Abstract
DNMT1 is recruited to substrate sites by PCNA and UHRF1 to maintain DNA methylation after replication. The cell cycle dependent recruitment of DNMT1 is mediated by the PCNA-binding domain (PBD) and the targeting sequence (TS) within the N-terminal regulatory domain. The TS domain was found to be mutated in patients suffering from hereditary sensory and autonomic neuropathies with dementia and hearing loss (HSANIE) and autosomal dominant cerebellar ataxia deafness and narcolepsy (ADCA-DN) and is associated with global hypomethylation and site specific hypermethylation. With functional complementation assays in mouse embryonic stem cells, we showed that DNMT1 mutations P496Y and Y500C identified in HSANIE patients not only impair DNMT1 heterochromatin association, but also UHRF1 interaction resulting in hypomethylation. Similar DNA methylation defects were observed when DNMT1 interacting domains in UHRF1, the UBL and the SRA domain, were deleted. With cell-based assays, we could show that HSANIE associated mutations perturb DNMT1 heterochromatin association and catalytic complex formation at methylation sites and decrease protein stability in late S and G2 phase. To investigate the neuronal phenotype of HSANIE mutations, we performed DNMT1 rescue assays and could show that cells expressing mutated DNMT1 were prone to apoptosis and failed to differentiate into neuronal lineage. Our results provide insights into the molecular basis of DNMT1 dysfunction in HSANIE patients and emphasize the importance of the TS domain in the regulation of DNA methylation in pluripotent and differentiating cells.
Introduction
Epigenetic mechanisms are crucial for the regulation of gene expression during embryonic development and cell differentiation. Tissue-specific DNA methylation patterns are established during embryogenesis by the de novo DNA methyltransferases DNMT3A and DNMT3B, whereas the propagation of these marks to future somatic cell generations is based on the maintenance DNA methyltransferase 1 (DNMT1) (1–5). The catalytic activity of DNMT1 is attributed to its C-terminal domain (CTD), however enzyme regulation, targeting and activation are mediated by the N-terminal domain (NTD) harboring distinct subdomains (6). During S phase DNMT1 localization at sites of DNA replication is mediated by the PCNA-binding domain (PBD) while heterochromatin binding during late S and G2 is mediated by the targeting sequence (TS) domain, both of which contribute to proper maintenance of DNA methylation patterns (7–9).
A key factor in the regulation of DNMT1 is Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1, also known as 95 kDa mouse nuclear protein (Np95)). By binding to hemimethylated DNA via its SET and RING associated (SRA) domain (10–14), UHRF1 targets DNMT1 to its substrate sites (10). In addition, UHRF1 binds to methylated H3K9 via its tandem Tudor domain (TTD) and to H3R2 via its plant homeodomain (PHD) (15–18).
By cooperative binding of repressive H3K9me3 marks and hemimethylated DNA, UHRF1 targets DNMT1 to newly synthesized DNA in heterochromatin after replication (19). In addition, the UHRF1 RING domain ubiquitinates H3 tails on K18 (K23 in Xenopus), which is specifically recognized by the ubiquitin interacting motif (UIM) in the TS domain of DNMT1 and required for DNA methylation (20,21). Besides intermolecular protein–protein binding, also intramolecular protein interactions serve as a prerequisite for DNMT1 activation. Firstly, in complex with unmethylated DNA, the linker between the zinc finger (CXXC) domain and the bromo-adjacent homology domain 1 (BAH1) blocks the access of DNA to the catalytic center (22). Secondly, the crystal structure of DNMT1 reveals that, in absence of DNA, the TS domain is inserted in the DNA-binding pocket of the CTD thereby inhibiting enzymatic activity (23). These two autoinhibitory mechanisms have to be overcome by structural changes before the methylation reaction can occur. Interaction of UHRF1 with DNMT1 releases the TS domain and enables catalytic activity of the CTD (24).
In addition to enzyme targeting and activation, protein stability also contributes to the regulation of maintenance DNA methylation. Stability and abundance of DNMT1 during the cell cycle is governed by UHRF1 dependent ubiquitination and deubiquitination by the ubiquitin specific peptidase 7 (USP7, also known as herpes virus associated ubiquitin specific protease (HAUSP)) which protects against proteasomal degradation (25,26). While Tip60 mediated acetylation promotes ubiquitination by UHRF1 and thereby marks DNMT1 for proteasomal degradation, the corresponding deacetylation by histone deacetylase 1 (HDAC1) contributes to the stabilization of DNMT1 (25,26).
Despite its well-known replication-coupled function as maintenance DNA methyltransferase in proliferating cells, DNMT1 is highly expressed in embryonic and adult postmitotic neurons, especially in the central nervous system (CNS) (27,28). Remarkably, DNA methylation is required in adult neurogenesis and its misregulation was described to be involved in the pathophysiology of neurodegenerative disorders (29,30). Several medical studies have reported heterozygous DNMT1 mutations causing DNA hypomethylation in patients suffering from hereditary sensory and autonomic neuropathy type IE (HSANIE, OMIM 614116) or autosomal dominant cerebellar ataxia deafness and narcolepsy (ADCA-DN, OMIM 604121). Strikingly, all causative mutations described to date affect a genomic region in DNMT1, which codes for the TS domain (31–38). Clinical manifestations of HSANIE and ADCA-DN include a broad phenotypic spectrum with sensory impairment, hearing loss and dementia as hallmarks of the disease (38). In particular, HSANIE is characterized by late onset neurologic disorders of the CNS and the peripheral nervous system that manifest in progressive degeneration predominantly of sensory and autonomic neurons (39). However, the pathophysiological trajectories underlying these neurological disorders caused by DNMT1 mutations remain mostly unknown.
In this study, we investigate the effect of DNMT1 mutations identified in HSANIE patients on the function of the TS domain in embryonic stem cells and neuronal progenitor cell differentiation. With functional complementation assays, we show that HSANIE associated mutations in mouse DNMT1 disrupt binding of UHRF1 and lead to defects in maintenance DNA methylation. We show that the TS point mutations affect the association with late replicating chromatin, catalytic complex formation at methylation sites and cell cycle dependent protein stability.
Importantly, ESCs expressing HSANIE mutants fail to differentiate into the neuronal lineage and are prone to undergo apoptosis. These results emphasize the importance of the complex interplay of UHRF1 with the TS domain in regulating DNMT1 function and DNA methylation in neurogenesis.
