Abstract
Establishing quantitative relationships between molecular structure and broad biological effects has been a longstanding challenge in science. Currently, no method exists for forecasting broad biological activity profiles of medicinal agents even within narrow boundaries of structurally similar molecules. Starting from the premise that biological activity results from the capacity of small organic molecules to modulate the activity of the proteome, we set out to investigate whether descriptor sets could be developed for measuring and quantifying this molecular property. Using a 1,567-compound database, we show that percent inhibition values, determined at single high drug concentration in a battery of in vitro assays representing a cross section of the proteome, provide precise molecular property descriptors that identify the structure of molecules. When broad biological activity of molecules is represented in spectra form, organic molecules can be sorted by quantifying differences between biological spectra. Unlike traditional structure–activity relationship methods, sorting of molecules by using biospectra comparisons does not require knowledge of a molecule's putative drug targets. To illustrate this finding, we selected as starting point the biological activity spectra of clotrimazole and tioconazole because their putative target, lanosterol demethylase (CYP51), was not included in the bioassay array. Spectra similarity obtained through profile similarity measurements and hierarchical clustering provided an unbiased means for establishing quantitative relationships between chemical structures and biological activity spectra. This methodology, which we have termed biological spectra analysis, provides the capability not only of sorting molecules on the basis of biospectra similarity but also of predicting simultaneous interactions of new molecules with multiple proteins.
Keywords: biospectra, proteome, structure–function relationships
Organic molecules have the intrinsic capacity of both storing and transmitting information. This ability provides the link between chemical scaffold design and biological activity. Identification of structure features that allow differentiation between effect and side-effect profiles of medicinal agents is currently rate limiting in drug discovery (1). Current understanding of structure–activity relationship (SAR) components evolved from “lock and key” models of protein–ligand interactions (2, 3). Each protein family has its own sets of rules, which depend on dynamic and structural aspects of ligand and ligand-binding site, for identifying molecular properties that provide specific interactions with proteins (4, 5). Current drug discovery methods estimate biological response of potential medicinal agents by constructing independent and linear models. Although these models provide a link between specific biological targets and therapeutic effects, the properties of natural signals are too complex to expect that an independent set of descriptors would be capable of forecasting broad biological responses (6). Hence refinement of traditional SAR methods hinges on development of nonlinear, multivariant models and availability of molecular property descriptors that take into account chirality and dynamic aspects not only of ligands but also of receptor targets (7). Obviously, these models must also factor in the variance of molecular property and biological response descriptors based on differences in biological environments (8). Considering the complexity of this requirement, computational solutions that precisely link molecular structure to broad biological response are currently not possible (9, 10). We report here an approach to structure–function studies that is based on measurements of the capacity of molecules to interact with the proteome (11).
Translation of Chemical Property Information into Biological Activity Spectra
What emerges from investigations on the complexity of biological response regulation is the obvious role that modularity plays at every level, from genes to proteins to functions. Modular structure design within protein families by using conserved structure elements in the active site (11), for example, forms the basis for 3D models, which drive SAR studies within protein families. This strategy even succeeds in protein families whose members have low sequence homology such as the cytochrome P450 superfamily, which consists of >2,000 members distributed across different organisms (12–15).
The investigation described below takes advantage of the modularity concept for linking biological response and structure design of molecules. Conservation of active site residues within protein families is the reason that, at high ligand concentration, the capacity of molecules to interact selectively with protein targets is lost. Consequently, determination of percent inhibition values for a molecule at single high drug concentrations not only measures its interaction with a single target but also provides information on a molecule's broad interaction potential with a large number of proteins represented in a gene family. Examining the ability of ligands to interact with proteins representing a wide range of gene families of the “drugable” proteome (16) under identical ligand concentration therefore provides an effective strategy for determining the interaction capacity of molecules with this important section of the proteome (17–19).
