100 nM BAC-C10; 100 nM BAC-C16; or 100 nM AY9944, a known inhibitor of cholesterol biosynthesis. After 48-h of treatment, cells were harvested and pelleted by centrifugation. Cell pellets were lysed by sonicating cold in 150 µL of 1× PBS for 30 min. The experiments were performed in triplicate for each condition. Protein weight was measured with the BioRad-DC Protein Assay Kit. The average protein weight of the Neuro2a cell pellets was 0.76 ± 0.10 mg with no statistical significance between treatment groups (Student’s t-test, P ≥ 0.1). To extract the lipids, we added 1 ml of NaCl aqueous solution (0.9%) and 4 ml of Folch solution (2:1, chloroform: methanol, containing 1 mM BHT and 1 mM PPh3) to the cell lysates. The mixture was briefly vortexed and centrifuged for 5 min. The lower organic phase was recovered and dried using the speed vacuum (Thermo Fisher Savant). Dried extracts were redissolved in 300 µL of methylene chloride. For analysis, 100 µL of the lipid extract was transferred to an autosampler vial, dried under Ar, and reconstituted with 200 µL of buffer B.
Liquid chromatography and ion mobility-mass spectrometry
Chromatographic separation was performed using a Waters Acquity FTN ultraperformance liquid chromatography (Waters Corp., Milford, MA) with a hydrophilic interaction column (HILIC; Phenomenex Kinetex, 2.1 × 100 mm, 1.7 µm) maintained at 40°C. The solvent system consisted of 50% acetonitrile/50% water with 5 mM ammonium acetate (buffer A) and 95% acetonitrile/5% water with 5 mM of ammonium acetate (buffer B) (32). Using a 0.5 ml/min flow rate, the linear gradient conditions were 0–1 min, 100% B; 4 min, 90% B; 7–8 min, 70% B; and 9–12 min, 100% B. Injection volumes were 5 and 15 μL for positive and negative mode analyses, respectively.
IM-MS analysis was performed on a Waters Synapt G2-Si HDMS (Waters Corp., Milford, MA) equipped with an ESI source. ESI capillary voltages of +3.0 and −2.0 kV were used for positive- and negative-mode analyses, respectively. Other ESI conditions were as follows: sampling cone, 40 V; extraction cone, 80 V; source temperature, 100°C; desolvation temperature, 350°C; cone gas, 10 L/h; and desolvation gas, 1000 L/h. Mass calibration was performed with sodium formate for the range of m/z 50–1200. Collision cross-section calibration was performed using a set of PC and PE CCS standards as previously described (36). Briefly, PC (positive mode) standards covering the range m/z 454–980 and 213–330 Å2 and PE (negative mode) standards covering the range m/z 410–786 and 199–274 Å2 were used to fit a curve of the form Ω′ = A(t′d + t0)B, where Ω′ is the corrected CCS value, t′d is the corrected drift time value, and A, t0, and B are fit parameters. For the analysis of cell extracts, IM separation was performed with a traveling wave velocity of 550 m/s and height of 40 V. Additional experiments with standard lipid extracts were performed at 500 m/s and 40 V in triplicate over three days. Untargeted MS/MS was performed with a collision energy ramp of 35–45 eV applied to the transfer region of the instrument. Data were acquired over m/z 50–1200 with 1 s scan time. Leucine enkephalin was acquired as a lockspray signal for postacquisition correction of m/z and drift time.
Data analysis
Data alignment, peak detection, and normalization were performed in Progenesis QI (Nonlinear Dynamics). The chromatographic region from 0.4 to 9 min was considered for peak detection. Adducts considered for deconvolution included [M+H]+, [M+Na]+, [M+H−H2O]+, [M+H−2H2O]+, and [M+NH4]+. Features with deconvoluted adducts are denoted with n, whereas features that could not be assigned a neutral mass are denoted with m/z. The reference sample for alignment was automatically selected by Progenesis QI, and data were normalized to all compounds. The normalization was performed by a median and mean absolute deviation approach on all detected abundances, which effectively corrects for technical variation (such as sample loading size) and minimizes influence by noise in the data and bias in the calculation. Features corresponding to the m/z of AY9944, BAC C10, and BAC C16 were excluded from the analysis. The resulting data set was filtered by ANOVA P value ≤ 0.05 and fold-change ≥ 1.5. Additional statistical analysis was performed in EZInfo (Umetrics). Identifications were made against the METLIN and LipidMAPS databases within 15 ppm mass accuracy. CCS values for lipid standard extracts were obtained using the DriftScope (v2.8; Waters Corp.) chromatographic peak detection algorithm with lock mass correction.
RESULTS
HILIC-IM-MS method for comprehensive lipidomics
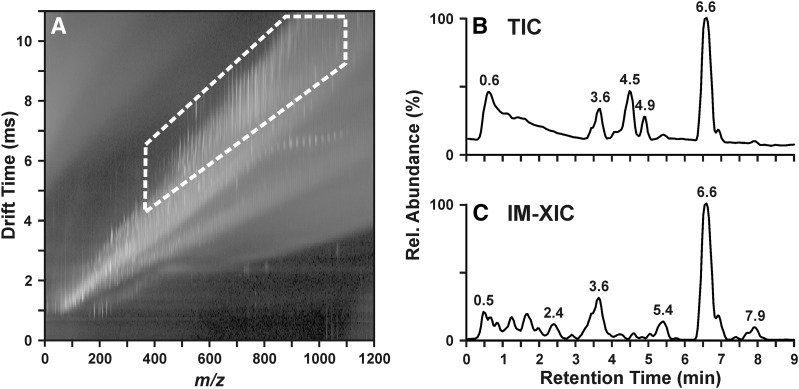
IM-MS has become a widely used platform for the analysis of biomolecules such as lipids because of its ability to resolve same-mass ions on the basis of structure. For the analysis of complex samples, this structural separation affords improvements in analyte specificity, i.e., signals corresponding to lipids can be isolated from other classes of biomolecules and chemical noise because of the low gas-phase density of lipid ions (24). Figure 1 demonstrates the improvement in analyte specificity and signal-to-noise ratio in the analysis of the lipids of Neuro2a neuroblastoma cells by LC-IM-MS. The lipid signals, outlined in Fig. 1A, occupy a distinct region of m/z-drift time space covering m/z ∼400–1050 and drift time 4–11 ms. The total ion chromatogram (TIC) from the LC dimension, shown in Fig. 1B, reveals several peaks but suffers from a high background in the early portion of the chromatogram. By excising the region containing the lipid signals (highlighted in Fig. 1A) away from the remaining signals, the background in the TIC (Fig. 1B) is visibly reduced, as shown in Fig. 1C. A number of peaks observed in Fig. 1B, such as those at 4.5 and 4.9 min, are significantly reduced in intensity in the ion mobility-extracted ion chromatogram (IM-XIC) (Fig. 1C), which indicates that the species at these retention times are not present in the highlighted region of Fig. 1A and are not lipids. In regions of the chromatogram where lipids are known to elute, such as 2.4, 5.4, and 7.9 min, no peak is clearly visible in Fig. 1B. The presence of such peaks is apparent only upon the removal of chemical noise by extracting out the lipid region of the IM-MS data, as shown in Fig. 1C.
