Abstract
Detection of viruses by flow cytometry is complicated by their small size. Here, we characterized the ability of a standard (FACSAria II) and a sub-micron flow cytometer (A50 Micro) to resolve HIV-1 viruses. The A50 was superior at resolving small particles but did not reliably distinguish HIV-1, extracellular vesicles, and laser noise by light scatter properties alone. However, single fluorescent HIV-1 particles could readily be detected by both cytometers. Fluorescent particles were sorted and retained infectivity, permitting further exploration of the functional consequences of HIV-1 heterogeneity. Finally, flow cytometry had a limit of detection of 80 viruses/ml, nearly equal to PCR assays. These studies demonstrate the power of flow cytometry to detect and sort viral particles and provide a critical toolkit to validate methods to label wild-type HIV-1; quantitatively assess integrity and aggregation of viruses and virus-based therapeutics; and efficiently screen drugs inhibiting viral assembly and release.
Keywords: Flow cytometry, flow virometry, HIV, extracellular vesicles, virus sorting, quantification
Introduction
Flow cytometry is a common method of analyzing cell types and states based on membrane and intracellular protein expression and complex parameters such as viability, DNA content, or proliferation history. Knowledge gained with the use of flow cytometry over the past few decades has greatly advanced our understanding of cell biology and immunology, and has revealed previously unappreciated cellular heterogeneity. For example, over 20 subsets of T cells have been identified (Perfetto et al., 2004), including specialized subcategories such as CD4+ and CD8+ T memory stem cells (TSCM) (Gattinoni et al., 2011; 2009; Zhang et al., 2005) and circulating memory follicular helper T(TFH) cells (Förster et al., 1994; Locci et al., 2013; Morita et al., 2011). Coupled with the advent of fluorescence-activated cell sorting (FACS), flow cytometry has enabled the purification of subpopulations of immune cells, facilitating further characterization of subsets through downstream biochemical and functional assays.
Flow cytometry has also contributed greatly to our understanding of viral infection of cells. Viruses have been studied indirectly by identifying cell subsets susceptible to infection and characterizing host and viral protein expression during the viral life cycle (Cavrois et al., 2002; McSharry, 2000). Immune responses to viruses can be characterized by proliferation assays, MHC tetramers, and cytokine and chemokine production (reviewed in (De Rosa, 2012)). However, few studies describe direct observation of viruses by flow cytometry due to inherent difficulties in observing small particles. These challenges include (1) minimal levels of light scattering from particles smaller than the wavelengths of light used in flow cytometers, (2) particle volumes approximately one million times smaller than that of a standard cell, with concomitant reductions in antigen binding sites, (3) the relatively low optical resolution of standard flow cytometers, and (4) contamination with extracellular microvesicles with similar size, density, composition, and light scatter characteristics (Lacroix et al., 2010; Nolan, 2015; Stoner et al., 2016).
These challenges are not insurmountable; over 30 years ago Howard Shapiro and colleagues demonstrated the ability of custom flow cytometers to detect viral particles (Hercher et al., 1979). In recent years, spurred in part by developments in the microvesicle field, flow cytometric evaluation of viruses – or “flow virometry” – has begun to yield information on the heterogeneity of viral particles. For example, analysis of HIV-1 viruses detected by fluorescent nanoparticles demonstrated the presence of distinct viral subpopulations characterized by differential incorporation of the host proteins LFA-1 and HLA-DR (Arakelyan et al., 2013). Dengue virus preparations were also found to be heterogeneous based on maturation state, and the ratio of mature to immature particles varied with the cell line used to produce virus (Zicari et al., 2016). Analysis of Nipah virus particles demonstrated three populations based on size characteristics with different incorporation levels of the viral glycoprotein NiV-F and NiV-G (Landowski et al., 2014). Finally, a sophisticated analysis of Junin virus particles demonstrated the presence of two viral subpopulations that differed in size (Gaudin and Barteneva, 2015). These subpopulations could be sorted by FACS and demonstrated differences in infectivity that correlated with glycoprotein and nucleic acid content, indicating the flow virometry not only enables detection of viral subpopulations but also downstream analysis in functional infectivity assays. Finally, flow virometry has recently been extended to analyze therapeutic preparations such as the homogeneity and aggregation characteristics of oncolytic vaccinia viruses (Tang et al., 2016) or the antigen expression on individual particles during human cytomegalovirus vaccine development (Vlasak et al., 2016).
In this study, we sought to build upon these pioneering flow virometry reports by assessing the capabilities of a high-performance standard flow cytometer (Becton Dickinson FACSAria II Special Order Research Project (SORP)) and a cytometer dedicated to submicron particle analysis (Apogee A50 Micro) for the detection and sorting of HIV-1 particles. The A50 was superior at detecting extracellular vesicles (EVs) and unlabeled HIV-1 viruses by light scatter alone but could not reliably distinguish between them and laser noise. These particles were entirely below the light scattering thresholds on the FACSAria. However, using fluorescently labeled viruses and detection by fluorescent threshold rather than light scatter, both machines readily identified single viruses and could distinguish them from EVs and machine noise. These labeled viruses will serve as useful tools to develop methods to specifically purify and label wild-type viruses and to optimize viral preparation, concentration, and storage for research and therapeutic purposes. Furthermore, we successfully sorted fluorescent HIV-1 viruses and demonstrated that these particles maintained the ability to infect target cells even when bearing HIV Env proteins, indicating that Env protein stability on viral particles is more robust than previously appreciated and opening the door for downstream functional analysis of sorted HIV-1 subpopulations. Finally, we determined that flow virometry has a limit of detection of ≤ 80 particles/ml, ~1250 times lower than ultrasensitive p24 ELISAs (~100,000 particles/ml) and nearly as sensitive as PCR-based viral load assays. These studies demonstrate the power of flow virometry to detect and sort individual, functional viruses and provide a toolkit for the systematic investigation of techniques to characterize viruses and to distinguish them from contaminating EV populations.
Materials and Methods
Cells
This study was conducted according to the principles specified in the Declaration of Helsinki and under local ethical guidelines (Case Western Reserve University Institutional Review Board (IRB)). Normal donor samples were de-identified and obtained from leukapheresis from ALLCELLS, LLC. All donors were negative for HBV, HCV, and HIV. CD4+ T cells were isolated by adding additional autologous red blood cells to leukapheresis samples and RosetteSep CD4+ T cell enrichment kit antibodies (STEMCELL Technologies) prior to ficoll gradient separation. Cells were cryopreserved and treated with benzonase upon thawing, prior to infection.
Biosafety
Flow cytometric analysis of infectious HIV-1 particles poses potential safety risks to machine operators. In particular, the use of flow cytometers to sort infectious viruses can result in the production of aerosols that pose a biosafety hazard. For sorting experiments, we utilized a FACSAria contained in a negative-pressure BioBubble (Propel Labs) containment system equipped with a 0.3 μm HEPA filtration at 200 cfm airflow. The cytometry core monitors system integrity using a “Glo-germ” procedure. All equipment and procedures are in compliance with the proposed NIH / International Society for the Advancement of Cytometry standards and approved by the Case Western Reserve University Environmental Health Safety Office. For all other experiments where recovery of infectious virus was not required, viruses were inactivated by fixation with 2% paraformaldehyde. These studies were approved by the Case Comprehensive Cancer Center Cytometry Core (FACSAria) and the Cleveland Clinic Flow Cytometry Core (A50 Micro).
