Abstract
The esterification of alcohols with fatty acids is a universal mechanism to form inert storage forms of sterols, di- and triacylglycerols, and retinoids. In ocular tissues, formation of retinyl esters is an essential step in the enzymatic regeneration of the visual chromophore (11-cis-retinal). Acyl-CoA wax alcohol acyltransferase 2 (AWAT2), also known as multifunctional O-acyltransferase (MFAT), is an integral membrane enzyme with a broad substrate specificity that has been shown to preferentially esterify 11-cis-retinol and thus contribute to formation of a readily available pool of cis retinoids in the eye. However, the mechanism by which this promiscuous enzyme can gain substrate specificity is unknown. Here, we provide evidence for an allosteric modulation of the enzymatic activity by 11-cis retinoids. This regulation is independent from cellular retinaldehyde-binding protein (CRALBP), the major cis-retinoid binding protein. This positive-feedback regulation leads to decreased esterification rates for 9-cis, 13-cis, or all-trans retinols and thus enables preferential synthesis of 11-cis-retinyl esters. Finally, electron microscopy analyses of the purified enzyme indicate that this allosteric effect does not result from formation of functional oligomers. Altogether, these data provide the experimental basis for understanding regulation of AWAT2 substrate specificity.
Keywords: multifunctional O-acyltransferase, vitamin A, 11-cis-retinol, cone photoreceptor, visual cycle
In vertebrates, vision depends on photosensitive pigments composed of 11-cis-retinal bound to protein scaffolds (opsins), which are localized in the outer segments of specialized photoreceptor cells, rods and cones (1). Light captured by these photoreceptors photo-isomerizes 11-cis-retinylidene to its all-trans configuration, induces a conformational change in the protein scaffold, and enables G-coupled protein-mediated signal transduction events that lead to the perception of light (2–4). Following the photo-isomerization, all-trans-retinal is released from the opsins. In order to restore photosensitivity, the visual pigments need to be regenerated with the new molecule of 11-cis-retinal. To ensure a proper supply of 11-cis-retinal, vertebrates evolved a metabolic pathway called the visual or retinoid cycle (4). This canonical visual pathway depends on lecithin:retinol acyltransferase (LRAT) and retinoid isomerase (RPE65) and takes place in photoreceptors and retinal pigmented epithelium (RPE) cells (5–8). There is accumulating evidence that the canonical retinoid cycle fails to explain major differences between rod and cone chromophore regeneration. Although these two types of photoreceptors share common structural design (9), they differ in the dynamic range of light intensity to which they can adapt (10–12). Rods are characterized by single photon sensitivity that results in saturation of their signaling capacity under relatively low light intensities (11, 13). Contrary to rods, cone responses have a very high photon saturation threshold, which enables them to function in photopic conditions (14, 15). Nevertheless, the quantum efficiency of photon capturing by rod and cone visual pigments is similar (16–18). Consequently, cones depend on rapid regeneration of the visual pigment that cannot be fully achieved by the relatively slow canonical visual cycle (15, 19, 20).
To support cone pigment regeneration, an alternate metabolic pathway has been proposed for cis retinoid production (20, 21). The principles of this noncanonical visual cycle are based on retinal-specific enzymatic activities and differences in retinoid composition between diurnal and nocturnal animals. In contrast to rats and mice, retinas and RPE cells of zebrafish, chicken, ground squirrel, macaque, and human contain a substantial amount of 11-cis-retinyl esters (22). This storage form of 11-cis retinoids can be readily converted into 11-cis-retinal to support cone function. Although most of the enzymes involved in the cone visual cycle remain to be uncovered, two enzymes, including dihydroceramide desaturase 1 (DES1) (23) and acyl-CoA wax alcohol acyltransferase 2 (AWAT2), also known as multifunctional O-acyltransferase (MFAT), (24) were recently proposed to be involved in this metabolic pathway. In the current model, DES1 catalyzes equilibrium isomerization of retinol yielding a mixture containing 9-cis, 11-cis, 9,13-di cis, and 13-cis-retinol, and all-trans-retinol in a ratio that closely resembles a thermodynamic equilibrium (23). Interestingly, 11-cis-retinol accounts for less than 1% of total retinol isomers in this mixture, yet only 11-cis-retinyl esters are accumulated in the ocular tissue. The enzyme that preferentially esterifies 11-cis-retinol has been proposed to be AWAT2 (24). Surprisingly, this integral membrane enzyme exhibits broad substrate specificity. In addition to vitamin A and its isomers, AWAT2 esterifies aliphatic fatty alcohols of various chain lengths (25–27). Based on the published kinetic parameters of human AWAT2 and the cis-retinol composition resulting from DES1 activity, one would expect significant accumulation of retinyl ester isomers different than 11-cis, which is not observed in vivo.
To scrutinize the function of AWAT2 in isomer-specific retinol esterification, we evaluated the enzymatic activity of mouse AWAT2 toward retinol isomers and examined the factors that affect substrate specificity of the enzyme. We show that AWAT2 enzymatic activity is allosterically modulated by 11-cis retinoids. This regulatory mechanism leads to decreased esterification rates for 9-cis, 13-cis, or all-trans retinols. Furthermore, by using isotope-labeled substrates, we demonstrate that esterification of 11-cis-retinol does not depend on cellular retinaldehyde-binding protein (CRALBP). These data provide a mechanism by which AWAT2 gains substrate specificity as needed for the preferential accumulation of 11-cis-retinyl esters in the eye.
MATERIAL AND METHODS
Materials
The all-trans-retinal, deuterated all-trans-retinal (5D-all-trans-retinal), all-trans-retinol, 9-cis-retinal, 11-cis-retinal, and 13-cis-retinal, as well as 9-cis-retinyl palmitate and palmityl myristate, were purchased from Toronto Research Company. Alternatively, cis retinoids and their esters were synthetized and purified as described below. All-trans-retinyl palmitate, lauryl chloride, palmitoyl chloride, dodecanol, and tetradecanol were obtained from Sigma-Aldrich, whereas acyl-CoAs were purchased from Avanti Polar Lipids. HPLC grade solvents were purchased from Fisher Scientific.
Chemical synthesis and purification of retinoids
To obtain geometric isomers of retinal or deuterated retinal, a quartz cuvette containing 0.5 ml of 40 mM all-trans-retinal in acetonitrile was placed on ice and exposed to light emitted by a 250 watt halogen bulb equipped with UV light filter. The lamp was positioned 25 cm above the cuvette. After 15 min, the retinoids were extracted with 5 ml of hexane. Collected hexane fraction was washed twice with a saturated solution of NaCl and dried under a stream of nitrogen. The residual mixture of retinal isomers was resolubilized in 0.5 ml of hexane. Separation of the geometric isomers was achieved by HPLC using an Agilent 1100 series system (Agilent Technologies) equipped with a diode-array detector. Retinals were eluted from a Luna silica column (250 × 21.2 mm, 10 μm) (Phenomenex) with an isocratic flow of 10% ethyl acetate in hexane (v/v) at the flow rate of 5 ml/min. The elution profile of retinoids was monitored at 360 nm. Chromatographic peaks that corresponded to 9-cis, 11-cis, 13-cis, and all-trans isomers were collected into separate tubes and the organic solvents were evaporated in a SpeedVac (Eppendorf). Purified retinals were redissolved in 0.5 ml of ethanol and stored at −80°C. Concentrations of the ethanolic stock solutions of retinals were determined spectrophotometrically using the following molar extinction coefficients: 36,100, 24,935, 35,500, and 42,880 M−1cm−1 for 9-cis, 11-cis, 13-cis, and all-trans isomers, respectively (28, 29). This procedure yielded 9-cis, 11-cis, 13-cis, and all-trans isomers of retinal in 10.9/26.1/17.5/45.5 ratio.
Retinals were reduced to the corresponding retinols by reacting them with ∼4 molar excess of sodium borohydride in 2 ml of ice-cold ethanol. Progress of these reactions was monitored by recording blue shift of the maximum absorbance of the reaction mixture from ∼360 to ∼325 nm, depending on the isomer. Upon completion, 2 ml of water were added to the reactions and retinoids were extracted with 4 ml of hexane. The organic phase was washed with 4 ml of saturated NaCl solution and dried in a SpeedVac. To verify purity of the final products, small aliquots were injected onto a Luna PREP silica column (250 × 4.6 mm, 10 μm) (Phenomenex) and eluted with 10% ethyl acetate in hexane (v/v) at the flow rate of 2 ml/min. Purified retinols were redissolved in 0.5 ml of ethanol and stored at −80°C.
Retinyl esters were prepared by the reaction between retinols and 2 molar excess of either acetic anhydride, palmitoyl chloride, or lauryl chloride in anhydrous dichloromethane in the presence of N,N-dimethylaminopyridine at 4°C for 4 h. The reaction was terminated by the addition of water and the retinoids were extracted with hexane. Completeness of the reaction and purity of retinyl esters were checked chromatographically using a Luna PREP silica column (250 × 4.6 mm, 10 μm). Retinyl esters and their geometric isomers were separated in 1% ethyl acetate in hexane (v/v), whereas retinols were eluted by increasing ethyl acetate concentration to 10% (v/v). In both cases, the flow rate was 2 ml/min. The isomers were identified based on their characteristic shape and maxima of the absorbance spectra (4).
