Significance
Regulator of the gabTD operon and its own gene (GabR) is an intriguing case of molecular evolution, displaying the evolutionary lineage between a pyridoxal-5′-phosphate (PLP)-dependent aminotransferase and a regulation domain of a transcription regulator. Here, PLP’s native function is not a catalytic coenzyme, but an effector of transcription regulation. The chemical species of GabR-PLP–GABA, which is responsible for GabR-mediated transcription activation, has been revealed as a stable external aldimine formed between PLP and GABA by a crystal structure with further support from results in mechanistic crystallography, NMR spectroscopy, and biological assays using both GABA and a GABA analog, (S)-4-amino-5-fluoropentanoic acid (AFPA), as a molecular probe. Our results provide mechanistic insights for a currently understudied Rhizobium meliloti rhizopine catabolism regulator (MocR)/GabR subfamily of bacterial transcription regulators.
Keywords: MocR, GabR, PLP, external aldimine, transcription activation
Abstract
The Bacillus subtilis protein regulator of the gabTD operon and its own gene (GabR) is a transcriptional activator that regulates transcription of γ-aminobutyric acid aminotransferase (GABA-AT; GabT) upon interactions with pyridoxal-5′-phosphate (PLP) and GABA, and thereby promotes the biosynthesis of glutamate from GABA. We show here that the external aldimine formed between PLP and GABA is apparently responsible for triggering the GabR-mediated transcription activation. Details of the “active site” in the structure of the GabR effector-binding/oligomerization (Eb/O) domain suggest that binding a monocarboxylic γ-amino acid such as GABA should be preferred over dicarboxylic acid ligands. A reactive GABA analog, (S)-4-amino-5-fluoropentanoic acid (AFPA), was used as a molecular probe to examine the reactivity of PLP in both GabR and a homologous aspartate aminotransferase (Asp-AT) from Escherichia coli as a control. A comparison between the structures of the Eb/O–PLP–AFPA complex and Asp-AT–PLP–AFPA complex revealed that GabR is incapable of facilitating further steps of the transamination reaction after the formation of the external aldimine. Results of in vitro and in vivo assays using full-length GabR support the conclusion that AFPA is an agonistic ligand capable of triggering GabR-mediated transcription activation via formation of an external aldimine with PLP.
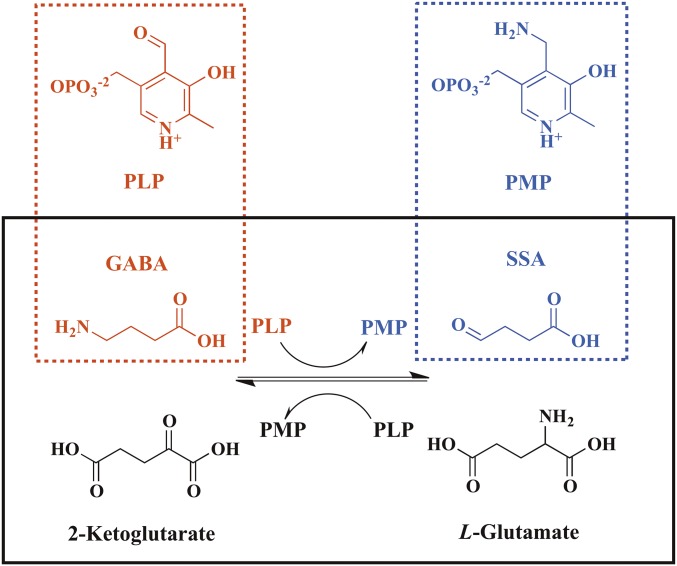
Bacterial transcription regulators are often directly responsive to the changes in metabolite concentrations (1, 2). The regulator of the gabTD operon and its own gene (GabR) responds to the increase of γ-aminobutyric acid (GABA) concentration in bacterial cells (3) and up-regulates glutamate regeneration from GABA. Belonging to the Rhizobium meliloti rhizopine catabolism regulator (MocR)/GabR subfamily in the repressor of the Bacillus subtilis gluconate operon (GntR) family of bacterial transcription regulators (2–5), GabR-type regulators characteristically have a large C-terminal effector-binding/oligomerization (Eb/O) domain, which is homologous to the type I aminotransferases (6), and a small N-terminal “winged-helix–turn–helix” (wHTH) DNA-binding domain (7). In B. subtilis, GabR represses its own transcription (3, 8). With both pyridoxal-5′-phosphate (PLP) and GABA bound, GabR activates the transcription of two genes, gabT and gabD, that encode the enzymes GABA aminotransferase (GABA-AT) and succinic semialdehyde (SSA) dehydrogenase, respectively, forming a pathway for glutamate regeneration from GABA (3, 8).
Previously, we have reported the crystal structures of full-length GabR as a “head-to-tail” dimer (9), in which electron density supported the existence of PLP bound to K312 of the Eb/O domain in the form of an internal aldimine. Although the PLP-binding site of GabR is largely conserved compared with the PLP-binding site of type I aminotransferases, no aminotransferase activity has been detected for GabR (9). To our knowledge, the existence of PLP in MocR/GabR-type transcription regulators is the only case known in nature where PLP functions as an effector in transcription regulation, including other reports of the MocR/GabR-type regulators DdlR, PdxR, and TauR (10–15). A previous report on GabR has shown that a second metabolite pair of pyridoxamine-5′-phosphate (PMP) and SSA could also cause transcription activation upon binding to GabR (8). In fact, the first half of the “Ping-Pong” transamination catalyzed by GABA-AT (Scheme 1 and Scheme S1) converts PLP and GABA to PMP and SSA reversibly (16). It was hypothesized that certain step(s) of the aminotransferase reaction might be required to trigger GabR-mediated transcription activation (8). Such a hypothesis has not been thoroughly proven, and the chemical identity of the GabR-PLP–GABA complex that results in transcription activation was most recently proposed as an external aldimine based solely on an observed spectroscopic signal and chemical logic (17), but no structural evidence is known. In the same paper, the dissociation constant of PLP is reported as 1.20 μM and the dissociation constant of GABA in the presence of PLP was reported as 2.6 mM. Using X-ray crystallography, NMR spectroscopy, and biological assays, an external aldimine species is reported here that is formed between PLP and GABA when both are bound to the effector-binding site of GabR and is apparently responsible for triggering GabR-mediated transcription activation.
Scheme 1.
GabR effectors and the GABA-AT reaction that is under GabR-mediated regulation are illustrated. The effectors for GabR-mediated transcription activation are shown in dashed boxes. The GABA-AT reaction in a “Ping-Pong” mechanism is shown in the solid box.
Scheme S1.
The mechanism of the first half of the GABA-AT reaction.
Results and Discussion
Protein Purification.
