Significance
In mammals, AMPA-type glutamate receptors (GluAs) are expressed ubiquitously in the central nervous system and play critical roles in synaptic plasticity, learning, and memory. Here we examined GluAs in the ascidian, Ciona intestinalis, and determined that they are expressed in a limited subset of cells during early development. We further find that GluAs are required for development of the ocellus, a photoreceptive organ used during the swimming stage, and for tail resorption and body axis rotation during metamorphosis. These functions require ion influx through GluAs. This is a demonstration of an in vivo requirement for GluAs in organ formation and morphogenesis. GluAs are also expressed during mammalian development, suggesting that developmental roles of GluAs may be functionally conserved.
Keywords: AMPA-type glutamate receptor, morphogeneis, ascidian, development
Abstract
AMPA-type glutamate receptors (GluAs) mediate fast excitatory transmission in the vertebrate central nervous system (CNS), and their function has been extensively studied in the mature mammalian brain. However, GluA expression begins very early in developing embryos, suggesting that they may also have unidentified developmental roles. Here, we identify developmental roles for GluAs in the ascidian Ciona intestinalis. Mammals express Ca2+-permeable GluAs (Ca-P GluAs) and Ca2+-impermeable GluAs (Ca-I GluAs) by combining subunits derived from four genes. In contrast, ascidians have a single gluA gene. Taking advantage of the simple genomic GluA organization in ascidians, we knocked down (KD) GluAs in Ciona and observed severe impairments in formation of the ocellus, a photoreceptive organ used during the swimming stage, and in resorption of the tail and body axis rotation during metamorphosis to the adult stage. These defects could be rescued by injection of KD-resistant GluAs. GluA KD phenotypes could also be reproduced by expressing a GluA mutant that dominantly inhibits glutamate-evoked currents. These results suggest that, in addition to their role in synaptic communication in mature animals, GluAs also have critical developmental functions.
Glutamate has been proposed to regulate neural development via nonsynaptic mechanisms, either by directly activating ionotropic and/or metabotropic glutamate receptors expressed on neural progenitors or indirectly by inducing neighboring cells to secrete molecules regulating neurogenesis, such as neurotropic factors (1–3). The early onset of AMPA-type glutamate receptor (GluA) expression in developing embryos also raises the possibility of other currently unidentified roles. For example, mouse gluA transcripts are detectable by embryonic day 10, and in zebrafish, zygotic gluA transcripts are expressed as early as the midblastula stage (4–10). These observations, combined with in vitro results, suggest a role for GluAs in neural cell proliferation (11, 12). In vertebrates, GluAs form tetramers consisting of various combinations of four subunits (GluA1–GluA4) (13–15). The presence of four gluA genes has prevented efficient generation of mice with all four genes deleted. Therefore, the precise role of GluAs in early development of neural and nonneural tissue has not yet been determined in vivo.
Here, we instead used the ascidian chordate Ciona intestinalis, the closest living relative to vertebrates (16), which is thought to have just a single gluA gene (17). Ciona larvae are transparent, enabling easy identification of cells expressing a target gene and detection of morphological phenotypes in the tissues and organs. The Ciona life cycle consists of two main phases: a short larval phase with a vertebrate-like notochord, which lasts until about 27 h postfertilization (hpf), and a nonmotile adult phase, which occurs after metamorphosis and lasts about 3–6 mo. After rapid embryogenesis, hatched larvae are able to swim for a few hours (17.5–24 hpf) (18, 19). During this stage, Ciona have a photoreceptive organ, the ocellus, which allows them to respond to light. Decreasing light induces swimming, whereas increasing light causes larva to stop swimming and settle onto substrates (20, 21). During the larval period, larvae prepare for the onset of metamorphosis, which usually begins with adhesion (24–27 hpf) (18, 19), followed, in turn, by tail regression into the trunk region (27–29 hpf), body-axis rotation (30–60 hpf), and the formation of adult organs including gill slits, the endostyle, and the digestive tract (22).
In this study, taking advantage of the simple genomic organization of ascidians, we provided convincing evidence that GluAs are essential for normal body development in vivo, including the formation of sensory organs and metamorphosis. We also indicated a critical role for ionotropic glutamate receptors in early development requiring ion flux through GluAs.
Results
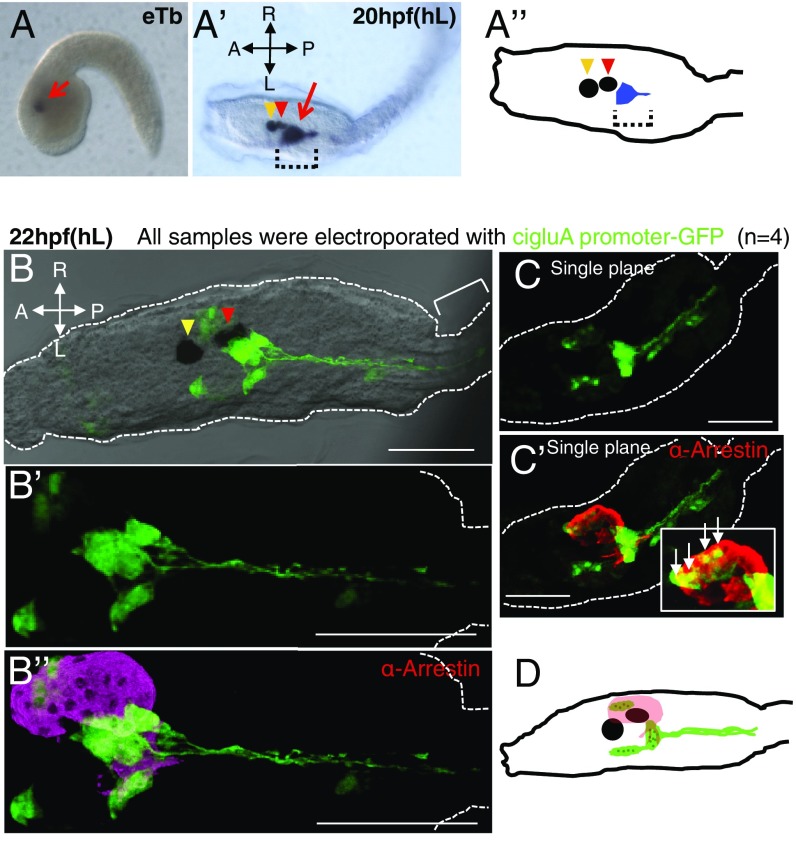
Analysis of the Ciona genome database and molecular cloning confirmed that Ciona has just a single GluA subunit with two splice variants at the C terminus (Fig. S1). Notably, clear zygotic expression of cigluA and the glutamate aspartate transporter (glast) starts at the neurula stage (7 hpf), whereas the expression of the vesicular glutamate transporter (vGlut) starts at the mTb (midtailbud) (11 hpf) stage. The neurula stage is when neuronal differentiation occurs. Compared with GluAs from other invertebrate species, the ciGluA sequence has the highest homology to mouse GluAs (Fig. 1B). In situ hybridization revealed ciGluA transcripts located in a restricted region behind the ocellus from the early tailbud stage and the hatching larval stage (Fig. 2 A–A”, arrow and brackets). To further characterize the identity and morphology of ciGluA-expressing cells, we created a reporter plasmid expressing GFP under the control of the cigluA promoter (cigluAp−GFP) (Fig. 1 C–C”’). Electroporation of cigluAp-GFP into Ciona fertilized eggs resulted in expression of GFP in specific cell populations. In vertebrates, GluA expression is observed throughout the CNS (23–25). Intriguingly, among the ∼100 neurons present in the CNS of Ciona (26), only about 10 were GFP-positive (Fig. 2 B–D). GFP signals showed patterns similar to gluA mRNA expression patterns in hatched larvae. The morphology of GFP-positive cells behind the ocellus indicated that they are neurons, with axons extending toward the motor ganglion (located between the head/trunk and tail) (Fig. 2 B–D). A few other GFP-positive cells colocalized with arrestin-positive photoreceptor cells in the ocellus (Fig. 2 C, C’, and D). Arrestin is a key protein for termination of the rhodopsin active state (27).
Fig. S1.