Fig. 1.
Lipids occupy a distinct region of IM-MS conformation space, outlined with the white dashed line in A. The high background in the total ion chromatogram (TIC) of a Neuro2a lipid extract in B is reduced, and more peaks become visible when the lipid region of the IM-MS plot (A) is extracted to yield an ion mobility-extracted ion chromatogram (IM-XIC) (C). Rel, relative.
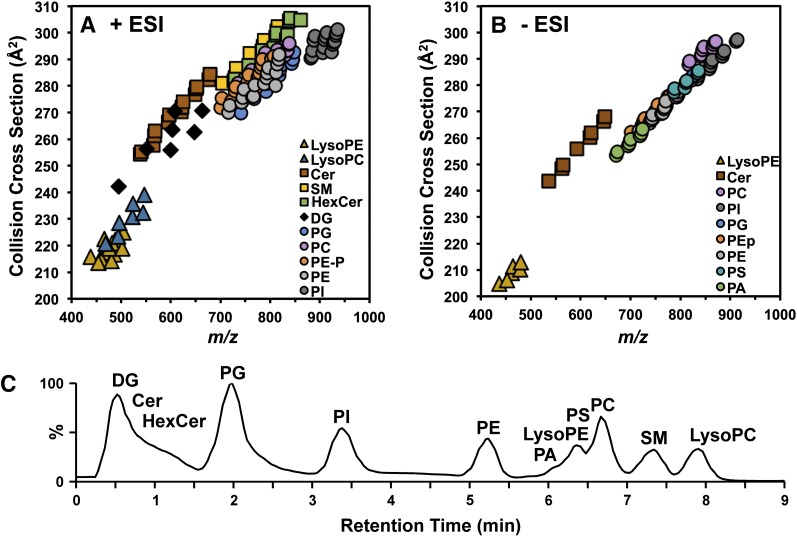
Using the lipid CCS calibrants we established recently (36), we generated over 250 lipid CCS values (positive, n = 165; negative, n = 93; relative standard deviation (RSD) ≤ 0.5% CCS) from a mixture containing 12 lipid classes: LysoPE, LysoPC, 1,3-DG, Cer, HexCer, PA, PC, PE (including plasmalogen PE, PEp), PG, PI, PS, and SM (Fig. 2A, B; supplemental Tables S2, S3). In addition to the broad resolution of lipid species from chemical noise and nonlipid species, IM-MS is also able to resolve class-specific differences in lipid species. Sphingolipids (square data points in Fig. 2A), such as ceramides, sphingomyelins, and hexosyl ceramides, tended to have larger CCS values than did glycerophospholipids (circle data points in Fig. 2A). The trendline of each class of lipids was obtained by fitting to power function, which shows the order of packing efficiencies of different lipid classes in the gas phase (supplemental Fig. S1, supplemental Table S1). Within the sphingolipid classes, CCS trend is dependent upon the presence and type of headgroup such that the trendlines lowered in the order of Cer > SM > HexCer. CCSs among the phospholipids were likewise dependent upon the type of headgroup. In positive mode, glycerophospholipid CCS trendlines were observed to decrease in the order of PC > PEp > PE > PG > PI. Two additional classes were observed only in negative mode (PAs and PSs), where CCS trendlines decreased in the order of PEp ≈ PS > PE ≈ PG > PA > PI. We suggest that these trendlines can be used to estimate the CCS of unknown lipid species in order to facilitate their identification (see discussion below regarding Fig. 3).
Fig. 2.
Collision cross-section measurements of 12 species of sphingolipids (squares), glycerolipids (diamonds), glycerophospholipids (circles), and lysoglycerophospholipids (triangles) in positive ionization mode (A) and negative ionization mode (B). (C) Ion mobility-extracted ion chromatogram from the HILIC separation of the 12-lipid mixture.
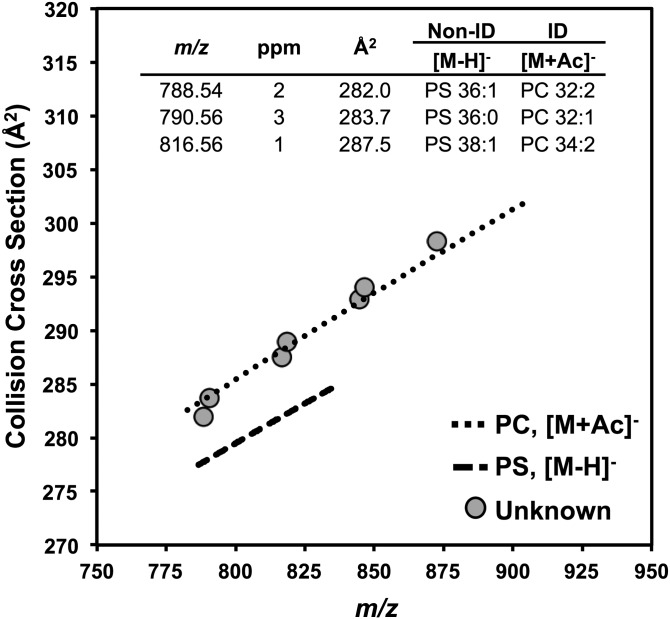
Fig. 3.
In negative ionization mode, PC [M+Ac]− and PS [M−H]− species are isobaric, as shown in the table within the figure. Unknown lipid signals (gray circles) from 6–7 min in the HILIC chromatogram were plotted onto trendlines fit to CCS values of known PS (long dashed line) and PC (short dashed line) species. The unknown lipids were determined to be PCs because their CCS values fall onto the PC trendline.
As shown in Fig. 2A and B, the region of the IM-MS plot from m/z 600–950 and 250–310 Å2 is densely occupied with several distinct classes of lipid species. Although there are large differences in CCS between several lipid classes (e.g., ∼2.5% difference between similar-mass SM and PC), the majority of lipid classes are more narrowly resolved. We observed several examples of lipids from different classes that were close in m/z and in CCS, particularly in negative mode where most ions were observed as [M−H]−. Several nominally isobaric PA and PG species observed in negative mode were not well resolved by CCS, such as PG 32:1 (m/z 719.49 and 262.5 Å2) and PA 38:6 (m/z 719.47 and 260.9 Å2), which differ in drift time by 0.05 ms and in CCS by 0.62%. Other examples include PE and PG species, such as PE 38:3 (m/z 768.5 and 271.7 Å2) and PG 36:4 (m/z 769.5 and 271.3 Å2), which differ by −0.5 Å2 (0.2%) in negative mode.
To improve our ability to accurately identify a large number of lipid species, we have chosen to perform chromatographic separation by HILIC prior to IM-MS analysis (32). Figure 2C shows the IM-XIC from the analysis of a mixture of the 12 lipid classes shown in Fig. 2A and B. HILIC conditions are similar to normal-phase chromatography in that lipids are separated on the basis of their headgroup polarity, such that the order of elution is as follows: DGs ≈ Cer < HexCer < PG < PI < PE < PA < LysoPE < PS ≈ PC < SM < LysoPC. As shown in Fig. 2C, the lipid species that were prone to similar-mass and similar-CCS overlap (i.e., PGs, PAs, PEs) are well separated in the chromatographic dimension with retention times of 2.1, 5.2, and 6.0 min, respectively. We also observed separation by fatty acid composition within a chromatographic peak, such that longer and unsaturated fatty acids elute prior to shorter or fully saturated fatty acids, respectively. For example, PE 34:2 elutes at 5.45 min, and PE 38:4 elutes at 5.30 min (supplemental Table S2).
