Significance
We present a genome-wide study of the signals responsible for the early induction of the body axis in the following experimental conditions: β-catenin morpholino; Wnt, Siamois, and Cerberus mRNAs; LiCl treatment; and dorsal-ventral regenerating half-embryos bisected at gastrula. Comparing 46 RNA-seq libraries, we uncovered the genetic networks that initiate dorsal-ventral patterning and Spemann’s organizer formation. We defined an early β-catenin signature that has only minor overlap with recently published late zygotic Wnt signatures. The relation of these early steps of development to endomesodermal germ layer induction was studied by overexpressing the growth factor antagonist Cerberus. This study offers a rich resource for understanding the earliest inductive events in the body plan of a model vertebrate embryo.
Keywords: embryonic induction, Wnt signaling, embryonic patterning, Cerberus, Spemann organizer
Abstract
The earliest event in Xenopus development is the dorsal accumulation of nuclear β-catenin under the influence of cytoplasmic determinants displaced by fertilization. In this study, a genome-wide approach was used to examine transcription of the 43,673 genes annotated in the Xenopus laevis genome under a variety of conditions that inhibit or promote formation of the Spemann organizer signaling center. Loss of function of β-catenin with antisense morpholinos reproducibly reduced the expression of 247 mRNAs at gastrula stage. Interestingly, only 123 β-catenin targets were enriched on the dorsal side and defined an early dorsal β-catenin gene signature. These genes included several previously unrecognized Spemann organizer components. Surprisingly, only 3 of these 123 genes overlapped with the late Wnt signature recently defined by two other groups using inhibition by Dkk1 mRNA or Wnt8 morpholinos, which indicates that the effects of β-catenin/Wnt signaling in early development are exquisitely regulated by stage-dependent mechanisms. We analyzed transcriptome responses to a number of treatments in a total of 46 RNA-seq libraries. These treatments included, in addition to β-catenin depletion, regenerating dorsal and ventral half-embryos, lithium chloride treatment, and the overexpression of Wnt8, Siamois, and Cerberus mRNAs. Only some of the early dorsal β-catenin signature genes were activated at blastula whereas others required the induction of endomesoderm, as indicated by their inhibition by Cerberus overexpression. These comprehensive data provide a rich resource for analyzing how the dorsal and ventral regions of the embryo communicate with each other in a self-organizing vertebrate model embryo.
With the recent completion of the Xenopus laevis genome (1), it has now become practical to study global transcriptional changes at the earliest stages of development in this classical model organism: for example, by analyzing the transcriptomes of the dorsal and ventral regions of the gastrula embryo (2). Recent studies analyzed the zygotic Wnt pathway in X. laevis and Xenopus tropicalis embryos, defining late gene signatures transcriptionally activated by endogenous Wnt8 expressed in the ventro-lateral marginal zone (3, 4). This zygotic Wnt signal occurs during mid- to late-gastrulation, promotes the formation of ventral-posterior tissues, and inhibits head development. For example, when Wnt8 DNA constructs that are expressed only at gastrula are injected into embryos (5), head structures are reduced. Reciprocally, overexpression of Wnt antagonists such as Dickkopf1 (Dkk1) (6) and Frzb1 (7) results in enlarged head structures.
The opposite phenotype is elicited by microinjection of Wnt mRNAs during early cleavage, generating embryos with spectacular secondary axes containing complete heads when injected into ventral blastomeres (8). When injected radially into embryos, Wnt8 mRNA leads to a dorsalized phenotype consisting entirely of head structures without trunks and a radial Spemann organizer (9–11). Similar dorsalizing effects are obtained by incubating embryos in lithium chloride (LiCl) solution at the 32-cell stage (12). LiCl mimics the early Wnt signal by inhibiting the enzymatic activity of glycogen synthase kinase 3 (GSK3) (13), an enzyme down-regulated by the canonical Wnt signal (14, 15). The molecular nature of the endogenous dorsal signal in Xenopus eggs remains elusive, but it is clear that its main effect is to cause the stabilization of β-catenin during cleavage on the dorsal side of the embryo (16, 17). A β-catenin antisense morpholino (β-CatMO) is one of the most potent and reliable reagents in X. laevis embryology, and its microinjection results in embryos lacking Spemann organizer tissue and dorsal development (18).
Dorsal β-catenin stabilization requires a cortical rotation of the egg cortex toward the animal pole and the displacement of maternal cytoplasmic determinants containing Dishevelled (Dvl) toward the animal pole (19, 20). Interestingly, the endogenous cytoplasmic determinants are refractory to inhibition by Dkk1 and Frzb mRNAs, which, however, readily inhibit microinjected Wnt mRNA (21, 22). The combination of maternal β-catenin, which induces the homeobox transcriptional activator Siamois/Twin (10), with high zygotic Nodal/TGF-β (called Xnr1/2/5/6 in Xenopus) signaling on the dorsal side induces formation of Spemann organizer tissue (23). The organizer in turn secretes BMP antagonists such as Chordin and Noggin, Wnt antagonists such as Dkk1, Frzb1 and Crescent, and multivalent inhibitors such as Cerberus (24). These proteins shape dorsal-ventral (D-V) and anterior-posterior (A-P) patterning (25, 26). Therefore, understanding the signaling pathways that induce the transcriptional activation of Spemann organizer tissue is of fundamental importance for understanding vertebrate morphogenesis.
In the present study, we applied genome-wide transcriptome analyses to investigate the induction of the Spemann organizer by the maternal Wnt/β-catenin signal. We used β-catenin MO–, Wnt8 mRNA-, Siamois mRNA-, and Cerberus mRNA-injected embryos and LiCl-treated embryos. We also performed RNA-seq of uninjected embryos bisected into dorsal and ventral fragments at midblastula and allowed to develop for 5 h before isolating RNA at early gastrula. This procedure maximizes dorsal-ventral (D-V) differences and, in addition, tests the regenerative potential of the embryo (26). A total of 46 RNA-seq libraries were examined, comparing levels of the expression of 43,673 transcripts at late blastula (stage 9) and early gastrula (stage 10.5) of X. laevis. The results define an early β-catenin signature that has very little overlap with the zygotic Wnt signature and generate a comprehensive genome-wide catalog of the earliest developmental transcriptional changes in a vertebrate embryo.
Results
Defining Spemann Organizer Induction by β-Catenin.
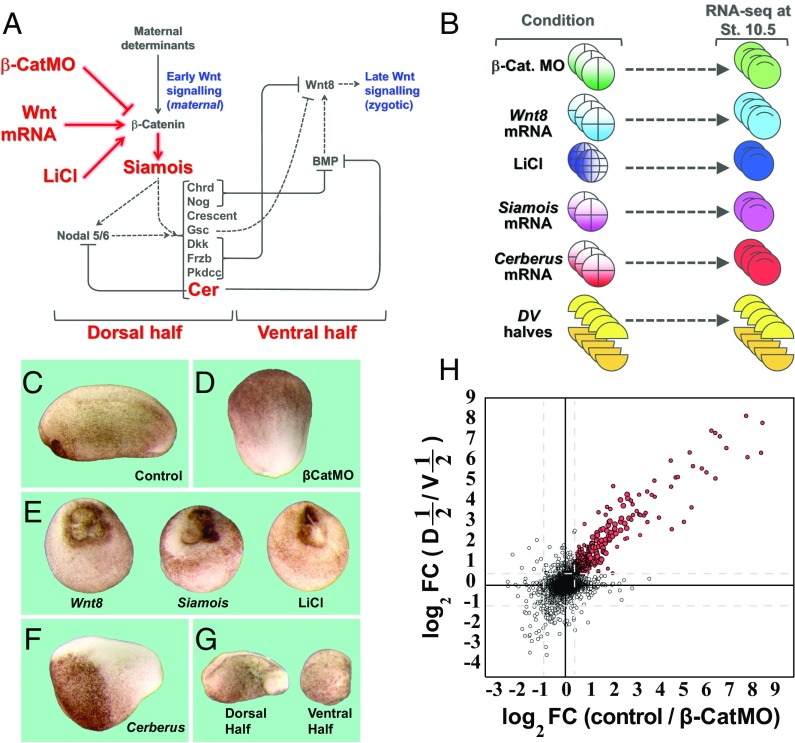
As shown in Fig. 1A, the dorsal maternal determinant signal can be blocked by microinjection of β-CatMO at the two- to four-cell stage. This signal can be mimicked by microinjection of Wnt8 mRNA, LiCl treatment, or microinjection of mRNA for the downstream effector Siamois. The early β-catenin signal results in the induction of the TGF-β–related molecules Xnr5/6, and a mixture of organizer growth factor inhibitors on the dorsal side that oppose signals provided by BMP and the zygotic (late) Wnt signaling pathways on the ventral side of the embryo (24, 25) (Fig. 1A).
Fig. 1.
