Here, Andreyeva et al. show that linker histone H1 is required for the underreplicated phenomenon in Drosophila salivary glands, in which tissues undergo endoreplication without cell division, and the latest replicating regions occasionally fail to complete endoreplication, resulting in underreplicated domains of polytene chromosomes. They demonstrate that H1 directly interacts with the suppressor of underreplication (SUUR) protein and is required for SUUR binding to chromatin in vivo and that the localization of H1 in chromatin changes profoundly during the endocycle.
Keywords: linker histone H1, suppressor of underreplication, polytene chromosomes, endoreplication, replication timing, intercalary heterochromatin
Abstract
Eukaryotic DNA replicates asynchronously, with discrete genomic loci replicating during different stages of S phase. Drosophila larval tissues undergo endoreplication without cell division, and the latest replicating regions occasionally fail to complete endoreplication, resulting in underreplicated domains of polytene chromosomes. Here we show that linker histone H1 is required for the underreplication (UR) phenomenon in Drosophila salivary glands. H1 directly interacts with the Suppressor of UR (SUUR) protein and is required for SUUR binding to chromatin in vivo. These observations implicate H1 as a critical factor in the formation of underreplicated regions and an upstream effector of SUUR. We also demonstrate that the localization of H1 in chromatin changes profoundly during the endocycle. At the onset of endocycle S (endo-S) phase, H1 is heavily and specifically loaded into late replicating genomic regions and is then redistributed during the course of endoreplication. Our data suggest that cell cycle-dependent chromosome occupancy of H1 is governed by several independent processes. In addition to the ubiquitous replication-related disassembly and reassembly of chromatin, H1 is deposited into chromatin through a novel pathway that is replication-independent, rapid, and locus-specific. This cell cycle-directed dynamic localization of H1 in chromatin may play an important role in the regulation of DNA replication timing.
In eukaryotic organisms, DNA replication does not occur synchronously throughout the genome but, rather, in discrete domains at different stages of S phase (Nakamura et al. 1986; Zink 2006). This asynchrony in replication timing has been well documented in Drosophila. During embryogenesis, the Drosophila genome initially replicates rapidly and synchronously. The cell cycle then lengthens, and replication becomes asynchronous, resulting in segregation of early and late replicating regions. As in other eukaryotes, active euchromatin replicates in early S phase, whereas repetitive DNA and other sequences assembled into condensed transcriptionally inert heterochromatin are copied later in S phase (Shermoen et al. 2010; Duronio 2012; Farrell et al. 2012). Following embryogenesis, asynchronous DNA replication continues in the imaginal tissues that will eventually form adult structures as well as in the differentiated tissues comprising the larval body.
Drosophila is a powerful model to study asynchronous replication timing owing to the polyploid nature of the larval tissues. The cells of larval tissues undergo endoreplication, resulting in multiple copies of the genome present in each cell. The homologous regions of each chromosome align and form giant polytene chromosomes that can be visualized by DNA staining to reveal characteristic patterns of intensely stained bands and lightly stained interbands. For instance, salivary gland polytene chromosomes consist of >1000 genome copies per cell by the end of the third instar (Zhimulev et al. 2004; Edgar et al. 2014). During endoreplication, heterochromatic regions that replicate near the end of S phase fail to replicate fully, leading to underreplicated loci that have a reduced ploidy compared with other regions of the polytene chromosomes. Underreplication (UR) of intercalary heterochromatin (IH) in the chromosome arms produces physical constrictions or “weak points” with an increased incidence of chromosome breaks, while UR of pericentric heterochromatin (PH) yields condensed unstructured chromatin that coalesces in a single chromocenter (Zhimulev et al. 2004).
The mechanisms underlying late replication and UR are still not fully understood. However, the Suppressor of UR (SuUR) gene has been shown to be essential for the establishment of UR (Belyaeva et al. 1998). SuUR encodes a protein with no known mammalian homolog but moderate similarity in its N terminus to the ATPase domain of the SNF2/SWI2 family (Makunin et al. 2002). Its mutation by a 6-kb insertion in the last exon (SuURES) results in full polytenization of IH. Cytological bands of normal ploidy are formed instead of the “weak points,” and chromosome break frequency is reduced (Belyaeva et al. 1998; Belyakin et al. 2005; Yarosh and Spradling 2014). Additionally, a portion of PH proximal to each of the chromosome arms forms a structured banding pattern similar to normal polytenized chromatin (Belyaeva et al. 1998). SUUR protein exhibits a highly dynamic spatial and temporal distribution pattern in polytene chromosomes. Whereas SUUR associates with PH throughout the endocycle, it emerges at IH only during the late endocycle S (endo-S) phase, when it colocalizes with the replication fork (Kolesnikova et al. 2013). Several recent studies suggest that SUUR may promote the formation of UR by inhibiting replication fork progression. The occupancy of ORC2, a component of the origin recognition complex (ORC), is reduced at sites of UR in salivary glands. It is not increased at underreplicated sites in SuURES mutants, indicating that enhanced replication at these regions is not due to the increased appearance of initiation sites; rather, SuUR mutation increases the rate of DNA replication (Belyaeva et al. 2012; Sher et al. 2012; Nordman et al. 2014). Despite significant advances in mechanistic understanding of the functions of SUUR, it is still unclear how SUUR interacts with the replication machinery to reduce the local rate of DNA replication or how it is able to localize specifically to late replicating/underreplicated regions of the genome during the endocycle.
Recent categorization of the Drosophila genome into different chromatin states using protein occupancy profiles (Filion et al. 2010; Kharchenko et al. 2011) has provided insight into the composition of underreplicated loci. A repressive chromatin state was identified that encompasses more than half of the genome, replicates late, and contains nearly all IH. This state is enriched for several proteins, including LAM, D1, IAL, and, notably, SUUR and linker histone H1. Intriguingly, H1 is the most highly enriched protein within this chromatin fraction (Kharchenko et al. 2011). Furthermore, SUUR and H1 were proposed to exhibit positive genome targeting interactions (van Steensel et al. 2010). H1 has also been implicated in suppression of replication initiation by several in vitro and in vivo studies (Lu et al. 1998; Thiriet and Hayes 2009; for review, see Flickinger 2015). Thus, similar to SUUR, H1 may negatively regulate DNA endoreplication. The localization of both SUUR and H1 to underreplicated chromatin suggests that they may work in concert to maintain its underreplicated status in Drosophila larvae.
The paradigmatic view of linker histone H1 as a global regulator of higher-order chromatin structure was derived principally from in vitro studies and the fact that, after core histones, H1 is the next most abundant chromatin protein. Earlier studies demonstrated that H1 condenses chromatin, protects the linker DNA, and regulates the spacing between nucleosome particles (Wolffe 1997; Woodcock et al. 2006). However, more recent work has shown that H1 also has locus-specific effects that are rendered through highly specific interactions between H1 and a variety of transcriptional and epigenetic regulatory proteins. For example, Drosophila H1 has been shown to be critical for the structural integrity of PH (Lu et al. 2009) by directly interacting with and recruiting the histone methyltransferase (HMT) Su(var)3-9, thereby promoting dimethylation of H3K9 at PH and transposon repression (Lu et al. 2013). Likewise, mammalian H1b has been shown to interact with the muscle-specific transcription factor Msx1, leading to repression of MyoD expression and inhibition of myogenesis (Lee et al. 2004). Several mammalian H1s were also shown to interact directly with DNA methyltransferases DNMT1 and DNMT3b to promote DNA methylation at the H19 and Meg3 (Gtl2) imprinting control regions (Yang et al. 2013). Even though functional consequences have not yet been established, physical interactions between linker histones and numerous other chromatin-bound proteins have been reported (for review, see McBryant et al. 2010), suggesting that H1 may partner with a variety of additional factors to regulate a wide variety of transactions occurring within chromatin.
In this study, we show that H1 is essential for the formation of underreplicated domains in both IH and PH of larval polytene chromosomes. H1 and SUUR exhibit a direct physical interaction, and, when H1 is depleted, the abundance of SUUR in salivary gland chromatin is greatly reduced. Importantly, H1 occupancy in polytene chromosomes exhibits a striking cell cycle-dependent temporal pattern. H1 protein is rapidly and strongly loaded into late replicating (including underreplicated) loci at the onset of endo-S phase, prior to the arrival of replication machinery, and is gradually redistributed as DNA replication progresses. Our findings provide evidence that H1 facilitates the recruitment of SUUR to late replicating and underreplicated chromatin and plays a critical role in the regulation of the timing of DNA endoreplication.