Only a few regions of the chromatogram contain multiple lipid species, namely, the early portion of the chromatogram containing DGs, ceramides, and hexosyl ceramides and the portion from 6–7 min containing PAs, LysoPEs, PSs, and PCs. In positive mode, these lipids mostly occur in distinct m/z regions, with LysoPEs having the smallest masses and PSs have the largest masses. In negative mode it is possible to observe isobaric PS and PC species in the form of [M−H]− and [M+CH3COO]−, respectively, but these lipid classes are well separated in the IM dimension. PCs tend to have the largest CCSs in positive mode; thus the addition of the acetate adduct further increases PC CCS values in relation to other glycerophospholipids such as PSs. Figure 3 demonstrates the ability to distinguish PS [M−H]− species from PC [M+CH3COO]− in a Neuro2a lipid extract. The data points in Fig. 3 represent lipid species found in the Neuro2a lipid extracts that could be either PS or PC species with the same mass accuracy. For example, the data point at m/z 788.5 and 282.0 Å2 returned database matches to PS (18:0/18:1) and PC (16:1/16:1) with 2 ppm mass accuracy. The overlay of the unknown data points onto trendlines fit to the CCSs of PC and PS standards (see Fig. 2, supplemental Fig. S1) indicated that these species are in fact PCs rather than PSs because the CCSs of the unknown lipids fall on the CCS trendline of known PCs. For example, the CCS value for m/z 816.56 in the Neuro2a sample (Fig. 3) was 287.5 Å2, and the CCS value of PC 34:2 (m/z 816.58) in the lipid mix was 287.7 Å2 (−0.07% different), which are within the accepted window of error (2% different) for positive identifications by CCS. Although the HILIC method does not fully resolve all lipid classes in the chromatographic dimension, enough separation is achieved in the IM dimension to successfully identify isobaric lipid classes, and all major lipids species are resolved in at least one of the two orthogonal dimensions of separation.
The effects of BAC on the Neuro2a lipidome
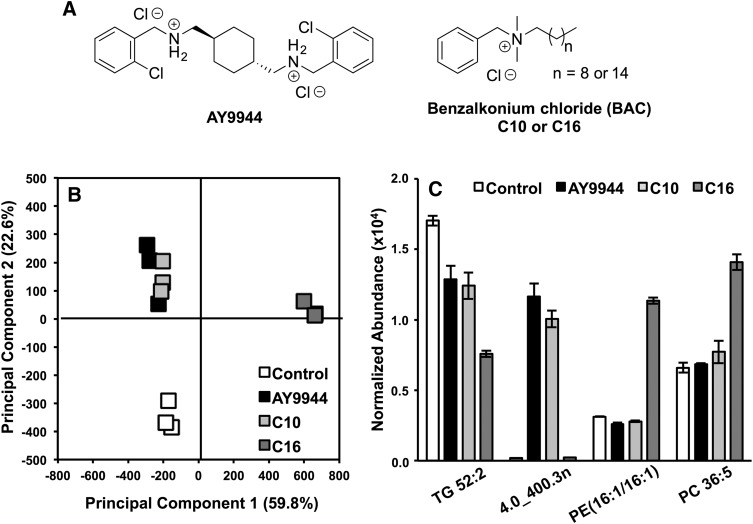
The HILIC-IM-MS method described above was used to characterize alteration in the lipid profiles of Neuro2a cells exposed to 100 nM of AY9944 or BACs with different alkyl chain lengths: BAC-C10 or BAC-C16 (Fig. 4A). AY9944 is a known inhibitor of 3β-hydroxysterol-Δ7-reductase (DHCR7), the last step of cholesterol biosynthesis (46). We recently found that BACs, a class of environmental toxins, also inhibit cholesterol biosynthesis, but their activities appear to vary with the length of the alkyl chain (47). Specifically, BAC-C10 acts as a potent inhibitor of DHCR7, whereas BAC-C16 does not, but BAC-C16 induced significantly more changes to genes associated with lipid homeostasis (47). Here we aim to elucidate the effects of these compounds on the lipidome of Neuro2a cells. Figure 4 summarizes the results of the lipid-profiling experiment. As seen in the principal components analysis (PCA) of the data, the largest difference was observed in the BAC-C16 treated samples, which separated from the control, AY9944, and BAC-C10 samples along principal component 1 (accounting for ∼60% of variance). As expected, the Neuro2a cells responded similarly to treatment with AY9944 and BAC-C10, with no clear delineation between the treatments in the PCA (Fig. 4B). However, clear separation was observed between the control group and the AY9944 and C10 group along principal component 2, which accounted for 22.6% of the variance in the dataset.
Fig. 4.
Results from lipidomic analysis of Neuro2a cells exposed to AY9944, BAC-C10 (C10), and BAC-C16 (C16) (A). (B) In the PCA, treatment groups separate from controls along PC2, and BAC-C16 is separated from AY9944, BAC-C10, and controls along PC1. (C) Group separation in the PCA is the result of molecular features such as TAG 52:2, PE (16:1/16:1), PC 36:5, and an unknown feature 4.0_400.3n. N = 3 for each condition.
Significantly changed lipid species were identified by retention time, m/z, and CCS (Table 1). All identified lipid species display CCS values that are well within 2% error of our collection of CCS values from lipid standards (if available) (Table 1). Figure 4C highlights several features that contributed to the group separations observed in the PCA. The most significant features by ANOVA P value were 4.0_398.3n and 4.0_400.3n (P = 1.4 × 10−11 and P = 9.6 × 10−12, respectively), which were greater than 30-fold more abundant in AY9944 and BAC-C10 samples than were the controls and BAC-C16 samples. Several triacylglycerol (TAG) species, including TAG 52:2, were decreased in all treatments in relation to the control. Among the treatments, BAC-C16 treatment had a greater effect (0.4-fold, P = 5.3 × 10−6) on TAG 52:2 levels than AY9944 (0.8-fold, P = 4.72 × 10−3) or BAC-C10 (0.7-fold, P = 2.9 × 10−3) treatment. Similar fold-change decreases were observed for TAG 54:3, TAG 54:2, and TAG 56:3, as well as a number of DG species including DGs 34:1 and 36:2 (Table 1). Sphingolipid levels also appeared to be affected by treatment with AY9944 and the BACs. Ceramides (d18:1/16:0) and (d18:1/24:1) were both decreased in the treatment groups in relation to controls (Table 1). Sphingomyelins of the same composition showed similar fold-change decreases across the treatment groups in relation to the control. Of interest, no significant change was observed in SM (d18:1/18:1) between the control and BAC-C16 treatment groups, but levels were decreased in AY9944 and BAC-C10 treatment (Table 1).
TABLE 1.