Expression of mouse AWAT2 in yeast
cDNA of mouse AWAT2 (NCBI accession number NM_177746) was purchased from OriGene Technologies. To prepare an expression vector, the cDNA was amplified by PCR using the following primer pair: forward, GCAGATACTAGTGTTTAATTATCAAACAATATCAATAATGTTCTGGCCCACCAAGAAGGACC and reverse, CGTCTAGACGCGTTCAAACTATCACCAGCTCCTGGGTCTCTGAGATGCC. The PCR product was digested with SpeI and MluI restriction enzymes and sub-cloned into the YepM vector (30). Saccharomyces cerevisiae strain BJ5457 (ATCC) was transfected by alkali-cation yeast transformation kit (MP Biomedicals) and the cells were plated on a leucine-deficient selection medium (−Leu) (MP Biomedicals). Colonies of yeast were picked up from the plate and inoculated into 25 ml of −Leu medium that contained 10% glycerol (v/v). The culture was incubated at 30°C for 16 h prior to transfer into 2 l of fresh −Leu/glycerol medium. Yeasts were grown until OD600 reached 1.2–1.4. The cells were harvested by centrifugation (6,000 g, 15 min), resuspended in 40 mM Tris/HCl (pH 8.0) with 250 mM sucrose, and disrupted by microfluidization at 100 psi (five cycles). Cell homogenate was spun to remove large cellular debris at 12,000 g for 20 min. The resulting supernatant was then centrifuged at 120,000 g for 1 h to collect the microsomal fraction, which was subsequently resuspended in 50 mM Tris/HCl (pH 8.0) and 250 mM sucrose, aliquoted, and stored at −80°C. Expression of AWAT2 in transfected yeasts was confirmed by Western blot analysis with anti-AWAT2 polyclonal antibody (NBP1-91574; Novus Biologicals).
Expression and purification of GST-AWAT2 fusion protein
To generate glutathione S-transferase (GST)-AWAT2 construct, cDNA of mouse AWAT2 was first subcloned in pGex-2T vector (GE Healthcare) using the following primers: forward, GCAGATGGATCCTTCTGGCCCACCAAGAAGGACC; reverse, CGTCTAGAATTCTCAAACTATCACCAGCTCCTGGGTCTCTGAGATGC; and EcoRI/BamHI restriction sites. Then, cDNA encoding GST-AWAT2 fusion protein was amplified by PCR with the following primers: forward, GCAGATACTAGTGTTTAATTATCAAACAATATCAATAATGTTCTGGCCCACCAAGAAGGACC and reverse, CGTCTAGACGCGTTCAAACTATCACCAGCTCCTGGGTCTCTGAGATGCC. After digestion with SpeI and MluI restriction enzymes, the PCR product was ligated into YepM vector. Expression of the fusion protein and purification of the yeast microsomal fraction was conducted as described above.
To purify GST-AWAT2, the yeast microsomes were incubated with 20 mM n-dodecyl-β-D-maltoside (DDM) in 5.4 mM sodium phosphate dibasic, 1.3 mM potassium phosphate monobasic, 137 mM NaCl, and 2.7 mM KCl solution (PBS) for 16 h at 4°C and spun at 140,000 g for 1 h at 4°C. The supernatant was diluted with PBS to lower the concentration of DDM to 2 mM and subsequently incubated with glutathione-Sepharose (GE Healthcare) for 3 h at 4°C. The resin was then placed in a chromatography column and washed with 10 column volumes of PBS. GST-AWAT2 was eluted with 10 mM reduced glutathione in PBS. Fractions containing the protein were pooled together, concentrated in a Centricon (Amicon) cutoff of 100 kDa, and loaded onto a Superdex 200 Increase 10/300 GL size exclusion column (GE Healthcare) equilibrated with 2 mM DDM in PBS. The protein was eluted with isocratic flow of the equilibration buffer (1 ml/min).
Negative stained single-particle transmission electron microscopy analysis
A purified GST-AWAT2 sample at 0.2 mg/ml concentration was applied onto a glow-discharged carbon-coated copper Quantifoil R2/1 grid (Quantifoil Micro Tools; 400 mesh) and stained using 2% (w/v) uranyl acetate. Briefly, the grid was blotted with purified sample and washed twice with distilled water prior to uranyl acetate staining (1 min). Micrographs were collected on an FEI Tecnai F20 TWIN microscope with a Tietz TemCam-F416 complementary metal-oxide semiconductor (CMOS)-based camera (4k × 4k).
The negative stained micrographs were processed with RELION 1.4 software (31). Briefly, 1,603 particles were manually picked to generate reference-free 2D classes (32). After expanding the dataset to 4,509 particles, different trials of 3D classification were performed with multiple initial models of GST and GST-AWAT2 without imposing symmetry restraints. The 3D classes revealed the particle size distribution of the negative stained dataset. The 3D classification trial with reasonable 3D volume classes (using volume/mass conversion of 0.81 Da/Å) was used to reconstruct a 3D structure. All 3D maps were visualized in UCSF Chimera (33). A larger dataset of the same negative stain conditions (12,846 particles) was used to confirm the particle size distribution.
Expression and purification of human apo-CRALBP
A clone of human CRALBP (NCBI accession number P12271.2) incorporated into pET19b vector was obtained from Dr. J. W. Crabb (Cole Eye Institute) (34). Bl21(DE3) Escherichia coli competent cells (Invitrogen) were transfected with this construct and a single colony was used to inoculate 25 ml of LB medium (Thermo Fisher) containing 50 μM ampicillin. Bacterial culture was grown overnight at 37°C and subsequently transferred to 2 l of fresh medium. The culture was shaken at 30°C for around 4 h until OD600 reached 0.4–0.6. Then, the temperature was lowered to 25°C and expression of CRALBP was induced by adding isopropyl-1-thio-D-galactopyranoside to a final concentration of 0.15 mM. At the same time, ampicillin concentration was increased to 100 μM. Four hours later, bacteria were harvested by centrifugation at 6,000 g for 15 min at 4°C. Bacteria pellet was resuspended in water and frozen at −80°C. The cells were lysed by osmotic shock (35) and sonicated (three times for 1 min). The resulting crude lysate was spun down at 36,000 g for 20 min at 4°C. The soluble fraction, after adjustment of its buffer composition to 50 mM Tris/HCl (pH 8.0), 250 mM NaCl, and 5 mM imidazole, was loaded onto a HisTrap HP, 5 ml column (GE Healthcare) equilibrated with loading buffer [50 mM Tris/HCl (pH 8.0) that contained 250 mM NaCl and 5 mM imidazole]. After loading, the column was washed with 10 column volumes of the loading buffer and CRALBP was eluted with gradient of imidazole in the loading buffer (5–300 mM, flow rate 1 ml/min for 30 min). Fractions containing protein were pooled together, concentrated on a Centricon (Amicon) cutoff 10 kDa to about 5 ml, diluted 10-fold with 10 mM Tris/HCl (pH 8.0), and loaded onto a UNO Q1 (BioRad) ion exchanger column. CRALBP was eluted with gradient of NaCl (0–0.5 M) over 30 min at a flow rate of 0.5 ml/min. Collected fractions were examined by SDS-PAGE, and those containing purified apo-CRALBP were pooled together, concentrated to 3 mg/ml, and 0.2 ml aliquots were stored at −80°C.
Formation of holo-CRALBP
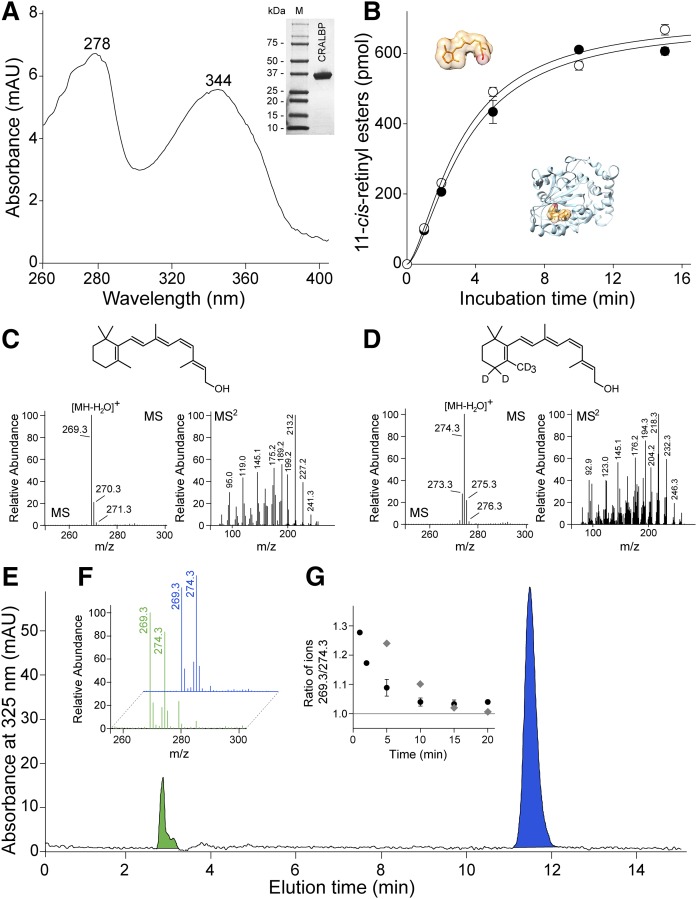
The holo-CRALBP was prepared by incubation 2 mg of the apolipoprotein with 2 molar excess of 11-cis-retinol delivered in DMSO (2%, v/v) for 30 min on ice in 10 mM Tris/HCl (pH 8.0), 10% glycerol (v/v). To remove unbound retinoid, the protein solution was diluted 10-fold with 10 mM Tris/HCl (pH 8.0), centrifuged at 36,000 g for 15 min at 4°C, and holo-CRALBP was repurified on a UNO Q1 column, as described above. The effectiveness of holo-CRALBP formation was verified spectrophotometrically by recording UV/Vis absorbance spectrum of the purified protein sample. The complex of CRALBP with 11-cis-retinol revealed a maxima absorbance ratio A344/A278 of 0.9 that corresponded to molar ratio of retinoid to the protein of 0.87, as determined by HPLC-based quantification of 11-cis-retinol extracted from holo-CRALBP.