Results
His1 is a suppressor of UR at IH and PH
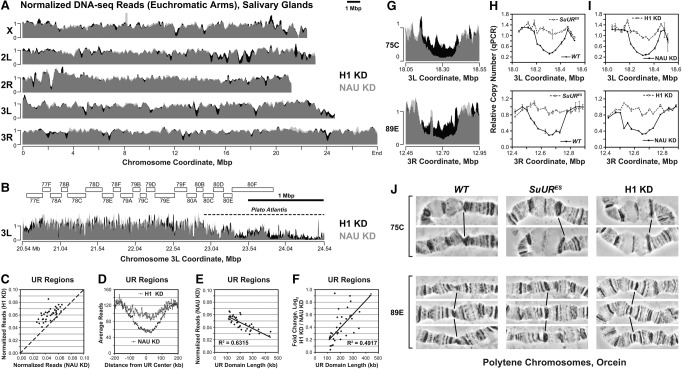
It has been suggested previously that Drosophila H1 and SUUR are enriched in repressive “black” chromatin (Filion et al. 2010) in cultured cells frequently used as a model for diploid cells (Lee et al. 2014). Since underreplicated regions of polytene chromosomes are predominantly located in this chromatin type (Yarosh and Spradling 2014) and since SUUR is required for the establishment and/or maintenance of UR (Belyaeva et al. 1998), we examined whether H1 is involved in promoting UR. To this end, we depleted salivary glands of wandering L3 larvae of H1 protein (to ∼15% of wild-type level) (Lu et al. 2009) by specific RNAi of the His1 gene and subjected the DNA to high-throughput sequencing (DNA-seq). For comparison, we knocked down a control gene, Nautilus (Wei et al. 2007). In the DNA of control salivary glands, several regions in the euchromatic arms exhibit a substantially lower DNA copy number than that of the flanking regions (Fig. 1A), consistent with their UR. Most underreplicated domains exhibit a gradual and symmetrical reduction of DNA copy number, with the minimum copy number at the center, as observed previously (Yarosh and Spradling 2014). Thus, the underreplicated domains that we identified by DNA-seq likely correspond to regions of UR that were determined previously by microarray (Sher et al. 2012) and whole-genome sequencing (Supplemental Fig. S1A; Yarosh and Spradling 2014). However, when H1 is depleted, the DNA copy number is substantially increased at nearly all major underreplicated regions (Fig. 1A,B).
Figure 1.
H1 is required for UR in salivary gland polytene chromosomes. (A) Genome-wide analyses of DNA copy number in Drosophila salivary gland cells. DNA from L3 salivary glands was subjected to high-throughput sequencing. DNA copy numbers (normalized to chromosome arm average) are shown across the entire mapped Drosophila genome. (X, 2L, 2R, 3L, and 3R) Chromosome arms. Genomic coordinates (in megabase pairs) are indicated at the bottom. H1 knockdown trace (H1 KD) is shown in black in the background, and the control trace (NAU KD) is shown in semitransparent light gray in the foreground; their overlap appears as dark gray. (B) Close-up view of a representative genomic region (proximal 3L). (Open rectangles) Polytene cytological bands; (dotted line) the region corresponding to “Plato Atlantis” (Belyaeva et al. 1998; Andreyeva et al. 2007). (C) Suppression of UR in H1-depleted salivary gland cells. For each identified underreplicated region, reads (normalized by the underreplicated region length and total read count) under H1 knockdown (Y-axis) are plotted against reads under control knockdown (X-axis). The dotted line represents equal DNA copy numbers for both conditions. (D) The average extent of UR and its suppression by H1 knockdown across underreplicated regions. Average read counts (normalized to total read count) were calculated across all identified underreplicated regions for control and H1 knockdowns as indicated (Y-axis). The distance from the underreplicated region center (in kilobases) is indicated on the X-axis. (E) Dependence of the extent of UR on the underreplicated region length. For each underreplicated region, normalized read counts in the control knockdown (Y-axis) are plotted against the length of the region (X-axis). (F) Dependence of the extent of H1 knockdown-dependent suppression of UR on the underreplicated region length. The log2 fold change of read counts in H1 knockdown relative to control (Y-axis) is plotted against the length of each underreplicated region (X-axis). (G) Close-up views of DNA copy number (from high-throughput sequencing, normalized to chromosome arm average) are shown for H1 (black in the background) and control (semitransparent light gray in the foreground) knockdowns. (75C and 89E) Corresponding cytological regions. Genomic coordinates (in megabase pairs) are indicated at the bottom. (H) Change of DNA copy number in homozygous SuURES mutant salivary glands versus the wild type (Oregon R; WT) was determined by quantitative PCR (qPCR). Copy numbers were calculated relative to embryonic DNA and normalized to a control intergenic region. The X-axis shows chromosome positions (in megabase pairs) of target amplicons. (I) Same as in H, except DNA copy numbers were compared between H1 knockdown (H1 KD) and the control (NAU KD). (J) Representative cytological images (orcein staining and phase contrast) of polytene chromosome fragments flanking cytological regions 75C and 89E in wild-type (Oregon R; WT), homozygous SuURES, and H1 knockdown (H1 KD) salivary glands. Slanted lines indicate 75C or 89E bands.
To quantify the effect of H1 depletion on DNA copy number, we determined the coordinates of underreplicated regions in the control data set and calculated the fold change of DNA copy number under H1 knockdown conditions for each region (see the Materials and Methods; Supplemental Fig. S1B). We identified 46 underreplicated domains, 39 of which represent IH, while seven lie outside of the euchromatic–heterochromatic transition regions and represent regions of PH (Supplemental Table S1; Riddle et al. 2011; Hoskins et al. 2015). Of the 46 underreplicated domains, most (33 out of 46) exhibit an increase (≥1.2-fold) of copy number upon H1 knockdown (Fig. 1C). On average, the depletion of H1 results in an ∼50% increase in the copy number when measured at the center of each domain (Fig. 1D), with several regions showing a nearly twofold increase (Supplemental Table S1). Consistent with a previous report (Yarosh and Spradling 2014), we found that DNA copy number is inversely correlated with the UR domain length (Fig. 1E). In addition, the fold change in copy number upon H1 knockdown is positively correlated with the UR domain length (Fig. 1F), indicating that longer underreplicated domains with a higher degree of UR are more susceptible to H1 depletion than shorter UR domains with less UR. Notably, the same relationships exist between the sizes of underreplicated domains and their susceptibility to SuUR mutation, as determined by analysis of DNA-seq data (Yarosh and Spradling 2014) for control and SuURES salivary glands (Supplemental Fig. S1C–F).
Two particular genomic regions (cytological bands 75C and 89E) represent “classical” sites of UR (Belyaeva et al. 1998). We observed that the decreased DNA copy number due to UR of these loci is strongly reversed by H1 knockdown (Fig. 1G). To validate our findings from DNA-seq, we isolated genomic DNA from control salivary glands and those strongly depleted of H1 (to <5% of the normal level) (Lu et al. 2009) and measured DNA copy numbers relative to wild-type embryonic DNA by quantitative PCR (qPCR) at ∼30-kb intervals across the entire 75C and 89E loci. As a control, we also compared DNA copy numbers at these loci using wild-type and SuURES salivary glands. In agreement with our DNA-seq data, the DNA copy numbers in wild-type and control salivary glands exhibit a gradual and symmetrical reduction at both regions compared with their flanking regions (Fig. 1H,I). In contrast and as expected, the DNA copy numbers in salivary glands from SuURES mutants across both loci are comparable with those in flanking regions, indicative of suppression of UR (Fig. 1H). Similarly, H1 depletion also results in a strong reversal of UR (Fig. 1I). Thus, upon extensive depletion of H1, the observed suppression of UR is almost complete and comparable with that produced by mutation of SuUR.
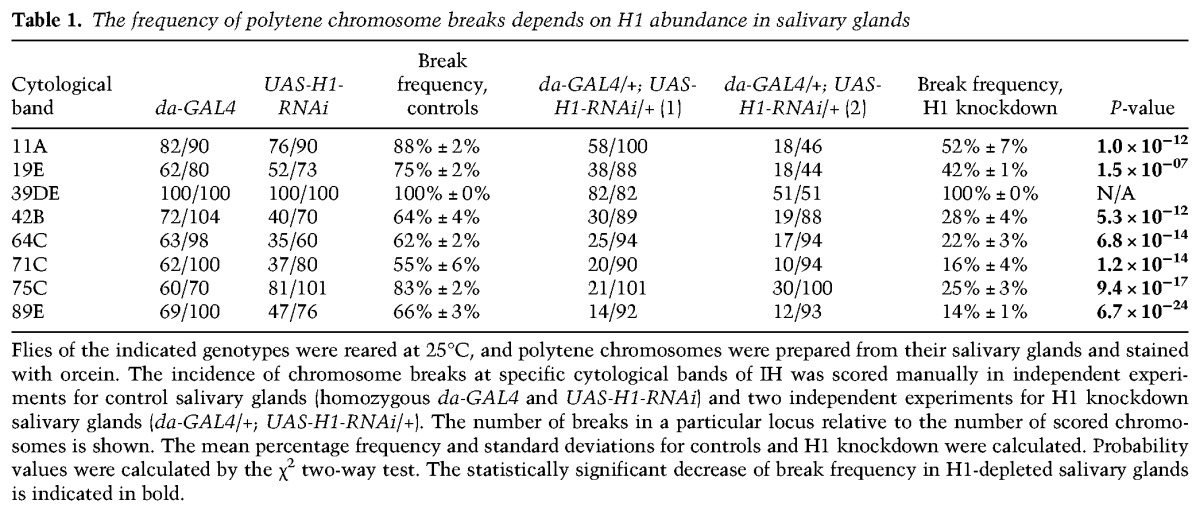
The phenomenon of UR was first observed cytologically and manifests itself as a constriction of corresponding polytene bands and increased incidence of chromosome breaks (Belyaeva et al. 1998; Zhimulev et al. 2004). We therefore evaluated the effect of H1 knockdown on the formation of chromosome breaks in polytene chromosomes. Representative examples of chromosome breaks in cytological regions 75C and 89E are shown in Figure 1J. Under control conditions (Fig. 1J, left panels), regions of UR are readily observable as bands with a pinched appearance relative to the flanking fully polytenized loci. As reported previously (Belyaeva et al. 1998), in SuURES mutants, the incidence of chromosome breaks is strongly decreased. Importantly, this effect is phenocopied by a moderate depletion of H1 (to ∼30% of the normal level). We calculated the chromosome break frequency at eight well-characterized underreplicated loci in animals of various genotypes and found that at nearly all of them, the break frequency was significantly reduced upon the abrogation of H1 expression (Table 1). Taken together, our observations demonstrate that, similar to SUUR, the linker histone H1 is essential for the establishment of UR in IH.