RNA-seq libraries for transcriptome analysis of early β-catenin targets. (A) Diagram showing the early β-catenin–dependent gene network on the dorsal side and the zygotic Wnt8 signal that arises later on the ventral side. The embryonic manipulations that were subjected to RNA-seq in the present study are indicated in red. (B) For RNA-seq, embryos were injected with β-catenin MO, Wnt8, Siamois, or Cerberus mRNA, treated with LiCl, or cut into dorsal and ventral halves at stage 8. All embryos were harvested at stage 10.5 for RNA extraction and RNA-seq. Some embryos were also analyzed at blastula stage 9. (C–G) Phenotypes of embryos subjected to manipulations in this study. Note that β-CatMO–injected embryos lacked all traces of a dorsal axis whereas Wnt8, Siamois mRNA microinjected, and LiCl-treated embryos were all radially dorsalized. Cerberus mRNA–microinjected embryos lacked endomesoderm and consisted mostly of anterior neural tissue and cement gland. Ventral half-embryos lacked dorsal axes whereas dorsal half-embryos developed a complete axis. (Magnification: 8×.) (H) Scatter plot comparing the log2 of fold change (FC) in RPKM upon β-catenin knockdown with that of dorsal halves/ventral halves. Shown here are 40,157 transcripts; red dots indicate transcripts decreased by β-catenin MO (by at least 1.4-fold in average of three experiments) and enriched in dorsal halves compared with ventral halves (by at least 1.5-fold in average of five experiments). Full datasets are provided in Dataset S1. Cer, Cerberus; D, dorsal half; St., stage; V, ventral half.
As indicated in Fig. 1B, the effect of β-catenin loss of function or Wnt8 mRNA gain of function was analyzed at early gastrula by RNA-seq in three independent experiments. In addition, inhibition of GSK3 by LiCl at the 32-cell stage and overexpression of Siamois mRNA were analyzed in duplicate, and the effect of blocking endomesoderm induction with Cerberus mRNA was analyzed in triplicate (Fig. 1B). As controls, we used uninjected embryos because previous work indicates that they behave like mock-injected embryos (27). We also analyzed transcripts from dorsal and ventral half-embryos (bisected at stage 8 and allowed to regenerate for 5 h until stage 10.5) in quintuplicate (Fig. 1B). In all experiments, sibling embryos were allowed to develop until tailbud stage, and gastrula stage 10.5 RNA samples were used for RNA-seq only when completely dorsalized or ventralized phenotypes, such as those in Fig. 1 C–G, were obtained. The RPKM (reads per kilobase per million mapped reads) for all annotated X. laevis genes are provided in Dataset S1, and the raw data for all 46 libraries are available at the GEO repository (accession no. GSE93195).
β-Catenin binds to thousands of gene promoters during early Xenopus development (3, 4). However, the initial accumulation of β-catenin is dorsal (16). In agreement with this finding, a scatter plot of the fold change (FC) of transcripts inhibited by β-CatMO correlated strongly with dorsal/ventral enrichment at early gastrula (Fig. 1H, red dots). Overall, many more transcripts were inhibited by β-catenin loss of function than were activated (Fig. S1A). We conclude that β-catenin knockdown provides an excellent method for D-V gene discovery.
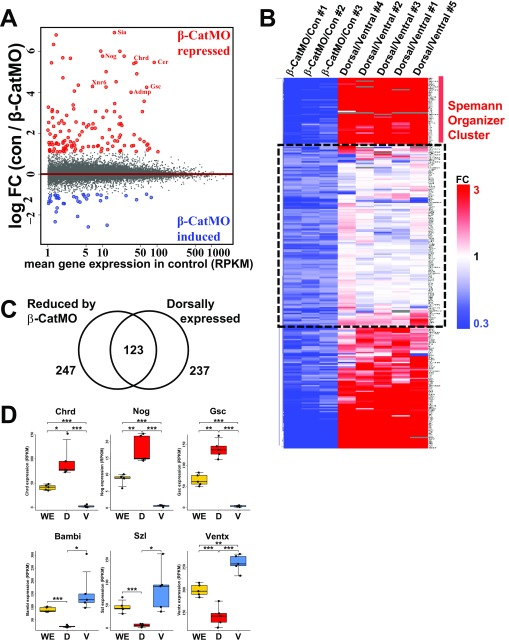
Fig. S1.
Transcriptome analysis of β-CatMO–injected embryos and dorsal and ventral halves. (A) MA-plot comparing gene expression between uninjected control and β-catenin MO–injected embryos including 16,729 transcripts. The average (three independent experiments) log2 fold change (FC) in expression of transcripts in control (con) embryos over β-CatMO–injected embryos (con/β-CatMO) is plotted on the ordinate; the average mean gene expression (in RPKM) in control embryos is represented on the abscissa. Red indicates β-CatMO–repressed transcripts (1.4-fold minimum decrease); blue indicates β-CatMO–induced transcripts (1.4-fold increased); gray dots indicate all other transcripts. Note that β-CatMO represses more genes than it activates, which is consistent with the widespread role of β-catenin as a transcriptional coactivator. (B) Heat map showing all transcripts that were repressed 1.4-fold by β-catenin MO in three independent pairs of libraries, and the dorsal to ventral (D/V) FC in five independent dorsal and ventral half libraries bisected at midblastula stage 8 and allowed to regenerate for 5 h until gastrula. Rows indicate FC of transcripts as indicated in the scale bar at the right of the panel. The various conditions are indicated at the top of the columns. Hierarchical clustering of rows clustered classical Spemann organizer genes on the top region. Genes within the dashed line box were repressed by β-CatMO but not enriched in the dorsal half; these were removed from our early β-catenin signature because they represent genes that require β-catenin independently of the maternal dorsal signal. (C) Venn diagram illustrating overlap of genes between β-CatMO–inhibited genes and dorsally enriched genes. Of 247 genes reduced by β-catenin MO and 237 genes enriched in dorsal side, 123 genes overlapped and defined our early dorsal β-catenin signature. In these stringent conditions, which require results above a certain threshold in all three independent experiments, some β-catenin–regulated genes may miss the cutoff. The complete stage 10.5 RPKM data are presented in Dataset S1, and, from this dataset, any genes that narrowly missed the cutoff can be retrieved. (D) Box plots of RPKMs of whole embryo (WE) and dorsal (D) and ventral (V) halves in five independent RNA-seq experiments. Note that, as expected, Chrd, Nog, and Gsc were significantly enriched in regenerating dorsal halves and that Bambi, Szl, and Ventx were enriched in ventral halves, which supports the quantitative nature of the RNA-seq analyses.
Defining the Early Dorsal β-Catenin Transcriptional Signature.
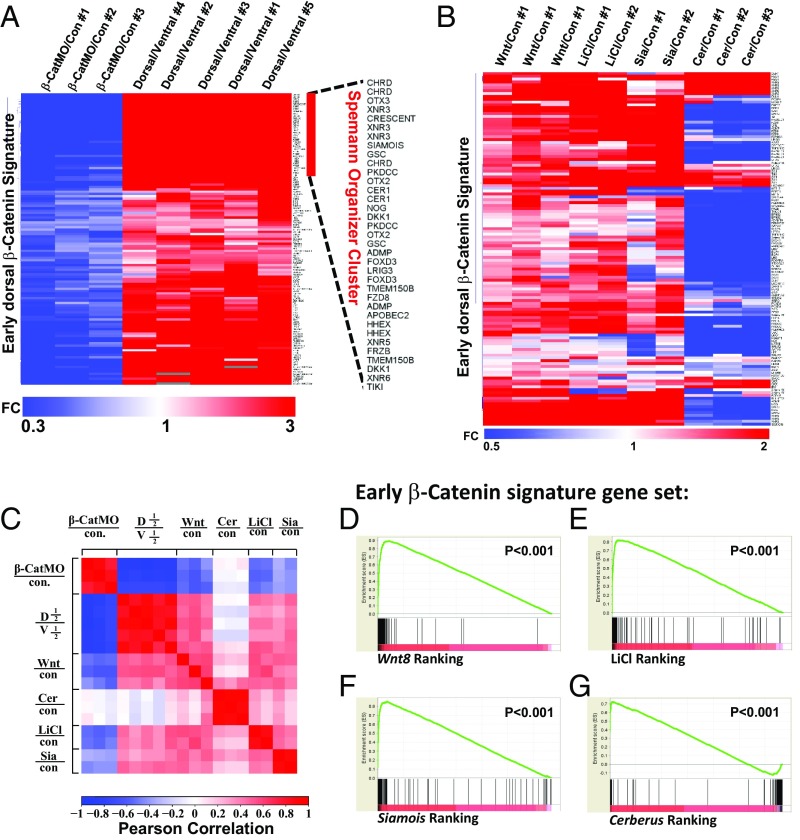
Among the genes repressed by β-CatMO, we found that 247 were reproducibly inhibited in each of all three experiments at stage 10.5 (Fig. S1B). About half of those genes were equally expressed in dorsal and ventral regenerating half-embryos, indicating that, although they required β-catenin, they were not dependent on the early dorsal maternal determinant signal (Fig. S1B, dashed box). Once genes without dorsal enrichment were removed, we were left with a set of 123 transcripts (Fig. S1C) that not only were decreased by β-CatMO in three independent replicates, but also were reproducibly enriched dorsally in four of five D-V experiments. The D-V quintuplicate samples identified most of the known Spemann organizer genes, as well as the classical ventrally enriched genes (Fig. S1D).