GluA amino acid sequence alignment (related to Fig. 1). (A) Green highlighting indicates the degree of conservation of amino acid residues between species (darker green, more conserved). The horizontal red arrows indicate the predicted and conserved transmembrane domains (TMDs). The horizontal blue arrows show the predicted and conserved glutamate binding regions (74, 75). The vertical red arrow indicates the residue subject to Q/R editing. Cin, C. intestinalis; Mmu, M. musculus. (B) Splice variants of the Ciona C-terminal sequence. A dash (-) indicates a gap.
Fig. 1.
Characterization of Ciona GluA. (A) Quantification of CiGluA, ciGLAST, and ci vGlut transcripts at the indicated developmental stage measured by quantitative real-time RT PCR. Equal amounts of total RNA were used for reverse transcription. More than 100 embryos were used for the analysis at each developmental stage. hpf, hours postfertilization; mTb, midtailbud stage. Data are expressed as the average of three wells plus the SE. (B) A maximum-likelihood tree of the various GluA proteins (cel, C. elegans; cin, C. intestinalis; dme, D. melanogaster; mmu, Mus musculus). The red arrow marks the position of ciGluA protein. Scale indicates 0.10 substitutions per position. (C) Schematic diagram of the cigluA gene. (C’–C’’’) Constructs used in this study. (C’) Schematic of the cigluA-promoter-GFP (cigluA-GFP) reporter construct. (C’’) Schematic of the ciGluA expression construct (cigluAp-GluA). (C’’’) Schematic of the MO-resistant ciGluA expression construct (cigluA-GluA*). Red bars indicate the MO target site that is disrupted in cigluA-GFP reporter construct (C’). The cross represents three silent mutations incorporated in the MO-resistant construct.
Fig. 2.
CiGluA is expressed in only a subset of neurons in the CNS. (A and A’) ciGluA expression analyzed using in situ hybridization. eTb, early tailbud stage, about 8.45 hpf (n = 26); hL, hatched larva at about 20 hpf (n = 31). The red arrow in A and the dotted brackets in A’ and A’’ indicate cigluA mRNA expression, the yellow arrowheads in A’ and A’’ indicate the otolith, and the red arrowheads in A’ and A’’ indicate the ocellus. (A’’) A schematic diagram illustrating the spatial relationship between cigluA mRNA-expressing cells and the otolith and ocellus in hL. (B–C’) Hatched larvae were electroporated with cigluA-GFP. (B) 3D reconstruction images showing merged DIC and GFP staining. The yellow arrowhead indicates the otolith, and the red arrowhead indicates the ocellus. The bracket indicates part of the tail. (B’) Higher magnification image of the GFP staining pattern in B. (B’’) Images costained for GFP and arrestin (a mature photoreceptor marker). (C) An independent sample expressing GFP and (C’) costained for arrestin. The white box in C’ is a higher magnification image of the photoreceptor region with arrows indicating cells that coexpress GFP and arrestin. (D) Schematic illustrating the spatial relationship between arrestin-positive photoreceptor cells (pink) and ciGluA-promoter–induced GFP-positive cells (green) in hL. The black circle and ellipse represent the otolith and the ocellus, and the dark green dots within the light green areas indicate cell nuclei. (Scale bars, 50 μm.)
Fig. S2.
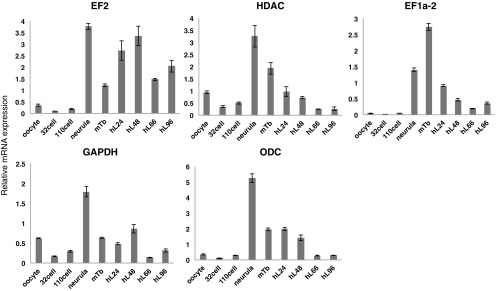
Relative transcript amounts for representative internal controls in Ciona (related to Fig. 1). Shown are CiEF2, ciHDAC, ciEF1a-2, ciGAPDH, and ciODC transcripts at each developmental stage measured with quantitative real-time PCR. The same amount of total RNA was used for reverse transcription. More than 100 embryos were used for the analysis at each developmental stage. Hpf, hours postfertilization; mTb, midtailbud stage. Data are expressed as average of three wells plus SE. EF1 or 2, elongation factor 1 or 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDAC, histone deacetylase; ODC, ornithine decarboxylase.
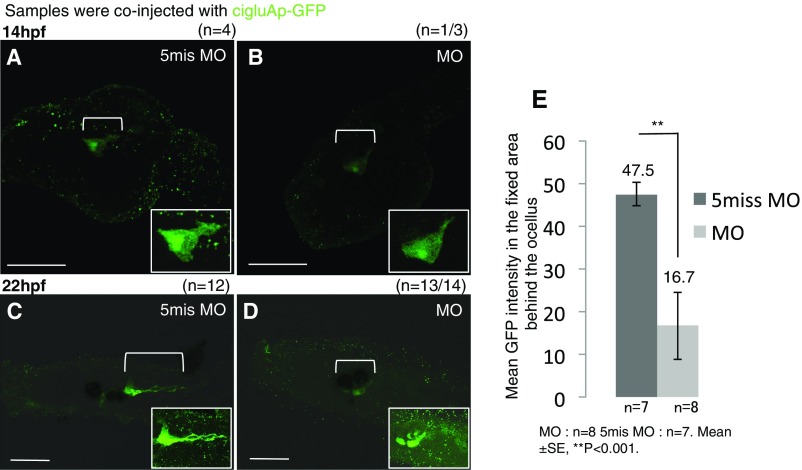
To explore the function of GluAs during development, we knocked down (KD) ciGluA expression using a morpholino oligonucleotide (MO), a stable oligonucleotide that inhibits mRNA translation starting from the fertilized egg stage. No morphological differences in ciGluA-expressing cells were observed at 14 hpf (Fig. S3 A and B). However, 8 h later (22 hpf), axonal extension from ciGluA-expressing cells had been arrested (Fig. S3 C and D), and arrestin-positive photoreceptor cells were absent from the KD larvae (Fig. 3 A and B). The larval ascidian ocellus consists of melanin-containing pigment cells and vertebrate-type photoreceptor cells (28, 29). Loss of ciGluAs did not influence the development of pigment cells (Fig. 3 A and B). We also measured intact GFP intensity before immunostaining in 20 hpf larvae and detected about 2.8-fold weaker intensity in MO-injected larvae than in control larvae (Fig. S3E).
Fig. S3.
GluA knockdown leads to abnormal differentiation of GluA-expressing cells (related to Figs. 2 and 3). (A and C) Samples injected with control 5mis MO. (B and D) MO against ciGluA. (A and B) Late tailbud stage larval images with GFP immunostaining. (C and D) Hatched larval DIC images merged with GFP immunostaining. The white boxes in A–D are higher magnification images of the bracket regions. (Scale bars, 50 μm.) (E) Mean intact GFP intensity in the bracketed region measured by ImageJ at 20 hpf. Mean ± SE, **P < 0.001.
Fig. 3.