Table 1.
The frequency of polytene chromosome breaks depends on H1 abundance in salivary glands
PH is also underreplicated in polytene chromosomes and forms a compact structure coalescing into a single chromocenter (Leach et al. 2000; Zhimulev et al. 2004). In the SuURES mutant, UR of some regions of PH, such as the “Plato Atlantis” of the third chromosome, is suppressed, and they form band–interband patterns similar to those of euchromatin (Belyaeva et al. 1998; Semeshin et al. 2001; Andreyeva et al. 2007). Whole-genome sequencing indicates that some of the mapped pericentric regions are severely underreplicated in the control, and the ploidy is partially increased in the absence of H1 (Fig. 1B). An extensive analysis of PH by computational alignment is difficult due to the lack of reliable mapping (Hoskins et al. 2002, 2007) and the large amounts of repetitive DNA, including transposable elements (TEs), in these regions (Smith et al. 2007; He et al. 2012; Hoskins et al. 2015). To better understand the role of H1 in PH, we evaluated UR of TE sequences independent of their genomic location (see the Materials and Methods). To validate this approach, we used sequencing data from wild-type and SuURES mutant salivary glands (Yarosh and Spradling 2014). We found that the mutation of SuUR results in an increase (≥1.2-fold) of copy numbers for most TEs (97 out of 124), with many (30) showing a twofold or greater increase (Supplemental Fig. S1G; Supplemental Table S2). Similarly, more than half of all TEs (67 out of 124) show an increased copy number upon H1 depletion, although the magnitude of the effect is somewhat weaker than that of SuUR mutation (Supplemental Fig. S1H). Thus, similar to SuUR mutation, H1 knockdown suppresses UR in both IH and PH regions of the genome. Together, our observations strongly indicate that abrogation of His1 expression in larvae phenocopies the mutation of SuUR and that both corresponding proteins function in the establishment of UR.
H1 is an upstream effector of UR and facilitates binding of SUUR to chromatin in vivo
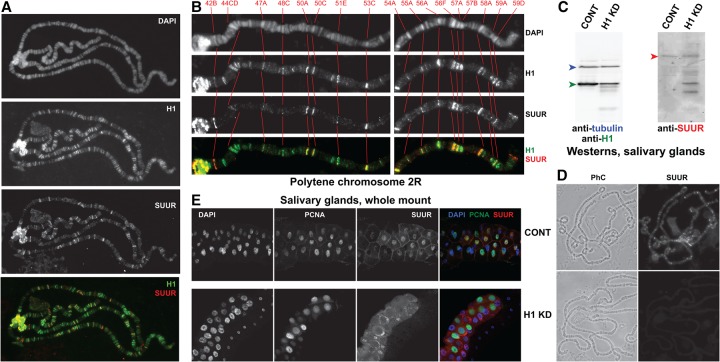
Our results indicate that H1 and SUUR have similar effects on DNA UR in salivary gland polytene chromosomes. Since H1 and SUUR colocalize throughout the genome in Kc167 cells (Filion et al. 2010), we determined their localization patterns in polytene chromosomes. Indirect immunofluorescence (IF) costaining revealed a substantial degree of overlap between H1 and SUUR in multiple bands within euchromatic arms and at the chromocenter (Fig. 2A). SUUR distribution in polytene chromosomes is known to vary depending on the stage of the endocycle (Kolesnikova et al. 2013). Thus, we simultaneously stained the polytene spreads for PCNA (data not shown) to determine the cell cycle phase as described (Kolesnikova et al. 2013) and selected polytene spreads in late endo-S phase, when the genomic occupancy of SUUR is maximal in IH and overlaps with that of PCNA. We observed strong colocalization of H1 and SUUR in late endo-S phase (Fig. 2A).
Figure 2.
Polytene chromosome loading and nuclear localization of SUUR depend on H1. (A) Colocalization of SUUR and H1 proteins in wild-type polytene chromosomes. Localization patterns of H1 and SUUR in larval polytene chromosomes were analyzed by indirect IF staining. H1 (green) and SUUR (red) signals overlap extensively in heterochromatin and euchromatic arms of polytene chromosomes. The polytene spread corresponds to a cell in late endo-S phase (PCNA staining is not shown). DAPI staining shows the overall chromosome morphology. (B) Detailed view of SUUR and H1 colocalization in polytene chromosome arm 2R during late endo-S phase. H1 (green) is abundant in all SUUR-positive (red) loci and in additional sites. DAPI staining shows the overall chromosome morphology and was used for an alignment of cytological positions. Red numbers at the top and corresponding cytological bands in all panels are connected with red lines. (C) Depletion of H1 protein by RNAi in salivary glands. Immunoblot analyses of H1 (left panel) and SUUR (right panel) in control (CONT; Oregon R) and H1 RNAi-depleted (H1 KD) salivary glands. H1 is strongly depleted (green arrowhead), whereas expression of the full-length SUUR (red arrowhead) is not substantially abrogated, although the protein stability appears to be compromised (note the truncated SUUR polypeptides in the H1 KD lane). Both blots were equally loaded; the left panel was additionally probed with antibodies to β-tubulin (loading control; blue arrowhead). (D) Decreased abundance of SUUR protein in H1-depleted polytene chromosomes. Polytene chromosomes were prepared from control and H1-depleted salivary glands as in C and stained with SUUR antibodies. IF signal for SUUR is not detectable in H1 knockdown. (Left panels) Phase contrast (PhC) images. (E) Abnormal subcellular distribution of SUUR protein in H1-depleted salivary glands. Control and H1-depleted salivary glands (as in C,D) were fixed and whole-mount-stained with DAPI (blue), PCNA (green), and SUUR (red) antibodies. Whereas SUUR is mostly nuclear in the control, it is released into cytoplasm upon H1 depletion.
At higher magnification (Fig. 2B), it is apparent that the distribution of SUUR in polytene chromosomes is limited to a subset of H1-positive loci. Thus, it is possible that H1 directs the genomic distribution of SUUR in salivary glands. To test this hypothesis, we compared polytene chromosome localization of SUUR in wild-type animals with that in animals depleted of H1 by RNAi. To avoid a complete disruption of polytene morphology (Lu et al. 2009; Kavi et al. 2016), H1 was only moderately depleted (to ∼30% of normal) (Fig. 2C; see the Supplemental Material). In H1-depleted polytene chromosomes in all stages of the endo-S, SUUR is not detectable in chromosome arms and persists only weakly (at just above the detection limit) in the chromocenter (Fig. 2D). Since SUUR is most readily detectable in polytene chromosomes of late endo-S phase cells, we additionally verified that H1 knockdown abrogates the binding of SUUR to chromatin in late endo-S. To this end, polytene spreads were stained with PCNA antibodies, and the nuclei were staged as described above. We observed that even in the late endo-S, the moderate depletion of H1 abolishes SUUR tethering to polytene chromosome arms (Supplemental Fig. S2A). In reciprocal experiments, we found that the mutation of SuUR does not appreciably affect the localization of H1 in polytene chromosomes (Supplemental Fig. S2B). Thus, the precise spatial distribution pattern of SUUR in the genome requires H1, whereas SUUR appears largely dispensable for H1 localization.
Finally, whole-mount immunostaining of isolated salivary glands (Fig. 2E) indicates that the predominantly nuclear localization of SUUR in control cells (Fig. 2E, top panels) is disrupted upon H1 depletion (Fig. 2E, bottom panels). Therefore, H1 is also required for nuclear localization of SUUR. Importantly, although a moderate H1 depletion brings about an almost complete removal of SUUR from polytene chromosomes, it does not appear to reduce the expression level of SUUR in salivary glands (Fig. 2C, also apparent in whole-mount staining of salivary glands in E). However, whereas only a single band of full-length SUUR is observed in the immunoblot of control cells, H1-depleted cells contain several smaller immunoreactive species, suggesting possible degradation of SUUR protein under these conditions. Therefore, in addition to its role in promoting localization of SUUR in chromatin, H1 seems to protect SUUR from degradation in vivo. In H1-depleted cells, SUUR appears to accumulate in the cytoplasm (Fig. 2E, bottom), where it may be unstable. Collectively, our findings indicate that SUUR depends on H1 for its localization in chromatin and that it functions in the nucleus downstream from H1.