Results
Deletions and TS domain mutations found in HSANIE patients affect DNMT1 activity in vivo
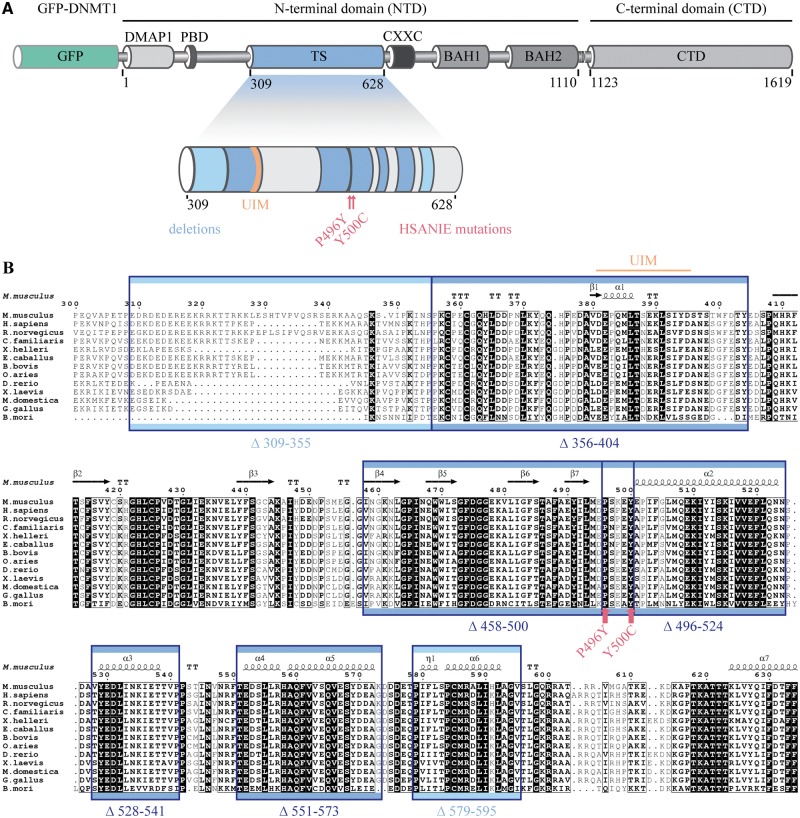
Although the catalytic activity of DNMT1 resides in its CTD, the NTD of the enzyme is indispensable for proper maintenance DNA methylation (6). Within the NTD the TS domain spans from amino acid 309 to 628 in mouse isoform 2 and is highly conserved among different species (Fig. 1). To map regions required for methylation activity, we generated a systematic set of DNMT1 deletion constructs and performed rescue experiments expressing the deletion constructs in Dnmt1−/− mouse embryonic stem cells (ESCs). Site specific DNA methylation analysis revealed that not only the regions between amino acid (aa) 356 to 404 and 458 to 500 (21), but also regions from 496 to 573 are functionally relevant to maintain methylation patterns at pericentromeric heterochromatin (Fig. 2A, shown in dark blue). DNMT1 either lacking the flexible, less conserved N-terminal region of the TS domain (aa 309 to 355), or the C-terminal region of the TS domain (aa 579 to 595) was able to restore methylation at major satellite repeats (Fig. 2A, shown in light blue).
Figure 1.
Domain structure of mouse DNMT1 and conservation of the TS domain among different species. (A) Domain structure of GFP-DNA methyltransferase 1 (DNMT1) with illustration of deletions and HSANIE associated point mutations in the targeting sequence (TS) domain. The large regulatory N-terminal domain (NTD) of DNMT1 is comprised of a DNA methyltransferase associated protein 1 (DMAP1)-binding domain, a proliferating cell nuclear antigen (PCNA)-binding domain (PBD), the TS domain harboring a ubiquitin interacting motif (UIM (21)), a zinc finger (CXXC) domain, two bromo-adjacent homology (BAH1 and BAH2) domains and a catalytically active C-terminal domain (CTD). (B) Primary sequence alignment of TS domains from different species. The secondary structure of the mouse TS domain is indicated (PDB: 3AV4 (23)). Highly conserved residues are shaded black. Deleted regions are indicated by blue rectangles and hereditary sensory and autonomic neuropathy type IE (HSANIE) associated point mutations by red arrows.
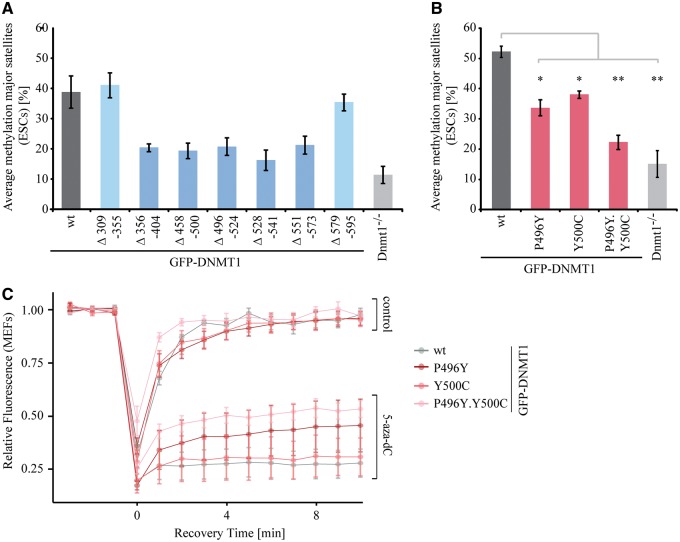
Figure 2.
TS domain deletions and HSANIE associated mutations affect the DNA methylation activity of DNMT1 in vivo. (A and B) Site-specific DNA methylation analysis at major satellite repeats of mouse Dnmt1−/− ESCs expressing transiently GFP-DNMT1 wild-type (wt) and deletions (A) or HSANIE associated point mutant constructs (B). DNA methylation levels of untransfected Dnmt1−/− ESCs are displayed for comparison. Shown are mean values ± standard deviations (SDs) from two to six (A) or (B) three independent biological replicates (average from eight CpG sites, respectively). (B) Two-sample t-tests were performed that assume equal variances. Asterisks represent significant differences: *P < 0.05, **P < 0.001. (C) Quantitative evaluation of FRAP data showing mean curves. Error bars represent standard deviations. FRAP of S-phase mouse embryonic fibroblasts (n = 5) expressing GFP-DNMT1 and GFP-DNMT1 harboring HSANIE mutations without drug treatment (upper panel) and after treatment with 30 µM 5-aza-dC for 30 min (lower panel).
Interestingly, mutations of two highly conserved amino acids in the TS domain (Fig. 1B) were linked to a neurodegenerative disease described as HSANIE (31,39). As the HSANIE associated mutations are located within a functionally relevant central region of the TS domain (Fig. 2A), we investigated the DNA methylation activity of GFP-DNMT1 harboring HSANIE mutations. To this end, we cloned HSANIE associated mutations P496Y (human: D490E.P491Y) and Y500C (human: Y495C) in mouse DNMT1 expression constructs and reintroduced the mutant proteins in Dnmt1−/− ESCs (Supplementary Material, Fig. S6). In line with previous findings (31), we found that transiently and stably expressed mutated DNMT1 is unable to rescue DNA methylation levels in vivo (Fig. 2B, Supplementary Material, Fig. S1A and S1C). Methylation levels at imprinted and unmethylated H19 promoter remained unchanged (Supplementary Material, Fig. S1B).
To investigate a potential targeting defect of DNMT1 harboring HSANIE associated mutations, we made use of a DNA methyltransferase trapping assay in living cells. In this assay, the cytosine analogue 5-aza-2′-deoxycytidine (5-aza-dC) is incorporated into DNA during replication and forms an irreversible covalent complex with the active methyltransferase at the C6 position of the cytosine residue thereby immobilizing the catalytically active enzyme at DNA replication foci (40). Trapping was measured by fluorescence recovery after photobleaching (FRAP). We found that DNMT1 harboring HSANIE mutations can be trapped, but exhibit a larger mobile fraction than DNMT1 wild-type (wt) indicating a lower targeting to substrate sites in living cells (Fig. 2C). In summary, we could show that the TS domain of DNMT1 is essential for maintenance DNA methylation. Deletions in the TS domain between amino acid 356 to 573 as well as point mutations found in HSANIE patients lead to decreased methylation activity in vivo.
