Significance
Glioblastoma (GBM) is the most lethal brain malignancy lacking effective treatment. In this study, we demonstrate that BCL6 is a prognostic marker and a targetable GBM-promoting factor. Silencing of BCL6 inhibits the malignant phenotype of GBM cells and triggers cellular senescence. We also identify AXL as an important BCL6 transcriptional target, the expression of which is also regulated positively by NCoR, a BCL6 cofactor. Either silencing of BCL6 or targeted disruption of the BCL6/NCoR complex diminishes AXL expression and inhibits GBM growth. This study elucidates a crucial BCL6-mediated signaling pathway in GBM biology. More importantly, our results highlight the promise and merit of targeting BCL6 for treating this deadly disease.
Keywords: ZBTB, BCL6, AXL, NCoR, glioblastoma multiforme
Abstract
ZBTB transcription factors orchestrate gene transcription during tissue development. However, their roles in glioblastoma (GBM) remain unexplored. Here, through a functional screening of ZBTB genes, we identify that BCL6 is required for GBM cell viability and that BCL6 overexpression is associated with worse prognosis. In a somatic transgenic mouse model, depletion of Bcl6 inhibits the progression of KrasG12V-driven high-grade glioma. Transcriptome analysis demonstrates the involvement of BCL6 in tumor protein p53 (TP53), erythroblastic leukemia viral oncogene homolog (ErbB), and MAPK signaling pathways. Indeed, BCL6 represses the expression of wild-type p53 and its target genes in GBM cells. Knockdown of BCL6 augments the activation of TP53 pathway in response to radiation. Importantly, we discover that receptor tyrosine kinase AXL is a transcriptional target of BCL6 in GBM and mediates partially the regulatory effects of BCL6 on both MEK-ERK (mitogen-activated protein/extracellular signal-regulated kinase kinase–extracellular signal-regulated kinase) and S6K-RPS6 (ribosomal protein S6 kinase–ribosomal protein S6) axes. Similar to BCL6 silencing, depletion of AXL profoundly attenuates GBM proliferation both in vitro and in vivo. Moreover, targeted inhibition of BCL6/nuclear receptor corepressor 1 (NCoR) complex by peptidomimetic inhibitor not only significantly decreases AXL expression and the activity of MEK-ERK and S6K-RPS6 cascades but also displays a potent antiproliferative effect against GBM cells. Together, these findings uncover a glioma-promoting role of BCL6 and provide the rationale of targeting BCL6 as a potential therapeutic approach.
Human ZBTB family genes consist of 49 members who encode transcription factors that modulate the transcription of genes involved in cell fate decision and lineage commitment (1). BCL6, also known as ZBTB27, is a master regulator of germinal center response and a proto-oncogene in diffuse large B-cell lymphoma (DLBCL) (2–4). Recent studies also unveiled the critical functions of BCL6 in pre–B-cell development, acute lymphoblastic leukemia, and chronic myeloid leukemia (5–7). In addition to hematopoietic cell lineages, BCL6 is involved in mammary epithelial differentiation and neurogenesis (8, 9). To execute its transcriptional activity, BCL6 requires homodimerization and forming a complex with cofactors including BCL6 corepressor (BCoR), nuclear receptor corepressor 1 (NCoR), and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) (10–12). BCL6 inhibitors are capable of blocking the interaction between BCL6 and BCoR/NCoR/SMRT and selectively killing BCL6-addicted cancer cells (13, 14), thus holding a promise for BCL6-targeted therapy. Notably, aberrant genomic or expressional changes of BCL6 have been reported in solid tumors, including breast cancer, ovarian cancer, and glioblastoma (GBM) (15–17). Intriguingly, although BCL6 confers growth advantages to breast cancer and ovarian cancer, it was found to suppress the development of medulloblastoma (18). Therefore, more efforts are required to decipher the lineage/tissue-specific networks orchestrated by BCL6.
GBM remains the most aggressive brain malignancy, with little improvement in prognosis or therapy for decades. Genomic analysis of GBM highlights the dysregulations of receptor tyrosine kinase (RTK), retinoblastoma protein (RB), and tumor protein p53 (TP53) pathways (19, 20). AXL is an oncogenic RTK that co-overexpresses frequently with its ligand GAS6 in glioma (21), yet the mechanism promoting its overexpression is unclear. As an important effector of many RTKs, the RAS pathway is hyperactivated in GBM. In addition, driver mutations of KRAS and NRAS have been identified in human diffuse gliomas (22). Of note, BCL6 mitigates Ras-induced senescence in mouse embryonic fibroblasts and human B cells (23). However, the functional relevance of BCL6 in GBM biology remains unknown. Here, with comprehensive molecular, cellular, and animal studies, we revealed the biological role of BCL6 in gliomagenesis and resolved a complex and critical BCL6-mediated signaling network in GBM cells.
Results
Identification of BCL6 as a Progrowth Factor in GBM.
