Abstract
We have devised and tested a new strategy for selectively delivering molecules to tumor cells. Cellular association of polyarginine-based, cell-penetrating peptides (CPPs) is effectively blocked when they are fused to an inhibitory domain made up of negatively charged residues. We call these fusions activatable CPPs (ACPPs) because cleavage of the linker between the polycationic and polyanionic domains, typically by a protease, releases the CPP portion and its attached cargo to bind to and enter cells. Association with cultured cells typically increases 10-fold or more upon linker cleavage. In mice xenografted with human tumor cells secreting matrix metalloproteinases 2 and 9, ACPPs bearing a far-red-fluorescent cargo show in vivo contrast ratios of 2-3 and a 3.1-fold increase in standard uptake value for tumors relative to contralateral normal tissue or control peptides with scrambled linkers. Ex vivo slices of freshly resected human squamous cell carcinomas give similar or better contrast ratios. Because CPPs are known to import a wide variety of nonoptical contrast and therapeutic agents, ACPPs offer a general strategy toward imaging and treating disease processes associated with linker-cleaving activities such as extracellular proteases.
Keywords: cancer, molecular imaging, polycation, transduction
Molecular imaging and therapy in patients would greatly benefit from generic, rational mechanisms to target contrast agents and therapeutic drugs to diseased tissues, especially tumors (1). Currently, the main strategies are based on antibodies against surface markers or ligands for receptors preferentially expressed in the target tissue (2). Although antibodies have occasionally been successful in targeting tumors (3), their irreducible bulk hinders penetration of solid tumors and excretion of unbound reagent (4), and elaborate reengineering is required to minimize immunogenicity (5, 6). A few small molecule ligands (2 kDa or less) for endogenous receptors have been preliminarily explored, but robust tumor specificity is rare or nonexistent (4). A fundamental limitation of simple antibody or ligand binding is the lack of amplification, where each target molecule (typically of low abundance) can bind at most one probe. Some amplification can be achieved by incorporating the probe into polymers or nanoparticles, but the increase in bulk worsens access to diseased tissue and removal from healthy organs. None of these approaches help get drugs across the plasma membrane into the cytoplasm and nucleus of diseased cells, the most desirable loci for modifying signal transduction or triggering cell death.
Certain polycationic sequences [variously dubbed cell-penetrating peptides (CPPs), membrane-translocating sequences, or protein transduction domains] can bring covalently attached payloads into mammalian cells without requiring specific receptors. CPPs were first discovered within a domain from Antennapedia homeobox protein and the tat protein from HIV-1 (7, 8). A variety of multicationic oligomers, including guanidinium-rich sequences, as simple as 6-12 consecutive arginines are now known to be equally or more effective (9-11). D-Amino acids are at least as good as natural L-amino acids and possibly better because the unnatural isomers resist proteolysis (10-12). Cargoes ranging in size from metal chelates and fluorescent dyes (13, 14) to iron oxide nanoparticles (15) and liposomes (16) can be imported, although the detailed mechanisms and subcellular localizations remain poorly understood and may differ, depending on cargo size, cell type, CPP sequence, and other experimental variables (17, 18). Initial attachment of the polycations to the cell surface is avid, rapid, and probably mediated by electrostatic attraction for anionic phospholipids and glycosaminoglycans. Much of the subsequent internalization probably occurs by endocytosis, because delivery of bioactive cargoes to the cytosol and nucleus can be enhanced by inclusion of sequences known for acidification-dependent disruption of endosomes (19, 20).
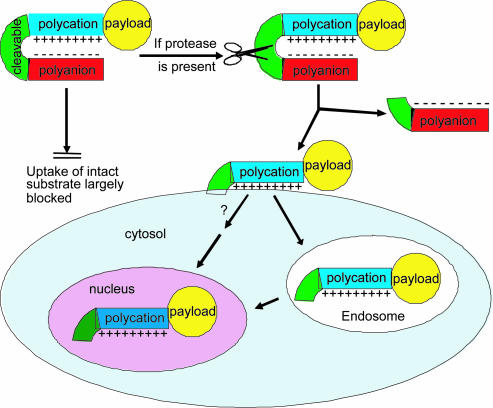
We now demonstrate a generic targeting mechanism based on selective local unleashing of CPPs, as schematized in Fig. 1. Cellular uptake of CPPs can be largely blocked by fusing them by means of cleavable linkers to polyanionic sequences, which neutralize the polycations by forming intramolecular hairpins of ≈2-3 kDa. We call such constructs activatable CPPs (ACPPs), because cleavage of the linkers dissociates the inhibitory polyanions, releasing the polycationic peptides and their cargo to attach to and enter cells. The mechanism (Fig. 1) is a flexible, modular, amplifying strategy to concentrate imaging and therapeutic agents on and within cells in the immediate vicinity of extracellular cleavage activities, such as matrix metalloproteinases (MMPs) in tumors. We chose MMP-2 and MMP-9 as our primary initial targets because they are the best characterized proteases overexpressed by tumors (21). Currently, at least 26 members of the MMP family have been identified. They play a crucial role in extracellular matrix degradation, tissue invasion, and metastasis (21-26).
Fig. 1.
Schematic diagram of activatable CPPs. Cellular uptake induced by a cationic peptide is blocked by a short stretch of acidic residues attached by a cleavable linker. Once the linker is cleaved, the acidic inhibitory domain drifts away, and the cationic CPP is free to carry its cargo into cells.
