Significance
Regeneration of the liver after injury is essential for proper hepatic function. Endothelial cells contribute to liver homeostasis via paracrine interaction with hepatocytes. Here we show that endothelial-derived activin A, as a transcriptional target of Krüppel-like factor 2 (KLF2), negatively regulates hepatocyte proliferation and liver regeneration. This study highlights a mechanism by which endothelial cells control liver regeneration.
Keywords: KLF2, activin A, vascular niche
Abstract
Endothelial cells (ECs) not only are important for oxygen delivery but also act as a paracrine source for signals that determine the balance between tissue regeneration and fibrosis. Here we show that genetic inactivation of flow-induced transcription factor Krüppel-like factor 2 (KLF2) in ECs results in reduced liver damage and augmentation of hepatocyte proliferation after chronic liver injury by treatment with carbon tetrachloride (CCl4). Serum levels of GLDH3 and ALT were significantly reduced in CCl4-treated EC-specific KLF2-deficient mice. In contrast, transgenic overexpression of KLF2 in liver sinusoidal ECs reduced hepatocyte proliferation. KLF2 induced activin A expression and secretion from endothelial cells in vitro and in vivo, which inhibited hepatocyte proliferation. However, loss or gain of KLF2 expression did not change capillary density and liver fibrosis, but significantly affected hepatocyte proliferation. Taken together, the data demonstrate that KLF2 induces an antiproliferative secretome, including activin A, which attenuates liver regeneration.
The liver has a central role in controlling metabolic activity and homeostasis. The importance of the liver is underlined by its ability to rapidly regenerate, which is achieved by proliferation of mature liver cells. The close interaction between the parenchymal cells (about 80%; hepatocytes) and nonparenchymal cells (about 20%; endothelial, Kupffer, and stellate cells) plays a major role in the regeneration process (1). Blood vessels and the liver sinusoidal endothelial cells (LSECs) lining these vessels deliver metabolites and oxygen to the tissue and ensure the export of waste products. In addition to these established functions, LSECs also provide angiocrine signaling, thereby determining the multicellular crosstalk leading to liver regeneration versus liver fibrosis (2). For example, it was shown that angiopoetin-2 (Ang-2) down-regulation leads to the release of paracrine factors, resulting in the inhibition of transforming growth factor-β (TGF-β) production and thus allowing hepatocyte proliferation in the early stages of liver regeneration (3). Moreover, stromal-derived factor-1 (SDF) and its receptors CXCR4 and CXCR7 control the balance between liver regeneration and fibrosis. CXCR7 signaling plays a crucial role in the release of hepatocyte growth factor (HGF), which is required for hepatocyte proliferation, whereas CXCR4 signaling leads to the release of TGF-β and subsequently to more liver fibrosis (4, 5).
Activin A, a dimer of two activin βA-subunits, is a member of the TGF-β family. Activin A signaling leads to the activation and nuclear translocation of SMAD proteins, which regulate target gene expression. Recent studies have highlighted the importance of activin A in controlling hepatocyte proliferation by inhibiting DNA synthesis and increasing the production of extracellular matrix from stellate cells, resulting in hepatic fibrosis (6, 7). Carbon tetrachloride (CCl4)-induced liver injury also up-regulates activin A, which results in liver fibrosis (8).
The phenotype of endothelial cells is regulated by mechanical activation of the cells induced by the blood flow. We hypothesized that the downstream signaling elicited by blood flow controls angiocrine signaling and liver regeneration. Krüppel-like factor 2 (KLF2) is a shear stress-inducible transcription factor, which is involved in critical regulation of endothelial gene expression. Endothelial-restricted deletion of KLF2 is embryonically lethal, characterized by severe loss of vascular tone, bleeding, and cardiac dysfunction induced by vascular complications (9). KLF2 was shown to control blood vessel maturation through smooth-muscle-cell migration (10) and to reduce endothelial activation in response of proinflammatory stimuli (11). Other studies have demonstrated that KLF2 restores the neovascularization capacities of aged circulating proangiogenic cells in vivo (12).
Because flow and KLF2 control endothelial activation, we aimed to elucidate the role of endothelial KLF2 in mediating hepatic regeneration upon liver damage.
Results
Loss of Endothelial KLF2 Expression Reduces Liver Damage.
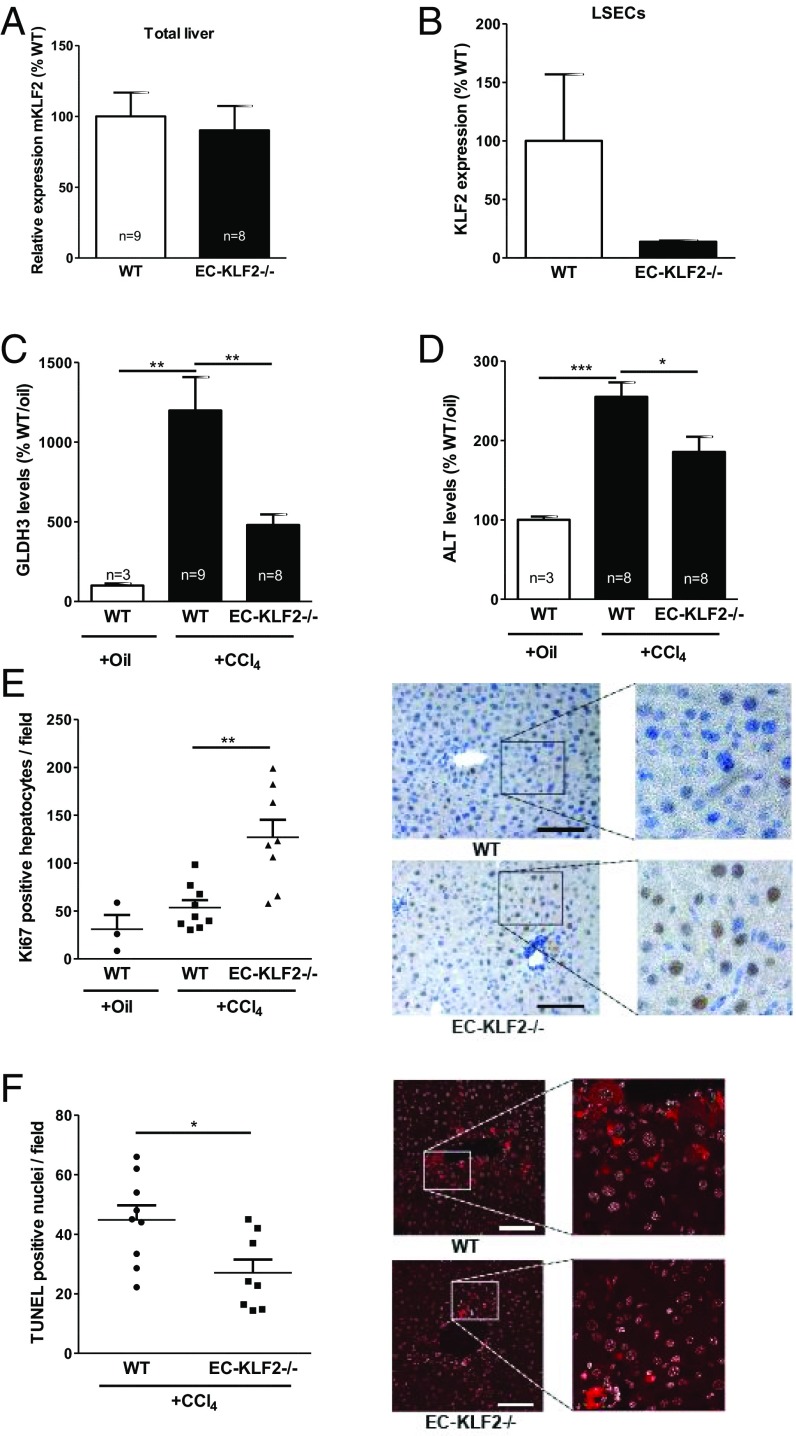
Flow hemodynamics controls the phenotype of endothelial cells (ECs) and may thereby influence liver regeneration. To study if the release of angiocrine factors from LSECs controlled liver regeneration and fibrosis, we analyzed the role of the flow-responsive transcription factor KLF2 in liver regeneration. Toward this end, we used endothelial specifically inducible KLF2-deficient mice [Cdh5-CreERT2;KLF2fl/fl (13), abbreviated EC-KLF2−/−] and wild-type (KLF2fl/fl, abbreviated WT) littermates and subjected these mice to CCl4 for 3 wk to induce chronic liver injury. CCl4 induces liver damage through different mechanisms including the formation of free radicals and disturbance of hepatocyte calcium homeostasis (14, 15). Whereas global KLF2 mRNA expression in the liver was not significantly affected (Fig. 1A), KLF2 mRNA expression in isolated LSECs was strongly reduced in EC-KLF2−/− mice compared with WT (Fig. 1B). Liver injury was determined by measuring serum levels of glutamate dehydrogenase (GLDH3) and alanine aminotransferase (ALT), which were significantly elevated in CCl4-treated mice in comparison with the oil-treated control group (Fig. 1 C and D), whereas aspartate aminotransferase (AST) and alkaline phosphatase (AP) were not changed. Interestingly, there was a significant reduction of the serum levels of GLDH3 and ALT in CCl4-treated EC-KLF2–deficient mice compared with WT mice (Fig. 1 C and D). Hepatocyte proliferation was significantly elevated in CCL4-treated EC-KLF2–deficient mice compared with WT mice (Fig. 1E). Moreover, CCl4-treated EC-KLF2–deficient mice displayed significantly less hepatocyte cell death compared with WT mice (Fig. 1F). These data indicated that loss of endothelial KLF2 expression reduced liver damage in a model of chronic liver injury.
Fig. 1.