Acyl-CoA-dependent retinol esterification assay
To test substrate specificity of AWAT2 toward geometric isomers of retinol, 9-cis, 11-cis, 13-cis, or all-trans-retinol was added to a reaction mixture containing 20 mM Tris/HCl (pH 7.0), 0.4 mM acyl-CoA, and 2 μl of microsomes isolated from yeast expressing AWAT2 (8 μg of total protein). The retinoids were delivered in 2 μl of N,N-dimethylformamide to a final concentration of 100 μM. The total volume of the reaction mixture was 0.2 ml.
Although C12:0-, C16:0-, and C18:1-CoA species were tested, myristoyl-CoA (C14:0) was chosen as a primary source of acyl moiety in most of the experiment because of the highest AWAT2 activity in the presence of this acyl-CoA as well as better HPLC separation of 9-cis-retinyl myristate from 11-cis-retinyl palmitate that was used in further experiments. The enzymatic reaction was carried out at 37°C for up to 150 min and quenched with 0.2 ml of ethanol. The retinoids were extracted with 0.3 ml of hexane. The retinoid composition was examined by normal phase HPLC using a Luna PREP silica column (250 × 4.6 mm, 10 μm) (Phenomenex) with a step gradient of 1% (v/v) ethyl acetate in hexane over 10 min followed by 20 min of 10% (v/v) ethyl acetate in hexane at a flow rate of 2 ml/min. Retinyl esters were detected at 325 nm and quantified by correlating peak areas with known quantities of synthetic standards.
To determine the respective Km and Vmax values for each retinol isomer, the initial velocity of the enzymatic reaction was calculated in the presence of increasing substrate concentration. The substrate concentrations ranged between 1 and 140 μM. The reaction was carried out for 5 min for 11-cis-retinol and 10 min for each of the other retinol isomers. GraphPad software was used to calculate the kinetic parameters by fitting the experimental data to the Hill model (equation 1) via nonlinear regression.
| (Eq. 1) |
To evaluate the substrate preference of AWAT2 in the presence of a mixture of the different retinol isomers, equimolar concentrations of isomers (25 μM each) were added to the same reaction. The reaction was incubated for 30 min prior to the retinoid extraction. All other conditions and analytical procedures were as described above.
Determination of mode of inhibition and inhibition constant
The initial velocity of AWAT2-dependent esterification was measured in the presence of 0, 2, 4, 6, or 10 μM of 11-cis-retinyl palmitate and variable concentrations of 9-cis-retinol. The inhibition constant (Ki) values were estimated by calculating the x-intercept from the equation resulting from the linear regression of the Dixon (36) and Cornish-Bowden (37) plots. To further evaluate the mode of inhibition and Ki values, the experimental data were subjected to the nonlinear regression analysis using a mixed inhibition model (equations 2, 3, and 4) (38) available in GraphPad software package.
| (Eq. 2) |
| (Eq. 3) |
| (Eq. 4) |
Acyl-CoA-dependent aliphatic alcohol esterification assay
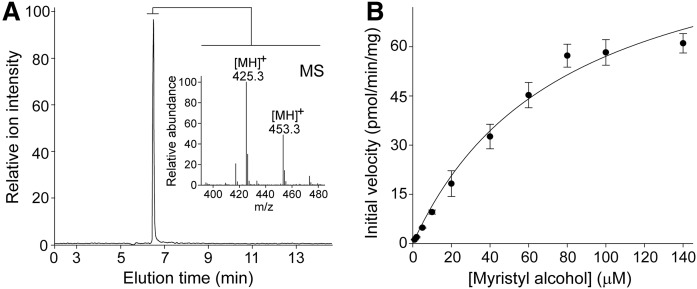
The kinetic parameters for the acyltransferase activity of AWAT2 were examined by incubating microsomes isolated from yeast cells expressing AWAT2 (8 μg of total protein) with tetradecanol at a concentration between 1 and 140 μM and 0.4 mM myristoyl-CoA in 20 mM Tris/HCl buffer (pH 7.4). The alcohols were delivered in 2 μl of N,N-dimethylformamide added to 0.2 ml of the reaction mixture. The enzymatic reaction was carried out at 37°C with moderate shaking in an Eppendorf thermomixer (600 rpm) for 10 min and terminated by the addition of 0.2 ml of ethanol. To enable LC-MS-base quantification of the fatty acid esters, 200 pmol of an internal standard (palmityl myristate) were added to each sample prior to extraction with 0.3 ml of hexane. After 2 min centrifugation at 18,000 g, the organic phase was collected from which 0.1 ml was injected onto a Luna PREP silica column (250 × 4.6 mm, 10 μm). The wax esters were eluted with an isocratic flow of 2% ethyl acetate (v/v) in hexane at a flow rate of 1.4 ml/min. The eluent was directed into an LXQ linear ion trap mass spectrometer (Thermo Scientific) via an atmospheric pressure chemical ionization source working in the positive mode. To ensure adequate sensitivity, the mass spectrometer parameters were tuned for palmityl myristate. Areas under ion intensity peaks for the internal standard and product of the enzymatic reaction were calculated using Xcalibur 2.2 software (Thermo Scientific).
Examination of role of CRALBP in 11-cis-retinol esterification
To verify whether 11-cis-retinol bound to CRALBP is preferentially esterified by AWAT2, the enzymatic reaction was conducted in the presence of equimolar concentrations (50 μM) of holo-CRALBP and isotopically labeled 5D-11-cis-retinol. The reaction was terminated by the addition of 0.2 ml of ethanol at different time points and retinoids were extracted with 0.3 ml of hexane. The isotopic composition of 11-cis-retinyl esters was examined by LC-MS. The hexanoic fraction was injected onto a Luna PREP silica column (250 × 4.6 mm, 10 μm) and retinyl esters were separated from unreacted 11-cis-retinol by a step gradient of ethyl acetate in hexane (2% for 10 min, followed by 20% for the next 10 min) at the flow rate of 1.4 ml/min. The eluent was directed into an LXQ linear ion trap mass spectrometer as described above. The portion of deuterated retinyl esters was calculated by comparing relative ion intensities at m/z 269.3 [MH-H2O]+ and m/z 274.3 [MH-H2O]+ that corresponded to unlabeled moiety and deuterated retinoid moiety, respectively.
The back exchange between CRALBP and free retinoid was examined by incubating holo-protein with 5D-11-cis-retinol in conditions equivalent to those used in the enzymatic assay. The only difference was that the yeast microsomal fraction was isolated from cells that did not express AWAT2. The samples were incubated for 5, 10, 15, or 20 min and then loaded onto a HiTrap Q HP column (5 ml) (GE Healthcare) equilibrated with 10 mM Tris/HCl (pH 8.0). CRALBP was eluted with a gradient of NaCl (0–0.5 M) over 30 min at the flow rate of 1 ml/min. Fractions containing repurified protein were pooled together, concentrated to 0.2 ml, and the 11-cis-retinol was extracted by addition of 0.2 ml ethanol and 0.3 ml of hexane. The isotopic composition of 11-cis-retinol was examined by LC-MS.
Statistical analysis
Data are represented as the mean ± SD. For the statistical analysis, results of at least three independent experiments were repeated in duplicate. The significance between two groups was determined by unpaired Student’s t-test, whereas the difference between multiple groups was analyzed using one-way ANOVA. SigmaPlot 11.0 (Systat Software) was used to perform the statistical analysis.