SUUR and H1 proteins interact physically
Because H1 is required for SUUR binding to chromatin, it is possible that H1 interacts directly with SUUR, as shown previously for another nuclear enzyme, the HMT Su(var)3-9 (Lu et al. 2013). To test this hypothesis, we performed in vitro pull-down assays using GST-tagged SUUR and extracts from wild-type adult Drosophila ovaries as a source of the endogenous native H1. We compared GST, full-length GST-SUUR, and three GST-tagged truncated forms of the SUUR polypeptide (Fig. 3A). The three fragments of SUUR were shown previously to play distinct functional roles in the establishment of UR (Kolesnikova et al. 2005; Pindyurin et al. 2008). All recombinant fusion proteins were loaded in the binding reactions at approximately equimolar concentrations (Fig. 3B), and their association with the ovarian H1 was probed by immunoblotting with an H1-specific antibody (Fig. 3C). These experiments indicate that H1 specifically interacts with the full-length SUUR and its central fragment (residues 371–578). Similar results were obtained in experiments in which native H1 from ovarian extracts was substituted with purified recombinant H1 (Supplemental Fig. S3). Thus, the physical interaction between H1 and the central fragment of SUUR is direct. Importantly, an overlapping fragment of SUUR (361–599) has been shown previously to be required for the localization of SUUR to chromatin, including IH, in vivo (Kolesnikova et al. 2005; Yurlova et al. 2009). Therefore, the central region of SUUR, which encompasses a positively charged domain and the nuclear localization signal (NLS), is sufficient to mediate a strong physical interaction with H1 and may be involved in tethering SUUR to H1-containing chromatin in vivo.
Figure 3.
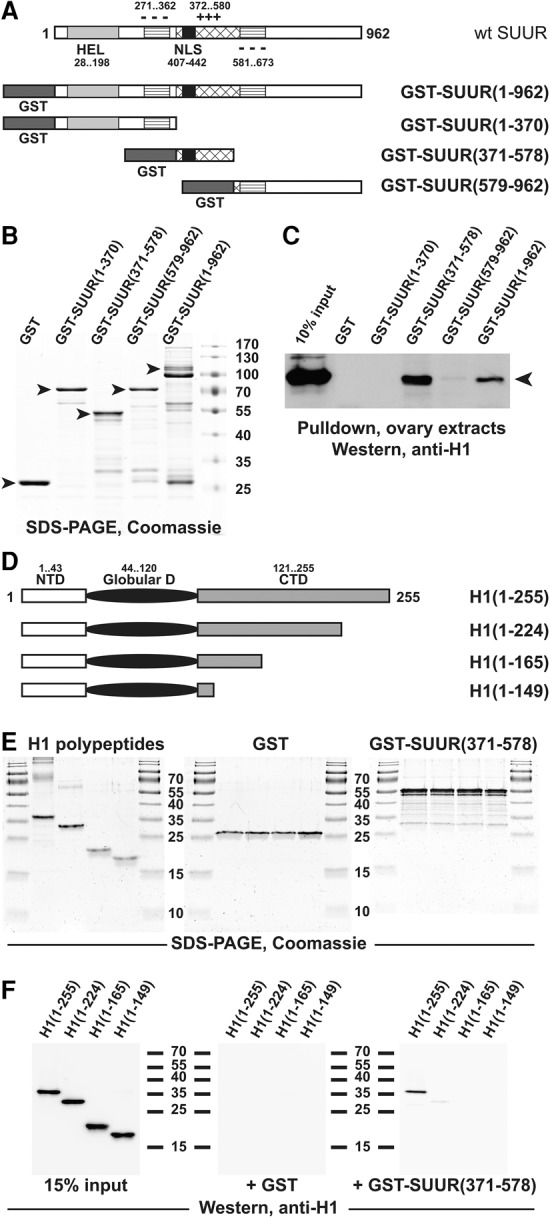
H1 and SUUR proteins exhibit direct physical interactions. (A) Schematic representation of GST-SUUR fusion expression constructs used for the analyses. The major structural domains of wild-type SUUR protein (open rectangle) are represented by light-gray (ATPase/helicase domain [HEL]), striped (negatively charged domain [−−−]), cross-hatched (positively charged domain [+++]), and black (NLS) boxes. Numbers indicate amino acid residues. N-terminal GST (dark-gray boxes) fusion constructs were prepared with full-length and the indicated truncations of SUUR. (B) Recombinant GST-SUUR fusion polypeptides. GST and GST fusion proteins (as in A) were expressed and purified from Escherichia coli, incubated with ovarian extracts, and analyzed by SDS-PAGE and Coomassie staining. Arrowheads indicate full-length polypeptide products; molecular mass marker sizes (in kilodaltons) are shown at the right. (C) SUUR-dependent GST pull-downs of endogenous H1 from Drosophila ovarian extracts. Whole-cell extracts from adult Drosophila ovaries were incubated with GST fusion proteins (B), and pull-down products were analyzed by H1-specific immunoblot along with the 10% input control. Endogenous native H1 (arrowhead) strongly interacts with full-length and the middle fragment (amino acids 371–578) of recombinant SUUR. (D) Schematic representation of recombinant H1 polypeptides used for the analyses. The three major structural domains of Drosophila H1 are represented by an open rectangle (N-terminal domain [NTD]), a filled oval (GD), and a shaded rectangle (C-terminal domain [CTD]). Numbers indicate amino acid residues. Recombinant untagged full-length H1 and its indicated C-terminal truncations were expressed and purified from E. coli. (E) Recombinant polypeptides used for in vitro GST pull-down experiments. Recombinant GST (middle panel) and GST-SUUR(371–578) fusion protein (right panel) were analyzed by SDS-PAGE and Coomassie staining; the protein loading is equivalent to that in F. (Left panel) Recombinant H1 polypeptides were analyzed similarly; the protein loading is approximately sevenfold higher than that in the corresponding panel of F. Molecular mass marker sizes (in kilodaltons) are shown between the panels. (F) SUUR-dependent in vitro GST pull-downs of recombinant H1 polypeptides. The indicated H1 polypeptides were incubated with GST and GST-SUUR(371–578) fusion proteins, and pull-down products were analyzed by H1-specific immunoblot along with the 15% input control. Full-length H1 but not any of its C-terminal truncations strongly interacts with SUUR(371–578).
We demonstrated previously that H1 exerts its multiple biological functions through independent biochemically separable activities of its three structural domains (Kavi et al. 2016). Moreover, different segments of the H1 protein are required for direct physical interactions with its several different biochemical partners. For instance, the central region of the H1 C-terminal domain (CTD) is required for the interaction with Su(var)3-9 in vitro and for H1's functions in heterochromatin in vivo. To determine whether the CTD is also involved in the physical interaction with SUUR, we compared the association in vitro between GST-SUUR(371–578) and full-length and several C-terminally truncated recombinant H1 polypeptides (Fig. 3D); GST itself was used as a negative control. Equimolar loading of all recombinant proteins in the binding reactions was confirmed by Coomassie staining (Fig. 3E). We found that full-length H1 can interact with the H1-binding domain of SUUR but that truncation of the distal 25% of the H1 CTD (or longer truncations) renders the polypeptide incapable of associating with GST-SUUR(371–578) (Fig. 3F). These experiments indicate that H1 and SUUR proteins bind each other directly through specific structural domains within their respective sequences.
The loading of H1 into chromatin is dynamically altered in the course of the endo-S phase
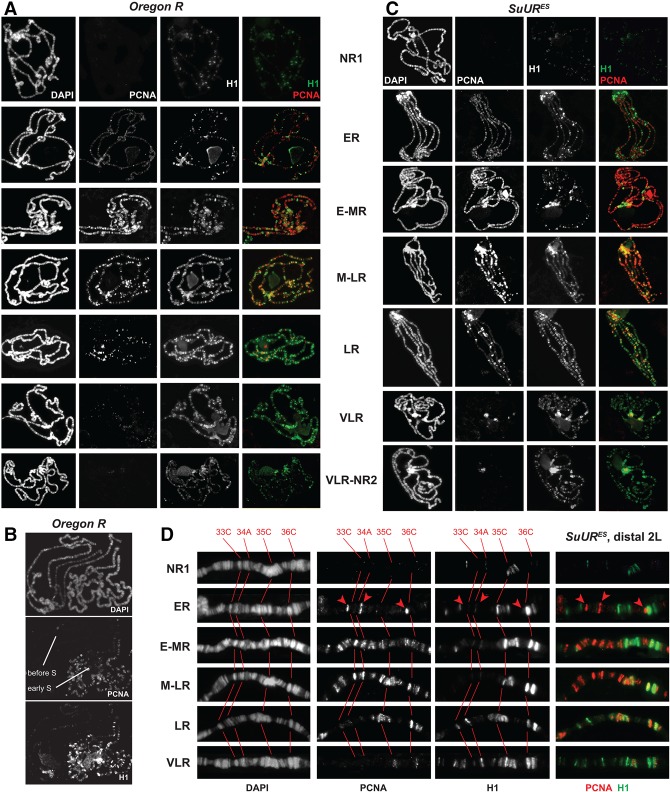
The apparent regulatory roles that H1 plays in endoreplication, particularly of the underreplicated IH, strongly suggest that H1 may exhibit a variable genomic distribution during the endocycle. Also, H1 interacts physically with SUUR, which is known to have endocycle-dependent chromosome localization (Kolesnikova et al. 2013). Thus, we analyzed the patterns of H1 distribution in polytene chromosomes in various stages of the endo-S phase. To this end, we used a previously described method (see the Materials and Methods; Kolesnikova et al. 2013). We collected wandering L3 larvae that were undergoing or approaching the final cycle of endoreplication of salivary gland DNA (Ashburner and Berendes 1978). Polytene chromosome spreads were then costained with H1 and PCNA antibodies, and the endocycle stages were assigned based on the PCNA staining patterns.