Enhanced hepatocyte proliferation after CCl4-induced chronic liver injury in endothelial-specific KLF2-deficient mice. (A) Relative KLF2 expression was measured by qRT-PCR in WT and EC KLF2-/- mice 21 d after CCl4-induced liver damage in total liver tissue. Data are expressed as mean ± SEM (WT-CCl4, n = 9; EC-KLF2, n = 8). (B) KLF2 expression was measured by qRT-PCR in WT and EC-KLF2-/- mice after CCl4-induced liver damage in isolated LSECs. Data are expressed as mean ± SEM (WT, n = 2; EC-KLF2, n = 2). (C and D) Serum GLDH3 and ALT enzyme activity after 21 d of CCl4 treatment were measured in WT and EC-KLF2-/- mice. Data are expressed as mean ± SEM (WT-Oil, n = 3; WT-CCl4, n = 9; EC-KLF2, n = 8). (E) Hepatocyte proliferation in liver sections after 3 wk of CCl4 treatment was analyzed by Ki67 staining. Representative images are shown in the Right panels. Data are expressed as mean ± SEM (WT-Oil, n = 3; WT-CCl4, n = 9; EC-KLF2, n = 8). (Scale bar: 100 µm.) (F) Hepatocyte death after 21 d of CCl4 treatment was determined by quantification of TUNEL-positive nuclei. Representative images are shown in the Right panels. Data are expressed as mean ± SEM (WT-CCl4, n = 9; EC-KLF2, n = 8). (Scale bar: 100 µm.) *P < 0.05, **P < 0.01, ***P < 0.001.
Overexpression of KLF2 in LSECs Reduces Hepatocyte Proliferation.
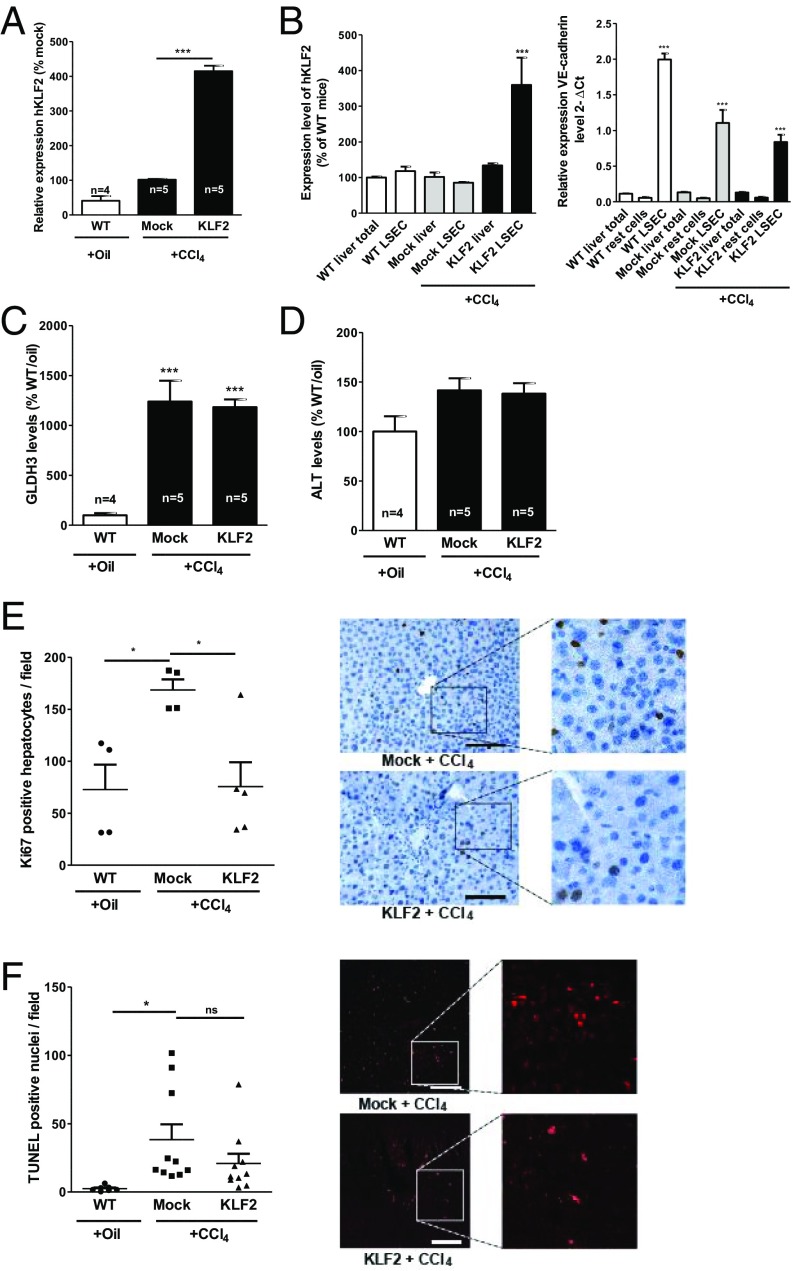
Next, we determined if the overexpression of KLF2 in endothelial cells affected the response to liver injury in WT mice using the same liver injury model (CCl4 treatment). To this end, we generated lentiviral constructs to specifically overexpress human KLF2 in LSECs (Fig. S1A). Selective overexpression of human KLF2 (hKLF2) in endothelial cells was demonstrated upon in vitro (Fig. S1B) and in vivo administration of the lentiviral vector (Fig. 2A). When analyzing cell fractions collected from the liver of hKLF2-treated mice, hKLF2 was specifically enriched in isolated LSECs (Fig. 2B). KLF2 overexpression did not affect CCl4-induced increase of GLDH3 and ALT serum levels compared with mock control groups (Fig. 2 C and D). However, overexpression of KLF2 significantly decreased hepatocyte proliferation (Fig. 2E). TUNEL staining showed significantly increased levels of cell death in CCl4-treated mock controls but not in KLF2-overexpressing mice; however, a direct comparison of the mock- versus hKLF2-treated group failed to show statistical significance (Fig. 2F). Together, these data showed that endothelial-specific overexpression of KLF2 reduced hepatocyte proliferation in vivo.
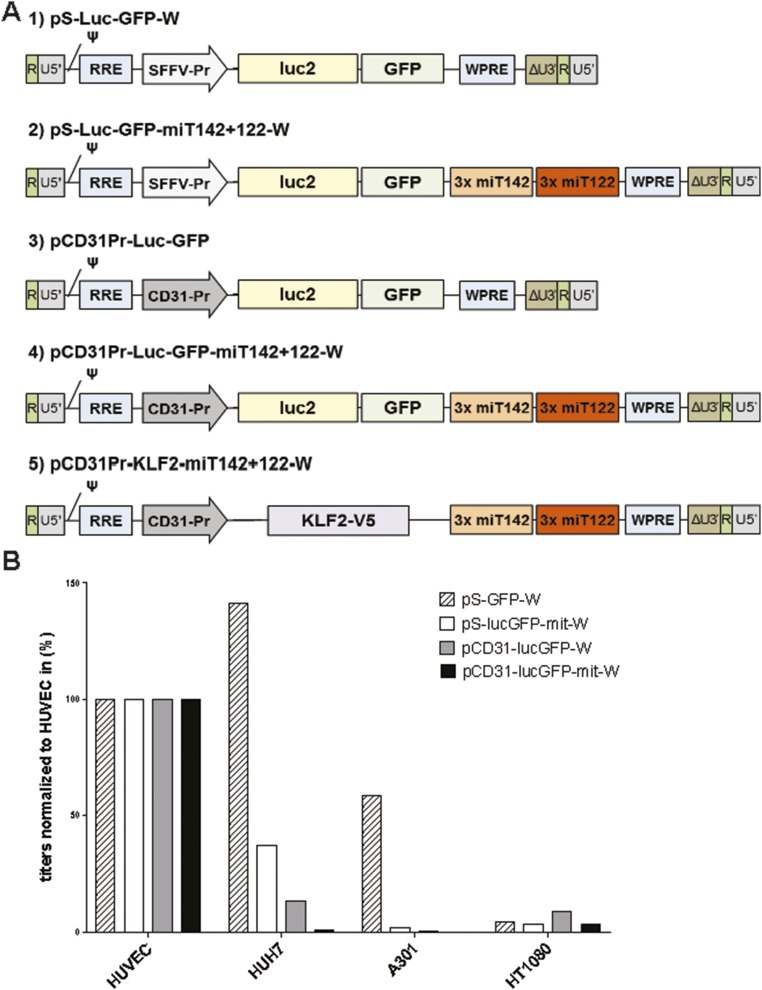
Fig. S1.
Generation of LSEC-specific vectors. (A) Schematic drawing of the transfer vector expression cassettes. Starting from the SFFV promoter containing plasmid pS-luc2-GFP-W (1), three copies of the target sequences for the liver-specific miR122 and the lymphocyte-specific miR142 were inserted into the 3′-UTR of the expression cassette (2). In addition, the ubiquitously expressed SFFV promoter was exchanged against the endothelial-cell–specific, murine CD31 promoter (3). In the third step, both elements were combined to achieve a highly LSEC-specific lentiviral vector (4). In the final step, the reporter construct luc2-GFP was exchanged against human KLF2 containing a C-terminal V5-tag (5). (B) Titers of vector stocks having packaged constructs: pS-GFP-W, pS-lucGFP-mit-W, pCD31-lucGFP-W, and pCD31-lucGFP-mit-W on different cell types. Serial dilutions of concentrated vectors were applied to HUVECs, HUH7 (human hepatoma cells), A301 (human T cells), and HT1080 (human fibrosarcoma). Three days post transduction cells were analyzed by flow cytometry for GFP expression. For each stock the dilutions within the linear range were identified and used to calculate titers in transducing units per mL.
Fig. 2.
