Abstract
Background:
The transcription factor 7-like 2 gene (TCF7L2) is an element of the Wnt signaling pathway. There is lack of evidence if TCF7L2 has a functional role in lipid metabolism and regulation of the components constitutes the metabolic syndrome (MetSyn). The aims of this study were to evaluate whether the risk allele of TCF7L2 gene polymorphism is associated with dyslipidemia and MetSyn.
Materials and Methods:
The MetSyn subjects were participated only based on the National Cholesterol Education Program – Third Adult Treatment Panel criteria. In this case–control study, the DNA from MetSyn patients without (n = 90) and with type 2 diabetes (T2D) (n = 94) were genotyped.
Results:
The results show that the genotype-phenotype for CC, CT/TT of TCF7L2 gene polymorphism correlated with body mass index and waist circumference in MetSyn and MetSyn + T2D subjects (r = −0.949 and r = −0.963, respectively). The subjects that only possess MetSyn but are not diabetics show the 2 h postprandial glucose and fasting blood glucose, glycated hemoglobin significantly lower (P < 0.05) than those subjects have both abnormality. The level of triglyceride in CT/TT carriers in MetSyn was higher than CC carriers (P = 0.025). A comparison with the controls subjects, the frequencies of the T allele in the groups of MetSyn (46.66%) and MetSyn + T2D (47.34%) show significantly different (P < 0.05). The odds ratios for T allele in (MetSyn)/(normal), (MetSyn + T2D)/(normal), and in (MetSyn + T2D)/(MetSyn) were 3.59 (95% confidence interval [CI], 1.33–9.67, P = 0.0093), 3.76 (95% CI, 1.40–10.07, P = 0.0068), and 1.08 (95% CI: 0.55– 2.11, P = 0.834), respectively.
Conclusion:
The results revealed the important insights essential for the role of TCF7L2 that the T allele of TCF7L2 plays a significant role in the susceptibility to dyslipidemia, MetSyn, and T2D.
Keywords: Metabolic syndrome, polymorphism, transcription factor 7-like 2 rs7903146, type 2 diabetes
INTRODUCTION
The metabolic syndrome (MetSyn) is a main clinical and public health challenge worldwide. It is considered as the major cause of morbidity and mortality due to genetics, unbalanced diet, and inactive lifestyle. In general, it is defined as a disorder of metabolism (energy utilization and storage). It is suggested that the MetSyn could be presented into clinical practice but with clear and distinct risk factors for health.[1] In recent years, several organizations have defined the MetSyn as interrelated risk conditions for increasing type 2 diabetes (T2D), stroke, and cardiovascular disease (CVD).[1,2,3]
Nowadays, it is believed that there is evidence for the existence and characteristics of the MetSyn and it is sufficiently criteria developed to support its inclusion in clinical practice. Different definitions for MetSyn in adults were presented by the international scientific communities. The organizations that developed clinical criteria for MetSyn are the International Diabetes Federation, World Health Organization (WHO), the European Group for the Study of Insulin Resistance, and the National Cholesterol Education Program – Third Adult Treatment Panel (NCEP-ATP-III).[4,5,6,7] All groups agreed on the core components of the MetSyn which are insulin resistance (elevated fasting plasma glucose), hypertension (elevated diastolic and systolic blood pressure [SBP]), dyslipidemia (high serum triglycerides (TGs) and low high-density lipoprotein levels), and obesity (abdominal or central obesity). However, they evaluate different clinical criteria to identify MetSyn. The ATP-III definition, unlike the other definitions, does not require impaired glucose regulation or insulin resistance as an essential component.