All proteins, including Escherichia coli aspartate aminotransferase (Asp-AT), were expressed and purified with supplemented PLP (∼1 mM), giving a yellow color resulting from the internal aldimine formed between PLP and a conserved lysine.
Crystallography of GabR Eb/O Domain and Asp-AT Complexed with Ligands.
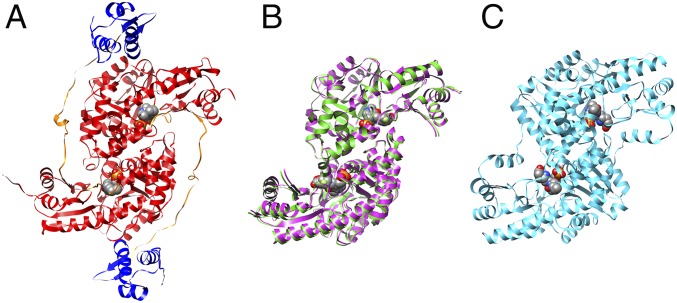
Crystallization of full-length GabR in the presence of GABA was not successful, so a truncated version (residues 88–479) lacking the helix–turn–helix domain was produced. This Eb/O domain was expressed, purified, and cocrystallized with PLP and GABA. Crystals of three complexes with PLP-ligand adducts bound, the Eb/O domain with a PLP-GABA adduct, a PLP–(S)-4-amino-5-fluoropentanoic acid (AFPA) adduct, and Asp-AT with a PLP-AFPA adduct, were obtained by cocrystallization. X-ray crystal structures of all three complexes were determined, refined, and converged with statistics shown in Table S1. In all three cases, a Schiff base adduct was formed and an appropriate model was built into the electron density maps (Fo-Fc) in the final rounds of refinement. The formed adduct was confirmed by the simulated annealing omit difference maps (Fo-Fc) generated in Phenix (18). The final models for the Eb/O–PLP–GABA, Eb/O–PLP–AFPA, and Asp-AT–PLP–AFPA complexes have been deposited in the Protein Data Bank (PDB) with ID codes 5T4J, 5T4K, and 5T4L, respectively. The Eb/O–PLP–GABA and Eb/O–PLP–AFPA complex structures were solved with molecular replacement and refined at 2.23 Å resolution and 2.25 Å resolution, respectively. The structure of the Asp-AT–PLP–AFPA complex was also solved with molecular replacement and refined at 1.53 Å resolution. The overall structure of the Eb/O domain retains the same head-to-tail homodimer architecture as in the full-length GabR and type I aminotransferases (Fig. 1).
Table S1.
Data processing and refinement statistics
| Protein | GabR EBO/GABA | GabR EBO/AFPA | Asp-AT/AFPA |
| PDB ID code | 5T4J | 5T4K | 5T4L |
| Data processing | |||
| Space group | C2221 | C2221 | C2221 |
| Cell dimension | |||
| α, β, γ; ° | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| a, b, c; Å | 92.4, 128.9, 65.1 | 96.6, 127.5, 67.6 | 84.9, 154.9, 79.3 |
| Resolution, Å | 49.2–2.23 | 50.8–2.25 | 29.0–1.53 |
| Rmerge,* % | 7.5 (46.4)† | 8.3 (64.4)† | 6.8 (68.7)† |
| I/σ, I | 29.3(4.7)† | 12.6(1.2)† | 34.0(1.7)† |
| Completeness, % | 99.8(97.9)† | 96.2(78.4)† | 98.2(83.4)† |
| Multiplicity | 5.5 (4.9)† | 3.1 (2.5)† | 12.2 (5.6)† |
| No. of reflections | 104,767 | 59,025 | 941,856 |
| No. of unique reflections | 19,166 | 19,311 | 77,111 |
| Refinement | |||
| Rwork‡/Rfree,§ % | 17.4/21.0 | 18.8/23.7 | 14.8/16.5 |
| No. of atoms | |||
| Protein | 2,921 | 2,292 | 3,465 |
| Ligand | 22 | 24 | 32 |
| Water | 136 | 182 | 510 |
| B-factors, Å2 | |||
| Wilson B | 35.1 | 30.7 | 20.2 |
| Protein | 40.4 | 42.2 | 23.7 |
| Ligand | 28.6–41.1 | 24.8–43.9 | 13.0–27.8 |
| Solvent | 44.9 | 42.9 | 39.41 |
| rmsd | |||
| Bond length, Å | 0.005 | 0.003 | 0.007 |
| Bond angle, ° | 0.86 | 0.60 | 1.13 |
| Ramachandran plot | |||
| Most favored, % | 96.7 | 96.4 | 97.2 |
| Allowed, % | 3.0 | 3.3 | 2.6 |
| Outliers, % | 0.3 | 0.3 | 0.2 |
Rmerge = Σ|Iobs − Iavg|/ΣIavg.
Values for the highest resolution bins are in parentheses.
Rwork = Σ|Fobs − Fcalc|/ΣFobs.
Five percent of the reflection data were selected at random as a test set, and only these data were used to calculate Rfree.
Fig. 1.
Head-to-tail dimers of the full-length GabR (PDB ID code 4N0B), Eb/O domain of GabR, and Asp-AT. Bound PLPs are shown in spheres. (A) In the full-length GabR, the Eb/O domains are in red, the flexible linker regions are in orange, and the DNA-binding domains are in blue. (B) Overlay of two dimeric structures of the GabR Eb/O domain in complexes with GABA (magenta) and AFPA (green). (C) Dimeric structure of E. coli Asp-AT (PDB ID code 1SFF, cyan).
Details of the Effector Binding Site in the GabR Eb/O Domain and Schiff Base Formation Between PLP and GABA.
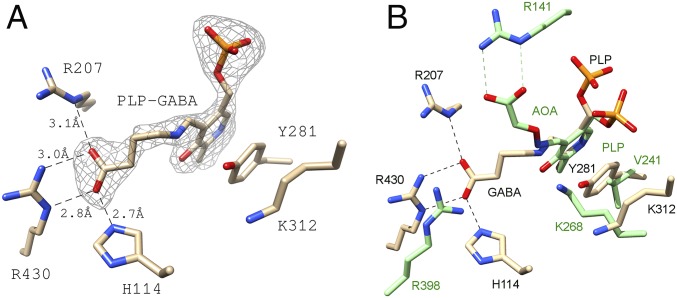
In the effector-binding pocket of the Eb/O domain, the omit map difference electron density (Fo-Fc) revealed the existence of a Schiff base formed between PLP and GABA (Fig. 2A). The ligands are refined with occupancies of 1 in each effector-binding site, indicating a stoichiometry of 1 for each monomer. Two basic residues, R207 and R430, along with H114 from the same subunit, interact with the carboxylate of the GABA moiety. In the previously published structure of the GABA-AT–PLP–aminooxyacetate (AOA) complex (Fig. 2B), the PLP-AOA adduct mimics the PLP-GABA external aldimine, whose carboxylate interacts with one of the conserved arginine residues (R141) (19). Compared with the homologous type I aminotransferases, such as GABA-AT (Fig. 2B), GabR is missing one (R141 in GABA-AT) of the two conserved arginines, which are involved in binding to dicarboxylic acid substrates (19, 20), such as l-glutamate or 2-ketoglutarate, in the second half of a typical transamination reaction (Scheme 1). This observed structural difference suggests that GabR has evolved to recognize monocarboxylic acids, such as GABA, preferentially and to orient the ligand in favor of forming a Schiff base with PLP.