RESULTS
Mouse AWAT2 preferentially esterifies 11-cis-retinol
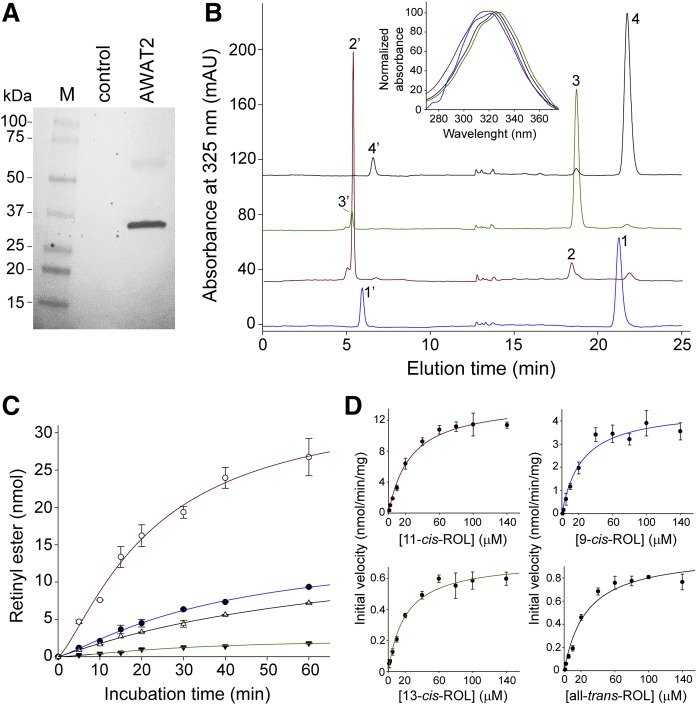
The high catalytic efficiency of 11-cis-retinol esterification by human AWAT2 (24) contrasts with the very broad substrate specificity of the enzyme (25, 26) and its mouse ortholog (27). In addition to retinols, AWAT2 accepts a variety of long-chain alcohols leading to formation of wax esters (25–27). To verify whether 11-cis specificity is characteristic for AWAT2 from cone-rich species, such as humans, or whether this phenomenon represents a more general property of AWAT2 from rod-dominant animals, we expressed mouse AWAT2 in yeast cells and tested its enzymatic activity toward geometric isomers of retinol. Because AWAT2 is an integral membrane protein localized in the endoplasmic reticulum, we used isolated microsomal fractions as a source for the enzymatic activity (Fig. 1A). The advantage of using yeast for expression of vitamin A-processing enzymes is the lack of endogenous retinoid metabolism. Suitably, the microsomes isolated from control yeasts did not demonstrate any detectable retinyl ester formation. Also, AWAT2-dependent esterification did not occur in the absence of externally added acyl-CoAs, indicating that endogenous acyl-CoAs were not copurified with the microsomal fractions.
Fig. 1.
Enzymatic activity of AWAT2 toward geometric isomers of retinol. A: Western blot detection of mouse AWAT2 in microsomal fraction isolated from yeast cells transfected with YepM containing the enzyme’s cDNA. The control sample represents membranes from the cells transfected with empty vector. B: HPLC chromatograms show separation of retinols and products of their AWAT2-dependent esterification. Peaks 1 and 1’ represent 9-cis-retinol and 9-cis-retinyl myristyl, respectively; peaks 2, and 2’ represent 11-cis-retinol and the corresponding ester, respectively; peaks 3 and 3’ represent 13-cis-retinol and its ester, respectively; and peaks 4 and 4’ represent all-trans-retinol and all-trans-retinyl ester, respectively. The inset shows characteristic UV/Vis spectra for each of the isomers. C: Comparison of the time courses of retinyl ester formation from retinol and its geometric isomers (blue line, 9-cis-retinol isomer; red line, 11-cis-retinol isomer; green line, 13-cis-retinol isomer; black line, all-trans-retinol isomer). D: Determination of kinetic parameters for the enzymatic reaction in the presence of retinol and its isomers. The curves represent dependence of the initial velocity of retinyl myristate formation on the substrate concentrations. The kinetic parameters of the reaction were determined by fitting the experimental points to the Michaelis-Menten model.
Incubation of vitamin A or its 9-, 11-, and 13-cis isomers with the microsomes containing AWAT2 resulted in the formation of corresponding retinyl esters (Fig. 1B). Notably, efficiency of the product formation strongly depended on the geometric isomer, as evidenced by comparison of the reaction time courses (Fig. 1C). Esterification of 11-cis-retinol progressed much faster than 9-cis, all-trans, or 13-cis isomers. These differences were reflected in kinetic parameters determined for each of the substrates (Fig. 1D, Table 1). The initial rate of the reaction, as a function of retinoid concentration, followed a typical steady state enzymatic kinetic. Thus, the kinetic parameters were calculated by fitting the experimental data to the Michaelis-Menten model. Maximum velocity (Vmax) for the esterification of 11-cis-retinol was over three times higher than for 9-cis-retinol and more than 14 times greater than for all-trans- and 13-cis-retinol, reaching 14.5 nmol/min/mg of total protein. Interestingly, the apparent Michaelis-Menten constants (Km) calculated for each of the isomers ranged between 20.9 and 26.7 μM; however, the differences between them held no statistical significance. Moreover, they were comparable to Km values determined for tetradecanol, a nonretinoid long-chain alcohol (Fig. 2A, B; Table 1). Similarly to human AWAT2 (24), these kinetic data indicate favorable esterification of 11-cis over other isomers of retinol for the mouse enzyme. However, there is an important deviation in how this substrate preferentiality is achieved. Unlike the human enzyme, which was characterized by approximately four times lower Km for 11-cis-retinol than for other isomers and a Vmax similar to that calculated for the 9-cis isomer, mouse AWAT2 revealed no differences in Km across tested substrates, but rather possessed a much higher Vmax value for the 11-cis isomer.
TABLE 1.
Kinetic parameters of AWAT2-dependent esterification of retinol isomers
| Substrate | Km (μM) | Vmax (nmol/min/mg) |
| 9-cis-retinol | 23.76 ± 6.24 | 4.55 ± 0.36 |
| 11-cis-retinol | 26.22 ± 3.95 | 14.52 ± 0.69 |
| 13-cis-retinol | 35.18 ± 7.82 | 1.06 ± 0.08 |
| all-trans-retinol | 23.59 ± 3.06 | 0.66 ± 0.04 |
| tetradecanol | 48.98 ± 17.56 | 1.2 ± 0.13 |
Fig. 2.
AWAT2 activity toward long-chain alcohol. A: LC-MS-based detection of myristate ester. The extracted ion chromatogram represents elution of product of the enzymatic reaction (myristyl myristate) detected at m/z 425.3 [MH]+. In addition to these ions, the MS spectrum averaged across this chromatography peak displays ions corresponding to palmityl myristate (m/z 453.3 [MH]+) that served as an internal standard for the purpose of quantification. B: Rate of myristyl myristate formation as a function of concentration of myristyl alcohol. By analogy to production of retinyl esters, the experimental points were used to determine the kinetic parameters for the enzymatic wax formation.
CRALBP-bound and free 11-cis-retinol as substrates for AWAT2
It is entirely plausible that the kinetic parameter of AWAT2 may not play a decisive role in determining 11-cis-retinol specificity of the enzyme. Alternatively, the substrate specificity of AWAT2 could be achieved by the interaction of AWAT2 with a specialized 11-cis-retinol binding protein (CRALBP). CRALBP is expressed in the RPE and Müller glia cells and facilitates the intracellular transport of cis retinoids (39, 40) (Fig. 3A). To test this hypothesis, we compared the enzymatic activity of AWAT2 in the presence of 11-cis-retinol delivered to the reaction mixture in an organic solvent (free form) or prebound to CRALBP. As shown in Fig. 3B, if tested separately, these two routes of substrate delivery did not affect progress of the esterification reaction. To further investigate the dependence of esterification on the carrier protein, the reaction was performed in the presence of equimolar concentrations of holo-CRALBP and free 11-cis-retinol labeled with five nonexchangeable deuterium atoms (Fig. 3C, D). Upon extraction with hexane, the isotopic composition of the esterification products was examined by LC-MS (Fig. 3E, F). A chromatography peak that corresponded to 11-cis-retinyl esters revealed presence of both m/z 269.2 and m/z 274.2 ions. These peaks correspond to the loss of acyl moiety from the unlabeled and labeled molecules upon ionization. Although the higher intensity of m/z 269.3 ions at early time points may suggest preferential esterification of the substrate bound to CRALBP (Fig. 3F, G), contribution of free deuterated 11-cis-retinol was very substantial. At the 1 min time point, deuterated 11-cis-retinyl esters represented 44% of total products. At this time point, less than 5% of the substrate was converted to the product, thus accumulated apo-CRALBP that could be reloaded in situ with free deuterated 11-cis-retinol had only minor influence on the obtained results.
Fig. 3.
AWAT2-dependent esterification of 11-cis-retinol in the presence of holo-CRALBP. A: The apo-CRALBP expressed in E. coli cells was purified to homogeneity (inset) and preincubated with 11-cis-retinol to form holo-protein. After repurification, the efficiency of ligand binding was verified by recording UV/Vis absorbance spectra. The spectrum indicates two maxima at 278 nm corresponding to the protein scaffold and at 344 nm that is representative of bound 11-cis-retinol. B: The time course of 11-cis-retinol esterification in the presence of either free (open circle) or CRALBP-bound (filled circle) substrate. C, D: Verification of the isotopic composition of 11-cis-retinol (C) and its deuterated form (D). C: The MS spectrum of unlabeled retinoid reveals the main isotopic peak at m/z 269.3 that corresponds to ionization-driven loss of water from the parent ion. The conforming tandem spectrum (MS2) shows the fragmentation pattern of the retinoid moiety. D: The main peak for heavy atom-labeled 11-cis-retinol is at m/z 274.3 that indicates five deuterium atoms incorporated into the molecule. The ion at m/z 274.3 that corresponds to the molecules labeled with only four heavy atoms represents less than 20% of the mixture. Importantly, the unlabeled retinoid was not detectable. E: HPLC separation of 11-cis-retinol (blue peak) and its esterified form (green peak) after 2 min incubation of AWAT2 with holo-CRALBP and free 5D-11-cis-retinol. The isotopic composition of the substrates (blue) and products (green) are shown in (F), whereas (G) represents temporal changes in the ratio between labeled and unlabeled 11-cis-retinyl esters during the enzymatic reaction (filled circle) and the time course of ligand exchange between holo-CRALBP and free 11-cis-retinol (gray diamonds).