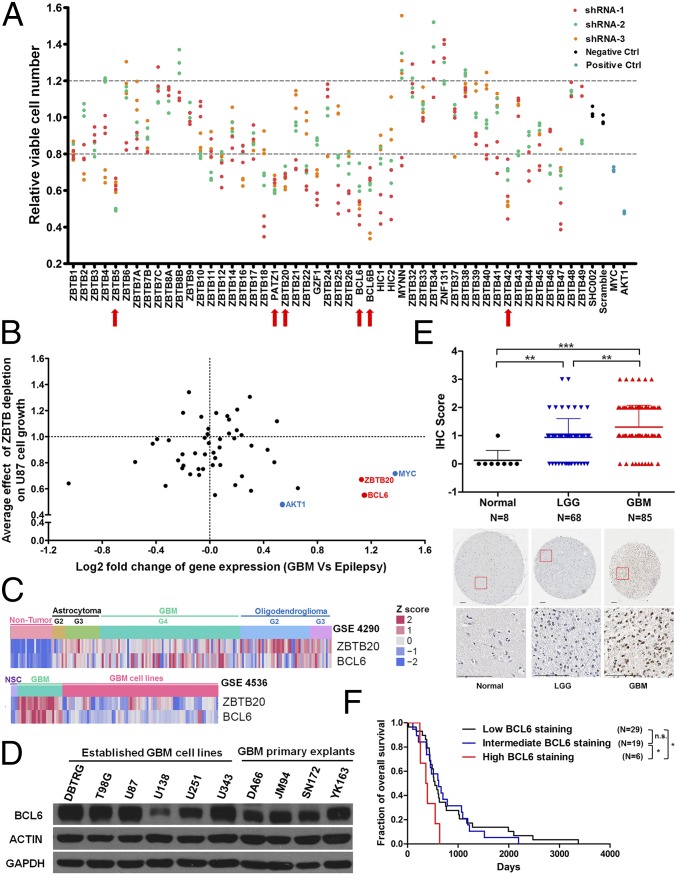
To explore the role of ZBTB family genes in GBM cell growth, a customized shRNA library was constructed to silence the expression of 49 ZBTB genes (SI Appendix, Table S1). This screen identified a list of progrowth genes including ZBTB5, PATZ1, ZBTB20, BCL6, BCL6B, and ZBTB42 (Fig. 1A). Among those, BCL6 and ZBTB20, both indispensable for cell viability, stood out as two top hits, as they were also upregulated in GBM (Fig. 1 B and C and SI Appendix, Fig. S1 A and B and Table S2) (19, 24–27). However, only BCL6 protein, not ZBTB20, was low or undetectable in normal brain tissues (SI Appendix, Fig. S1 C and D) (28). The expression of BCL6 protein was verified in both established GBM cell lines and primary explants (Fig. 1D) (29). Of note, GBM cells expressed a similar level of BCL6 protein as BCL6-positive breast cancer cells, albeit lower than BCL6-dependent DLBCL cells (SI Appendix, Fig. S1E). Next, we sought to examine the potential involvement of BCL6 in glioma progression. Through immunohistochemistry (IHC) analysis of 153 primary glioma specimens and 8 normal brain samples, BCL6 expression was robustly elevated in tumor samples and positively correlated with glioma pathological grade (Fig. 1E). Moreover, high BCL6 expression strongly predicted a worse prognosis (Fig. 1F). Hence, these results uncover BCL6 as a glioma-promoting gene and a biomarker whose up-regulation correlates with clinical grades.
Fig. 1.
Identification of BCL6 as a progrowth factor in glioma. (A) Relative viable number of U87 GBM cells after infection with lentiviral particles encoding indicated shRNAs. SHC002 and pLKO-scramble were used as negative controls (Negative Ctrl), and shRNAs targeting MYC and AKT1 were used as positive controls (Positive Ctrl). Candidate genes (marked by red arrows) were selected only if all independent shRNAs and all repeats showed consistent increase or decrease (>20%) in relative viable cell number compared with negative controls. (B) Plot showing the correlation between average effect of ZBTB depletion on U87 cell growth and differential gene expression (GBM vs. epilepsy; GSE4290). BCL6 and ZBTB20 were highlighted in red as candidates for growth-promoting genes. MYC and AKT1 were marked in blue as positive controls for oncogenes. (C) Heat map showing the expression of BCL6 and ZBTB20 in nontumor brain tissues (epilepsy), GBM cell lines, and primary GBM samples. G2, G3, and G4 denote World Health Organization grade II, III, and IV, respectively. NSC, neural stem cells. (D) Western blot result showing the endogenous BCL6 expression among 6 established GBM cell lines and 4 primary GBM explants. (E) IHC results and representative staining images showing BCL6 expression in normal brain, LGG (lower-grade glioma), and GBM samples. (Scale bars, 100 μm). IHC scores were compared using a nonparametric statistical test. (F) Survival curves for patients with GBM with differential BCL6 expression (Low, IHC score 0 or 1; Intermediate, IHC score 2; High, IHC score 3). Log-rank test was applied.
BCL6 Expression Is a Functional Requisite for Glioma Cell Growth.
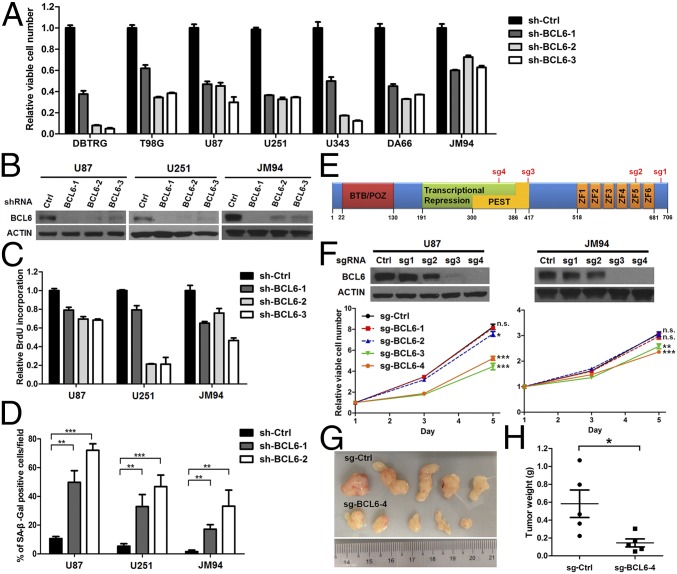
To verify the involvement of BCL6 in human GBM cell growth, we first rescreened the effects of BCL6 knockdown in five established GBM cell lines and two primary explant lines by shRNAs (Fig. 2 A and B). Depletion of endogenous BCL6 markedly reduced GBM cell viability, BrdU incorporation, and colony-forming ability (Fig. 2 A–C and SI Appendix, Fig. S2 A and B). Meanwhile, BCL6 knockdown triggered cellular senescence and G0/G1 arrest, but did not induce either sub-G1 cell population or caspase 3/7 activity (Fig. 2D and SI Appendix, Fig. S2 and C and D). Moreover, both siRNA- and CRISPR/Cas9-mediated BCL6 silencing showed similar antiproliferation effects (Fig. 2 E and F and SI Appendix, Fig. S2 E–G) (30). Importantly, depletion of endogenous BCL6 in GBM cells powerfully suppressed xenograft tumor growth in immunodeficient mice (Fig. 2 G and H). Collectively, our loss-of-function analyses strongly suggest that BCL6 is required for human GBM cell growth, both in vitro and in vivo.