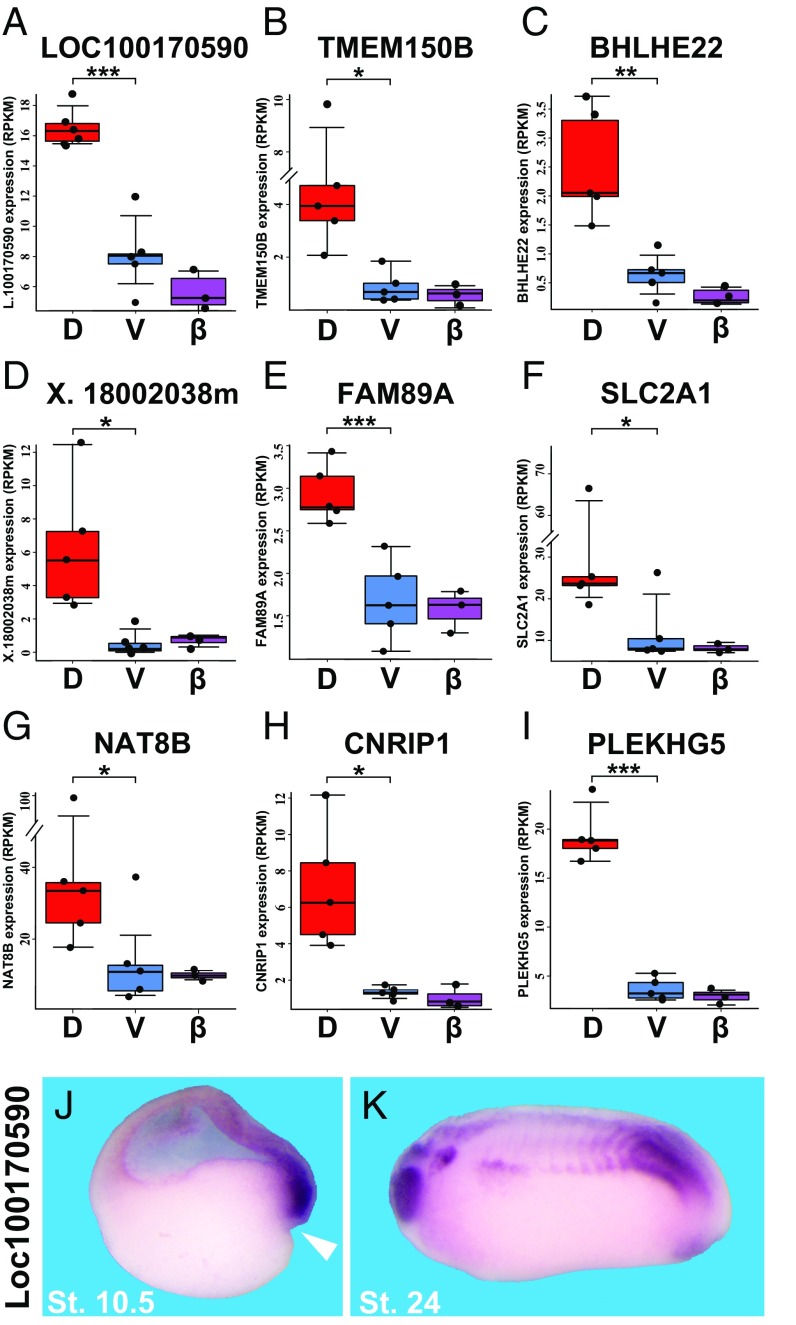
The early dorsal β-catenin signature identified by these experiments is listed in Fig. 2A. Of these 123 dorsal genes, 86 were reported to be bound by β-catenin in X. tropicalis (4) (Dataset S2). There were also 124 genes that were decreased by β-CatMO but not dorsally enriched, of which 34 bound β-catenin in ChIP experiments (Dataset S3) (4). The early dorsal signature includes many secreted factors and transcription factors known to be expressed in Spemann’s organizer (2, 24), as well as many previously undescribed dorsal genes. As shown in Fig. 3, nine of these genes were enriched dorsally in a statistically significant way, and included PLEKHG5 (a RhoA GEF also known as SYX); TMEM150B (a transmembrane protein); BHLHE22 (a class E basic helix–loop–helix protein); FAM89A (Family with Sequence Similarity 89 Member A); Xlaev18002038m (unknown); CRNRIP1 (cannabinoid receptor interacting protein); NAT8B (probable N-acetyl transferase 8B); and LOC100170590.L (unknown Xenopus ORF MGC115585). In situ hybridization for LOC100170590.L confirmed that it was indeed expressed in the Spemann organizer at stage 10.5 whereas, at tailbud (stage 24), it was localized in the eye, brain, somites, and proctodeum (Fig. 3 J and K). The requirement for β-catenin and dorsal enrichment for LOC100170590.L were validated through qRT-PCR (Fig. S2A). This gene has a possible homolog in coelacanth fishes, but not in mammals (Fig. S2B).
Fig. 2.
The early dorsal β-catenin gene signature. (A) Genes were categorized as follows: secreted factors, transcriptional factors and regulators, transmembrane protein and receptors, enzymes/kinases, and others and unknowns. Human gene symbols and full names are shown (2), except when Xenopus-specific names exist. Because the subtetraploid X. laevis has long and short homologs for many genes, duplicate names have been removed from this list. For unknown genes, the gene IDs from the JGI9 X. laevis genome are indicated. (B) Venn diagram showing that the early β-catenin gene signature is distinct from the late zygotic Wnt signature (3), with only three genes in common.
Fig. 3.
The early β-catenin signature contains previously undescribed dorsal transcripts. (A–I) Box plots indicate dorsal and ventral gene expression levels (in RPKM) in five independent regenerating half-embryo experiments. These genes have either not been previously characterized or have been only superficially investigated. Box plots illustrate the reproducibility of replicate experiments. Each dot represents the expression in RPKM of the corresponding gene in a distinct experiment. Genes above the whisker are considered outliers according to the interquartile range (IQR) calculated for each box plot. The difference between dorsal and ventral expression was statistically significant for all nine genes, with all genes surpassing the 95% confidence interval mark at a minimum. The β-CatMO RPKMs (β) were similar to those of ventral halves. Statistical significance was determined through two-tailed pairwise P values. Significant differences in expression values (RPKM) are indicated as *≤0.05, **≤0.01, and ***≤0.005. (J) In situ hybridization validating that unknown Loc100170590 is indeed expressed in Spemann organizer mesoderm at gastrula. White arrowhead indicates the dorsal blastopore lip. (Magnification: 16×.) (K) At tailbud stage 24, Loc1001170590 is expressed in eye, central nervous system, somites, and proctodeum. (Magnification: 14×.) D, dorsal; St., stage; V, ventral.
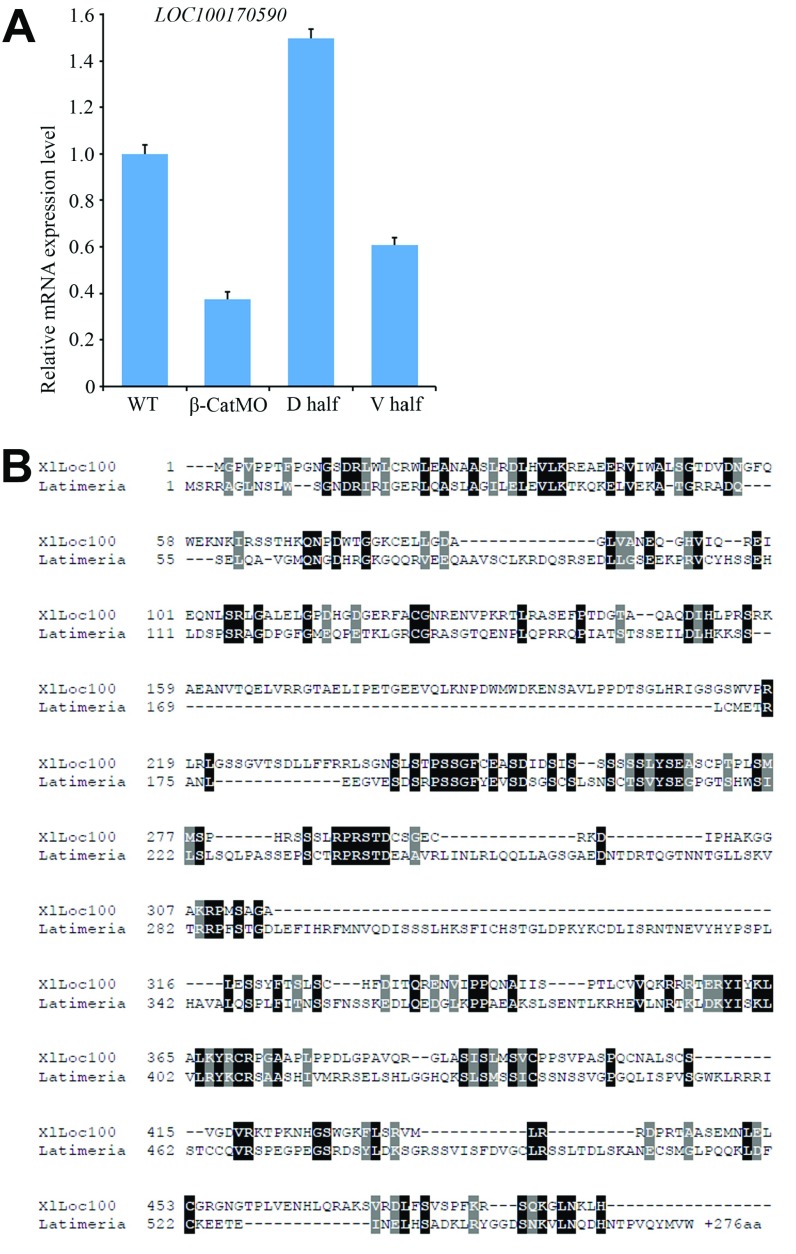
Fig. S2.
qRT-PCR validation and sequence homology of LOC100170590. (A) β-catenin–dependent regulation and D-V enrichment of LOC100170590 was validated by qRT-PCR analysis. β-CatMO decreased LOC100170590 mRNA levels compared with uninjected WT controls (WT), and expression in dorsal half-embryos (D half) was significantly higher than in ventral half-embryos (V half). (B) Amino acid sequence comparison between Xenopus laevis Loc100170590 and Latimeria chalumnae XP_005988953.1 (a coelacanth fish) revealed a 27% degree of identity. The coelacanth gene has been annotated as possible Dapper homolog-like 2, but XlLoc100170590 lacks critical Dapper-like features, such as a PDZ domain and a leucine zipper. The sequence alignment was generated by using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) followed by BoxShade (embnet.vital-it.ch/software/BOX_form.html). Black and gray shaded boxes indicate identical and similar amino acid residues, respectively. There were no significant homologies with mammalian genomes.