For characterizing the interaction capacity of molecules with the proteome, percent inhibition values are treated as a continuum of data points and not as independent observations (9, 20–22). From this perspective, individual percent inhibition values are not interpreted in terms of biological response significance but instead are used to describe the interaction capacity of a molecule with the proteome. Representation of this molecular property as a biological activity spectrum has several important consequences. First, biospectra provide a uniform description of this molecular property, which integrate SAR determinants of both ligand and ligand-binding sites. A second feature is that biospectra derived from percent inhibition values at single ligand concentration are normalized and directly comparable. As a result, spectroscopic methods can be used to compare interaction capacities of molecules with the proteome across a wide range of structure classes. Biospectra similarity between molecules is measured by using similarity measures such as Euclidean distance, cosine correlation, city block distance, or Tanimoto coefficient. For the SAR study presented below, biospectra similarity was determined by using cosine correlation. The other similarity measures provided similar results.
Methods
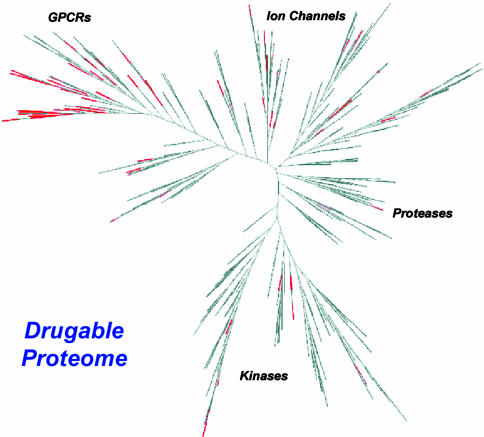
To probe the utility of biological activity spectra as an intrinsic molecular property descriptor, we selected a portion of the BioPrint database‡ of Cerep (Rueil-Malamaison, France) (23) for our investigation. We used 1,567 structurally diverse molecules and 92 ligand-binding assays for constructing a data set containing complete percent inhibition values at 10 μM ligand concentration. Primary screening at 10 μM was carried out in duplicate. Additional screening was carried out at 10 μM ligand concentration if results varied by more than 20%. The 92 assays were selected to represent a cross section of the drugable proteome as shown in Fig. 1 (16–19). The 1,567-compound data set represented the complete list of molecules in the BioPrint database that had percent inhibition values for all of the 92 assays. The compounds and assays used for this investigation are listed in Data Sets 1 and 2, which are published as supporting information on the PNAS web site. decisionsite 7.2 (Spotfire, Somerville, MA) was used for hierarchical clustering and profile similarity searches.
Fig. 1.
A cross section of the drugable proteome. Proteins are clustered on the basis of sequence homology. Proteins in close proximity in this dendrogram are members of the same gene family and share sequence similarity and structure similarity in regulatory and ligand-binding domains. Cerep's BioPrint database‡ (23) consists of >100 in vitro assays. Forty-two of the 92 assay constituents used in our studies (shown in red) are G-protein-coupled receptors (GPCRs). The rest encompass a wide range of functionally diverse proteins representing a number of protein superfamilies.
Results and Discussion
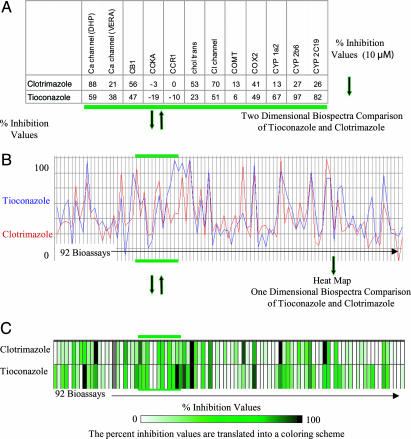
To ensure that direct structure–activity information would not, in any way, affect the outcome of our assessment of the general utility of biospectra as molecular property descriptors, the biological activity spectra of clotrimazole (1) and tioconazole (2) (Fig. 2) were selected as a starting point for our investigation because sterol 14α-demethylase (CYP51), the putative drug target (15) of these antifungal agents, is not part of the 92-protein bioassay suite. Fig. 2 shows the biological activity profiles for clotrimazole and tioconazole by using the suite of in vitro assays described above and listed in Data Set 2. Biological activity data for these two molecules can be represented in several ways: (i) a table listing the percent inhibition values of 1 and 2 in each of the assays (Fig. 2 A); (ii) a two-dimensional spectra representation of percent inhibition values, e.g., percent inhibition values for each molecule in the 92 bioassays (Fig. 2B); (iii) a one-dimensional comparison of percent inhibition values by using a heat map representing individual percent inhibition values with a coloring scheme (Fig. 2C).