Therefore, by NCEP-ATP-III criteria, individuals were defined as having MetSyn, if they have at least three of the following factors abnormal: (1) waist circumference >102 cm in men and >88 cm in women, (2) blood pressure; SBP to diastolic blood presser ≥130/85 mmHg, (3) serum TG ≥150 mg/dL (1.7 mmol/L), 4) high-density lipoprotein (HDL) cholesterol <40 mg/dL (1.0 mmol/L) in men and <50 mg/dL (1.3 mmol/L) in women, and (5) fasting blood glucose (FBG) >110 mg/dL (6.1 mmol/L).[4,7]
The prevalence of MetSyn reported in the America's and European population are about 25% and 15%, respectively in.[8,9] The prevalence of MetSyn in Iran, according to the NCEP-ATP-III definition was estimated to be 34.7%, which is clearly higher than the average rate mentioned in the USA and Europe.[10] Several studies have shown that people with MetSyn, suffered from heart attacks more than those who have not MetSyn between three and five times.[11] The etiology of MetSyn is complex and determined by the interaction of environmental factors and genetic. Based on the thrifty genotype hypothesis, genes involved in efficiently storing and saving energy could predispose to MetSyn.
Several specific genes are involved in metabolism, such as genes in the systems of energy balance, lipid and insulin metabolism, lipolysis, thermogenesis, fuel oxidation, and glucose uptake in adipocyte tissue and skeletal muscle. Many of these genes have been associated with MetSyn in various ethnic populations. These candidate genes include but are not limited to peroxisome proliferator-activated receptor γ, adiponectin, CD36, β-adrenergic receptors, insulin receptor substrates, 11 β-hydroxysteroid dehydrogenase type 1, C-reactive protein, tumor necrosis factor-α, calpain-10, upstream transcription factor-1, and skeletal muscle glycogen synthase-1.[12,13,14]
Transcription factor 7-like 2 gene (TCF7L2), previously known as TCF-4, is a member of the high mobility group box transcription factors. TCF7L2 is a major transcription factor of the canonical Wnt signaling pathway known as β-cat/TCF. This bipartite transcription factor is formed by free β-catenin (β-cat) and a member of the TCF protein family, affecting preadipocyte differentiation, inflammatory status, pancreatic beta-cell function, and cortisol/aldosterone secretion.[15] Impaired Wnt signaling has an important role in generation of ectopic fat, insulin resistance, and elevated plasma lipids.[16]
The network of Wnt signaling pathways and β-cat/TCF7L2 bipartite transcription factor in obesity, insulin resistance, insulin secretion, lipoprotein metabolism, and inflammation is important to understand for prevention and treatment of MetSyn, diabetes, and CVD.[17] Many studies confirmed the association between the single-nucleotide polymorphism (SNP) (rs7903146) in the TCF7L2 and T2D in population of different ethnic origins and also Persian.[18]
Involvement of the TCF7L2 rs7903146 (C/T) gene polymorphism in regulating essential or major components of the MetSyn such as hypertension, dyslipidemia, insulin sensitivity, and obesity is not well understood.[19,20,21] Researchers are still investigating to find the relationship between the frequency of a number of genes and their association with increased risk of T2D and MetSyn components. There is lack of evidence if TCF7L2 has a functional role in lipid metabolism and has a direct functional role in the regulation of the components constituted the MetSyn. Therefore, the aims of this study were to evaluate if the risk allele of TCF7L2 (rs7903146C/T) gene is associated with dyslipidemia and MetSyn and T2D.
MATERIALS AND METHODS
Study subjects
The patient population consist of a total of 184 subjects, MetSyn (n = 90) and MetSyn + T2D (n = 94). All participants were well informed the purposes of the study, and hopefully, all were happy to make positive contribution. Written informed consent from patients and participants in the study were also obtained to register for a confidential barcode. All subjects with self-reported history of disease and registered data for treatments that confound the study were excluded from the study. A group of unrelated healthy subjects from healthy population was used as control to test TCF7L2 genotypes for departure from Hardy–Weinberg equation (HWE). All peripheral blood samples of patients were collected from patients from the Diabetes Clinic Centers/Isfahan City, Iran. The MetSyn subjects diagnosed based on the definition provided by the NCEP-ATP-III.[4,7] The MetSyn subjects were encounter in this study if they had the four previously mentioned criteria of the NCEP-ATP-III definition. The MetSyn + T2D were included in this study according to the NCEP-ATP-III to include MetSyn plus WHO criteria to enclose the T2D subjects.