Functional relevance of UHRF1 domains on the regulation of DNMT1 enzymatic activity
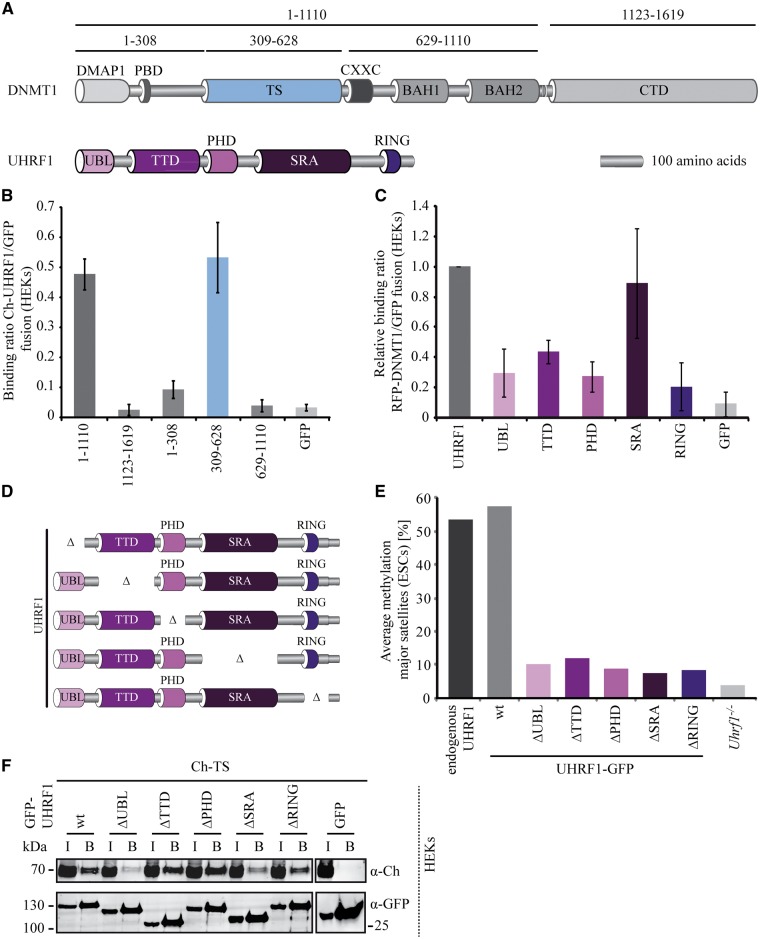
UHRF1 is required for maintaining DNA methylation patterns after replication by direct interaction with DNMT1 (10). To elucidate which protein domains are involved in the interaction of DNMT1 and UHRF1 (Fig. 3A), we generated a systematic set of constructs of the NTD and CTD of DNMT1 as well as single domains of UHRF1 and performed a binding assay. We used a semi-quantitative fluorescence protein–protein interaction assay, in which GFP-tagged proteins are immobilized on a GFP-multiTrap plate, incubated with RFP/Ch-tagged proteins and binding ratios are determined by fluorescence readout (41). Mapping studies on UHRF1 showed that the SRA domain interacted with DNMT1 (Fig. 3C). In terms of DNMT1, the TS mediated the binding to UHRF1 (Fig. 3B). To analyze domain contribution to the TS domain interaction in the UHRF1 protein context, we tested the binding of Ch-TS to UHRF1-GFP single domain deletions (Fig. 3D). We generated constructs with single domain deletions of UHRF1 and found that deletions of the TTD, the PHD and the RING domain had little to no effect on the interaction, whereas deletions of the UBL and the SRA domain reduced the binding of Ch-TS to UHRF1-GFP indicating that the TS domain of DNMT1 can interact with two domains of UHRF1, the UBL and the SRA domain of UHRF1 (Fig. 3F). In our experimental set-up, the UBL domain alone, however, did not interact with DNMT1 suggesting a cooperative binding mode involving the UBL and SRA domain of UHRF1.
Figure 3.
DNMT1 TS domain interaction with UHRF1 is necessary for DNA methylation. (A) Schematic outline of DNMT1 and the ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) expression constructs used for protein–protein interaction mapping studies. UHRF1 harbors an ubiquitin-like domain (UBL) followed by a tandem Tudor domain (TTD), a plant homeodomain (PHD), a SET and RING associated (SRA) domain and a really interesting new gene (RING) domain. (B) Mapping and relative quantification of the interaction GFP-DNMT1 with Ch-UHRF1 by fluorescence protein–protein interaction assay in vitro. Ratios of Ch-UHRF1 over GFP fusion proteins are shown as mean values ± standard error of the mean (SEM) of three to six biological replicates. (C) Mapping and relative quantification of the interaction GFP-UHRF1 with RFP-DNMT1 by fluorescence protein–protein interaction assay in vitro. Ratios of RFP-DNMT1 over GFP fusion proteins are shown as mean values ± SEM of three biological replicates normalized to the binding ratio of the GFP-UHRF1 full length protein. (D) Schematic outline of different UHRF1-GFP single domain deletion (Δ) constructs used for rescue experiments. (E) Local methylation analysis at major satellite repeats. CpG methylation levels in Uhrf1−/− ESCs stably expressing UHRF1-GFP wt or single domain deletions were analyzed. DNA methylation levels of untransfected Uhrf1−/− ESCs are shown for comparison. Values represent means from eight CpG sites. (F) Coimmunoprecipitation of UHRF1-GFP wt and single domain deletion mutants and Ch-TS. GFP and Ch fusion constructs were coexpressed in HEK293T cells and cell lysates were used for immunoprecipitation with the GFP-Trap. Bound fractions were detected by immunoblotting with anti-GFP and anti-Ch antibodies. GFP was used as negative control. I = Input, B = Bound.
To examine which domains of UHRF1 are functionally relevant for regulation of DNMT1 enzymatic activity, we investigated the ability of UHRF1 single domain deletions in mediating DNA methylation in vivo. All Uhrf1−/− ESC lines stably expressing UHRF1-GFP with single domain deletions displayed low methylation levels suggesting a DNMT1 recruitment defect (Fig. 3E). Consequently, we investigated the dependence of DNMT1 subnuclear localization on UHRF1 domains. In the wt UHRF1-GFP cell line, DNMT1 showed late S phase-specific heterochromatin association that was abolished in all single domain UHRF1 deletion cell lines displaying diffuse nuclear localization of DNMT1 comparable to Uhrf1−/− ESCs (Supplementary Material, Figs S3 and S4). In conclusion, all UHRF1 domains are required for recruitment of DNMT1 for maintenance DNA methylation.
HSANIE associated mutations in TS domain of DNMT1 affect the interaction with UHRF1 and lead to dissociation from heterochromatin
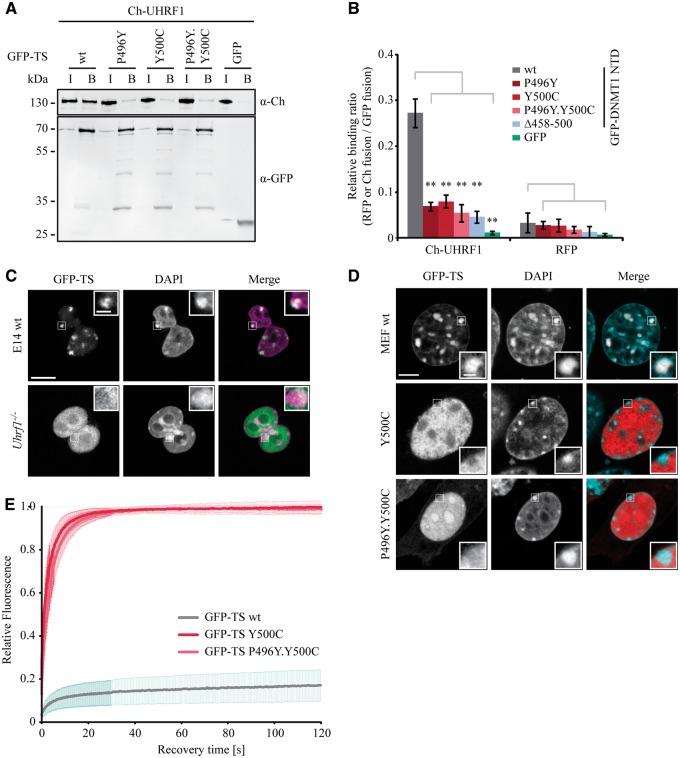
As HSANIE associated mutations in the TS domain of DNMT1 lead to hypomethylation at pericentric heterochromatin and the TS mediates the interaction with UHRF1, we investigated the effects of HSANIE mutations on UHRF1 interaction. With coimmunoprecipitation experiments, we found that mutated TS was defective in binding UHRF1 (Fig. 4A), which was confirmed by a semiquantitative fluorescence protein–protein interaction assay (Fig. 4B). Expression of GFP-TS in ESCs revealed that while the TS domain was tightly associated with heterochromatin in wt cells, enrichment of the TS domain at chromocenters was lost and the signal was diffusely distributed in the nucleus in UHRF1 depleted cells (Uhrf1−/−) (Fig. 4C). A similar delocalization was found in UHRF1 binding deficient TS domain mutants (Fig. 4D, Supplementary Material, Fig. S5). With half nucleus FRAP we investigated the mobility of TS domain fusions in MEFs and found that HSANIE mutations caused fast protein mobility indicating chromatin binding defects (Fig. 4E). Notably, the interaction of DNMT1 with the replication protein PCNA was not altered by the HSANIE mutations arguing against severe misfolding of the mutant proteins (Supplementary Material, Fig. S2). In conclusion, our findings suggest that the TS domain is crucial for mediating the interaction of DNMT1 with UHRF1 and that this interaction is affected by the HSANIE associated mutations located in this region.