KLF2 overexpression in LSECs reduces hepatocyte proliferation after CCl4-induced chronic liver injury. After CCl4-induced liver damage, mice were injected with control virus (mock), KLF2 virus (KLF2), or left untreated (WT). After 3 wk, mice were sacrificed and livers removed. (A) Relative human KLF2 expression was determined by qPCR. Data are expressed as mean ± SEM (WT, n = 3; mock-CCl4, n = 5; KLF2-CCl4, n = 5). (B) Human KLF2 as well as VE-cadherin mRNA expression levels in isolated LSECs compared to those of nonendothelial cells (flow through). (C and D) Serum GLDH3 and ALT enzyme activity expressed as mean ± SEM. (WT-Oil, n = 4; Mock-CCl4, n = 5; KLF2-CCl4, n = 5). (Scale bar: 100 µm.) (E) Hepatocyte proliferation in liver sections was analyzed by Ki67 staining. Representative images are shown in the Right panels. Data are expressed as mean ± SEM (WT-Oil, n = 4; Mock-CCl4, n = 4; KLF2-CCl4, n = 5). (Scale bar: 100 µm.) (F) Hepatocyte death was determined by quantifying TUNEL-positive nuclei. Representative images are shown in the Right panels. Data are expressed as mean ± SEM (WT-Oil, n = 7; Mock-CCl4, n = 10; KLF2-CCl4, n = 10). (Scale bar: 100 µm.) *P < 0.05, ***P < 0.001.
Loss or Gain of KLF2 Expression Does Not Change Capillary Density and Liver Fibrosis.
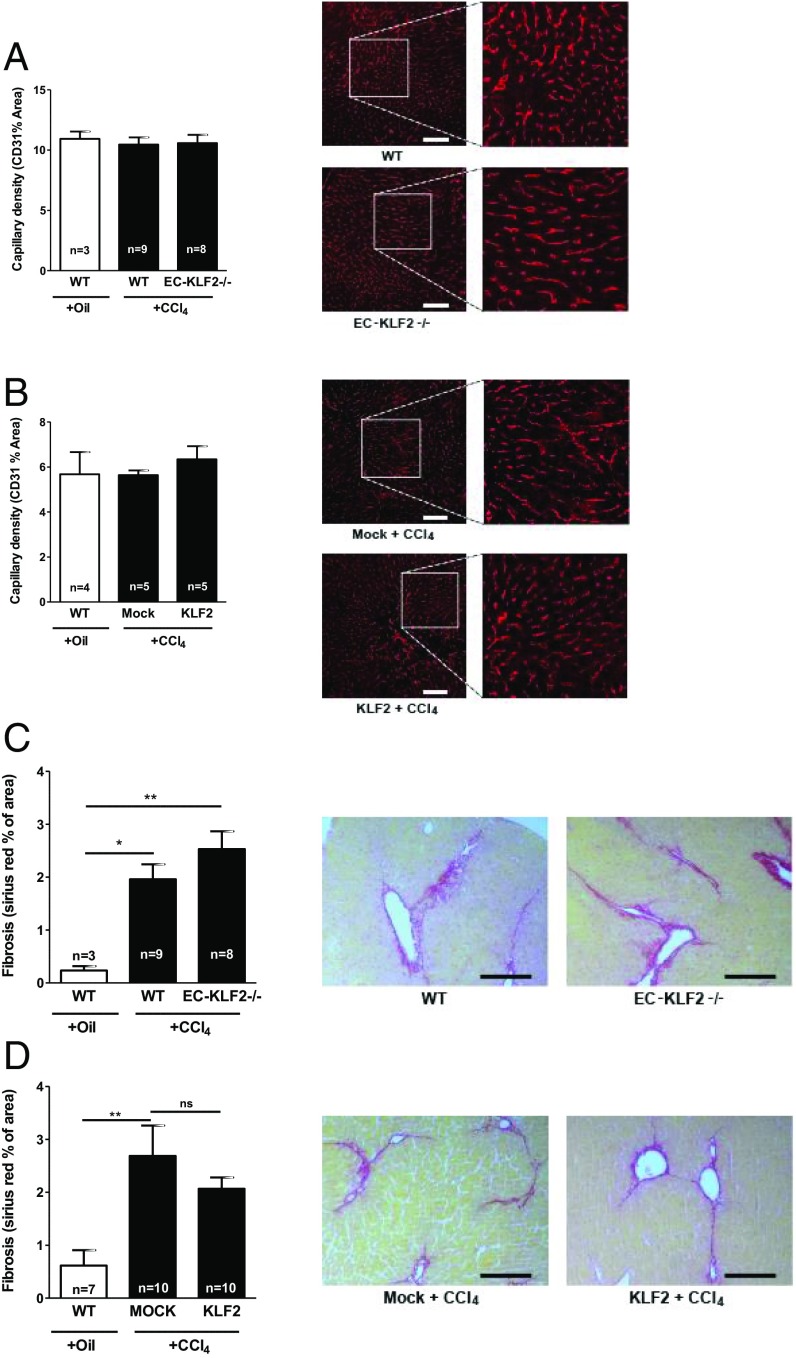
Globally KLF2 heterozygous mice show an increase in capillary density (16). Consistently, adenoviral KLF2 overexpression inhibits angiogenesis (17). We therefore determined whether endothelial-specific inducible KLF2 deletion or overexpression affected vessel density in the liver (Fig. 3 A and B). However, endothelial-specific gain- and loss-of-function of KLF2 did not affect liver microvascular density, suggesting that endothelial KLF2 controlled liver regeneration independently of angiogenesis. One of the hallmarks of liver injury is fibrosis. Therefore, we next assessed liver fibrosis by Sirius red staining. Chronic CCl4 treatment significantly induced liver fibrosis in WT mice and in EC-KLF2–deficient mice without significant differences between the groups (Fig. 3C). CCl4 treatment also significantly induced fibrosis in mock-treated mice, whereas no significant augmentation was detected in CCl4-treated KLF2 transduced mice; however, the differences between mock- and KLF2-treated mice failed to achieve statistical significance (Fig. 3D). These data indicated that endothelial KLF2 did not change endothelial cell density and had no significant effect on liver fibrosis but predominantly affected hepatocyte proliferation.
Fig. 3.
No change in capillary network density and liver fibrosis. (A and B) Capillary density was detected by CD31 staining (indicated by red) in liver sections after 21 d of CCl4 treatment of WT and EC-KLF2-/- mice (A) or (B) WT mice treated with control virus (Mock) or KLF2 virus. Data are expressed as mean ± SEM (WT-Oil, n = 3; WT-CCl4, n = 9; EC-KLF2, n = 8; WT-Oil, n = 4; mock-CCl4, n = 5; KLF2-CCl4, n = 5). (Scale bars: 100 μm.) (C) Liver fibrosis (Sirius red staining) was measured in liver sections after 21 d of CCl4 treatment of WT and EC-KLF2-/- mice. Data are expressed as mean ± SEM (WT-Oil, n = 3; WT-CCl4, n = 9; EC-KLF2, n = 8). (Scale bar: 100 μm.) (D) Liver fibrosis (Sirius red staining) was measured in liver sections after 21 d of CCl4 treatment of WT mice treated with control virus (mock) or KLF2 virus. Data are expressed as mean ± SEM (WT-Oil, n = 7; Mock-CCl4, n = 10; KLF2-CCl4, n = 10). (Scale bar: 100 μm.) Representative images are shown in the Right panels of A–D. *P < 0.05, **P < 0.01.
KLF2 Induces the Secretion of Activin A from Endothelial Cells.