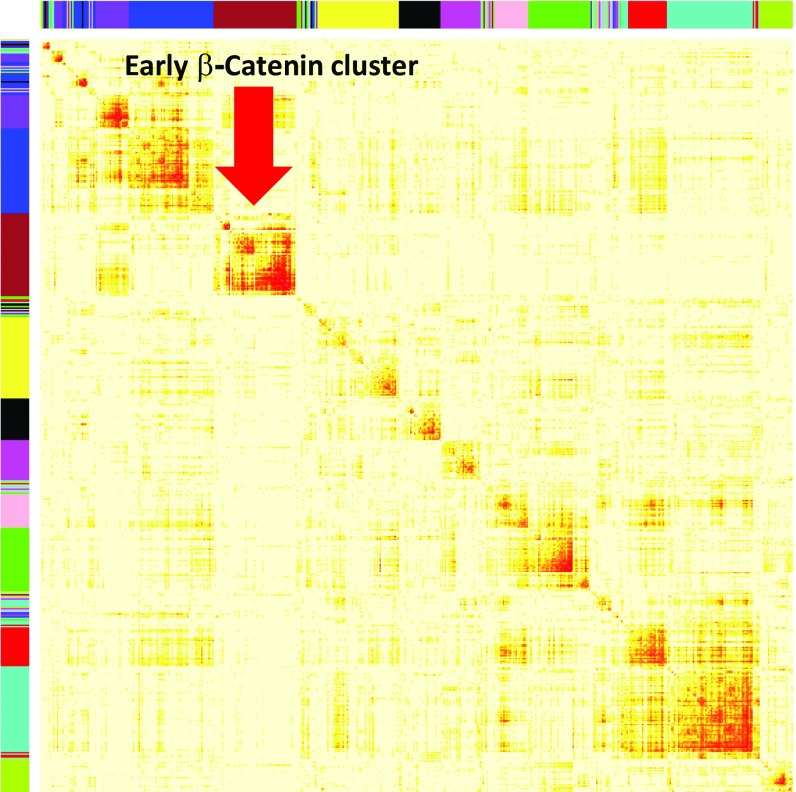
In generating our early β-catenin signature, several arbitrary cutoff criteria were applied (RPKM above 1, decrease by 1.4-fold in three β-CatMO experiments, and dorsal enrichment above 1.5-fold in four libraries). We therefore sought an independent method to validate this signature. To this end, we applied Weighted Gene Coexpression Network Analysis (WGCNA) to 29 libraries sequenced at stage 10.5 (28). This method, which detects modules of highly correlated genes, generated a cluster of 233 genes that contained 106 of the 123 transcripts (86%) in the early β-catenin signature (Fig. S3 and Dataset S4), which suggests that our early β-catenin signature is independent of subjective cutoff points.
Fig. S3.
Weighted Gene Coexpression Network Analysis (WGCNA) of stage 10.5 libraries. WGCNA unsupervised hierarchical clustering identified a distinct cluster (brown cluster, red arrow) that contained 106 of the 123 genes that compose the early β-catenin signature. Hierarchical clustering of marbling score-related genes and visualization of gene modules was performed. The colored bars are directly consistent with the module (color) for the clusters of genes.
The signature presented here defines 123 X. laevis dorsal transcripts that require the early β-catenin signal. These results should be compared with those recently obtained by Kjolby and Harland (3), who used embryos microinjected with the Wnt antagonist Dkk1 to identify a late Wnt signature of 82 genes that constitute zygotic Wnt/β-catenin transcriptional targets at midgastrula (stage 11.5). Remarkably, only 3 genes (IRX1, MYF5, and SEBOX) overlapped between the early and late Wnt signatures (Fig. 2B). We conclude that a very specific early dorsal β-catenin gene signature can be defined by combining a requirement for β-catenin and dorsal expression.
The Early β-Catenin Gene Signature Is Activated by LiCl and by Wnt8 and Siamois mRNAs.
The repression by β-CatMO and dorsal enrichment of the 123 transcripts that constitute the early β-catenin signature can be visualized in Fig. 4A. Unsupervised hierarchical grouping clustered the classical Spemann organizer genes together, such as Chordin, Xnr3, Crescent, Siamois, Goosecoid, PKDCC, OTX2, Cerberus, Noggin, Dkk1, ADMP, and others (Fig. 4A, sidebar).
Fig. 4.
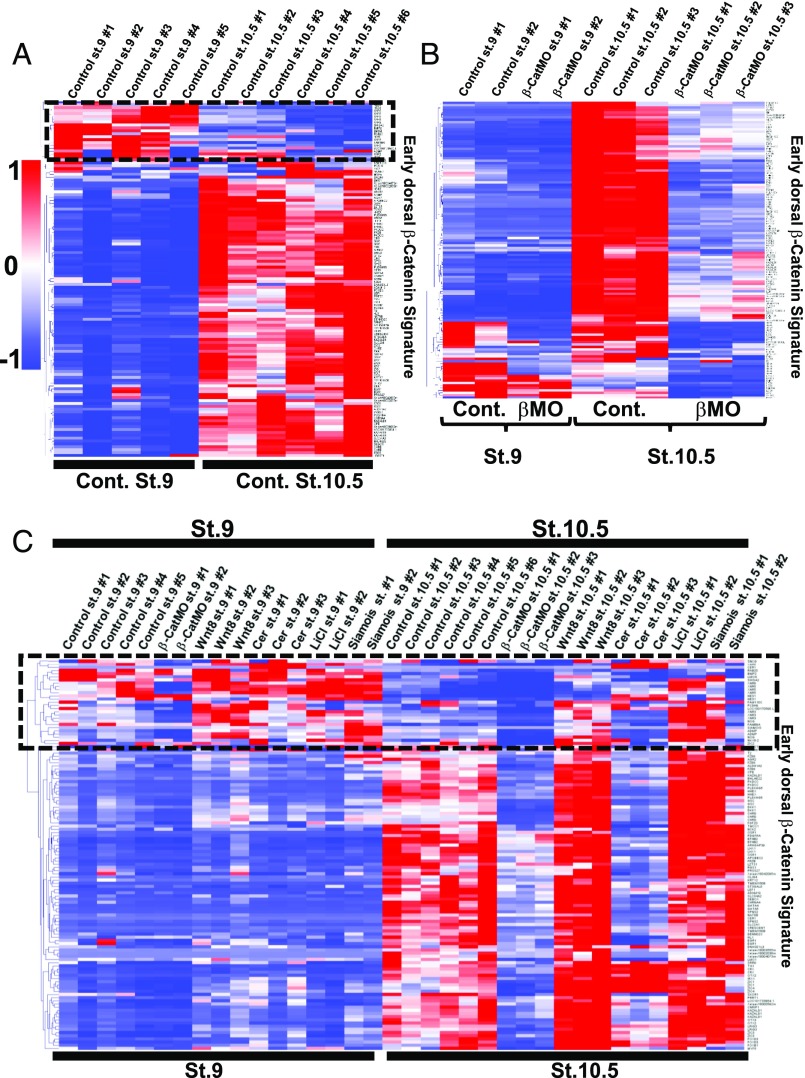
Correlation of the early dorsal β-catenin gene signature with Wnt8, LiCl, Siamois-induced genes, and Cerberus-repressed genes. (A) Heat map showing the fold change of early β-catenin signature genes in three β-catenin MO and five dorsal halves with respect to control and ventral half-embryo libraries, respectively. Fold changes (FC) over controls were used as inputs, and unsupervised hierarchical clustering of columns and rows was performed. Note that hierarchical clustering of rows identified most classic known Spemann organizer genes on the top of the heat map. (B) Heat map displaying regulation of genes composing the early β-catenin signature by Wnt8 mRNA, LiCl, and Siamois or Cerberus mRNAs. (C) Correlation matrix of β-CatMO–, Wnt8-, Cerberus-, and Siamois-injected embryos and LiCl-treated embryos with the early dorsal β-catenin signature. Correlation scores were calculated as Pearson correlation coefficients (PCCs) and were color-coded as shown in the scale bar on the bottom of the panel. Note that the β-CatMO (in blue) anticorrelates with all conditions except Cerberus mRNA-injected embryos. These results show that the early β-catenin signature obtained via RNA-seq is reproducible and readily identifies dorsalizing and ventralizing conditions. (D–G) Gene Set Enrichment Analysis (GSEA) showed that the early β-catenin gene set was significantly enriched at the top of the Wnt8-, LiCl-, and Siamois-induced gene rankings, and at the bottom of the Cerberus-induced ranking. Cer, Cerberus; con, control; D1/2, dorsal half; Sia, Siamois; V1/2, ventral half.