Figure 4.
HSANIE associated point mutations in the TS domain reduce the interaction of DNMT1 with UHRF1. (A) Coimmunoprecipitation of GFP-TS wt, P496Y, Y500C and P496Y.Y500C mutant constructs and Ch-UHRF1. GFP and Ch fusion constructs were coexpressed in HEK293T cells and cell lysates were used for immunoprecipitation with the GFP-Trap. Bound fractions were analyzed by immunoblotting with anti-GFP and anti-Ch antibodies. GFP was used as negative control. I = Input, B = Bound. (B) Fluorescence protein–protein interaction assay of the GFP-DNMT1 NTD with Ch-UHRF1. After one-step purification of the GFP-DNMT1 NTD wt and mutant constructs in a GFP-multiTrap plate, the binding of Ch-UHRF1 expressed in HEK293T cells was determined by fluorescence readout. GFP and RFP were used as negative control. Shown are mean relative binding ratios ± SEM of Ch-UHRF1 or RFP over GFP fusion proteins from four to six biological replicates. Two-sample t-tests were performed that assume equal variances. Significance compared to the relative binding ratio of GFP-DNMT1 NTD wt are indicated: *P < 0.05, **P < 0.001. (C) Confocal midsections of fixed mouse ESCs (wt, Uhrf1−/−) transiently expressing GFP-TS and DNA was counterstained with DAPI. In the merged image, DAPI is depicted in magenta. Scale bar 10 µm; enlargements: 3-times magnification, scale bar 2 µm. (D) Confocal midsections of fixed MEF cells transfected with GFP-TS wt or GFP-TS Y500C and P496Y.Y500C constructs. In the merged image, GFP-TS is depicted in red and DAPI in magenta. Scale bar 5 µm; enlargements: 3-times magnification, scale bar 1 µm. (E) Protein mobility of GFP-TS wt and HSANIE associated GFP-TS Y500C and P497Y.Y500C mutants in living MEF cells (n = 13) determined by half nucleus fluorescent recovery after photobleaching (FRAP) analysis. Curves represent mean ± SEM.
The destabilization of HSANIE associated GFP-DNMT1 TS mutants is cell cycle-dependent
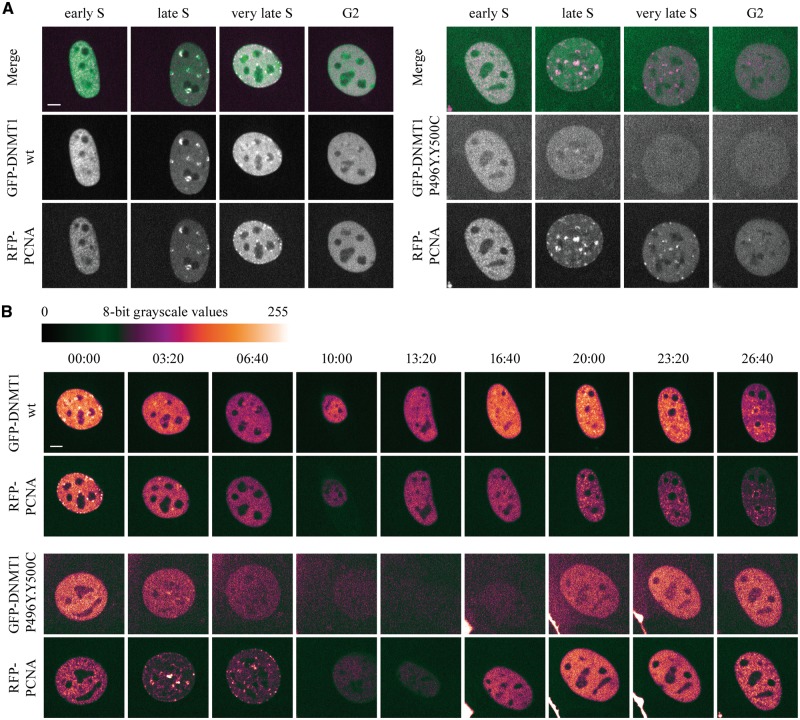
As the TS domain is essential for late S phase-specific localization and mobility of DNMT1 (42), we tested the effect of HSANIE mutations on subcellular localization of GFP-DNMT1 on a single cell level throughout the cell cycle. We imaged living cells coexpressing GFP-DNMT1 wt or GFP-DNMT1 P496Y.Y500C with RFP-PCNA as a cell cycle marker. While the localization in early to mid S phase was comparable to wt, the GFP signal of mutant DNMT1 dropped when cells entered late S phase. In addition, the double point mutant displayed only weak late S phase-specific association with heterochromatin and did not display the characteristic, prolonged TS domain mediated heterochromatin association in G2 (Fig. 5A). Interestingly, the signal of GFP-DNMT1 P496Y.Y500C recovered in G1 phase indicating a cell cycle dependency of this defect (Fig. 5B). To investigate a potential stability defect of mutant DNMT1, we performed cycloheximide experiments, which showed that GFP-DNMT1 abundance and stability was reduced by the HSANIE mutations (P496Y, Y500C or P496Y.Y500C) (Supplementary Material, Fig. S6). These results provide evidence for a cell cycle-dependent destabilization of GFP-DNMT1 harboring HSANIE mutations beginning in late S phase that might be caused by insufficient targeting to late replicating heterochromatin.
Figure 5.
The destabilization of the DNMT1 HSANIE associated mutations is cell cycle dependent. (A and B) Spinning disk confocal midsections of MEF cells transiently coexpressing GFP-DNMT1 wt or P496Y.Y500C mutant and RFP-PCNA as a cell cycle marker. (A) Selected frames from live cell series are shown of GFP-DNMT1 wt (left panel) and GFP-DNMT1 P496Y.Y500C (right panel). Cells were tracked starting from early S until G2 phase. In the merged image, RFP-PCNA is depicted in magenta. (B) Live cell series of MEF cells shown in (A) transiently coexpressing GFP-DNMT1 wt (upper panel) or P496Y.Y500C (lower panel). Starting from very late S phase (wt) or mid S phase (P496Y.Y500C) images were taken every 200 min. White represents the highest and black the lowest intensity. Scale bar 5 µm.