Materials and Methods
Peptide Synthesis and Fluorophore Labeling. Peptides were synthesized on an automatic peptide synthesizer by using standard protocols for fluorenylmethoxycarbonyl solid-phase synthesis. Detailed information regarding peptide synthesis, fluorophore labeling, and poly(ethylene glycol) attachment (PEGylation) can be found in the supporting information, which is published on the PNAS web site.
Peptide Cleaved by MMP-2 (PLGLAG). MMP-2 proenzyme (5 μg in 80 μl of 50 mM Tris·HCl buffer) was activated with 2.5 mM 4-aminophenylmercuric acetate at 37°C for 2 h. Afterward, we added 32 μl of 0.5 mM peptide stock solution and incubated the mixture for 1 hr at room temperature. Enzyme cleavage progress was monitored by HPLC. The HPLC chromatograms showed that near complete cleavage was accomplished after 30 min of incubation. The new peak was collected, and its mass was determined by mass spectroscopy. The mass spectrum indicated that the enzyme was cut between glycine and leucine residues of the MMP-2 substrates as predicted, giving products such as NH2-eeeeee-ahx-PLG and LAG-rrrrrrrrr-ahx-c(Fl)-CONH2.
FACS Analysis and Microscopy. Jurkat cells were cultured in RPMI medium 1640 plus 10% (vol/vol) FBS to a density of 0.5-1 × 106 cells per ml. The media was refreshed 1 day before the assay of ACPPs. Cells were washed with Hanks' balanced salt solution (HBSS) buffer three times, resuspended in HBSS at 0.5-1 × 106 cells per ml, stained with 1 μM peptide in HBSS at room temperature for 10 min, washed three times with cold HBSS, and analyzed by flow cytometry at 530-nm emission for fluorescein-labeled peptides or at 675 nm for Cy5-labeled peptides. We collected 10,000 events from cells judged to be healthy by their forward and side scatter. Peptide association with HT-1080 cells was similarly quantified by flow cytometry after release from adherence with trypsin. For microscopic imaging, HT-1080 cells grown to 70% confluency were washed with HBSS three times, stained with 1.25 μM peptide and 1 μg/ml Hoechst 33258 (a nuclear stain), rinsed twice, trypsinized, replated on polylysine-coated dishes, and imaged for Cy5 content (excitation, 625-645 nm; emission, 665-695 nm) and Hoechst 33258 (excitation, 375-385 nm; emission, 420-460 nm).
Xenografts in Mice. Nude mice (age, 4-6 weeks) were injected s.c. with ≈106 HT-1080 cells. Once the tumors had reached ≈5-7 mm in size (typically 1-2 weeks later), animals were anesthetized with 100 mg/kg ketamine and 5 mg/kg midazolam), weighed, and injected with ≈100 μl of 60 μM peptide through the tail vein. Animals were then imaged at various times by using a Nikon f/1.2 camera lens in front of a cooled charge-coupled device camera (SenSys, Photometrics, Tucson, AZ). For longer lasting imaging studies, animals were allowed to wake up after 2 h of anesthesia and were reanesthetized at ≈4 and 6 hr for further data collection. Plasma half-lives were determined by the decrease in fluorescence intensity of ≈5-μl blood samples withdrawn periodically into heparinized capillaries. After imaging was ended, animals were killed with halothane, and organs of interest were harvested and weighed. For frozen sectioning, tissues were added to OCT cryopreservative and frozen on dry ice and hexane. Samples were stored at -80°C and cut into 5-μm sections at -20°C by using a cryotome. Cy5 fluorescence was imaged as described above. To measure standardized uptake values (SUVs), 30 mg of each tissue was added to 100 μl of a buffered 1% SDS mixture (pH 7.6) and protease inhibitor mixture (Roche Diagnostics). The tissue was then homogenized, heated to 70°C for 15 min, microwaved for 15 sec, centrifuged at 20,500 × g for 15 min, then imaged on the same system used for whole mice. Two sets of standards (liver and kidney) were used to calibrate fluorescence intensity in terms of peptide concentration. From this calibration, the quantity of peptide in 30 mg of tissue for each organ was calculated. SUVs were calculated as the molality of peptide in the tissue divided by the total injected dose as mol/kg of body weight.
Squamous Cell Carcinoma Specimens. Human squamous cell carcinoma specimens from planned resections of neoplasms were collected postoperatively according to a protocol with institutional review board approval. The specimens were in ice-cold normal saline for 30 min during transport back to the laboratory, where they were cut by hand to ≈1-mm-thick slices, added to 1 ml of 1 μM peptide for 15 min at room temperature, rinsed five times for 2 min in 1 ml of HBSS, cryosectioned, and imaged as described above.