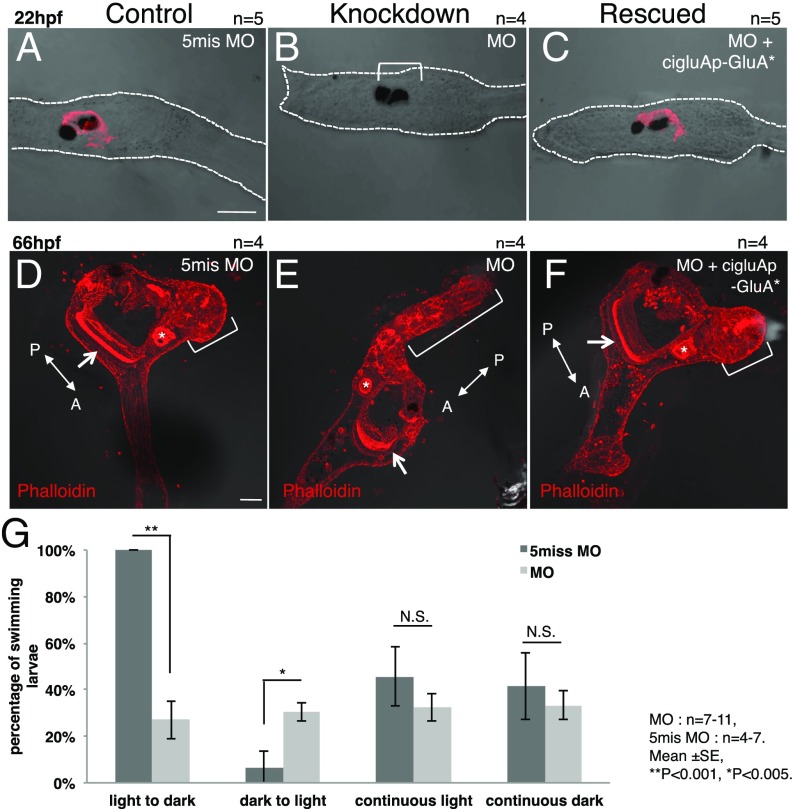
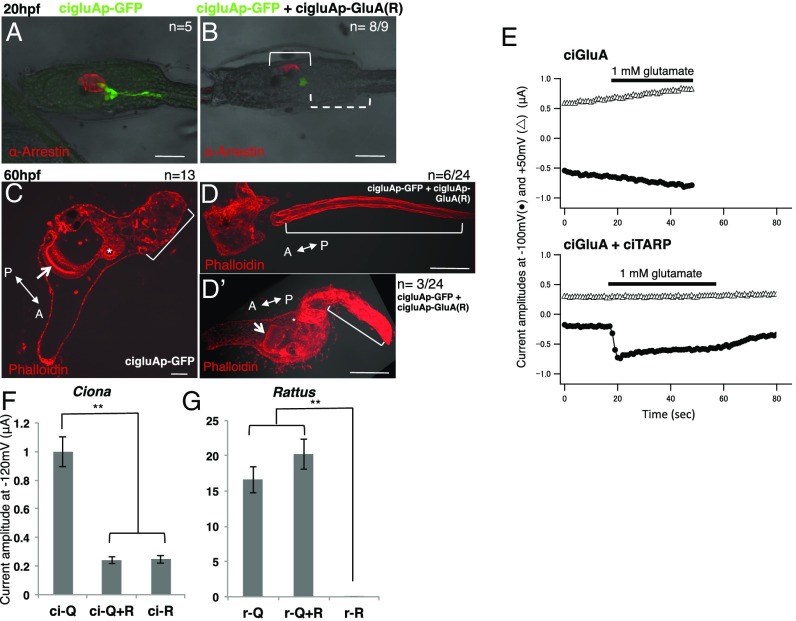
GluA knockdown leads to defects in sensory-organ differentiation and metamorphosis. (A and D) Samples injected with control 5mis MO. (B and E) MO against ciGluA. (C and F) MO against ciGluA rescued with the MO-resistant ciGluA construct. (A–C) Hatched larval DIC images merged with arrestin immunostaining. The arrestin signal was almost completely absent in MO-injected larvae (bracket in B). (D–F) Juvenile images showing merged DIC and phalloidin staining (F-actin). The arrow and asterisk in D–F indicate the endostyle and digestive tract, respectively, showing that internal organs developed normally. The bracket in E indicates the tail that failed to shrink. The brackets in D and F indicate normal, shrunken tails. [Scale bars, 50 μm (A and D).] (G) Percentage of larvae that swam in indicated light conditions. Mean ± SE, **P < 0.001, *P < 0.005.
At 66 hpf, the process of metamorphosis was severely impaired, resulting in tails that failed to shrink and defective body-axis rotation (Fig. 3 D and E). These developmental defects were completely rescued by injecting a MO-resistant ciGluA construct, suggesting a specific, rather than off-target, action of the MO (Fig. 3 C and F). To confirm that disappearance of the photoreceptor marker, arrestin, was accompanied by functional defects, we evaluated shadow-response behavior, which is active swimming caused by an acute loss of light (20, 21). ciGluA KD larvae showed reduced swimming behavior in response to a dark stimulus (Fig. 3G). We have demonstrated that GluAs are crucial for normal differentiation of GluA-expressing cells in vivo. Moreover, this is evidence for the indispensability of GluAs in organ developmental and behavioral phenotypes, including the formation of sensory organs, sensory-guided swimming behavior, and metamorphosis.
We used the Xenopus oocyte expression system to confirm the physiological properties of ciGluAs. CiGluAs did not induce clear inward currents triggered by glutamate (Fig. 4E, upper traces). This is not unexpected, as Caenorhabditis elegans and Drosophila melanogaster GluAs require coinjection of transmembrane GluA regulatory proteins (TARPs) to achieve a glutamate response (30–32). TARPs facilitate the membrane targeting and channel function of GluAs (33, 34). Therefore, we isolated the cDNA for Ciona TARP (ciTARP) (Fig. S4) and coexpressed it with ciGluA in Xenopus oocytes (Fig. 4E, lower traces). The amplitude of glutamate-evoked responses was relatively small, but this may be explained either by the requirement of additional auxiliary proteins or by the intrinsic ion permeability of the ciGluAs (Fig. 4 F and G, left column).
Fig. 4.
Artificial Ciona R-type GluAs dominantly inhibit activity of Q-type GluAs. (A and B) DIC images of hatched larvae merged with GFP and arrestin staining patterns. In B, the dotted bracket indicates GFP-positive cells with defective axons and the solid bracket indicates partial loss of arrestin-positive photoreceptor cells in an R-type ciGluA electroporated larva. (C–D’) DIC images merged with F-actin labeling with phalloidin in juveniles. The arrow and asterisk in C and D’ indicate the endostyle and digestive tract, respectively, showing that the internal organs developed normally. The bracket in D indicates a cut tail. The bracket in D’ indicates a tail that failed to shrink. The bracket in C indicates a normal, shrunken tail. In control samples, all samples (n = 13) showed normal regression. In the R-type ciGluA electroplated samples (n = 22), 32% of samples were normal (n = 7), and 68% (n = 15) showed abnormal tail regression. Among these, seven showed impaired regression, and eight showed no regression. Seven of the eight larvae with no regression had cut tails. [Scale bars, 50 μm (A–C) and 100 μm (D and D’).] (E) Current flow induced by glutamate application in Xenopus oocytes voltage clamped at –100 mV (filled circles) and +50 mV (open triangles). Upper plots currents in oocytes injected with cRNA for ciGluA alone, and Lower plots currents in oocytes injected with both cRNA for ciGluA and ciTARP. Black bars indicate the timing of glutamate application. (F and G) The average peak current amplitude after glutamate application in Xenopus oocytes injected with only Q-type or R-type, or mixed subunit types (Q/R-hetero). All data were obtained in the absence of extracellular Ca2+, and TARP was coexpressed in all groups. r, rat. n = 6–11. Mean ± SE. **P < 0.001.
Fig. S4.
TARP amino acid sequence alignment (related to Fig. 4). Red highlighting indicates the degree of conservation of amino acid residues between species (dark red, conserved in all five species; red, conserved in 4/5 species; pink, conserved in 3/5 species). Black arrows indicate the boundaries of the predicted transmembrane domains (TMDs) of mouse stargazin. Cin, C. intestinalis; Cle, C. elegans; Dme, D. melanogaster; Mmu, M. musculus; Xla, X. laevis. Dashes (-) indicate gaps.
Mammalian GluAs can be divided into two classes based on of Ca2+ permeability. Ca2+ permeability depends on the substitution of a single amino acid residue in the ion channel pore from glutamine (Q) to arginine (R) by RNA editing (called Q/R editing) (1–3, 35–38). GluAs that include the R-type GluA2 subunit lack Ca2+ permeability (13, 39). In very early development, most mammalian GluAs are Ca2+-permeable GluAs (Ca-P GluAs), but the Ca2+-impermeable GluAs (Ca-I GluAs) population increases rapidly by birth, due to intensive Q/R RNA editing of the GluA2 subunit (40–44). Defects in this process cause serious problems (45–49). Decreased efficiency in Q/R editing is observed in, for example, patients with amyotrophic lateral sclerosis (ALS), leading to increasing Ca2+ influx and cell death in motor neurons (45, 46). Mice with genetic manipulations that inhibit Q/R conversion of GluA2 subunits die within a few weeks of birth from epilepsy and excess Ca2+ influx into neurons (49). Thus, the presence of R-type receptors is crucial for mammals to survive. To test whether similar Q/R editing and cooperation of Ca-P GluAs and Ca-I GluAs occur in ascidians, we first assessed the Q/R editing status in Ciona. No Q/R editing was detected at any of the three developmental stages we selected, suggesting that Ciona is likely to have only Q homomultimers (ci-Q/Q) (Fig. S5A). We next investigated the electrophysiological properties of ci-Q/Q using Xenopus oocytes, which have endogenous Ca2+-activated Cl− channels. Thus, Ca2+ permeability of GluAs can be monitored as a transient outward current through the Cl− channels (Materials and Methods for details). We demonstrated that ciGluAs are Ca2+-permeable with inward rectification (Fig. S5B). The ciGluA Ca2+-permeability index was almost the same as that for rat GluA (Fig. S5C). Thus, Ciona has only Ca-P GluAs throughout its life span because ciGluAs are the sole AMPA-type receptor in this species. Other invertebrates, such as nematodes and flies, have also been reported to have only Q-type GluAs (50). The requirement for Ca-I GluAs in mammals appears to be evolutionarily unique and may be related to the role of GluAs in synaptic transmission acquired during the evolution of vertebrates.