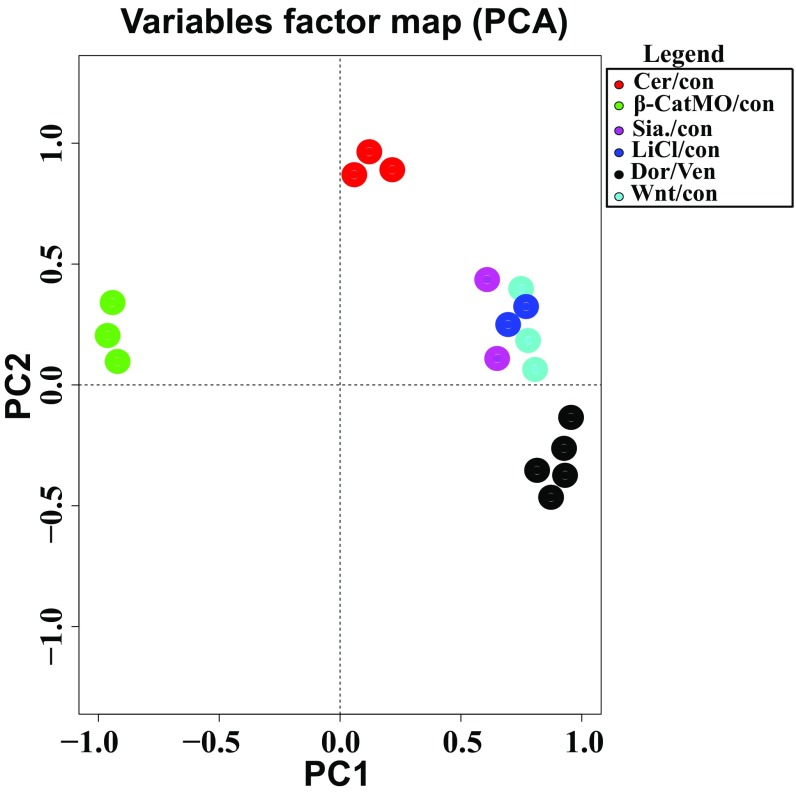
To investigate the relationship between the early β-catenin signature and the Wnt pathway, we microinjected embryos radially with Wnt8 mRNA in triplicate. Heat map analysis indicated that most early dorsal β-catenin signature genes were activated by Wnt8 mRNA (Fig. 4B). Similarly, LiCl treatment and Siamois mRNA microinjection (in duplicate) induced most genes in this signature (Fig. 4B). The expression of the early β-catenin signature across the different conditions sequenced was reproducible, as indicated by Pearson correlation matrix analysis (Fig. 4C). In addition, principal component analysis (PCA) showed that replicates and Spemann organizer-inducing conditions clustered together, confirming reproducible expression of the early β-catenin signature (Fig. S4). Dataset S2 provides the RPKM levels for the early β-catenin signature genes in each library.
Fig. S4.
The expression of the 123-gene early dorsal β-catenin signature examined via principal component analysis (PCA) to analyze dimensionality in six experimental conditions: Cerberus/control (Cer/con), β-catenin morpholino/control (β-CatMO/con), Siamois/control (Sia/con), lithium chloride/control (LiCl/con), dorsal/ventral regenerating halves (Dor/Ven), and Wnt8/control (Wnt/con). Each axis represents a principal component (PC), with the first one having the most variation. PCA analysis confirmed the reproducibility of the early β-catenin signature by clustering replicate conditions together and distinguishing ventralizing (β-catenin depletion) and dorsalizing treatments. The Cerberus mRNA-injected condition was located between the β-CatMO and the rest of the sequenced conditions due to its ability to inhibit both Nodal and BMP target genes.
Analysis of the early β-catenin signature enrichment among Wnt8 mRNA-, LiCl-, Siamois mRNA-, and Cerberus mRNA-induced genes was performed using Gene Set Enrichment Analysis (GSEA) (29). All genes with RPKM values above 1 (in corresponding control embryos, ∼16,000 genes) were ranked according to their average fold change. The early β-catenin gene set was greatly enriched within the genes ranked according to their levels of induction by Wnt8 mRNA (indicated as vertical black lines in Fig. 4D). Similarly, LiCl- and Siamois-induced gene ranking showed enrichment of this gene set at the top of the list (Fig. 4 E and F). We conclude that the dorsal β-catenin gene set closely mirrors early Wnt, LiCl, and Siamois signaling in the early embryo.
Cerberus Inhibits Most Endomesodermal Organizer Genes.
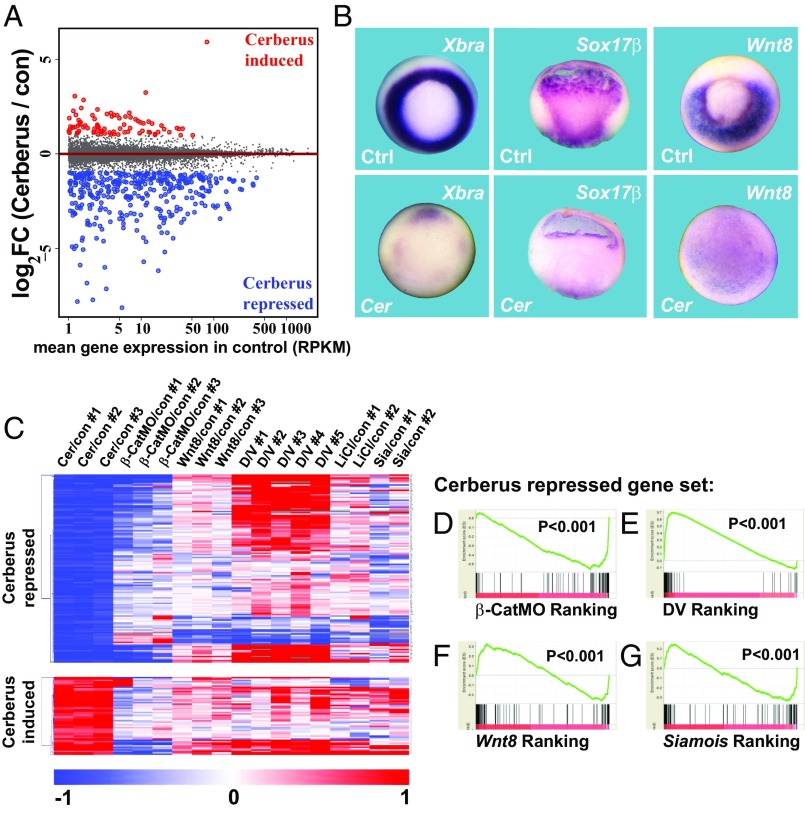
Cerberus is a protein secreted by the head organizer that functions as a multivalent growth factor antagonist by inhibiting the Nodal/TGFβ, BMP, and Wnt pathways (30, 31). To investigate the convergence of these pathways with the early β-catenin signature, we overexpressed Cerberus mRNA in X. laevis embryos, and RNA-seq libraries were prepared at early gastrula in three independent experiments. Most genes within the early dorsal β-catenin signature were inhibited, presumably indicating a requirement for endomesodermal differentiation (Fig. 4B, in blue). However, a few genes in the signature were increased by Cerberus mRNA, including Xnr5, Xnr6, and many neural genes such as Hes1, Zic1, Zic4, Otx2, Irx1, Nkx6-2, and Tiki/Trabd2a (Fig. 4B). GSEA analysis confirmed that many components of the β-catenin gene set were inhibited by Cerberus mRNA although some transcripts still ranked highly in the list (Fig. 4G).
When the entire transcriptome of Cerberus mRNA-injected embryos was examined, many more genes were significantly repressed than were induced (Fig. 5A). This observation was consistent with the inhibition of mesodermal and endodermal differentiation marked by Xbra and Sox17β, respectively (Fig. 5B). It is noteworthy that expression of xWnt8, the major Wnt that signals from ventrolateral mesoderm during Xenopus gastrulation, is essentially eliminated in Cerberus mRNA-injected embryos (Fig. 5B and Dataset S1). Cerberus overexpression represses, and induces, a wide range of genes of both dorsal and ventral origin by stage 10.5 (Fig. 5C and Datasets S5 and S6). Previous RNA-seq studies used the Nodal/TGF-β receptor inhibitor SB431542 to define Nodal target genes (27). We observed that Cerberus, a Nodal antagonist, strongly repressed 107 genes, of which 49 overlapped with genes repressed by SB431542 (Dataset S5). In GSEA analyses, a set of 107 genes repressed by Cerberus were found predominantly at the bottom of the correlation of those inhibited by β-CatMO (Fig. 5D). Cerberus-repressed genes also correlated with genes arranged according to their D-V enrichment (Fig. 5E). When the Cerberus-repressed gene set was probed against rankings of transcripts induced by Wnt8 or Siamois, they appeared at either the top or bottom of the ranked list, presumably reflecting the dual inhibition of Nodal and BMP pathways by this multivalent antagonist (Fig. 5 F and G). We conclude that many, but not all, of the early β-catenin–induced genes require endomesoderm germ layer specification by stage 10.5.