Results
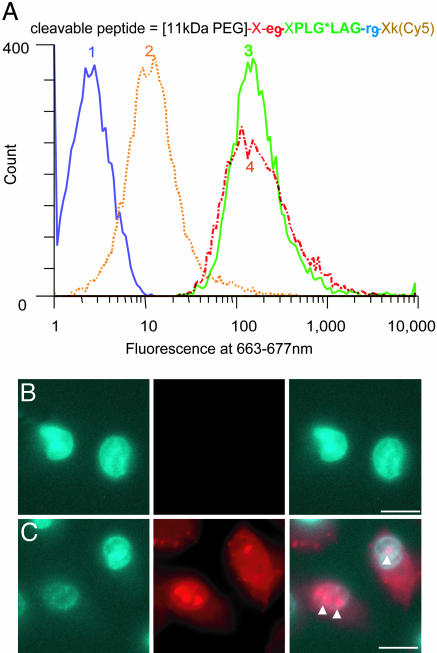
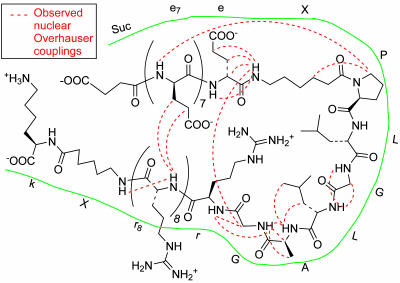
Until Cleaved Off, Polyanionic Sequences Inhibit Association of CPPs with Cells. Given that the initial binding of CPPs to cells is electrostatic, we asked whether association with cells could be prevented by appending polyanionic sequences to give the polycations intramolecular diversions. Fluorescently labeled peptides were synthesized with nine arginine residues fused by means of cleavable linkers to six to nine consecutive acidic residues, usually glutamate. We incubated these peptides, either intact or with linkers precleaved, with Jurkat lymphocytes or HT-1080 fibrosarcoma cells and assessed cell fluorescence by flow cytometry and fluorescence microscopy of the live unfixed cells after washing away unbound peptides. Fig. 2 shows results with HT-1080 cells and an ACPP cleavable by MMP-2. The intact peptide showed 18-fold less uptake than the equimolar mixture of the two fragments resulting from linker cleavage, which in turn was similar to a control CPP with only the polycation. The flow cytometric histograms showed that fluorescence on or in healthy cells was unimodal and reasonably homogeneous (Fig. 2 A). Single cell microscopy (Fig. 2B) confirmed that cargo uptake was far greater after linker cleavage and indicated that a significant fraction reached the nucleus, as judged by accumulation of fluorescence in the nucleoli, similar to results previously reported for polycation-mediated transduction (17). Analogous cleavage-dependent association with cells was observed with a variety of ACPPs containing different numbers of arginine residues, different polyanionic sequences, and linkers cleavable by a variety of proteases, including enterokinase, MMP-2, MMP-9, and urokinase plasminogen activator, or even by simple reduction of a disulfide bond (Table 1). In the best case, cell labeling increased >100-fold when the polyanion was cut off from the polycation. Both the arginine residues and the acidic residues could be D-amino acids, as desirable to restrict in vivo proteolysis to the central linker between the two domains. Greater contrast was obtained when the polycationic, not the polyanionic, region was closer to the C terminus. We hypothesize that this preference is because the new amino terminus created by proteolytic cleavage would reinforce the polycationic charge, whereas, if the polycation is at the N terminus, proteolysis would append a negatively charged carboxylate to the polycation. Cleavage-dependent contrast was equally observable with fluorescein or the far-red fluorophore Cy5 as cargo and with or without a PEG tail (Table 1). Such PEGylation increases solubility and slows in vivo excretion but is not necessary to block CPP activity. ACPPs Adopt a Hairpin Conformation Before Cleavage. Polyanion inhibition of polycation uptake would be most easily understood if the oppositely charged segments zippered together as shown in Fig. 1. Direct evidence for such a hairpin structure was obtained by homonuclear two-dimensional NMR analysis (see supporting information for methods). Fig. 3 shows the observed nuclear Overhauser effect correlations, which reflect proton-proton proximities. The observed short-range couplings within the PLGLAG linker indicate a turn conformation (see supporting information for more detail). In addition, the numerous nuclear Overhauser effects between the strings of D-arginine and D-glutamate clearly indicate pairings that would stabilize the hairpin turn. Taken together, the data indicate that the presence of a hairpin structure, although they are not sufficient to define a complete atomic-level structure owing to chemical shift overlap.
Fig. 2.
Association of ACPPs with live HT-1080 cells depends on cleavage by MMP-2, as demonstrated by FACS analysis (A) and microscopy (B and C). (A) Trace 1 (blue) shows untreated cells. Traces 2 (orange) and 3 (green) show cells incubated for 10 min with 1 μM uncleaved or precleaved peptide, respectively. Cells incubated with 1 μMr9k(Cy5) are shown in red (Trace 4). (B) HT-1080 cells were incubated with 1 μg/ml Hoechst 33258 (Left) and 1.25 μM uncleaved peptide (Center) and imaged at Hoechst or Cy5 wavelengths (overlaid at Right). (C) Results from a similar experiment with cleaved peptide. The arrowheads indicate possible nucleoli.