Flow cytometers
Volumetric iGFP data were obtained with the use of an A50Micro (Apogee Flow Systems, Hertfordshire, UK), a flow cytometer dedicated to nanoparticle analysis, located at the Cleveland Clinic Foundation. The machine is equipped with a blue (488nm) 50mW laser and 525/50, 575/30, 680/35, and 740 nm emission filters. The A50 Micro used in this study was equipped with a large angle light scatter (LALS) detector that collects light scattered at a 15–150° angle and a small angle light scatter (SALS) detector that collects light scattered at 0.5–5° angles in relation to the sample stream. A threshold of LALS 12 was used to analyze all samples at 259V SALS, 320V LALS and 470V FL1 (Green PMT). The 18-color BD FACSAria II SORP sorter equipped with a powerful 355nm (60mW), 405nm (100mW), 488nm (100mW), 532nm (150mW), 640nm (100mW) lasers, PMT-SSC detector and 1.0 FSC ND filter was set to 250V SSC, 10V FSC for all experiments to reduce the laser noise from the FSC channel. The FACSAria uses a side scatter (SSC) detector positioned at 90° relative to the stream and a forward scatter (FSC) detector that collects light at ~5° angle. Therefore, the LALS and SALS parameters on the A50 Micro are roughly equivalent to the SSC and FSC detectors, respectively, on the FACSAria. Sheath fluid was pre-filtered at 0.2 μm pore for all experiments.
Beads
Cytometer performance was measured using a set of green fluorescent polystyrene bead standards (Invitrogen #F13839) in 20nm, 40nm, 100nm, 200nm, 500nm, 1 μm, and 1.9 μm diameters. The beads were sonicated, vortexed, and diluted to approximately 50,000/ml. Detection method used was the same as virus detection strategy. On the Aria, beads were examined using a threshold of 200 in the B515 channel (Blue 488nm laser excitation, 515 ± 10 nm emission detection), which allowed for visualization of beads on FSC x SSC cytograms in spite of low detectable scattered light in those two detectors by small (20nm–100nm) beads. On A50M, a threshold of LALS12 was used.
Production and quantification of HIV-based lentiviral particles
Lentiviral particles were produced as previously described (Tilton et al., 2014). Briefly, HEK293 T cells were seeded in 10cm dishes and cultured overnight prior to transfection using calcium phosphate methods (Pear et al., 1993) with 10.0 μg of wild-type or fluorescent HIV core (constructs described in detail below). As indicated in the text, 6.0 μg of CXCR4-tropic HIV Env JOTO.TA1.2247 (Wilen et al., 2011) (obtained from Drs. Beatrice Hahn and George Shaw) and 7.5 μg β-lactamase-Vpr plasmid were added to the HIV core to determine Env-dependent aggregation and fusogenicity of sorted particles. The wild-type HIV-1 core consisted of pNL43-deltaE-EGFP (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3-delta-E-EGFP (Cat #11100) from Drs. Haili Zhang, Yan Zhou, and Robert Siliciano), which expresses EGFP in cells but does not incorporate EGFP into particles and therefore produces non-fluorescent HIV virions. The fluorescent cores consisted of pNL4-3-deltaE-Gag-iGFP (Hübner et al., 2009)–obtained from Dr. Benjamin Chen–and Gag-iGFP-MS2SL, or Gag-iKOκ-MS2SL. The -MS2SL constructs were created in our laboratory using a previously described non-fluorescent Gag-MS2SL construct (Dilley et al., 2011) as a template. Briefly, this viral construct contains loss-of-function deletions in pol, vif, vpr, vpu, and env, while gag, nef, tat, rev and all cis-acting sequences are preserved. For the Gag-iGFP-MS2SL construct, we amplified the 5′ LTR-p6 region of the Gag-iGFP construct from Dr. Chen (Hübner et al., 2009) and 3 PCR products spanning the remainder of a non-fluorescent ΔPol-pNL4-3 construct obtained from Dr. Wei-Shau Hu (Dilley et al., 2011). We constructed a final plasmid using Gibson assembly. For the Gag-iKOκ-MS2SL virus, we used the recently created Gag-iGFP-MS2SL virus as a template and replaced the egfp gene with mKOκ, derived from the Chicken Mermaid S188 plasmid, a gift from Vincent Pieribone (Addgene plasmid # 53617) (Han et al., 2013) using Gibson assembly. Particles were harvested 72 hours after transfection, filtered through a 0.2 μm filter (unless indicated otherwise), and concentrated by ultracentrifugation through a 20% sucrose cushion according to published protocols (Kutner et al., 2009). A p24 ELISA (Cell Biolabs, Inc.) was used to correlate p24 content with FVM concentration data.
A50M speed of analysis varied from 1.5 μl/min at the highest sample concentrations to 101 μl/min at the low sample concentrations. The total respective analyzed volumes varied from 1.5μl to 303μl, which is near the maximum capacity of the A50M sample-loading syringe. For the FACSAria, we used the lowest analysis speed to minimize the probability of coincidence detection; all samples are representative 1-minute analyses unless indicated otherwise. We determined over multiple datasets that analysis of more than ~6,000 particles/second on the FACSAria resulted in considerable coincidental detection of viral particles. Generally, the lowest speed of analysis was therefore used, unless sample density was well below this number.
Virus sorting and infection assay
Virus dilution falling within single-particle detection range (1000x dilution, unless otherwise noted) was visualized at 10V FSC, 250V SSC and the appropriate fluorescent channel PMT voltage was first adjusted based on a 0.1μm PBS−/− control to show <10 events/second at lowest speed. We used 488 nm excitation and 515/20 nm emission channel for GFP and/or 532 nm excitation with a 575/26 filter for KOκ detection. This value was contained between 450V and 530V in our setup. For each sort we used a 70μm tip and 70 psi sheath pressure. For infection, about 1.6×107 iGFP/pNL events were sorted and concentrated at 1×105xg for 2 hours at 4°C, then resuspended in 200μl of 0.1μm filtered PBS−/− overnight at 4°C. Unsorted virus concentration was normalized by addition of filtered PBS using calculated iGFP counts/sec based on a 10-fold dilution of the concentrated resuspension. 1×106 enriched primary CD4+ T cells resuspended at 2×107 cells/ml in RPMI with 10% FBS, 1% L-glutamine and were overlayed with 60 μl of each virus suspension, or PBS, and spinoculated and stained as described before (Tilton et al., 2014). Cells were incubated with CCF2-AM (Invitrogen) in CO2-independent media (Gibco) for 1hour in accordance with manufacturer instructions, washed, and resuspended overnight in CO2-independent media containing 10% FBS and 2.5 mM probenicid. The cells were then washed, stained for 30 minutes at 4°C using anti-human CD3 Brilliant Violet 650 (BioLegend), CD4 APC (eBioscience), and live/dead fixable blue viability dye (Invitrogen). The cells were then washed, resuspended in 1% paraformaldehyde and analyzed for viral fusion on an LSRII analytical flow cytometer (Becton Dickinson). At least 50,000 events were collected for each sample, and each infection condition was performed in triplicate.
Data analysis and statistics
Flow cytometry data were analyzed with FlowJo v10.0.8 (FlowJo, LLC, Ashland, OR). Unless otherwise specified, data in figures represent mean values and standard error of the mean. All p values were corrected for multiple comparisons where appropriate. Statistical analyses were performed using the paired T-test using GraphPad Prism v7.0.