Fig. 2.
Biological activity spectra of antifungal agents clotrimazole and tioconazole. These spectra were constructed by using 92 bioassay data points from Cerep's BioPrint array.‡ The bioassay proteins, listed in Data Set 2, are located on the x axis. Associated percent inhibition values (A), determined at 10 μM drug concentrations for each compound, are described in the two-dimensional spectra view shown in B and as a heat map shown in C, which presents the same information and layout for the x axis and uses a coloring scheme for expressing percent inhibition values. A white to green to black gradient expresses values between 0% and 100% inhibition. This coloring scheme is applied to all heat maps shown in this publication.
Biospectra Similarity. The capacity of clotrimazole and tioconazole to interact with the proteome is reflected in the biological activity spectrum constructed from individual percent inhibition values for these 92 bioassays obtained at 10 μM drug concentration (Fig. 2). Experience with physicochemical spectra teaches that spectra similarity parallels structure similarity. Hence it is generally accepted that molecules with identical structure have identical spectra and that spectra similarity between molecules decreases with decreasing structure similarity. There are two methods commonly used for spectra comparison. One method for establishing biospectra similarity between molecules is based on direct biospectra profile comparison. Operationally, this method rank orders compounds in the database by using similarity scores [similarity to profile (SP) scores], obtained by comparing the biospectra of every molecule in the database with the biospectrum of a reference compound. The second method, which is not biased, identifies spectra similarity [by using the confidence in cluster similarity (CCS) scale] based on hierarchical clustering. Hierarchical clustering identifies the best possible matches between compound pairs in the entire database (optimal overall fit), whereas profile searching identifies the compound that is most similar to the reference profile (best reference fit). Conducting a profile search using biospectra of clotrimazole (1) (Fig. 2) as entry point revealed three biospectra of greatest similarity in the entire 1,567-compound database: tioconazole (2) with a similarity (SP) rating of 0.79 and two other related azoles, 3 and 4, the former with a similarity rating of 0.81 and the latter with a rating of 0.79. Remarkably, this biospectra similarity search identifies molecules that all have antifungal activity and hence identifies molecules with similar biological response capacity.
A second profile search, using the biospectra of tioconazole (2) (Fig. 2) as entry point, revealed the following additional molecules (azoles) with high similarity ranking: 3 [similarity (SP) ranking of 0.92], 4 (0.90), 5 (0.84), 6 (0.86), 7 (0.78), 8 (0.79), and 9 (0.87). The results obtained in these two biospectra profile searches were compared with results obtained by hierarchical clustering using the unweighted pair-group method (UPGMA). The same similarity measure was used for profile searching and hierarchical clustering. Comparison between these independent spectra comparison methods (comparing SP and CCS values) indicates that biospectra spectra similarity parallels structure similarity and biological response similarity. Accordingly, if the spectra profile similarity [profile (SP) similarity values] ranking between molecules is >0.8, then the molecules retrieved in a profile search are structurally and pharmacologically closely aligned. Likewise, hierarchical clustering identifies structurally and pharmacologically closely related molecules, if molecules in a cluster exhibit a confidence in cluster similarity ranking (CCS) of >0.8. The results of profile searching and hierarchical clustering merge, e.g., identify molecules as most similar if profile search ranking or CCS value are >0.8 (see Fig. 4A). Examination of the biospectra similarity of these and other molecules in the database reveals that structure similarity relationships (visual inspection) erode when similarity (SP) rankings or CCS values are <0.80.