Fig. 2.
PLP and GABA bound to the truncated Eb/O domain. (A) Simulated annealing omit map (Fo-Fc) is shown around the PLP-GABA Schiff base at 3σ. (B) Eb/O–PLP–GABA complex in comparison to GABA-AT with AOA bound to PLP mimicking the PLP-GABA external aldimine (green, PDB ID code 1SFF).
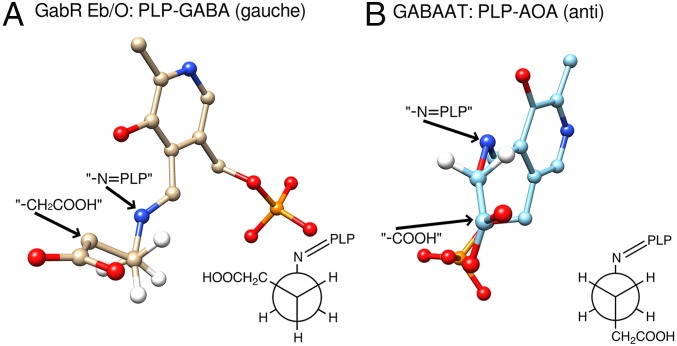
Interestingly, the conserved basic residues in GabR are not aligned with GABA-AT residue R141, which had been proposed to interact with the GABA carboxylate (19, 21). Although the AOA-PLP adduct, which mimics the GABA-PLP in the E. coli GABA-AT structure, binds to the active site in an anticonformation, the PLP-GABA Schiff base in GabR is in a gauche conformation (Fig. 3). The dihedral torsional angle of C2-C1-O-N in the PLP-AOA “structural mimic” is measured to be 157° (23° off from an ideal anticonformation) in UCSF Chimera. In the Eb/O–PLP–GABA complex structure (Fig. 3A), the dihedral torsional angle of Cα-Cβ-Cγ-N in the PLP-GABA Schiff base was measured to be 77.3° (17.3° off from an ideal gauche conformation) in UCSF Chimera. Compared with the anticonformation, the gauche conformation is of higher energy and a relatively minor conformation for GABA molecules in an aqueous environment in the cell.
Fig. 3.
GABA moiety displays a Gauche conformation. (A) GABA bound in the Eb/O domain is in a gauche conformation. The PLP-GABA external aldimine is shown in stick and ball form. The Cγ atom is placed (hidden) behind the Cβ atom, in the same orientation as the Newman projection for the dihedral torsional angle Cα-Cβ-Cγ-N in PLP-GABA, shown in the gauche conformation. (B) AOA-PLP imine bound in E. coli GABA-AT (PDB ID code 1SFF) mimicking the PLP-GABA Schiff base is shown in ball and stick form. The O atom is placed (hidden) behind the C1 atom in the same orientation as the Newman projection for the dihedral torsional angle C2-C1-O-N in PLP-AOA, shown in the anticonformation.
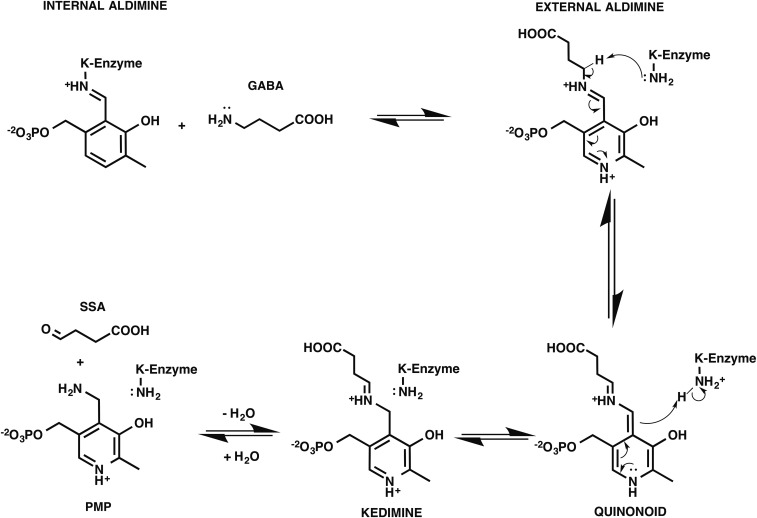
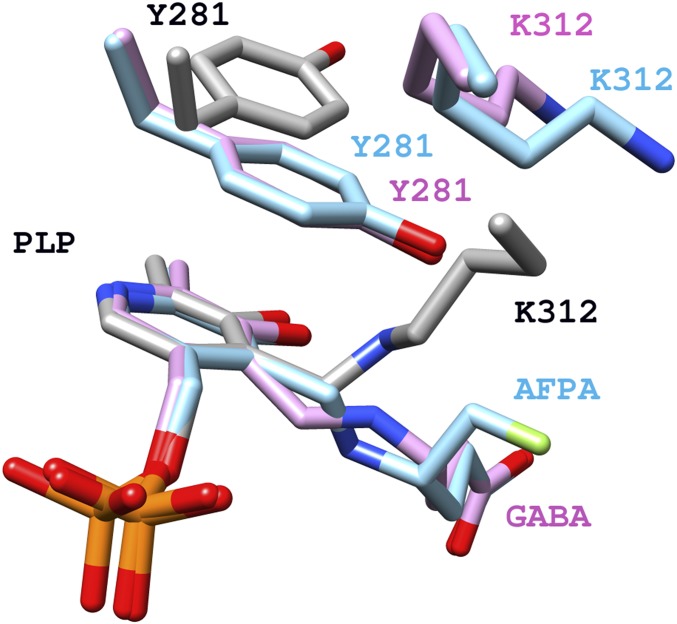
In all PLP-dependent enzymes, a conserved lysine forms an internal aldimine with PLP in its resting form and acts as a key catalytic residue during catalytic reactions, such as in the first half of the “Ping-Pong” transamination (6) (Scheme S1). The conserved K312 (9) (Fig. 4) was previously proven to be essential for GabR’s function, although the catalytic capacity to facilitate transamination has never been detected for GabR (8). In the Eb/O–PLP–GABA complex structure (Fig. 2A), Y281 is observed to block K312 from accessing the PLP-GABA Schiff base, preventing reformation of the internal aldimine as well as further transamination-like catalysis, in which the freed catalytic lysine would deprotonate the carbon adjacent to the nitrogen atom of the GABA-PLP adduct (19, 22) (Scheme S1). As a result, GabR is unlikely to catalyze the first half of the transamination reaction shown in Scheme 1. As we previously reported (9), in the absence of GABA, Y281 is found in a different orientation, allowing internal aldimine formation (Fig. 4). The observed obstruction by Y281 supersedes the previous speculation that GabR’s inability to stabilize the external aldimine might be the reason for the lack of observed transaminase activity (9).