Fig. S5.
Functional characterization of the constructs used in this study (related to Fig. 4). (A) Amino acid sequence alignments for Ciona GluA, rat GluA1, and rat GluA 2, around the Q/R editing site (arrow). (B) Representative current recordings in response to glutamate application in the presence or absence of Ca2+. Bars at the top indicate the timing of application of glutamate and Ca2+. Current amplitudes were measured at the points before glutamate application (black arrows), at the current peak in 0 mM Ca2+ (blue arrows), and in 5 mM Ca2+ (red arrows). (C) The Ca2+ permeability index was calculated by normalizing [current increase (at +60 mV in 5 mM Ca2+)] by [current increase (at –120 mV in 0 mM Ca2+)]. This is to normalize for the GluA expression level. N.S., not significant; r, rat.
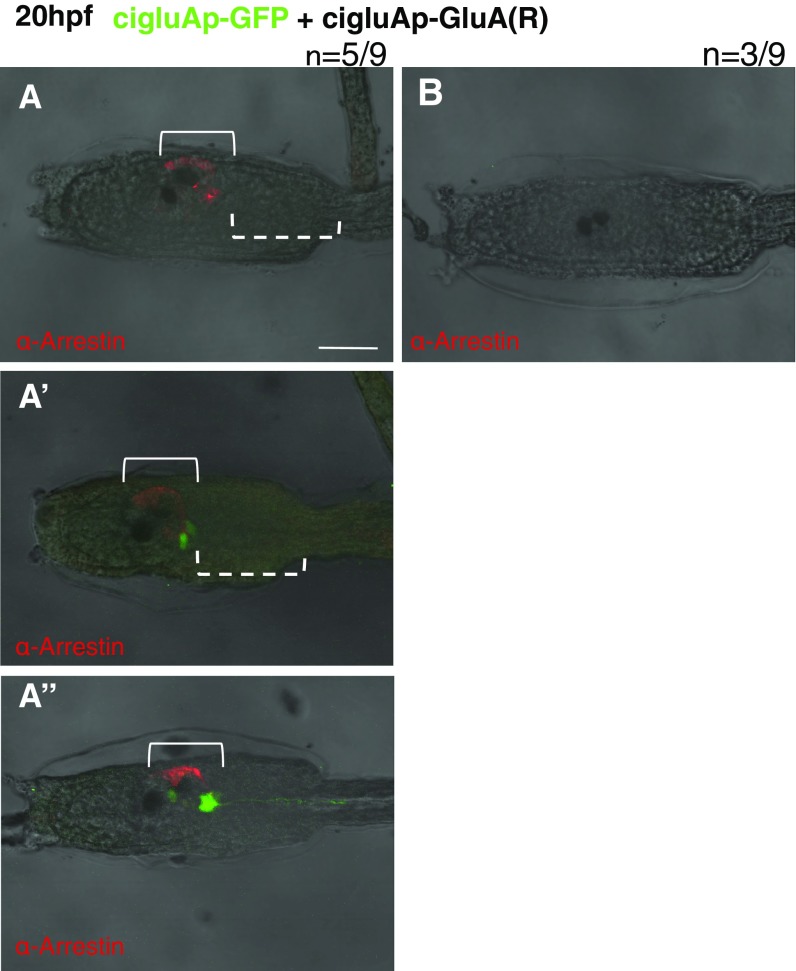
In vertebrates, a single amino acid substitution alters the calcium permeability of GluAs. Does an identical substitution artificially created in ciGluAs similarly alter the properties of the ion channel? To test this, we created a ciGluA construct that was artificially edited at the Q/R site and studied its electrophysiological properties. In the course of characterization of heteromeric AMPA receptors, we uncovered distinct properties of Ciona receptors compared with mammalian receptors. Unexpectedly, the current amplitude of Q- and R-hetero multimers (ci-Q/R) was significantly smaller than that of ci-Q/Q multimers and at a similar level to that of ci-R/R multimers (Fig. 4F). This is in clear contrast with mammals, where r-R/R is almost nonfunctional and the r-Q/R current is similar to that of r-Q/Q (Fig. 4G), as shown previously by Hollmann et al. (13). These results indicate that requirement of Ca2+ permeability of ciGluAs during development cannot be examined, whereas the artificial R-type ciGluA subunit functions as a dominant-negative subunit. This enabled us to discriminate between the possible functional and structural roles of GluAs during early development. Specifically, we electroporated an R-type (ci-R) expression construct into fertilized Ciona oocytes, which endogenously express only Q-type ciGluAs, expecting the dominant-negative effect of ci-R to result in suppression of GluA-mediated cation influx. Expression of ci-R under the cigluA promoter led to a phenotype similar to, but slightly milder than, that produced by ciGluA KD by MO. At 20 hpf, seven larvae out of the nine showed loss of axonal extension (Fig. S6). Five out of nine larvae electroporated with R-type GluAs had a partial loss of arrestin-positive cells, with three out of those nine larvae showing a loss of arrestin-positive cells resembling the loss in MO experiments (Fig. 4 A and B and Fig. S6). At 60 hpf, we found two types of defects in metamorphosis. One was similar to defects observed in MO-injected larvae, with defective tail regression but normally developed adult organs (Fig. 4D’), and the other consisted of larvae with cut tails and defective adult organs (Fig. 4D). In addition, body-axis rotation did not occur in ci-R electroporated larvae (Fig. 4 D and D’). The observed developmental defects from ci-R induction are caused by reduced ion influx through GluAs (Fig. 4F) and not by loss of GluA function as a signal transducer and/or adhesion molecule, which is independent of ion permeability. The fact that vertebrate-like single amino acid substitution of ciGluAs induced dramatic developmental phenotypes in ascidian larvae illustrates the indispensable role of ciGluAs as a glutamate-sensitive ion channel during early development.
Fig. S6.
Various phenotypes seen in R-type GluA electroporated larvae (related to Fig. 4). (A–A’’) Representative images of larvae with partial loss of arrestin-positive photoreceptors. The solid brackets in A–A’’ show that R-type ciGluA-electroporated larvae have photoreceptor cells with partial loss of arrestin-positive cells. The dotted brackets show the region where the GFP signal disappeared (A) and the GFP-positive cells without their axons (A’). (Scale bar, 50 μm.) (B) Representative image of an R-type GluA-electroporated larva lacking expression of GFP and arrestin.
Discussion
In this study, we uncovered a unique expression pattern of ciGluAs. In contrast to mammals where GluAs are expressed throughout the CNS (51), expression in Ciona is restricted to a small population of neural cells (Fig. 2 A–D). This suggests a possibility that GluAs may have originally functioned in development and later transitioned to become a predominant mechanism for synaptic neurotransmission. Consistent with this possibility, we demonstrate that ciGluAs are required for organ formation and dynamic body transformation. Vertebrates have two types of visual systems: lateral paired eyes and parietal or pineal eyes. The Ciona ocellus has been reported to be homologous with the pineal eye. First, the ocellus originates from the same part of the neural plate as the vertebrate pineal eye. Both are derived from the lateral part of the neural plate and have a final location in a dorsal part of the anterior brain (52, 53). Second, the ascidian ocellus serves as a photoreceptive organ, necessary for the shadow response (54, 55). Pineal eyes in larval amphibians have the same function (56). In our studies, lack of ciGluAs caused loss of arrestin immunoreactivity and a reduction in shadow-response behavior (Fig. 3 A, B, and G), which suggests that ciGluAs are required for pineal eye-like organ formation and function. Supporting this idea, GluA expression in the pineal body has been reported in macaques and in the developing rat (57, 58). Further experiments, for example, with specific marker antibodies, are needed to confirm whether the ascidian ocellus is indeed homologous with the pineal eye.