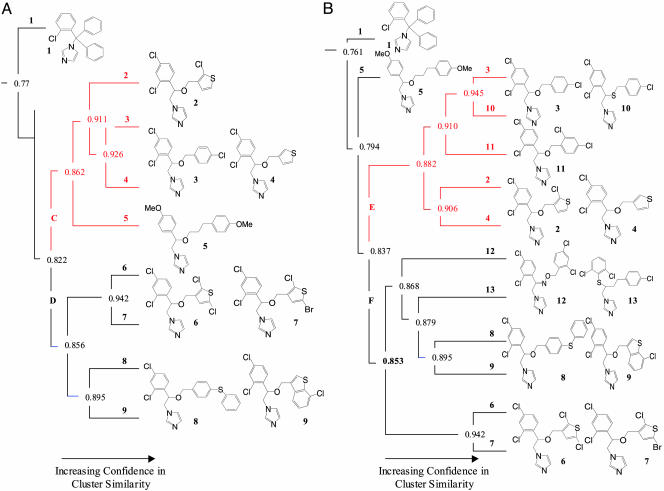
Fig. 4.
Hierarchical clustering of biospectra provides the azole section of this linkage map using 1,567 molecules in hierarchical clustering (see Fig. 3) (A) and the new y-axis dendrogram section containing azole derivatives 1–13 and using 1,571 molecules in hierarchical clustering, resulting from the addition of 10–13 to the database (B).
Hierarchical Clustering of the 1,567 Biospectra. The hierarchical clustering of 1,567 compounds and 92 bioassays is shown in Fig. 3A. Hierarchical clustering of the biospectra of 1,567 molecules listed in Cerep's BioPrint database, using the UPGMA algorithm and cosinus correlation as similarity measurement, provides two SAR dendrograms. The dendrogram depicted on the y axis provides similarity assessments of molecules based on their capacity to interact with the proteome (biospectra similarity). The “biological activity” similarity between these molecules can be ascertained by considering the organization of the y-axis dendrogram. This organization captures the biological response similarity of these molecules. Accordingly, molecules on proximate branches of the y-axis dendrogram in Fig. 3 have closely related biological activity spectra and closely related pharmacology. The x-axis dendrogram, on the other hand, clusters proteins on the basis of ligand-binding site similarity. This property determines the appearance and organization of biological activity spectra. Biological activity spectra are organized on the basis of the capacity of ligand-binding domains to differentiate between ligands (25). The ligand-binding domain similarity between the 92 proteins is objectively ascertained through hierarchical clustering. Hierarchical clustering organizes the layout of the x-axis dendrogram and provides an unbiased mean for biological spectra comparisons between organic molecules.
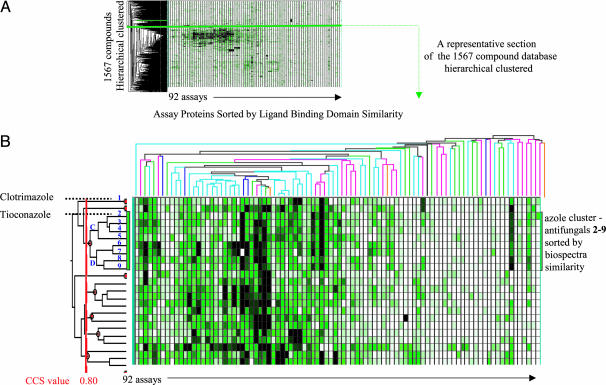
Fig. 3.
Hierchical clustering of 1,567 compounds by using percent inhibition values. (A) A heat map and x-axis and y-axis dendrograms obtained for the complete SAR matrix. Over 140,000 data points with a dimension 92 × 1,567 (assays × molecules) resides in the heat map. (B) A portion (23 molecules in 92 assays) of the heat map containing clotrimazole (1) and tioconazole (2), which were described in Fig. 2. The data are organized by using two classification schemes (dendrograms): one with horizontal orientation on top (x-axis dendrogram) and the other with vertical orientation on the left side (y-axis dendrogram). Receptors appearing in the x-axis dendrogram are color coded according to memberships in designated protein superfamilies: blue, G-protein-coupled receptors; pink, enzymes; green, ion channels; purple, transporters; orange, receptors; black, steroid receptors. Providing an unbiased organization of biospectra of individual molecules (shown on the y axis). The x-axis dendrogram clusters proteins into groups based on interaction profile similarity between proteins by using the percent inhibition values of 1,567 molecules as the measure. Proteins with similar percent inhibition value distribution (similar ligand-binding domain characteristics) appear on proximate branches of the x-axis dendrogram. The y-axis dendrogram, on the other hand, clusters molecules on the basis of similarity ranking obtained by comparing biospectra by using the UPGMA algorithm (molecule comparison). Biospectra similarity between clusters and individual molecules is measured by using confidence in CCS values. Clusters to the right of the red line of the y-axis dendrogram have CCS values >0.80. A similar scoring method for comparing molecular structure based on IR spectra similarity has been described by Varmuza et al. (24).