Altered lipids in Neuro2a cells after treatment with AY9944 (AY), BAC-C10 (C10), or BAC-C16 (C16)
| Annotationa | Retention Time (min) | m/z | Mass Error (ppm) | CCS (Å2) | ΔCCS (%)c | Abundance in Control | Fold-changee | f | ||||
| AY | C10 | C16 | AY | C10 | C16 | |||||||
| TAG 52:2 | 0.4 | 876.802 | 1.7 | 318.6 | — | 17024 ± 346 | 0.8 | 0.7 | 0.4 | 4.7 × 10−3 | 2.9 × 10−3 | 5.3 × 10−6 |
| TAG 54:3 | 0.4 | 902.819 | −1.6 | 323.4 | — | 8865 ± 192 | 0.8 | 0.7 | 0.4 | 5.8 × 10−4 | 0.01 | 3.3 × 10−6 |
| TAG 54:2 | 0.4 | 904.831 | 2.3 | 326.3 | — | 9611 ± 246 | 0.8 | 0.7 | 0.5 | 0.01 | 3.7 × 10−3 | 3.7 × 10−5 |
| DG 34:1 | 0.4 | 577.520 | −0.9 | 256.6 | 13257 ± 317 | 0.8 | 0.8 | 0.6 | 0.01 | 4.1 × 10−4 | 4.8 × 10−4 | |
| DG 36:2 | 0.4 | 603.535 | 1.0 | 260.8 | −1.0 | 8231 ± 129 | 0.8 | 0.7 | 0.5 | 5.0 × 10−3 | 4.1 × 10−3 | 2.5 × 10−5 |
| Cer(d18:1/16:0) | 0.5 | 520.510 | −0.7 | 252.7 | — | 11200 ± 2419 | 0.5 | 0.4 | 0.5 | 0.03 | 0.02 | 0.03 |
| Cer(d18:1/24:1) | 0.5 | 630.619 | 0.1 | 273.7 | −1.1 | 6701 ± 1216 | 0.7 | 0.6 | 0.5 | 0.08 | 0.04 | 0.02 |
| PG 32:1 (−) | 2.5 | 743.484 | 0.7 | 261.6 | −0.3 | 1862 ± 120 | 1.4 | 1.1 | 2.7 | 0.01 | 0.29 | 8.8 × 10−5 |
| PG 34:2 (−) | 2.4 | 769.522 | 2.0 | 266.7 | −0.2 | 4401 ± 515 | 1.3 | 1.0 | 4.2 | 0.42 | 0.94 | 3.0 × 10−3 |
| 4.0_400.3n | 4.0 | 383.331 | — | 204.4 | — | 189 ± 20 | 61.7 | 53.3 | 1.3 | 6.7 × 10−5 | 2.1 × 10−5 | 0.03 |
| 4.0_398.3n | 4.0 | 381.317 | — | 201.8 | — | 435 ± 28 | 40.1 | 32.6 | 1.3 | 1.0 × 10−4 | 6.3 × 10−5 | 2.0 × 10−3 |
| PE 36:3 | 5.4 | 742.539 | −0.9 | 264.7 | −0.7 | 10377 ± 444 | 0.8 | 0.7 | 1.5 | 0.01 | 3.2 × 10−3 | 4.6 × 10−4 |
| PE 16:1-18:2b | 5.4 | 714.508 | −1.0 | 265.4 | — | 2740 ± 47 | 0.8 | 0.9 | 3.2 | 3.4 × 10−4 | 3.1 × 10−3 | 1.5 × 10−6 |
| PE 16:1-16:1b | 5.5 | 688.494 | −3.4 | 261.2 | −1.0d | 3128 ± 25 | 0.8 | 0.9 | 3.6 | 3.3 × 10−3 | 1.7 × 10−3 | 8.3 × 10−7 |
| PC 36:5 | 6.4 | 780.554 | 0.2 | 281.7 | −0.8 | 6598 ± 361 | 1.0 | 1.2 | 2.1 | 0.40 | 0.10 | 8.9 × 10−5 |
| PC 36:3 | 6.4 | 784.586 | 1.1 | 288.5 | 1.0 | 1012 ± 91 | 0.9 | 1.1 | 1.8 | 0.35 | 0.57 | 2.4 × 10−3 |
| SM(d18:1/16:0) | 7.4 | 703.574 | 0.7 | 279.5 | −0.6 | 12784 ± 3775 | 0.3 | 0.3 | 0.5 | 0.04 | 0.03 | 0.08 |
| SM(d18:1/18:1) | 7.3 | 729.590 | 0.2 | 281.5 | −0.6 | 1487 ± 136 | 0.6 | 0.5 | 1.0 | 0.02 | 0.00 | 0.72 |
| SM(d18:1/24:1) | 7.1 | 813.682 | 3.5 | 299.7 | −0.3 | 939 ± 235 | 0.4 | 0.3 | 0.5 | 0.04 | 0.02 | 0.06 |
| LysoPE 16:1 (−) | 6.6 | 450.262 | −0.9 | 207.5 | — | 631 ± 133 | 0.7 | 0.8 | 1.7 | 0.12 | 0.20 | 0.01 |
| LysoPC 16:1 | 7.9 | 494.326 | 3.0 | 223.8 | 0.2 | 3931 ± 262 | 1.0 | 1.3 | 1.7 | 0.59 | 0.31 | 1.6 × 10−3 |
Long dashes in cells indicate not available.
Results are from positive-mode analysis unless otherwise specified by (−).
Fatty acid composition determined from additional MS/MS experiments.
Relative to CCS values in supplemental Tables S2 and S3.
Relative to drift tube CCS from Ref. 36.
Fold-change relative to control.
Student’s t-test against control.
The separation of the BAC-C16 treatment group away from the other treatment groups and controls is mainly attributed to the elevated phospholipid levels in the BAC-C16 group. Species from several classes of phospholipids were affected by BAC-C16 treatment. Figure 4C shows examples of the altered phospholipid levels for PE 16:1-16:1 (confirmed by MS/MS) and PC 36:5. The levels of PE 16:1-16:1 were slightly decreased with AY9944 or BAC-C10 treatment in relation to the control (0.8- and 0.9-fold), whereas a 3.6-fold increase in abundance was observed with BAC-C16 treatment (P = 8.3 × 10−7). A similar pattern in abundance profiles was observed for PE 16:1-18:2, as shown in Table 1. However, the fold-change increase in BAC-C16 observed for PE 36:3 was nearly half those of PE 16:1-16:1 and PE 16:1-18:2 (1.5-fold vs. >3-fold). Eleven additional PE species were elevated in BAC-C16 treatment in relation to controls with fold-changes ≥ 1.5 (supplemental Table S4). On the basis of these observations, we examined the expression profiles of LysoPEs in the Neuro2a cells to determine whether the increased levels of PEs was due to decreased metabolism of PEs to LysoPEs (supplemental Table S5). However, we found that LysoPEs are consistently higher in BAC-C16-treated cells than in control cells.