Table 1. Effect of different linkers and acidic inhibitory domains on ACPP association with Jurkat and HT-1030 cells assayed by flow cytometry.
| Sequence | Uptake before cleavage | Uptake after cleavage (reagent) | Increase caused by cleavage |
|---|---|---|---|
| EEEEEDDDDK*AXRRRRRRRRRXC(Fl) | 0.18 | 2.4 (EK) | 13 |
| EEEEEDDDDK*ARRRRRRRRRXC(Fl) | 0.07 | 1.2 (EK) | 17 |
| EDDDDK*AXRRRRRRRRRXC(Fl) | 0.30 | 2.3 (EK) | 8 |
| EEDDDDK*ARXRRXRRXRRXRRXC(Fl) | 0.015 | 0.11 (EK) | 7 |
| DDDDDDK*ARRRRRRRRRXC(Fl) | 0.05 | 0.77 (EK) | 16 |
| EEDDDDK*AXrrrrrrrrrXC(Fl) | 0.07 | 1.2 (EK) | 17 |
| eeeeeeXPLG*LAGrrrrrrrrrXc(Fl) 10 min | 0.086, 0.034 | 1.3, 1.3 (MMP-2) | 16, 39 |
| eeeeeeXPLG*LAGrrrrrrrrrXc(Fl) 60 min | 0.11 | 2.1 (MMP-2) | 19 |
| UeeeeeeeeXPLG*LAGrrrrrrrrrXk(Fl) | 0.006 | 0.74 (MMP-2) | 123 |
| eeeeeeXPLG*LAGrrrrrrrrrXc(Cy5) | nc | nc (MMP-2) | 36 |
| UeeeeeeXPLG*LAGrrrrrrrrrXc(Cy5) | nc | nc (MMP-2) | 20 |
| UeeeeeeeeXPLG*LAGrrrrrrrrrXk(Cy5.5) | nc | nc (MMP-2) | 17 |
| [11-kDa PEG]XeeeeeeeeeXPLG*LAGrrrrrrrrrXk(Cy5) | 0.012, 0.10 | 0.82, 2.1 (MMP-2) | 68, 21 |
| 0.10 | 1.87 (MMP-9) | 18 | |
| [11-kDa PEG]XeeeeeeeeeXLALGPGrrrrrrrrrXk(Cy5)† | 0.019 | 0.021 (MMP-2) | 1.13 |
| — | 0.020 (MMP-9) | 1.07 | |
| Fl-XrrrrrrrrrXPLG*LAGeeeeeeee-βAla | 0.004 | 0.06 (MMP-2) | 16 |
| Fl-XrrrrrrrrrXSGRS*Aeeeeeeee-βAla | 0.012 | 0.05 (uPA) | 4 |
| eeeeeeXSGRS*AXrrrrrrrrrXc(Cy5) | nc | nc (uPA) | 11 |
| Fl-rrrrrrrrrc-*-ceeeeee‡ | 0.092 | 0.72 (reduction) | 8 |
For sequences, lowercase characters indicate d-amino acid. All peptides were amidated at C terminus. Values represent the results of triplicate experiments performed on the same day. Some entries have two values because the triplicate experiments were repeated on another day.
Cleavage site; U, succinoyl; X, 6-aminohexanoyl; Fl, fluorescein. Uptake before and after cleavage was measured by FACS, normalized to Fl-GGRRRRRRRRRR or rrrrrrrrrk(Cy5), except for some measurements not calibrated (nc) with respect to either reference peptide. EK, enterokinase; uPA, urokinase plasminogen activator.
This scrambled control should be uncleavable, so the rightmost column refers to increase due to enzyme exposure rather than cleavage.
Disulfide-linked. In “ceeeeee,” the N terminus is d-glu and the amidated C terminus is d-cys.
Fig. 3.
Nuclear Overhauser effects observed in two-dimensional NMR of a simple ACPP, succinyl-e8-XPLGLAG-r9-Xk, where X denotes 6-aminohexanoyl. Dashed red lines indicate observed nuclear Overhauser effects, and the green line highlights the peptide outline for clarity.
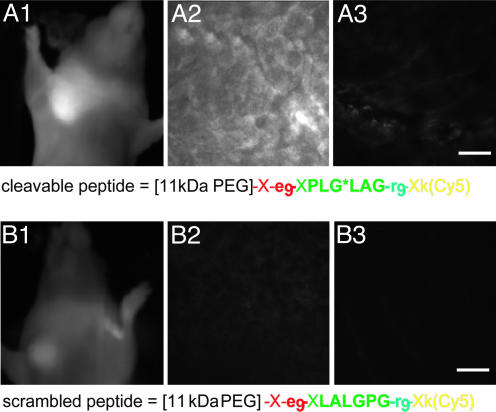
MMP-2 Cleavable ACPPs Concentrate in Human Tumors Xenografted into Mice. We next tested whether ACPPs could light up protease-expressing human tumor xenografts in whole mice. We chose HT-1080 tumors in the axilla of nude mice because these tumors express both MMP-2 and MMP-9 and have been used to test other MMP-2 cleavable contrast agents (22, 26). Adding a PEG tail to the peptide proved helpful to prevent excessively rapid excretion; PEGs of 5, 11, and 21 kDa gave plasma half-lives of ≈5, 15, and 38 min, respectively, consistent with trends reported in ref. 27. Anesthetized mice were injected through the tail vein with either an MMP-2 cleavable ACPP, an isomeric scrambled version verified not to be a substrate for MMP-2 or MMP-9, or an all-D-amino acid version. All peptides had Cy5 attached to permit in vivo imaging of the far-red fluorescence through the skin. Fig. 4A1 shows that the tumor is the brightest fluorescence visible in the live animal injected with the MMP-2-cleavable ACPP, whereas Fig. 4B1 shows much less tumor contrast from a different animal injected with the scrambled analog. Similar cleavage-dependent contrast was seen in frozen sections at higher magnifications (Fig. 4 A2, A3, B2, and B3). To quantitate the results, we measured the contrast index defined as (fluorescence intensity of tumor - autofluorescence)/(fluorescence of normal contralateral region - autofluorescence). This index was 2.1 ± 0.17 (mean ± SE, n = 6) for the cleavable ACPP, which was modestly but significantly higher (P < 0.02, two-tailed t test) than the values obtained for both the scrambled isomer (1.3 ± 0.16, n = 2) and the all-D-amino acid control (1.5 ± 0.11, n = 4). The latter values may differ from 1.0 because of the phenomenon of enhanced permeability and retention, whereby macromolecules passively accumulate in tumors because their vasculature is leakier than that of healthy tissue (28). Nevertheless, the amount of cleavable ACPP that accumulates in the tumor is significantly more than can be accounted for by the enhanced-permeability-and-retention effect, arguing for local unmasking of the CPP by enzymes secreted by the tumor.