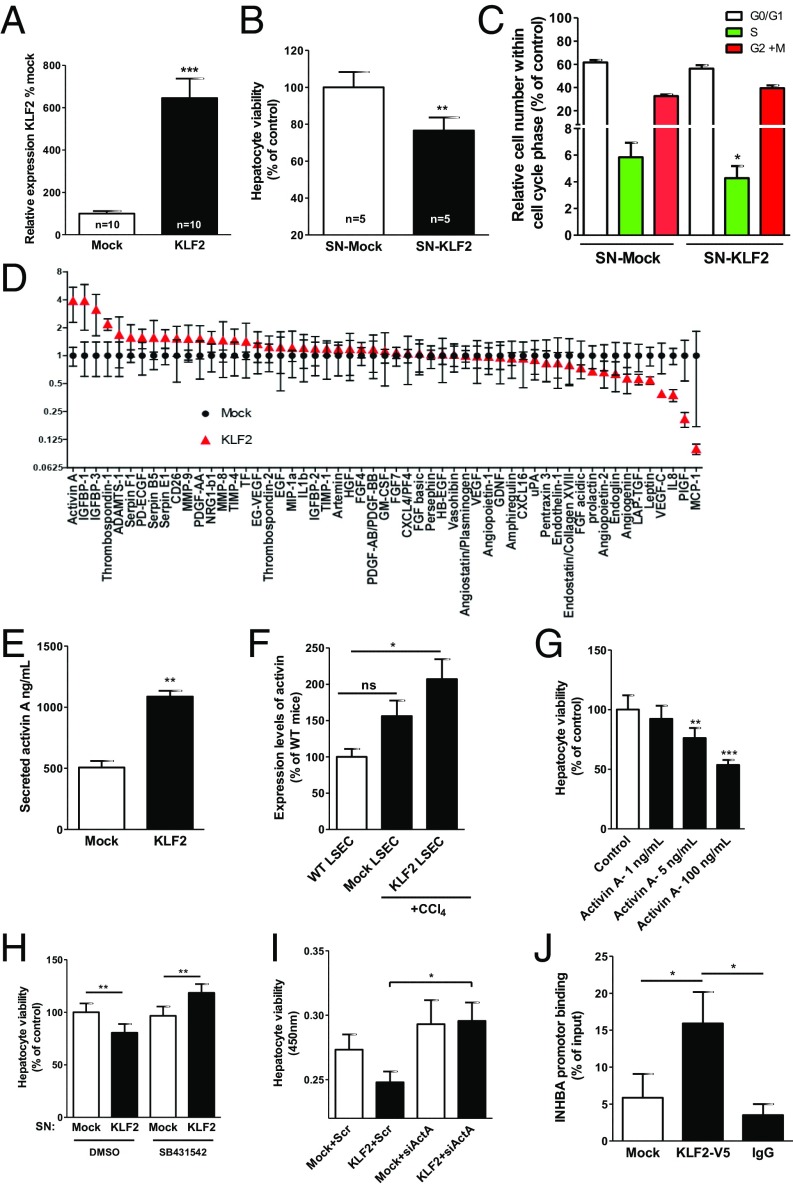
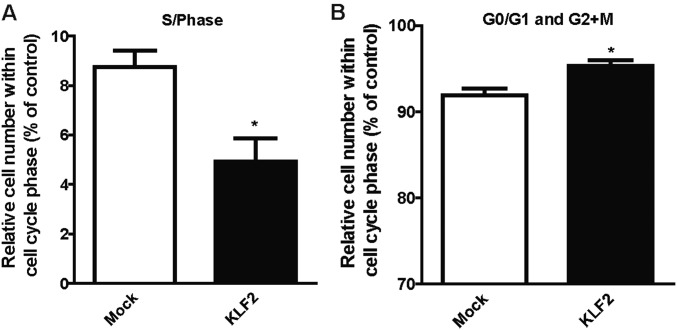
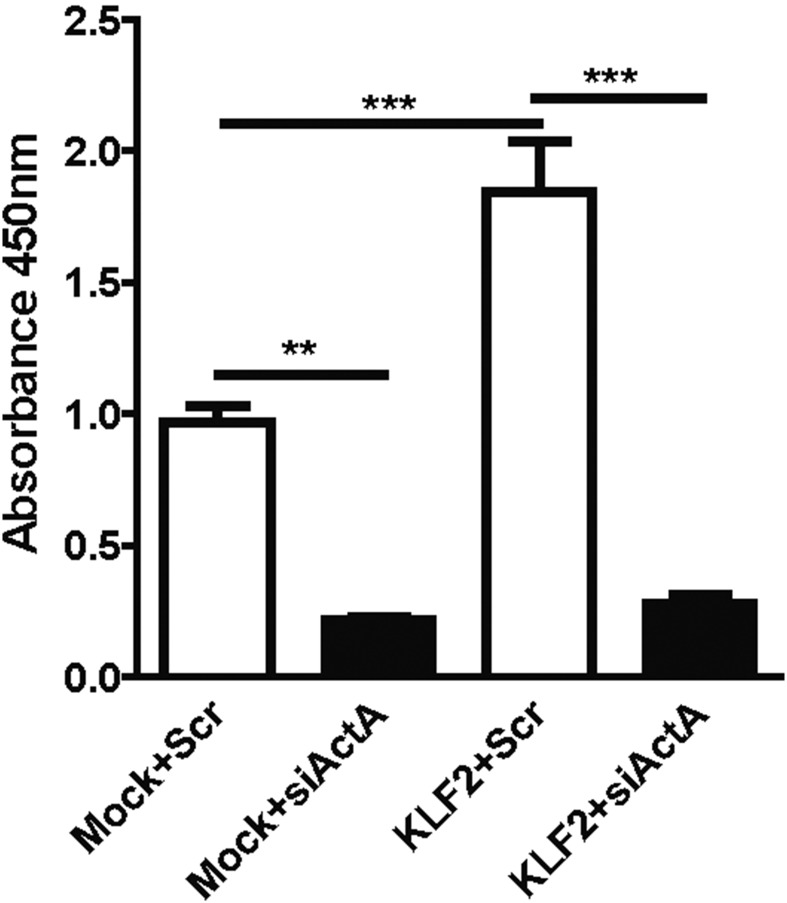
To obtain insight into the mechanisms of endothelial KLF2 control of hepatocyte proliferation, we determined whether endothelial cells communicate in a paracrine manner with hepatocytes. Therefore, we overexpressed KLF2 by using lentiviral vectors (18) in ECs in vitro (Fig. 4A) and determined whether supernatants (SNs) of endothelial cells overexpressing KLF2 regulated hepatocyte proliferation. Indeed, SNs from KLF2-overexpressing cells reduced hepatocyte cell number (Fig. 4B). This was confirmed by a BrdU incorporation assay, which showed that SNs from KLF2-overexpressing cells reduced the S-phase and concomitantly increased the overall number of hepatocytes in the G0/G1 and G2/M phases (Fig. 4C and Fig. S2 A and B). To determine how KLF2 reduced hepatocyte proliferation, we next analyzed angiocrine factors in SNs derived from ECs overexpressing KLF2, using an angiocrine factor antibody array. Intriguingly, several factors were significantly affected by KLF2 overexpression in comparison with mock controls (Fig. 4D). We validated these results by measuring secretion of selected factors with a potential role in liver regeneration, such as activin A, IGFBP-3, EGF, VEGF-C, Ang-2, and endoglin in SNs of KLF2 overexpression ECs by ELISA (Fig. S3).
Fig. 4.
KLF2 regulates activin A, and activin A inhibits the proliferation of hepatocyte. (A) Relative expression of KLF2 in HUVECs after lentiviral KLF2 transduction/overexpression. Data are expressed as mean ± SEM (Mock, n = 10; KLF2, n = 10). (B) Hepatocyte proliferation was measured by a Cell Counting Kit-8 (CCK-8) kit after incubation with SNs from HUVEC-overexpressing KLF2 for 18 h. Data are expressed as mean ± SEM (Mock-SN, n = 5; KLF2-SN, n = 5). (C) Hepatocyte cell-cycle analysis was detected by BrdU staining upon treatment with SN from HUVEC-overexpressing KLF2 for 18 h. Data are expressed as mean ± SEM (Mock-SN, n = 4; KLF2-SN, n = 4). (D) Angiocrine factor secretion was measured in SNs from HUVEC-overexpressing KLF2 and mock-transfected cells. Data are expressed as mean ± SEM (Mock-SN, n = 3; KLF2-SN, n = 3). (E) Activin A was measured in supernatant from endothelial cells overexpressing KLF2 and mock. Data are expressed as mean ± SEM (Mock, n = 4; KLF2, n = 4). (F) Relative mRNA expression of activin A detected in LSECs isolated of WT mice, CCL4-treated WT mice additionally injected with control virus (mock), or KLF2 virus. Data are expressed as mean ± SEM (WT, n = 3; mock-CCl4, n = 5; KLF2-CCl4, n = 5) (G) Hepatocyte proliferation was dose-dependently inhibited by recombinant activin A. Data are expressed as mean ± SEM (n = 14). (H) Hepatocyte proliferation was measured by the CCK-8 kit after simultaneous treatment with SNs from KLF2-overexpressing HUVECs and the Alk5 receptor inhibitor (SB431542, 10 µM), which blocks activin A signaling. Data are expressed as mean ± SEM (n = 4). (I) Hepatocyte proliferation was measured by the CCK-8 kit after simultaneous treatment with SNs from KLF2-overexpressing HUVECs and siRNA against activin A. Data are expressed as mean ± SEM (n = 4). (J) Chromatin immunoprecipitation experiments revealed direct binding of V5-tagged KLF2 to the activin A promoter. Data are expressed as mean ± SEM (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. S2.
KLF2 inhibits the proliferation of hepatocytes. Hepatocyte cell-cycle analysis was detected by BrdU staining after treatment with SNs from HUVEC-overexpressing KLF2 for 18 h. (A) Hepatocytes in S-phase. (B) Hepatocytes in G0/G1 and G2+M phases. Data are expressed as mean ± SEM (Mock-SN, n = 4; KLF2-SN, n = 4). *P < 0.05.
Fig. S3.
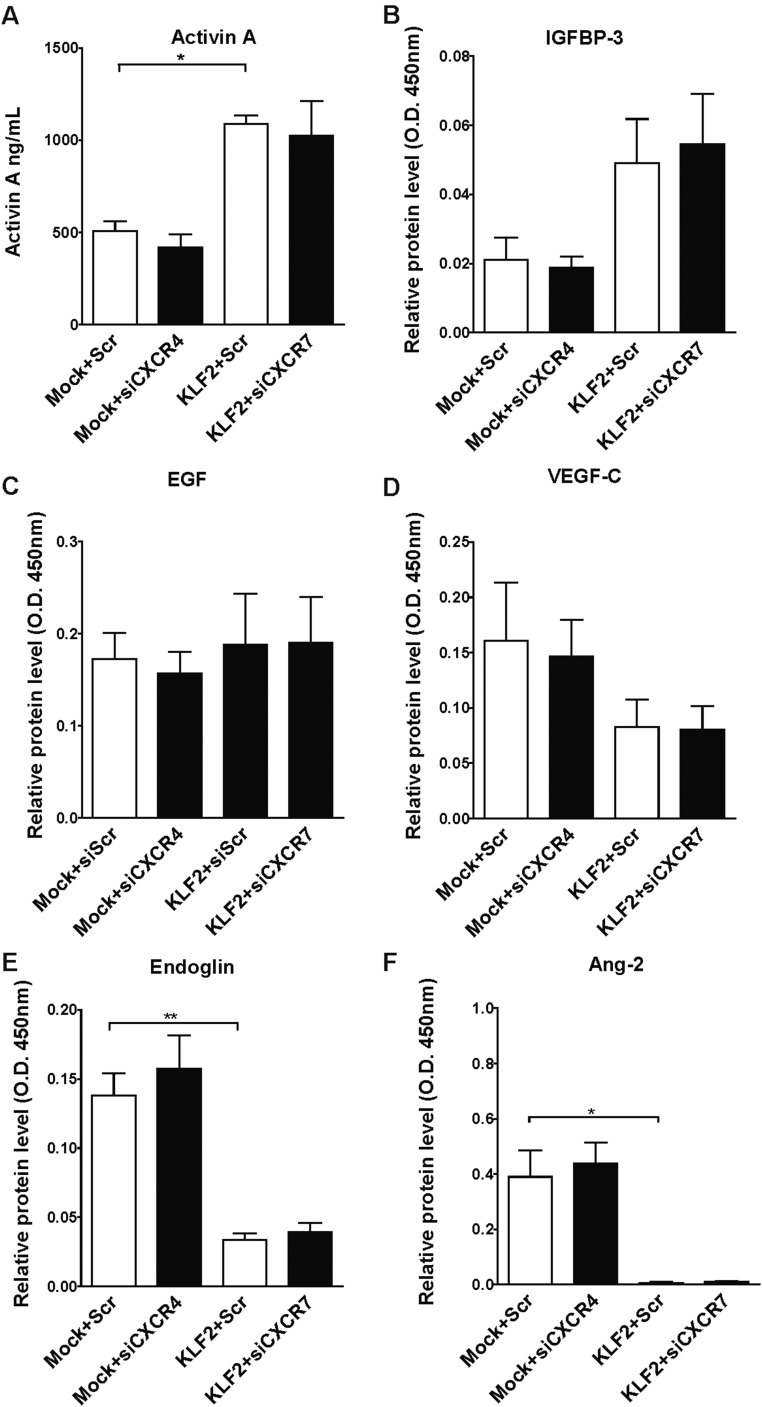
KLF2 affects the secretion of angiocrine factors. (A–F) HUVECs were transduced with lentivirus to overexpress KLF2 or with Mock lentivirus. At least 4 d after transduction, cells were transfected with siRNA to silence CXCR4, CXCR7, or nontargeting control siRNAs. Two days after transfection, the supernatants of these cells were analyzed by ELISA to determine levels of (A) activin A, (B) IGFBP3, (C) EGF, (D) VEGFC, (E) endoglin, and (F) Ang2. Data are expressed as mean ± SEM; n = 3–4 per group. *P < 0.05, **P < 0.01.