Genotyping of single-nucleotide polymorphism rs7903146 (C/T) polymorphism
Peripheral blood samples of the patient were collected in ethylenediaminetetraacetic acid anticoagulated tubes. Then, DNA was extracted from whole-blood samples using the QIAGEN DNA purification kit (QIAGEN Ltd., UK). DNA integrity was checked by ultraviolet spectrophotometer at 260/280 nm and by agarose electrophoresis to visualize the purity. The genotyping was performed by mismatched polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The following primers were used for the experiment.
Forward: 5’-TTA GAG AGC TAA GCA CTT TTT AGG TA-3’
Reverse: 5’-AGA GAT GAA ATG TAG CAG TGA AGT G-3’
The above specific PCR primers amplified a 201-bp fragment, in which there is a specific restriction site to determine the allele differences of the rs7903146 variant. The PCR was carried out on an Eppendorf thermal cycler (AG2231, Eppendorf, Germany) under the following conditions for PCR-RFLP: 94°C for 4 min by 35 cycles of 94°C for 30 min, 61.1°C for 1 min, 72°C for 20 s, and a final extension of 72°C for 5 min.
Finally, the PCR-RFLP products were then digested with 0.5 unit of RsaI restriction enzyme (Promega UK Ltd.). After 4-h incubation at 37°C, the digestive products were visualized by 3% agarose and/or polyacrylamide gel electrophoresis.
Statistical analysis
SPSS for Windows, version 22 (SPSS, Chicago, IL, USA), was used for statistical analysis. For all statistical analyses, P < 0.05 was considered as statistically significant. All quantitative values are expressed as mean ± standard deviation HWE for the distribution of genotypes was estimated by the Chi-square test. The odds ratios (ORs) with 95% confidence interval (CI) to understand the different association were calculated by multiple logistic regressions with confounders determined by a backward conditional elimination method to find a significance level below 0.05. The logistic regression analysis was also performed with and without adjustment for age and sex as well as body mass index (BMI). STATA/SE 10 program was used for power calculation. The mean of clinical and biochemical characteristics was compared by t-test or between groups by post hoc and Mann–Whitney tests.