Fig. 4.
Comparison of the external aldimine vs. internal aldimine in GabR. External aldimine Schiff bases (magenta and cyan), internal aldimine Schiff base with K312 (gray), and Y281 are shown in stick form. Labels and carbon atoms for internal aldimine (PDB code ID 4N0B) are in black and gray, respectively. Labels and carbon atoms for the Eb/O–PLP–GABA complex are in magenta. Labels and carbon atoms for the Eb/O–PLP–AFPA complex are in cyan.
External Aldimine as the Final State of GabR-Mediated Reaction.
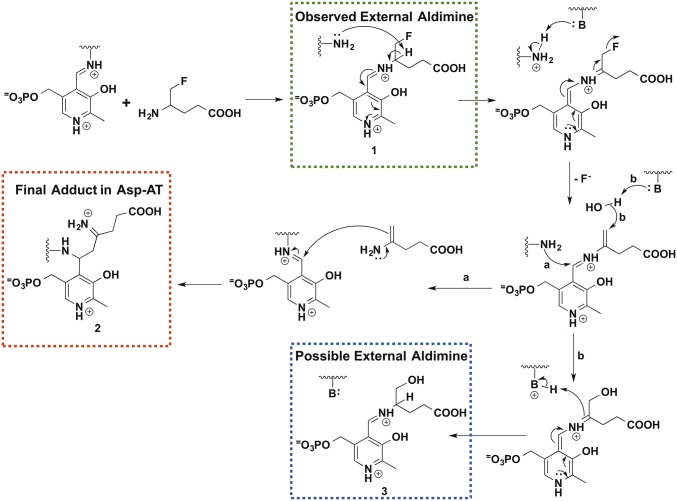
The Eb/O–PLP–GABA complex structure at 2.23 Å resolution could not allow us to distinguish between external aldimine and ketimine (Scheme S1) for the chemical identity of the PLP-GABA Schiff base observed. Therefore, we used a mechanism-based inactivator, AFPA, which was designed to inactivate PLP-dependent aminotransferases irreversibly (23), as a molecular probe. A previously proposed inactivation mechanism of an aminotransferase, mammalian GABA-AT, by AFPA is shown in Scheme 2. Once a typical aminotransferase reaction progresses beyond the external aldimine 1 stage, the AFPA moiety releases the fluoride ion, resulting in a reactive intermediate. Eventually, AFPA inactivates the PLP-dependent aminotransferase via a ternary adduct, in which the AFPA moiety is covalently linked to both PLP and the catalytic lysine, or alternatively forms external aldimine 3. The catalytic lysine freed after the formation of the external aldimine 1 plays a critical role in assisting the irreversible inactivation of the PLP-dependent enzyme. The formation of the proposed ternary adduct should provide evidence that the reaction between AFPA and PLP can progress beyond the external aldimine 1, reaching quinonoid or ketimine intermediates (Scheme 2 and Scheme S1). In contrast, the intact fluorine group of AFPA should indicate that the reaction could not proceed beyond the formation of the external aldimine 1. Due to the overall desirable crystallographic behavior of E. coli Asp-AT, we chose it as a control for the cocrystallization experiment with AFPA. E. coli Asp-AT, a homolog of the GabR Eb/O domain, has previously been used successfully as a model enzyme to study mechanism-based inactivators of PLP-dependent enzymes (24–26).
Scheme 2.
Proposed reaction mechanism of AFPA. The green dashed box (1) shows the final adduct in the GabR Eb/O domain, the red dashed box (2) shows the expected adduct for Asp-AT, and the blue dashed box (3) indicates an alternative external aldimine species.
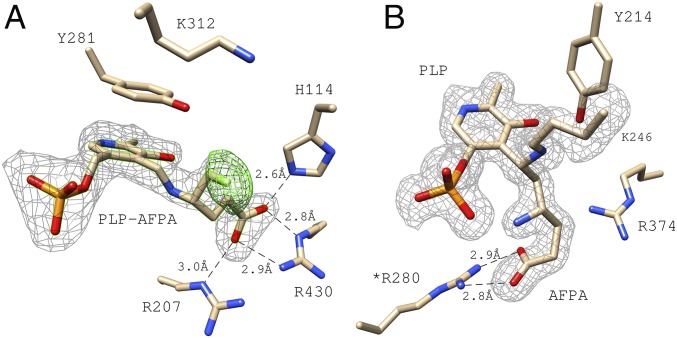
Both the GabR Eb/O domain and Asp-AT were cocrystallized with PLP and AFPA. The structure of the Eb/O–PLP–AFPA complex (refined at 2.25 Å resolution) confirms the formation of an external aldimine with the fluorine atom intact in the final adduct (Fig. 5A). To validate the existence of the fluorine atom, extra rounds of refinement were done after deletion of the fluorine atom; the omit map difference electron density (Fo-Fc) strongly supports the presence of the fluorine atom in the complex. Residue Y281 is observed to be in the same position as in the Eb/O–PLP–GABA complex structure (Fig. 4), preventing the access of K312 to the PLP-AFPA Schiff base, which could otherwise potentially push the reaction further along the proposed inactivation pathway (Scheme 2).
Fig. 5.
AFPA as a molecular probe in the Eb/O domain and in Asp-AT (as a control). (A) PLP-AFPA external aldimine is bound to the EB/O domain. Two sets of simulated annealing electron density omit maps (Fo-Fc) are shown. Both are shown at 3.5 σ. The gray map was generated omitting the complete ligand adducts, confirming the existence of the ligands. The green map was generated omitting only the fluorine atom, confirming the existence of fluorine in AFPA. (B) Electron density omit map of the E. coli Asp-AT–AFPA–PLP complex, generated by omitting AFPA, PLP, and lysine 246 from the calculation. *Indicates that R280 is from a different subunit of the Asp-AT dimer.