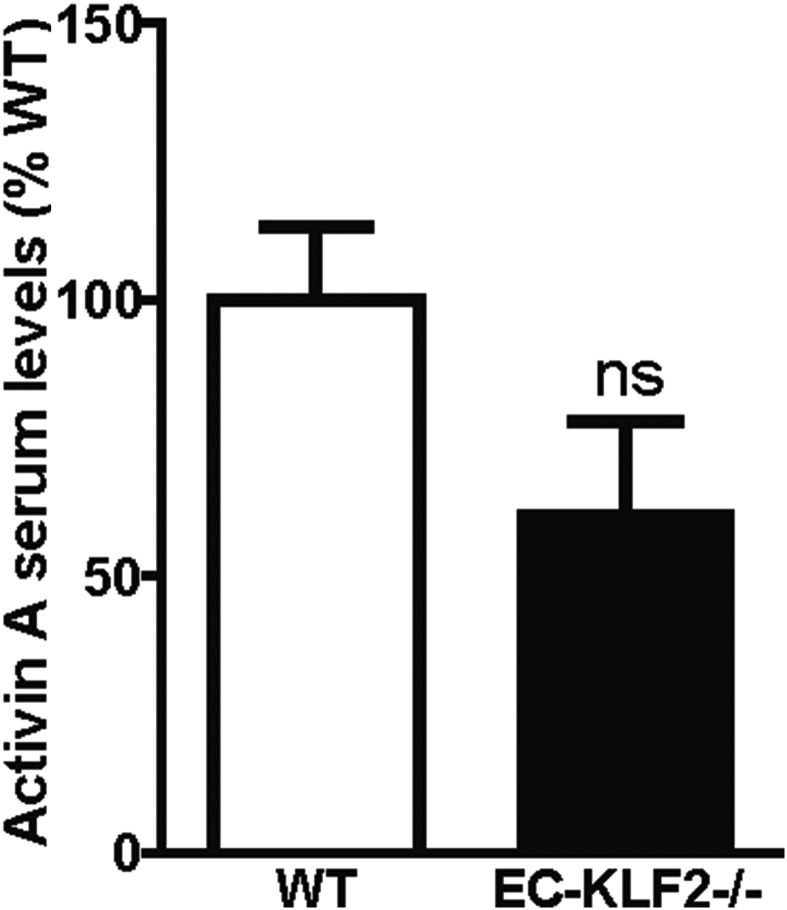
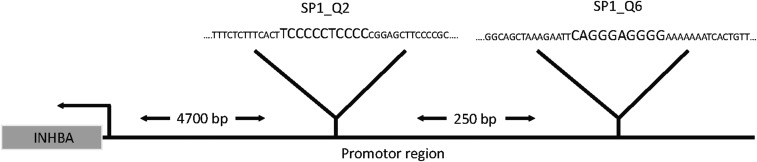
The most highly up-regulated factor after KLF2 overexpression was activin A (Fig. 4E). Because activin A has been shown to regulate hepatocyte proliferation and liver regeneration (19), we selected this factor for further analysis. Consistent with the regulation of activin A by KLF2 in vitro, activin A levels were significantly increased in the LSECs isolated from KLF2-transduced mice (Fig. 4F), but no significant reduction of activin A plasma levels was detectable in EC-KLF−/− mice (Fig. S4). To assess whether activin A was responsible for KLF2-dependent regulation of hepatocyte proliferation, we determined the effect of recombinant activin A on hepatocyte proliferation in vitro. Indeed, hepatocyte proliferation was diminished by activin A (Fig. 4G). Next, we determined whether blocking activin A signaling in hepatocytes reversed the inhibitory effects of the supernatant of KLF2-overexpressing cells on hepatocyte proliferation. Because activin A is part of the TGF-β–signaling pathway, we used the pharmacological receptor blocker for ALK5 SB431542 (20) to inhibit activin A downstream signaling. Inhibition of activin A activity prevented the inhibitory effect of the SNs of KLF2-overexpressing ECs on hepatocyte proliferation (Fig. 4H). These results were confirmed by siRNA-mediated silencing of activin A (Fig. 4I and Fig. S5). To determine how KLF2 induced activin A secretion, we analyzed the activin A promoter region and found conserved KLF2-binding sites (Fig. S6). Subsequent chromatin immunoprecipitation experiments with V5-tagged KLF2 showed that KLF2 indeed bound directly to the activin A promotor (Fig. 4J). In summary, these data showed that KLF2 induced activin A expression and secretion from endothelial cells, which inhibited hepatocyte proliferation.
Fig. S4.
KLF2 induces the secretion of activin A in vivo. Serum activin A levels were measured in WT and EC-KLF2−/− mice by ELISA. Data are expressed as mean ± SEM (WT, n = 6; EC-KLF2, n = 6).
Fig. S5.
Activin A-dependant hepatocyte proliferation. ELISA measurement of activin A in supernatant from HUVECs transduced with mock or KLF2 vector and additionally transfected with siRNA for activin A or scramble control as indicated. Data are expressed as mean ± SEM (n = 4) **P < 0.01, ***P < 0.001.
Fig. S6.
KLF2-binding sides at the activin A promotor. Schematic drawing of conserved KLF2-binding sites in the activin A promotor region.
Endothelial KLF2 Contributes to Liver Damage via Activin A Secretion.
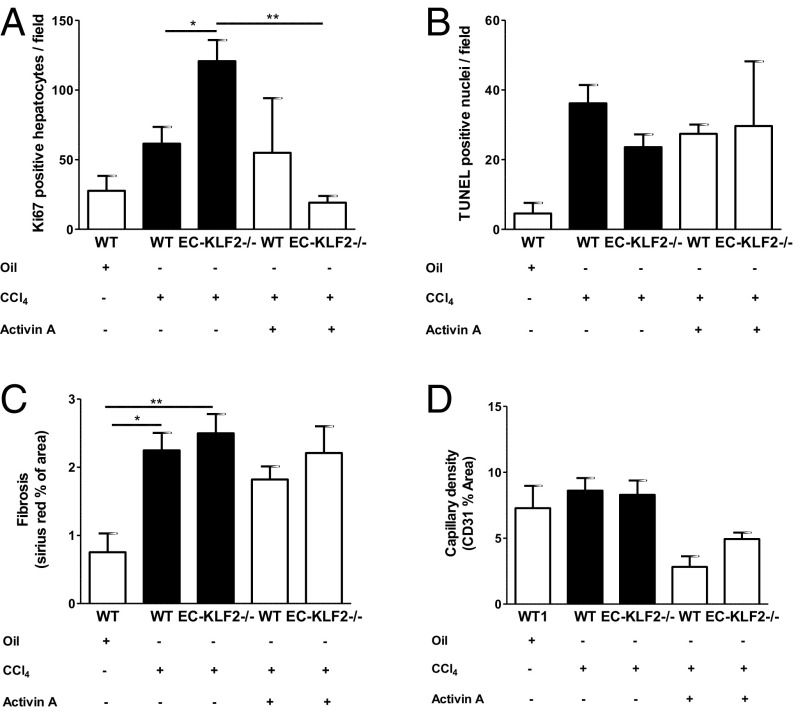
After demonstrating that KLF2-mediated induction of activin A regulated hepatocyte proliferation in vitro, we aimed to address the in vivo relevance of these findings. We administered recombinant activin A in combination with CCl4 for 3 wk and measured the effects on hepatocyte proliferation and cell death in EC-KLF2−/− compared with WT mice. Exogenous activin A blocked the induction of hepatocyte proliferation in EC-KLF2–deficient mice (Fig. 5A), whereas hepatocyte death remained largely unaffected (Fig. 5B). Furthermore, activin A did not affect liver fibrosis (Fig. 5C), but tended to reduce capillary density (Fig. 5D). Together, the results indicated that KLF2 induced an antiproliferative secretome, including activin A, which attenuated liver regeneration.
Fig. 5.