The temporal changes that H1 distribution in the genome undergoes during the endocycle are striking. The H1 staining pattern is extremely dynamic, and its association with particular loci depends strongly on their replication status (Fig. 4A). Before the onset of endo-S, H1 is present at a low abundance in most polytene bands. Upon initiation of endoreplication (in very early S), H1 is strongly loaded into thick, late replicating bands. This loading takes place very rapidly, since all early endo-S nuclei contain a uniformly elevated amount of H1 in their thick bands. The intensity of the IF signal at these loci gradually dissipates in the course of the endo-S, but, at the same time, the signal gradually increases at early replicating loci (upon completion of their replication by late endo-S). Consequently, throughout the endo-S phase (with a possible exception of the mid- to late endo-S) (Fig. 4A, fourth row from the top), the staining patterns of H1 and PCNA remain mutually exclusive. By the end of the endo-S phase (when PCNA staining is limited to pericentric regions), the distribution of H1 becomes more uniform. Interestingly, the average H1 staining intensity after endo-S appears to rise compared with the intensity before endo-S (Fig. 4A, cf. first and seventh rows), consistent with H1 de novo synthesis and deposition during the S phase of the cell cycle (Jackson and Chalkley 1985; Guglielmi et al. 2013). Higher-resolution images further confirm these observations (Supplemental Fig. S4A,B and legend).
Figure 4.
H1 distribution in polytene chromosomes is altered dynamically during endo-S phase. (A) Stage-dependent distribution of H1 during endo-S phase in wild-type polytene chromosomes. H1 (green) and PCNA (red) genome-wide distribution patterns were examined by IF staining of polytene chromosomes. The stages of endo-S phase were established based on PCNA distribution in the wild type according to Kolesnikova et al. (2013). H1 and PCNA exhibit mostly mutually exclusive patterns throughout the endocycle. DAPI staining shows the overall chromosome morphology. (NR1) No replication (before the onset of the endo-S phase); (ER) early replication (early endo-S phase); (E-MR) early to mid-replication; (M-LR) mid- to late replication; (LR) late replication; (VLR) very late replication; (NR2) no replication (after the completion of the endo-S phase). (B) Differential loading of H1 in polytene chromosomes before and at the onset of endo-S phase. Polytene chromosomes were stained and analyzed as in A. Two adjacent polytene spreads corresponding to “before” and early endo-S phase exhibit a dramatic difference in the level of H1 loading into chromosomes. (C) Stage-dependent distribution of H1 during endo-S phase in SuUR mutant polytene chromosomes. Polytene chromosomes were prepared from homozygous SuURES mutant larvae and analyzed as in A. (D) Stage-dependent distribution of H1 during endo-S phase in SuUR mutant distal polytene chromosome arm 2L. Detailed view of fragments of polytene chromosomes from (or similar to) those presented in C. DAPI staining shows the overall chromosome morphology and was used for an alignment of cytological positions. Red numbers at the top and corresponding cytological bands in grayscale panels are connected with red lines. (Red arrowheads) Early replicating interbands.
The observed quantitative differences of H1 occupancy during different stages of endoreplication are not subjective, since all analyzed nuclei were processed, stained, and exposed in identical conditions, frequently on the same slide. In fact, these differences were confirmed by visualizing polytene spreads in different stages when they are adjacent to each other. For instance, Figure 4B presents two adjacent nuclei, one of which exhibits no discernable PCNA staining (before endo-S), and the other exhibits a PCNA-staining pattern characteristic of the early endo-S. The many-fold difference of H1 occupancy between the two polytene spreads is immediately apparent, with the majority of the strong H1-specific signal in the “early endo-S” polytene spread limited to late replicating regions.
Although the distribution patterns of PCNA and H1 are largely mutually exclusive, we also observed a partial overlap of their IF signals in certain late replicating polytene loci during the middle to late stages of endo-S (e.g., 50A) (Supplemental Fig. S4B). This observation may reflect a genuine colocalization of the proteins or may stem from a relatively low resolution of the method. The late replicating genomic sites, especially underreplicated IH loci, are not fully polytenized and have a compressed cytological appearance. However, their morphology is substantially improved in SuUR mutants because of the DNA copy number increase (Belyaeva et al. 1998). Since the mutation of SuUR does not strongly affect the distribution of H1 in polytene chromosomes (Supplemental Fig. S2C), it afforded us an opportunity to examine H1 endocycle-dependent distribution in SuURES polytene chromosomes to achieve a better spatial resolution. Indeed, SuURES mutant polytene chromosomes exhibit the same dynamic model of H1 deposition during the progression of endo-S phase (Fig. 4C). Importantly, owing to the suppression of UR, the strong and rapid deposition of H1 during early endo-S into nonreplicating loci, including underreplicated domains, becomes even more readily evident. In higher-resolution images, these chromosome regions (e.g., 35C and 36C) (Fig. 4D) accumulate high levels of H1 protein. The levels of H1 protein in these regions are reduced by late endo-S, when they undergo replication. Early replicating regions (red arrowheads in Fig. 4D) do not contain detectable H1 during early endo-S, and only minimal amounts of H1 are loaded in them as the endo-S phase progresses. The regions that undergo replication during the middle of endo-S (33C and 34A) do not contain high amounts of H1 during early S, and moderate amounts of H1 are deposited into these loci only after their replication is completed.
The increased spatial resolution in polytene chromosomes of SuURES larvae also allowed us to assess a putative “colocalization” of H1 and PCNA during late endo-S. At the level of fine microscopic resolution, it is obvious that although the IF signals for H1 and PCNA are closely spaced with respect to each other in late replicating regions, they are in fact separated, and an increase of PCNA occupancy invariably correlates with a decrease of H1 occupancy and vice versa (Supplemental Fig. S4C,D).
Collectively, our results indicate (1) that H1 exhibits a highly dynamic occupancy in polytene chromosomes as they undergo endoreplication and (2) that the replication machinery appears to be distributed in a mutually exclusive pattern with H1. These mutually exclusive distribution patterns likely contribute to the regulation of endoreplication by H1.
Discussion
The role of Drosophila H1 in the establishment of UR in salivary gland cells
In this study, we demonstrate that virtually all major sites of UR throughout the Drosophila genome exhibit a substantial increase in salivary gland DNA copy number upon depletion of the linker histone H1 (Fig. 1; Supplemental Table S1), thus implicating H1 in the regulation of endoreplication.
In our control knockdown salivary glands, we identified 46 underreplicated domains (Supplemental Fig. S1A; Supplemental Table S1). While these regions are in general agreement with previous efforts to map underreplicated domains by less sensitive microarray analyses (Belyakin et al. 2005; Sher et al. 2012), we identified fewer underreplicated sites than a recent report that used high-throughput sequencing of salivary gland DNA (Yarosh and Spradling 2014). Notably, the underreplicated domains that our analyses failed to detect represent sites with the weakest degree of UR. One possible source of variation is the distinct technical approach that we used compared with Yarosh and Spradling (2014), as simultaneous sequencing of a nonpolytenized (embryonic) genome as a means to normalize the reads from underrepresented sequences in polytenized tissues (Yarosh and Spradling 2014) likely provides additional sensitivity. Another potential explanation could lie in the relative sequencing depth of our respective assays (approximately fourfold lower in our study), considered crucial for the analyses of next-generation sequencing data (Jung et al. 2014; Sims et al. 2014). However, this explanation is less likely, as subsampling of our reads to much lower depths (see the Supplemental Material) yielded no appreciable difference in the number and location of identified underreplicated sites or the change in copy number upon H1 knockdown.
On average, a moderate knockdown of H1 led to an ∼50% copy number gain at the center of underreplicated domains in IH (Fig. 1D). The copy number is not restored to the same degree as that in a SuUR genetic mutant (Supplemental Fig. S1C–G; Yarosh and Spradling 2014). The difference is likely attributable to the incomplete depletion of H1. In fact, in an independent biological validation experiment that resulted in an ∼95% depletion of H1, an almost complete restoration of copy number was observed (Fig. 1, cf. G and I). The observation of an almost complete reversal of UR in cells depleted of H1 (but still wild type for SuUR) strongly suggests an epistatic mechanism of action in which both H1 and SUUR act together in the same biochemical pathway.
