Abstract
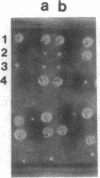
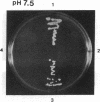
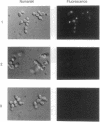
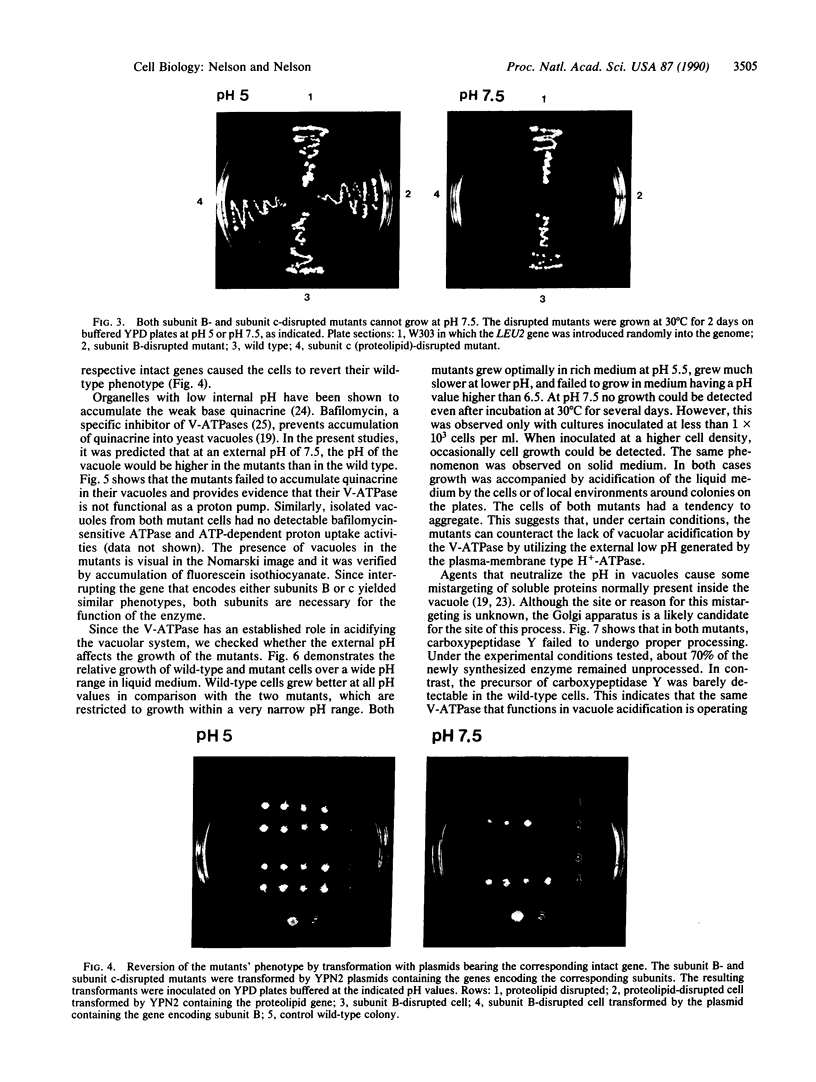
The main function of vacuolar H(+)-ATPases in eukaryotic cells is to generate proton and electrochemical gradients across the membranes of the vacuolar system. The enzyme is composed of a catalytic sector with five subunits (A-E) and a membrane sector containing at least two subunits (a and c). We disrupted two genes of this enzyme, in yeast cells, one encoding a subunit of the membrane sector (subunit c) and another encoding a subunit of the catalytic sector (subunit B). The resulting mutants did not grow in medium with a pH value higher than 6.5 and grew well only within a narrow pH range around 5.5. Transformation of the mutants with plasmids containing the corresponding genes repaired the mutations. Thus failure to lower the pH in the vacuolar system of yeast, and probably other eukaryotic cells, is lethal and the mutants may survive only if a low external pH allows for this acidification by fluid-phase endocytosis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Al-Awqati Q. Proton-translocating ATPases. Annu Rev Cell Biol. 1986;2:179–199. doi: 10.1146/annurev.cb.02.110186.001143. [DOI] [PubMed] [Google Scholar]
- Banta L. M., Robinson J. S., Klionsky D. J., Emr S. D. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988 Oct;107(4):1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. D., Ljungdahl P. O., Trumpower B. L. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. I. Construction of a RIP1 deletion strain and isolation of temperature-sensitive mutants. J Biol Chem. 1989 Mar 5;264(7):3713–3722. [PubMed] [Google Scholar]
- Bowman B. J., Bowman E. J. H+-ATPases from mitochondria, plasma membranes, and vacuoles of fungal cells. J Membr Biol. 1986;94(2):83–97. doi: 10.1007/BF01871190. [DOI] [PubMed] [Google Scholar]
- Bowman E. J., Siebers A., Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denda K., Konishi J., Oshima T., Date T., Yoshida M. The membrane-associated ATPase from Sulfolobus acidocaldarius is distantly related to F1-ATPase as assessed from the primary structure of its alpha-subunit. J Biol Chem. 1988 May 5;263(13):6012–6015. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma Y., Ohsumi Y., Anraku Y. Properties of H+-translocating adenosine triphosphatase in vacuolar membranes of SAccharomyces cerevisiae. J Biol Chem. 1981 Nov 10;256(21):10859–10863. [PubMed] [Google Scholar]
- Mellman I., Fuchs R., Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- Moriyama Y., Nelson N. Cold inactivation of vacuolar proton-ATPases. J Biol Chem. 1989 Feb 25;264(6):3577–3582. [PubMed] [Google Scholar]
- Moriyama Y., Nelson N. Lysosomal H+-translocating ATPase has a similar subunit structure to chromaffin granule H+-ATPase complex. Biochim Biophys Acta. 1989 Apr 14;980(2):241–247. doi: 10.1016/0005-2736(89)90405-7. [DOI] [PubMed] [Google Scholar]