CiGluAs are expressed in a subpopulation of photoreceptor cells, and loss of GluA expression or activity results in reduced amounts of arrestin (Figs. 2 A–D and 3 A and B). How does GluA in a subpopulation affect all arrestin-positive cells? The loss of GluAs in photoreceptor cell progenitors in the early stages of development may prevent proliferation of these cells, leading to the absence or reduction of arrestin-positive cells. We observed zygotic expression of ciGluA transcripts at the neurula stage (Fig. 1A), whereas arrestin expression starts later, during the early tailbud stage (59). Weak GFP signals in GluA KD cells probably reflect the immaturity of those cells (Fig. S4 A–E). In rodents, functional GluAs are expressed in proliferative neural precursor cells in addition to mature neurons (60, 61). However, prior in vitro studies have not identified which cells expressing GluAs are required for progenitor differentiation (11, 12). In the present study, we demonstrated that GluAs are crucial for the development of GluA-expressing cells, as shown by the partial arrestin expression and arrested axonal extension of GluA-expressing cells in promoter-induced ci-R electroporated larvae (Fig. 4 A and B and Fig. S6). In the course of metamorphosis, only tail regression and body-axis rotation were inhibited by decreased functionality of ciGluAs in our experiments (Figs. 3 D and E and 4 C–D’). This suggests that the other metamorphosis steps are controlled by independent mechanisms and that only tail regression and body-axis rotation are on the same cascade. Several previous studies with mammalian cells in vitro have demonstrated that GluAs contribute to axonal development and cell morphology (62–65). It is possible that the axons that vanished with GluA KD or ci-R electroporation may transmit the signal to start tail regression and body-axis rotation.
Quantitative real-time PCR results (Fig. 1A) indicated that gluA is expressed at the neurula stage, whereas expression of vGlut, which is required for synaptic transmission, begins later at the mTb stage (11 hpf). Furthermore, GLASTs, which remove extracellular glutamate independently from synaptic function (66–68), are also expressed at the neurula stage. These data suggest that the functions of ciGluA during early development may be independent from GluA’s roles in synaptic transmission. Our identification of ascidian GluA suggests that a prototypical gluA gene encoded a calcium-permeable channel and may have been associated with calcium signaling involved during development (69, 70). This gene likely later evolved into the four different gluA genes (GluA1, GluA2, GluA3, and GluA4) present in mammals that function to depolarize cells through mainly sodium influx.
Materials and Methods
cigluAp-GFP and cigluAp-ciGluA Coding Sequence Construction and Mutagenesis.
Following dephosphorylation, a pSP eGFP vector (from Takeo Horie) was cut out by digestion with Pvu II. The ∼8 kbp amplified PCR fragment, including the cigluA regulatory sequence (RS) and the ATG start codon, was phosphorylated and cloned into the pSP vector above (termed “pSP cigluA RS vector”). The 8 kbp RS was amplified by PCR with the primers CGATCTTCGGTATCGTAAGAGATCA (Fwd), TTGAAATTTTACCTGGTGTGC (Rev), TTTTACTTCTTTTGTACTCCCGTA (Nested Fwd), and GCGGTGTGCAAAATCGTACT (Nested Rev).
To insert the GFP and cigluA coding sequences (CDSs) under the cigluA RS, restriction site-generating mutagenesis was performed at the start codon site of the RS vector, leading to inhibition of the transcription of exons contained within the 8 kbp RS sequence. The mutagenesis primers were gatcgtcgacCCGGTATGTCCACAGCATTC (Fwd) and gtcAggAtccCACTTGTTGTTACTGCATAA (Rev) (lowercase letters in the primer sequences indicate mismatched nucleotides introduced to generate the restriction site for XbaI).
After digestion with XbaI, blunting, and dephosphorylation, eGFP and cigluA CDS fragments were fused into the pSP cigluA RS sequence. For eGFP, we inserted the Pvu-II–digested eGFP fragments described above.
To make the MO-resistant construct, we used the following primers to generate silent mutations in the cigluA-RS–induced ciGluA CDS construct: GAATGTCTCGtaGgACTATAAAAATAG (Fwd) and CGAAATTGCATTTGAAACTAT (Rev). Construction images are presented in Figs. 1C and 2C”’.
Behavioral Test for Light Sensitivity.
We evaluated light-sensing behavior in fully matured swimming larvae (22 hpf, st.28 according to FABA2: chordate.bpni.bio.keio.ac.jp/faba2). To produce the dark and light stimuli, we alternately covered and uncovered the larvae with a black plastic sheet. Swimming behavior (movement of the tail from side to side) toward the dark stimulus was recorded with a digital camera (Olympus CAMEDIA SP-350) and scored manually.
Electrophysiology in Xenopus Oocytes.
After linearization with NheI (ciGluA and ciTARP in the pGEMHE vector) or XhoI [ratGluA2 in the pBlueScript II SK(–) vector], cRNAs were transcribed with a T8 RNA transcription kit (Invitrogen). Rat GluA2Q and 2R vectors were gifts from J. Boulter, University of California, Los Angeles. Xenopus laevis were anesthetized in water containing 0.15% tricaine, and oocytes were collected through a surgical procedure, as described previously (71). The isolated oocytes were treated with collagenase (type 1, 2 mg/mL; Sigma) for 6 h and injected with 50 nl of cRNA solution. The amounts of injected cRNA were as follows: ciGluA (25 ng) and ciTARP (0 ng or 25 ng) (Fig. 4E), Q [17 ng (Q-only, Q+R) or 0 ng (R-only)], R [0 ng (Q-only) or 17 ng (Q+R, R-only)], and ciTARP [17 ng (all)] (Fig. 4 F and G and Fig. S5 B and C). After incubation at 17 °C for 3–4 d in frog Ringer’s solution [88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.3 mM Ca(NO3)2, 0.41 mM CaCl2, and 0.82 mM MgSO4, pH 7.6, with 0.1% penicillin–streptomycin solution], oocytes were placed in a chamber of a volume of ∼160 μL. Macroscopic glutamate-induced currents were recorded under two-electrode voltage clamp using an OC-725C (Warner Instruments), Digidata 1322A, and pClamp 9.2 (Molecular Devices). Intracellular microelectrodes were filled with 3 M K-acetate and 10 mM KCl and had resistances between 0.2 MΩ and 0.5 MΩ in the standard bath solution, ND96 (98 mM NaCl, 2 mM KCl, 3 mM MgCl2, and 5 mM Hepes, pH 7.4). In the experiments with bath Ca2+ (Fig. S4 B and C), 5 mM CaCl was simply added to the ND96. Oocytes were clamped at −120 mV, and 40 μL of a 5× concentrated stock of glutamate solution was added directly to the bath (160 μL) by pipette. Washout of glutamate was achieved by bath perfusion.
Xenopus oocytes have endogenous Ca2+-activated Cl− channels (72–74). Thus, Ca2+ permeability of GluA can be monitored in oocytes with a high sensitivity. Upon addition of 5 mM Ca2+ to the bath, glutamate-evoked Ca2+ influx through Q-type ciGluAs activates the Cl− channels, especially at +60 mV, where Ca2+-induced Cl− current can be detected better (Fig. S5B). The data were analyzed offline in pClamp 9.2, Igor Pro (Wavematrix) and Excel (Microsoft). All animal experiments involving X. laevis in this study conformed to the guidelines of the Animal Care and Use Committee of the National Institute for Physiological Sciences (Okazaki, Japan) and were performed with the approval of the committee.
MO Injection.
To inhibit ciGluAs expression, we used the MO technique (Gene Tools). The MO antisense sequence was TAGTACGGCGAGACATTTTAACTGG, and the control MO sequence with five nucleotide mismatches was TAcTAgGGCGAcAgATTTTAACTcG. On the basis of previous studies, 10 fmoles MO in distilled water was injected into intact fertilized eggs (75).
Estimation of Tail Regression.
We estimate the regression of tail by the criteria using the ratio of tail length and trunk length; normal regression, 0–0.5; impairment regression, 0.5–2 (shown as a representative in Fig. 4D’); no regression, 2< (shown as a representative in Fig. 4D). Based on the criteria, in control samples, 100% of samples (n = 13) are normal regression. The ratio of 12 samples is 0 and only one sample is 0.1.
Sequence Alignment and Molecular Phylogeny.