RESULTS
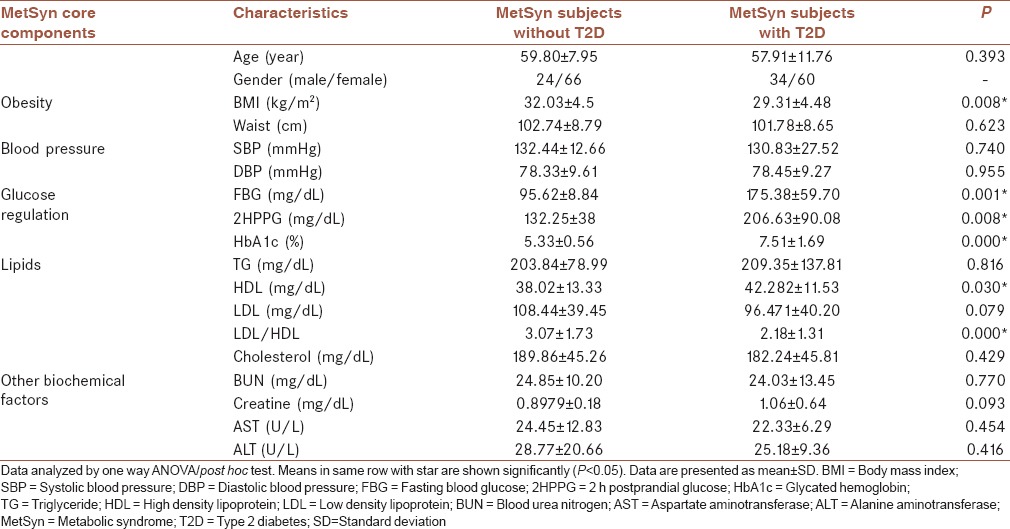
The biochemical factors of the subjects were measured to distinguish the MetSyn and diabetics conditions in individuals. Subjects were characterized and grouped according to the essential components for MetSyn and T2D. The main features of subjects with regard to cutoff levels provided by NCEP-ATP-III and WHO to distinguish the MetSyn + T2D from the MetSyn were measured. The characteristics in subjects were measured and the results are summarized in Table 1. The biochemical factors in the MetSyn were compared with the MetSyn + T2D subjects. The comparison revealed that there are significant differences (P < 0.05) in BMI, HDL, and low-density lipoprotein/HDL ratio. As shown in Table 1, FBG, 2 h postprandial glucose (2HPPG), and glycated hemoglobin (HbA1c) in the MetSyn subjects which have T2D are significantly higher (P < 0.05) than those subjects without T2D.
Table 1.
The clinical and biochemical characteristics of the metabolic syndrome subject
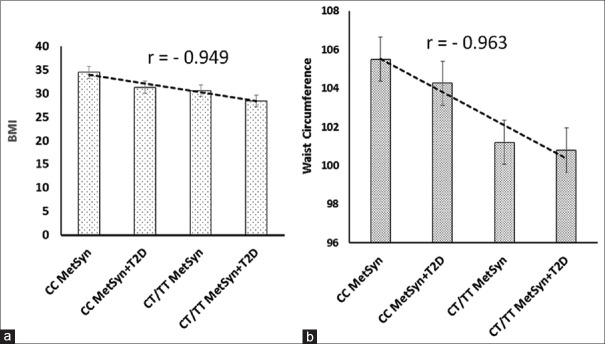
The clinical and biochemical characteristics of the subjects based on the rs7903146(C/T) genotypes (CC and CT/TT) were analyzed, and the results are shown in Table 2. The results revealed that there is a genotype-phenotype correlation as shown in Figure 1. The CC, CT/TT genotype frequencies in MetSyn and MetSyn + T2D with BMI, and waist circumference were correlated with coefficient correlations of r = −0.949 and r = −0.963, respectively. TG in MetSyn group with CT/TT genotype is higher than the CC genotype (P > 0.05), [Table 2].
Table 2.
The clinical and biochemical characteristics of the subjects based on rs7903146 (C/T) genotypes
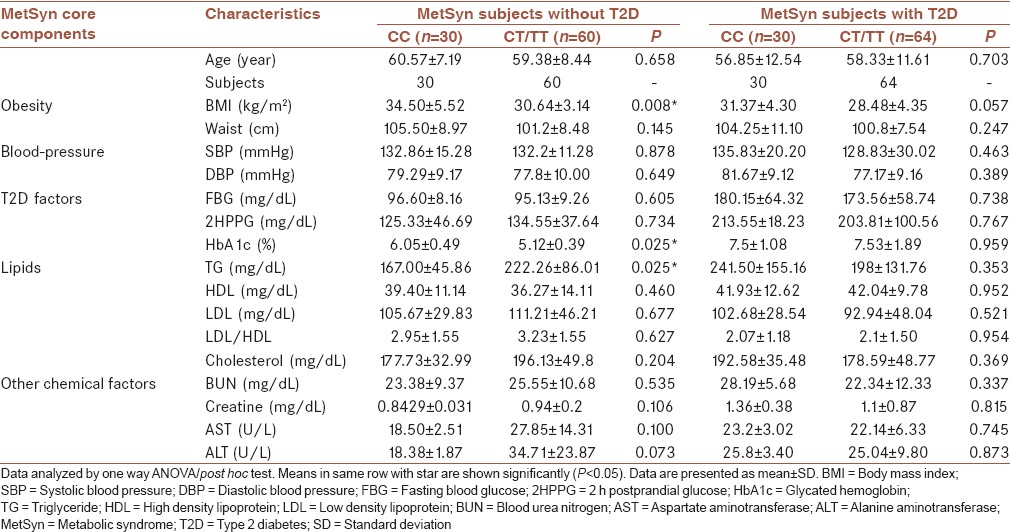
Figure 1.