Assessment of Biological Activity Profile Similarity. The dendrogram flanking the y axis of Fig. 3A shows the binning of 1,567 molecules into clusters based on biospectra similarity. The biospectra similarity between molecules is measured with CCS values resulting from hierarchical clustering. Inspecting the structures of molecules grouped in the y-axis dendrogram in Fig. 3A reveals that the clustering of compounds based on biospectra similarity measurements separates these molecules into distinct structural series and pharmacology classes. Accordingly, molecules on proximate branches of the y-axis dendrogram in Fig. 3A exhibiting a CCS ranking of ≥0.8 are structurally closely related. This empirically determined CCS value is in agreement with biospectra similarity rankings obtained through direct spectra comparison (profile searching) studies shown above. Fig. 3A organizes 73 clusters containing 317 compounds with a score of ≥0.80 confidence range. Each of these clusters identifies structurally and pharmacologically closely related molecules. Thirty-three clusters in this database contain 109 compounds with CCS values ≥0.90, indicating molecules whose biospectra profiles are even more closely aligned.
A portion (23 compounds) of the entire data set, containing clotrimazole (1), tioconazole (2), and related molecules 2–9, is shown in Fig. 3B. Compounds 2–9 are antifungal agents with similar structural motifs (Fig. 4A). The cluster of molecules 2–9 (“azole” cluster) shown in Figs. 3B and 4 is prototypical of the other clusters in the 1,567-compound database that have a confidence in cluster similarity value of ≥0.80. Inspection of Fig. 4A reveals that dendrogram sections C and D resolve structurally less similar molecules into subgroups containing molecules 2–5 and another containing 6–9, and these are further resolved into groupings providing tightly associated structures. The closer the confidence in similarity value approaches unity in these subgroups, the greater the structural similarity. Accordingly, in the azole cluster shown in Fig. 4A, the biospectra of molecules 6 and 7 have a CCS value of 0.94. This is the highest biospectra similarity score within this group of antifungal imidazole derivatives and identifies two structures that differ by only a single atom: chlorine versus a bromine atom in the thiophene moiety of the azole nucleus. A CCS score of only 0.77 was obtained in the biospectra comparison of clotrimazole (molecule 1) and tioconazole (molecule 2). These values indicate that these molecules have significant structural differences.
The fact that high CCS values correlate tightly with molecular structure similarity has obvious implications. A major one is the ability of approaching structure–function studies from either direction (predicting structure from function or predicting function from structure). Experience with physicochemical spectroscopic methods such as NMR, IR, or mass spectrometry teaches that similarity relationships between molecules and spectra are exchangeable and, as a result, chemical structure can be assigned by using spectra as input and spectra can be predicted by using molecular structure information as input.
Predicting Biospectra by Using Molecular Structure Information. For testing the inverse process, using chemical structure information for determining biospectra of molecules, four compounds, 10–13, were added to the 1,567-compound database to assess the forecasting capabilities of this methodology. Compounds 10–13 were omitted in the first analysis, shown in Fig. 4A, because they did not have a complete data set: they lacked percent inhibition values for either bradykinin B2 or the enzyme P55fyn receptors in the 92-protein assay suite. Visual comparison of the structures of 10–13 with molecules 2–9 (shown in Fig. 4) reveals a strong structural similarity between these molecules. Based on this assessment and the observations described above, the biospectra profiles of molecules 10–13 are expected to be more closely aligned to those of molecules 2–9 than any of the other 1,559 remaining compounds in the database. For this reason, the CCS values for 10–13 and compounds 2–9 in the azole cluster are projected to exceed CCS values of 0.8 and hence only one of the 73 clusters with CCS values ≥0.8 in Fig. 4 should include molecules 2–13.