In addition to PC 36:5 (Fig. 4C; 2.1-fold, P = 8.9 × 10−5), 10 other PCs were significantly elevated in BAC-C16 samples, including PCs 36:4, 36:3, p34:2, p34:1, 32:2, and p32:1 (supplemental Table S6). The fold-change increases for PCs ranged from 1.7- to 3.2-fold in relation to controls. Among the LysoPCs, LysoPC 16:1 was increased in BAC-C16 treatment (1.7-fold, P = 1.6 × 10−3), whereas no significance (P > 0.05) was found between AY9944 or BAC-C10 treatment in relation to the control. A slight decrease was observed for the most abundant LysoPC, LysoPC 18:1, in the BAC-C16-treated cells (0.8-fold, P = 0.019) in relation to the controls (supplemental Table S6). Altered PG levels were also observed in the BAC-C16 group (Table 1, supplemental Table S7). Eleven PGs were increased in relation to controls, ranging from 1.5- to 4.2-fold. The elevated PGs had fatty acid compositions similar to those of the elevated PEs and PCs, such as PG 32:2 and PG 34:3.
DISCUSSION
Untargeted lipidomics by HILIC-IM-MS
Structural separations by ion mobility are highly informative in the analysis of lipids from complex samples, where many isobaric or nearly isobaric lipids are observed. A number of studies, including our own data presented in Fig. 2, have shown that the largest structural differences in lipids are found between sphingolipids (i.e., Cer, HexCer, SM) versus glycerophospholipids (PC, PE, PA, etc.) because of the different backbones of the lipid, i.e., sphingosine versus acylglycerol (25–27, 29, 48–50). IM can also resolve additional lipid classes within each group on the basis of their different headgroups. There is strong consensus between the literature and our results that PCs have larger gas-phase conformations than other phospholipids such as PEs, likely due to the bulky nature of the quaternary ammonium in the choline headgroup in relation to the smaller and more linear ethanolamine headgroup (49, 50). Within the PEs, we observe a clear separation between PEs with two ester linkages and PEs with one vinyl ether linkage (plasmalogen PEs, PEp), with the plasmalogen PEs having slightly larger CCS values than PEs with the same fatty acid compositions in positive mode (supplemental Table S1) and slightly smaller CCS values in negative mode (supplemental Table S2). The decrease in CCS we observed between PEs and plasmalogen PEs in negative mode is consistent with the observations of Paglia et al. (50), who reasoned that the decrease in CCS was the result of the additional double bond in the PEp. This reduction in CCS with the addition of double bonds is a common phenomenon in all lipid classes, with each double bond contributing a 1–5% decrease in CCS (28, 49).
The formation of lipid ions by electrospray ionization is largely determined by the lipid backbone headgroup, or both, which may be neutral, basic, or acidic. We found very clear preferences in the formation of adducts between various lipid classes in positive ionization mode. PIs and PGs, which do not have any amines and are rich in oxygens, tend to form sodium adducts or appear as water loss fragments in positive mode. PCs and SMs, both of which contain a quaternary amine in the choline headgroup, predominately form protonated adducts. The influence of adduct formation on lipid CCS values has been described by Jackson et al. (25), who reported that sodiated lipids display more compact gas-phase structure than do protonated lipids. Interestingly, we observed that ion mobility resolution of phospholipid classes is lower in negative mode than in positive mode (Fig. 2A, B). We attribute this phenomenon to the diversity of adducts (i.e., protonated, sodiated, water loss) formed in positive mode, whereas negative-mode lipid ions are predominately formed by the loss of a proton. This less efficient separation of phospholipid classes in negative-mode IM-MS analysis can make it challenging to use CCS alone to distinguish similar-mass lipid species of different classes in this mode, such as the similar-mass PGs and PAs described above.
Conventionally, identification of unknown lipid species relies heavily on MS/MS experiments in both positive and negative modes. Although there are many unique lipid species, the fragments they yield may not be unique, given the common fragmentation pathways shared within a lipid class or among all lipids. For example, in negative-mode MS/MS analysis, two different lipids may yield the same fatty acid fragments, even though they belong to distinct lipid classes. Although it has become common to use the IM dimension as a filter in untargeted MS/MS analysis, the large number of similar-mass lipid precursors within a narrow range of drift times may lead to convoluted assignments without an additional separation to resolve different lipid classes (32). We have demonstrated here that the addition of HILIC chromatography prior to IM-MS achieves resolution of at least 12 lipid classes. The overlap of similar-mass lipid precursors and fragments belonging to different lipid classes is minimized, and the few lipid classes that coelute and occupy similar mass ranges, such as ceramides and DGs, as well as PCs and PSs, can be easily resolved in the IM dimension, as demonstrated in Fig. 3. We note that the workflow developed here is more suited for untargeted discovery-type experiments. Adaptation of the method for absolute quantitation is possible, given that internal standards for each class of lipids were added at the beginning of the sample processing, and the data were normalized by the combination of the internal standards. However, for the most sensitive and accurate quantitation, a targeted analysis would still be preferred after the initial discovery experiments.
CCS measurements of lipids by TWIM-MS calibration
We have recently demonstrated that the choice of calibrant has a significant impact on the CCS values generated for phospholipids on TWIM-MS platforms (36). The lipid CCS values reported here were generated from lipid standard CCS calibrants describe previously (36). The difference between the calibrated CCSs of PEs and PCs identified in the lipid mix and the drift tube CCS of the corresponding PE and PC calibration standards was less than 1% in both positive and negative modes. Table 1 contains the CCS values for lipids identified in the Neuro2a cells, which are up to 1.1% different from the same lipid species identified from the 12-lipid mix in supplemental Tables S2 and S3. However, our CCS values tended to be smaller (average of -3.7% and -4.4% different in positive and negative modes, respectively) than previously published lipid CCS values calibrated with nonlipid calibrants, such as poly-DL-alanine (50, 51). This observation is consistent with trends we observed for calibration with lipid standards versus calibration with poly-DL-alanine (36).
The calibration approach of determining CCS values is the most rapid method for obtaining structural data for large datasets such as those encountered in untargeted lipidomics regardless of the IM platform (i.e., traveling wave or drift tube). The ability to rapidly determine CCS values presents an opportunity to build comprehensive databases of CCS measurements. However, the 3–4% variability in calibrated CCS values obtained from calibrants other than lipids presents a challenge for the use of CCS values in lipidomics databases. For example, the difference in CCS between two different types of glycerophospholipids in positive mode is on the order of 1–2% for species with similar mass and unsaturation, such as PC 36:3 and PEp 40:2 (m/z 784.6; 285.5 Å2 and 290.0 Å2, respectively). In addition, similar-mass PCs and HexCers, such as PC 38:4 and HexCer (d18:1/24:1), tend to be 2.5% different in CCS despite belonging to different classes of lipids. In both instances, the CCS difference between lipid classes is smaller than the calibration errors using poly-DL-alanines, which prevents the use of these values as discriminating data for identification. It is critically important that calibrated CCS values approach those of drift tube measurements as closely as possible in order to develop comprehensive lipid CCS databases that can be used for assignment of identifications in untargeted lipidomics, and our lipid CCS calibrants most closely satisfy these criteria.