We found that H1 and SUUR are also involved in UR of PH. For instance, both the mapped pericentric regions (Fig. 1B) and TE sequences (Supplemental Figure S1H; Supplemental Table S2), which are highly abundant in pericentric regions (Hoskins et al. 2002; Kaminker et al. 2002), exhibit an increase of DNA copy number upon H1 knockdown. We show that the SuURES mutation also results in a robust loss of UR at PH, as measured by changes in DNA copy number at TEs. The abrogation of H1 expression gives rise to a somewhat weaker effect on the UR of PH than that of IH (Supplemental Fig. S1), which is consistent with an almost complete elimination of SUUR protein from polytene chromosome arms in salivary glands depleted of H1 by RNAi but the persistence of residual SUUR at their PH (Fig. 2D). The role of H1 in maintaining the underreplicated state of PH may be relevant to its important regulatory functions in constitutive heterochromatin, where it recruits Su(var)3-9, facilitates H3K9 methylation, and maintains TEs in a transcriptionally repressed state (Lu et al. 2013). Recently, it was proposed that TE repression in ovarian somatic cells involves an H3K9 methylation-independent process through recruitment of H1 by Piwi–piRNA complexes, resulting in reduced chromatin accessibility (Iwasaki et al. 2016). Our results also implicate UR of TE sequences in polytenized cells as yet another putative mechanism that contributes to regulation of their expression. Interestingly, it was shown previously that double mutants encompassing both the Su(var)3-9 and SuUR mutant alleles exhibit a synthetically increased predominance of novel band–interband structures at PH compared with the mutation of SuUR alone (Andreyeva et al. 2007). While the evidence suggests a relationship between UR and transcriptionally repressive epigenetic states, such as H3K9 methylation, the nature of this relationship remains largely speculative (Posukh et al. 2015).
Dissection of the physical interaction between H1 and SUUR
Additionally, we demonstrate that SUUR protein physically interacts with H1 in both a complex mixture of whole-cell extracts that contain endogenous native H1 (Fig. 3C) and recombinant purified H1 polypeptides (Fig. 3F; Supplemental Fig. S3). Furthermore, we delineate the particular structural domains of the two proteins that are required for the interaction. SUUR protein contains several sequence features (Fig. 3A) that have been implicated in regulation of UR and binding to specific proteins. Although SUUR possesses a putative bromodomain (Tchurikov et al. 2004), it contains no identifiable DNA-binding domain, so the mechanism that allows SUUR to exhibit a preference for specific genomic underreplicated loci is unknown. The positively charged central region is both necessary and sufficient to interact with heterochromatin protein 1a (HP1a) (Pindyurin et al. 2008), which suggests a possible involvement of HP1a in tethering SUUR to H3K9me2/3-rich PH. However, the specific localization of SUUR to underreplicated IH, which is not enriched for H3K9me2/3 (Filion et al. 2010), remains enigmatic. We now demonstrate that the central region of SUUR is also sufficient for binding directly to H1 in vitro. Considering that the central region of SUUR is essential for the faithful localization of the protein to chromatin in vivo, including underreplicated IH (Kolesnikova et al. 2005; Yurlova et al. 2009), it seems likely that H1 directly mediates the tethering of SUUR to chromatin in underreplicated regions.
The tripartite structure of H1 provides multiple binding interfaces for interacting proteins and thus allows H1 to mediate several biochemically separable functions in vivo (Kavi et al. 2016). For instance, the globular domain and proximal 25% of the CTD are required for H1 loading into chromatin, while the proximal 75% of the CTD is needed for normal polytene morphology, H3K9 methylation, and physical interactions with Su(var)3-9. In this study, we discover a previously unknown function for the distal 25% of the H1 CTD, which we show to be essential for binding to SUUR. Deletion of this region of H1 results in a near-complete loss of the interaction with SUUR (Fig. 3F). Thus, in addition to its critical functions in heterochromatin structure and activity, the CTD of H1 is likely also important in facilitating UR.
Dynamic locus-specific distribution of H1 in the chromatin of endoreplicating cells
One of the most striking findings in this study is the observation that the genomic occupancy of H1 undergoes profound changes during the endoreplication cycle. It also remains largely mutually exclusive with that of DNA polymerase clamp loader PCNA (Fig. 4), which is consistent with the observed depletion of H1 in nascent chromatin compared with mature chromatin (Alabert et al. 2014).
H1 is heavily loaded into late replicating loci at the onset of replication (when these loci are silent for replication). Combined, our observations indicate that the chromosome distribution of H1 during the endocycle is governed by at least three independent processes (Supplemental Fig. S5). Two of them—replication-dependent (RD) eviction of H1 and RD deposition of H1 after the passage of replication fork—are related to the well-recognized obligatory processes of chromatin disassembly and reassembly during replication (Budhavarapu et al. 2013; MacAlpine and Almouzni 2013). The third pathway, which directs early deposition of H1 into late replicating loci, has not been described previously. This process is (1) replication-independent (RI); (2) locus-specific, with a strong preference for late replicating sites; and (3) apparently more rapid than the RD deposition of H1, since very high levels of H1 occupancy are observed in all nuclei immediately after the initiation of endo-S. It is possible that the RI pathway of H1 loading into chromatin is mediated by a selective recruitment of H1 based on epigenetic core histone modification-dependent mechanisms. For instance, mammalian H1.2 was reported to recognize H3K27me3 (Kim et al. 2015), and this modification is very abundant in IH (Sher et al. 2012).
Also, the RI mechanism for deposition of H1 probably does not involve de novo nucleosome assembly, as H1 is known to exhibit a mutually exclusive distribution with RI core histone variants (Braunschweig et al. 2009), and there is no known nuclear process during early S phase that requires core histone turnover. In the future, it will be interesting to further confirm that RI nucleosome assembly does not take place during early replication in salivary gland polytene chromosomes. Finally, the locus-specific RI deposition of H1 in early endo-S chromatin may be conserved in the normal S phase of diploid tissues, and it will require independent experimentation with sorted mitotically dividing cells to confirm this possibility.
Regulation of SUUR distribution and function by H1 in vivo
We also provide cytological evidence that the functions of H1 and SUUR are biochemically linked. Specifically, we demonstrate that SUUR localizes to a subset of H1-positive bands (Fig. 2A,B) and requires H1 for its precise distribution in polytene chromosomes (Fig. 2D), nuclear localization (Fig. 2E), and stability in salivary gland cells (Fig. 2C). These observations implicate H1 as an upstream effector of SUUR functions in vivo and an essential component of the biological pathway that maintains loci of reduced ploidy in polytenized cells. Importantly, this finding adds to a growing list of biochemical partners of H1 that mediate their chromatin-directed functions in an H1-dependent fashion (Lu et al. 2013; Yang et al. 2013; Xu et al. 2014; Iwasaki et al. 2016).
Interestingly, even a moderate depletion of H1 (to ∼30% of normal) results in a complete removal of SUUR from chromosome arms (Fig. 2D; Supplemental Fig. S2A). Thus, H1-dependent localization of SUUR requires high concentrations of the linker histone in chromatin. This conclusion is also consistent with SUUR colocalization with polytene loci that are the most strongly stained for H1 (Fig. 2A,B). In contrast, elimination of the H3K9me2 mark from polytene spreads requires very extensive depletion of H1 (Lu et al. 2009, 2013), whereas the moderate depletion of H1 does not strongly affect H3K9 dimethylation in the chromocenter or polytene arms (Supplemental Fig. S6A). Therefore, the robust effect of even moderate H1 depletion on SUUR localization in chromatin is unlikely to be mediated indirectly through disorganization of heterochromatin structure.
Unexpectedly, the cell cycle-dependent temporal pattern of H1 localization (Fig. 4) is not identical to that of SUUR. In contrast to H1, SUUR protein (1) is only weakly present in IH during early endo-S phase, (2) achieves the maximal occupancy at IH loci only in the late endo-S, and (3) colocalizes with PCNA at certain sites (Kolesnikova et al. 2013). The observations made in this study and in previous works can be summarized in the following model for H1-mediated regulation of SUUR association with chromatin (Supplemental Fig. S5). The initiation of the deposition of SUUR in chromosomes is strongly dependent on H1. More specifically, SUUR is preferentially localized to chromatin domains that are highly enriched for H1. For instance, the tremendously elevated concentration of H1 in IH of early endo-S cells promotes and nucleates the initiation of deposition of SUUR into these regions. However, the pattern of SUUR occupancy at these sites does not occur temporally in parallel with that of H1. Initially, the exceptionally high abundance of H1 in late replicating loci during early endo-S is not paralleled by a simultaneous comparable increase of SUUR occupancy (Fig. 4B, cf. Supplemental Fig. S6B). Rather, loading of SUUR into these sites lags significantly behind H1 occupancy. Thus, the rate of SUUR localization to H1-rich IH appears to be much slower than that of the RI deposition of H1 into these loci. After the initial recruitment, further loading of SUUR does not require H1, and SUUR continues (in a slower fashion) to accumulate at IH throughout the endo-S phase even when H1-enriched domains dissipate in the course of DNA endoreplication (Fig. 4). The additional loading of SUUR in chromatin is likely facilitated by its self-association through dimerization of the N terminus (Kolesnikova et al. 2005) and physical interactions with the replication fork (Kolesnikova et al. 2013), as proposed previously. In this fashion, SUUR achieves its maximal concentration in IH loci by the late endo-S (Kolesnikova et al. 2013).