Fig. 5.
The Cerberus gene signature identifies early dorsal β-catenin genes that require endomesodermal induction. (A) MA-plot comparing gene expression of 16,729 mRNAs between control and Cerberus mRNA-injected embryos. The average log2 fold change in expression of transcripts in Cerberus mRNA-injected embryos over control embryos is plotted on the y axis; the average gene expression in RPKM of uninjected control embryos is on the x axis. This MA-plot confirms the proper normalization of the RNA-seq data because the horizontal nature of the line across the zero baseline shows that there is no systematic bias. (B) Cerberus overexpression greatly inhibits Xbra, xSox17β, and xWnt8 mRNA expression when examined by in situ hybridization. (Magnification: 10×.) (C) Heat map showing how gene signatures repressed or induced by Cerberus mRNA were differentially regulated by various dorsalizing or ventralizing conditions. Note that a large block of Spemann organizer genes strongly activated in D/V regenerating half-embryos (in red, top of the heat map) are repressed by Cerberus-mediated endomesodermal inhibition. (D–G) GSEA analysis of the Cerberus-repressed gene set. Approximately 16,000 transcripts were ranked according to β-CatMO, D/V expression, and Wnt8 and Siamois mRNA induction and were examined for the ranking of Cerberus-repressed genes (vertical lines). Cer, Cerberus; con, control; Ctrl, control; D/V, dorsal/ventral ratio; Sia, Siamois.
The Early β-Catenin Signature Is Established at Gastrula.
In this study, we concentrated on the analysis of genes expressed at early gastrula because it is at this stage that Spemann organizer tissue is fully functional. We also analyzed gene expression at the earlier stage 9 (blastula, about 3.5 h before harvesting RNA from siblings used for gastrula studies) in some experiments and provide the complete RPKM data for stage 9 libraries in Dataset S7. As shown in Fig. 6A, most of the 123 genes in the early β-catenin signature were not activated until gastrula (shown as blue at stage 9 in the RPKM heat map). However, a subset of β-catenin targets had higher expression at blastula (in red) relative to gastrula stage 10.5 (Fig. 6A, boxed on top). Accordingly, most genes in the early β-catenin signature were unaffected by β-CatMO at stage 9 (Fig. 6B). Results from all experimental conditions tested at blastula and gastrula (control, β-CatMO, Wnt8 mRNA, Cerberus mRNA, LiCl, and Siamois mRNA) were allowed to cluster hierarchically by row according to RPKM expression as shown in Fig. 6C. At blastula stage 9, most genes in the signature were unaffected by β-catenin depletion or dorsalizing treatments (Fig. 6C, in blue). However, a group of genes did respond to Wnt-mimicking agents and were inhibited by β-CatMO at stage 9 (boxed at top of Fig. 6C and listed in Dataset S8), which included the classical Wnt targets Siamois (10) and Xnr3 (32). In addition, many organizer-specific genes were induced at stage 9, such as Xnr5, Xnr6, Admp, Cerberus, Noggin, and Zic2. The relative delay in expression of the majority of the early β-catenin signature genes expressed at gastrula is likely explained by the need of the early β-catenin signal to activate Siamois as well as Xnr5/6 expression to achieve full organizer expression (33, 34), as indicated in the pathway outlined earlier in Fig. 1A.
Fig. 6.
Comparison of the expression of the early dorsal β-catenin gene signature at blastula and gastrula stages. (A) Heat map showing that the majority of the early dorsal β-catenin gene signature had higher expression levels (in red) in WT control (Cont.) embryos at stage 10.5 rather than at blastula stage 9. RPKMs of the genes constituting the early β-catenin signature genes at blastula stage 9 or gastrula stage 10.5 (results from five and six sequenced libraries, respectively) were normalized by row. (B) The majority of genes in the early β-catenin signature were unaffected by β-CatMO at stage 9; the effect of β-catenin MO becomes evident at stage 10.5. (C) Heat map comparing RPKM levels of the early dorsal β-catenin signature in 46 libraries at blastula and gastrula stages. Only a minority of the β-catenin signature were inhibited by β-CatMO or activated by Wnt-like signals at blastula stage 9. RPKM values of all libraries were allowed to cluster automatically according to rows. The cluster delimited by a box at the top indicates transcripts that are capable of being induced or repressed by dorsalizing or ventralizing treatments at blastula stage 9. A list of these genes is provided in Dataset S8 and includes many known early Wnt targets, such as Siamois, Xnr3, Xnr5, Xnr6, Noggin, and ADMP. The scale bar at the bottom indicates normalized RPKM values. In conclusion, most genes in the early β-catenin signature become sensitive to β-catenin depletion only at gastrula stage, presumably indicating the requirement of the endodermal and mesodermal germ layer induction for Spemann organizer development. Cer, Cerberus; Cont, control; St., stage.
Discussion
In Xenopus, fertilization triggers a cortical rotation that transports Dvl-containing cytoplasmic organelles, called maternal dorsal determinants, along microtubules toward prospective dorsal regions (19, 35). The rotation brings cytoplasmic determinants from vegetal cytoplasm in contact with the marginal zone of the egg before first embryonic cleavage (36, 37). This process determines the site at which the dorsal side of the embryo is formed by stabilizing maternal β-catenin protein, which accumulates in dorsal nuclei as early as the two- to four-cell stage (16, 17). The full molecular composition of the cytoplasmic determinant and the mechanism by which β-catenin is stabilized remain unknown although the participation of maternal Wnt11 and Dvl proteins, perhaps in combination with the formation of multivesicular bodies, has been proposed (22, 38). It is likely that the elusive cytoplasmic determinant is protein in nature because recent RNA-seq analyses of early cleavage stages (eight-cell) of X. laevis or X. tropicalis embryos have not detected any asymmetrically expressed mRNAs along the D-V axis (39, 40). The early β-catenin stabilization, in combination with zygotic Siamois and Nodal signaling, induces Spemann organizer formation at the gastrula stage (11). There has been considerable progress in identifying the components of the Spemann organizer (2, 41–43). However, a global examination of the transcriptomic landscapes elicited by the early nuclear β-catenin signal and associated pathways that induce formation of Spemann organizer tissue was lacking.
The Early β-Catenin Dorsal Gene Set.
In this study, we set out to determine the transcriptional output of the early maternal β-catenin signal. To this end, we sequenced the transcriptome of β-CatMO–injected embryos at gastrula (stage 10.5) in three independent experiments and compared them with their uninjected controls. We used β-CatMO as a loss-of-function reagent because it blocks dorsal development with 100% penetrance (18). Secreted Wnt antagonists (such as Dkk1 and Frzb) are ineffective at blocking the endogenous early β-catenin cytoplasmic determinant, yet inhibit the zygotic Wnt8 ventral-lateral signal that mediates A-P patterning (24). We found 247 transcripts that reproducibly required β-catenin, but, of these, to our surprise, only about half were dorsal genes, which suggested that many β-catenin transcriptional targets were not dependent on the dorsal cytoplasmic determinant signal. In fact, recent ChIP-seq studies indicate that endogenous β-catenin binds to 5,000 genes in the X. tropicalis genome, but only a small fraction of these are regulated by Wnt at any one stage (4).
To specifically focus on the genetic program activated by β-catenin on the dorsal side, we sequenced libraries from regenerating dorsal and ventral half-embryos bisected at midblastula and cultured until early gastrula in five independent experiments. This approach resulted in much higher D-V differences than those observed in previous studies using dissected dorsal and ventral blastopore lips (2) because the BMP and late Wnt8 signals in the ventral half were unopposed by diffusing organizer-secreted antagonists (26). Transcripts were filtered so that only those decreased by β-catenin knockdown and dorsally enriched resulted in a set of 123 genes, which were named the early dorsal β-catenin signature (Fig. 2A).
This filtering approach was validated by WGCNA (28), a conventional clustering method that grouped 86% of the early β-catenin signature within a single cluster. The early β-catenin signature included many Spemann organizer genes. In addition, it identified many other genes that have been previously undescribed (such as LOC100170590.L) (Fig. 3 A, J, and K), unknown to be dorsally enriched, or, in one case, only superficially described (PLEKHG5) (Fig. 3I) (43). Studies are needed to dissect the function of these dorsal genes in D-V patterning. A likely explanation for why these genes had been missed in our previous studies (2) is that dorsal and ventral half-embryos were allowed to regenerate, maximizing differences as a result of unopposed BMP and Wnt8 signaling in ventral halves.
A point of interest is that the Wnt8 gene itself is highly increased in ventral half-embryos and, conversely, has one of the highest fold decreases in Siamois-injected and LiCl-treated embryos (Dataset S1). Wnt8 is responsible for most of the late Wnt signature in Xenopus and is known to be induced by BMP4 (4, 44). The genes inhibited by β-catenin but not dorsally enriched also constitute an interesting group, in particular in light of the recent finding that β-catenin is bound to thousands of gene loci in Xenopus (4). Of these 124 genes, 34 bound β-catenin in ChIP assays (4) (Dataset S3).
A Rich Genome-Wide Embryonic Resource.