Fig. 4.
HT-1080 tumor xenografts visualized with activatable CPPs. HT-1080 tumors were implanted into the mammary fat pad of nude mice and allowed to grow until they reached 5-7 mm in diameter. (A1) A live anesthetized animal imaged 50 min after injection with 6 nmol of cleavable peptide. (A2 and A3) Tumor and muscle histology from a different animal killed 30 min after injection. (B1-B3) A similar experiment with the scrambled peptide. (Scale bars, 30 μm.)
Although Fig. 4A1 shows that tumors become visible in intact live animals, such fluorescence images are highly biased in favor of superficial tissues, skin > s.c. tumors > deep organs. To measure the true distribution of the peptides unbiased by anatomical depth, postmortem tissue samples from different organs were homogenized in detergent to release the labeled probe, clarified by centrifugation, and quantified by Cy5 fluorescence relative to tissue standards spiked with known amounts of dye. SUVs, defined as (moles of recovered peptide/weight of tissue sample)/(moles injected into animal/total body weight), are compiled in Table 2, comparing a cleavable ACPP with its all-D-amino acid control. Although the kidney and liver have the highest absolute SUVs, as typical for peptides, the tumors gave a higher ratio of SUVs between the cleavable and control peptide: 3.1. Also, of the tissues with appreciable uptake, only in the tumors did the difference between the two peptides attain statistical significance (P < 0.05, two-tailed t test).
Table 2. SUVs (mean ± SD) 1 hr after injection of peptide into mice.
| Organ | Cleavable peptide | All-d-amino acid control peptide |
|---|---|---|
| Skin | 0.6 ± 0.4 | 0.2 ± 0.3 |
| Muscle | 0.2† | 0.0† |
| Brain | 0.2† | 0.1 ± 0.1 |
| Intestine | 0.6 ± 0.3 | 0.4 ± 0.4 |
| Liver | 1.2 ± 0.5 | 0.6 ± 0.1 |
| Spleen | 0.2† | 0.0† |
| Colon | 0.9 ± 0.3 | 0.7 ± 0.2 |
| Lung | 0.1 ± 0.1 | 0.0 ± 0.1 |
| Plasma | 1.4 ± 0.1 | 1.7 ± 0.2 |
| Salivary | 0.4 ± 0.1 | 0.3 ± 0.1 |
| Kidney | 5.3 ± 0.6 | 3.2 ± 0.9 |
| Heart | 0.2 ± 0.1 | 0.0† |
| Pancreas | 0.7 ± 0.2 | 0.2† |
| Tumors (P < 0.05) | 1.3 ± 0.4 | 0.4 ± 0.3 |
The cleavable peptide was [11-kDa PEG]-X-e9-XPLG*LAG-r9-Xk(Cy5), and the uncleavable peptide was [11-kDa PEG]-X-e9-Xplglag-r9-Xk(Cy5).
Standard deviation was <0.05 in these samples.
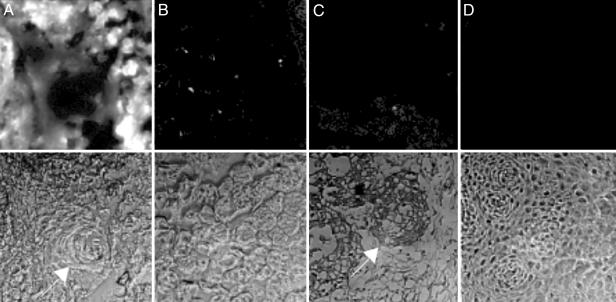
ACPPs Light Up Human Squamous Cell Carcinomas. Although human tumor cell lines xenografted into immunodeficient mice are popular cancer models, they fail to mimic many aspects of real human tumors. To get a preliminary indication whether ACPPs would work on clinically relevant neoplasms, we applied ACPPs to coarse sections cut from tissue freshly resected from patients undergoing surgery for squamous cell carcinoma of the aerodigestive tract. These surgical samples contained both neoplastic and normal tissue, distinguishable by cell morphology and histological staining. The ACPP, whose covalently attached PEG was reduced to 5 kDa to facilitate diffusion, consistently stained tumor tissue more brightly than normal tissue, whereas the scrambled peptide or the ACPP coadministered with N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (a Zn2+ chelator and broad-spectrum MMP inhibitor), showed no such consistent pattern. In Fig. 5, which shows our best result so far, A Upper-D Upper are Cy5 fluorescence images displayed at a uniform gain, whereas A Lower-D Lower are transmitted light views of the same fields. The squamous cell carcinoma tumor tissue exposed to cleavable peptide (Fig. 5A) was much more fluorescent than normal tissue exposed to cleavable peptide (Fig. 5B) or either tissue exposed to scrambled peptide (Fig. 5 C and D). Contrast, defined as (tumor tissue fluorescence - autofluorescence)/(normal tissue fluorescence - autofluorescence), was almost eight in this example. Contrast tended to be greatest where the tumor tissue had a high histologic grade of malignancy. An example in Fig. 5A is that the keratin pearl, characteristic of differentiated squamous epithelium (29), was less fluorescent than the surrounding tumor. The contrast averaged 2.7 ± 0.2 (mean ± SD) from two patients with relatively differentiated oral cavity/oropharynx tumors (low to moderate histologic grade of malignancy), whereas two high-grade laryngeal tumors gave more contrast, 6.5 ± 3.4. Also, lymphocytic granulation tissue was nearly as bright as the tumors themselves, possibly because of the release of MMPs from lymphocytes. Normal tissue immediately adjacent to tumor tissue was noticeably brighter than more remote normal tissue, possibly because of the presence of immune cells or to diffusion of the soluble proteases.