Results
Detection of small particles by flow cytometry
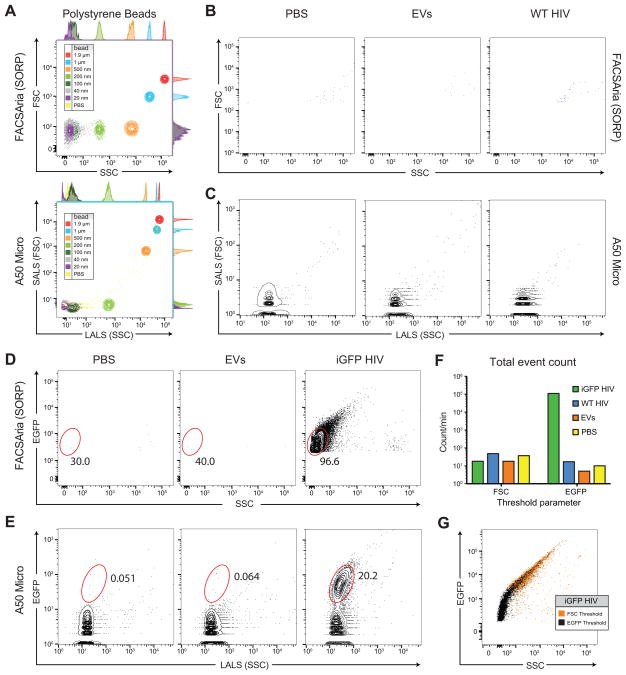
As a first step in assessing the ability of flow cytometers to detect single HIV virions, we evaluated the sensitivity of a high-end conventional flow cytometer (BD FACSAria SORP) and a cytometer specifically designed for detection of sub-micron particles (Apogee A50 Micro) using green fluorescent polystyrene beads with defined sizes. Beads were detected by fluorescence and their relative forward scatter (FSC) and side scatter (SSC) light parameters evaluated. We found that the FACSAria was able to resolve 1.9 μm, 1 μm, 500 nm, and 200 nm beads distinctly by FSC and SSC (Figure 1A) whereas beads of 100 nm, 40 nm, and 20 nm were below machine thresholds for FSC and SSC and were only detectable due to their fluorescence. In agreement with previous reports, the photomultiplier tube (PMT) on the SSC parameter allowed for greater resolution than the photodiode detector on the FSC parameter (van der Pol et al., 2012): 200 nm beads were distinguishable from smaller beads by SSC but not by FSC. In contrast to the Aria, the A50 Micro has features incorporated specifically for the detection of submicron particles including a photomultiplier tube for FSC detection, a smaller flow volume at the point of laser interrogation of the sample, and enhanced optics (Lacroix et al., 2010). Consistent with this design, the A50 Micro demonstrated improved FSC resolution, distinguishing between 500 nm and 200 nm beads by both FSC and SSC (Figure 1A) whereas the Aria could only distinguish them by SSC. However, 100 nm beads were still poorly detected by the A50 and formed an overlapping population with 40 and 20 nm beads and laser noise by light scatter alone. Low-range resolution improves when green fluorescence is used to supplement light scatter: although 20 and 40 nm beads partially overlap, they are more distinct from each other as well as from the 100 nm beads and the PBS control (Supplemental Figure 1).
Fig 1.
Detection of small particles and HIV-1 using flow cytometry. (A) Analysis of polystyrene beads of defined sizes by forward (FSC) and side (SSC) light scatter properties on the FACSAria II (SORP) and A50 Micro. (B,C) Analysis of filtered PBS and supernatants from HEK293T cells not transfected (EVs) or transfected with a ΔEnv wild-type HIV core plasmid (WT-HIV) analyzed by the (B) FACSAria II or (C) A50 Micro. (D,E) Analysis of PBS, untransfected HEK293 supernatants, or supernatants from cells transfected with a ΔEnv Gag-iGFP core plasmid (iGFP-HIV) on the (D) FACSAria II or (E) A50 Micro. (F) Comparison of events/second of PBS, EVs, WT-HIV, or iGFP-HIV on the FACSAria II using SSC or fluorescent thresholds. (G) iGFP events detected using a FSC or EGFP threshold differ substantially in B515 and SSC parameters as analyzed on the FACSAria II.
Detection of HIV particles and extracellular vesicles
HIV particles are approximately 120–150 nm in diameter (Briggs et al., 2006; 2003; Hanne et al., 2016; Pang et al., 2014); however HIV has a much lower refractive index (~1.42) (Pang et al., 2016) than do polystyrene beads (1.605) (van der Pol et al., 2012; 2014) and therefore would be expected to scatter less light than beads of a similar size. For example, 100 nm polystyrene bead scatter similar levels of light as 160–200 nm biological vesicles (van der Pol et al., 2013). Indeed, very small biological particles such as extracellular vesicles (EVs) or viruses with a diameter below 500 nm scatter very little light, especially in the forward direction. To systematically test the ability of the FACSAria and A50 Micro to resolve HIV particles, we generated virus from transfected HEK293T cells with a plasmid encoding an HIV core and harvested supernatants after 72 hours. For initial experiments, a plasmid encoding an Env protein was deliberately omitted to reduce the presence of viral aggregates. As a control for EVs, which consist of (1) exosomes, 40–100 nm vesicles of endocytic origin, (2) ectosomes or shedding microvesicles, 50 nm–1 μm vesicles derived from the plasma membrane, (3) and apoptotic blebs, 50 nm–5 μm particles released by dying cells (reviewed in (Mathivanan et al., 2010)), supernatants from un-transfected HEK293T cells were also collected. All samples were filtered through a 0.45 μm membrane prior to analysis. On the FACSAria, using a forward scatter (FSC) threshold of 200, the EV control supernatant and the supernatants containing HIV did not significantly differ from the media control, with very few detectable events in all conditions (Figure 1B). On the A50, a highly abundant, low-FSC low-SSC population present in the PBS control was observed, which corresponds to laser noise due to the very low SSC (LALS) threshold required to observe very small changes at the instrument’s limits of light scatter detection. Compared to the PBS control, the EV samples contained a small positive shift in SSC and a concomitant reduction in relative numbers of collected noise events. The wild-type (WT) HIV sample was characterized by even larger shifts in SSC and reduction in machine noise (Supplemental figure 2); however, machine noise, EVs, and WT HIV form largely overlapping populations by light scatter on the A50 (Figure 1C). These data indicate that, as expected, standard cellular flow cytometers are very poor at detecting viruses. Even the A50, a machine dedicated to small particle detection, displayed only nominal vesicle detection by light scatter alone, whether in filtered EV samples or with WT HIV.
To increase our confidence in detecting HIV particles, we generated fluorescently labeled virions using the HIV Gag-iGFP virus developed by Benjamin Chen’s laboratory that incorporate a GFP molecule between the matrix and capsid domains of Gag (Dale et al., 2011; Hübner et al., 2009) and a minimal Gag backbone developed by Wei-Shau Hu’s lab (Dilley et al., 2011). We confirmed virus presence in the supernatants by western blot and confocal microscopy (data not shown). Based on prior reports, we hypothesized that these bright, engineered particles could be used in conjunction with a fluorescence threshold rather than a FSC threshold to increase sensitivity and specificity of virus detection (Brussaard et al., 2000; Brussaard, 2004). As expected, filtered PBS and EV controls that were not transfected with the Gag-iGFP clone showed virtually no events on the FACSAria using a B515 (488 nm blue laser with a 515/20 nm detection filter) threshold (Figure 1D). In contrast, the supernatants containing Gag-iGFP HIV particles revealed the presence a GFP-bright population almost entirely (96.6%) contained within the 0–102 SSC interval, beneath the lowest setting for a standard SSC threshold (Figure 1D). Similar results were observed with control and Gag-iGFP containing supernatants on the A50 Micro, with a fluorescence parameter allowing clear distinction between viral particles and unlabeled EVs or machine noise (Figure 1E). Using FSC or SSC thresholds, event counts for PBS, EVs, WT HIV, and iGFP HIV were between 10–100 events/min (Figure 1F). The number of events dropped slightly for PBS, EVs, and WT HIV when a fluorescence threshold was applied; however, iGFP-HIV detection exceeded 105 events/min, an approximately 4-log increase in event frequency. Viral concentrations, as determined by p24 ELISA, were similar between WT HIV and iGFP HIV, suggesting that standard light scatter thresholds only detect a small fraction of viruses–and likely only a fraction of EVs as well. Importantly, the abundant iGFP population detected using an EGFP threshold differs markedly in B515 and SSC parameters compared with the iGFP population detected using a FSC threshold (Figure 1G). With a fluorescent threshold enabled, the iGFP population is predominant EGFP-dim with low SSC values. In contrast, using the FSC threshold, nearly all detected iGFP events were EGFP-bright and generated an elongated “tail” in the SSC channel that we believe correspond to viral aggregates, as described in detail below.