The calculator for finding the correlation coefficient (r) values which represent the relationship between (a) body mass index and (b) waist circumference with genotypes. Pearson's formula is used for this calculation
The genotype distribution at this SNP did not obey the HWE within both groups of the MetSyn and MetSyn + T2D subjects (P > 0.05).
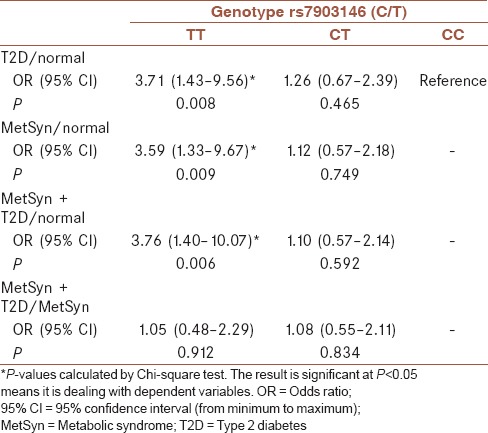
The genotype and allele frequency data are shown in Table 3. The analysis of the data shows that the frequency of the T risk allele was approximately equal in two groups, i.e., MetSyn (46.66%) and MetSyn + T2D (47.34%). In other word, the frequency of T allele is not meaningfully different (P = 0.884). The genotypes frequency of CC, CT, and TT in MetSyn subjects are 31.11%, 44.44%, and 24.44%, respectively. That is likely similar to the genotypes frequency in MetSyn + T2D subjects for CC, CT, and TT which are 29.79%, 45.74%, and 24.47%, respectively. The ORs of the presence of T allele frequency together with the presence of T2D and MetSyn are presented in Table 4. Logistic regression analysis and Chi-square test indicated that the OR for the T risk allele for (rs7903146C/T) in (MetSyn)/(normal), (MetSyn + T2D)/(normal), and (MetSyn + T2D)/(MetSyn) are 3.59 (95% CI, 1.33–9.67 P = 0.0093), 3.76 (95% CI, 1.40–10.07 P = 0.0068), and 1.08 (95% CI: 0.55–2.11 P = 0.834), respectively, as shown in Table 4.
Table 3.
Frequency of the rs7903146 (C/T) polymorphism and association of this variant with metabolic syndrome and type 2 diabetes in the study subjects
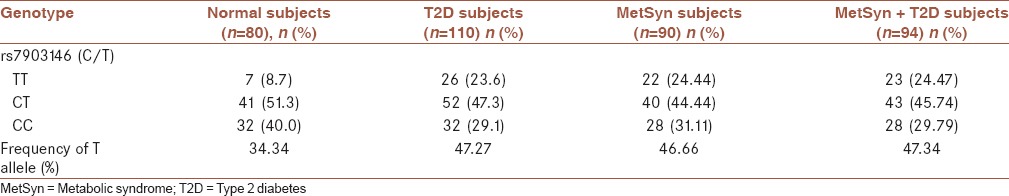
Table 4.