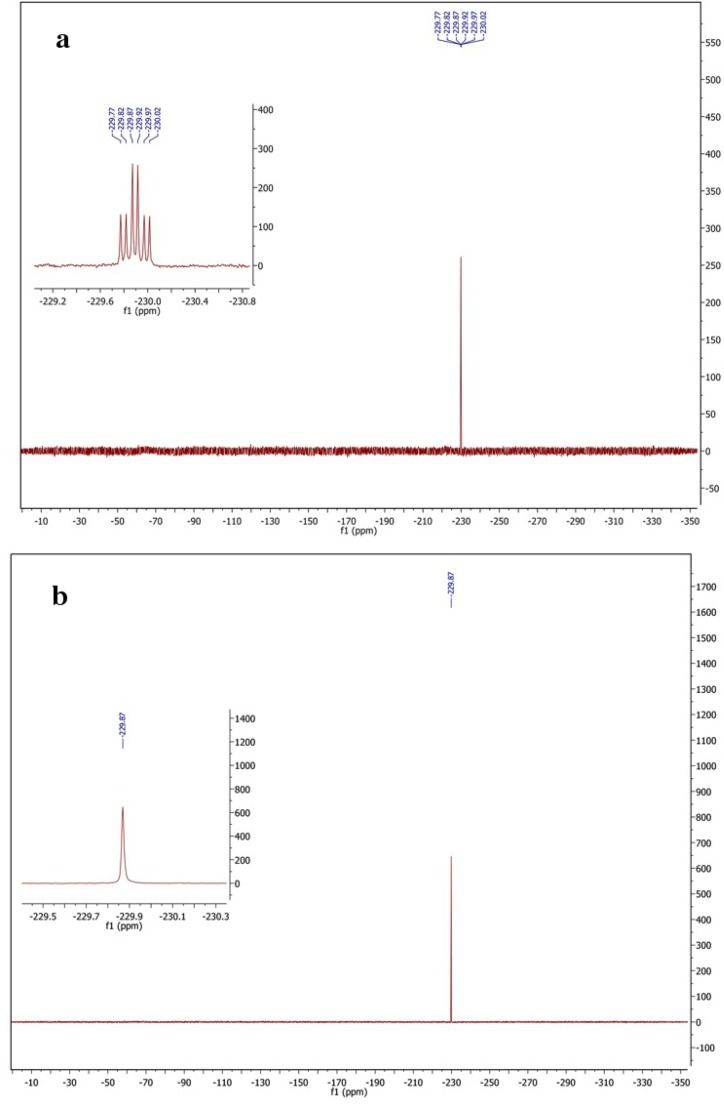
This electron density could also be fitted with another atom, so additional validation was used to establish its identity. It is chemically plausible that a hydroxyl or water could react with the reactive intermediate and replace the eliminated fluorine group; the resulting adduct 3 would be structurally similar to the external aldimine proposed here (adduct 1 in Scheme 2). At the 2.25 Å resolution obtained for the Eb/O–PLP–AFPA complex structure, it is impossible to distinguish adduct 3 from adduct 1 even with the omit map difference electron density (Fo-Fc) validation shown (as green mesh) in Fig. 5A. Pathway b in Scheme 2 and adduct 3 have never previously been proven in studies of AFPA inactivation. Nevertheless, the AFPA-treated protein sample was subjected to fluorine NMR spectroscopy to eliminate the possibility of an alternative adduct (adduct 3 in Scheme 2, via pathway b in Scheme 2). The results are shown in Fig. S1. Fig. S1A is the fluorine NMR spectrum that shows a triplet of doublets (J1 = 25.3 Hz, J2 = 21.4 Hz), because the fluorine atom is split by its two neighboring hydrogen atoms and then split again by another hydrogen atom on the adjacent carbon in the external aldimine form. Fig. S1B represents the proton-decoupled fluorine NMR spectrum that shows a singlet, because the fluorine atom and the protons are decoupled. As concluded, the fluorine atom remains intact in the same environment as intact AFPA, even after treatment with a saturated amount of GabR protein.
Fig. S1.
Fluorine NMR spectra. (A) Fluorine NMR showing a triplet of doublets (J1 = 25.3 Hz, J2 = 21.4 Hz). (B) Proton-decoupled fluorine NMR spectroscopy that shows a singlet.
As a positive control, the structure of the Asp-AT–PLP–AFPA complex (refined at 1.53 Å resolution) showed that the proposed ternary adduct 2 has formed via pathway a in Scheme 2 (Fig. 5B), indicating that the catalytic actions of the conserved lysine have occurred in Asp-AT but not in the GabR Eb/O domain. On the basis of structural information and NMR results, it was concluded that the external aldimine 1 is the final chemical species when both PLP and AFPA are bound to the GabR Eb/O domain. The obtained results using AFPA as a molecular probe suggest that the observed Schiff base in the Eb/O–PLP–GABA complex structure is also an external aldimine.
For aminotransferases in general, the external aldimine intermediate can be transiently observed spectroscopically (27). However, mutation of a catalytic residue (28) or chemical modification of the ligands, such as the amino acid (19) or PLP (29, 30), was required to trap a stable external aldimine in an aminotransferase. GabR is a protein reported to be capable of forming a stable external aldimine between PLP and GABA for its function in transcription regulation. Our results are consistent with the ability of another group to detect the external aldimine of GabR using spectroscopic approaches (17). Spectroscopic changes, consistent with the formation of external aldimine, were also reported for DdlR (11). Only mild global conformational changes are introduced in the PLP-bound Eb/O domain of GabR upon binding to GABA and formation of the external aldimine. A least squares structural comparison revealed that the Eb/O domains of the GabR-PLP dimer (PDB ID code 4N0B) aligned well with the Eb/O domains of the Eb/O–PLP–GABA complex or the Eb/O–PLP–AFPA complex, with rmsd values for Cα atoms around 1.12 Å and 1.13 Å, respectively. Conformational changes could be more pronounced in the linker region between the Eb/O and wHTH domains, and within the wHTH domains in the full-length dimeric GabR; these conformational changes likely contribute to the triggering of transcription activation.
Functional Significance of External Aldimine Formation.