These results indicate that neither the FACSAria nor the A50 Micro can distinguish between unlabeled viruses and EVs–the FACSAria because these particles are below light scatter thresholds for detection and the A50 Micro because viruses and EVs form overlapping populations by FSC and SSC parameters that are largely indistinguishable from the noise, falling below the light scatter sensitivity of the instrument. While the FACSAria can detect fluorescently labeled viruses, this population approaches the fluorescence threshold value and some of the virus population may not be detected. In contrast, the entire fluorescent virion population on the A50 Micro is clearly defined and suggests that this dedicated sub-micron flow cytometer indeed detects fluorescence signals from HIV-1 particles more sensitively and quantitatively.
Detection of single HIV particles by flow cytometry
Two major factors complicate the ability to detect single viral particles by flow cytometry. First, viruses can form aggregates, which we believe correspond to the “tail” of events in the 102–105 SSC range in Figure 1G. A second complication of detecting single viruses is that due to the small size of viral particles compared to the volume of sheath fluid at the interrogation point of the lasers on standard flow cytometers, there is a concern of coincidental detection–or swarm detection–in which multiple viruses pass by the laser simultaneously and are recorded as a single event. Of note, the A50 Micro contains a smaller interrogation volume and offers lower collection speeds than the FACSAria to reduce swarm detection.
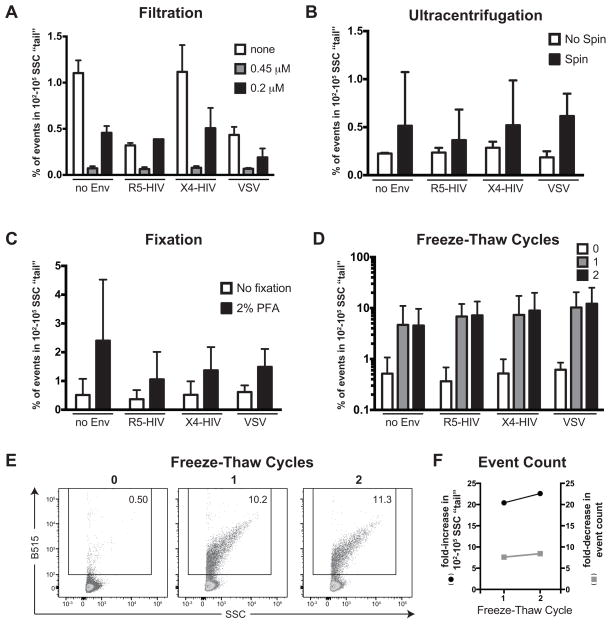
To investigate whether the SSC-high “tail” of iGFP events reflected viral aggregates, we performed a series of experiments to determine whether this population was affected by factors thought to influence aggregation including filtration, centrifugation, freeze/thaw cycles, fixation with 2% paraformaldehyde, or Env protein incorporation. To assess the effects of filtration, iGFP HIV-containing supernatants were centrifuged at 500xg for 5 minutes to remove cellular debris and then either left unfiltered or passed through a 0.45 μm or 0.2 μm filter prior to flow cytometric analysis. In the absence of filtration, the frequency of events in the SSC-high “tail” ranged from ~0.3–1.1% of the total events, with the highest levels observed in the no Env and X4-HIV conditions (Figure 2A). Filtration through a 0.45 μm filter virtually eliminated this population for all conditions (p<0.05 for all conditions); however, filtration through a 0.2 μm filter unexpectedly resulted in higher levels of SSC-high events compared to the 0.45 μM filter. Next, we investigated the effects of ultracentrifugation of viruses at 25,000xg through a 20% sucrose cushion and fixation with 2% paraformaldehyde (PFA) on the presence of SSC-high events. Although ultracentrifugation increased SSC-high event frequencies in all Env conditions, this enhancement was highly variable (Figure 2B) and did not reach statistical significance. Similar results were observed with 2% PFA fixation (Figure 2C). In contrast, freeze-thaw cycles resulted in a dramatic and highly significant (p<0.001 for all conditions) increase in SSC-high event frequency (Figure 2D, 2E). The increase in the SSC-high “tail” upon freeze-thawing could be due either to the de novo formation of viral aggregates from the single virus population or due to a selective destruction of single viruses that results in an enrichment of SSC-high events. Indeed, freeze-thaw cycles do result in a destruction of particles as measured by fold-decrease in event count. However, the fold-decrease in event count is far lower than would be required to explain the fold-increase in the SSC-high population (Figure 2F), providing compelling evidence that this population indeed reflects viral aggregate formation from single particles. This hypothesis is further supported by the strong reduction of the SSC-high population upon filtration with a 0.45 μm filter. We speculate that the unexpected presence of the SSC-high population following passage through a 0.2 μm filter may reflect de novo aggregate formation from viruses interacting with each other as they pass through the narrow filter pore. Interestingly, we did not observe substantial differences in aggregate formation depending on the presence or absence of HIV or VSV Env proteins.
Fig 2.
Assessment of factors influencing viral aggregation. Percentage of SSC-high “tail” events in viral supernatant samples (A) left unfiltered or filtered through 0.45 or 0.2 μm filters, (B) centrifuged at 25,000xg for 2 hours through a 20% sucrose cushion, (C) fixed with 2% paraformaldehyde, or (D) after 0, 1, or 2 freeze-thaw cycles. (E) Histogram of single viral particles and larger SSC-high events prior to and after freeze-thaw cycles using particles bearing an R5-tropic HIV Env. (F) Fold-increase in the SSC-high “tail” compared to fold-reduction in event count with freeze-thaw cycles using viruses bearing an R5-tropic HIV Env. Similar results were observed with the no Env, X4-tropic HIV Env, and VSV-G Env conditions.
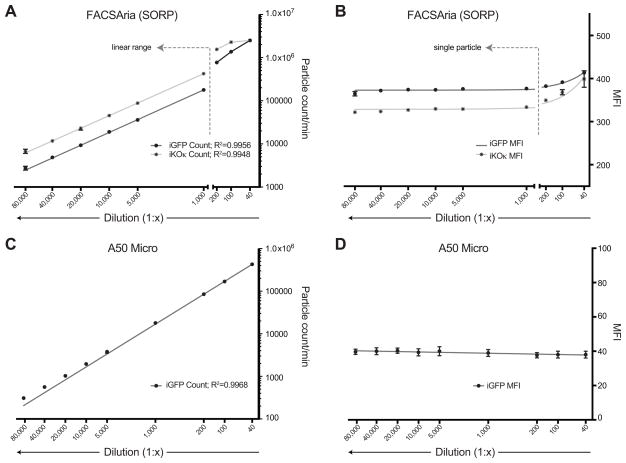
To assess whether coincidental detection or swarm detection was occurring in our samples, we utilized a well-established method for identifying single particles by monitoring event counts and mean fluorescent intensity (MFI) as the sample is diluted (Nolan, 2015; van der Pol et al., 2012). For single particles, a linear relationship is expected between the dilution factor and the number of events detected per minute, whereas MFI should remain constant. Non-linear relationships between dilution and event count or increasing MFI at higher concentrations of virus are indicative of coincidental detection. Viruses were prepared as above, with the exception that we generated a second fluorescent virus, termed Gag-imKOκ, that incorporates a bright, orange-emitting fluorescent protein called mKOκ (Tsutsui et al., 2008) between matrix and capsid in place of GFP. The resuspended viral stocks were subsequently diluted from frozen 40x–80,000x and run in triplicate on the FACSAria and A50 Micro. On the FACSAria, a strong linear relationship between dilution factor and event count was observed for both Gag-iGFP particles (R2=0.9956, p<0.0001) and Gag-imKOκ particles (R2=0.9948, p<0.0001)(Figure 3A) for 1,000x–80,000x dilutions, whereas more concentrated samples (40x, 100x, and 200x dilutions) deviated from this relationship. Additionally, we found that the MFI of events in dilutions between 1,000x–80,000x were stable whereas MFI increased with more concentrated viruses (Figure 3B). Together, these data strongly suggest that single particles are being detected at dilutions of 1:1,000 to 1:80,000 whereas in more concentrated samples swarm detection is occurring. A similar linear relationship between Gag-iGFP virus event counts and dilutions was observed with the A50 Micro (R2=0.9968, p<0.0001); however only the Gag-iGFP particles could be examined based on the lack of an appropriate laser to excite mKOκ (Figure 3C). Additionally, the MFI was constant over the entire dilution range from 1:40–1:80,000, with no evidence of swarm detection at higher concentrations (Figure 3D). The improved performance of the A50 Micro is likely the result of design features such as a smaller interrogation volume to reduce coincidental detection, and adjustable sample flow at rates as low as 0.75 μl/min.