HSANIE associated point mutations affect neuronal progenitor differentiation
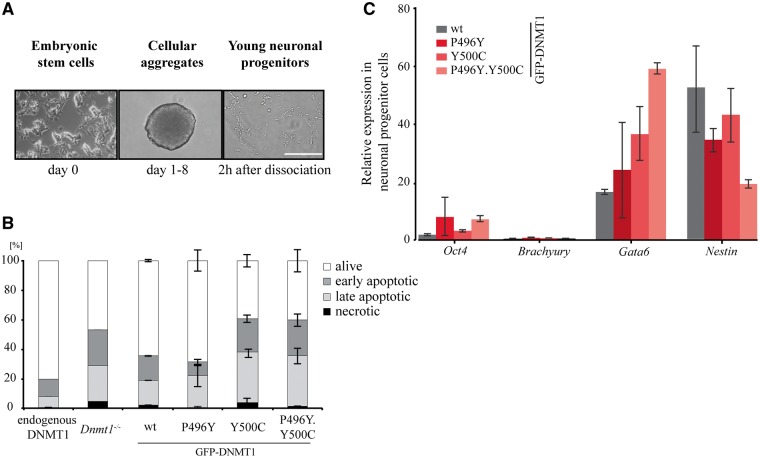
As HSANIE associated mutations in DNMT1 lead to a neuronal restricted phenotype in HSANIE patients (31), we investigated the potential of cells expressing DNMT1 with HSANIE associated mutations to differentiate into the neuronal lineage. During neuronal progenitor cell (NPC) differentiation (Fig. 6A), we examined cell viability of wt, Dnmt1−/− and different ESC lines carrying HSANIE associated mutations (Supplementary Material, Fig. S7). In the pluripotent state and on day 8 of differentiation (Supplementary Material, Fig. S8), wt, Dnmt1−/− and mutant cell lines exhibited a similar viability degree. Strikingly, young neuronal progenitors generated from Dnmt1−/− cells, as well as stably expressing GFP-DNMT1 Y500C and double mutants P496Y.Y500C did not fully develop into differentiated NPCs, but instead were prone to undergo apoptosis 2 h after dissociation from cellular aggregates (Fig. 6B). Furthermore, we compared mRNA expression levels in NPCs of pluripotency gene Oct4 and Brachyury, Gata6 and Nestin for early mesodermal, primitive endodermal and ectodermal lineage, respectively. GFP-DNMT1 Y500C and double mutant P496Y.Y500C NPCs had lower expression levels of developmental marker Nestin, a marker for neuronal progenitor differentiation and elevated expression levels of pluripotency marker Oct4 indicating an uncompleted differentiation in NPCs (Fig. 6C). The results highlight the importance of proper DNMT1 function during neurogenesis and indicate that the limited differentiation potential of DNMT1 deficient cells cannot be rescued by DNMT1 harboring HSANIE associated mutations.
Figure 6.
HSANIE associated point mutations in the TS domain show different survival rate and differentiation potential during neuronal progenitor differentiation. (A) Mouse embryonic stem cells are differentiated into young neuronal progenitor cells (NPCs). Scale bar: 200 µm. (B) Flow cytometric analysis of alive, early apoptotic, late apoptotic and necrotic cells. Two hours after dissociation of cellular aggregates, young neuronal progenitors were analyzed using Annexin V and propidium iodide staining. Bar graphs represent mean values ± standard deviation (SD) from two to three biological replicates. (C) RNA expression profiles of NPCs in biological duplicates (wt) or triplicates (P496Y, Y500C, P496Y.Y500C) of pluripotency factor Oct4, lineage specific markers Brachyury, Gata6 and Nestin for mesodermal, endodermal and ectodermal lineage, respectively. All ddCt values are normalized to wt Oct4 expression.
Discussion
For long, DNA methylation has been regarded as a stable epigenetic mark set by the de novo DNA methyltransferases DNMT3A and DNMT3B during development and maintained after each round of DNA replication by DNMT1 (1–5). This simple view of DNA methylation, however, cannot explain why DNMT1 is expressed and required in postmitotic cells (27,28). Despite or because of the central and ubiquitous role in DNA methylation only very few human diseases were linked to heterozygous DNMT1 mutations that are mostly restricted to the TS domain within the regulatory NTD of DNMT1 (43). So far, two neurodegenerative diseases HSANIE and ADCA-DN have been linked to mutations in the TS domain of DNMT1 (31,32) but the underlying molecular mechanism remained elusive.
To investigate how simple amino acid exchanges outside the catalytic domain of DNMT1 may lead to global hypomethylation and neurodegenerative diseases, we transferred these mutations to a controlled ESC differentiation system and assayed for cellular defects. As UHRF1 plays a central role in the regulation of DNMT1, we first investigated whether the HSANIE mutations affect this interaction. Our coimmunoprecipitation experiments and deletion analyses show an interaction of the TS domain with the SRA domain of UHRF1, which is consistent with previous yeast two-hybrid screens (44). Interestingly, the HSANIE mutations are located within this part of the TS domain and indeed weaken this protein–protein interaction. We could demonstrate that mutated DNMT1 showed weaker association with heterochromatin in late S phase and failed to maintain DNA methylation in ESC, which is consistent with the previous observation that UHRF1 is required for recruitment of DNMT1 (10). The interaction of UHRF1 with DNMT1 was also shown to be essential for allosteric activation of the enzyme to enable binding of substrate DNA to the CTD (23,24,45–47). The weakened interaction could explain the reduced activity of mutated DNMT1 pull-downs in vitro (31) and fits well with our observation that DNMT1 harboring HSANIE mutations are impaired in catalytic complex formation as measured with our trapping assay in living cells. In addition to defects in enzyme activation and targeting, we show that HSANIE mutations also cause proteasomal degradation of DNMT1 in late S and G2 phase, likely as a result of failed heterochromatin binding, which is consistent with previous studies (31). Similar protein destabilization was shown for chromatin unbound DNMT3A and DNMT3B (48,49).
Besides harboring a binding site for UHRF1, the TS domain of DNMT1 also contains a UIM that recognizes H3K18 ubiquitinated by UHRF1 (21). Although this UIM is well separated in primary and tertiary structure from the sites mutated in HSANIE patients, allosteric interference with H3K18ub binding cannot be ruled out. In addition to UHRF1 and histone binding, DNMT1 is subjected to further protein interactions and multiple posttranslational modifications, which likely contribute to proper regulation of DNA methylation (50). Several of these interactions may not be absolutely required but may enhance local or global efficiency and thereby contribute to the fine tuning DNMT1 activity as we found for the interaction with PCNA, which 2-fold enhances DNMT1 efficiency in living cells (51,52). While it is unclear which of these interactions are affected by the HSANIE mutations in different cell types, we could clearly show that ESCs carrying these mutations are impaired in neuronal differentiation and prone to apoptosis.
Curiously, although HSANIE TS domain mutations are present in all patient tissues, they mostly affect the neuronal lineage causing both central and peripheral neurodegeneration. Although it is not clear yet how dynamic the changes in DNA methylation could be in postmitotic neurons, the turnover and change of DNA methylation has emerged as one possible modulator of neuronal plasticity in response to external or internal stimuli (53–55). This fits with the observation that neuronal tissues are characterized by strong expression of ten-eleven translocation (TET) genes and high hmC levels (56–59). Therefore, DNA modification might be more dynamic in postmitotic neurons than previously thought so that even small changes in protein–protein interactions and activity might unbalance the equilibrium of DNA modifications. But it remains unclear why neuronal tissues are specifically affected by these DNMT1 mutations in HSANIE patients.
In summary, we show that HSANIE mutations affect DNMT1 interaction with the essential cofactor UHRF1, cause cell cycle dependent degradation of DNMT1 and impair neuronal differentiation. These data add to our understanding of the role and regulation of DNMT1 during differentiation and help to understand DNMT1 dysfunction and hypomethylation in the pathogenesis of this neurodegenerative disease.