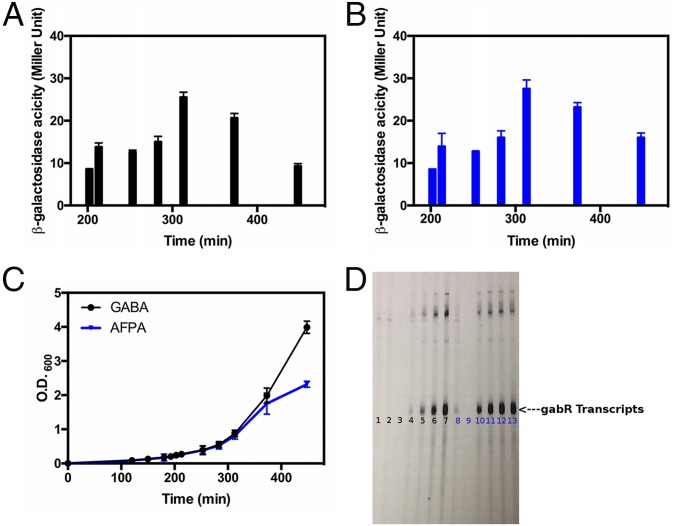
To extrapolate the functional interpretation from the GabR Eb/O domain to the full-length GabR, both in vivo and in vitro assays were conducted to compare the biological effects of GABA and AFPA. When examining GabR-mediated activation in vivo using a gabT/lacZ fusion (3) (Fig. 6A), both GABA and AFPA triggered GabR-mediated transcription activation in B. subtilis cells. The time course of transcription activation was measured and plotted as the calculated Miller units over a 450-min period after addition of the effectors (Fig. 6 A and B). The growth of the culture was also plotted at the same time points as the Miller unit measurements (Fig. 6C). The results clearly demonstrate that both AFPA and GABA cause transcription activation at 200 min and beyond. Although the culture grew better after 300 min in the presence of GABA than it did in the presence of AFPA, the gene activation level was actually more pronounced for AFPA than for GABA after 300 min.
Fig. 6.
Biological effects of GABA and AFPA. (A) Expression of gabT/lacZ is indicated by calculated Miller units over time. The calculated Miller units of the culture without addition of GABA or AFPA were no higher than 2. (B) Self-developed film indicating the newly synthesized gabT mRNA. (C) Culture growth (indicated with O.D.600 values) while supplemented with GABA or AFPA. (D) Lane 1: control without GABA or AFPA. Lanes 2–7: GABA added at concentrations of 0.625, 1.25, 2.5, 5, 10, and 20 mM, respectively. Lanes 8–13: AFPA added with concentrations of 0.625, 1.25, 2.5, 5, 10, and 20 mM, respectively.
In vitro, GabR, PLP, RNA polymerase, and the gabT promoter-containing DNA fragments were mixed with radioactively labeled nucleotides as described previously (8). The gabT transcripts, which are indicated as the appearance of the newly synthesized P32-labeled bands, were formed upon addition of either GABA or AFPA (Fig. 6D) proportional to the amount of the effector added. AFPA triggered the GabR-mediated transcription activation in vitro with a higher potency than did GABA. The GabR-PLP–AFPA complex may be a better transcriptional activator than the GabR-PLP–GABA complex. This interpretation could explain the long-lasting activation effect caused by AFPA shown in the in vivo experiments after 300 min (Fig. 6 A–C). Overall, we conclude that AFPA activates the transcription of the gabT promoter via the same chemical principle, which is the formation of the external aldimine, as GABA does in B. subtilis. The activation of transcription by GabR induced by GABA has also been reported in Bacillus thuringiensis, indicating that this function is not unique to B. subtilis (31).
Conclusions
Using crystallography, we have shown that PLP bound to the GabR Eb/O domain reacts with the effector GABA to form a stable external aldimine, which apparently triggers the transcription from the gabT promoter in B. subtilis. This conclusion is consistent with the published spectroscopic analysis of GabR (17) and offers an explanation for the previous results that the effector pair of PMP and SSA can also trigger GabR-mediated transcription activation in vitro (8). Through a reverse transamination-like reaction (Scheme S1), SSA and PMP can potentially form a Schiff base, presenting a structure similar to the PLP-GABA external aldimine (8). However, SSA and PMP may not work as an actual effector pair in vivo. With an evolutionarily modified PLP-GABA–binding site, the GabR Eb/O domain is capable of preferentially binding to GABA and forming an external aldimine. To perform the role of a transcription regulator, GabR employs Y281 to subdue the Eb/O domain’s catalytic potential by stabilizing the external aldimine formed and preventing its further modification.
Materials and Methods
Materials.
Chemicals for the assays were purchased from Fisher Scientific and Sigma–Aldrich. Cloning vectors were obtained from Integrated DNA Technologies. E. coli BL21 (DE3) cells were purchased from New England BioLabs. Bacterial growth media and antibiotics were obtained from Fisher Scientific and Sigma–Aldrich. Crystallization screen solutions and other crystallization supplies were purchased from Hampton Research and Emerald Bio. All chemicals were of the highest quality available.
Truncation and Cloning of GabR Eb/O Domain.
The sequence coding for the GabR Eb/O domain was amplified by PCR using previously cloned full-length GabR (in pETite vector from Lucigen) as the template (9), the specific oligonucleotides as primers (forward primer designed as GAAGGAGATATACATATGATCGACCAGAGCGATTGGATA and reverse primer designed as GTGATGGTGGTGATGATGATCCCCTGTAACGGGGATTTT), and Phusion HF DNA polymerase, following the manufacture’s instructions. The thermocycler program used included an initial denaturation for 120 s at 95 °C; 35 cycles at 95 °C for 30 s, 50 °C for 60 s, and 72 °C for 60 s; and a final extension at 72 °C for 5 min. The PCR product was inserted into a pETite vector for recombinant expression of GabR Eb/O domain protein with a C-terminal His6-tag.
Protein Expression and Purification.
Full-length GabR protein and l-aspartate aminotransferase from E. coli were expressed and purified as described (8). The GabR Eb/O domain was overexpressed in E. coli and purified using a similar method. Transformed E. coli BL21 (DE3) cells were grown in 1 L of LB supplemented with 50 μg/mL kanamycin at 37 °C and 250 rpm using a MaxQ6000 shaker bath incubator (Thermo Fisher Scientific) until OD600 nm reached ∼0.6. Protein expression was induced by the addition of 0.5 mM isopropyl-beta-d-1-thiogalactopyranoside. Cells were incubated at 25 °C and harvested after 16 h by centrifuging using a JA10 rotor (Beckman) at 4,800 rpm and 4 °C for 10 min. The cell paste was resuspended in the lysis buffer containing 1× PBS and 10 mM imidazole (pH 8.0), followed by sonication. The resulting suspension was centrifuged twice using a JA20 rotor (Beckman) at 16,500 rpm and 4 °C for 20 min, and the soluble fraction was loaded onto a 10-mL His-Trap column (GE Life Sciences) containing Ni2+ and previously equilibrated with lysis buffer. Elution of the retained proteins was achieved with a linear imidazole gradient (25-column volume, 10–485 mM). Fractions containing the truncated EB/O domain were pooled, concentrated to 2 mL, and loaded onto a 16/60 Superdex 200 size exclusion column (GE Life Sciences) that was previously equilibrated with 84% elution buffer. Fractions containing enzyme were pooled, concentrated, and used immediately after exchanging into suitable buffers for crystallization and biological assays. The wild-type Asp-AT was overexpressed and purified with the protocol used by Onuffer and Kirsch (32).