An area that could be improved in this work, as in the lipid CCS field in general, is to annotate the lipid species with detailed fatty acid composition and positional isomers, which is largely limited by the available of authentic standards at the moment. In the cases in which we obtained the acyl chain identity based on standards or MS fragmentation spectra, the acyl composition of each lipid species was mostly a single combination of fatty acids (known fatty acid compositions are noted in Table 1 and supplemental Tables S2, S3). However, the positional isomers were not differentiated in this work. Previous work by Baker and coworkers (49) reported that positional isomers of phospholipids could be differentiated by IM separation, but the difference between the drift times of each pair of isomers is less than 1%, which is beyond the resolution of our instrument. In another study by Groessl et al. (52), the effect of E/Z isomers on the CCS of PC(18:1/18:1) was also found to be less than 0.5%.
BAC exposure alters lipid homeostasis
The lipidomic results on BAC-exposed Neuro2a cells are consistent with our previous studies on the biological activities of individual BACs (47). Because BAC-C10 acts as a potent inhibitor of DHCR7, we anticipated that the effects of AY9944 and BAC-C10 treatments on the Neuro2a lipidome would be similar. We indeed observed tight clustering of the AY9944 and BAC-C10 groups in the PCA, which was the result of highly elevated (fold-change > 10) levels of species at 4.0 min in the chromatogram that were not observed in the control or BAC-C16 group. These features consisted of two pairs of signals at m/z 381.3 and 383.3 (4.0_398.3n and 4.0_400.3n in Table 1) and m/z 363.3 and 365.3 differing by 18 Da, which are similar to the pattern of water loss fragments (i.e., [M+H−H2O]+ and [M+H−2H2O]+, respectively) observed for oxysterols (46, 53). The prominent biochemical features of DHCR7 inhibition are decreased cholesterol levels and elevated levels of 7-DHC, 7-dehydrodesmosterol (7-DHD), as well as the 7-DHC-derived oxysterols 4β-OH-7-DHC, 4α-OH-7-DHC and DHCEO (46, 53, 54). We have previously shown that 7-DHC, 7-DHD, and DHCEO are significantly elevated in AY9944- and BAC-C10-treated Neuro2a cells (47). It is likely that the observed oxysterols are 7-DHC- or 7-DHD-derived oxysterols, given that the mass difference between the tentative oxysterols is the same as the mass difference between 7-DHC and 7-DHD (i.e., 384.3 Da and 382.3 Da, respectively). However, their appearance at 4.0 min in the chromatogram is inconsistent with oxysterols, which are not retained (elute in the solvent front) under HILIC conditions. It is plausible that the oxysterols were conjugated to a larger, more polar compound, which resulted in the uncharacteristic retention time. For example, glucuronidation and sulfation of oxysterols have been observed (55, 56). On the basis of our results, the addition of a carbohydrate unit (i.e., Cer to HexCer) results in a 0.2 min increase in HILIC retention time (Fig. 2). A larger increase in retention time may be likely depending on the polarity and size of the modification and its location on the oxysterol.
We observed alterations in sphingolipid levels across all treatment groups. The ∼2-fold reduction in ceramide levels observed in AY9944-, BAC-C10-, and BAC-C16-treated Neuro2a cells is of particular interest given the importance of ceramides, and sphingolipids in general, in brain development and function (56–58). We observed similar decreases in sphingomyelins, which are synthesized from ceramides, with the same fatty acid composition. However, SM (d18:1/18:1) was an exception in that no significant reduction was observed in BAC-C16 treatment and no change was observed for Cer (d18:1/18:1) in any of the treatment groups. Collectively, these results suggest that the alteration in sphingolipid homeostasis is dependent upon both the individual BAC and the sphingolipid composition.
Treatment of Neuro2a cells with BAC-C16 had a large effect on glycerophospholipid metabolism resulting in elevated levels of several classes of lipids, including PEs, LysoPEs, PCs, and PGs, with the largest effect observed for PEs (supplemental Tables S4–S7). These same lipid classes were only mildly affected by AY9944 and BAC-C10. Changes in glycerolipid levels also appeared to correlate with BAC length, for which BAC-C16 treatment resulted in a greater fold-change reduction in TAGs and DGs than did BAC-C10 treatment (Table 1). In the context of elevated phospholipid levels, the decreased DG levels in BAC-C16 treatment suggest increased glycerophospholipid biosynthesis from DGs rather than decreased DG synthesis. In our previous study, gene expression analysis revealed that fatty acid synthase was significantly upregulated in BAC-C16-treated Neuro2a cells (47), which is consistent with the elevated phospholipid levels observed in BAC-C16 treatment.
BACs have been widely used as disinfectants in products ranging from cleaning products to medical products for more than 50 years (59). Their mechanism of disinfection stems from the ability of the positively charged quaternary ammonium headgroup to disrupt the negatively charged lipid membranes of microbes. Several adverse health effects related to BAC exposure have been reported, including occupational asthma and ocular diseases (60, 61). We demonstrated here that treatment of neuroblastoma cells with BAC-C16 and BAC-C10 resulted in major revisions to lipid homeostasis affecting sterol, sphingolipid, glycerolipid, and phospholipid metabolism. The effects of BAC exposure appear to be moderated by their alkyl chain length, with BAC-C16 and BAC-C10 resulting in alterations to distinct lipid metabolism pathways. The effects of BAC-C10 on lipid homeostasis are similar to those of AY9944, a known inhibitor of cholesterol biosynthesis, which recapitulates the biochemical defects of Smith-Lemli-Opitz syndrome in rats (46). Many of the lipid species affected, such as PEs and SMs, play significant roles in neurological development and function (62, 63). These results in conjunction with our previous findings, suggest that BAC exposure could lead to or contribute to the pathogenesis of developmental disorders if exposure occurs during developmental stages (47).
CONCLUSIONS
We have developed a comprehensive approach for lipidomics by coupling HILIC chromatographic separation with ion mobility-mass spectrometry. This method successfully resolved similar-mass lipid species, a major challenge in lipidomics analysis, on the basis of polar headgroups and fatty acid composition in the HILIC dimension and by lipid backbone and headgroup in the IM dimension. CCS values, calibrated by our previously established lipid calibrants, and retention times are reported for over 250 lipid signals from the analysis of 12 lipid classes, with RSDs ≤ 0.5% CCS for triplicate analyses. The method reported here, combining lipid CCS calibrants, HILIC chromatography, and ion mobility-mass spectrometry, represents a comprehensive approach to the resolution and structural analysis of lipids from tissues and cells. We applied HILIC-IM-MS to characterize the effects of benzalkonium chloride exposure on the lipidome of the Neuro2a neuroblastoma cell line. We found that lipid homeostasis was differentially altered in BAC-C10 and BAC-C16 exposure, and this disruption of lipid metabolism pathways at noncytotoxic levels of BAC exposure could have significant implications in human environmental health.