Fig. 2.
BCL6 expression is a functional requisite for GBM cell growth. (A) Effect of shRNA-mediated depletion of BCL6 on GBM cell viability. In each cell line, sh-BCL6 groups were significantly different from the control (sh-Ctrl) group. (B) Western blot results showing the efficiency of BCL6 knockdown. (C and D) Effect of shRNA-mediated depletion of BCL6 on BrdU incorporation (C) and senescence-associated β-galactosidase (SA-β-Gal) activity (D). (E) Schematic representation of potential BCL6 editing sites by four individual sgRNAs. (F) Western blot results showing the efficiency of each sgRNA against BCL6 and the effect of BCL6 silencing by CRISPR/Cas9 system on cell viability. (G and H) Effect of BCL6 depletion by CRISPR/Cas9 system on tumor growth. U87 cells stably expressing either sg-BCL6-4 or sg-Ctrl were s.c. injected into NOD scid gamma mice. Tumors were harvested 30 d after implantation and weighed.
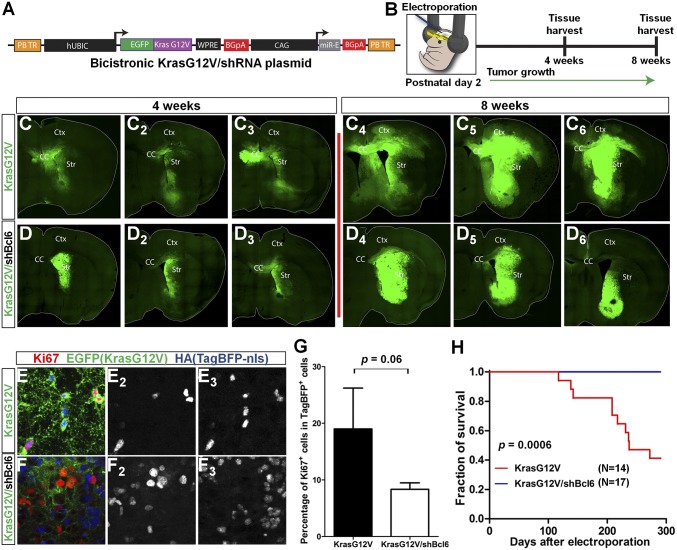
Next, to investigate the role of Bcl6 during gliomagenesis, we used a murine model of postnatal electroporation coupled with PiggyBac transposition for stable transgenesis of KrasG12V and shRNA targeting Bcl6 (31–33). Wild-type CD-1 mice were electroporated at postnatal day 2 with plasmids harboring either EGFP-KrasG12V (control) or EGFP-KrasG12V/miR-E–based shRNA against Bcl6 #275 (shBcl6.275) (Fig. 3 A and B and SI Appendix, Fig. S3 A–D). A TagBFP2 blue fluorescent protein with a hemagglutinin (HA) epitope tag (TagBFP-HA)-nuclear localization sequence reporter and pBase were coelectroporated to facilitate counting of tumor cells and for transposition, respectively. Neural precursor cells along the rostro-caudal axis of the left lateral ventricle where plasmids were delivered were susceptible to electroporation and subsequently transformed by KrasG12V (31, 33). Mouse brains were analyzed at 4 and 8 wk postelectroporation to monitor tumor growth. Mice electroporated with KrasG12V/shBcl6 showed a clear reduction in glioma size and constricted tumor margins compared with the KrasG12V control group (Fig. 3 C and D and SI Appendix, Fig. S3 E and F). In accordance, tumor cells from the KrasG12V/shBcl6 group tended to be less proliferative, as evidenced by the decreased Ki67 positivity at the ventral margin of tumors (Fig. 3 E–G). There was a clear trend toward increased tumor cell apoptosis in KrasG12V/shBcl6 mice relative to KrasG12V control, although because of intertumor heterogeneity, this result did not achieve statistical significance (SI Appendix, Fig. S3 G–I). Most importantly, KrasG12V/shBcl6 animals survived much longer than KrasG12V control group animals (Fig. 3H; P = 0.0006), strongly suggesting that Bcl6 promotes the progression of KrasG12V-driven glioma.
Fig. 3.