Crystallization of the GabR Eb/O and X-Ray Diffraction Data Collection.
After the initial crystallization screen and optimization, the GabR Eb/O domain was crystallized via hanging drop methods at 25 °C. The hanging drops were prepared with 1 μL of 12 mg/mL preincubated protein with inhibitor solution and 1 μL of the reservoir solution that contained 0.1 M Tris (pH 8.5) buffer and 20% ethanol. Crystals were harvested in about 10 d. For cryocooling, crystals were transferred to the reservoir solution with 20% glycerol as a cryoprotectant, and were flash-cooled by plunging in liquid nitrogen. The cocrystallization of Asp-AT and AFPA was conducted according to previous studies (24, 25). Diffraction data were collected at GM/CA@APS beamlines 23ID-B and 23ID-D at the Advanced Photon Source (Argonne National Laboratory). The wavelength used for data collection was 1.0332 Å, and diffraction images were recorded on 4 × 4 tiled CCD detectors (Rayonix) with 300 × 300-mm2 sensitive areas. All data were indexed, integrated, scaled, and merged using HKL2000 (33).
Phasing, Model Building, and Refinement.
Diffraction data were phased with molecular replacement using the program Phaser (34) in the CCP4 software suite (35). A previously solved full-length GabR structure (9), truncated to contain residues 107–479, was used as the search model. The previously solved AspAT model 2Q7W (25) was used as the search model for the E. coli Asp-AT–PLP–AFPA structure. Rigid body refinement followed by restrained refinement was carried out in Refmac (36). Model building was conducted using the program Coot (37). Iterative rounds of model building and refinement generated the final coordinates. The external aldimine ligands, PLP-GABA or PLP-AFPA, were removed from the model; the model without a ligand was refined in Phenix using simulated annealing refinement. The starting temperature was 5,000 K, the final temperature was 300 K, and the annealing process was conducted in 50 steps with a cooling rate of 100° per step. The SA OMIT map was then generated after the above refinement.
In Vivo and in Vitro Assays for GabR-Dependent Transcription Activation.
The in vivo assay was conducted as previously described (8), using GABA or AFPA as an effector (0.625–20 mM). The expression signal was detected using ONPG (ortho-nitrophenyl-β-galactoside) as a substrate for the β-galactosidase (lacZ gene product) generating a spectroscopic signal at 420 nm. The time course was measured and plotted over a 200-min period. Each point is the average value of triplicate measurements. For each measurement, the Miller units were calculated using the following equation: . In vitro GabR-dependent RNA polymerase reactions were performed as previously described (8). The PCR fragment (50 nM) contained the gabR/gabT regulatory region. The total reaction was carried in transcription buffer containing 40 mM Tris⋅HCl; 100 mM KCl; 10 mM MgCl2; 5% glycerol; 0.1 mM EDTA; 1 mM DTT; 0.1 mg/mL BSA with 4 units of RNasin (ribonuclease inhibitor); 150 μM ATP, CTP, and GTP; 20 μM UTP; 0.5–1 μL/Ci 32P-labeled UTP; and 0.02–0.04 unit of E. coli RNA polymerase. The reaction was incubated at 37 °C for 15 min and terminated by additional of 4 μL of the 20 mM EDTA, 95% formamide dye solution. The samples were subsequently heated at 80 °C for 5 min. The samples were then analyzed without further purification using 7 M urea/5–6% polyacrylamide DNA sequencing gels. GABA was purchased from Sigma. AFPA was synthesized according to a previous publication (38).
NMR Spectroscopy.
The protein (100 mg) was treated with AFPA. The molar ratio of the protein and AFPA was 2:1 to minimize free AFPA in the solution. The sample was concentrated to a volume of 0.4 mL by centrifugation. Deuterium oxide (0.15 mL) was added, and the solution was transferred to a 5-mm standard NMR tube. Fluorine NMR and proton-decoupled fluorine NMR spectra were taken at the Northwestern Integrated Molecular Structure Education and Research Center facility on an Agilent DD2 500-MHz spectrometer with an Agilent 5-mm HFX probe at 26 °C [19F NMR δ 229.87 (td, J1 = 25.3 Hz, J2 = 21.4 Hz), proton-decoupled 19F NMR δ 229.87 (s)].
Structural Analysis and Figure Making.
All structural analyses were conducted in Coot (37) and UCSF Chimera (39). Structure comparison of the PLP/amino acid-binding site was conducted in two steps (Figs. 2B and 6): First, the overall structural comparison/overlap was conducted in UCSF Chimera using the Needleman–Wunsch algorithm. Second, manual adjustments were done by overlapping the pyridine ring of the PLP to achieve better pattern matching for the conserved key residues in the PLP/amino acid-binding site for comparison. All structural figures were made in UCSF Chimera.
Acknowledgments
We thank Dr. Miguel Ballicora for helpful discussions and NIH Grant 1R15GM113229-01 (to D.L.) and Loyola University Chicago (D.L.) for funding. The GM/CA@APS has been funded in whole or in part with federal funds from the National Cancer Institute (Grant ACB-12002) and the National Institute of General Medical Sciences (Grant AGM-12006). R.B.S. is funded by the National Institute on Drug Abuse (Grant DA030604). Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. Support for the NMR spectrometer funding was provided by the International Institute of Nanotechnology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5T4J, 5T4K, and 5T4L).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703019114/-/DCSupplemental.