The odds ratio of the presence of T allele frequency associated with the presence of type 2 diabetes and metabolic-syndrome
DISCUSSION
The association between a common variant of TCF7L2 polymorphisms (rs7903146) and T2D mellitus has been widely investigated in Iranian and many other populations.[18] Up to now, no significant function for TCF7L2 is being well documented. The MetSyn with several biochemical components causes many chronic diseases such as diabetes and CVD. There is no report related to the association between dyslipidemia and MetSyn with TCF7L2 polymorphisms. As it was expected, subjects with MetSyn + T2D had FBG, 2HPPG, and HbA1c significantly higher (P < 0.05) than those subjects without T2D. This was consistence with WHO definition for T2D. In this study, there is a genotype-phenotype correlations. The CC, CT/TT genotype frequencies in MetSyn and MetSyn + T2D with BMI and waist circumference with coefficient correlations of r = −0.949 and r = −0.963, respectively. Therefore, in this investigation, the genotyping in the MetSyn subjects which carrying the rs7903146 T allele (risk allele) was significantly associated with decreasing BMI and waist circumference.
The study on the MetSyn + T2D subjects carrying the rs7903146 T allele, the BMI, and waist circumference in this group was also slightly decreased but was not significantly different. It is known that the TCF7L2 plays a critical role in the Wnt signaling pathway to regulate adipose cell differentiation and adipogenesis.[19,20,21,22] This phenomena could be associated with minor increase in TCF7L2 expression in adipose tissue.[23] HbA1c is formed by long-term exposure of normal hemoglobin to plasma glucose in a nonenzymatic glycation pathway. Therefore, HbA1c level is proportional to average blood glucose, and it is a useful factor for diabetes treatment monitoring. HbA1c in MetSyn is significantly lower than MetSyn + T2D. The results were expected to be associated to phenotype. HbA1c levels in the MetSyn groups with the rs7903146 C allele are higher than T allele carriers. There are no differences between the HbA1c levels in both allele in the MetSyn + T2D. Although the C allele is good factor to prevent MetSyn, HbA1c concentration might not be correlated to TCF7L2 genotype. There is one report suggest that there is an association between the T risk allele and the levels of HbA1c.[24]
MetSyn subjects with rs7903146 T risk allele possess TG higher than that of rs7903146 C allele. These results are consistent with the previous study that the TCF7L2 polymorphisms could be associated with MetSyn components such as dyslipidemia, waist circumference, and BMI.[21] The TCF7L2 gene polymorphism might be regulated by TG or free fatty acids in plasma. This result is consistence with the previous observation that the T2D patients with the C allele carriers are likely to have MetSyn features.[25] Further experiments require to find the real fact.
The variations in TCF7L2 gene polymorphism particularly rs7903146 could be a genetic risk factor for T2D.[26,27] The beta-cell dysfunction is also present in MetSyn patients, affecting dynamic pathways of glucose metabolism.[28] The alterations in glucose metabolism cause by these gene variations have been investigated largely. The T allele of TCF7L2 is a risk factor regulates insulin secretion. The TCF7L2 is transcription factor of Wnt signaling involves in several biological processes, from the differentiation of adipocytes, and the regulation of adipokines to pancreatic beta cell function.[29] The presence of the T allele of TCF7L2 rs7903146 gene variant could be linked to other diseases such as the incidence of cancer or the severity of coronary artery disease.[30] The OR and the logistic regression analysis for T allele indicated that the common variant of TCF7L2 ra7903146 could be associated with MetSyn and the T allele of TCF7L2 ra7903146 gene polymorphism is a risk allele for MetSyn as well as type-2 diabetes mellitus. The variants of the TCF7L2 gene polymorphism could likely predict the development of T2D mellitus and abnormalities introduce MetSyn components such as dyslipidemia.
CONCLUSIONS
The present study demonstrated that TCF7L2 plays a significant role in the regulation of lipid and glucose metabolism, wherein the changes in its biological function could cause alteration in the susceptibility to dyslipidemia, MetSyn, and T2D. Our finding related to T allele, which plays as a risk allele in diabetes and metabolic syndrome, openes a narrow window to find the etiology of these diseases with important insights necessary for evaluating the role of TCF7L2 polymorphism in metabolic pathways.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
AUTHORS’ CONTRIBUTIONS
AP designed and contributed in the conception of the work, conducting the study, revising the draft, approval of the final version of the manuscript, and agreed for all aspects of the work.