Bcl6 silencing inhibits glioma progression in mice. (A) Schematic of bicistronic KrasG12V/shBcl6.275 miR-E knockdown construct. (B) Postnatal day 2 electroporation of wild-type CD-1 mice targeting left lateral ventricle of the brain with pBase, TagBFP-HA-nuclear localization sequence, and EGFP-KrasG12V/shBcl6 (or EGFP-KrasG12V) expressing plasmids. (C and D) Stitched images of tumors at 4- and 8-wk times in KrasG12V and KrasG12V/shBcl6 mice (n = 3 per time). CC, Ctx, and Str denote corpus callosum, cerebral cortex, and striatum, respectively. (E and F) Coronal section of 8-wk tumors from KrasG12V (E) and KrasG12V/shBcl6 (F) mice stained with Ki67, EGFP, and HA (TagBFP). (E2 and E3) Colocalization of Ki67 (E2) and TagBFP signals (E3). (F2 and F3) Ki67+ cells (F2) largely did not colocalize with TagBFP+ cells (F3). (G) Quantification of percentage of Ki67+ cells among the TagBFP-expressing cells at the ventral margin of tumors from indicated groups. Data represent means ± SEM (n = 3). (H) Kaplan-Meier survival curves for KrasG12V (n = 14) and KrasG12V/shBcl6 (n = 17) mice. Log-rank test was applied.
BCL6 Represses the TP53 Pathway.
To dissect the GBM-associated roles of BCL6, a cDNA microarray analysis was performed in U87 cells. About 800 genes were differentially expressed following BCL6 depletion (SI Appendix, Table S3, false discovery rate ≤ 0.05) (34, 35). Remarkably, pathway enrichment analysis of these genes identified TP53, erythroblastic leukemia viral oncogene homolog (ErbB), and MAPK pathways among the top hits (SI Appendix, Fig. S4A), prompting us to investigate the functional relevance of BCL6 in these pathways.
First, we examined the effect of BCL6 on the TP53 pathway. In BCL6-silenced U87, DBTRG, and U343 cells (harboring wild-type TP53 and CDKN2A deletion), both transcript and protein levels of TP53 (p53), CDKN1A (p21), and CDKN1B (p27) were markedly elevated (SI Appendix, Fig. S4 B and C). Not surprisingly, p53 protein level in U251 and T98G cells (harboring mutant TP53 and CDKN2A deletion) was not altered after BCL6 knockdown (SI Appendix, Fig. S4D). In addition, while ectopic expression of BCL6 suppressed the TP53 expression, doxycycline-induced depletion of BCL6 in U87 cells increased p53 protein (SI Appendix, Fig. S4 E–G) (36). Further, TP53 knockdown alleviated cell cycle dysregulation caused by BCL6 depletion (SI Appendix, Fig. S4 H and I), suggesting the involvement of TP53 pathway as a downstream effector of BCL6 in controlling cell cycle progression.
Next, we explored the effect of BCL6-p53 axis on radiation response. BCL6 counteracted the p53 expression after radiation treatment (SI Appendix, Fig. S5 A–E). Six-Gray radiation robustly induced p-p53S15, ac-p53K382, and p21, which could be elevated further by BCL6 silencing (SI Appendix, Fig. S5 B–D). Simultaneous knockdown of TP53 in BCL6-depleted cells abolished p21 level (SI Appendix, Fig. S5E), confirming that p21 expression is dependent on p53. Moreover, BCL6 silencing exhibited an additive effect with radiation in suppressing the viability of GBM cells, which was partially reversed by TP53 knockdown (SI Appendix, Fig. S5F), suggesting that BCL6-p53 axis plays a role in GBM radiotolerence.
Identification of AXL as a Target of BCL6.
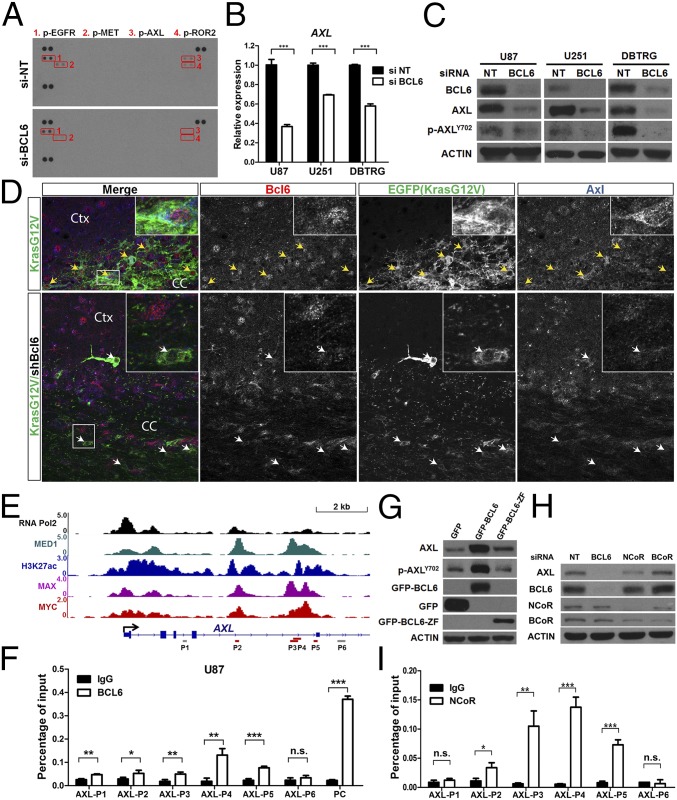
In addition to TP53 inactivation, dysregulation of RTK network is another major driver in GBM. The strong growth dependence of GBM on BCL6 expression prompted us to examine whether BCL6 has a potential role in RTK signaling. Through a human phospho-RTK antibody array, we observed a marked down-regulation of AXL, MET, ROR2, and EGFR phosphorylation in BCL6-depleted cells (Fig. 4A). Notably, high AXL expression was associated with worse overall survival of patients with GBM (SI Appendix, Fig. S6A), indicating its crucial function in GBM progression. Further analysis revealed that mRNA and protein levels of AXL, as well as its phosphorylation, were all decreased following BCL6 silencing (Fig. 4 B and C and SI Appendix, Fig. S6 B and C and Table S3). BCL6 level was positively correlated with both p-AXLY702 and AXL in BCL6-disrupted isogenic clones derived from JM94 cells (SI Appendix, Fig. S6D). Moreover, glioma populations in KrasG12V/shBcl6 mice showed diminished Bcl6 and Axl expression compared with those in the KrasG12V control group (Fig. 4D and SI Appendix, Fig. S6E). Chromatin immunoprecipitation (ChIP), coupled with quantitative real-time PCR (qPCR), revealed further a selective recruitment of BCL6 to the intron 4 region of the AXL locus where ChIP-seq signals of MED1, MYC, MAX, and H3K27ac were highly enriched (Fig. 4 E and F and SI Appendix, Fig. S6F) (37). Only wild-type BCL6, but not its Zinc-finger domain (BCL6-ZF), was able to induce AXL expression (Fig. 4G), suggesting that this regulation requires BCL6 cofactors. BCL6N21K H116A, a mutant defective in recruitment of NCoR and BCoR (12), exhibited a lower activity in promoting AXL expression than its wild-type counterpart (SI Appendix, Fig. S6G). Depletion of NCoR, rather than BCoR, partially resembled the effect of BCL6 knockdown on AXL expression (Fig. 4H). Indeed, NCoR and BCL6 co-occupied the intron 4 region of AXL locus (Fig. 4I), and their concordant expression was significantly associated with AXL level in GBM samples (SI Appendix, Fig. S6H). Collectively, these data suggest AXL as a downstream target of BCL6 in GBM.