References
- 1.Resch M, Schiltz E, Titgemeyer F, Muller YA. Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA. Nucleic Acids Res. 2010;38(7):2485–2497. doi: 10.1093/nar/gkp1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem. 2002;277(15):12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- 3.Belitsky BR, Sonenshein AL. GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol Microbiol. 2002;45(2):569–583. doi: 10.1046/j.1365-2958.2002.03036.x. [DOI] [PubMed] [Google Scholar]
- 4.Rossbach S, Kulpa DA, Rossbach U, de Bruijn FJ. Molecular and genetic characterization of the rhizopine catabolism (mocABRC) genes of Rhizobium meliloti L5-30. Mol Gen Genet. 1994;245(1):11–24. doi: 10.1007/BF00279746. [DOI] [PubMed] [Google Scholar]
- 5.Haydon DJ, Guest JR. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;63(2-3):291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 6.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 7.Bramucci E, Milano T, Pascarella S. Genomic distribution and heterogeneity of MocR-like transcriptional factors containing a domain belonging to the superfamily of the pyridoxal-5′-phosphate dependent enzymes of fold type I. Biochem Biophys Res Commun. 2011;415(1):88–93. doi: 10.1016/j.bbrc.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Belitsky BR. Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator. J Mol Biol. 2004;340(4):655–664. doi: 10.1016/j.jmb.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Edayathumangalam R, et al. Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT. Proc Natl Acad Sci USA. 2013;110(44):17820–17825. doi: 10.1073/pnas.1315887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tramonti A, et al. Molecular mechanism of PdxR – a transcriptional activator involved in the regulation of vitamin B6 biosynthesis in the probiotic bacterium Bacillus clausii. FEBS J. 2015;282(15):2966–2984. doi: 10.1111/febs.13338. [DOI] [PubMed] [Google Scholar]
- 11.Takenaka T, Ito T, Miyahara I, Hemmi H, Yoshimura T. A new member of MocR/GabR-type PLP-binding regulator of D-alanyl-D-alanine ligase in Brevibacillus brevis. FEBS J. 2015;282(21):4201–4217. doi: 10.1111/febs.13415. [DOI] [PubMed] [Google Scholar]
- 12.Jochmann N, Götker S, Tauch A. Positive transcriptional control of the pyridoxal phosphate biosynthesis genes pdxST by the MocR-type regulator PdxR of Corynebacterium glutamicum ATCC 13032. Microbiology. 2011;157(Pt 1):77–88. doi: 10.1099/mic.0.044818-0. [DOI] [PubMed] [Google Scholar]
- 13.Belitsky BR. Role of PdxR in the activation of vitamin B6 biosynthesis in Listeria monocytogenes. Mol Microbiol. 2014;92(5):1113–1128. doi: 10.1111/mmi.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Qaidi S, Yang J, Zhang JR, Metzger DW, Bai G. The vitamin B6 biosynthesis pathway in Streptococcus pneumoniae is controlled by pyridoxal 5′-phosphate and the transcription factor PdxR and has an impact on ear infection. J Bacteriol. 2013;195(10):2187–2196. doi: 10.1128/JB.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiethaus J, Schubert B, Pfänder Y, Narberhaus F, Masepohl B. The GntR-like regulator TauR activates expression of taurine utilization genes in Rhodobacter capsulatus. J Bacteriol. 2008;190(2):487–493. doi: 10.1128/JB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, et al. Kinetic and crystallographic analysis of active site mutants of Escherichia coli gamma-aminobutyrate aminotransferase. Biochemistry. 2005;44(8):2982–2992. doi: 10.1021/bi048657a. [DOI] [PubMed] [Google Scholar]
- 17.Okuda K, et al. Role of the aminotransferase domain in Bacillus subtilis GabR, a pyridoxal 5′-phosphate-dependent transcriptional regulator. Mol Microbiol. 2015;95(2):245–257. doi: 10.1111/mmi.12861. [DOI] [PubMed] [Google Scholar]
- 18.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 19.Liu W, et al. Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase. Biochemistry. 2004;43(34):10896–10905. doi: 10.1021/bi049218e. [DOI] [PubMed] [Google Scholar]
- 20.Storici P, et al. Structures of gamma-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5′-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with gamma-ethynyl-GABA and with the antiepilepsy drug vigabatrin. J Biol Chem. 2004;279(1):363–373. doi: 10.1074/jbc.M305884200. [DOI] [PubMed] [Google Scholar]
- 21.Storici P, Qiu J, Schirmer T, Silverman RB. Mechanistic crystallography. Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid as elucidated by crystallography. Biochemistry. 2004;43(44):14057–14063. doi: 10.1021/bi0487185. [DOI] [PubMed] [Google Scholar]
- 22.Miyahara I, Hirotsu K, Hayashi H, Kagamiyama H. X-ray crystallographic study of pyridoxamine 5′-phosphate-type aspartate aminotransferases from Escherichia coli in three forms. J Biochem. 1994;116(5):1001–1012. doi: 10.1093/oxfordjournals.jbchem.a124620. [DOI] [PubMed] [Google Scholar]
- 23.Silverman RB, Levy MA. Mechanism of inactivation of gamma-aminobutyric acid-alpha-ketoglutaric acid aminotransferase by 4-amino-5-halopentanoic acids. Biochemistry. 1981;20(5):1197–1203. doi: 10.1021/bi00508a022. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Pozharski E, Fu M, Silverman RB, Ringe D. Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid. Biochemistry. 2010;49(49):10507–10515. doi: 10.1021/bi101325z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu D, et al. Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals “a tale of two mechanisms”. Biochemistry. 2007;46(37):10517–10527. doi: 10.1021/bi700663n. [DOI] [PubMed] [Google Scholar]
- 26.Lepore BW, et al. Chiral discrimination among aminotransferases: inactivation by 4-amino-4,5-dihydrothiophenecarboxylic acid. Biochemistry. 2010;49(14):3138–3147. doi: 10.1021/bi902052x. [DOI] [PubMed] [Google Scholar]
- 27.Schnackerz KD, et al. Identification and spectral characterization of the external aldimine of the O-acetylserine sulfhydrylase reaction. Biochemistry. 1995;34(38):12152–12160. doi: 10.1021/bi00038a008. [DOI] [PubMed] [Google Scholar]
- 28.Griswold WR, Castro JN, Fisher AJ, Toney MD. Ground-state electronic destabilization via hyperconjugation in aspartate aminotransferase. J Am Chem Soc. 2012;134(20):8436–8438. doi: 10.1021/ja302809e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griswold WR, Fisher AJ, Toney MD. Crystal structures of aspartate aminotransferase reconstituted with 1-deazapyridoxal 5′-phosphate: Internal aldimine and stable L-aspartate external aldimine. Biochemistry. 2011;50(26):5918–5924. doi: 10.1021/bi200436y. [DOI] [PubMed] [Google Scholar]
- 30.Griswold WR, Toney MD. Role of the pyridine nitrogen in pyridoxal 5′-phosphate catalysis: Activity of three classes of PLP enzymes reconstituted with deazapyridoxal 5′-phosphate. J Am Chem Soc. 2011;133(37):14823–14830. doi: 10.1021/ja2061006. [DOI] [PubMed] [Google Scholar]
- 31.Peng Q, et al. Activation of gab cluster transcription in Bacillus thuringiensis by γ-aminobutyric acid or succinic semialdehyde is mediated by the Sigma 54-dependent transcriptional activator GabR. BMC Microbiol. 2014;14:306. doi: 10.1186/s12866-014-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onuffer JJ, Kirsch JF. Redesign of the substrate specificity of Escherichia coli aspartate aminotransferase to that of Escherichia coli tyrosine aminotransferase by homology modeling and site-directed mutagenesis. Protein Sci. 1995;4(9):1750–1757. doi: 10.1002/pro.5560040910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Silverman RB, Andruszkiewicz R, Nanavati SM, Taylor CP, Vartanian MG. 3-Alkyl-4-aminobutyric acids: The first class of anticonvulsant agents that activates L-glutamic acid decarboxylase. J Med Chem. 1991;34(7):2295–2298. doi: 10.1021/jm00111a053. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]