We used CLC Main Workbench software for amino acid alignment. For molecular phylogenetic analysis, we used MEGA7. Amino acid sequences of the glutamate receptors were aligned by MUSCLE, and a maximum-likelihood tree with 100 replicates of bootstrap analysis was constructed from a dataset of gap-free 350 amino acid alignment covering the S1-TMD4 region.
Animals and handling, quantitative real-time PCR, cloning, Q/R editing status, immunohistochemistry, whole-mount in situ hybridization, accession numbers, and statistics were as described in SI Materials and Methods.
SI Materials and Methods
Animals and Handling.
C. intestinalis type A (Ciona robusta) was collected from the Maizuru and Misaki regions in Japan, and their eggs and sperm were handled as described previously (76). Embryos at the one-cell stage were electroporated with 40 μg of plasmid DNA as previously described (77). After MO injection and electroporation, the embryos were maintained in agar-coated dishes with Millipore-filtered seawater containing 50 μg/mL streptomycin sulfate at 18 °C. Embryos were fixed at room temperature (RT) for 30 min in 4% (wt/vol) paraformaldehyde dissolved in 0.5 M NaCl, 0.1 M Mops, pH 7.5. Fluorescence was observed with an FV1000 confocal microscope (Olympus). All MO injection and electroporation experiments were repeated independently at least three times.
Quantitative Real-Time PCR.
We obtained the total RNA from over 100 embryos at each developmental stage using RNeasy Mini kits, according to the manufacturer’s protocol (Qiagen). At each stage, 0.5 μg of total RNA was reverse transcribed with ReverTra Ace kits (Toyobo), following the manufacturer’s instructions. KAPA SYBER FAST qPCR Master Mix kits (Kapa Biosystems) and an ABI PRISM 7300 Real-Time PCR System were used for target amplification and detection. The primers were as follows: for ciGluA, caacatcccatggattac (Fwd) and tccttccatcccaacttg (Rev); ci-GLAST, gtgttgttcttggcatcgtg (Fwd) and cgataagcggcagaatcatc (Rev); ci-vGlut, cggggtggccttctgtattt (Fwd) and gttggcaacgataatcgccc (Rev); Ci-EF-1a, cacttggtcgtttcgctgt (Fwd) and cttcttgccgactttcttgg (Rev); Ci-EF2, gggcacgttgatttctcatct (Fwd) and tcgcaatacggtctcggttt (Rev); ci-HDAC, agatacaccacgggcatagc (Few) and ttgggtcgttaaccagcttc (Rev); ci-GAPDH, gcgatcaagactgcaatgaa (Fwd) and aaaaatgctgcttcgcttgt (Rev); and ci-ODC, ttccgcacgtggatgtaata (Fwd) and gaggagttttggcgtcacat (Rev).
Cloning.
To isolate candidate CiGluA cDNA, we searched predicted ciGluA sequences using online databases [Joint Genome Institute (JGI), C. intestinalis ver.1 (JGI ver.1), genome.jgi.doe.gov/ciona4/ciona4.home.html]. We selected the single gene predicted by a homology search (model “ci0100143594”). We followed the same procedure for ciTARP, referring to the registered gene model (model “ci0100133946”) in JGI ver.2 (genome.jgi.doe.gov/Cioin2/Cioin2.home.html).
On the basis of the predicted partial sequences, we performed 5′, 3′ gene RACE using Gene Racer kits (Life Technology) to determine the entire cDNA sequences using the total RNA pool from hatched larvae (at about 22 hpf). The full-length ciGluA and ciTARP sequences obtained by PCR were phosphorylated and ligated with linearized, blunted, and dephosphorylated pGEMHE vectors for electrophysiology experiments and inserted into pBluescript II SK (−) vectors for in situ hybridization. We used Ligation High ver.2 (Toyobo) for the ligation reaction, T4 Nucleotide Kinase (Takara) for DNA phosphorylation, Blunting High (Toyobo) to blunt the DNA, and E. coli Alkaline Phosphatase (Toyobo) for dephosphorylation of the linearized vector.
Q/R Editing Status.
We cloned the ciGluA pore region, including the Q/R editing site, from cDNA at three developmental stages (before gastrula, 4 hpf; hatched larva, 22 hpf; and young adult, 66 hpf). The primers for PCR were GTTGGAGTAAGTGTGGTCCTC (Fwd) and CACAGTAAGAAAAGCTGCAAG (Rev). Fragments cloned into the EcoRV site of the pBlueScripts II SK (−) vector were sequenced with T7 general primers to confirm the Q/R editing status.
Immunohistochemistry.
Immunostaining of whole-mount specimens was carried out as described previously (78, 79). The Ciona larvae were fixed with 4% paraformaldehyde dissolved in 0.5 M NaCl, 0.1 M Mops, pH 7.5, overnight at 4 °C. They were then washed five times for 10 min each time in 0.2% Triton-X in PBS (TPBS) before blocking in 1 mg/mL goat serum in TPBS, all day or overnight at 4 °C. They were incubated with anti-ciArr antiserum (1:1,000; kindly provided by T. Horie, University of Tsukuba, Shimoda, Japan and M. Nakagawa, University of Hyogo, Kamigori, Japan) and anti-GFP antibody (1:1,000; Nacalai Tesque) in the blocking buffer overnight and washed with TPBS at 4 °C for 8 h. Subsequently, they were incubated with Alexa 546-conjugated goat anti-mouse IgG (1:1,000; Molecular Probes, Invitrogen) in TPBS at 4 °C overnight. Alexa 488-conjugated rat anti-Phalloidin antibody was used as above as a secondary antibody. After the specimens were rinsed several times with TPBS, they were mounted in 50% (vol/vol) glycerol and observed under an FV1000 microscope (Olympus).
Whole-Mount in Situ Hybridization.
CiGluA cDNA in a pBluescript II SK (−) vector was linearized by XhoI digestion. The antisense probes were synthetized with a digoxigenin-labeling mix (Roche) and T3 RNA polymerase, according to standard methods. Embryos were fixed in 4% paraformaldehyde in 0.1 M Mops (pH 7.5) and 0.5 M NaCl at 4 °C overnight. The specimens were washed in 30% (vol/vol) ethanol and then twice in 80% (vol/vol) ethanol, before storage in 80% ethanol at −30 °C. After washing once with 30% ethanol and twice with PBT (PBS containing 0.1% Tween20), the fixed specimens were partially digested with Proteinase K (3 µg/mL for tailbud embryos, 6 µg/mL for late tailbud larva and hatched larva) in PBT for 40 min at 37 °C (shaken every 5–10 min). After washing twice in PBT, the specimens were postfixed with 4% paraformaldehyde in PBT (pH 7.5) for 1 h at RT (shaken intermittently) and then washed three more times with PBT. After washing with a hybridization buffer for 10 min at RT, the specimens were prehybridized for 2 h at 42 °C. The specimens were heated for 5 min at 80 °C before hybridization with digoxigenin-labeled probes at 50 °C for 16 h. The hybridization buffer contained 50% (vol/vol) formamide, 5× saline–sodium citrate buffer (SSC), 0.1 mg/mL yeast tRNA, 5× Denhart’s solution, 1% Tween20, and the DIG-labeled DNA probe. After hybridization, the specimens were washed twice for 15 min in the wash solution (2× SSC, 50% formamide, 0.1% Tween20) at 50 °C twice, and then washed with solution A [0.5 M NaCl, 10 mM Tris–Cl (pH 8.0), 5 mM EDTA, 1% Tween20] three times. They were soaked in solution A containing 20 µg/mL RNaseA at 37 °C for 30 min and then washed with solution A. Next, the specimens were soaked in 2× SSC, 50% formamide, and 0.1% Tween20 at 50 °C for 20 min and then twice in 5× SSC, 50% formamide, and 0.1% Tween20 at 50 °C for 15 min. The specimens were then washed twice with PBT and blocked with 0.5% (wt/vol) blocking reagent (Roche Diagnostics) in PBT for 30 min, before overnight incubation with 1:2,000 alkaline–phosphatase-conjugated anti-digoxigenin antibody (Fab fragment, Roche). The specimens were washed with PBT (>0.4 mL) on a shaker at RT for 30 min four times and then washed with PBT (>0.4 mL) on a shaker at RT for 2 h or at 4 °C overnight. This was followed by washing with buffer III [III-a contained 200 mM Tris–Cl (pH 9.5), 200 mM NaCl; III-b contained 100 mM MgCl2, 1:1] twice. For signal detection, the specimens were incubated with NBT/BCIP/alkaline phosphatase buffer at RT in the dark. The reaction was stopped in PBT and then observed in PBT or in 50–80% glycerol.