Materials and Methods
Mammalian expression constructs and antibodies
Mouse fusion constructs were generated using enhanced green fluorescent protein (GFP), monomeric red fluorescent protein (RFP) or monomeric cherry (Ch). The expression constructs for RFP-DNMT1, GFP-DNMT1 wt, GFP-DNMT1Δ356-404, GFP-DNMT1Δ458-500, GFP-NTD, Ch-TS, GFP-DNMT1 (1-308), GFP-TS, GFP-DNMT1 (629-1110) and GFP-CTD have been described previously (9,21,26,40,60,61). GFP-DNMT1 deletion and HSANIE point mutants as well as UHRF1-GFP deletion expression constructs were derived from the corresponding wt constructs by overlap extension PCR (62). The GFP-UHRF1, UHRF1-GFP, Ch-UHRF1 and GFP-UHRF1 single domain constructs have been reported before (17,51,63,64). GFP, RFP and RFP-PCNA have been reported before (9,25,65,66). All constructs were verified by DNA sequencing. The following monoclonal antibodies were used for immunoblotting: rat anti-RFP/Ch (5F8, Chromotek; (67), rat anti-GFP (3H9, Chromotek). In dependence on the expected intensity of the signals, secondary antibodies either conjugated to horseradish peroxidase (HRP) (anti-rat (Dianova)) or conjugated to fluorescent dyes (anti-rat Alexa Fluor 488, Alexa Fluor 594 or Alexa Fluor 647 (Invitrogen)) were applied. For detection of HRP-conjugated antibodies an ECL Plus reagent (GE Healthcare, Thermo Scientific) was used.
Cell culture, transfection and immunofluorescence staining
HEK293T cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 50 µg/ml gentamicin. MEF cells were cultured in Dulbecco’s modified Eagle medium supplemented with 15% fetal bovine serum, non-essential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol (Gibco-BRL), 100 U/ml penicillin and 100 µg/ml streptomycin (PAA Laboratories GmbH). Mouse ESCs were cultured as published (51) with the exception that the medium was supplemented with 2i inhibitors (1 µM MEK inhibitor PD and 3 µM GSK-3 inhibitor CHIR) (2i, Axon Medchem) (63). To analyze the DNA methylation level in stably expressing UHRF1-GFP ESC lines, we cultured the cell lines in medium supplemented with 1,000 U/ml recombinant mouse leukemia inhibitory factor LIF (Millipore). Mouse J1 Dnmt1−/− ESCs are homozygous for the c null allele and have been described before (65). Mouse E14 wt and Uhrf1−/− cells (M. Muto and H. Koseki) as well as J1 triple knockout cells (Masaki Okano) have been reported previously (68). HEK293T cells were transfected with polyethylenimine (Sigma). Mouse ESCs and MEF cells were transfected with Lipofectamin (Invitrogen). Fixation, DAPI counterstaining and image acquisition cells was performed as described before (17).
Live cell microscopy, DNA methyltransferase trapping assay and fluorescence after photobleaching analysis
Live cell imaging and the trapping assay were performed as described previously (40,42). Briefly, mouse embryonic fibroblasts stably expressing RFP-PCNA were seeded on multi-well imaging slides (ibidi) and transfected using Lipofectamine 3000 (Life Technologies) following the manufacturer’s protocol. Trapping assay was performed on an Ultraview-Vox spinning disk system (Perkin-Elmer) equipped with an EMCCD camera (Hamamatsu, Japan) a microirradiation system, and an environmental chamber kept at 37 °C with 5% CO2, using a 63X 1.4 NA Plan-Apochromat oil-immersion objective (Zeiss). Imaging was performed with 488 nm and 561 nm solid-state laser diodes, using minimal gain and 2×2 camera binning for a final pixel size of 220 nm. Cells in late S-phase were visually identified for photobleaching experiments. For each photobleaching experiment, three pre-bleach images were acquired, before using the microirradiation system with the 488 nm laser to photobleach a small area (<1 µm). After bleaching, images were acquired for both fluorophores every 10 s for up to 5 min.
Data extraction and FRAP normalization was performed in Fiji. Briefly, images were background subtracted and registered to correct for xy drift and cell motion artifacts. ROIs corresponding to the bleached area and to the entire nucleus were manually selected, and mean fluorescence intensities were extracted for each timepoint. From these raw intensities, two normalization steps were performed. First each data point was normalized to its corresponding average pre-bleach intensity, and then each data point from the bleached area was normalized to the total nuclear fluorescence at its corresponding timepoint to correct for acquisition photobleaching. Normalized recovery data was then imported into R (69).
Generation of stable ESC lines and DNA methylation analysis
Forty-eight hours after transfection with GFP tagged constructs, GFP positive ESCs were separated using a fluorescence activated cell sorting (FACS) Aria II instrument (Becton Dikinson) and the cells were subsequently grown in selective medium containing 10 µg/ml blasticidin (GFP-DNMT1 cell lines) or normal medium (UHRF1-GFP cell lines). After expansion, cells were again FACS sorted one or two more times until at least 90% of the population was GFP positive. Furthermore, the GFP-DNMT1 cell lines were single cell sorted and clones with low expression levels were chosen for further analysis. The GFP-DNMT1 ESC line has been reported before (26). Genomic DNA isolation, bisulfite conversion, Primer sets and PCR conditions were described before (61,70). All PCR products were analyzed by pyrosequencing (Varionostic).
Neuronal progenitor cell (NPC) differentiation
The differentiation of pluripotent ESCs into NPCs was based on a protocol described before (71).
Analysis of cell viability
In order to analyze cell viability, we stained apoptotic cells with annexin V and necrotic cells with propidium iodide. To test the efficiency of the staining, apoptosis was induced by treatment of the cells with 5 µM staurosporine for 2 h (‘apoptotic control’) (Supplementary Material, Fig. S8). For the staining 200,000 cells were resuspended in 100 µl annexin binding buffer (100 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). Cells were stained with 5 µl Alexa Fluor 350 conjugated annexin V (Thermo Fisher) for 15 min at room temperature protected from light. 400 µl annexin binding buffer were added and necrotic cells were stained by addition of 20 μg/ml propidium iodide solution (Sigma Aldrich) shortly before analysis by flow cytometry using a FACS Aria II instrument (Becton Dickinson). Quantification of alive, necrotic, early and late apoptotic cells was performed using the single cell analysis software FlowJo.
RNA isolation, cDNA synthesis and real-time PCR
Total RNA was isolated from cells using a nucleospin triprep kit (Macherey-Nagel). 500 ng of total RNA was reverse transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s instructions. Real-time PCR was conducted using LightCycler® 480 SYBR Green I Master on a LightCycler® 480 Instrument II (Roche). PCR efficiency and primer pair specificity was examined using a standard curve of serially diluted cDNA and melting curve, respectively. After normalization to the transcript level of glyceraldehyde phosphate dehydrogenase, data were analyzed based on the 2-ΔΔCT method (72). A detailed list of primers used in the real-time PCR is shown in Supplementary Material, Table S1.