CRALBP was shown to exist in two conformational states that affect the lipid-exchange loop region (41–43). Dynamic transitions from open to closed conformations affect access to the ligand-binding cavity and also influence stability of the ligand-bound form of the protein. Thus, to assess the rate of back-exchange of the ligand, holo-CRALBP was incubated with deuterated 11-cis-retinol in conditions analogous to those used for the enzymatic assay with the exception of the microsomes that were isolated from cells not expressing AWAT2. The carrier protein was subsequently repurified on a HiTrap Q HP ion exchange column and the isotopic composition of 11-cis-retinol associated with the protein fractions was examined by LC-MS. Addition of equimolar amounts of deuterated 11-cis-retinol to holo-CRALBP resulted in time-dependent enrichment in isotopically labeled ligand bound to the protein. Nearly 80% of the ligand was exchanged within 7 min of incubation and the equilibrium was achieved after 15 min (Fig. 3G). Although, the rate of exchange definitely confirmed the dynamic nature of CRALBP interaction with the ligand, this phenomenon did not affect the overall interpretation of the data. Formation of retinyl esters from labeled and unlabeled retinols became detectable at very early time points, whereas the time required for back-exchange was significantly longer. Thus, both the carrier protein-bound and free 11-cis-retinol served as effective substrates for AWAT2, indicating that the CRALBP-dependent delivery of the ligand is not a major factor that determines substrate specificity of the enzyme.
Activity of AWAT2 in the presence of a mixture of retinol isomers
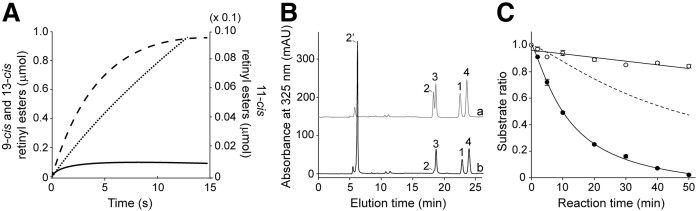
DES1 enzymatic activity produces a mixture of retinol isomers, which contains less than 1% of the 11-cis isomer (23). Although effective concentrations of retinol in the retina are difficult to estimate, it is legitimate to assume that, as with most enzymes, AWAT2 operates in substrate concentrations far below the Km values. By knowing the enzymatic parameters and ratio between the substrates, we can model the progress of the product formation. Such analysis clearly indicates that differences in Km or Vmax do not compensate for the over 100-fold higher concentration of 9- and 13-cis-retinol (Fig. 4A). In fact, 9- and 13-cis-retinyl esters constitute the main products formed even at the earliest time points. Although lack of accumulation of 13-cis-retinyl esters can be explained by the absence of an acceptor for 13-cis retinoid in vivo, 9-cis-retinal would be incorporated into the opsins. Thus, kinetic parameters for either human or mouse enzyme do not explain the exclusive accumulation of 11-cis-retinyl esters in the eyes.
Fig. 4.
Substrate selectivity of AWAT2 in the presence of a mixture of retinol isomers. A: Theoretical progress of the esterification reaction. Accumulation of 9-, 11-, and 13-cis-retinyl esters shown as dashed line, solid line, and dotted line, respectively, was simulated based on the kinetic parameters summarized in Table 1. The initial concentrations of the substrates resembled thermodynamic equilibrium between retinol isomers and were set below the Km values at 0.1, 1.0, and 2.3 μM for 11-, 9-, and 13-cis isomers, respectively. B: Representative chromatograms indicate changes in the composition of retinol isomers upon incubation with AWAT2. Trace (a) corresponds to separation of 9-cis-, 11-cis-, 13-cis-, and all-trans-retinol (peaks 1, 2, 3, and 4, respectively) from an equimolar mixture. Trace (b) shows the composition of retinoids extracted after 30 min of incubation in the standard condition for AWAT2 enzymatic assay. Disproportional (compared to other isomers) esterification of 11-cis-retinol is evident by diminishing of peak 2 and concomitant appearance of peak 2’ corresponding to 11-cis-retinyl myristate. C: The time course of the enzymatic reaction conducted in the presence of mixture of retinol isomers. Because formation of retinyl esters other than 11-cis was not sufficient for reliable quantification, the data are represented as ratios between substrates 11-cis- or 9-cis-retinol (filled circles) and 13-cis-retinol (open circles). The dashed line corresponds to the expected rate of 9-cis-retinyl myristate formation based on the kinetic parameters determined solely in the presence of 9-cis-retinol (Fig. 1A, C, D; Table 1). There is an apparent discrepancy between the efficiency of 9-cis-retinol esterification in the presence and absence of other retinol isomers.
In the standard enzymatic assay, AWAT2 was incubated with a single type of substrate. However, in physiological settings, the enzyme operates in the presence of several retinol isomers. To imitate this condition, the esterification reaction was performed with equimolar concentrations of 9-, 11-, and 13-cis-retinol, and all-trans-retinol. HPLC analysis showed that the predominant chromatography peak corresponded to 11-cis-retinyl myristate (Fig. 4B). Surprisingly, this analysis revealed the absence of 9-cis-, 13-cis-, or all-trans-retinyl esters. Based on the kinetic parameters described above, lack of evident 9-cis-retinyl esters was particularly astonishing. Thus, to better characterize this phenomenon, we followed the reaction progress over time. Because the amount of retinyl esters other than 11-cis was not sufficient for reliable quantification, the disappearance of corresponding alcohol substrates was used to monitor the progress of the reaction. Additionally, to correct for thermal conversion of 9- or 11-cis-retinol into 13-cis-retinol upon prolonged incubation, we represented the data as ratios between these isomers. This analysis showed that, as compared with the reaction progress estimated based on Vmax value, esterification of 9-cis-retinol was dramatically attenuated in the presence of other isomers (Fig. 4C).
The 11-cis-retinoids modulate isomer specificity of AWAT2
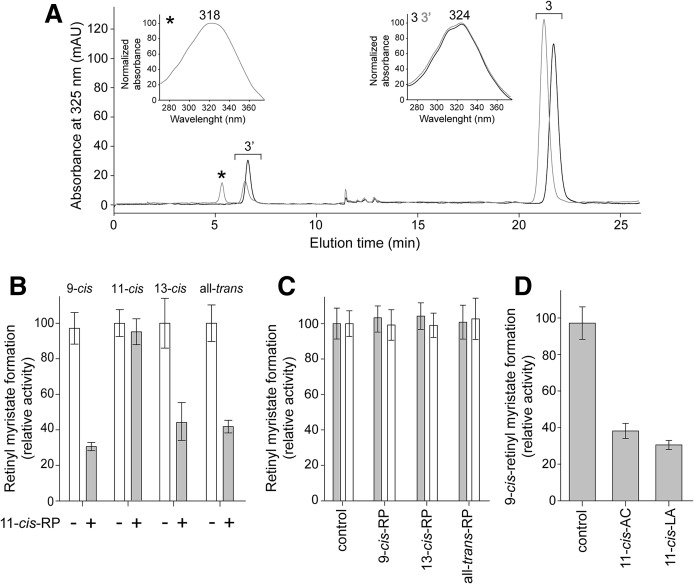
To investigate the origin of the altered enzymatic activity of AWAT2 in the presence of a mixture of retinol isomers, we examined to determine whether reaction products exhibited any influence on the enzymatic reaction. Under the applied experimental conditions, addition of 9-cis-, 13-cis-, or all-trans-retinyl palmitates did not have any effect on formation of 9-cis- or 11-cis-retinyl myristate. In contrast, the presence of 10 μM 11-cis-retinyl palmitate significantly reduced the amount of 9-cis-retinyl ester produced by AWAT2 (Fig. 5A–C). The 11-cis-retinyl palmitate also inhibited 13-cis- and all-trans-retinol esterification, but, importantly, had no effect on formation of 11-cis-retinyl myristate (Fig. 5B). To determine whether the inhibitory effect was related to the acyl moiety carried by the retinyl ester, we examined 11-cis-retinyl laurate and acetate in addition to 11-cis-retinyl palmitate. All of these derivatives showed comparable reduction in esterification of 9-cis-retinol (Fig. 5D).
Fig. 5.
The effect of 11-cis-retinyl esters on AWAT2 activity. A: Inhibition of 9-cis-retinol esterification in the presence of 11-cis-retinyl palmitate. The enzymatic reaction was carried out in the presence of 6 μM 11-cis-retinyl ester and 100 μM 9-cis-retinol. After 20 min of incubation, the retinoid composition was examined by HPLC. The analysis indicated a lower amount of 9-cis-retinyl esters (peak 3’) produced in the presence of 11-cis-retinyl esters (gray trace) as compared with a control reaction (black trace). The chromatography peaks were identified based on the elution time and characteristic UV/Vis spectra (insets). The 11-cis-retinyl ester and the corresponding spectrum with maximum absorbance at 318 nm are marked with asterisks. Peak 3 and confirming UV/Vis spectra with maximum at 324 nm (inset) indicate 9-cis-retinol. B: The 11-cis-retinyl palmitate (11-cis-RP) inhibited esterification of 9-cis-, 13-cis-, or all-trans-retinol, but not 11-cis-retinol. Ten micromoles of selected retinyl esters were preincubated with AWAT2 containing microsomes prior to the addition of the alcohol substrate. C: The 9-cis-, 13-cis-, or all-trans-retinyl palmitates had no effect on esterification of retinol isomers. D: The inhibitory effect of 11-cis-retinyl esters was not dependent on the length of the acyl moiety. Results of the enzymatic assays performed in the presence of 11-cis-retinyl palmitate, laureate (LA), or acetate (AC) revealed a comparable level of inhibition for each of the tested compounds.