- Nelson H., Mandiyan S., Nelson N. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J Biol Chem. 1989 Jan 25;264(3):1775–1778. [PubMed] [Google Scholar]
- Nelson H., Nelson N. The progenitor of ATP synthases was closely related to the current vacuolar H+-ATPase. FEBS Lett. 1989 Apr 10;247(1):147–153. doi: 10.1016/0014-5793(89)81259-1. [DOI] [PubMed] [Google Scholar]
- Nelson N. Structure, Function, and Evolution of Proton-ATPases. Plant Physiol. 1988 Jan;86(1):1–3. doi: 10.1104/pp.86.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. Structure, molecular genetics, and evolution of vacuolar H+-ATPases. J Bioenerg Biomembr. 1989 Oct;21(5):553–571. doi: 10.1007/BF00808113. [DOI] [PubMed] [Google Scholar]
- Nelson N., Taiz L. The evolution of H+-ATPases. Trends Biochem Sci. 1989 Mar;14(3):113–116. doi: 10.1016/0968-0004(89)90134-5. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980 Aug;21(1):205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y., Anraku Y. Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J Biol Chem. 1981 Mar 10;256(5):2079–2082. [PubMed] [Google Scholar]
- Orci L., Malhotra V., Amherdt M., Serafini T., Rothman J. E. Dissection of a single round of vesicular transport: sequential intermediates for intercisternal movement in the Golgi stack. Cell. 1989 Feb 10;56(3):357–368. doi: 10.1016/0092-8674(89)90239-0. [DOI] [PubMed] [Google Scholar]
- Preston R. A., Murphy R. F., Jones E. W. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7027–7031. doi: 10.1073/pnas.86.18.7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G. A. Pulse labeling of yeast cells and spheroplasts. Methods Enzymol. 1983;97:324–329. doi: 10.1016/0076-6879(83)97144-6. [DOI] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985 Apr;40(4):1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Rothblatt J., Schekman R. A hitchhiker's guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. doi: 10.1016/s0091-679x(08)61165-6. [DOI] [PubMed] [Google Scholar]
- Rothman J. H., Yamashiro C. T., Kane P. M., Stevens T. H. Protein targeting to the yeast vacuole. Trends Biochem Sci. 1989 Aug;14(8):347–350. doi: 10.1016/0968-0004(89)90170-9. [DOI] [PubMed] [Google Scholar]
- Serrano R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta. 1988 Feb 24;947(1):1–28. doi: 10.1016/0304-4157(88)90017-2. [DOI] [PubMed] [Google Scholar]
- Stevens T. H., Rothman J. H., Payne G. S., Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986 May;102(5):1551–1557. doi: 10.1083/jcb.102.5.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida E., Ohsumi Y., Anraku Y. Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1090–1095. [PubMed] [Google Scholar]
- Weisman L. S., Wickner W. Intervacuole exchange in the yeast zygote: a new pathway in organelle communication. Science. 1988 Jul 29;241(4865):589–591. doi: 10.1126/science.3041591. [DOI] [PubMed] [Google Scholar]