Supplementary Material
Footnotes
Abbreviations:
- BAC
- benzalkonium chloride
- CCS
- collision cross section
- Cer
- ceramide
- DG
- diacylglycerol
- DHCR7
- 3β-hydroxysterol-Δ7-reductase
- DMS
- differential mobility spectrometry
- DTIM
- drift tube ion mobility
- HexCer
- hexosyl ceramide
- HILIC
- hydrophilic interaction liquid chromatography
- IM-MS
- ion mobility-mass spectrometry
- IM-XIC
- ion mobility-extracted ion chromatogram
- LysoPC
- lysophosphatidylcholine
- LysoPE
- lysophosphatidylethanolamine
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PCA
- principal components analysis
- PE
- phosphatidylethanolamine
- PEp
- plasmalogen phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phospatidylinositol
- PS
- phosphatidylserine
- TAG
- triacylglycerol
- TIC
- total ion chromatogram
- TWIM
- traveling wave ion mobility
Financial support for this work was provided by the National Institutes of Health Grant R00 HD073270 (L.X.), a pilot grant from the University of Washington Center for Exposures, Diseases, Genomics, and Environment (NIH P30ES007033), and the startup fund to L.X. from the Department of Medicinal Chemistry in the School of Pharmacy at the University of Washington. J.H. acknowledges support from a National Institutes of Environmental Health Sciences training grant (T32 ES007032). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Yeagle P. L. 1989. Lipid regulation of cell membrane structure and function. FASEB J. 3: 1833–1842. [PubMed] [Google Scholar]
- 2.Wymann M. P., and Schneiter R.. 2008. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9: 162–176. [DOI] [PubMed] [Google Scholar]
- 3.Berliner J. A., Leitinger N., and Tsimikas S.. 2009. The role of oxidized phospholipids in atherosclerosis. J. Lipid Res. 50(Suppl): S207–S212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musso G., Gambino R., and Cassader M.. 2009. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog. Lipid Res. 48: 1–26. [DOI] [PubMed] [Google Scholar]
- 5.Gross R. W., and Han X.. 2007. Lipidomics in diabetes and the metabolic syndrome. Methods Enzymol. 433: 73–90. [DOI] [PubMed] [Google Scholar]
- 6.Di Paolo G., and Kim T. W.. 2011. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat. Rev. Neurosci. 12: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos C. R., and Schulze A.. 2012. Lipid metabolism in cancer. FEBS J. 279: 2610–2623. [DOI] [PubMed] [Google Scholar]
- 8.Sounni N. E., Cimino J., Blacher S., Primac I., Truong A., Mazzucchelli G., Paye A., Calligaris D., Debois D., De Tullio P., et al. 2014. Blocking lipid synthesis overcomes tumor regrowth and metastasis after antiangiogenic therapy withdrawal. Cell Metab. 20: 280–294. [DOI] [PubMed] [Google Scholar]
- 9.Han X., and Gross R. W.. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 10.Schmelzer K., Fahy E., Subramaniam S., and Dennis E. A.. 2007. The lipid maps initiative in lipidomics. Methods Enzymol. 432: 171–183. [DOI] [PubMed] [Google Scholar]
- 11.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis E. A., Deems R. A., Harkewicz R., Quehenberger O., Brown H. A., Milne S. B., Myers D. S., Glass C. K., Hardiman G., Reichart D., et al. 2010. A mouse macrophage lipidome. J. Biol. Chem. 285: 39976–39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quehenberger O., and Dennis E. A.. 2011. The human plasma lipidome. N. Engl. J. Med. 365: 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwudke D., Liebisch G., Herzog R., Schmitz G., and Shevchenko A.. 2007. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol. 433: 175–191. [DOI] [PubMed] [Google Scholar]
- 15.Merrill A. H. Jr., Sullards M. C., Allegood J. C., Kelly S., and Wang E.. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 36: 207–224. [DOI] [PubMed] [Google Scholar]
- 16.Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., and Brown H. A.. 2007. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432: 21–57. [DOI] [PubMed] [Google Scholar]
- 17.Han X. 2007. Neurolipidomics: challenges and developments. Front. Biosci. 12: 2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harkewicz R., and Dennis E. A.. 2011. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 80: 301–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemmer D. E., Hudgins R. R., and Jarrold M. F.. 1995. Naked protein conformations: cytochrome c in the gas phase. J. Am. Chem. Soc. 117: 10141–10142. [Google Scholar]
- 20.von Helden G., Wyttenbach T., and Bowers M. T.. 1995. Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science. 267: 1483–1485. [DOI] [PubMed] [Google Scholar]
- 21.McLean J. A., Ruotolo B. T., Gillig K. J., and Russell D. H.. 2005. Ion mobility-mass spectrometry: a new paradigm for proteomics. Int. J. Mass Spectrom. 240: 301–315. [Google Scholar]
- 22.Kanu A. B., Dwivedi P., Tam M., Matz L., and Hill H. H. Jr. 2008. Ion mobility-mass spectrometry. J. Mass Spectrom. 43: 1–22. [DOI] [PubMed] [Google Scholar]
- 23.Fenn L. S., and McLean J. A.. 2008. Biomolecular structural separations by ion mobility-mass spectrometry. Anal. Bioanal. Chem. 391: 905–909. [DOI] [PubMed] [Google Scholar]
- 24.Fenn L. S., Kliman M., Mahsut A., Zhao S. R., and McLean J. A.. 2009. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal. Bioanal. Chem. 394: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson S. N., Ugarov M., Post J. D., Egan T., Langlais D., Schultz J. A., and Woods A. S.. 2008. A study of phospholipids by ion mobility TOFMS. J. Am. Soc. Mass Spectrom. 19: 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kliman M., May J. C., and McLean J. A.. 2011. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta. 1811: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May J. C., Goodwin C. R., Lareau N. M., Leaptrot K. L., Morris C. B., Kurulugama R. T., Mordehai A., Klein C., Barry W., Darland E., et al. 2014. Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal. Chem. 86: 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H. I., Kim H., Pang E. S., Ryu E. K., Beegle L. W., Loo J. A., Goddard W. A., and Kanik I.. 2009. Structural characterization of unsaturated phosphatidylcholines using traveling wave ion mobility spectrometry. Anal. Chem. 81: 8289–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridenour W. B., Kliman M., McLean J. A., and Caprioli R. M.. 2010. Structural characterization of phospholipids and peptides directly from tissue sections by MALDI traveling-wave ion mobility-mass spectrometry. Anal. Chem. 82: 1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shvartsburg A. A., Isaac G., Leveque N., Smith R., and Metz T.. 2011. Separation and classification of lipids using differential ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 22: 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lintonen T. P., Baker P. R., Suoniemi M., Ubhi B. K., Koistinen K. M., Duchoslav E., Campbell J. L., and Ekroos K.. 2014. Differential mobility spectrometry-driven shotgun lipidomics. Anal. Chem. 86: 9662–9669. [DOI] [PubMed] [Google Scholar]