Fig. 5.
ACPP staining of surgically resected human squamous cell carcinoma tissue. Fresh tumor tissue was sliced in 1-mm slices and incubated in 1 μM cleavable (A) or uncleavable (B) peptide for 15 min, washed, and frozen. Sections (5 μm) were taken for fluorescence microscopy by using a 10× objective, and tissue type was verified by hematoxylin/eosin stain. (A) The arrow indicates a differentiated keratin pearl. As a control, histologically normal tissue from the same patient was treated similarly with MMP-2 cleavable peptide (C) or scrambled peptide (D). (C) The arrow indicates a ring of invading tumor cells.
Discussion
We believe the selective activation of CPPs has the following advantages: (i) It should be adaptable to a wide variety of imaging and therapeutic modalities, including radioactivity, because the payload or cargo need not have any particular spectroscopic properties. CPP-mediated uptake has already been demonstrated with gamma-ray emitters and MRI contrast agents as well as potential therapeutic agents (30). Close integration between imaging and therapy would thus be facilitated. Demonstration of ACPPs with nonoptical cargoes will be an important next step. (ii) Catalytic amplification is inherent; i.e., each protease molecule can cleave multiple substrate molecules, whereas each epitope can only bind one antibody at a time. (iii) ACPPs help deliver the cargo not just to the surface of the target cell but inside and to the nucleus, which is important for therapeutic payloads if not for in vivo imaging. (iv) Molecular masses can be varied over a wide range from quite small (≈18 aa or ≈2 kDa) up to nanoparticles of several nanometers in diameter (15, 16, 31). Depending on whether polymers are appended to the polyanionic versus polycationic portion, one can choose whether they are discarded or retained after linker cleavage. Excessive molecular mass has the disadvantage of decreasing penetration into solid tumors, particularly when they have high interstitial fluid pressure (29). (v) The highly modular substrates are synthesized by standard methods of peptide synthesis and bioconjugation, without requiring fermentation or high-level expression systems, yet they contain enormous scope for rational or combinatorial variation. (vi) The high content of D-amino acids should reduce immunogenicity, although this remains to be tested experimentally. Other guanidinium-decorated nonpeptidic backbones, such as carbamates and peptoids, are known to be competent for cell uptake (32) and should be modulatable in analogy to the peptides discussed above. (vii) Extracellular proteases are mechanistically important in cancer (33), particularly in angiogenesis and metastasis, unlike many tumor antigens of unknown function. In principle, tumor cells that try to become resistant by down-regulating their proteases are likely to become less aggressive and metastatic. Also, there is some hope that multiple subtypes of cancers may converge upon up-regulating a relatively limited repertoire of proteases, giving each successful substrate a wider range of clinical indications. (viii) Proteases that are or can become extracellular are crucial to many other disease processes, including thrombosis, congestive heart failure, inflammation, neurodegeneration, and infectious pathogens (34-37). We are not even limited to proteases: Any conditions that sever the vetoing polyanion from the polycation (e.g., agents that reduce disulfide bonds) offer potential mechanisms for localization.
Our initial tests in vivo have all been with substrates for soluble proteases, such as MMP-2 and MMP-9, mainly because these MMPs have well established roles in metastasis and angiogenesis, clear substrate preferences, and commercial sources for in vitro testing. However, soluble proteases have the potential disadvantage of gradually leaking from the tumor into the general circulation, where they would contribute to background signal and reduced contrast. MMPs have been detected in the plasma and urine of cancer patients at levels that show positive correlations with the severity of metastatic disease (23), although the relative enzyme activities in tumors versus blood do not seem to be known. To circumvent diffusion of soluble MMPs, we are building substrates for membrane-bound MMPs, such as MT1-MMP (24, 25). Other membrane-bound proteases including members of the ADAM (a disintegrin and metalloprotease) family (38) are potential alternatives.
Up to now, we have avoided incorporating additional contrast mechanisms, such as fluorescence dequenching (22, 26) or enhanced permeability and retention of adequately large polymers (28), within tumors with leaky vasculature. When maximum contrast and sensitivity become more important than mechanistic purity, it may be advantageous to attach ACPPs to nanoparticles or large polymers to harness the enhanced-permeability-and-retention effect. In the case of fluorescence, crowding fluorophores together on a polymer or nanoparticle (22, 26, 39, 40) or including a quencher on the end of the polyanion might improve contrast by suppressing fluorescence of the uncleaved substrate.