MR carried out the molecular biology experiments and participated in revising the draft and agreed for all aspects of the work, read and approved the final draft of manuscript.
HK participated in molecular biology experiments.
MF agreed for all aspects of the work.
Acknowledgments
The authors acknowledge the Isfahan University of Medical Sciences and the Molecular Biology and Genetic Laboratory to help us to do the study. This study was approved in Isfahan Payame Noor University (Grant number: 18035).
REFERENCES
- 1.Kahn R, Buse J, Ferrannini E, Stern M. American Diabetes Association; European Association for the Study of Diabetes. The metabolic syndrome: Time for a critical appraisal: Joint statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2005;28:2289–304. doi: 10.2337/diacare.28.9.2289. [DOI] [PubMed] [Google Scholar]
- 2.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: A joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–5. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 3.Alberti KG, Zimmet P, Shaw J IDF Epidemiology Task Force Consensus Group. The metabolic syndrome – A new worldwide definition. Lancet. 2005;366:1059–62. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 4.Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–97. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 5.Balkau B, Charles MA. Comment on the provisional report from the WHO consultation. European Group for the Study of Insulin Resistance (EGIR) Diabet Med. 1999;16:442–3. doi: 10.1046/j.1464-5491.1999.00059.x. [DOI] [PubMed] [Google Scholar]
- 6.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 7.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. American Heart Association; National Heart, Lung, et al. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109:433–8. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 8.Beltrán-Sánchez H, Harhay MO, Harhay MM, McElligott S. Prevalence and trends of metabolic syndrome in the adult U.S. population, 1999-2010. J Am Coll Cardiol. 2013;62:697–703. doi: 10.1016/j.jacc.2013.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu G, Qiao Q, Tuomilehto J, Balkau B, Borch-Johnsen K, Pyorala K DECODE Study Group. Prevalence of the metabolic syndrome and its relation to all-cause and cardiovascular mortality in nondiabetic European men and women. Arch Intern Med. 2004;164:1066–76. doi: 10.1001/archinte.164.10.1066. [DOI] [PubMed] [Google Scholar]
- 10.Delavari A, Forouzanfar MH, Alikhani S, Sharifian A, Kelishadi R. First nationwide study of the prevalence of the metabolic syndrome and optimal cutoff points of waist circumference in the Middle East: The national survey of risk factors for noncommunicable diseases of Iran. Diabetes Care. 2009;32:1092–7. doi: 10.2337/dc08-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Te Morenga LA, Levers MT, Williams SM, Brown RC, Mann J. Comparison of high protein and high fiber weight-loss diets in women with risk factors for the metabolic syndrome: A randomized trial. Nutr J. 2011;10:40. doi: 10.1186/1475-2891-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groop L. Genetics of the metabolic syndrome. Br J Nutr. 2000;83(Suppl 1):S39–48. doi: 10.1017/s0007114500000945. [DOI] [PubMed] [Google Scholar]
- 13.Pollex RL, Hegele RA. Genetic determinants of the metabolic syndrome. Nat Clin Pract Cardiovasc Med. 2006;3:482–9. doi: 10.1038/ncpcardio0638. [DOI] [PubMed] [Google Scholar]
- 14.Hoteit M, Arabi A, Habib R, Mahfouz R, Baddoura R, Halaby G, et al. Estrogen receptor a is not a candidate gene for metabolic syndrome in Caucasian elderly subjects. Metabolism. 2014;63:50–60. doi: 10.1016/j.metabol.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Schinner S. Wnt-signalling and the metabolic syndrome. Horm Metab Res. 2009;41:159–63. [Google Scholar]