Repeating the hierarchical clustering process as described before but, in this instance, using 1,571 instead of 1,567 molecules (by including molecules 10–13) creates a new complete linkage map. The azole section of the new linkage map shown in Fig. 4B contains, as anticipated, molecules 2–13. Remarkably, not only is the inclusion of 10–13 in the dendrogram of Fig. 4B predictable, but in addition, where molecules 10–13 specifically reside in dendrogram 4B and how closely these molecules are associated with other members of the cluster can also be rationalized. For projecting the evolution of the dendrogram shown Fig. 4B, one matches up molecules 10–13 with those most similar in structure (differing by the least number of atoms) in Fig. 4A (26). Predictions of bioactivity profiles, dendrogram relationships, and CCS values are based on competition experiments involving molecules 10–13 and compounds 2–9. The starting point for these calculations is the dendrogram section in Fig. 4A derived from the y-axis dendrogram shown in Fig. 3B. Displacement of molecules from this (and other) clusters takes place if a new hierarchical cluster is projected to yield a higher CCS value in biospectra similarity competition.
Predicting the Evolution of the Dendrogram in Fig. 4B. Accordingly, to project how the added molecules 10–13 integrate with the molecules residing in Fig. 4A, the following individual structure comparisons were made. Molecules 10 and 11 were first singled out because of their structural similarity to molecule 3 in dendrogram section C of Fig. 4A. In fact, by examining the structures of molecules residing in this section, one recognizes that molecule 3, which is aligned with 4 in Fig. 4A, is actually structurally more similar to molecules 10 and 11 than to molecule 4. This is because molecules 10 and 3 differ by only a single atom; molecule 10 has sulfur instead of an oxygen atom in the ether linkage, whereas molecules 3 and 4 (clustered in Fig. 4A with a CCS of 0.93) have greater structural diversity (a chlorophenyl group in 3 is replaced by a thiophene moiety in 4). Likewise, molecule 11 is structurally more similar to molecule 3 than to molecules 2, 4, 5, and 10. Once more, molecule 11 differs from molecule 3 by only a single atom (a chlorine atom is substituted for a hydrogen atom on the phenyl ring of the benzyl group) and 11, in turn, differs only by two atoms from 10 (oxygen for a sulfur atom and chlorine for a hydrogen atom). These comparisons establish a close structural relationship between molecules 3, 10, and 11. Inspection of the CCS values in Fig. 4A indicates that the displacement of molecule 4 by either molecule 10 or molecule 11 in the cluster pair of 3 and 4 shown in Fig. 4A requires a CCS value that exceeds 0.93 because that value serves as the standard for the association of 3 and 4. Whether 10 or 11 is more closely aligned with 3 is not obvious; however, the assessment of structural similarity predicts a cluster for molecule 3, 10, and 11 (with 4 being displaced) and that one of the molecule pairs 3 and 10 or 3 and 11 has a CCS value >0.93. In addition, because the structure comparisons made above indicate that molecules 10 and 11 are structurally less similar to each other than they are to molecule 3, the prediction would be that molecule 3, 10, and 11 have a dendrogram relationship where one molecule pair, 3 and 10 or 3 and 11, is projected to have a CCS value of ≥0.93 and the remaining, either 10 or 11, a CCS value of ≥0.91 because this value will compete with the CCS value given for the biospectra comparison between structurally less similar molecules (2 and molecules 3 and 4) in Fig. 4A.
Furthermore, by examining the structural similarity of molecule 2 in relation to molecules 3, 4, 5, 10, and 11, one recognizes that molecule 2 is structurally more similar to molecule 4 than to molecules 3, 5, 10, or 11. Again, molecules 2 and 4 differ by only one atom (a chlorine versus a hydrogen atom in the thiophene moiety of molecule 4). Fig. 4A lists a CCS value of 0.91 for these molecules. The CCS value of 0.91 should remain intact. In addition, the CCS value of 0.86 for the clustering of 2, 3, and 4 with 5 dictates the dendrogram relationship between clusters containing molecule pair 2 and 4 and molecules 3, 10, and 11. The CCS value separating these clusters is projected to be ≥0.86 to compete successfully with the value obtained for the biospectra comparison of cluster 3, 10, and 11 with structurally less similar molecule 5 in Fig. 4A.