Fig. 4.
Identification of AXL as a target of BCL6. (A) Phospho-RTK array result showing the down-regulation of phosphorylated AXL in U87 cells with depletion of BCL6. Red boxes indicate the chemiluminescent signals in duplicate for p-EGFR (1), p-MET (2), p-AXL (3), and p-ROR2 (4). (B and C) qPCR (B) and Western blot (C) results showing the down-regulation of AXL after BCL6 depletion in GBM cell lines. RNA and protein samples were harvested 36 and 72 h posttransfection, respectively. Data represent means ± SD (n = 3). (D) Immunofluorescence staining showing the correlated expression between Bcl6 and Axl in EGFP+ tumor cells (indicated by arrows) at the margin of tumors in KrasG12V (Upper) and KrasG12V/shBcl6 (Lower). (E) The ChIP-seq signals of RNA Pol2, MED1, H3K27ac, MAX, and MYC in U87 AXL locus were visualized using Integrative Genomics Viewer. Y axis represents the value of reads per million. Gray and red bars indicate the template regions amplified by respective ChIP-qPCR primers. (F) ChIP-qPCR results showing the enrichment of BCL6 binding in AXL locus. Cells were fixed 2 h after the addition of fresh medium. The 5′ UTR region of BCL6 was applied as a positive control (PC). (G) Effect of either GFP-BCL6 or GFP-BCL6-ZF on AXL expression. (H) Effect of BCL6, NCoR, and BCoR silencing on AXL expression. (I) ChIP-qPCR results showing the enrichment of NCoR binding in AXL locus.
AXL Partially Mediates the BCL6 Effect on both MEK-ERK and S6K-RPS6 Cascades.
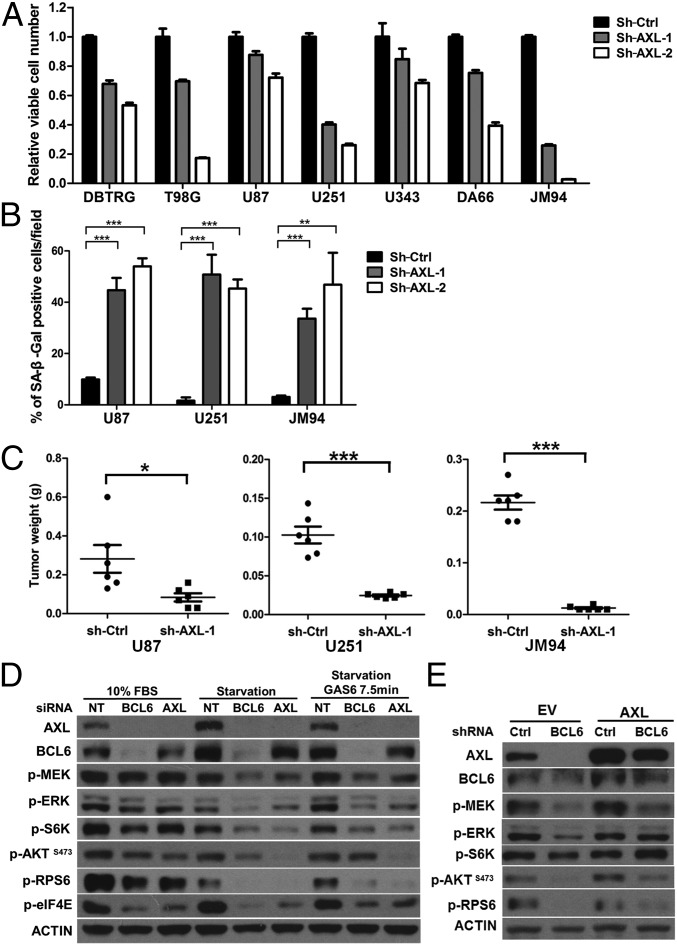
In the following study, we sought to characterize the role of BCL6-AXL axis in GBM. Similar to BCL6 silencing, AXL depletion reduced cell viability and anchorage-independent growth of GBM cells with concurrent induction of senescence and G0/G1 arrest (Fig. 5 A and B and SI Appendix, Fig. S7 A–E). Moreover, AXL depletion potently inhibited the growth of GBM xenografts (Fig. 5C). Notably, dominant negative inhibition of AXL (AXL-DN) via overexpression of its kinase-defective counterpart (38) resembled, albeit to a lesser extent, the effects of AXL knockdown in GBM cells, both in vitro and in vivo (SI Appendix, Fig. S7 F–I).