Endothelial KLF2 contributes to liver damage via activin A secretion in vivo. (A) Hepatocyte proliferation in liver sections after 3 wk of simultaneous CCl4 and recombinant activin A treatment was analyzed by Ki67 staining in WT and EC-KLF2-/- mice. Data are expressed as mean ± SEM (WT-Oil, n = 6; WT-CCl4, n = 12; EC-KLF2, n = 12; WT-CCl4+activin A, n = 4; EC-KLF2–CCl4+activin A, n = 4). (B) Hepatocyte death was detected by TUNEL staining in liver sections after 3 wk of simultaneous CCl4 and recombinant activin A treatment. Data are expressed as mean ± SEM (WT-Oil, n = 3; WT- CCl4, n = 13; EC-KLF2, n = 11; WT-CCl4+activin A, n = 4; EC-KLF2–CCl4+activin A, n = 4) (C) Liver fibrosis was determined by Sirius red staining in liver sections after 3 wk of simultaneous CCl4 and recombinant activin A treatment in WT and EC-KLF2-/- mice. Data are expressed as mean ± SEM (WT-Oil, n = 6; WT-CCl4, n = 13; ECKLF2, n = 12; WT-CCl4+activin A, n = 4; EC-KLF2–CCl4+activin A, n = 4). (D) Capillary density (CD31) was examined in liver sections after 3 wk of simultaneous CCl4 and recombinant activin A treatment in WT and EC-KLF2-/- mice. Data are expressed as mean ± SEM (WT-Oil, n = 6; WT-CCl4, n = 13; EC-KLF2, n = 12; WT-CCl4+activin A, n = 4; EC-KLF2–CCl4+activin A, n = 4).
Discussion
In this study, we aimed to investigate the role of the flow-responsive transcription factor KLF2 in liver regeneration. We show that endothelial cell-specific deletion of KLF2 in mice reduces liver damage and augments hepatocyte proliferation in a chronic CCl4-mediated liver injury model. We further demonstrate that KLF2 overexpression in vitro induces activin A expression and secretion from endothelial cells, which inhibits hepatocyte proliferation. Taken together, the results indicate that KLF2 induces an antiproliferative secretome, including activin A, which attenuates liver regeneration. These findings are further supported by endothelial-specific overexpression of KLF2. Although we cannot rule out that KLF2 is also expressed in some nonendothelial cells, the vector preferentially transduced liver endothelial cells, resulting in reduced hepatocyte proliferation in vivo. However, KLF2 overexpression did not affect the CCl4-induced damage as measured by the release of GLDH3 and ALT as well as TUNEL staining. We hypothesize that endogenous endothelial KLF2 is required to protect hepatocytes against cell death, but overexpression of KLF2 in endothelial cells cannot further augment this cell protective effect.
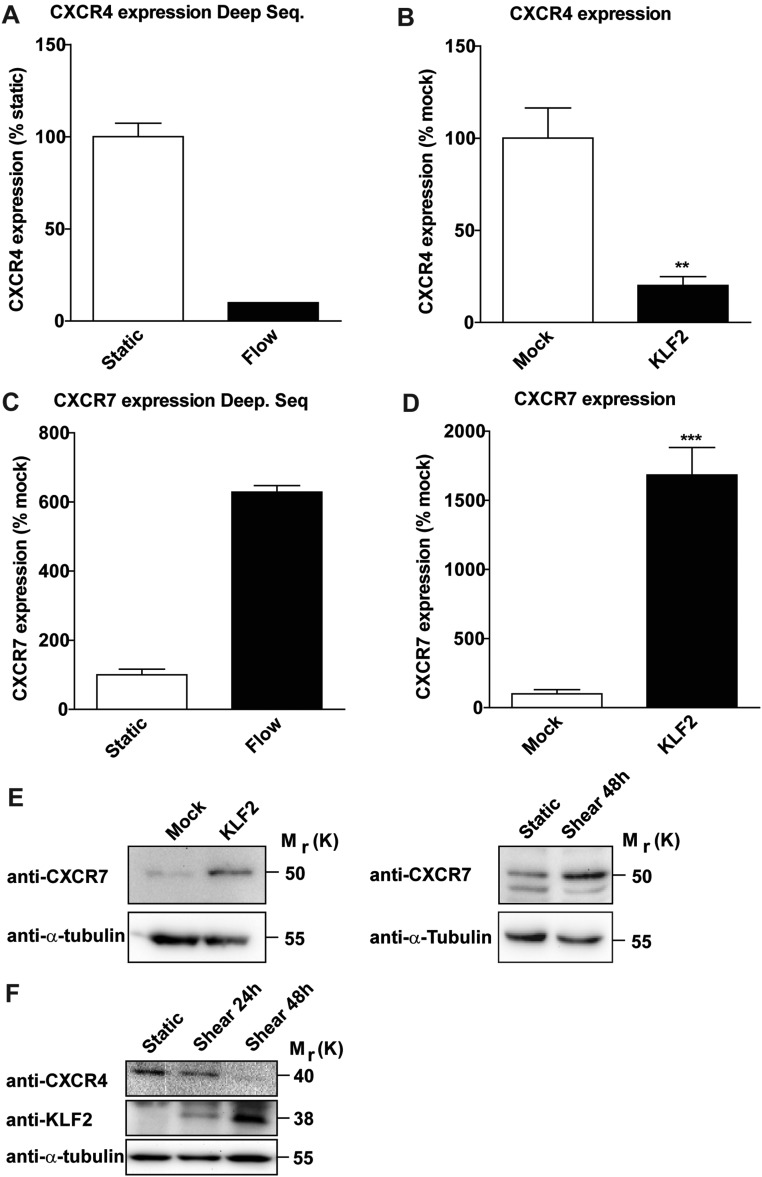
Endothelial cells not only are important for oxygen delivery, but also act as a paracrine source for signals that determine tissue regeneration and fibrosis (5). Because vascular density was not changed either upon endothelial-specific overexpression or deletion of KLF2, we hypothesized that KLF2 does not affect angiogenesis or endothelial cell survival, but regulates liver regeneration by controlling the endothelial secretome. Endothelial-mediated paracrine regulation of liver regeneration was shown to be critically controlled by the SDF-1 receptors CXCR4, which mediates the release of profibrotic cytokines, and CXCR7, which was shown to provide a proregenerative niche (4). Interestingly, we demonstrate that in vitro overexpression of KLF2 in ECs reciprocally regulates CXCR4 and CXCR7 expression on mRNA as well as protein level (Fig. S7). However, analysis of angiocrine factor secretion after silencing of CXCR4 or CXCR7 did not reveal significant differences in the secretome profile (Fig. S3), indicating that the CXCR4/CXCR7 balance may not play a major role in KLF2-mediated secretion of paracrine active factors by endothelial cells, at least in vitro.
Fig. S7.
KLF2 regulates CXCR4 and CXCR7 expression in ECs. (A) RNA sequencing data of CXCR4 expression after exposure of HUVECs to high shear stress at 20 dynes/cm2 for 72 h compared with static control (n = 2). (B) Relative CXCR4 expression in endothelial cells overexpressing KLF2 using lentivirus or Mock-transduced controls. Data are expressed as mean ± SEM (Mock, n = 7; KLF2, n = 7). (C) Deep sequencing data of CXCR7 expression after exposure of HUVECs to high shear stress at 20 dynes/cm2 compared with static control (n = 2). (D) Relative CXCR7 expression in endothelial cells overexpressing Mock and KLF2. Data are expressed as mean ± SEM (Mock, n = 7; KLF2, n = 7). (E and F) Total cell protein lysates from mock- and KLF2-transduced cells as well as HUVECs exposed to laminar shear stress at 20 dynes/cm2 were analyzed by Western blot and probed with antibodies against (E) CXCR7, (F) CXCR4, and α-tubulin, which served as a loading control. **P < 0.01, ***P < 0.001.
KLF2 overexpression in vitro regulated the secretion of various cytokines, and one of the most prominent up-regulated cytokines was activin A. Activin A has an important role during liver regeneration. It was shown that activin A inhibits hepatocyte proliferation, and administration of the activin A antagonist follistatin led to acceleration of liver regeneration after partial hepatectomy (21). During liver regeneration, activin A and its receptors display a characteristic time-dependent expression: activin A receptors are down-regulated immediately after partial hepatectomy. However, activin A’s expression is normalized during the subsequent 72 h, which may allow for the response to mitogenic stimuli (7). After CCl4-induced liver injury, activin A expression is elevated, which results in liver fibrosis (8). Blocking the activity of activin A with anti-activin A antibodies after CCl4 treatment reduces the necrotic area and the secretion of serum transferases (22). However, activin A was also shown to induce the renewal of liver architecture by inducing collagen production in hepatic stellate cells (HSC) and tubulogenesis of LSECs (23, 24). However, consistent with conflicting reports showing no effect of activin A on collagen production (24), we also did not observe significant effects of KLF2 on fibrosis (Fig. 3 C and D) or collagen mRNA expression.