Comparing dendrogram section C (Fig. 4A) with dendrogram section E (Fig. 4B) indicates that the predictions made above are accurate: not only are the anticipated alignments of molecules in the new azole cluster correct but so too are the CCS values obtained for individual clusters. Accordingly, confidence in cluster similarity values in combination with chemical structure similarity assessments accurately predicts biological activity profiles and dendrogram relationships of previously not-tested molecules (27). This analysis can be extended to molecules 12 and 13, whose structural similarity with molecules 1–11 is less pronounced. Comparing biospectra and CCS values of structurally related molecules (biospectra comparison of molecules with CCS values >0.80 in Fig. 3A), provides a wealth of information for drug design. Determination of differences in CCS values (delta CCS, or differences between other similarity measures) in biospectra competition experiments provides, for example, qualitative and quantitative assessment of “bioequivalence” of structure fragments (bioisosteres).
The experiments described above indicate that the information content stored in biospectra matches the information content of physicochemical descriptors and that biospectra are precise descriptors of molecular properties capable of differentiating molecules on the basis of single atom pair differences. The accurate differentiation of structurally similar molecules is based on standardized descriptions of the capacity of molecules to interact with the proteome and not on information on the putative drug target. This approach in structure–function studies enables consideration of wide response determinants and does not require representation of response as a linear combination of independent relationships. Moreover, simple rules provide accurate projections for establishing response–structure relationships, based on y-axis dendrogram clusters of structurally related molecules in Fig. 3A. As a result, biological spectra analysis permits precise assessment of molecular properties relevant to structure–function relationships not only for individual molecules but also for clusters of compounds embedded in chemical series.
Summary. Comparing biological activity profiles of molecules assayed at a single ligand concentration provides an unbiased means for establishing quantitative relationships between chemical structure and broad biological effects without using information on affinities for putative drug targets. Being able to measure and quantify structural similarity of organic molecules by using biological molecular property descriptors has significant implications in many areas of science. For medicinal chemists, understanding the biological consequence of structural changes in a molecular series is essential to the drug discovery process. For combinatorial chemists, understanding the relationship between new synthetic libraries and existing molecules is crucial. Herein we have shown a paradigm that uses the unique structural properties of the proteome to order organic molecules. This method is flexible and is capable of incorporating diverse databases of molecules and proteins. Biological spectra analysis (biological spectroscopy) has significant implications for systems biology and drug discovery. Biological activity profiles are precise indicators not only of molecular properties such as molecular structure but also of the biological response capacity of molecules.
Supplementary Material
Author contributions: A.F.F., W.T.L., P.F.T., and R.A.V. designed research; A.F.F., W.T.L., P.F.T., and R.A.V. performed research; A.F.F., W.T.L., P.F.T., and R.A.V. contributed new reagents/analytic tools; A.F.F., W.T.L., P.F.T., and R.A.V. analyzed data; and A.F.F. and R.A.V. wrote the paper.
Abbreviations: SAR, structure–activity relationship; SP, similarity to profile; CCS, confidence in cluster similarity.
Footnotes
BioPrint is a commercial database, which is available to subscribers and contains data on >2,500 drugs and drug-like molecules.