We chose far-red fluorescence as our first imaging modality for three reasons: the cyanine dyes are stable and easy to conjugate, the imaging equipment is relatively simple to use and cheap, and its spatial resolution spans the full range from subcellular to whole animal. In mice, fluorescence imaging can reach a significant fraction of the intact animal, especially when aided by tomographic techniques (40). In larger animals and in patients, the few-millimeters-deep penetration restricts the utility of fluorescence to (i) the most superficial dermatological tumors, (ii) the retina, (iii) tumors near the surface of a body cavity accessible by endoscopy (41), and (iv) the margins of a surgical resection. Real-time molecular imaging of the margins of a resection while the patient is still on the operating table would be of great value to the surgeon to decide whether any invasive carcinoma tissue remained lurking at or just beyond the tissue just removed. Instrumentation for infrared image-guided surgery has been described (42). Topical application of the contrast agent would be ideal, but i.v. infusion is another possibility.
The ability of a polyanionic peptide domain to inhibit binding and entry of a closely apposed polycationic CPP is functionally reminiscent of intramolecular fluorescence resonance energy transfer, in which an acceptor chromophore quenches the fluorescence of a nearby donor fluorophore. In each case, if the linker is cleaved and the inhibitory moiety diffuses away, the active partner (the CPP or the donor fluorophore) is unmasked. The unmasking of CPPs has a completely different underlying mechanism and a much slower time scale but much broader range of useful imaging modalities and cargoes than fluorescence resonance energy transfer.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Baird for helpful discussions and technical assistance with frozen section preparation, Dr. Richard Lin for sharing expertise on xenografts and mouse imaging, Coyt Jackson for technical assistance with FACS analysis, Paul Steinbach for maintaining the imaging setups, and Dr. Larry Gross for assisting with mass spectrometry. This work was supported by the Howard Hughes Medical Institute, the Keck Institute, National Institutes of Health Grants DK54441 and GM54038 (to P.A.J.) and NS27177 (to R.Y.T.), and Department of Energy Grant DE-FG03-01ER63276 (to R.Y.T.).
Author contributions: T.J., E.S.O., Q.T.N., M.R., P.A.J., and R.Y.T. designed research; T.J., E.S.O., and M.R. performed rersearch; Q.T.N. contributed new reagents/analytic tools; T.J., E.S.O., M.R., P.A.J., and R.Y.T. analyzed data; and T.J., E.S.O., M.R., P.A.J., and R.Y.T. wrote the paper.
Abbreviations: CPP, cell-penetrating peptides; ACPP, activatable CPP; MMP, matrix metalloproteinase; SUV, standardized uptake value; PEG, polyethylene glycol.
References
- 1.Etzioni, R., Urban, N., Ramsey, S., McIntosh, M., Schwartz, S., Reid, B., Radich, J., Anderson, G. & Hartwell, L. (2003) Nat. Rev. Cancer 3, 243-252. [DOI] [PubMed] [Google Scholar]
- 2.Weissleder, R. & Mahmood, U. (2001) Radiology 219, 316-333. [DOI] [PubMed] [Google Scholar]
- 3.Harris, M. (2004) Lancet Oncol. 5, 292-302. [DOI] [PubMed] [Google Scholar]
- 4.Winnard, P., Jr., & Raman, V. (2003) J. Cell Biochem. 90, 454-463. [DOI] [PubMed] [Google Scholar]
- 5.Olafsen, T., Tan, G. J., Cheung, C. W., Yazaki, P. J., Park, J. M., Shively, J. E., Williams, L. E., Raubitschek, A. A., Press, M. F. & Wu, A. M. (2004) Protein Eng. Des. Sel. 17, 315-323. [DOI] [PubMed] [Google Scholar]
- 6.Sundaresan, G., Yazaki, P. J., Shively, J. E., Finn, R. D., Larson, S. M., Raubitschek, A. A., Williams, L. E., Chatziioannou, A. F., Gambhir, S. S. & Wu, A. M. (2003) J. Nucl. Med. 44, 1962-1969. [PMC free article] [PubMed] [Google Scholar]
- 7.Vives, E., Brodin, P. & Lebleu, B. (1997) J. Biol. Chem. 272, 16010-16017. [DOI] [PubMed] [Google Scholar]
- 8.Richard, J. P., Melikov, K., Vives, E., Ramos, C., Verbeure, B., Gait, M. J., Chernomordik, L. V. & Lebleu, B. (2003) J. Biol. Chem. 278, 585-590. [DOI] [PubMed] [Google Scholar]
- 9.Rothbard, J. B., Kreider, E., Vandeusen, C. L., Wright, L., Wylie, B. L. & Wender, P. A. (2002) J. Med. Chem. 45, 3612-3618. [DOI] [PubMed] [Google Scholar]