Accession Numbers.
The CDS sequences of ciGluA and ciTarp were deposited in DDBJ/EMBL/GenBank under accession numbers LC070681 (Ciona glutamate receptor variant 1), LC070682 (Ciona glutamate receptor variant 2), and LC152057 (ciTARP).
Statistics.
Statistical analyses used Student’s paired or unpaired t tests.
Acknowledgments
We give special thanks to Dr. Takeo Horie (University of Tsukuba, Shimoda, Japan), who gave us the pSP eGFP plasmid and critical advice. The anti-arrestin antibody was a kind gift from Dr. Masashi Nakagawa (University of Hyogo, Kamigori, Japan) and Dr. Horie (University of Tsukuba, Shimoda, Japan). We thank Dr. Hiroki Takahashi (National Institute for Basic Biology, Okazaki, Japan) for providing the ascidian samples for the real-time PCR and in situ hybridization. We also thank Dr. Yoshimichi Murata (Tohoku University) for the pGEMHE plasmid. Rat GluA2 cDNA plasmid was kindly gifted by Dr. James Boulter (University of California, Los Angeles). Dr. Kotaro Oka (Keio University) and Dr. Junjiro Horiuch (Tokyo Metropolitan Institute of Medical Science, Tokyo) kindly gave us helpful discussion. This work was supported by Japan Society for the Promotion of Science KAKENHI Grants 26830033 (to S.H.), 16K07426 and 16H01451 (to K.H.), 25117006 and 26250014 (to S.O.), and 16K14569 and 26290016 (to H.O.) and also by Japan Science and Technology Agency CREST Grant 14529570 (to S.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in DDBJ/EMBL/GenBank [accession nos. LC070681 (Ciona glutamate receptor variant 1), LC070682 (Ciona glutamate receptor variant 2), and LC152057 (Ciona TARP)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612943114/-/DCSupplemental.
References
- 1.Haydar TF, Wang F, Schwartz ML, Rakic P. Differential modulation of proliferation in the neocortical ventricular and subventricular zones. J Neurosci. 2000;20(15):5764–5774. doi: 10.1523/JNEUROSCI.20-15-05764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlett K. Glutamate as a modulator of embryonic and adult neurogenesis. Curr Top Med Chem. 2006;6(10):949–960. doi: 10.2174/156802606777323665. [DOI] [PubMed] [Google Scholar]
- 3.Brazel CY, Nuñez JL, Yang Z, Levison SW. Glutamate enhances survival and proliferation of neural progenitors derived from the subventricular zone. Neuroscience. 2005;131(1):55–65. doi: 10.1016/j.neuroscience.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Sugden SG, Zirpel L, Dietrich CJ, Parks TN. Development of the specialized AMPA receptors of auditory neurons. J Neurobiol. 2002;52(3):189–202. doi: 10.1002/neu.10078. [DOI] [PubMed] [Google Scholar]
- 5.Luján R, Shigemoto R, López-Bendito G. Glutamate and GABA receptor signalling in the developing brain. Neuroscience. 2005;130(3):567–580. doi: 10.1016/j.neuroscience.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 6.Lin WH, Wu CH, Chen YC, Chow WY. Embryonic expression of zebrafish AMPA receptor genes: Zygotic gria2α expression initiates at the midblastula transition. Brain Res. 2006;1110(1):46–54. doi: 10.1016/j.brainres.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 7.Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21(5):1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Métin C, Denizot JP, Ropert N. Intermediate zone cells express calcium-permeable AMPA receptors and establish close contact with growing axons. J Neurosci. 2000;20(2):696–708. doi: 10.1523/JNEUROSCI.20-02-00696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoppmann V, Wu JJ, Søviknes AM, Helvik JV, Becker TS. Expression of the eight AMPA receptor subunit genes in the developing central nervous system and sensory organs of zebrafish. Dev Dyn. 2008;237(3):788–799. doi: 10.1002/dvdy.21447. [DOI] [PubMed] [Google Scholar]
- 10.Bettler B, et al. Cloning of a novel glutamate receptor subunit, GluR5: Expression in the nervous system during development. Neuron. 1990;5(5):583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- 11.Luk KC, Sadikot AF. Glutamate and regulation of proliferation in the developing mammalian telencephalon. Dev Neurosci. 2004;26(2-4):218–228. doi: 10.1159/000082139. [DOI] [PubMed] [Google Scholar]
- 12.Martins RA, Linden R, Dyer MA. Glutamate regulates retinal progenitors cells proliferation during development. Eur J Neurosci. 2006;24(4):969–980. doi: 10.1111/j.1460-9568.2006.04966.x. [DOI] [PubMed] [Google Scholar]
- 13.Hollmann M, Hartley M, Heinemann S. Ca2+ permeability of KA-AMPA--gated glutamate receptor channels depends on subunit composition. Science. 1991;252(5007):851–853. doi: 10.1126/science.1709304. [DOI] [PubMed] [Google Scholar]
- 14.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280(5369):1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 15.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462(7274):745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439(7079):965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 17.Okamura Y, et al. Comprehensive analysis of the ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol Genomics. 2005;22(3):269–282. doi: 10.1152/physiolgenomics.00229.2004. [DOI] [PubMed] [Google Scholar]
- 18.Cloney RA. Ascidian larvae and the events of metamorphosis. Am Zool. 1982;22:817–826. [Google Scholar]
- 19.Degnan BM, Souter D, Degnan SM, Long SC. Induction of metamorphosis with potassium ions requires development of competence and an anterior signalling centre in the ascidian Herdmania momus. Dev Genes Evol. 1997;206(6):370–376. doi: 10.1007/s004270050066. [DOI] [PubMed] [Google Scholar]
- 20.Svane IB, Young CM. The ecology and behavior of ascidian larvae. Oceanogr Mar Biol Annu Rev. 1989;27:45–90. [Google Scholar]
- 21.Tsuda M, Kawakami I, Shiraishi S. Sensitization and habituation of the swimming behavior in ascidian larvae to light. Zoolog Sci. 2003;20(1):13–22. doi: 10.2108/zsj.20.13. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama-Ishimura A, Chambon JP, Horie T, Satoh N, Sasakura Y. Delineating metamorphic pathways in the ascidian Ciona intestinalis. Dev Biol. 2009;326(2):357–367. doi: 10.1016/j.ydbio.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 23.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 24.Schwenk J, et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014;84(1):41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 25.Belachew S, Gallo V. Synaptic and extrasynaptic neurotransmitter receptors in glial precursors’ quest for identity. Glia. 2004;48(3):185–196. doi: 10.1002/glia.20077. [DOI] [PubMed] [Google Scholar]
- 26.Nicol D, Meinertzhagen IA. Cell counts and maps in the larval central nervous system of the ascidian Ciona intestinalis (L.) J Comp Neurol. 1991;309(4):415–429. doi: 10.1002/cne.903090402. [DOI] [PubMed] [Google Scholar]
- 27.Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu Rev Pharmacol Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 28.Kusakabe T, et al. Ci-opsin1, a vertebrate-type opsin gene, expressed in the larval ocellus of the ascidian Ciona intestinalis. FEBS Lett. 2001;506(1):69–72. doi: 10.1016/s0014-5793(01)02877-0. [DOI] [PubMed] [Google Scholar]
- 29.Kusakabe T, Tsuda M. Photoreceptive systems in ascidians. Photochem Photobiol. 2007;83(2):248–252. doi: 10.1562/2006-07-11-IR-965. [DOI] [PubMed] [Google Scholar]
- 30.Walker CS, et al. Conserved SOL-1 proteins regulate ionotropic glutamate receptor desensitization. Proc Natl Acad Sci USA. 2006;103(28):10787–10792. doi: 10.1073/pnas.0604520103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, et al. Evolutionary conserved role for TARPs in the gating of glutamate receptors and tuning of synaptic function. Neuron. 2008;59(6):997–1008. doi: 10.1016/j.neuron.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker CS, et al. Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc Natl Acad Sci USA. 2006;103(28):10781–10786. doi: 10.1073/pnas.0604482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311(5765):1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- 34.Ziff EB. TARPs and the AMPA receptor trafficking paradox. Neuron. 2007;53(5):627–633. doi: 10.1016/j.neuron.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Jonas P, Burnashev N. Molecular mechanisms controlling calcium entry through AMPA-type glutamate receptor channels. Neuron. 1995;15(5):987–990. doi: 10.1016/0896-6273(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 36.Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991;252(5013):1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- 37.Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253(5023):1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- 38.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 39.Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17(1):58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nutt SL, Kamboj RK. Differential RNA editing efficiency of AMPA receptor subunit GluR-2 in human brain. Neuroreport. 1994;5(13):1679–1683. doi: 10.1097/00001756-199408150-00034. [DOI] [PubMed] [Google Scholar]