Fig. 5.
AXL contributes partially to the GBM-promoting effects of BCL6. (A–C) Effect of AXL depletion by shRNAs on cell viability (A), cellular senescence (B), and xenograft growth (C) in NOD scid gamma mice. (A) In all cell lines, sh-AXL groups were significantly different from the corresponding sh-Ctrl group. (D) Effect of siRNA-mediated silencing of BCL6 and AXL on downstream targets in U87 cells under complete medium (10% FBS), serum starvation (48 h), or GAS6 stimulation (100 ng/mL). (E) Effect of ectopic AXL expression on BCL6-depleted cells. U251 GBM cells expressing either ectopic AXL or empty vector (EV) were infected with indicated shRNAs and serum starved for 48 h before harvest.
We next explored signaling pathways regulated by BCL6-AXL axis in GBM. Under serum starvation, AXL contributed to the maintenance of basal phosphorylation of mitogen-activated protein/extracellular signal-regulated kinase kinase (MEK), extracellular signal-regulated kinase (ERK), and S6 kinase (S6K), and S6 ribosomal protein (RPS6) (Fig. 5D and SI Appendix, Fig. S7 J–L), all of which could be activated further by AXL ligand, GAS6 (Fig. 5D), suggesting that both MEK-ERK and S6K-RPS6 axes are the faithful downstream targets of AXL. Notably, these AXL downstream molecules were suppressed markedly following BCL6 silencing under both 10% FBS and serum starvation conditions (Fig. 5D and SI Appendix, Fig. S7L). siRNAs targeting either BCL6 or AXL diminished AXL expression to a similar extent and dampened the activation of both MEK-ERK and S6K-RPS6 axes upon GAS6 stimulation (Fig. 5D). Importantly, reexpression of ectopic AXL could compensate partially for this deficiency caused by BCL6 depletion (Fig. 5E). Together, these results suggest that AXL partially mediates the effect of BCL6 on promoting MEK-ERK and S6K-RPS6 cascades in GBM cells.
Targeting BCL6 in GBM.
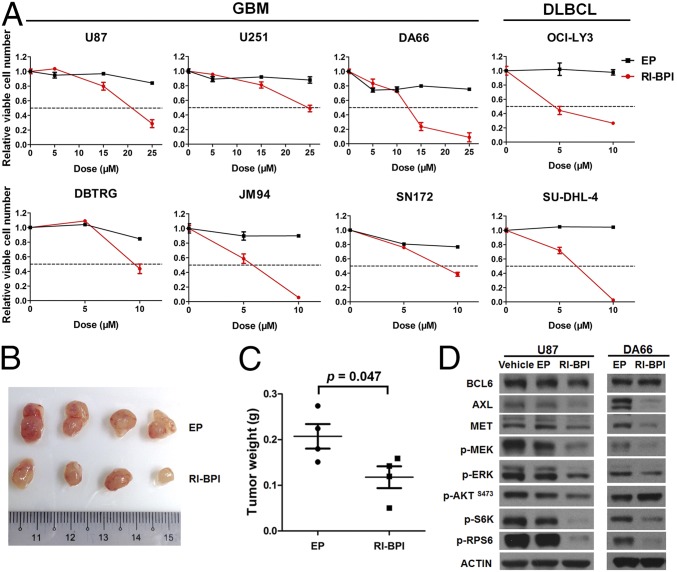
Considering the glioma-promoting function of BCL6 described earlier, targeted inhibition of BCL6 serves as an attractive therapeutic strategy. To test this, we examined the anti-GBM effects of a peptidomimetic inhibitor (RI-BPI), which specifically blocks the interaction between BCL6 and its cofactors (13). RI-BPI treatment potently suppressed GBM cell growth both in vitro and in vivo (Fig. 6 A–C). Importantly, about half of GBM cells displayed similar response to RI-BPI compared with BCL6-dependent DLBCL cells (OCI-LY3 and SU-DHL-4), which are known to be highly sensitive to this inhibitor (Fig. 6A). RI-BPI also substantially reduced the levels of AXL, MET, p-MEK, p-ERK, p-S6K, and p-RPS6 in GBM cells compared with empty peptide or vehicle control (Fig. 6D), echoing the effects of BCL6 depletion described earlier. In addition, isogenic JM94 cells that were deficient for BCL6 expression showed improved responses to EGFR inhibitors (gefitinib and erlotinib), but moderate or unchanged sensitivity to cisplatin or temozolomide treatment (SI Appendix, Fig. S8), suggesting that BCL6 inhibition may provide additional benefit for combinational targeted therapy. Collectively, our data demonstrate that BCL6 can serve as a therapeutic target in GBM.
Fig. 6.
Targeting BCL6 in glioma. (A) Effect of RI-BPI on GBM cell viability. GBM cells were treated with indicated concentration of either RI-BPI or empty peptide (EP) for 72 h before analysis. OCI-LY3 and SU-DHL-4 cells were used as positive controls. (B and C) Effect of RI-BPI treatment on JM94 cell growth in vivo. Peptides were administrated intraperitoneally (50 mg⋅kg−1⋅day−1 for 4 d). (D) Western blot results showing the effect of RI-BPI (20 µM, 24 h) on BCL6 downstream targets.