Protein–protein interaction assay and coimmunoprecipitation
For protein–protein interaction assays and coimmunoprecipitation GFP and RFP or Ch fusion constructs were expressed in HEK293T cells and 2 days after transfection cells were harvested in ice cold PBS. Cell pellets from one to two 10 cm dishes were lysed in 200 µl lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM MgCl2, 0.1% NP-40, 1× protease inhibitor, 2 mM PMSF, 1 mg/ml DNaseI (AppliChem)) and a protein–protein interaction assay in GFP-multiTrap plates (Chromotek) was performed as described (41) with the following adaptations: GFP extracts were equalized to a concentration of 60 nM in immunoprecipitation buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA) prior to one step purification in blocked (3% milk) GFP-multiTrap plates. After stringent washing (wash buffer; 20 mM Tris–HCl pH 7.5, 300 mM NaCl, 0.5 mM EDTA), purified GFP fusion proteins were incubated with crude protein extracts of RFP or Ch fusion proteins at a concentration of 1.1 to 2.1 µM diluted in IP buffer (excess of amount RFP or Ch fusion proteins in relation to GFP fusions: 18–35 times). Bound fractions were quantified by fluorescence intensity measurements with a Tecan Infinite M1000 plate reader (Tecan). For coimmunoprecipitation assays, the GFP and RFP or Ch fusion constructs were coexpressed in HEK293T cells, protein extracts were equalized and depending on the expression amounts of 5–30 pmol GFP-fusion protein were applied for the coimmunoprecipitation with the GFP-Trap (Chromotek). Note that the plasmid amount of GFP fusion construct and RFP fusion constructs used for transfection was adapted in a way to have at least a 3-fold excess of the molar RFP or Ch fusion protein amount in relation to GFP fusions. Bound fractions were firstly detected by fluorescence intensity measurements and secondly by immunoblotting using specific antibodies.
Statistical Analysis
Results were depicted as mean values ± standard deviations (SDs) or as mean values ± standard errors of the mean (SEM) from the number of biological replicates indicated in the corresponding figure legend. The difference between two mean values was analyzed by Student’s t-test and was considered as statistically significant in case of P < 0.05 (*) and highly significant for P < 0.001 (**).
Sequence Alignments
Alignments were prepared using ClustalW2 (73) and ESPript (74).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to the following colleagues for providing ESCs and somatic cells: Masahiro Muto and Haruhiko Koseki for mouse wt E14 and Uhrf1−/− ESCs; En Li and Taiping Chen for mouse J1 wt and Dnmt1−/− ESCs; Masaki Okano for J1 TKO ESCs; Thomas Jenuwein for wt MEF cells.
Conflict of Interest statement. None declared.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft [grant number SFB 646/B10, and SFB1064/A17 to H.L]. M.S. and S.L. are fellows of the Integrated Research Training Group (IRTG) of the SFB1064. S.L. was funded by an award of Lehre@LMU of the Ludwig-Maximilians Universität Munich. P.W. was a fellow of the Graduate School Life Science Munich (LSM). K.S. was and J.R. is supported by the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS-LS). J.R. is supported by a doctoral fellowship from Fonds de recherche du Québec - Santé. W.Q. was supported by the China Scholarship Council (CSC). Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft [grant number SFB1064/A17 to H.L].
References
- 1. Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 2. Goll M.G., Bestor T.H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem., 74, 481–514. [DOI] [PubMed] [Google Scholar]
- 3. Cheng X., Blumenthal R.M. (2008) Mammalian DNA methyltransferases: a structural perspective. Structure, 16, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Law J.A., Jacobsen S.E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet., 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith Z.D., Meissner A. (2013) DNA methylation: roles in mammalian development. Nat. Rev. Genet., 14, 204–220. [DOI] [PubMed] [Google Scholar]
- 6. Margot J.B., Aguirre-Arteta A.M., Di Giacco B.V., Pradhan S., Roberts R.J., Cardoso M.C., Leonhardt H. (2000) Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J. Mol. Biol., 297, 293–300. [DOI] [PubMed] [Google Scholar]
- 7. Leonhardt H., Page A.W., Weier H.U., Bestor T.H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell, 71, 865–873. [DOI] [PubMed] [Google Scholar]
- 8. Chuang L.S., Ian H.I., Koh T.W., Ng H.H., Xu G., Li B.F. (1997) Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science, 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- 9. Easwaran H.P., Schermelleh L., Leonhardt H., Cardoso M.C. (2004) Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep., 5, 1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bostick M., Kim J.K., Estève P.O., Clark A., Pradhan S., Jacobsen S.E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science, 317, 1760–1764. [DOI] [PubMed] [Google Scholar]
- 11. Sharif J., Muto M., Takebayashi S.I., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature, 450, 908–912. [DOI] [PubMed] [Google Scholar]
- 12. Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature, 455, 818–821. [DOI] [PubMed] [Google Scholar]
- 13. Avvakumov G.V., Walker J.R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C.H., Dhe-Paganon S. (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature, 455, 822–825. [DOI] [PubMed] [Google Scholar]
- 14. Qian C., Li S., Jakoncic J., Zeng L., Walsh M.J., Zhou M.M. (2008) Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem., 283, 34490–34494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Citterio E., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P.P., Bonapace I.M. (2004) Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol., 24, 2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karagianni P., Amazit L., Qin J., Wong J. (2008) ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol., 28, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rottach A., Frauer C., Pichler G., Bonapace I.M., Spada F., Leonhardt H. (2010) The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res., 38, 1796–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng J., Yang Y., Fang J., Xiao J., Zhu T., Chen F., Wang P., Li Z., Yang H., Xu Y. (2013) Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem., 288, 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., Wong J. (2013) UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun., 4, 1563. [DOI] [PubMed] [Google Scholar]
- 20. Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F., et al. (2013) Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature, 502, 249–253. [DOI] [PubMed] [Google Scholar]
- 21. Qin W., Wolf P., Liu N., Link S., Smets M., La Mastra F., Forné I., Pichler G., Hörl D., Fellinger K., et al. (2015) DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res., 25, 911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song J., Rechkoblit O., Bestor T.H., Patel D.J. (2010) Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science, 331, 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takeshita K., Suetake I., Yamashita E., Suga M., Narita H., Nakagawa A., Tajima S. (2011) Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. USA, 108, 9055–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bashtrykov P., Jankevicius G., Jurkowska R.Z., Ragozin S., Jeltsch A. (2014) The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J. Biol. Chem., 289, 4106–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du Z., Song J., Wang Y., Zhao Y., Guda K., Yang S., Kao H.Y., Xu Y., Willis J., Markowitz S.D., et al. (2010) DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signa.l, 3, ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin W., Leonhardt H., Spada F. (2011) Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem., 112, 439–444. [DOI] [PubMed] [Google Scholar]
- 27. Goto K., Numata M., Komura J.I., Ono T., Bestor T.H., Kondo H. (1994) Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation, 56, 39–44. [DOI] [PubMed] [Google Scholar]
- 28. Inano K., Suetake I., Ueda T., Miyake Y., Nakamura M., Okada M., Tajima S. (2000) Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J. Biochem., 128, 315–321. [DOI] [PubMed] [Google Scholar]
- 29. Zhao C., Deng W., Gage F.H. (2008) Mechanisms and functional implications of adult neurogenesis. Cell, 132, 645–660. [DOI] [PubMed] [Google Scholar]
- 30. Braun S.M.G., Jessberger S. (2014) Adult neurogenesis and its role in neuropsychiatric disease, brain repair and normal brain function. Neuropathol. Appl. Neurobiol., 40, 3–12. [DOI] [PubMed] [Google Scholar]