We have compiled the RNA-seq results in complete RPKM files of expression levels for the 43,673 genes annotated in the Xenopus genome (1) comprising a total of 46 RNA-seq libraries. This rich resource is now accessible to the Xenopus community or anyone interested in the complete transcriptional repertoire of Spemann organizer-inducing signals in vertebrate embryos. The embryonic transcriptional response should be considered globally because, for each action in the organizer, there is a reaction in the ventral side (45).
The widespread dorsal gene activation by overexpression of the homeobox protein Siamois was particularly interesting because it implied that most of the early maternal β-catenin signal is mediated through this Xenopus-specific transcription factor (10, 46, 47). The X. laevis genome contains six active Siamois/Twin genes, which ensure a rapid response to the maternal β-catenin signal (48). We note that Siamois overexpression tended to generate more robust RPKM inductions than Wnt8 or LiCl, presumably because this transcription factor reached higher concentration levels after direct injection (Dataset S1), even though the dorsalized embryonic phenotypes were comparable (Fig. 1E). Microinjection of Cerberus mRNA repressed genes that require endoderm and mesoderm formation and increased neural genes within the early dorsal β-catenin signature. The Cerberus data also defined global gene signatures repressed and induced by this mRNA that may help decipher the physiological role of this growth factor antagonist.
We also examined the timing of the activation of the early β-catenin signature. By comparing the transcriptomes of Spemann organizer activating or inhibiting treatments at stage 9 (blastula) versus 10.5 (about 3.5 h later), we confirmed that, whereas all genes within the dorsal β-catenin signature are activated by stage 10.5, a small cluster of these genes were expressed at higher levels at stage 9 (Fig. 6). It is known that in Xenopus β-catenin bound to TCF-3 plays an important role in recruiting epigenetic regulators even before zygotic transcription (49). Nuclear β-catenin recruits Prmt2 (a protein-histone arginine methyltransferase) that causes an epigenetic mark called H3R8Me2 (dimethylation of histone H3 at Arginine 8), which deposits an epigenetic memory that poises promoters for transcription up to six cell cycles before Wnt target genes are first transcribed (49). The maternal β-catenin/Wnt signal, followed by zygotic Siamois and Nodal signaling, provides the initial step in the process that gives rise to the formation of the Spemann organizer in the dorsal marginal zone at the gastrula stage (11, 23). Spemann organizer tissue then orchestrates dorsal ventral patterning by secreting a mixture of growth factor antagonists (24).
Comparison of the Early and Late Wnt Signatures.
The cellular responses to Wnt ligands in early development are stage-dependent, which represents a difficulty when trying to assign a general transcriptional signature for the pathway. There are two distinct waves of Wnt signaling in Xenopus early development. Maternal Wnt/β-catenin signaling drives the formation of the Spemann organizer in dorsal regions whereas zygotic Wnt8, expressed at mid- to late-gastrula in ventro-lateral mesoderm, promotes the development of the posterior region of the embryo.
Recently, two studies have defined the late zygotic Wnt8 gene set using a combination of RNA-seq and ChIP-sEq (3, 4). Kjolby and Harland (3) used the Wnt inhibitor Dkk1 to define the transcriptome activated by Wnt signal at mid- to late-gastrula in X. laevis. A total of 82 genes constituted their late Wnt signature. Nakamura et al. (4) used X. tropicalis to screen for target genes repressed by a morpholino specifically targeting Wnt8. As controls, they rescued these effects with Wnt8 DNA and also selected only those genes bound by endogenous β-catenin at TCF/LEF sites. These more stringent criteria identified fewer genes (4), but all were included in the late signature from the other laboratory (3). Strikingly, it was found that β-catenin bound to 5,193 genes at gastrula, most of which are not regulated by Wnt at this stage; β-catenin recruitment does not imply transcriptional regulation, which must depend on additional developmental contexts (4).
That the early and late Wnt signatures would differ was expected (11). However, the extreme degree of divergence we found was surprising. We identified only 3 genes (Irx, Myf5, and Sebox) that overlapped between our early β-catenin signature of 123 genes and the late Wnt signature of 82 genes (3) (Fig. 2B). It is currently unknown what provides this “switch” from maternal to late β-catenin/Wnt signaling although epigenetic mechanisms or stage-specific TCF proteins may be involved (49, 50). It has been proposed that β-catenin recruitment may occur during early embryonic stages to DNA regulatory elements responsible for Wnt-regulated expression in other tissues at much later stages (4). In this regard, cross-talk between signaling pathways such as BMP, Nodal, and FGF may be important. In addition, as yet unknown lineage master regulators, as occurs with the recruitment of β-catenin in hematopoietic progenitor cells (51), might be involved in triggering differential transcriptional programs. It is generally accepted that the main role of β-catenin at gastrula is to signal dorsally and establish the Spemann organizer. However, in the present study it was found that many other genes require β-catenin for transcription but are not dorsally enriched, perhaps indicating as yet unknown transcriptional roles for β-catenin in early development. Studies are needed to investigate the effects of β-catenin/Wnt signaling at later embryonic stages: for example, to examine the deployment of the Hox complexes that are not yet active at the stages studied here.
Conclusion
This work presents an early dorsal β-catenin signature of 123 genes and provides the complete transcriptional landscape of treatments that massively stimulate or inhibit formation of the Spemann organizer in X. laevis. The genome-wide transcriptomic signatures associated with β-catenin loss of function, with dorsal and ventral half-embryos, and with Wnt8, LiCl, Siamois, and Cerberus phenotypes and the specific differences and similarities among them provide a rich substratum for investigations into the self-organizing nature of the earliest gene regulation events during development of a vertebrate embryo.
Materials and Methods
Embryo Manipulations, mRNA Injections, and Whole-Mount in Situ Hybridization.
X. laevis were purchased from Nasco. Embryos were generated through in vitro fertilization and cultured in 0.1× Marc's modified Ringer’s (MMR) and staged according to Nieuwkoop and Faber (52). A total of 24 ng of morpholino oligonucleotide against Xenopus β-catenin (18) was injected four times marginally at two-cell stage. For in vitro mRNA synthesis, pCS2-xWnt8, pCS2-xCerberus, and pCS2-xSiamois were linearized with NotI and transcribed with SP6 RNA polymerase using the Ambion mMessage mMachine kit. Synthetic mRNAs were injected into the marginal zone of each blastomere at the four-cell stage as follows: 3 pg of xWnt8; 100 pg of xCerberus; and 20 pg of xSiamois. All of the injected embryos were cultured until stage 9 or 10.5. For LiCl treatment, 32-cell stage embryos were treated with 0.3 M LiCl in 0.1× MMR for 7 min, washed three times with 0.1× MMR, and cultured until stage 9 or 10.5. For dorsal and ventral half-embryos, regularly cleaving stage 8 midblastula embryos were bisected into dorsal and ventral halves (44) and cultured until gastrula stage 10.5. For hybridization probe synthesis, pCS2-xWnt8 was linearized with BamH1 and transcribed with T3 RNA polymerase; pCS2-Xbra with Sal1 and transcribed with SP6 RNA polymerase; pCS2-xSox17β with Apa1 and transcribed with SP6 RNA polymerase. Whole-mount in situ hybridizations were performed as described at www.hhmi.ucla.edu/derobertis.
cDNA Library Preparation, RNA Sequencing, and Data Analysis.
Total RNA was isolated from two whole embryos or six half-embryos (dorsal or ventral) with the Absolutely RNA Miniprep Kit (Agilent). Libraries were constructed with the Illumina TruSeq RNA Library Preparation Kit V2 according to the manufacturer's protocol. One microgram of total RNA was used as input RNA to make each library, each of which was sequenced on an Illumina Hi-SEq. 2000 using standard methods to generate 100-bp single-end reads at the Broad Stem Cell Research Center at the University of California, Los Angeles (UCLA). RNA sequencing data were processed as described previously (2, 53, 54). All RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession No. GSE93195). Gene set enrichment analysis (GSEA) was performed using the GSEA software from the Broad Institute (software.broadinstitute.org/gsea/index.jsp) (29, 55). Statistical significance was measured with a permutation based Kolmogorov–Smirnoff nonparametric rank test (1000 permutations). For Weighted Gene Coexpression Network Analysis (WGCNA) (28), a gene coexpression network was constructed using the 2,000 transcripts that showed greatest variations upon microinjection of β-CatMO and their expression values throughout the 29 libraries sequenced at stage 10.5 (Dataset S1). To find modules of highly correlated genes, average linkage hierarchical clustering was performed, and modules were assigned by color codes.
Heat Maps.
Heat maps were generated in Multiexperiment Viewer (MeV) (56). For Fig. 6, RPKMs were used as inputs, and rows/genes were normalized using the mean and SD of the row to which the value belongs, using the following formula: Value = [(Value) – Mean(Row)]/[SD(Row)]; hierarchical clustering using Manhattan distance was performed. The heat map in Fig. 5 used fold change as input and was similarly normalized.
Mean Log Ratios-Average Plot and PCA Analysis.