Discussion
In this study, we identify a critical role of BCL6 in promoting glioma cell proliferation and glioma progression, as well as the prognostic value of BCL6 overexpression in patients with GBM. Our data suggest that BCL6 expression is progressively up-regulated from lower-grade glioma to GBM. Translocation of BCL6 was reported in more than 36% of patients with GBM (17); however, genomic rearrangement of BCL6 locus is surprisingly not identified in the Cancer Genome Atlas (TCGA) glioma cohort (n > 1,100) (22), indicating that BCL6 translocation in GBM remains elusive. In addition, copy number variation and somatic mutation of BCL6 are infrequent in GBM (19, 20). Therefore, nongenomic mechanism is likely involved in BCL6 overexpression in GBM. Notably, our analysis showed that primary GBM samples expressed a higher level of BCL6 than ex vivo cultured GBM cells (SI Appendix, Fig. S1A), indicating that GBM microenvironment or extrinsic factors may modulate BCL6 expression.
Although the mechanism of BCL6 overexpression in GBM is unclear, our loss-of-function studies strongly support a glioma-promoting role of BCL6 in both human GBM cell lines and mouse glioma models. Depletion of BCL6 in GBM cells induced G0/G1 arrest and reduced BrdU incorporation. In line with this observation, BCL6 overexpression promoted G1-S transition of ovarian cancer cells (16). Our data also suggest that both TP53 and AXL pathways are functionally involved in BCL6-mediated cell cycle regulation. Moreover, silencing of BCL6 induced senescence in GBM cells, which was phenocopied by AXL knockdown. Interestingly, this process appeared to be independent of known senescence regulators p53 and p16 (encoded by CDKN2A) (23, 39). In addition, GBM cells barely showed activation of apoptosis on BCL6 knockdown, in contrast to mammary epithelial cells (9, 15).
In a somatic transgenic glioma model, silencing of Bcl6 in neural precursor cells could suppress significantly, but not abolish, the formation of KrasG12V-driven glioma, suggesting that Bcl6 is critical to promote glioma progression but is dispensable for its initiation. Taking advantage of EGFP labeling of KrasG12V-transformed cells that accurately separated tumor from stroma (Fig. 3 C and D and SI Appendix, Fig. S3 E and F), we observed that glioma cells diffused extensively into the brain parenchyma in KrasG12V mice at 8 wk postelectroporation; in sharp contrast, all the brains from KrasG12V/shBcl6 animals at the same point displayed circumscribed tumor borders, little migration outside the tumor bulk, and virtually no cortical invasion. Thus, Bcl6 may also foster the invasive growth of glioma. Notably, we observed an increased Ki67 positivity in nonglioma cells in KrasG12V/shBcl6 glioma tissues compared with KrasG12V tumors, suggesting that Bcl6 may regulate glioma microenvironment.
TP53 pathway is frequently attenuated in GBM (19, 20). In this study, we report an additional layer of TP53 pathway repression through BCL6 activation in GBM. Similar observations have been reported in the context of germinal-center B cells, DLBCL, and chronic myeloid leukemia (4, 7), suggesting that the BCL6-TP53 regulatory route is not tissue-specific. Interestingly, RI-BPI could not de-repress TP53 pathway in GBM cells (SI Appendix, Fig. S9), suggesting that the formation of BCL6/BCoR or BCL6/NCoR complex is dispensable for TP53 repression in this context. Notably, after BCL6 silencing, the levels of ac-p53K382, p-p53S15, and p21 were augmented significantly in response to 6-Gy, but not 12-Gy, radiation, suggesting that the effect of BCL6 on p53 signaling may be dose-dependent of radiation, and that alternative pathways triggered by high-dose radiation may compensate for BCL6 loss. In addition, quantitative analysis showed that p21 level corresponded more closely to ac-p53K382 and total p53 levels than p-p53S15 upon BCL6 silencing and radiation (SI Appendix, Fig. S5B), indicating that acetylation of p53 at K382 potentiates p21 expression in BCL6-depleted GBM cells (40).
Overexpression of AXL is prevalent in glioma tissues and is associated with worse prognosis of patients with glioma (21). Although genomic amplification of AXL was observed in a small portion of GBM cases (19, 20), the mechanisms leading to AXL overexpression remain largely unknown. Here, we report that AXL is a transcriptional target of BCL6 in GBM. Both BCL6 and NCoR were recruited to the intron 4 region of AXL locus. However, P3 and P4 regions with high enrichment of both factors did not contain a consensus BCL6 binding motif, suggesting that binding of BCL6 over these segments may involve a noncanonical mechanism. Notably, both RI-BPI treatment and NCoR knockdown achieved partial suppression of AXL, whereas BCL6 knockdown completely abolished AXL expression, suggesting that BCL6-mediated induction of AXL involves a NCoR-independent mechanism. Nevertheless, NCoR contributes to the full induction of AXL by forming the BCL6/NCoR complex, supporting the notion that NCoR can also function to enhance transcription (41). Further, we demonstrated that both BCL6 and AXL positively regulated MEK-ERK and S6K-RPS6 cascades, and that the BCL6-dependent effect on these pathways was partially mediated by AXL. Interestingly, the effects of BCL6 and AXL on GBM cell growth were more pronounced in anchorage-independent condition and xenograft assay when compared with 2D monolayer culture, suggesting that microenvironmental factors may be involved in the progrowth function of BCL6 and AXL (42).