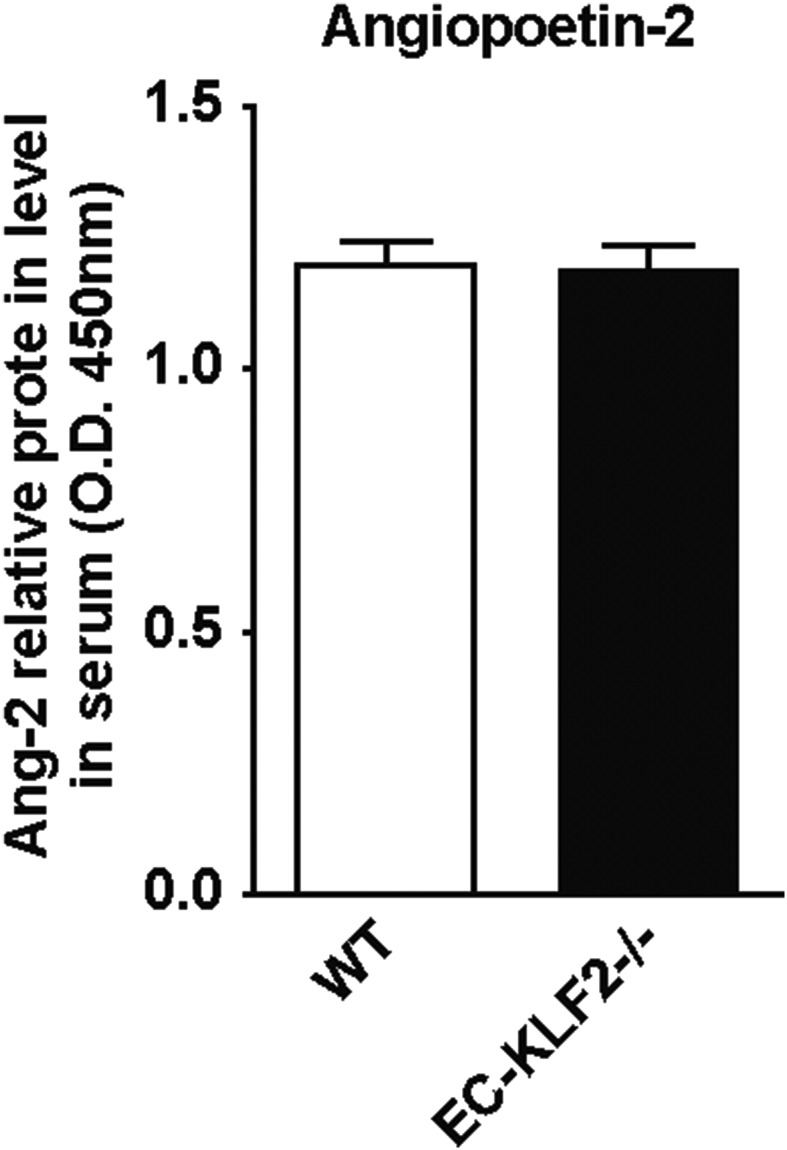
In endothelial-specific KLF2-deficient mice, we observed diminished levels of circulating activin A, which is associated with an increase in hepatocyte proliferation and a decrease in hepatocyte death in these mice. However, recombinant activin A treatment in combination with CCl4 for 3 wk only modestly reduced the increase in proliferating hepatocytes observed in EC-KLF2–deficient mice, and cell death was not affected. These findings suggest that activin A preferentially affects hepatocyte proliferation, which is in line with the biological effect of activin A on DNA synthesis (25, 26). Of note, addition of recombinant activin A only partially reversed the phenotype of EC-KLF2–deficient mice, suggesting that other factors contribute to the observed phenotype. Indeed, KLF2 regulated the secretion of many additional factors in endothelial cells in vitro, which may contribute to the regulation of liver regeneration in vivo. For example, Ang-2 expression plays a critical role during liver regeneration in vivo (3). Indeed, overexpression of KLF2 led to significant down-regulation of Ang-2 in vitro (Fig. S3F), but circulating Ang-2 levels remained unchanged in EC-KLF2–deficient mice (Fig. S8). Endoglin, another factor, which was down-regulated in KLF2-overexpressing ECs, has been shown to impact hepatic fibrosis by controlling TGF-β signaling through ALK-Smad pathways (27). Finally, overexpression of KLF2 leads to secretion of IGFBP-3, which is associated with fibrosis and steatosis of nonalcoholic fatty liver disease (28). All of these factors combined likely mediate the antiregenerative effects of endothelial KLF2 in the liver.
Fig. S8.
Endothelial-specific deletion of KLF2 does not affect Ang-2 serum levels. Serum Ang-2 levels were measured in WT and EC-KLF2−/− mice by ELISA. Data are expressed as mean ± SEM (WT, n = 6; EC-KLF2, n = 6).
Taken together, our results demonstrate the important role of liver endothelial cells as a source for paracrine factors, which contribute to the liver microenvironment that regulates liver regeneration and fibrosis. Notably, the findings of this study highlight a role for activin A as one major angiocrine factor released from endothelial cells, which controls liver regeneration by regulating hepatocyte proliferation and liver homeostasis. Although liver endothelial cells express higher levels of activin A compared with nonendothelial cells (Fig. S9), we cannot exclude that other cells also provide activin A. Finally, the data highlight a central role for the endothelial KLF2-activin A axis as a negative regulator of liver regeneration. Because KLF2 is mainly described as a flow-induced transcription factor, we hypothesize that absence of blood flow, in the case of liver injury, facilitates liver regeneration. Upon restoration of blood flow, the up-regulation of KLF2 and activin A inhibits hepatocyte proliferation to allow normal liver homeostasis.
Fig. S9.
Activin A expression levels (qPCR) in LSECs and non-LSEC liver cells. Activin A mRNA expression levels in isolated LSECs compared with those of nonendothelial cells (flow through). Data are expressed as mean ± SEM (WT, n = 3; mock-CCl4, n = 5; KLF2-CCl4, n = 5).
Materials and Methods
Cell Culture and in Vitro Assays.
Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and cultured in endothelial basal medium (EBM) (Lonza) supplemented with 10% FBS (Invitrogen) and EGM-SingleQuots (Lonza). The hepatocyte cell line AML-12 was cultured in DMEM nutrient mixture F-12 HAM medium (Sigma) with 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 5 ng/mL selenium, and 40 ng/mL dexamethasone, 90%, and FBS, 10%. For more details SI Materials and Methods.
Viral Vectors.
The LSEC-specific lentiviral vector CD31-KLF2-V5-miT122+142-LV (KLF2) was made LSEC-specific by a combination of transcriptional targeting and microRNA (miRNA)-detargeting, according to ref. 29. In detail, the spleen focus forming virus (SFFV) promoter of the pSEW (30) transfer vector plasmid was exchanged by the endothelial-cell–specific CD31 promoter and two triple miRNA-detargeting sequences matching miRNA142-3P (31) and miRNA122 target sites (29, 32) were inserted 3′ to the expression cassette (Fig. S1). After verification of specific expression in target cells (Fig. S1), the sequence of the reporter gene was exchanged by the human KLF2-V5 sequence (12). VSVG-pseudotyped vectors were produced by cotransfection in 293T cells and were concentrated by low-speed centrifugation (30). Long-term overexpression of KLF2 and Mock was done as previously described (33). Lentiviral particles were generated as previously described (12).
Human Angiogenesis Array.
Secreted proteins were quantified by the human angiogenesis array (Proteomic profilerTM; R&D) according to the manufacturer’s instructions. SNs from mock and KLF2 transduced cells were collected and concentrated by centrifugation (150,000 × g for 2 h) and stored at −80 °C.
Mouse Experiments and Immunohistochemistry.
Cdh5-CreERT2 mice (34) and KLF2 flox/flox (35) were described previously and kindly provided by R. Adams, Max Planck Institute for Molecular Biomedicine, Department of Tissue Morphogenesis, Muenster, Germany, and E. Sebzda, Vanderbilt University, Nashville, TN, respectively. Mice were administered with seven injections of tamoxifen (2 mg, Sigma) intraperitoneally over a period of 2 wk (five times in the first week and two times in the second week). Litter mates that do not have the Cdh5-CreERT2 transgene (KLF2fl/fl) received the same tamoxifen administrations and served as WT controls. The animals were then treated twice weekly with CCl4, 0.5 µg/g dissolved in peanut oil, for 3 wk to induce chronic liver injury after which they were killed. Recombinant activin A (100 µg/mouse; R&D) was additionally administered by an Alzet microosmotic pump for rescue experiments. LSEC-specific KLF2 overexpression was achieved by tail-vein injection of CD31-KLF2-V5-miT122+142-LV or Mock-LV in PBS in 8-wk-old C57BL/6 mice (Charles River). CCl4 treatment was initiated as described above 4 d post vector application. All mouse experiments were carried out in accordance with the principles of laboratory animal care as well as according to the German national laws. The studies were approved by the local ethical committee (FU/ 1026 Regierungspräsidium Darmstadt, Hessen, Germany). For more details, SI Materials and Methods.
Statistical Analysis.
Data are expressed as mean ± SEM. Microsoft Excel or GraphPad Prism 5 software was used to assess statistical significance. Two treatment groups were compared by Student’s t test. Multiple group comparisons were done by ANOVA using Tukey test. Results were considered statistically significant when *P < 0.05, **P < 0.01, and ***P < 0.001.
SI Materials and Methods
RNA Isolation and Real-Time Quantitative PCR.
RNA isolation was performed using miRNeasy Kit (Qiagen) according to the manufacturer’s instruction. cDNA was generated from 1 µg total RNA using MuLV reverse transcriptase (Life Technologies) and random hexamers (Thermo Scientific). For gene expression analysis, real-time qPCR was applied by using Fast SYBR Green (Applied Biosystems) in a StepOnePlus machine (Applied Biosystems). Gene expression was normalized to RPLP0.
Cell Viability.
Proliferation assays were performed with a CCK-8 kit (Dojindo Laboratories) according to the manufacturer’s protocol. This assay allows the determination of cell viability in proliferating cells by measuring dehydrogenase activity, which is directly proportional to the number of living cells. AML-12 cells were incubated with supernatant from mock- or KLF-overexpressing HUVECs for 24 h and 48 h at 37 °C and 5% CO2. At each indicated time point, 10 μL CCK-8 solution was added to the cells and incubated for 2 h at 37 °C. Measurement of formazan was performed with a plate reader at 450 nm at the above-mentioned time points (Synergy HT; BioTek Instruments).
Proliferation Assay.
Hepatocytes (AML-12) were incubated with 10 mM BrdU for 60 min and subsequently stained using the BrdU Flow Kit (BD Pharmingen). According to the manufacturer’s instruction, cells were stained with 2.5 µL of anti–BrdU-V450 for 20 min and 10 µL of 7-AAD for 10 min, both at room temperature, and were analyzed using FACS Canto II device (BD Biosciences).
ELISAs.
Levels of activin A, Ang-2, endoglin, IGFBP-3, EGF, and VEGF-C in serum or in SNs of KLF2-overexpressing endothelial cells were measured using Quantikine Activin A ELISA (R&D) according to the manufacturer’s instructions.
Chromatin Immunoprecipitation.