Fig 3.
Detection of single HIV-1 particles on the FACSAria II and A50 Micro. (A) Particle count/min of iGFP and iKOκ viruses by dilution factor on the FACSAria II. R2=0.9956 and 0.9948, respectively, p<0.0001 for both. (B) Mean fluorescent intensity (MFI) of iGFP and iKOκ viruses by dilution factor on the FACSAria II. (C) Particle count/min of iGFP viruses by dilution factor on the A50 Micro. R2=0.9968, p<0.0001. A lack of a laser with an appropriate excitation wavelength makes detection of iKOκ viruses impossible on the A50 machine used in this study. (D) Mean fluorescent intensity (MFI) of iGFP viruses by dilution factor on the A50 micro.
HIV-1 viruses can be sorted and maintain ability to infect target cells
While flow virometry has demonstrated the ability to detect heterogeneous viral subpopulations that differ in size, maturation state, host and viral protein incorporation, and nucleic acid content (Arakelyan et al., 2013; Gaudin and Barteneva, 2015; Landowski et al., 2014; Zicari et al., 2016), being able to functionally characterize these subpopulations is critical for assessing how heterogeneity influences viral transmission, replication, and immunity. For instance, in a recent study by Gaudin and Barteneva, Junin viruses were sorted into low and high fractions based upon size. Normalized for virus input, the high fraction demonstrated twofold enhanced infectivity compared with the low fraction, which subsequently correlated with the percentage of viruses expressing both envelope glycoproteins and viral nucleic acids (Gaudin and Barteneva, 2015).
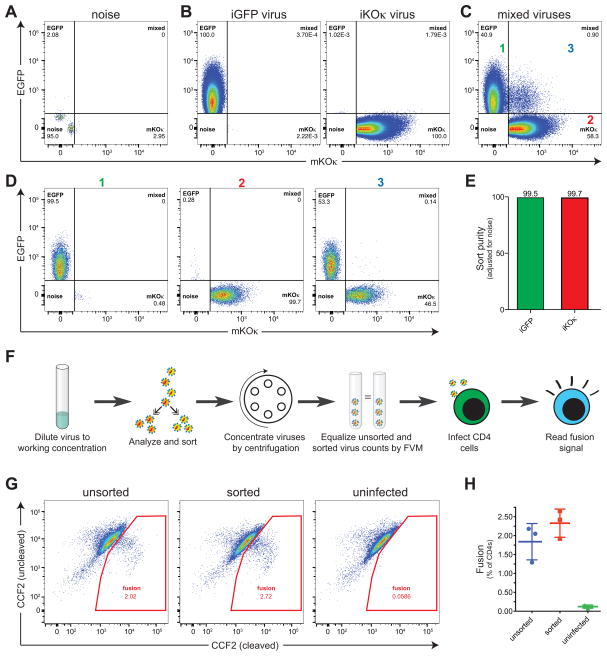
To determine whether HIV-1 particles could be efficiently sorted and maintain infectivity, we first analyzed PBS filtered through a 0.1 μM filter on the FACSAria and set gates to exclude ≥ 95% of machine noise (Figure 4A), while single-color controls show excellent signal specificity in each channel (Figure 4B). We then assessed a population of mixed EGFP and mKOκ viruses (pre-sort fraction) and found that the vast majority of particles were singly fluorescent for EGFP (40.9%) or mKOκ (58.3%), although some double-positive events were also recorded (0.90%) (Figure 4C). Following sorting based on these quadrant gates, viruses were reanalyzed on the FACSAria and the EGFP post-sort fraction found to contain 99.5% EGFP+ viruses while the mKOκ post-sort was 99.7% pure (Figure 4D & 4E). Interestingly, the double-positive population was found to contain only single-positive events after sorting, suggesting that the initial double-positive population represented either swarm detection or short-lived viral aggregates that were reversed upon sorting (Figure 4D).
Fig 4.
Sorting and infectivity of HIV-1 viruses. (A) 0.1 μm filtered PBS was analyzed on the FACSAria II and gates drawn that eliminated ≥ 95% of machine noise. (B) Analysis of single-positive iGFP or iKOκ viruses. (C) Mixed iGFP or iKOκ viruses demonstrating (1) EGFP-only events, (2) mKOκ-only events, and (3) EGFP and mKOκ double positive events. Sorting was performed on the FACSAria II using a 70 μm nozzle at 70 PSI. (D) Representative post-sort cytograms corresponding to gates 1, 2, and 3 are shown. The starting concentration of each virus was ~3000 events/sec and the final sorted concentrations were typically between 80 and 300 events/sec. (E) Chart showing the sort purity of iGFP and iKOκ viruses, adjusted for machine noise. (F) Schematic of the experiment analyzing the ability of sorted viruses to successfully infect cells. Briefly, virus was diluted, analyzed and sorted, and concentrated by centrifugation. Unsorted viruses were diluted to equivalent concentrations as determined by flow virometry and unsorted virus, sorted virus, or PBS used to infect cells. Fusion was determined by β-lactamase dependent CCF2 dye cleavage. (G) Analysis of CCF2 dye cleavage by unsorted and sorted viruses and PBS. Representative pseudocolor plots from of one of three replicate experiments. (H) Fusion in CD4+ T cells exposed to unsorted and sorted virus or PBS controls, 3 replicates.
To assess whether sorted HIV-1 viruses maintained their ability to enter cells, we generated maturation-competent EGFP+ viruses bearing a patient derived CXCR4-tropic HIV Env and packaging β-lactamase-Vpr. Upon fusion with host cells, these viruses release active β-lactamase that cleaves the FRET substrate CCF2, generating a colorimetric change to the cells that can be analyzed by flow cytometry (Cavrois et al., 2002). Viruses were sorted on the FACSAria using 70 psi sheath pressure and a 70 μm tip, then concentrated over a 20% sucrose cushion by ultracentrifugation at 100,000xg for 2 hrs at 4°C. Concentrated viruses were resuspended in filtered PBS and particles/minute determined by flow virometry. Unsorted viruses were diluted in PBS to equivalent concentrations, also verified by flow virometry. Sorted and unsorted fractions were then mixed with unstimulated, primary CD4+ T cells, spinoculated for 2h at 1200xg and analyzed for CCF2 dye cleavage 24h after infection (schematic shown in Figure 4F). To our surprise, sorting and concentration of viruses did not diminish infectivity of intact HIV-1 particles, with three sorted replicates demonstrating similar infectivity compared to unsorted fractions (sorted: 2.36 ± 0.41% fusion; unsorted: 1.71 ± 0.44% fusion, p=0.14) (Figure 4G, H). Both sorted and unsorted viruses demonstrated significantly higher CCF2 dye cleavage than uninfected controls (p=0.0006 and p=0.003, respectively).