References
- 1.Xue, L. & Bajorath, J. (2000) Comb. Chem. High Throughput Screen. 3, 363-372. [DOI] [PubMed] [Google Scholar]
- 2.Blow, D. M. (1976) Acc. Chem. Res. 9, 145-152. [Google Scholar]
- 3.Huber, R. & Bode, W. (1978) Acc. Chem. Res. 11, 114-122. [Google Scholar]
- 4.Yoshida, Y. & Aoyama, Y. (1987) Biochem. Pharmacol. 36, 229-235. [DOI] [PubMed] [Google Scholar]
- 5.Mottola, D. M., Laiter, S., Watts, V. J., Tropsha, A., Wyrick, S. D., Nichols, D. E. & Mailman, R. B. (1996) J. Med. Chem. 39, 285-296. [DOI] [PubMed] [Google Scholar]
- 6.Csete, M. E. & Doyle, J. C. (2002) Science 295, 1664-1669. [DOI] [PubMed] [Google Scholar]
- 7.Zou, X., Sun, Y. & Kuntz, I. D. (1999) J. Am. Chem. Soc. 121, 8033-8043. [Google Scholar]
- 8.Keinan, S. & Avnir, D. (1998) J. Am. Chem. Soc. 120, 6152-6159. [Google Scholar]
- 9.Poroikov, V. V., Filimonov, D. A., Ihlenfeldt, W.-D., Gloriozova, T. A., Lagunin, A. A., Borodina, Y. V., Stepanchikova, A. V. & Nicklaus, M. C. (2003) J. Chem. Inf. Comput. Sci. 43, 228-236. [DOI] [PubMed] [Google Scholar]
- 10.Gillet, V. J., Willett, P. & Bradshaw, J. (1998) J. Chem. Inf. Comput. Sci. 38, 165-179. [DOI] [PubMed] [Google Scholar]
- 11.Panchenko, A. R., Kondrashov, F. & Bryant, S. (2004) Protein Sci. 13, 884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., LeTrong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 13.Ji, H.-T., Zhang, W.-N., Zhou, Y.-J., Lu, J.-G., Zhu, J. & Zhu, J. (1998) Shengwu Huaxue Yu Shengwu Wuli Xuebao 30, 419-426. [Google Scholar]
- 14.Graham, S. E. & Peterson, J. A. (1999) Arch. Biochem. Biophys. 369, 24-29. [DOI] [PubMed] [Google Scholar]
- 15.Ji, H., Zhang, W., Zhou, Y., Zhang, M., Zhu, J., Song, Y., Lü, J. & Zhu, J. (2000) J. Med. Chem. 43, 2493-2505. [DOI] [PubMed] [Google Scholar]
- 16.Swindells, M. B. & Overington, J. P. (2002) Drug Discov. Today 7, 516-521. [DOI] [PubMed] [Google Scholar]
- 17.Mirzabekov, A. & Kolchinsky, A. (2002) Curr. Opin. Chem. Biol. 6, 70-75. [DOI] [PubMed] [Google Scholar]
- 18.Pazos, F. & Valencia, A. (2001) Protein Eng. 14, 609-614. [DOI] [PubMed] [Google Scholar]
- 19.Moore, M. N. (2002) Aquat. Toxicol. 59, 1-15. [DOI] [PubMed] [Google Scholar]
- 20.Horvath, D. & Jeandenans, C. (2003) J. Chem. Inf. Comput. Sci. 43, 680-690. [DOI] [PubMed] [Google Scholar]
- 21.Blower, P. E., Yang, C., Fligner, M. A., Verducci, J. S., Yu, L., Richman, S. & Weinstein, J. N. (2002) Pharmacogenomics J. 2, 259-271. [DOI] [PubMed] [Google Scholar]
- 22.Lewi, P. J. (1976) Arzneim.-Forsch. 26, 1295-1300. [PubMed] [Google Scholar]
- 23.Krejsa, C. M., Horvath, D., Rogalski, S. L., Penzotti, J. E., Mao, B., Barbosa, F. & Migeon, J. C. (2003) Curr. Opin. Drug Discov. Dev. 6, 470-480. [PubMed] [Google Scholar]
- 24.Varmuza, K., Karlovits, M. & Demuth, W. (2003) Anal. Chim. Acta 490, 313-324. [Google Scholar]
- 25.Kauvar, L. M., Higgins, D. L., Villar, H. O., Sportsman, J. R., Engqvist-Goldstein, A., Bukar, R., Bauer, K. E., Dilley, H. & Rocke, D. M. (1995) Chem. Biol. 2, 107-118. [DOI] [PubMed] [Google Scholar]
- 26.Barbosa, F. & Horvath, D. (2004) Curr. Top. Med. Chem. 4, 589-600. [DOI] [PubMed] [Google Scholar]
- 27.Raffa, R. B. (1999) Life Sci. 65, 967-980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.