- 32.Baker P. R., Armando A. M., Campbell J. L., Quehenberger O., and Dennis E. A.. 2014. Three-dimensional enhanced lipidomics analysis combining UPLC, differential ion mobility spectrometry, and mass spectrometric separation strategies. J. Lipid Res.; Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shvartsburg A. A., and Smith R. D.. 2008. Fundamentals of traveling wave ion mobility spectrometry. Anal. Chem. 80: 9689–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pringle S. D., Giles K., Wildgoose J. L., Williams J. P., Slade S. E., Thalassinos K., Bateman R. H., Bowers M. T., and Scrivens J. H.. 2007. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int. J. Mass Spectrom. 261: 1–12. [Google Scholar]
- 35.Giles K., Williams J. P., and Campuzano I.. 2011. Enhancements in travelling wave ion mobility resolution. Rapid Commun. Mass Spectrom. 25: 1559–1566. [DOI] [PubMed] [Google Scholar]
- 36.Hines K. M., May J. C., McLean J. A., and Xu L.. 2016. Evaluation of collision cross section calibrants for structural analysis of lipids by traveling wave ion mobility-mass spectrometry. Anal. Chem. 88: 7329–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pulfer M., and Murphy R. C.. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 38.Wakelam M. J., Pettitt T. R., and Postle A. D.. 2007. Lipidomic analysis of signaling pathways. Methods Enzymol. 432: 233–246. [DOI] [PubMed] [Google Scholar]
- 39.Spagou K., Tsoukali H., Raikos N., Gika H., Wilson I. D., and Theodoridis G.. 2010. Hydrophilic interaction chromatography coupled to MS for metabonomic/metabolomic studies. J. Sep. Sci. 33: 716–727. [DOI] [PubMed] [Google Scholar]
- 40.Spagou K., Wilson I. D., Masson P., Theodoridis G., Raikos N., Coen M., Holmes E., Lindon J. C., Plumb R. S., Nicholson J. K., et al. 2011. HILIC-UPLC-MS for exploratory urinary metabolic profiling in toxicological studies. Anal. Chem. 83: 382–390. [DOI] [PubMed] [Google Scholar]
- 41.Ivanisevic J., Zhu Z. J., Plate L., Tautenhahn R., Chen S., O’Brien P. J., Johnson C. H., Marletta M. A., Patti G. J., and Siuzdak G.. 2013. Toward ’omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 85: 6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng L., T’Kind R., Decuypere S., von Freyend S. J., Coombs G. H., and Watson D. G.. 2010. Profiling of lipids in Leishmania donovani using hydrophilic interaction chromatography in combination with Fourier transform mass spectrometry. Rapid Commun. Mass Spectrom. 24: 2074–2082. [DOI] [PubMed] [Google Scholar]
- 43.Fei F., Bowdish D. M. E., and McCarry B. E.. 2014. Comprehensive and simultaneous coverage of lipid and polar metabolites for endogenous cellular metabolomics using HILIC-TOF-MS. Anal. Bioanal. Chem. 406: 3723–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dodbiba E., Xu C. D., Payagala T., Wanigasekara E., Moon M. H., and Armstrong D. W.. 2011. Use of ion pairing reagents for sensitive detection and separation of phospholipids in the positive ion mode LC-ESI-MS. Analyst. 136: 1586–1593. [DOI] [PubMed] [Google Scholar]
- 45.Schwalbe-Herrmann M., Willmann J., and Leibfritz D.. 2010. Separation of phospholipid classes by hydrophilic interaction chromatography detected by electrospray ionization mass spectrometry. J. Chromatogr. A. 1217: 5179–5183. [DOI] [PubMed] [Google Scholar]
- 46.Xu L., Liu W., Sheflin L. G., Fliesler S. J., and Porter N. A.. 2011. Novel oxysterols observed in tissues and fluids of AY9944-treated rats—a model for Smith-Lemli-Opitz syndrome. J. Lipid Res. 52: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herron J., Reese R., Tallman K. A., Narayanaswamy R., Porter N. A., and Xu L.. 2016. Identification of environmental quaternary ammonium compounds as direct inhibitors of cholesterol biosynthesis. Toxicol. Sci. 151: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLean J. A. 2009. The mass-mobility correlation redux: the conformational landscape of anhydrous biomolecules. J. Am. Soc. Mass Spectrom. 20: 1775–1781. [DOI] [PubMed] [Google Scholar]
- 49.Kyle J. E., Zhang X., Weitz K. K., Monroe M. E., Ibrahim Y. M., Moore R. J., Cha J., Sun X., Lovelace E. S., Wagoner J., et al. 2016. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 141: 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paglia G., Angel P., Williams J. P., Richardson K., Olivos H. J., Thompson J. W., Menikarachchi L., Lai S., Walsh C., Moseley A., et al. 2015. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 87: 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., and Vertes A.. 2015. Energy charge, redox state, and metabolite turnover in single human hepatocytes revealed by capillary microsampling mass spectrometry. Anal. Chem. 87: 10397–10405. [DOI] [PubMed] [Google Scholar]
- 52.Groessl M., Graf S., and Knochenmuss R.. 2015. High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst. 140: 6904–6911. [DOI] [PubMed] [Google Scholar]
- 53.Xu L., Korade Z., Rosado D. A., Liu W., Lamberson C. R., and Porter N. A.. 2011. An oxysterol biomarker for 7-dehydrocholesterol oxidation in cell/mouse models for Smith-Lemli-Opitz syndrome. J. Lipid Res. 52: 1222–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L., Mirnics K., Bowman A. B., Liu W., Da J., Porter N. A., and Korade Z.. 2012. DHCEO accumulation is a critical mediator of pathophysiology in a Smith-Lemli-Opitz syndrome model. Neurobiol. Dis. 45: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng L. J., Griffiths W. J., Nazer H., Yang Y., and Sjovall J.. 1997. High levels of (24S)-24-hydroxycholesterol 3-sulfate, 24-glucuronide in the serum and urine of children with severe cholestatic liver disease. J. Lipid Res. 38: 926–934. [PubMed] [Google Scholar]
- 56.Bjorkhem I., Andersson U., Ellis E., Alvelius G., Ellegard L., Diczfalusy U., Sjovall J., and Einarsson C.. 2001. From brain to bile. Evidence that conjugation and omega-hydroxylation are important for elimination of 24S-hydroxycholesterol (cerebrosterol) in humans. J. Biol. Chem. 276: 37004–37010. [DOI] [PubMed] [Google Scholar]
- 57.van Echten-Deckert G., and Herget T.. 2006. Sphingolipid metabolism in neural cells. Biochim. Biophys. Acta. 1758: 1978–1994. [DOI] [PubMed] [Google Scholar]
- 58.Posse de Chaves E., and Sipione S.. 2010. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 584: 1748–1759. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert P., and Moore L. E.. 2005. Cationic antiseptics: diversity of action under a common epithet. J. Appl. Microbiol. 99: 703–715. [DOI] [PubMed] [Google Scholar]
- 60.Purohit A., Kopferschmitt-Kubler M. C., Moreau C., Popin E., Blaumeiser M., and Pauli G.. 2000. Quaternary ammonium compounds and occupational asthma. Int. Arch. Occup. Environ. Health. 73: 423–427. [DOI] [PubMed] [Google Scholar]
- 61.Pisella P. J., Pouliquen P., and Baudouin C.. 2002. Prevalence of ocular symptoms and signs with preserved and preservative free glaucoma medication. Br. J. Ophthalmol. 86: 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Posse de Chaves E. I. 2006. Sphingolipids in apoptosis, survival and regeneration in the nervous system. Biochim. Biophys. Acta. 1758: 1995–2015. [DOI] [PubMed] [Google Scholar]
- 63.Farooqui A. A., Horrocks L. A., and Farooqui T.. 2000. Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem. Phys. Lipids. 106: 1–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.