Flow virometry is orders of magnitude more sensitive than ELISA at detecting HIV particles
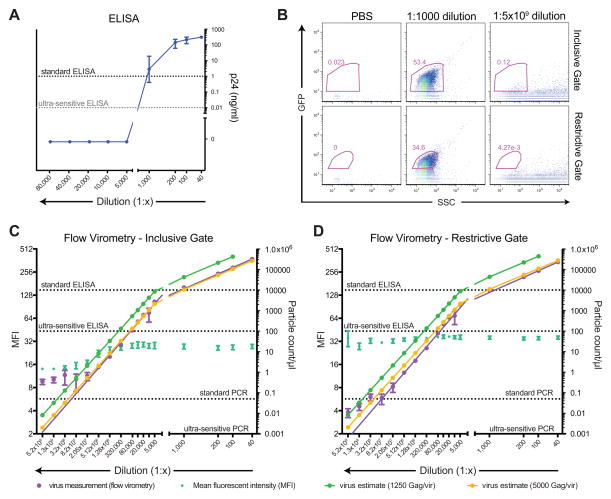
Multiple common HIV quantification standards exist, including measuring p24 protein by ELISA, commonly used to assess viruses produced in vitro, and PCR-based assays used to measure viral load in patient samples. The p24 ELISA has a limit of detection (LOD) of 1 ng p24 equivalents/ml for standard assays and ~10 pg p24/ml for ultra-sensitive assays, which corresponds to ~10,000,000 and 100,000 particles/ml, respectively, based upon estimates of ~2500 Gag molecules/virus (Carlson et al., 2008; Ganser-Pornillos et al., 2012). The PCR viral load assay is more sensitive, with an LOD of 50 copies/ml for the standard assay and 1 copy/ml for ultra-sensitive assays. To assess the LOD of flow virometry, a p24 ELISA was performed alongside flow virometry over an extended range of viral stock dilutions ranging from 1:40 to 1:5×109. The p24 ELISA successfully detected HIV at relatively high concentrations, but dilutions of 1:1000 and greater were at or below the limit of detection (LOD) of this standard assay (Figure 5A).
Fig 5.
Analysis of the limit of detection of viruses by flow virometry and comparison with ELISA and PCR assays. (A) A viral stock was diluted and assessed at the stated dilution factors for p24 concentration by standard (1 ng/ml sensitivity) p24 ELISA. All results are the mean of 6 replicates. (B) Analysis of PBS, concentrated HIV particles (1:1000 dilution), and extremely dilute HIV particles (1:5×109 dilution) on the A50 Micro. An inclusive gate (top row) was drawn based on PBS controls; however in very dilute samples analyzed for long periods of time (~3 minutes) on the A50 Micro, laser ‘noise’ with low EGFP fluorescence can be detected. A restrictive gate (bottom row) eliminates the vast majority of this noise while retaining approximately 70% of the viral events in the inclusive gate. (C) Analysis of particle count/μl and MFI of events in the inclusive gate by viral dilution on the A50 Micro. Using the p24 ELISA results from the 1:100 dilution, high and low estimates for viral particles were calculated based on 1,250 or 5,000 Gag molecules/virus, respectively, and mathematically adjusted for other dilutions to serve as a reference for flow virometry results. (D) Analysis of particle count/μl and MFI of events in the restrictive gate by viral dilution on the A50 Micro. High and low estimates of viruses based on p24 ELISA were calculated as described above. All dilutions for (C) and (D) were generated and analyzed in triplicate. Reference lines for ELISA and PCR: standard p24 ELISA (1 ng/ml sensitivity = 10,000,000 particles/ml based on 10,000 particles/pg p24 at 2,500 Gag molecules/virus); ultrasensitive p24 ELISA (10 pg/ml sensitivity = 100,000 particles/ml); standard PCR = 50 copies/ml; ultra-sensitive PCR = 1 copy/ml.
To determine the LOD of flow virometry, we used the A50 Micro that allows determination of absolute particle counts in volumes up to 300 μl. We employed two gating strategies: an inclusive gate based on PBS that included all GFP viruses but overlapped slightly with EVs and machine noise and a restrictive gate designed to eliminate the vast majority of EVs and noise (Figure 5B).
Using the inclusive gating strategy, flow virometry produced linear results from dilutions of 1:40 to 1:2×107 (R2=0.9931, p<0.0001), with 674 ± 166 particles/ml detected (Figure 5C). Using the p24 ng/ml concentration determined by ELISA at the 1:100 dilution–which was within the linear range of ELISA and had low CVs–high and low virus estimates were mathematically calculated based on Gag content of 1250 and 5000 molecules per virus (Briggs et al., 2004; Li et al., 2000) and plotted for the other dilutions to serve as a reference for the flow virometry measurements. At dilutions of 1:2×107 and greater, particle count stabilized at ~0.5 particles/μl. In addition, the MFI of viral particles began to decline at concentrations of 1:1.3×106 and higher. Together, these data suggest that EVs or machine noise in the inclusive gate are increasingly detected at low dilutions, stabilizing particle count and reducing GFP as fewer actual viral particles are detected. Overall, intact fluorescent particles, as observed by flow virometry, tracked well with the lower estimates of viral particles based on 5000 Gag molecules/virus; however it is important to recognize that ELISA measures p24 from both intact and disrupted viruses. Recent studies and reviews have converged on an estimate of roughly 2500 Gag molecules per virus (Carlson et al., 2008; Ganser-Pornillos et al., 2012); if these estimates are accurate they would indicate that ~50% of Gag as measured by ELISA was from intact particles, consistent with the loss of core integrity through ultracentriguation, cryopreservation, and thawing of the viral stocks.
Using the restrictive gating strategy, flow virometry generated linear results from dilutions of 1:40 to 1:8×107 (R2=0.9982, p<0.0001), reflecting detection of 54 ± 16 particles/ml (Figure 5D). As the restrictive gate captures ~70% of viral events, we conclude that these samples have ~80 particles/ml, making flow virometry at least 1,250x more sensitive than ultra-sensitive ELISAs and 125,000x more sensitive than standard ELISAs such as the one used in this study. Indeed, the LOD of flow virometry approaches that of the standard PCR assay for viral load (~50 copies/ml). Notably, the particle count/μl continued to decrease in dilutions down to 1:5×109, however the change in slope in dilutions of 1:3×108 and greater indicate that particle detection is unreliable at these concentrations due to loading capacity limitations of the A50 Micro that restrict the total volume of sample that can be analyzed. Further supporting the conclusion that individual viral particles are being detected–as opposed to EVs or machine noise–the MFI was stable over all dilutions from 1:40 to 1:5×109. Together, these results indicate that flow virometry is remarkably sensitive for detecting labeled HIV-1 particles and far exceeds the limits of detection of any current assay not dependent upon amplification of viral products.
Discussion
In this study, we sought to build upon previous flow virometry findings by systematically testing and assessing the capabilities of a high-end standard flow cytometer (BD FACSAria SORP) and a cytometer designed for detection of submicron particles (Apogee A50 Micro). Our observation that even cytometers specifically designed for submicron particle analysis cannot distinguish between unlabeled viruses and extracellular vesicles (EVs) is not surprising given the similar sizes and composition of these populations. Indeed, these similar characteristics of EVs makes them a common contaminant of HIV-1 preparations (Bess et al., 1997) that is difficult to remove by filtration or density centrifugation. Flotation of viruses in gradients such as sucrose or OptiPrep also co-purifies EVs unless their relative densities are altered, such by digestion of surface proteins by subtilisin (Ott, 2009) or treatment with cholesterol (Linde et al., 2013), both which likely affect viral infectivity. Therefore, most preparations of viruses would be expected to be contaminated by EV populations. This raises two important concerns. First, for therapeutic viral preparations–such as oncolytic viruses, vaccines, and gene therapy vectors–the effects of EVs on the patient will also need to be considered. Second, off-target binding of antibodies to EVs will likely confound flow virometry in the same way that non-specific staining complicates flow cytometry, and appropriate controls will be necessary for proper interpretation of results (Lacroix et al., 2010; Trummer et al., 2008).