Mean log ratios-average plots (MA-plots) were generated in R. For Fig. S1A, the log2 fold change (FC) in expression of transcripts between control embryos corresponding to β-CatMO embryos was plotted versus the average RPKM expression of transcripts in control embryos. In Fig. 4A, the log2 FC in expression of transcripts between Cerberus embryos to control embryos was plotted against the average RPKM levels of expression in control embryos. Transcripts with an average RPKM below 1 across all conditions were eliminated. Principal component analysis (PCA) in Fig. S4 was generated in R by comparing log2 FC in all libraries for transcripts identified in our early β-catenin signature.
Scatter and Box Plots.
The scatter plot matrix in Fig. 1H contains 40,157 transcripts from dorsal and ventral half RNA-seq libraries. The log2 FC in average expression of transcripts between dorsal and ventral half-embryos (five experiments) was plotted versus the log2 FC of control embryos compared with β-CatMO embryos (three experiments). Transcripts highlighted in red are those with fold changes higher than 1.5 and 1.4 in the dorsal/ventral (D/V) and con/β-CatMO conditions, respectively. Box plots in Fig. S1 and Fig. 3 were generated in R. The middle line in the box indicates the median, the box edges indicate the 25th/75th percentiles, and the whiskers indicate min and max values. The statistical significance of differences in gene expression levels between pairwise sets of genes was tested using the Mann–Whitney test and indicated as follows: *≤0.05, **≤0.01, and ***≤0.005.
Cloning and qRT-PCR.
To clone full-length Loc100170590, forward and reverse PCR primers were designed according to the genomic sequence deposited in the Xenbase database (www.xenbase.org/entry/). The oligos also contained upstream sequences for Gateway-mediated cloning. PCR was performed on cDNA obtained from gastrula stage X. laevis embryos, resulting in an amplification product migrating at the expected size (about 1.5 kb). The PCR product was purified, cloned in a pDonr221 vector and subsequently in a home-made Gateway-compatible pCS2 vector suitable for antisense probe and in vitro mRNA synthesis. Primers for cloning were as follows: Fwd, GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGTGTTCCGATGGGCCCTGTC; Rev, GGGGACCACTTTGTACAAGAAAGCTGGGTCATGAAGCTTATTCAACCCTTTTTGAC. Primers for qRT-PCR were as follows: Fwd, GCCCTTGAGTCTTCTTAT; Rev, CAGACATGAGAGAGATGG.
Supplementary Material
Acknowledgments
We thank Prof. Guoping Fan (UCLA) for suggestions in library preparation; and members of the E.M.D.R. laboratory for comments on the manuscript. We thank Dr. Pengpeng Liu for bioinformatics instruction. E.A.S. was supported by the MARC program (NIH T34 GM008563-21) and M.D.J.B was supported by NIH Training Grant R25 GM055052. This work was supported by the Norman Sprague Endowment and the Howard Hughes Medical Institute, of which E.M.D.R. is an investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE93195).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700766114/-/DCSupplemental.
References
- 1.Session AM, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538(7625):336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding Y, et al. Genome-wide analysis of dorsal and ventral transcriptomes of the Xenopus laevis gastrula. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kjolby RA, Harland RM. Genome-wide identification of Wnt/β-catenin transcriptional targets during Xenopus gastrulation. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura Y, de Paiva Alves E, Veenstra GJ, Hoppler S. Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules. Development. 2016;143(11):1914–1925. doi: 10.1242/dev.131664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christian JL, Moon RT. Interactions between Xwnt-8 and Spemann organizer signaling pathways generate dorsoventral pattern in the embryonic mesoderm of Xenopus. Genes Dev. 1993;7(1):13–28. doi: 10.1101/gad.7.1.13. [DOI] [PubMed] [Google Scholar]
- 6.Glinka A, et al. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391(6665):357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 7.Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell. 1997;88(6):747–756. doi: 10.1016/s0092-8674(00)81921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahon AP, Moon RT. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell. 1989;58(6):1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 9.Smith WC, Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67(4):753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- 10.Kessler DS. Siamois is required for formation of Spemann’s organizer. Proc Natl Acad Sci USA. 1997;94(24):13017–13022. doi: 10.1073/pnas.94.24.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hikasa H, Sokol SY. Wnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol. 2013;5(1):a007955. doi: 10.1101/cshperspect.a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao KR, Elinson RP. The entire mesodermal mantle behaves as Spemann’s organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol. 1988;127(1):64–77. doi: 10.1016/0012-1606(88)90189-3. [DOI] [PubMed] [Google Scholar]
- 13.Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93(16):8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Loh KM, van Amerongen R, Nusse R. Generating cellular diversity and spatial form: Wnt signaling and the evolution of multicellular animals. Dev Cell. 2016;38(6):643–655. doi: 10.1016/j.devcel.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Schneider S, Steinbeisser H, Warga RM, Hausen P. Beta-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57(2):191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 17.Larabell CA, et al. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136(5):1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: A novel antisense approach. Dev Biol. 2000;222(1):124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- 19.Rowning BA, et al. Microtubule-mediated transport of organelles and localization of beta-catenin to the future dorsal side of Xenopus eggs. Proc Natl Acad Sci USA. 1997;94(4):1224–1229. doi: 10.1073/pnas.94.4.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JR, Moon RT. Analysis of the signaling activities of localization mutants of beta-catenin during axis specification in Xenopus. J Cell Biol. 1997;139(1):229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taelman VF, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143(7):1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobrowolski R, De Robertis EM. Endocytic control of growth factor signalling: Multivesicular bodies as signalling organelles. Nat Rev Mol Cell Biol. 2011;13(1):53–60. doi: 10.1038/nrm3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agius E, Oelgeschläger M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127(6):1173–1183. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niehrs C. Regionally specific induction by the Spemann-Mangold organizer. Nat Rev Genet. 2004;5(6):425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- 26.Plouhinec JL, Zakin L, Moriyama Y, De Robertis EM. Chordin forms a self-organizing morphogen gradient in the extracellular space between ectoderm and mesoderm in the Xenopus embryo. Proc Natl Acad Sci USA. 2013;110(51):20372–20379. doi: 10.1073/pnas.1319745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu WT, et al. Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program. Development. 2014;141(23):4537–4547. doi: 10.1242/dev.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature. 1996;382(6592):595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 31.Piccolo S, et al. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397(6721):707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKendry R, Hsu SC, Harland RM, Grosschedl R. LEF-1/TCF proteins mediate wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev Biol. 1997;192(2):420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 33.Labbé E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci USA. 2000;97(15):8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid CD, Zhang Y, Sheets MD, Kessler DS. Transcriptional integration of Wnt and Nodal pathways in establishment of the Spemann organizer. Dev Biol. 2012;368(2):231–241. doi: 10.1016/j.ydbio.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weaver C, Kimelman D. Move it or lose it: Axis specification in Xenopus. Development. 2004;131(15):3491–3499. doi: 10.1242/dev.01284. [DOI] [PubMed] [Google Scholar]
- 36.Nieuwkoop PD. Origin and establishment of embryonic polar axes in amphibian development. Curr Top Dev Biol. 1977;11:115–132. doi: 10.1016/s0070-2153(08)60744-9. [DOI] [PubMed] [Google Scholar]
- 37.Miller JR, et al. Establishment of the dorsal-ventral axis in Xenopus embryos coincides with the dorsal enrichment of dishevelled that is dependent on cortical rotation. J Cell Biol. 1999;146(2):427–437. doi: 10.1083/jcb.146.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao Q, et al. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120(6):857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 39.De Domenico E, Owens ND, Grant IM, Gomes-Faria R, Gilchrist MJ. Molecular asymmetry in the 8-cell stage Xenopus tropicalis embryo described by single blastomere transcript sequencing. Dev Biol. 2015;408(2):252–268. doi: 10.1016/j.ydbio.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flachsova M, Sindelka R, Kubista M. Single blastomere expression profiling of Xenopus laevis embryos of 8 to 32-cells reveals developmental asymmetry. Sci Rep. 2013;3:2278. doi: 10.1038/srep02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hufton AL, Vinayagam A, Suhai S, Baker JC. Genomic analysis of Xenopus organizer function. BMC Dev Biol. 2006;6:27. doi: 10.1186/1471-213X-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faunes F, et al. Identification of novel transcripts with differential dorso-ventral expression in Xenopus gastrula using serial analysis of gene expression. Genome Biol. 2009;10(2):R15. doi: 10.1186/gb-2009-10-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popov IK, et al. Identification of new regulators of embryonic patterning and morphogenesis in Xenopus gastrulae by RNA sequencing. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoppler S, Moon RT. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech Dev. 1998;71(1-2):119–129. doi: 10.1016/s0925-4773(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 45.Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123(6):1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81(1):85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 47.Ishibashi H, et al. Expression of Siamois and Twin in the blastula Chordin/Noggin signaling center is required for brain formation in Xenopus laevis embryos. Mech Dev. 2008;125(1-2):58–66. doi: 10.1016/j.mod.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haramoto Y, et al. Identification and comparative analyses of Siamois cluster genes in Xenopus laevis and tropicalis. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 49.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19(2):220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roël G, et al. Lef-1 and Tcf-3 transcription factors mediate tissue-specific Wnt signaling during Xenopus development. Curr Biol. 2002;12(22):1941–1945. doi: 10.1016/s0960-9822(02)01280-0. [DOI] [PubMed] [Google Scholar]
- 51.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147(3):577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieuwkoop PD, Faber J. 1967. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis (Garland Publishing Inc., New York) (republished in 1994)
- 53.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 56.Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27(22):3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.