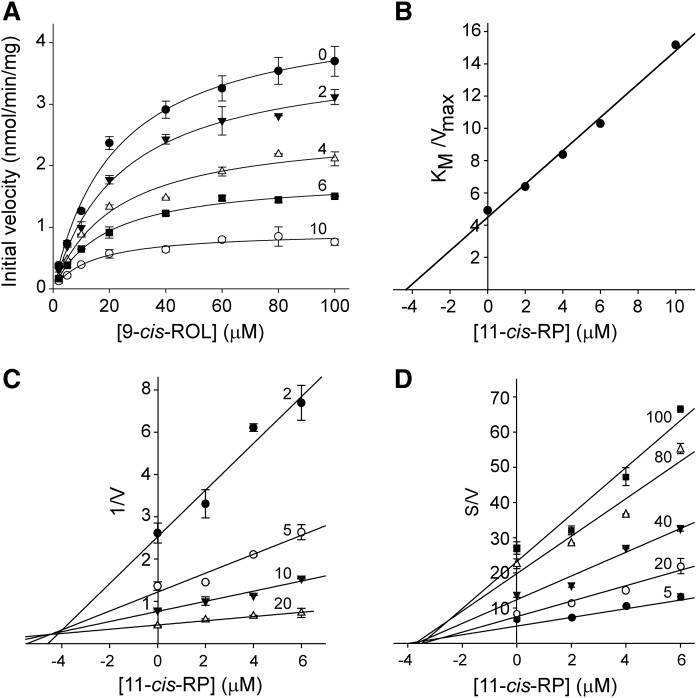
Following the identification of 11-cis-retinyl esters as modulators of AWAT2 activity, we determined the mode of inhibition by measuring changes in the initial rate of the enzymatic reaction in the presence of 2, 4, 6, or 10 μM of 11-cis-retinyl palmitate. The kinetic parameters were calculated by fitting the experimental data to the Hill model. Increasing the concentration of 11-cis-retinyl palmitate led to a dramatic reduction in the maximum rate of the reaction, as compared with the control experiment (Fig. 6A). Concomitantly, changes in Km were relatively modest, suggesting that 11-cis-retinyl palmitate acts predominantly in a noncompetitive fashion (Table 2). However, to better assess the mode of inhibition, the data were fitted into a mixed-model inhibition plot. Values of the parameter α, which determines the degree to which the binding of inhibitor changes the affinity of the enzyme for the substrate, were calculated to be 0.83 ± 0.23. Because α >> 1 indicates competitive inhibition and α ≈ 0 relates to a pure uncompetitive inhibitor, we concluded that the calculated values confirmed a largely noncompetitive mode of action. A subsequent calculation of Ki based on the noncompetitive model yielded a value of 4.5 ± 0.34 μM. A similar value of 4.2 μM was obtained when the Km/Vmax was plotted as a function of the inhibitor concentrations (Fig. 6B). These calculations were further confirmed by the Dixon plot that yielded Ki = 4.4 μM for the inhibitor binding to the free enzyme (Fig. 6C), whereas a slightly smaller value of 3.4 μM was obtained using a Cornish-Bowden plot for determining binding of the inhibitor to the enzyme-substrate complex (Fig. 6D).
Fig. 6.
Characterization of the mode of inhibition by 11-cis-retinyl palmitate. A: The effect of 11-cis-retinyl palmitate on the initial reaction rates. The enzymatic activities were measured at concentrations ranging from 0 to 10 μM as indicated by the numbers adjacent to each of the curves. The experimental points were fitted to the Hill equation (black lines) and the apparent values for Km and Vmax were calculated (Table 2). A significant decrease in Vmax values was not accompanied by major changes in Km, suggesting a noncompetitive mode of inhibition. Further confirmation of the type of inhibition and determination of the inhibition constants was achieved by analyzing the ratio between the substrate concentration for half-saturation and the limiting rate (Km/Vmax) (B), a Dixon plot showing the relationship between the reciprocal velocity of the reaction and the inhibitor concentration (C), and Cornish-Bowden plot where the ratio between the substrate concentration and the initial velocity was plotted against the inhibitor concentration (D). For panels C and D numbers adjacent to each of the curves indicate the substrate concentrations (μM).
TABLE 2.
Changes in the kinetic parameters of 9-cis-retinol esterification in the presence of 11-cis-retinyl palmitate
| Inhibitor 11-cis-RP (μM) | Km (μM) | Vmax (nmol/min/mg) |
| 0 | 22.25 ± 2.48 | 4.53 ± 0.17 |
| 2 | 24.32 ± 2.27 | 3.81 ± 0.12 |
| 4 | 21.87 ± 4.04 | 2.61 ± 0.16 |
| 6 | 18.74 ± 1.73 | 1.82 ± 0.05 |
| 10 | 15.26 ± 2.58 | 0.96 ± 0.05 |
11-cis-RP, 11-cis-retinyl palmitate.
These data indicate that 11-cis-retinyl esters affect the acyl transfer onto retinol isomers by impeding esterification of 9-cis, 13-cis, and all-trans retinols. Because the rate limiting step of the enzymatic reaction is the deprotonation of a substrate’s alcohol group, the lack of changes in Km values in the presence of 11-cis-retinyl esters suggests that the inhibition is not related to alterations in substrate affinity. Instead, this effect can most likely be attributed to allosterically induced changes in the active site of the enzyme, which impede the activation of the substrate and decrease Vmax.
The oligomeric state of AWAT2 examined by electron microscopy
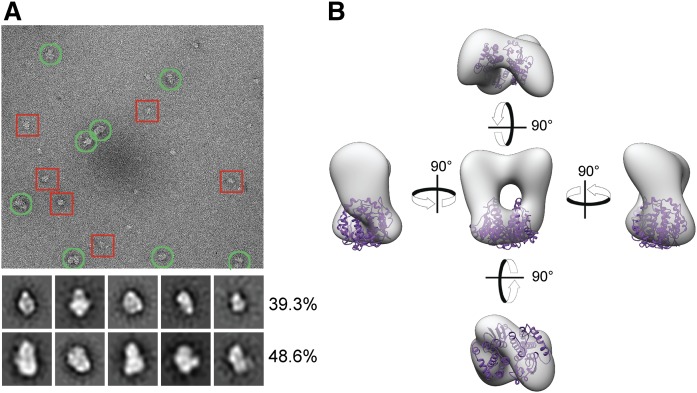
Allosteric modulation of enzymatic activities is often accomplished via functional oligomeric assembly of an enzyme. Therefore, we tested to determine whether AWAT2 forms ordered homo-oligomeric structures. Because our attempt to purify native AWAT2 from yeast microsomes did not yield sufficient quality protein sample, the enzyme was expressed and purified in a GST-fusion form. After solubilization with DDM, the enzyme was purified on a glutathione affinity resin followed by gel filtration in the presence of the detergent. The resulting GST-AWAT2 fusion protein was imaged by negative-stain electron microscopy (EM) that allowed for the visualization of individual protein particles. Separate protein particles with consistent size and shape were evident in the micrographs (Fig. 7A). These represented GST-AWAT2 monomers and more abundant homo-dimers. Larger heterogeneous particles, which are characteristic for nonspecific protein aggregates, were excluded from further analysis. Single-particle analysis was used to reconstruct a 3D EM map of GST-AWAT2 in the absence of enforced symmetry (Fig. 7B). The density for the well-ordered GST-AWAT2 fusion protein was clearly distinguishable in most of the 2D classes and in the reconstructed 3D map (Fig. 7B). Because GST forms an obligatory dimer on its own, the oligomeric state of GST-AWAT2 can be attributed predominantly to GST-GST interaction. We did not find evidence for higher order oligomers that involved contacts between AWAT2; thus, the monomer most likely constitutes a functional unit of the enzyme.
Fig. 7.
Evaluation of the oligomeric state of AWAT2 by EM. A: A representative cryo-electron micrograph of purified GST-AWAT2. The red squares indicate fusion protein monomers, whereas the green circles indicate fusion protein homodimers. The 2D classes shown were produced using the 1,603 particle dataset. The top row of classes represents fusion protein monomers, whereas the second row corresponds to homodimers. Each row is marked with the percentage distribution of each oligomeric state calculated from the 12,846 particle dataset. B: Three views of the 3D reconstructed density adjusted to a reasonable volume using the volume/mass conversion of 0.81 Da/Å. The crystal structure of GST (purple) is docked into the density using the Chimera fit to map function.