In addition to AXL, we found that BCL6 knockdown reduced the phosphorylation of EGFR, MET, and ROR2 in U87 cells under regular culture condition. However, upon acute EGF stimulation, BCL6 silencing either did not affect the induction of p-EGFRY1068 (U87 cells) or even promoted it (JM94 cells; SI Appendix, Fig. S8), indicating that BCL6 may contribute to maintaining the steady-state level of p-EGFRY1068, but its effect on p-EGFRY1068 during acute EGF response may vary according to cellular context. As AXL and MET have been shown to reinforce EGFR signaling and to mediate the resistance to EGFR tyrosine kinase inhibitors (43–45), down-regulation of AXL and MET by BCL6 inhibition may exert a more profound effect on RTK pathway and overcoming therapeutic resistance of EGFR inhibitors. In support of this notion, BCL6 depletion sensitized GBM cells to EGFR inhibitors (SI Appendix, Fig. S8). Together with our finding that targeting BCL6 by RI-BPI effectively inhibited GBM cell viability with concurrent down-regulation of AXL and MET proteins, these results not only identify BCL6 as a druggable candidate but also suggest a potential combinational therapeutic strategy for treating GBM.
Materials and Methods
Extended materials and methods are provided in SI Appendix. Either signed consent forms or exemptions have been obtained for all human samples. The study of human samples was approved by the University of California, Los Angeles, Institutional Review Board. All murine xenograft experiments were in compliance with ethical regulations of the Institutional Animal Care and Use Committee of National University of Singapore. Electroporation-based murine experiments were approved by the Institutional Animal Care and Use Committee of Cedars-Sinai Medical Center. Unless otherwise stated, two-tailed Student’s t test was used to analyze the potential statistical difference between two groups. n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001. Log-rank test was used for survival analysis.
Supplementary Material
Acknowledgments
We thank Sumiko Takao and Vaidehi Krishnan for help with γ-radiation experiments; Annouck Luyten, Yi-Ting Qiao, and Kol-Jia Yong for ChIP protocols and technical support; Hui-Min Geng, Christian Hurtz, Anand Jeyasekharan, and Shojiro Kitajima for reagent sharing; and Hazimah Binte Mohd Nordin for help with mouse breeding. This work is funded by the National Research Foundation Singapore under its Singapore Translational Research Investigator Award (to H.P.K.; NMRC/STaR/0021/2014), the Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2013-T2-2-150), the Singapore Ministry of Health’s National Medical Research Council Centre Grant awarded to National University Cancer Institute of Singapore, the National Research Foundation Singapore, and the Singapore Ministry of Education under its Research Centres of Excellence initiatives, and is additionally supported by philanthropic donations from the Melamed family and Blanche and Steven Koegler, as well as Tom Collier “Regatta for Hope” Foundation. We also acknowledge support from the Donna and Jesse Garber Awards for Cancer Research (D.-C.L.), the Samuel Oschin Comprehensive Cancer Institute Cancer Research Forum Award (J.J.B.), the Board of Governors RMI of Cedars-Sinai (J.J.B.), and National Institutes of Health Grant R33 CA202900 (to J.J.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE77053).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609758114/-/DCSupplemental.
References
- 1.Siggs OM, Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basso K, Dalla-Favera R. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 3.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 5.Duy C, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duy C, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurtz C, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208:2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiberi L, et al. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat Neurosci. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- 9.Logarajah S, et al. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 2003;22:5572–5578. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- 10.Hatzi K, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Reports. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghetu AF, et al. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad KF, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 13.Cerchietti LC, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113:3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerchietti LC, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker SR, et al. The transcriptional modulator BCL6 as a molecular target for breast cancer therapy. Oncogene. 2015;34:1073–1082. doi: 10.1038/onc.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YQ, et al. BCL6 is a negative prognostic factor and exhibits pro-oncogenic activity in ovarian cancer. Am J Cancer Res. 2014;5:255–266. [PMC free article] [PubMed] [Google Scholar]
- 17.Ruggieri S, et al. Translocation of the proto-oncogene Bcl-6 in human glioblastoma multiforme. Cancer Lett. 2014;353:41–51. doi: 10.1016/j.canlet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Tiberi L, et al. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing Sonic Hedgehog signaling. Cancer Cell. 2014;26:797–812. doi: 10.1016/j.ccell.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Brennan CW, et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin L, et al. Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutterer M, et al. Axl and growth arrest-specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:130–138. doi: 10.1158/1078-0432.CCR-07-0862. [DOI] [PubMed] [Google Scholar]
- 22.Ceccarelli M, et al. TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shvarts A, et al. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 2002;16:681–686. doi: 10.1101/gad.929302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 26.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.Yin D, et al. Proteasome inhibitor PS-341 causes cell growth arrest and apoptosis in human glioblastoma multiforme (GBM) Oncogene. 2005;24:344–354. doi: 10.1038/sj.onc.1208225. [DOI] [PubMed] [Google Scholar]
- 30.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breunig JJ, et al. Ets factors regulate neural stem cell depletion and gliogenesis in Ras pathway glioma. Cell Reports. 2015;12:258–271. doi: 10.1016/j.celrep.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunning MJ, Smith ML, Ritchie ME, Tavaré S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 37.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vajkoczy P, et al. Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc Natl Acad Sci USA. 2006;103:5799–5804. doi: 10.1073/pnas.0510923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu RY, et al. BCL-6 negatively regulates macrophage proliferation by suppressing autocrine IL-6 production. Blood. 2005;105:1777–1784. doi: 10.1182/blood-2004-08-3171. [DOI] [PubMed] [Google Scholar]
- 40.Barlev NA, et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 41.Meyer MB, Pike JW. Corepressors (NCoR and SMRT) as well as coactivators are recruited to positively regulated 1α,25-dihydroxyvitamin D3-responsive genes. J Steroid Biochem Mol Biol. 2013;136:120–124. doi: 10.1016/j.jsbmb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 43.Meyer AS, Miller MA, Gertler FB, Lauffenburger DA. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal. 2013;6:ra66. doi: 10.1126/scisignal.2004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jun HJ, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–3050. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.