The functional roles of H1 in regulation of DNA endoreplication
We demonstrate that H1 has a pivotal function in the establishment of UR of specific IH loci in polytenized salivary gland cells. Our findings that H1 interacts directly with SUUR in vitro and is required for SUUR localization to late replicating IH in polytene chromosomes in vivo strongly suggest that the H1-mediated recruitment of SUUR promotes UR by obstructing replication fork progression in its cognate underreplicated loci but does not affect replication origin firing (Nordman et al. 2014). However, the remarkable temporal pattern of H1 distribution in endoreplicating polytene chromosomes suggests that it may also play a direct SUUR-independent role in regulation of endoreplication. This is especially plausible considering that the temporal distribution patterns of SUUR and H1 are dissimilar.
In contrast to the role of SUUR in slowing down the replication fork progression during late endo-S phase, H1 (acting in the absence of SUUR during early endo-S) may function to repress the initiation of endoreplication, as proposed in several studies (Lu et al. 1998; Thiriet and Hayes 2009). Our DNA-seq analyses also suggest this mechanism. Compared with the relatively smooth, flat profiles of DNA copy numbers in SuURES mutant salivary glands (Supplemental Fig. S6C), the profiles in H1-depleted cells exhibit a jagged, uneven appearance (Fig. 1G), indicative of aberrant local initiation of replication. Unfortunately, our experimental system (cytological analyzes of salivary glands) cannot be used to further confirm this idea. First, an extensive depletion of H1 results in the loss of polytene morphology (Lu et al. 2009); second, since the staging of endo-S progression is based on PCNA staining, a spurious activation of ectopic replication origins would result in an incorrect calling of the stage. To further complicate these analyses, polytenized cells are not amenable to other methods of cell cycle staging, such as fluorescence-activated cell sorting (FACS). In the future, it will be important to examine the role of H1 in regulation of DNA replication timing in sorted Drosophila diploid cells.
Materials and methods
Genomic DNA for high-throughput sequencing
Libraries for high-throughput sequencing of chromatin from His1 and control (Nau) knockdown salivary glands of Drosophila melanogaster L3 larvae were prepared using standard protocols. See the Supplemental Material for details.
Sequencing alignment, identification of underreplicated regions, and quantification of their parameters
Sequencing was performed on an Illumina HiSeq2500. For identification of underreplicated regions in IH, single-end reads were aligned to the R5/dm3 release of the Drosophila genome using Bowtie (Langmead et al. 2009), yielding a total of 9,785,692 and 2,624,291 uniquely aligning reads for the control and H1 knockdown samples, respectively. The sequencing tracks were uploaded to Gene Expression Omnibus (accession number GSE95215). For the control knockdown, underreplicated regions were identified using a custom bash/R script. Briefly, aligned reads in the control knockdown were scanned along 5-kb windows, and regions were designated as underreplicated if 20 consecutive windows fell below the average per-chromosome read count.
The fold change between H1 and control knockdowns was calculated for each of the derived UR coordinates using HOMER annotatePeaks.pl (Heinz et al. 2010). Normalized reads for each underreplicated region were calculated (for both control and H1 knockdown) as the total aligned reads per million divided by the length of the underreplicated domain (in kilobases) identified in the control knockdown sample. Fold change was then calculated as reads per million per kilobase (H1 knockdown) divided by reads per million per kilobase (control knockdown).
To evaluate changes in DNA copy number at PH, reads were aligned to a custom “genome” containing the dm3 release with repeat sequences masked by RepeatMasker, combined with the canonical transposon sequence set (version 9.4.1, Berkeley Drosophila Genome Project). Uniquely aligned read counts for each TE were calculated using SAMtools (Li et al. 2009). Sequencing data for analysis of wild-type and SuURES polytenization (Yarosh and Spradling 2014) were obtained from the Short Read Archive (BioProject ID PRJNA244953).
Polytene chromosomes and DNA break frequency analyses
Larvae (UAS-His1-dsRNA-10-3/+; da-GAL4/+ or wild-type controls) were reared at 25°C. L3 larvae in puff stages 3–8 (Ashburner and Berendes 1978) were selected for salivary gland dissection. Polytene chromosomes were prepared and stained with acetic orcein by standard methods (Zhimulev et al. 1982). Briefly, the glands were dissected in PBS (137 mM NaCl, 3 mM KCl, 8 mM NaH2PO4, 2 mM KH2PO4), transferred to aceto-orcein solution (1% orcein in 45% acetic acid) for 10–15 min followed by 55% lactic acid for 1–2 min, and squashed. Phase contrast images were obtained on an Olympus BX51 microscope, DP52 camera, 100× lens with oil immersion. The frequency of chromosome breaks/weak points was calculated for chromosomes in the last two stages of polyteny (i.e., the thickest) as described (Zhimulev et al. 1982) in two independent experiments. In total, >70 chromosomes from 20 individual slides were analyzed for each genotype and every cytological region.
Indirect IF staining
For all cytological experiments, larvae were reared and collected at 25°C or 29°C (Fig. 2E). Salivary glands from wandering third instar larvae were dissected in PBS. Glands were transferred into a formaldehyde-based fixative (one ∼15-µL drop of 3% lactic acid, 45% acetic acid, 3.7% formaldehyde on a coverslip) for 2 min (Novikov et al. 2007), squashed, and frozen in liquid N2. The coverslips were removed, and slides were placed in 70% ethanol for 20 min and stored at −20°C. The slides were washed three times for 5 min in PBS + 0.1% Triton X-100 (PBST). Primary antibodies were incubated overnight at 4°C in PBST + 0.1% BSA and washed three times for 5 min each with PBST. Secondary antibodies were incubated for 2 h at room temperature in PBST + 0.1% BSA and washed three times for 5 min each with PBST, and squashes were mounted in VectaShield medium with 0.15 µg/mL DAPI. For secondary antibody combinations that contained Alexa 647 conjugates, DNA was stained with 0.1 µg/mL DAPI in PBST for 30 min, and squashes were mounted in Prolong Gold anti-fade mountant (Molecular Probes). IF images were obtained with a Zeiss Axio Observer.Z1, Axiocam 506 mono (D) microscopy camera using a 63×/1.40 plan apo lens with oil immersion using ZEN 2012 software. Phase contrast images were obtained with an Olympus BX51 microscope, DP52 camera using a 100×/1.30 Uplan FI Ph3 lens with oil immersion.
For whole-mount IF staining, L3 larvae were reared at 29°C, and salivary glands were dissected in PBS and fixed in 4% paraformaldehyde for 20 min at room temperature. The glands were washed three times in methanol and stored at −20°C if necessary. They were hydrated three times in PBST for 10 min and permeabilized for 30 min at 37°C in PBS + 0.3% Triton X-100. Blocking was performed for 30 min at 37°C in PBS supplemented with 15% fetal calf serum and 1% BSA. The glands were incubated with primary antibodies diluted in blocking solution for 16 h at 4°C, washed three times with PBST for 30 min, and incubated with secondary antibodies in blocking solution overnight at 4°C. The stained glands were washed three times with PBST for 30 min, stained with DAPI (0.1 µg/mL) for 30 min, and mounted in Prolong Gold. IF images were obtained on a Zeiss LSM 710 confocal microscope using a 20×/0.50 EC Plan-Neofluar lens.
Recombinant proteins and GST pull-down assays
Plasmids encoding GST-tagged full-length SUUR and its middle part (amino acids 371–578) were described previously (Makunin et al. 2002; Pindyurin et al. 2008). pGEX-4T-SUUR(1-370) and pGEX-4T-SUUR(579–962) expression constructs were produced by PCR and molecular cloning. Whole-cell extracts from wild-type Drosophila ovaries were used as a source of the native H1 protein for GST pull-downs. Ovarian extracts were prepared according to Bleichert et al. (2013). Alternatively, GST pull-downs were performed with recombinant H1 polypeptides (full-length and C-terminally truncated). The expression constructs and purification protocol have been described (Kavi et al. 2016). See the Supplemental Material for details of cloning, protein purification, and pull-down protocols.
Supplementary Material
Acknowledgments
We are grateful to Harald Saumweber and Giorgia Siriaco for antibodies and fly stocks. We thank Elena Vershilova for expert technical assistance, and Elena Belyaeva for help with chromosome break frequency scoring. We also thank Elena Belyaeva and members of the Pindyurin, Zhimulev, Skoultchi, and Fyodorov laboratories for critical reading of the manuscript and valuable suggestions. Antibodies were raised at the Center for Genetic Resources of Laboratory Animals, Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences (RFMEFI61914X0005 and RFMEFI61914X0010). This work was supported by grants from the National Institutes of Health (NIH) to A.I.S. (GM093190 and GM116143) and D.V.F. (GM074233), a Cancer Center Support grant from the NIH to Albert Einstein College of Medicine (CA013330), a grant from the Russian Fundamental Scientific Research Project to I.F.Z. (0310-2016-0005), a Russian Foundation for Basic Research grant to A.V.P. (16-04-01598), and a grant from the Russian Science Foundation to A.V.P (16-14-10288). T.J.B. was supported in part by NIH F32 Fellowship (GM115210) and the NIH Institutional Research and Academic Career Development Award/K12 training grant (GM1027779). S.H. and M.A.W. were supported in part by NIH F30 Fellowships (CA210539 and DK107182, respectively).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.295717.116.