DISCUSSION
In most tissues, retinol esterification and the subsequent accumulation of chemically inert retinyl esters in cytoplasmic lipid droplets provide a safe way to store excess vitamin A and control retinoid homeostasis. However, in the eye, retinyl esters are an integral part of a metabolic pathway that results in the regeneration of visual pigments. In the established canonical visual cycle, LRAT-dependent esterification of vitamin A provides a direct substrate for RPE65 to produce 11-cis-retinol (44, 45), whereas the cone-specific regeneration pathway may rely on a mass action-driven accumulation of 11-cis-retinyl esters facilitated by AWAT2 (24). Although the end product is identical, these two esterification reactions fundamentally differ. LRAT is a highly specialized enzyme with a relatively narrow substrate specificity that utilizes phosphatidylcholine as a donor of the acyl moiety (46–48). Alternatively, retinol esterification is accomplished in an acyl-CoA-dependent manner by an acyl-CoA diacylglycerol acyltransferase (DGAT)1 and several representatives of the DGAT2 protein family, including AWAT2. In contrast to LRAT, these enzymes utilize a broad spectrum of acyl acceptors and mediate the synthesis of di- and triacylglycerols or wax mono-esters in addition to retinyl esters (25, 26, 49, 50). Consequently, the elucidation of factors that determine substrate specificity of this class of enzymes becomes critical for understanding their biological functions.
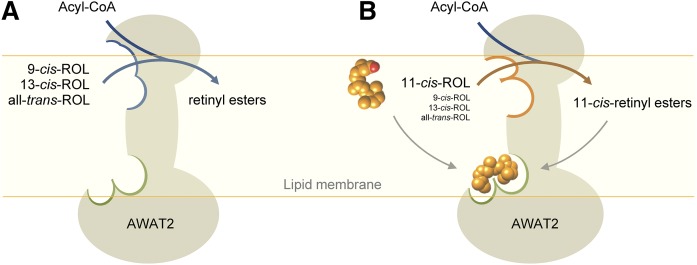
Because of the growing interest in the role that AWAT2 partakes in ocular vitamin A metabolism, we investigated the activity of AWAT2 toward retinol isomers and confirmed the enzyme’s supreme efficiency in esterifying 11-cis-retinol as compared with the 9-cis, 13-cis, or all-trans isomers. Interestingly, we noted that, in the presence of a mixture of different isomers, the rate of production of retinyl esters, other than 11-cis, was further suppressed, leading to the hypothesis that 11-cis-retinyl esters might be responsible for this effect. Kinetic studies provided experimental support for this postulate. The molecular mechanism by which 11-cis-retinyl esters influence AWAT2 activity is not entirely clear at present. The most plausible scenario is that the binding of 11-cis retinoids causes a ligand-induced structural rearrangement that makes the enzyme more efficient in 11-cis-retinol esterification (Fig. 8). The conformational change that creates this positive feedback system does not need to be extensive. A significant decrease in Vmax in the presence of 11-cis-retinyl esters was accompanied by a lack of major variations in Km values, suggesting that the rearrangement does not affect binding of the substrate, but rather is restricted to a few catalytic residues. The enzymatic activity of AWAT2 depends on a histidine residue that is a part of the conserved HHXXXDG motif (51). This histidine serves as a general base enabling activation of the substrate’s hydroxyl group. Because geometric isomers of retinol differ in spatial positioning of the functional group, even slight reorientation of the imidazole ring with the histidine residue in respect to the substrate’s hydroxyl group may impose profound effects on the efficiency of the enzymatic reaction. The overall measurable enzymatic effect of such changes would be an enhanced esterification of 11-cis-retinol accompanied by a suppression of the enzymatic activity toward other isomers.
Fig. 8.
A schematic representation of the modulation of AWAT2 enzymatic activity. A: In the absence of 11-cis-retinoids, AWAT2 reveals relaxed substrate selectivity allowing for esterification of a variety of geometric isomers of retinol. B: Interaction of the retinoid moiety in the 11-cis configuration with a putative allosteric site (marked with green boundary) leads to changes in the active site (blue and orange boundaries) that result in narrowing substrate specificity of the enzyme and selective production of 11-cis-retinyl esters.
It is important to note that the effect of 11-cis-retinyl esters does not exclude the possibility that 11-cis-retinol itself may be a physiological regulator of AWAT2. The lack of clear dependence of AWAT2 activation on the length of the acyl moiety in 11-cis-retinyl esters provides support for this hypothesis. Thus, although difficult to prove experimentally, the accelerated rate of 11-cis-retinol esterification may already be a result of enzyme activation by the substrate. This type of substrate activation is not unprecedented in acyltransferases. The representative example is ACAT1, which was shown to be allosterically activated by cholesterol and oxysterols in addition to the less effectual translational and posttranslational mechanisms (52).
Although the allosteric ligand-induced conformational changes may occur via interactions between multiple protomers, it is difficult to rationalize this model in light of the biochemical data. The absence of sigmoidal dependence of the reaction velocity on 11-cis-retinol concentration when the enzyme is incorporated into the phospholipid membrane and the lack of evidence for spontaneous AWAT2-induced oligomerization of purified GST-fused enzyme strongly suggest that the monomer of AWAT2 is the functional unit that is regulated. If this model turns out to be correct, it would be interesting to discover whether the 11-cis retinoid binding site, which causes modification of the enzymatic activity, is the same as or distinct from the catalytic site for esterification.
By investigating factors that might influence AWAT2 activity, we excluded the possibility that CRALBP serves as a special carrier that delivers 11-cis-retinol to the enzyme and, thus, affects substrate specificity. Consequently, 11-cis-retinol produced by DES1 is likely not channeled to AWAT2 via CRALBP in vivo. Furthermore, the experimental approach employed in this study provided evidence of a dynamic exchange between a ligand bound to CRALBP and external 11-cis-retinol. This finding coincides with previous molecular dynamics calculations as well as observations that CRALBP releases its retinoid cargo upon interaction with negatively charged phospholipids (53). These molecular properties of CRALBP clearly indicate that this carrier protein cannot serve as a storage pool of 11-cis retinoids inside the cell, but rather is involved in facilitating rapid transport of the ligand between cellular compartments or plasma membrane.
RPE cells and retina are the only tissues in which 11-cis retinoids are abundant and play important physiological functions. Thus, we believe that the regulatory mechanism described in this report principally applies to the eye. However, AWAT2 is not an eye-specific protein. In fact, it is expressed predominantly in the epidermis and sebaceous gland (25, 26). Moreover, its expression pattern within the ocular tissues is not limited to Müller cells or RPE, but spreads over other cell types (24). How might a broadly expressed and promiscuous enzyme contribute to the specific production of 11-cis-retinyl esters in the eye? To answer this question one needs to consider the multifunctionality of acyl-CoA-dependent acyltransferases attributed in their substrate ambiguity. Thus, the exact function of these enzymes depends mostly on the accessibility to the specific substrates or their composition, which in turn is determined by cell type. AWAT2 seems to be a perfect example illustrating this concept. While expressed in tissues lacking 11-cis-retinol, the function of AWAT2 is based on its catalytic activity toward long-chain alcohols or mono- and di-glycerols; whereas in cells containing robust retinoid metabolism, formation of retinyl esters might become dominant. Thus, in a larger sense, AWAT2 can be considered a case of gene sharing that describes different functions of a protein associated with similar molecular mechanisms, but different substrates, depending on the cell types.
In summary, biochemical analyses of AWAT2 led to the identification of a mechanism in which 11-cis retinoids act as modulators of AWAT2 activity by suppressing esterification of 9-cis, 13-cis, or all-trans retinols concurrently increasing the enzyme specificity toward 11-cis isomer. Our observations may provide an explanation for the mass action-driven selective accumulation of 11-cis-retinyl esters in the retinas that support regeneration of cone visual pigments.
Footnotes
Abbreviations:
- AWAT2
- acyl-CoA wax alcohol acyltransferase 2
- CRALBP
- cellular retinaldehyde-binding protein
- DDM
- n-dodecyl-β-D-maltoside
- DES1
- dihydroceramide desaturase 1
- DGAT
- acyl-CoA diacylglycerol acyltransferase
- EM
- electron microscopy
- GST
- glutathione S-transferase
- −Leu
- leucine-deficient selection
- LRAT
- lecithin:retinol acyltransferase
- MFAT
- multifunctional O-acyltransferase
- RPE
- retinal pigmented epithelium
- RPE65
- retinoid isomerase
This work was supported by National Eye Institute, National Institutes of Health Grant EY023948 (M.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflict of interest with the contents of this article.