- 10.Wright, L. R., Rothbard, J. B. & Wender, P. A. (2003) Curr. Protein Pept. Sci. 4, 105-124. [DOI] [PubMed] [Google Scholar]
- 11.Gammon, S. T., Villalobos, V. M., Prior, J. L., Sharma, V. & Piwnica-Worms, D. (2003) Bioconjugate Chem. 14, 368-376. [DOI] [PubMed] [Google Scholar]
- 12.Polyakov, V., Sharma, V., Dahlheimer, J. L., Pica, C. M., Luker, G. D. & Piwnica-Worms, D. (2000) Bioconjugate Chem. 11, 762-771. [DOI] [PubMed] [Google Scholar]
- 13.Bhorade, R., Weissleder, R., Nakakoshi, T. & Moore, A. T. C. H. (2000) J. Am. Chem. Soc. 11, 301-305. [DOI] [PubMed] [Google Scholar]
- 14.Bullok, K. E., Dyszlewski, M., Prior, J. L., Pica, C. M., Sharma, V. & Piwnica-Worms, D. (2002) Bioconjugate Chem. 13, 1226-1237. [DOI] [PubMed] [Google Scholar]
- 15.Lewin, M., Carlesso, N., Tung, C.-H., Tang, X.-W., Cory, D., Scadden, D. T. & Weissleder, R. (2000) Nat. Biotech. 18, 410-414. [DOI] [PubMed] [Google Scholar]
- 16.Torchilin, V. P. & Levchenko, T. S. (2003) Curr. Protein Pept. Sci. 4, 133-140. [DOI] [PubMed] [Google Scholar]
- 17.Potocky, T. B., Menon, A. K. & Gellman, S. H. (2003) J. Biol. Chem. 278, 50188-50194. [DOI] [PubMed] [Google Scholar]
- 18.Thoren, P. E., Persson, D., Isakson, P., Goksor, M., Onfelt, A. & Norden, B. (2003) Biochem. Biophys. Res. Commun. 307, 100-107. [DOI] [PubMed] [Google Scholar]
- 19.Lundberg, M., Wikstrom, S. & Johansson, M. (2003) Mol. Ther. 8, 143-150. [DOI] [PubMed] [Google Scholar]
- 20.Wadia, J. S., Stan, R. V. & Dowdy, S. F. (2004) Nat. Med. 10, 310-315. [DOI] [PubMed] [Google Scholar]
- 21.Talvensaari-Mattila, A., Paakko, P. & Turpeenniemi-Hujanen, T. (2003) Br. J. Cancer 89, 1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bremer, C., Bredow, S., Mahmood, U., Weissleder, R. & Tung, C. H. (2001) Radiology 221, 523-529. [DOI] [PubMed] [Google Scholar]
- 23.La Rocca, G., Pucci-Minafra, I., Marrazzo, A., Taormina, P. & Minafra, S. (2004) Br. J. Cancer 90, 1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratnikov, B. I., Deryugina, E. I. & Strongin, A. Y. (2002) Lab. Invest. 82, 1583-1590. [DOI] [PubMed] [Google Scholar]
- 25.Sounni, N. E., Janssen, M., Foidart, J. M. & Noel, A. (2003) Matrix Biol. 22, 55-61. [DOI] [PubMed] [Google Scholar]
- 26.Bremer, C., Tung, C. H. & Weissleder, R. (2001) Nat. Med. 7, 743-748. [DOI] [PubMed] [Google Scholar]
- 27.Yamaoka, T., Tabata, Y. & Ikada, Y. (1994) J. Pharm. Sci. 83, 601-606. [DOI] [PubMed] [Google Scholar]
- 28.Duncan, R. (2003) Nat. Rev. Drug Discovery 2, 347-360. [DOI] [PubMed] [Google Scholar]
- 29.Pierce, B. G., Shikes, R. & Fink, L. M. (1978) Cancer, A Problem of Developmental Biology (Prentice-Hall, Englewood Cliffs, NJ).
- 30.Franc, B. L., Mandl, S. J., Siprashvili, Z., Wender, P. & Contag, C. H. (2003) Mol. Imaging 2, 313-323. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, M., Kircher, M. F., Josephson, L. & Weissleder, R. (2002) Bioconjugate Chem. 13, 840-844. [DOI] [PubMed] [Google Scholar]
- 32.Wender, P. A., Mitchell, D. J., Pattabiraman, K., Pelkey, E. T., Steinman, L. & Rothbard, J. B. (2000) Proc. Natl. Acad. Sci. USA 97, 13003-13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egeblad, M. & Werb, Z. (2002) Nat. Rev. 2, 161-174. [DOI] [PubMed] [Google Scholar]
- 34.Spinale, F. G. (2002) Circ. Res. 90, 520-530. [DOI] [PubMed] [Google Scholar]
- 35.Close, D. R. (2001) Ann. Rheum. Dis. 60, Suppl. 3, 62-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Lampe, B., Barthel, B., Coupland, S. E., Riecken, E. O. & Rosewicz, S. (2000) Gut 47, 63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leake, A., Morris, C. M. & Whateley, J. (2000) Neurosci. Lett. 291, 201-203. [DOI] [PubMed] [Google Scholar]
- 38.Duffy, M. J., Lynn, D. J., Lloyd, A. T. & O'Shea, C. M. (2003) Thromb. Haemost. 89, 622-631. [PubMed] [Google Scholar]
- 39.Funovics, M., Weissleder, R. & Tung, C. H. (2003) Anal. Bioanal. Chem. 377, 956-963. [DOI] [PubMed] [Google Scholar]
- 40.Ntziachristos, V., Tung, C. H., Bremer, C. & Weissleder, R. (2002) Nat. Med. 8, 757-760. [DOI] [PubMed] [Google Scholar]
- 41.Funovics, M. A., Weissleder, R. & Mahmood, U. (2004) Radiology 231, 659-666. [DOI] [PubMed] [Google Scholar]
- 42.De Grand, A. M. & Frangioni, J. V. (2003) Technol. Cancer Res. Treat. 2, 553-562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.