References
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16: 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyeva EN, Kolesnikova TD, Demakova OV, Mendez-Lago M, Pokholkova GV, Belyaeva ES, Rossi F, Dimitri P, Villasante A, Zhimulev IF. 2007. High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3-9 double mutants. Proc Natl Acad Sci 104: 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Berendes HD. 1978. Puffing of polytene chromosomes. In The genetics and biology of Drosophila (ed. Ashburner M, Wright TRF), pp. 315–394. Academic Press, London. [Google Scholar]
- Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE. 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci 95: 7532–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Goncharov FP, Demakova OV, Kolesnikova TD, Boldyreva LV, Semeshin VF, Zhimulev IF. 2012. Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS One 7: e30035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF. 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci 102: 8269–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Balasov M, Chesnokov I, Nogales E, Botchan MR, Berger JM. 2013. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife 2: e00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Hogan GJ, Pagie L, van Steensel B. 2009. Histone H1 binding is inhibited by histone variant H3.3. EMBO J 28: 3635–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhavarapu VN, Chavez M, Tyler JK. 2013. How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ. 2012. Developing S-phase control. Genes Dev 26: 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. 2014. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Farrell JA, Shermoen AW, Yuan K, O'Farrell PH. 2012. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev 26: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger RA. 2015. Possible role of H1 histone in replication timing. Dev Growth Diff 57: 1–9. [DOI] [PubMed] [Google Scholar]
- Guglielmi B, La Rochelle N, Tjian R. 2013. Gene-specific transcriptional mechanisms at the histone gene cluster revealed by single-cell imaging. Mol Cell 51: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Caudy A, Parsons L, Rosebrock A, Pane A, Raj S, Wieschaus E. 2012. Mapping the pericentric heterochromatin by comparative genomic hybridization analysis and chromosome deletions in Drosophila melanogaster. Genome Res 22: 2507–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, Kaminker JS, Kennedy C, Mungall CJ, Sullivan BA, Sutton GG, et al. 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: RESEARCH0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, et al. 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Murano K, Ishizu H, Shibuya A, Iyoda Y, Siomi MC, Siomi H, Saito K. 2016. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol Cell 63: 408–419. [DOI] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. 1985. Histone synthesis and deposition in the G1 and S phases of hepatoma tissue culture cells. Biochemistry 24: 6921–6930. [DOI] [PubMed] [Google Scholar]
- Jung YL, Luquette LJ, Ho JW, Ferrari F, Tolstorukov M, Minoda A, Issner R, Epstein CB, Karpen GH, Kuroda MI, et al. 2014. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res 42: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3: RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi H, Emelyanov AV, Fyodorov DV, Skoultchi AI. 2016. Independent biological and biochemical functions for individual structural domains of Drosophila linker histone H1. J Biol Chem 291: 15143–15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kim K, Punj V, Liang G, Ulmer TS, Lu W, An W. 2015. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci Rep 5: 16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova TD, Makunin IV, Volkova EI, Pirrotta V, Belyaeva ES, Zhimulev IF. 2005. Functional dissection of the suppressor of underreplication protein of Drosophila melanogaster: identification of domains influencing chromosome binding and DNA replication. Genetica 124: 187–200. [DOI] [PubMed] [Google Scholar]
- Kolesnikova TD, Posukh OV, Andreyeva EN, Bebyakina DS, Ivankin AV, Zhimulev IF. 2013. Drosophila SUUR protein associates with PCNA and binds chromatin in a cell cycle-dependent manner. Chromosoma 122: 55–66. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach TJ, Chotkowski HL, Wotring MG, Dilwith RL, Glaser RL. 2000. Replication of heterochromatin and structure of polytene chromosomes. Mol Cell Biol 20: 6308–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. 2004. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304: 1675–1678. [DOI] [PubMed] [Google Scholar]
- Lee H, McManus CJ, Cho DY, Eaton M, Renda F, Somma MP, Cherbas L, May G, Powell S, Zhang D, et al. 2014. DNA copy number evolution in Drosophila cell lines. Genome Biol 15: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Sittman DB, Romanowski P, Leno GH. 1998. Histone H1 reduces the frequency of initiation in Xenopus egg extract by limiting the assembly of prereplication complexes on sperm chromatin. Mol Biol Cell 9: 1163–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. 2009. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev 23: 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Kavi H, Kim BJ, Guzzardo PM, Emelyanov AV, Xu N, Hannon GJ, Zavadil J, Fyodorov DV, et al. 2013. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science 340: 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Almouzni G. 2013. Chromatin and DNA replication. Cold Spring Harbor Persp Biol 5: a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF. 2002. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160: 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryant SJ, Lu X, Hansen JC. 2010. Multifunctionality of the linker histones: an emerging role for protein–protein interactions. Cell Res 20: 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C. 1986. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res 165: 291–297. [DOI] [PubMed] [Google Scholar]
- Nordman JT, Kozhevnikova EN, Verrijzer CP, Pindyurin AV, Andreyeva EN, Shloma VV, Zhimulev IF, Orr-Weaver TL. 2014. DNA copy-number control through inhibition of replication fork progression. Cell Rep 9: 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov DV, Kireev I, Belmont AS. 2007. High-pressure treatment of polytene chromosomes improves structural resolution. Nat Methods 4: 483–485. [DOI] [PubMed] [Google Scholar]
- Pindyurin AV, Boldyreva LV, Shloma VV, Kolesnikova TD, Pokholkova GV, Andreyeva EN, Kozhevnikova EN, Ivanoschuk IG, Zarutskaya EA, Demakov SA, et al. 2008. Interaction between the Drosophila heterochromatin proteins SUUR and HP1. J Cell Sci 121: 1693–1703. [DOI] [PubMed] [Google Scholar]
- Posukh OV, Maksimov DA, Skvortsova KN, Koryakov DE, Belyakin SN. 2015. The effects of SUUR protein suggest its role in repressive chromatin renewal during replication in Drosophila. Nucleus 6: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, et al. 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 21: 147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeshin F, Belyaeva S, Zhimulev F. 2001. Electron microscope mapping of the pericentric and intercalary heterochromatic regions of the polytene chromosomes of the mutant suppressor of underreplication in Drosophila melanogaster. Chromosoma 110: 487–500. [DOI] [PubMed] [Google Scholar]
- Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL. 2012. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res 22: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O'Farrell PH. 2010. Developmental control of late replication and S phase length. Curr Biol 20: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. 2014. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 15: 121–132. [DOI] [PubMed] [Google Scholar]
- Smith CD, Shu S, Mungall CJ, Karpen GH. 2007. The release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 316: 1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchurikov NA, Kretova OV, Chernov BK, Golova YB, Zhimulev IF, Zykov IA. 2004. SuUR protein binds to the boundary regions separating forum domains in Drosophila melanogaster. J Biol Chem 279: 11705–11710. [DOI] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ. 2009. Linker histone phosphorylation regulates global timing of replication origin firing. J Biol Chem 284: 2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Braunschweig U, Filion GJ, Chen M, van Bemmel JG, Ideker T. 2010. Bayesian network analysis of targeting interactions in chromatin. Genome Res 20: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Rong Y, Paterson BM. 2007. Stereotypic founder cell patterning and embryonic muscle formation in Drosophila require nautilus (MyoD) gene function. Proc Natl Acad Sci 104: 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. 1997. Histone H1. Int J Biochem Cell Biol 29: 1463–1466. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14: 17–25. [DOI] [PubMed] [Google Scholar]
- Xu N, Emelyanov AV, Fyodorov DV, Skoultchi AI. 2014. Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK–STAT signaling. Epigenetics Chromatin 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SM, Kim BJ, Norwood Toro L, Skoultchi AI. 2013. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci 110: 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W, Spradling AC. 2014. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev 28: 1840–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurlova AA, Makunin IV, Kolesnikova TD, Posukh OV, Belyaeva ES, Zhimulev IF. 2009. Conservation of domain structure in a fast-evolving heterochromatic SUUR protein in drosophilids. Genetics 183: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev IF, Vlassova IE, Belyaeva ES. 1982. Cytogenetic analysis of the 2B3-4–2B11 region of the X chromosome of Drosophila melanogaster. III. Puffing disturbance in salivary gland chromosomes of homozygotes for mutation l(1)pp1t10. Chromosoma 85: 659–672. [DOI] [PubMed] [Google Scholar]
- Zhimulev IF, Belyaeva ES, Semeshin VF, Koryakov DE, Demakov SA, Demakova OV, Pokholkova GV, Andreyeva EN. 2004. Polytene chromosomes: 70 years of genetic research. Int Rev Cytol 241: 203–275. [DOI] [PubMed] [Google Scholar]
- Zink D. 2006. The temporal program of DNA replication: new insights into old questions. Chromosoma 115: 273–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.