REFERENCES
- 1.Wald G. 1968. The molecular basis of visual excitation. Nature. 219: 800–807. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard R., and Wald G.. 1952. Cis-trans isomers of vitamin A and retinene in the rhodopsin system. J. Gen. Physiol. 36: 269–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., et al. . 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 289: 739–745. [DOI] [PubMed] [Google Scholar]
- 4.Kiser P. D., Golczak M., and Palczewski K.. 2014. Chemistry of the retinoid (visual) cycle. Chem. Rev. 114: 194–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin M., Li S., Moghrabi W. N., Sun H., and Travis G. H.. 2005. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 122: 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moiseyev G., Chen Y., Takahashi Y., Wu B. X., and Ma J. X.. 2005. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. USA. 102: 12413–12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redmond T. M., Poliakov E., Yu S., Tsai J. Y., Lu Z., and Gentleman S.. 2005. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. USA. 102: 13658–13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redmond T. M., Yu S., Lee E., Bok D., Hamasaki D., Chen N., Goletz P., Ma J. X., Crouch R. K., and Pfeifer K.. 1998. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 20: 344–351. [DOI] [PubMed] [Google Scholar]
- 9.Mustafi D., Engel A. H., and Palczewski K.. 2009. Structure of cone photoreceptors. Prog. Retin. Eye Res. 28: 289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yau K. W. 1994. Phototransduction mechanism in retinal rods and cones. The Friedenwald lecture. Invest. Ophthalmol. Vis. Sci. 35: 9–32. [PubMed] [Google Scholar]
- 11.Kefalov V. J. 2012. Rod and cone visual pigments and phototransduction through pharmacological, genetic, and physiological approaches. J. Biol. Chem. 287: 1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann L., and Palczewski K.. 2015. Advances in understanding the molecular basis of the first steps in color vision. Prog. Retin. Eye Res. 49: 46–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baylor D. A., Lamb T. D., and Yau K. W.. 1979. Responses of retinal rods to single photons. J. Physiol. 288: 613–634. [PMC free article] [PubMed] [Google Scholar]
- 14.Werblin F. S., and Copenhagen D. R.. 1974. Control of retinal sensitivity. 3. Lateral interactions at inner plexiform layer. J. Gen. Physiol. 63: 88–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paupoo A. A. V., Mahroo O. A. R., Friedburg C., and Lamb T. D.. 2000. Human cone photoreceptor responses measured by the electoretinogram a-wave during and after exposure to intense illumination. J. Physiol. 529: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Findlay J. B. C., and Pappin D. J. C.. 1986. The opsin family of proteins. Biochem. J. 238: 625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagins W. A. 1955. The quantum efficiency of bleaching of rhodopsin in situ. J. Physiol. 129: 22P–23P. [PubMed] [Google Scholar]
- 18.Hubbard R., and Kropf A.. 1958. The action of light on rhodopsin. Proc. Natl. Acad. Sci. USA. 44: 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht S., Haig C., and Chase A. M.. 1937. The influence of light adaptation on subsequent dark adaptation of the eye. J. Gen. Physiol. 20: 831–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mata N. L., Radu R. A., Clemmons R. S., and Travis G. H.. 2002. Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron. 36: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mata N. L., Ruiz A., Radu R. A., Bui T. V., and Travis G. H.. 2005. Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of all-trans-retinol to 11-cis-retinol. Biochemistry. 44: 11715–11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mustafi D., Kevany B. M., Bai X., Golczak M., Adams M. D., Wynshaw-Boris A., and Palczewski K.. Transcriptome analysis reveals rod/cone photoreceptor specific signatures across mammalian retinas. Hum. Mol. Genet. Epub ahead of print. August 9, 2016; doi:10.1093/hmg/ddw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaylor J. J., Yuan Q., Cook J., Sarfare S., Makshanoff J., Miu A., Kim A., Kim P., Habib S., Roybal C. N., et al. . 2013. Identification of DES1 as a vitamin A isomerase in Muller glial cells of the retina. Nat. Chem. Biol. 9: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaylor J. J., Cook J. D., Makshanoff J., Bischoff N., Yong J., and Travis G. H.. 2014. Identification of the 11-cis-specific retinyl-ester synthase in retinal Muller cells as multifunctional O-acyltransferase (MFAT). Proc. Natl. Acad. Sci. USA. 111: 7302–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen C. L., Brown C. H. t., Monetti M., and Farese R. V. Jr. 2005. A human skin multifunctional O-acyltransferase that catalyzes the synthesis of acylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turkish A. R., Henneberry A. L., Cromley D., Padamsee M., Oelkers P., Bazzi H., Christiano A. M., Billheimer J. T., and Sturley S. L.. 2005. Identification of two novel human acyl-CoA wax alcohol acyltransferases: members of the diacylglycerol acyltransferase 2 (DGAT2) gene superfamily. J. Biol. Chem. 280: 14755–14764. [DOI] [PubMed] [Google Scholar]
- 27.Miklaszewska M., Kawinski A., and Banas A.. 2013. Detailed characterization of the substrate specificity of mouse wax synthase. Acta Biochim. Pol. 60: 209–215. [PubMed] [Google Scholar]
- 28.Hubbard R., P. K. Brown, and Bownds D.. 1971. Methodology of vitamin A and visual pigments. Methods Enzymol. 18C: 615–653. [Google Scholar]
- 29.Robeson C. D., Blum W. P., Dieterle J. M., Cawley J. D., and Baxter J. G.. 1955. Chemistry of vitamin A. XXV. Geometrical isomers of vitamin A aldehyde and an isomer of its α-ionone analog. J. Am. Chem. Soc. 77: 4120–4125. [Google Scholar]
- 30.Figler R. A., Omote H., Nakamoto R. K., and Al-Shawi M. K.. 2000. Use of chemical chaperones in the yeast Saccharomyces cerevisiae to enhance heterologous membrane protein expression: high-yield expression and purification of human P-glycoprotein. Arch. Biochem. Biophys. 376: 34–46. [DOI] [PubMed] [Google Scholar]
- 31.Scheres S. H. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres S. H. 2015. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E.. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 34.Crabb J. W., Chen Y., Goldflam S., West K., and Kapron J.. 1998. Methods for producing recombinant human cellular retinaldehyde-binding protein. Methods Mol. Biol. 89: 91–104. [DOI] [PubMed] [Google Scholar]
- 35.Burger A., Berendes R., Voges D., Huber R., and Demange P.. 1993. A rapid and efficient purification method for recombinant annexin V for biophysical studies. FEBS Lett. 329: 25–28. [DOI] [PubMed] [Google Scholar]
- 36.Dixon M. 1953. The determination of enzyme inhibitor constants. Biochem. J. 55: 170–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornish-Bowden A. 1974. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 137: 143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copeland R. A. 2005. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. Methods Biochem. Anal. 46: 1–265. [PubMed] [Google Scholar]
- 39.Saari J. C., Nawrot M., Kennedy B. N., Garwin G. G., Hurley J. B., Huang J., Possin D. E., and Crabb J. W.. 2001. Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron. 29: 739–748. [DOI] [PubMed] [Google Scholar]
- 40.Xue Y., Shen S. Q., Jui J., Rupp A. C., Byrne L. C., Hattar S., Flannery J. G., Corbo J. C., and Kefalov V. J.. 2015. CRALBP supports the mammalian retinal visual cycle and cone vision. J. Clin. Invest. 125: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu T., Jenwitheesuk E., Teller D. C., and Samudrala R.. 2005. Structural insights into the cellular retinaldehyde-binding protein (CRALBP). Proteins. 61: 412–422. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z., Yang Y., Shaw N., Bhattacharya S., Yan L., West K., Roth K., Noy N., Qin J., and Crabb J. W.. 2003. Mapping the ligand binding pocket in the cellular retinaldehyde binding protein. J. Biol. Chem. 278: 12390–12396. [DOI] [PubMed] [Google Scholar]
- 43.Wu Z., Hasan A., Liu T., Teller D. C., and Crabb J. W.. 2004. Identification of CRALBP ligand interactions by photoaffinity labeling, hydrogen/deuterium exchange, and structural modeling. J. Biol. Chem. 279: 27357–27364. [DOI] [PubMed] [Google Scholar]
- 44.Deigner P. S., Law W. C., Canada F. J., and Rando R. R.. 1989. Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science. 244: 968–971. [DOI] [PubMed] [Google Scholar]
- 45.Kiser P. D., Zhang J., Badiee M., Li Q., Shi W., Sui X., Golczak M., Tochtrop G. P., and Palczewski K.. 2015. Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nat. Chem. Biol. 11: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J., Dong Z., Mundla S. R., Hu X. E., Seibel W., Papoian R., Palczewski K., and Golczak M.. 2015. Expansion of first-in-class drug candidates that sequester toxic all-trans-retinal and prevent light-induced retinal degeneration. Mol. Pharmacol. 87: 477–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golczak M., and Palczewski K.. 2010. An acyl-covalent enzyme intermediate of lecithin:retinol acyltransferase. J. Biol. Chem. 285: 29217–29222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ong D. E., Kakkad B., and MacDonald P. N.. 1987. Acyl-CoA-independent esterification of retinol bound to cellular retinol-binding protein (type II) by microsomes from rat small intestine. J. Biol. Chem. 262: 2729–2736. [PubMed] [Google Scholar]
- 49.Cheng J. B., and Russell D. W.. 2004. Mammalian wax biosynthesis. I. Identification of two fatty acyl-coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 279: 37789–37797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yen C. L., Monetti M., Burri B. J., and Farese R. V. Jr. 2005. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 1502–1511. [DOI] [PubMed] [Google Scholar]
- 51.Stöeveken T., Kalscheuer R., and Steinbüchel A.. 2009. Both histidine residues of the conserved HHXXXDG motif are essential for wax ester synthase/acyl-CoA:diacylglycerol acyltransferase catalysis. Eur. J. Lipid Sci. Technol. 111: 112–119. [Google Scholar]
- 52.Cheng D., Chang C. C. Y., Qu X. M., and Chang T. Y.. 1995. Activation of acyl-coenzyme A-cholesterol acyltransferase by cholesterol or by oxysterol in a cell-free system. J. Biol. Chem. 270: 685–695. [DOI] [PubMed] [Google Scholar]
- 53.Saari J. C., Nawrot M., Stenkamp R. E., Teller D. C., and Garwin G. G.. 2009. Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids. Mol. Vis. 15: 844–854. [PMC free article] [PubMed] [Google Scholar]