- 16.Song K, Wang S, Mani M, Mani A. Wnt signaling, de novo lipogenesis, adipogenesis and ectopic fat. Oncotarget. 2014;5:11000–3. doi: 10.18632/oncotarget.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almario RU, Karakas SE. Roles of circulating WNT-signaling proteins and WNT-inhibitors in human adiposity, insulin resistance, insulin secretion, and inflammation. Horm Metab Res. 2015;47:152–7. doi: 10.1055/s-0034-1384521. [DOI] [PubMed] [Google Scholar]
- 18.Palizban A, Nikpour M, Salehi R, Maracy MR. Association of a common variant in TCF7L2 gene with type 2 diabetes mellitus in a Persian population. Clin Exp Med. 2012;12:115–9. doi: 10.1007/s10238-011-0144-7. [DOI] [PubMed] [Google Scholar]
- 19.Sjögren M, Lyssenko V, Jonsson A, Berglund G, Nilsson P, Groop L, et al. The search for putative unifying genetic factors for components of the metabolic syndrome. Diabetologia. 2008;51:2242–51. doi: 10.1007/s00125-008-1151-4. [DOI] [PubMed] [Google Scholar]
- 20.Delgado-Lista J, Perez-Martinez P, García-Rios A, Phillips CM, Williams CM, Gulseth HL, et al. Pleiotropic effects of TCF7L2 gene variants and its modulation in the metabolic syndrome: From the LIPGENE study. Atherosclerosis. 2011;214:110–6. doi: 10.1016/j.atherosclerosis.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 21.Phillips CM, Goumidi L, Bertrais S, Field MR, McManus R, Hercberg S, et al. Dietary saturated fat, gender and genetic variation at the TCF7L2 locus predict the development of metabolic syndrome. J Nutr Biochem. 2012;23:239–44. doi: 10.1016/j.jnutbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 22.Franklin CS, Aulchenko YS, Huffman JE, Vitart V, Hayward C, Polašek O, et al. The TCF7L2 diabetes risk variant is associated with HbA1(C) levels: A genome-wide association meta-analysis. Ann Hum Genet. 2010;74:471–8. doi: 10.1111/j.1469-1809.2010.00607.x. [DOI] [PubMed] [Google Scholar]
- 23.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–3. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 24.Cauchi S, Choquet H, Gutiérrez-Aguilar R, Capel F, Grau K, Proença C, et al. Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity (Silver Spring) 2008;16:476–82. doi: 10.1038/oby.2007.77. [DOI] [PubMed] [Google Scholar]
- 25.Melzer D, Murray A, Hurst AJ, Weedon MN, Bandinelli S, Corsi AM, et al. Effects of the diabetes linked TCF7L2 polymorphism in a representative older population. BMC Med. 2006;4:34. doi: 10.1186/1741-7015-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 27.Helgason A, Pálsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–25. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 28.Serralde-Zúñiga AE, Guevara-Cruz M, Tovar AR, Herrera-Hernández MF, Noriega LG, Granados O, et al. Omental adipose tissue gene expression, gene variants, branched-chain amino acids, and their relationship with metabolic syndrome and insulin resistance in humans. Genes Nutr. 2014;9:431. doi: 10.1007/s12263-014-0431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilar JS, Begum AN, Alvarez J, Zhang XB, Hong Y, Hao J. Directed cardiomyogenesis of human pluripotent stem cells by modulating Wnt/ß-catenin and BMP signalling with small molecules. Biochem J. 2015;469:235–41. doi: 10.1042/BJ20150186. [DOI] [PubMed] [Google Scholar]
- 30.Sousa AG, Marquezine GF, Lemos PA, Martinez E, Lopes N, Hueb WA, et al. TCF7L2 polymorphism rs7903146 is associated with coronary artery disease severity and mortality. PLoS One. 2009;4:e7697. doi: 10.1371/journal.pone.0007697. [DOI] [PMC free article] [PubMed] [Google Scholar]