The fluorescent viral particles employed here that contain EGFP or mKOκ within the viral Gag protein will be valuable controls for flow virometry studies of HIV-1. By intrinsically labeling the viral structural proteins, EVs and viruses can be discriminated and the effectiveness of labeling strategies to distinguish viruses from EVs can be assessed on fluorescent viruses prior to use with wild-type viruses. Several potential strategies exist for labeling wild-type viruses: lipid dyes, nucleic acid stains, and antibodies. EVs also contain lipid bilayers and can package mRNA; however, there is evidence that viruses can be preferentially labeled with certain lipophilic tracers (Tang et al., 2016). Antibodies against proteins that are exclusively or preferentially localized to viral particles could prove an effective labeling strategy. The viral Env proteins are a logical target but are expressed at extremely low frequencies–an average of 9 to 15 trimers per particle (Magnus et al., 2009; Yang et al., 2006; 2005)–and therefore will require a combination of extremely bright fluorophores and machine optimizations to reduce background noise. The Env proteins are of particular interest because viral heterogeneity in Env conformation or density could contribute to the predominance on non-neutralizing antibodies against HIV if only a small fraction of viruses express fusion-competent proteins. Indeed, recent single-particle analysis of HIV-1 using optical tweezers demonstrated that Env glycoprotein incorporation differed by more than an order of magnitude between particles (Pang et al., 2014); this heterogeneity may play an important role in transmission, replication, and immune responses to HIV-1.
An alternative strategy to labeling viruses is to selectively deplete EVs. For instance, the CD45 antigen is present in western blot analysis of EVs from CD4+ T cells and macrophages, but HIV-1 viruses were not captured by immunoprecipitation with CD45 antibodies, suggesting this antigen is specifically excluded from viruses (Esser et al., 2001). This finding, in turn, gave rise to a method for removing EVs from HIV-1 preparations using CD45 immunodepletion (Ott, 2009). However, recent evidence suggests that EVs released by activated CD4+ T cells are heterogeneous (van der Vlist et al., 2012) and whether all EVs from T cells and macrophages express CD45 has not been definitively established using single-particle analysis techniques. Although selective depletion of EVs is a potentially viable strategy, specific markers would need to be identified for standard producer cell lines or other infectable cells–HEK293T cells, for instance, do not express CD45.
Overcoming the challenges to identifying or purifying viruses will be critical to take full advantage of flow virometry for studying viral heterogeneity and its consequences for HIV-1 infection. Importantly, our results suggesting that HIV particles can be sorted by FACS and retain the ability to fuse with target cells indicates that viral heterogeneity could subsequently be further characterized at a functional level and the infectivity of distinct subpopulations assessed. The finding that patient-derived HIV-1 Envs were as infectious following sorting as unsorted viruses was surprising given previous reports on the fragility of the Env glycoproteins (Papandreou et al., 1996), especially since a high sheath pressure of 70 psi was used to reduce the volume of the sort droplet to minimize the final output volume and maximize post-sort concentration. Viruses were lost during the sorting and ultracentrifugation steps, with the latter resulting in an approximately 10-fold reduction in total viral particle numbers. Nevertheless, the particles recovered from sorting and concentration were as infectious as unsorted particles. These results could have important consequences for vaccine development if certain subpopulations of HIV are non- or poorly-fusogenic as they could be excluded from vaccine preparations with a concomitant reduction in decoy antigens.
Finally, the observation that flow virometry is an ultra-sensitive method for detecting fluorescent HIV-1 has implications for assays to measure virus production in a variety of settings. Flow virometry with fluorescent viruses can be used to rapidly and cheaply gather data on optimal methods for virus production, concentration, and storage with quantitative monitoring of particle integrity and aggregation. The ease and sensitivity of measuring particle release into the supernatant makes flow virometry an attractive platform for evaluating drugs targeting HIV assembly and budding. If methods to selectively label or purify viruses are successful at eliminating EV populations, FVM could replace p24 ELISA for applications such as determining virus concentration, whole-virus immunoprecipitation, and viral outgrowth assays for detection of reactivated latent HIV-1 viruses. In addition to being far more sensitive than ELISA, flow virometry also has the advantage of measuring intact viral particles and therefore would be expected to correlate more accurately with viral infectivity. The simplicity of flow virometry and its low cost compared to current quantification methods make it a technically attractive alternative.
In conclusion, flow virometry represents an inexpensive, rapid, powerful and sensitive technology for the detection of HIV-1 virions but is currently complicated by extracellular vesicle populations with similar light scatter properties as unlabeled viruses. We have developed and characterized fluorescently labeled viruses that will allow assessment of particle integrity and aggregation and will serve as important specificity controls for methods to distinguish between unlabeled viruses and extracellular vesicles. The observation that HIV-1 particles can be sorted and maintain infectivity suggests that functional analysis of sorted viral subpopulations is possible and could provide critical insights for viral transmission, infection, and vaccine design.
Supplementary Material
Highlights.
Neither a conventional flow cytometer (FACSAria) or a flow cytometer dedicated to submicron particle analysis (A50 Micro) can reliably detect wild-type HIV virions using forward and side scatter signals.
Both cytometers can detect HIV-1 particles that are fluorescently labeled.
HIV-1 particles can be sorted by FACSAria without losing the ability to fuse with target cells.
The A50 Micro can quantify labeled viruses with a sensitivity rivaling that of PCR.
Fluorescent viruses coupled with flow virometry can rapidly and cheaply monitor drugs affecting assembly and budding, conditions affecting viral particle integrity and aggregation, and will serve as useful controls for methods to detect wild-type HIV virions.