- 31. Klein C.J., Botuyan M.V., Wu Y., Ward C.J., Nicholson G.A., Hammans S., Hojo K., Yamanishi H., Karpf A.R., Wallace D.C., et al. (2011) Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet., 43, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winkelmann J., Lin L., Schormair B., Kornum B.R., Faraco J., Plazzi G., Melberg A., Cornelio F., Urban A.E., Pizza F., et al. (2012) Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum. Mol. Genet., 21, 2205–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. David G., Gosal D., Ealing J., Mignot E. (2013) A mutation in the DNMT1 gene causing autosomal dominant ataxia with deafness and cataplexy. J. Neurol. Neurosurg. Psychiatry, 84, e2.44–e2.25346970 [Google Scholar]
- 34. Pedroso J.L., Povoas Barsottini O.G., Lin L., Melberg A., Oliveira A.S.B., Mignot E. (2013) A novel de novo exon 21 DNMT1 mutation causes cerebellar ataxia, deafness, and narcolepsy in a Brazilian patient. Sleep, 36, 1257–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yuan J., Higuchi Y., Nagado T., Nozuma S., Nakamura T., Matsuura E., Hashiguchi A., Sakiyama Y., Yoshimura A., Takashima H. (2013) Novel mutation in the replication focus targeting sequence domain of DNMT1 causes hereditary sensory and autonomic neuropathy IE. J. Peripher. Nerv. Syst., 18, 89–93. [DOI] [PubMed] [Google Scholar]
- 36. Moghadam K.K., Pizza F., La Morgia C., Franceschini C., Tonon C., Lodi R., Barboni P., Seri M., Ferrari S., Liguori R., et al. (2014) Narcolepsy is a common phenotype in HSAN IE and ADCA-DN. Brain, 137, 1643–1655. [DOI] [PubMed] [Google Scholar]
- 37. Sun Z., Wu Y., Ordog T., Baheti S., Nie J., Duan X., Hojo K., Kocher J.P., Dyck P.J., Klein C.J. (2014) Aberrant signature methylome by DNMT1 hot spot mutation in hereditary sensory and autonomic neuropathy 1E. Epigenetics, 9, 1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baets J., Duan X., Wu Y., Smith G., Seeley W.W., Mademan I., McGrath N.M., Beadell N.C., Khoury J., Botuyan M.V., et al. (2015) Defects of mutant DNMT1 are linked to a spectrum of neurological disorders. Brain, 138, 845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klein C.J., Bird T., Ertekin-Taner N., Lincoln S., Hjorth R., Wu Y., Kwok J., Mer G., Dyck P.J., Nicholson G.A. (2013) DNMT1 mutation hot spot causes varied phenotypes of HSAN1 with dementia and hearing loss. Neurology, 80, 824–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schermelleh L., Spada F., Easwaran H.P., Zolghadr K., Margot J.B., Cardoso M.C., Leonhardt H. (2005) Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods, 2, 751–756. [DOI] [PubMed] [Google Scholar]
- 41. Pichler G., Jack A., Wolf P., Hake S.B. (2012) Versatile toolbox for high throughput biochemical and functional studies with fluorescent fusion proteins. PLoS One, 7, e36967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schneider K., Fuchs C., Dobay A., Rottach A., Qin W., Wolf P., Álvarez-Castro J.M., Nalaskowski M.M., Kremmer E., Schmid V., et al. (2013) Dissection of cell cycle-dependent dynamics of Dnmt1 by FRAP and diffusion-coupled modeling. Nucleic Acids Res., 41, 4860–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bestor T.H., Edwards J.R., Boulard M. (2015) Notes on the role of dynamic DNA methylation in mammalian development. Proc. Natl. Acad. Sci. USA, 112, 6796–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Achour M., Jacq X., Rondé P., Alhosin M., Charlot C., Chataigneau T., Jeanblanc M., Macaluso M., Giordano A., Hughes A.D., et al. (2008) The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene, 27, 2187–2197. [DOI] [PubMed] [Google Scholar]
- 45. Syeda F., Fagan R.L., Wean M., Avvakumov G.V., Walker J.R., Xue S., Dhe-Paganon S., Brenner C. (2011) The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. J. Biol. Chem., 286, 15344–15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bashtrykov P., Rajavelu A., Hackner B., Ragozin S., Carell T., Jeltsch A. (2014) Targeted mutagenesis results in an activation of DNA methyltransferase 1 and confirms an autoinhibitory role of its RFTS domain. Chembiochem, 15, 743–748. [DOI] [PubMed] [Google Scholar]
- 47. Berkyurek A.C., Suetake I., Arita K., Takeshita K., Nakagawa A., Shirakawa M., Tajima S. (2014) The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J. Biol. Chem., 289, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong S., Liang G., Sharma S., Lin J.C., Choi S.H., Han H., Yoo C.B., Egger G., Yang A.S., Jones P.A. (2009) Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol. Cell. Biol., 29, 5366–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharma S., De Carvalho D.D., Jeong S., Jones P.A., Liang G. (2011) Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet., 7, e1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qin W., Leonhardt H., Pichler G. (2011) Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus, 2, 392–402. [DOI] [PubMed] [Google Scholar]
- 51. Schermelleh L., Haemmer A., Spada F., Rösing N., Meilinger D., Rothbauer U., Cardoso M.C., Leonhardt H. (2007) Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res., 35, 4301–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Spada F., Haemmer A., Kuch D., Rothbauer U., Schermelleh L., Kremmer E., Carell T., Längst G., Leonhardt H. (2007) DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol., 176, 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fan G., Beard C., Chen R.Z., Csankovszki G., Sun Y., Siniaia M., Biniszkiewicz D., Bates B., Lee P.P., Kuhn R., et al. (2001) DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci., 21, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Borrelli E., Nestler E.J., Allis C.D., Sassone-Corsi P. (2008) Decoding the epigenetic language of neuronal plasticity. Neuron, 60, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu N.K., Baek S.H., Kaang B.K. (2011) DNA methylation-mediated control of learning and memory. Mol. Brain, 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Globisch D., Münzel M., Müller M., Michalakis S., Wagner M., Koch S., Brückl T., Biel M., Carell T. (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One, 5, e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Szwagierczak A., Bultmann S., Schmidt C.S., Spada F., Leonhardt H. (2010) Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res., 38, e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science, 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fellinger K., Rothbauer U., Felle M., Längst G., Leonhardt H. (2009) Dimerization of DNA methyltransferase 1 is mediated by its regulatory domain. J. Cell. Biochem., 106, 521–528. [DOI] [PubMed] [Google Scholar]
- 61. Frauer C., Rottach A., Meilinger D., Bultmann S., Fellinger K., Hasenöder S., Wang M., Qin W., Söding J., Spada F., et al. (2011) Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One, 6, e16627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 63. Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature, 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. De Vos M., El Ramy R., Delphine Q., Patricia W., Fabio S., Najat M., Federica B., Valérie S., Heinrich L., Bonapace I.M., et al. (2014) Poly(ADP-ribose) Polymerase 1 (PARP1) Associates with E3 ubiquitin-protein ligase UHRF1 and modulates UHRF1 biological functions. J. Biol. Chem., 289, 16223–16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lei H., Oh S.P., Okano M., Jüttermann R., Goss K.A., Jaenisch R., Li E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- 66. Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rottach A., Kremmer E., Nowak D., Leonhardt H., Cardoso M.C. (2008) Generation and characterization of a rat monoclonal antibody specific for multiple red fluorescent proteins. Hybridoma, 27, 337–343. [DOI] [PubMed] [Google Scholar]
- 68. Meilinger D., Fellinger K., Bultmann S., Rothbauer U., Bonapace I.M., Klinkert W.E.F., Spada F., Leonhardt H. (2009) Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep., 10, 1259–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. R Core Team (2016) R: A Language and Environment for Statistical Computing. The R Foundation for Statistical Computing, Vienna, Austria.
- 70. Tucker K.L., Beard C., Dausmann J., Jackson-Grusby L., Laird P.W., Lei H., Li E., Jaenisch R. (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev., 10, 1008–1020. [DOI] [PubMed] [Google Scholar]
- 71. Bibel M., Richter J., Lacroix E., Barde Y.A. (2007) Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc., 2, 1034–1043. [DOI] [PubMed] [Google Scholar]
- 72. Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 73. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol., 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res., 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.