KLF2-V5– or mock-transduced HUVECs were washed twice with PBS before cross-linking with 37% formaldehyde for 10 min at room temperature. Cells were lysed with cytoplasmic lysis buffer, followed by nuclear lysis buffer. Extracts of nuclei were sonified using Bransons Diagenode for 45 cycles on high amplitude (30 s on followed by 30 s off) and 30 cycles on low amplitude (30 s on followed by 30 s off). Lysates were centrifuged at 10,000 × g for 10 min to remove cellular debris, and SNs were incubated with magnetic beads and anti-V5 antibody. Immunocomplexes were washed four times with low salt buffer, high salt buffer, LiCl buffer, and PBS. Cross-linking was reversed by Proteinase K digestion at 62 °C for 2 h, and DNA was isolated using phenol:chloroform:isoamyl alcohol extraction followed by qPCR (primer sequences were INHBA-for ACCAGCAAGGGCAACTTCTT and INHBA-rev AGATTCCCAGCTGCACTCAC).
Western Blot.
Cells were lysed in RIPA lysis buffer (Sigma) supplemented with protease inhibitors (Roche) for 15 min on ice. Cell lysates were centrifuged at 20,000 × g at 4 °C, and the protein concentration was determined by the Bradford method. Equal amounts of protein were loaded on SDS-polyacryamide gels and were separated. Proteins were blotted onto nitrocellulose membrane, and the following antibodies were used to analyze anti-V5 (1:1,000; Thermo Scientific): CXCR4 (1:1,000; Millipore), CXCR7 (1 µg/mL; Millipore), SEMA3B (1:5,000; Thermo Scientific), and α-tubulin (mouse monoclonal anti-human tubulin, 1:1,000; NeoMarkers). Secondary antibodies were purchased from Jackson ImmunoResearch.
Immunochemistry and Immunofluorescence Staining.
Liver samples were fixed overnight in 4% paraformaldehyde. Tissues were subsequently embedded in paraffin for additional analysis. Paraffin sections were used for different stainings with appropriate primary antibodies as follows: rat anti-CD31 (1:30, DIA-310; Dianova) and rabbit anti-Ki67 (1:100, Abcam). After incubation with the Alexa-conjugated secondary antibody (liver sections were counterstained with DAPI) or HRP antibody (liver sections were counterstained with H&E), apoptotic cell visualization was achieved using a TUNEL kit (Roche) according to the manufacturer’s instructions. Collagen deposition was visualized using a standard protocol for Sirius red (Sigma). Three representative images per mouse were acquired using confocal microscope LSM780 or Zeiss Z1 observer.
Liver Enzyme Measurements.
The liver enzymes GLDH, transaminases AST and ALT, and AP were measured in serum obtained from KLF2-deficient mice or WT mice. Measurements were performed in the central laboratory of Universitätsklinikum Frankfurt, Goethe-University, Frankfurt, Germany, by using standard procedures.
LSEC Isolation.
LSECs were isolated by a modified two-step collagenase perfusion method. Briefly, the mice were anesthetized by i.p. injection of a mixture of Ketamine (100 mg/kg) and Xylazine (10 mg/kg). Then the abdomens were exposed, and a 18G catheter was placed into the portal vein. The liver was first perfused at 5 mL/min with EGTA buffer at 37 °C for 5 min. Then, the liver was perfused with Liver Digest Medium (Invitrogen) for 8 min. The liver was then dissociated in suspension buffer and filtered with a 100-μm pore cell strainer. The cell suspension was centrifuged at 50 × g for 2 min twice to eliminate hepatocytes. The supernatant-containing LSECs were pelleted by centrifugation at 300 × g for 10 min. Fractions containing LSECs were first enriched by gradient centrifugation using 24% HistoDenz at 1,500 × g for 20 min without cessation. Final purification of LSECs was performed using CD146 magnetic beads according to the manufacturer’s instruction (Miltenyi Biotec).
Acknowledgments
We thank A. Fischer and M. Muhly-Reinholz for technical support. This work was supported by Deutsche Forschungsgemeinschaft Grants SFB/TR23-A10 (to S.D.), SFB/TR23-A3 (to H.G.A.), and SFB834 (to R.A.B.); by the Excellence Cluster Cardio-Pulmonary System (S.D.); and by the LOEWE Center for Cell and Gene Therapy, State of Hessen (T.A., C.J.B., R.A.B., and S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613392114/-/DCSupplemental.
References
- 1.Taub R. Liver regeneration: From myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 2.Manavski Y, Boon RA, Dimmeler S. Vascular niche controls organ regeneration. Circ Res. 2014;114:1077–1079. doi: 10.1161/CIRCRESAHA.114.303452. [DOI] [PubMed] [Google Scholar]
- 3.Hu J, et al. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science. 2014;343:416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- 4.Ding B-S, et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2014;505:97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding B-S, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y-G, Lui HM, Lin S-L, Lee JM, Ying S-Y. Regulation of cell proliferation, apoptosis, and carcinogenesis by activin. Exp Biol Med (Maywood) 2002;227:75–87. doi: 10.1177/153537020222700201. [DOI] [PubMed] [Google Scholar]
- 7.Date M, et al. Differential regulation of activin A for hepatocyte growth and fibronectin synthesis in rat liver injury. J Hepatol. 2000;32:251–260. doi: 10.1016/s0168-8278(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 8.Gold EJ, et al. Changes in activin and activin receptor subunit expression in rat liver during the development of CCl4-induced cirrhosis. Mol Cell Endocrinol. 2003;201:143–153. doi: 10.1016/s0303-7207(02)00417-3. [DOI] [PubMed] [Google Scholar]
- 9.Hergenreider E, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Bohanan CS, Neumann JC, Lingrel JB. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. J Biol Chem. 2008;283:3942–3950. doi: 10.1074/jbc.M707882200. [DOI] [PubMed] [Google Scholar]
- 11.SenBanerjee S, et al. KLF2 is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boon RA, et al. Kruppel-like factor 2 improves neovascularization capacity of aged proangiogenic cells. Eur Heart J. 2011;32:371–377. doi: 10.1093/eurheartj/ehq137. [DOI] [PubMed] [Google Scholar]
- 13.Monvoisin A, et al. VE-cadherin-CreERT2 transgenic mouse: A model for inducible recombination in the endothelium. Dev Dyn. 2006;235:3413–3422. doi: 10.1002/dvdy.20982. [DOI] [PubMed] [Google Scholar]
- 14.McCay PB, Lai EK, Poyer JL, DuBose CM, Janzen EG. Oxygen- and carbon-centered free radical formation during carbon tetrachloride metabolism. Observation of lipid radicals in vivo and in vitro. J Biol Chem. 1984;259:2135–2143. [PubMed] [Google Scholar]
- 15.Long RM, Moore L. Elevated cytosolic calcium in rat hepatocytes exposed to carbon tetrachloride. J Pharmacol Exp Ther. 1986;238:186–191. [PubMed] [Google Scholar]
- 16.Kawanami D, et al. Kruppel-like factor 2 inhibits hypoxia-inducible factor 1alpha expression and function in the endothelium. J Biol Chem. 2009;284:20522–20530. doi: 10.1074/jbc.M109.025346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharya R, et al. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the Kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- 18.Doddaballapur A, et al. Laminar shear stress inhibits endothelial cell metabolism via KLF2-mediated repression of PFKFB3. Arterioscler Thromb Vasc Biol. 2015;35:137–145. doi: 10.1161/ATVBAHA.114.304277. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, et al. Activin A induces growth arrest through a SMAD-dependent pathway in hepatic progenitor cells. Cell Commun Signal. 2014;12:18. doi: 10.1186/1478-811X-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boon RA, et al. KLF2 suppresses TGF-beta signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler Thromb Vasc Biol. 2007;27:532–539. doi: 10.1161/01.ATV.0000256466.65450.ce. [DOI] [PubMed] [Google Scholar]
- 21.Kogure K, et al. A single intraportal administration of follistatin accelerates liver regeneration in partially hepatectomized rats. Gastroenterology. 1995;108:1136–1142. doi: 10.1016/0016-5085(95)90212-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang D-H, et al. Role of activin A in carbon tetrachloride-induced acute liver injury. World J Gastroenterol. 2013;19:3802–3809. doi: 10.3748/wjg.v19.i24.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wada W, Kuwano H, Hasegawa Y, Kojima I. The dependence of transforming growth factor-beta-induced collagen production on autocrine factor activin A in hepatic stellate cells. Endocrinology. 2004;145:2753–2759. doi: 10.1210/en.2003-1663. [DOI] [PubMed] [Google Scholar]
- 24.Rodgarkia-Dara C, et al. The activin axis in liver biology and disease. Mutat Res. 2006;613:123–137. doi: 10.1016/j.mrrev.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Schwall RH, et al. Activin induces cell death in hepatocytes in vivo and in vitro. Hepatology. 1993;18:347–356. doi: 10.1016/0270-9139(93)90018-i. [DOI] [PubMed] [Google Scholar]
- 26.Hully JR, et al. Induction of apoptosis in the murine liver with recombinant human activin A. Hepatology. 1994;20:854–862. doi: 10.1002/hep.1840200413. [DOI] [PubMed] [Google Scholar]
- 27.Finnson KW, Philip A. Endoglin in liver fibrosis. J Cell Commun Signal. 2012;6:1–4. doi: 10.1007/s12079-011-0154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichikawa T, et al. Role of growth hormone, insulin-like growth factor 1 and insulin-like growth factor-binding protein 3 in development of non-alcoholic fatty liver disease. Hepatol Int. 2007;1:287–294. doi: 10.1007/s12072-007-9007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Annoni A, et al. In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood. 2009;114:5152–5161. doi: 10.1182/blood-2009-04-214569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anliker B, et al. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat Methods. 2010;7:929–935. doi: 10.1038/nmeth.1514. [DOI] [PubMed] [Google Scholar]
- 31.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 32.Qiao C, et al. Liver-specific microRNA-122 target sequences incorporated in AAV vectors efficiently inhibits transgene expression in the liver. Gene Ther. 2011;18:403–410. doi: 10.1038/gt.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dekker RJ, et al. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 34.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc. 2010;5:1518–1534. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
- 35.Lee JS, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]