Acknowledgments
This work was supported by the Case Western Reserve University/University Hospitals Centers for AIDS Research: NIH Grant Number: P30 AI036219 and by NIH Grant R21AI113148. M.B. designed and performed experiments and wrote the manuscript; J.C.T. supervised the project, edited and wrote the manuscript.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arakelyan A, Fitzgerald W, Margolis L, Grivel JC. Nanoparticle-based flow virometry for the analysis of individual virions. J Clin Invest. 2013;123:3716–3727. doi: 10.1172/JCI67042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bess JW, Gorelick RJ, Bosche WJ, Henderson LE, Arthur LO. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- Briggs JAG, Grünewald K, Glass B, Förster F, Kräusslich HG, Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Briggs JAG, Simon MN, Gross I, Kräusslich H-G, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- Briggs JAG, Wilk T, Welker R, Kräusslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CP, Marie D, Bratbak G. Flow cytometric detection of viruses. J Virol Methods. 2000;85:175–182. doi: 10.1016/s0166-0934(99)00167-6. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Briggs JAG, Glass B, Riches JD, Simon MN, Johnson MC, Müller B, Grünewald K, Kräusslich HG. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- Dale BM, McNerney GP, Hübner W, Huser TR, Chen BK. Tracking and quantitation of fluorescent HIV during cell-to-cell transmission. Methods. 2011;53:20–26. doi: 10.1016/j.ymeth.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC. Vaccine applications of flow cytometry. Methods. 2012;57:383–391. doi: 10.1016/j.ymeth.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley KA, Ni N, Nikolaitchik OA, Chen J, Galli A, Hu W-S. Determining the frequency and mechanisms of HIV-1 and HIV-2 RNA copackaging by single-virion analysis. J Virol. 2011;85:10499–10508. doi: 10.1128/JVI.05147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser MT, Graham DR, Coren LV, Trubey CM, Bess JW, Arthur LO, Ott DE, Lifson JD. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J Virol. 2001;75:6173–6182. doi: 10.1128/JVI.75.13.6173-6182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster R, Emrich T, Kremmer E, Lipp M. Expression of the G-protein--coupled receptor BLR1 defines mature, recirculating B cells and a subset of T-helper memory cells. Blood. 1994;84:830–840. [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Adv Exp Med Biol. 2012;726:441–465. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, Wang E, Douek DC, Price DA, June CH, Marincola FM, Roederer M, Restifo NP. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, Paulos CM, Muranski P, Restifo NP. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin R, Barteneva NS. Sorting of small infectious virus particles by flow virometry reveals distinct infectivity profiles. Nat Commun. 2015;6:6022. doi: 10.1038/ncomms7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Jin L, Platisa J, Cohen LB, Baker BJ, Pieribone VA. Fluorescent protein voltage probes derived from ArcLight that respond to membrane voltage changes with fast kinetics. PLoS ONE. 2013;8:e81295. doi: 10.1371/journal.pone.0081295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanne J, Göttfert F, Schimer J, Anders-Össwein M, Konvalinka J, Engelhardt J, Müller B, Hell SW, Kräusslich HG. Stimulated Emission Depletion Nanoscopy Reveals Time-Course of Human Immunodeficiency Virus Proteolytic Maturation. ACS Nano. 2016;10:8215–8222. doi: 10.1021/acsnano.6b03850. [DOI] [PubMed] [Google Scholar]
- Hercher M, Mueller W, Shapiro HM. Detection and discrimination of individual viruses by flow cytometry. J Histochem Cytochem. 1979;27:350–352. doi: 10.1177/27.1.374599. [DOI] [PubMed] [Google Scholar]
- Hübner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FYS, Li XD, Asmuth DM, Huser T, Chen BK. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Lacroix R, Robert S, Poncelet P, Dignat-George F. Overcoming limitations of microparticle measurement by flow cytometry. Semin Thromb Hemost. 2010;36:807–818. doi: 10.1055/s-0030-1267034. [DOI] [PubMed] [Google Scholar]
- Landowski M, Dabundo J, Liu Q, Nicola AV, Aguilar HC. Nipah virion entry kinetics, composition, and conformational changes determined by enzymatic virus-like particles and new flow virometry tools. J Virol. 2014;88:14197–14206. doi: 10.1128/JVI.01632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- Linde ME, Colquhoun DR, Ubaida Mohien C, Kole T, Aquino V, Cotter R, Edwards N, Hildreth JEK, Graham DR. The conserved set of host proteins incorporated into HIV-1 virions suggests a common egress pathway in multiple cell types. J Proteome Res. 2013;12:2045–2054. doi: 10.1021/pr300918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S International AIDS Vaccine Initiative Protocol C Principal Investigators. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. 2009;83:1523–1531. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- McSharry JJ. Analysis of virus-infected cells by flow cytometry. Methods. 2000;21:249–257. doi: 10.1006/meth.2000.1005. [DOI] [PubMed] [Google Scholar]
- Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JP. Flow Cytometry of Extracellular Vesicles: Potential, Pitfalls, and Prospects. Curr Protoc Cytom. 2015;73:13.14.1–16. doi: 10.1002/0471142956.cy1314s73. [DOI] [PubMed] [Google Scholar]
- Ott DE. Purification of HIV-1 virions by subtilisin digestion or CD45 immunoaffinity depletion for biochemical studies. Methods Mol Biol. 2009;485:15–25. doi: 10.1007/978-1-59745-170-3_2. [DOI] [PubMed] [Google Scholar]
- Pang Y, Song H, Cheng W. Using optical trap to measure the refractive index of a single animal virus in culture fluid with high precision. Biomed Opt Express. 2016;7:1672–1689. doi: 10.1364/BOE.7.001672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Song H, Kim JH, Hou X, Cheng W. Optical trapping of individual human immunodeficiency viruses in culture fluid reveals heterogeneity with single-molecule resolution. Nat Nanotechnol. 2014;9:624–630. doi: 10.1038/nnano.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou MJ, Idziorek T, Miquelis R, Fenouillet E. Glycosylation and stability of mature HIV envelope glycoprotein conformation under various conditions. FEBS Lett. 1996;379:171–176. doi: 10.1016/0014-5793(95)01505-1. [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- Stoner SA, Duggan E, Condello D, Guerrero A, Turk JR, Narayanan PK, Nolan JP. High sensitivity flow cytometry of membrane vesicles. Cytometry A. 2016;89:196–206. doi: 10.1002/cyto.a.22787. [DOI] [PubMed] [Google Scholar]
- Tang VA, Renner TM, Varette O, Le Boeuf F, Wang J, Diallo JS, Bell JC, Langlois MA. Single-particle characterization of oncolytic vaccinia virus by flow virometry. Vaccine. 2016;34:5082–5089. doi: 10.1016/j.vaccine.2016.08.074. [DOI] [PubMed] [Google Scholar]
- Tilton CA, Tabler CO, Lucera MB, Marek SL, Haqqani AA, Tilton JC. A combination HIV reporter virus system for measuring post-entry event efficiency and viral outcome in primary CD4+ T cell subsets. J Virol Methods. 2014;195:164–169. doi: 10.1016/j.jviromet.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trummer A, De Rop C, Tiede A, Ganser A, Eisert R. Isotype controls in phenotyping and quantification of microparticles: a major source of error and how to evade it. Thromb Res. 2008;122:691–700. doi: 10.1016/j.thromres.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Coumans F, Varga Z, Krumrey M, Nieuwland R. Innovation in detection of microparticles and exosomes. J Thromb Haemost. 2013;11(Suppl 1):36–45. doi: 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Coumans FAW, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, Sturk A, van Leeuwen TG, Nieuwland R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12:1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- van der Pol E, van Gemert MJC, Sturk A, Nieuwland R, van Leeuwen TG. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J Thromb Haemost. 2012;10:919–930. doi: 10.1111/j.1538-7836.2012.04683.x. [DOI] [PubMed] [Google Scholar]
- van der Vlist EJ, Arkesteijn GJA, van de Lest CHA, Stoorvogel W, Nolte-’t Hoen ENM, Wauben MHM. CD4(+) T cell activation promotes the differential release of distinct populations of nanosized vesicles. J Extracell Vesicles. 2012;1:3235. doi: 10.3402/jev.v1i0.18364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak J, Hoang VM, Christanti S, Peluso R, Li F, Culp TD. Use of flow cytometry for characterization of human cytomegalovirus vaccine particles. Vaccine. 2016;34:2321–2328. doi: 10.1016/j.vaccine.2016.03.067. [DOI] [PubMed] [Google Scholar]
- Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, Sherrill-Mix SA, Patro SC, Secreto AJ, Jordan APO, Lee G, Kahn J, Aye PP, Bunnell BA, Lackner AA, Hoxie JA, Danet-Desnoyers GA, Bushman FD, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. Engineering HIV-Resistant Human CD4+ T Cells with CXCR4-Specific Zinc-Finger Nucleases. PLoS Pathog. 2011;7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. 2006;80:4388–4395. doi: 10.1128/JVI.80.9.4388-4395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG. Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat Med. 2005;11:1299–1305. doi: 10.1038/nm1326. [DOI] [PubMed] [Google Scholar]
- Zicari S, Arakelyan A, Fitzgerald W, Zaitseva E, Chernomordik LV, Margolis L, Grivel JC. Evaluation of the maturation of individual Dengue virions with flow virometry. Virology. 2016;488:20–27. doi: 10.1016/j.virol.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.