- 41.Whitney NP, et al. Calcium-permeable AMPA receptors containing Q/R-unedited GluR2 direct human neural progenitor cell differentiation to neurons. FASEB J. 2008;22(8):2888–2900. doi: 10.1096/fj.07-104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahlstedt H, Daniel C, Ensterö M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19(6):978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venø MT, et al. Spatio-temporal regulation of ADAR editing during development in porcine neural tissues. RNA Biol. 2012;9(8):1054–1065. doi: 10.4161/rna.21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pachernegg S, Münster Y, Muth-Köhne E, Fuhrmann G, Hollmann M. GluA2 is rapidly edited at the Q/R site during neural differentiation in vitro. Front Cell Neurosci. 2015;9:69. doi: 10.3389/fncel.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427(6977):801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 46.Kwak S, Hideyama T, Yamashita T, Aizawa H. AMPA receptor-mediated neuronal death in sporadic ALS. Neuropathology. 2010;30(2):182–188. doi: 10.1111/j.1440-1789.2009.01090.x. [DOI] [PubMed] [Google Scholar]
- 47.Peng PL, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49(5):719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt HD, et al. ADAR2-dependent GluA2 editing regulates cocaine seeking. Mol Psychiatry. 2015;20(11):1460–1466. doi: 10.1038/mp.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brusa R, et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270(5242):1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 50.Chen YC, et al. Identifications, classification, and evolution of the vertebrate alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor subunit genes. J Mol Evol. 2001;53(6):690–702. doi: 10.1007/s002390010256. [DOI] [PubMed] [Google Scholar]
- 51.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 52.Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol. 1987;121(2):526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- 53.Eagleson GW, Harris WA. Mapping of the presumptive brain regions in the neural plate of Xenopus laevis. J Neurobiol. 1990;21(3):427–440. doi: 10.1002/neu.480210305. [DOI] [PubMed] [Google Scholar]
- 54.Kajiwara S, Yoshida M. Changes in behavior and ocellar structure during the larval life of solitary ascidians. Biol Bull. 1985;169:565–577. [Google Scholar]
- 55.Bone Q. On the locomotion of ascidian tadpole larvae. J Mar Biol Assoc U K. 1992;72:161–186. [Google Scholar]
- 56.Foster RG, Roberts A. The pineal eye in Xenopus laevis embryos and larvae: A photoreceptor with a direct excitatory effect on behavior. J Comp Physiol. 1982;145:413–419. [Google Scholar]
- 57.Mick G. Non-N-methyl-D-aspartate glutamate receptors in glial cells and neurons of the pineal gland in a higher primate. Neuroendocrinology. 1995;61(3):256–264. doi: 10.1159/000126847. [DOI] [PubMed] [Google Scholar]
- 58.Kaur C, Sivakumar V, Ling EA. Expression of N-methyl-D-aspartate (NMDA) and alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) GluR2/3 receptors in the developing rat pineal gland. J Pineal Res. 2005;39(3):294–301. doi: 10.1111/j.1600-079X.2005.00245.x. [DOI] [PubMed] [Google Scholar]
- 59.Horie T, Orii H, Nakagawa M. Structure of ocellus photoreceptors in the ascidian Ciona intestinalis larva as revealed by an anti-arrestin antibody. J Neurobiol. 2005;65(3):241–250. doi: 10.1002/neu.20197. [DOI] [PubMed] [Google Scholar]
- 60.Jansson LC, Wigren HK, Nordström T, Akerman KE. Functional α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors in differentiating embryonic neural progenitor cells. Neuroreport. 2011;22(6):282–287. doi: 10.1097/WNR.0b013e3283457b34. [DOI] [PubMed] [Google Scholar]
- 61.Maric D, et al. Functional ionotropic glutamate receptors emerge during terminal cell division and early neuronal differentiation of rat neuroepithelial cells. J Neurosci Res. 2000;61(6):652–662. doi: 10.1002/1097-4547(20000915)61:6<652::AID-JNR9>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 62.Hsu WL, et al. Glutamate stimulates local protein synthesis in the axons of rat cortical neurons by activating α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors and metabotropic glutamate receptors. J Biol Chem. 2015;290(34):20748–20760. doi: 10.1074/jbc.M115.638023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fannon J, Tarmier W, Fulton D. Neuronal activity and AMPA-type glutamate receptor activation regulates the morphological development of oligodendrocyte precursor cells. Glia. 2015;63(6):1021–1035. doi: 10.1002/glia.22799. [DOI] [PubMed] [Google Scholar]
- 64.Catsicas M, Allcorn S, Mobbs P. Early activation of Ca(2+)-permeable AMPA receptors reduces neurite outgrowth in embryonic chick retinal neurons. J Neurobiol. 2001;49(3):200–211. doi: 10.1002/neu.1075. [DOI] [PubMed] [Google Scholar]
- 65.Poluch S, et al. AMPA receptor activation leads to neurite retraction in tangentially migrating neurons in the intermediate zone of the embryonic rat neocortex. J Neurosci Res. 2001;63(1):35–44. doi: 10.1002/1097-4547(20010101)63:1<35::AID-JNR5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 66.Sutherland ML, Delaney TA, Noebels JL. Glutamate transporter mRNA expression in proliferative zones of the developing and adult murine CNS. J Neurosci. 1996;16(7):2191–2207. doi: 10.1523/JNEUROSCI.16-07-02191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furuta A, Rothstein JD, Martin LJ. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci. 1997;17(21):8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartfuss E, Galli R, Heins N, Götz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229(1):15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 69.Tosti E, Boni R, Gallo A. Ion currents in embryo development. Birth Defects Res C Embryo Today. 2016;108(1):6–18. doi: 10.1002/bdrc.21125. [DOI] [PubMed] [Google Scholar]
- 70.Rosenberg SS, Spitzer NC. Calcium signaling in neuronal development. Cold Spring Harb Perspect Biol. 2011;3(10):a004259. doi: 10.1101/cshperspect.a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakajo K, Nishino A, Okamura Y, Kubo Y. KCNQ1 subdomains involved in KCNE modulation revealed by an invertebrate KCNQ1 orthologue. J Gen Physiol. 2011;138(5):521–535. doi: 10.1085/jgp.201110677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miledi R, Parker I. Chloride current induced by injection of calcium into Xenopus oocytes. J Physiol. 1984;357:173–183. doi: 10.1113/jphysiol.1984.sp015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miledi R. A calcium-dependent transient outward current in Xenopus laevis oocytes. Proc R Soc Lond B Biol Sci. 1982;215(1201):491–497. doi: 10.1098/rspb.1982.0056. [DOI] [PubMed] [Google Scholar]
- 74.Barish ME. A transient calcium-dependent chloride current in the immature Xenopus oocyte. J Physiol. 1983;342:309–325. doi: 10.1113/jphysiol.1983.sp014852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamada L, et al. Morpholino-based gene knockdown screen of novel genes with developmental function in Ciona intestinalis. Development. 2003;130(26):6485–6495. doi: 10.1242/dev.00847. [DOI] [PubMed] [Google Scholar]
- 76.Hotta K, et al. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn. 2007;236(7):1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- 77.Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124(3):589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 78.Horie T, Nakagawa M, Orii H, Tsuda M. Whole structure of the photoreceptors in the ascidian larva visualized by an antibody against arrestin (Ci-Arr) J Photosci. 2002;19:272–274. [Google Scholar]
- 79.Tsuda M, Sakurai D, Goda M. Direct evidence for the role of pigment cells in the brain of ascidian larvae by laser ablation. J Exp Biol. 2003;206(Pt 8):1409–1417. doi: 10.1242/jeb.00235. [DOI] [PubMed] [Google Scholar]