Significance
The adaptive immune system forms our primary defense against bacteria and viruses. Key players of this system are antigen receptors, dimeric molecules formed by two different types of immunoglobulin domains. It is generally believed that these receptors evolved from an ancestral dimer formed by only a single type of immunoglobulin. Using laboratory evolution, we have recreated such homodimeric receptors and characterized their interactions by X-ray crystallography. Our findings provide molecular insights and support of long-held theories concerning the evolution of the adaptive immune system. They also provide a blueprint for the experimental reconstruction of ancestral proteins in the large number of cases in which evolution has obscured sequence similarities beyond recognition, and which cannot be analyzed using current sequence-based approaches.
Keywords: protein evolution, protein structure, directed evolution, antibody, homodimer
Abstract
Ancestral protein reconstruction allows the resurrection and characterization of ancient proteins based on computational analyses of sequences of modern-day proteins. Unfortunately, many protein families are highly divergent and not suitable for sequence-based reconstruction approaches. This limitation is exemplified by the antigen receptors of jawed vertebrates (B- and T-cell receptors), heterodimers formed by pairs of Ig domains. These receptors are believed to have evolved from an extinct homodimeric ancestor through a process of gene duplication and diversification; however molecular evidence has so far remained elusive. Here, we use a structural approach and laboratory evolution to reconstruct such molecules and characterize their interaction with antigen. High-resolution crystal structures of reconstructed homodimeric receptors in complex with hen-egg white lysozyme demonstrate how nanomolar affinity binding of asymmetrical antigen is enabled through selective recruitment and structural plasticity within the receptor-binding site. Our results provide structural evidence in support of long-held theories concerning the evolution of antigen receptors, and provide a blueprint for the experimental reconstruction of protein ancestry in the absence of phylogenetic evidence.
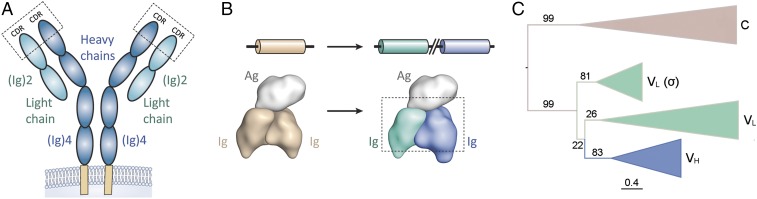
The adaptive immune system of jawed vertebrates provides specific responses to pathogens and forms long-lasting immunological memory of past encounters (1). Key components of this system are B and T lymphocytes and their cognate receptors: B-cell receptor (membrane-bound or secreted as antibodies) and T-cell receptor. Both receptors are heterodimeric molecules formed by the association of Ig domain building blocks, which then assemble into higher order complexes. This behavior is exemplified by the canonical Y-shaped IgG antibody molecule, composed of two heavy chains (each containing four Ig domains) and two light chains (each containing two Ig domains) (2). Within the N-terminal Ig domains of the receptors, hypervariable complementarity determining regions (CDRs) mediate contact with antigen (Fig. 1A). The diversity and specificity of these receptors is based on the genetic rearrangement of variable (V), diverse (D) and joining (J) gene segments, mediated by recombination-activating gene enzymes, with further diversity introduced through somatic hypermutation (3).
Fig. 1.
Evolution of rearranging Ig antigen receptors. (A) Chain structure of modern-day heterodimeric antigen receptors (IgG antibody isotype shown). The molecule is composed of two heavy chains (each containing four Ig domains, in blue) and two light chains (each containing two Ig domains, in cyan) (2). Hypervariable CDRs, which mediate contact with antigen, are highlighted. (B) Gene duplication and diversification events are believed to have converted a homodimeric Ig format (tan) to the heterodimeric format (cyan/blue) observed in the B lymphocyte-based (B-cell receptor, antibodies) and T lymphocyte-based (T-cell receptor) adaptive immune systems of jawed vertebrates. (C) Condensed maximum likelihood phylogeny of Ig heavy and light chains of vertebrates. Branches are labeled with bootstrap values from 100 replicates. (Scale bar: mean number of substitutions per site.) C, antibody constant.
Although the molecular mechanisms and structural features of rearranging Ig antigen receptors are well documented, only limited insights have been gained into their evolutionary origins (4, 5). Reconstructing the evolutionary history of antigen receptors using molecular phylogenetic analysis has proven difficult due to high levels of evolutionary divergence (6–8). Indeed, Ig receptor genes are completely absent from the genomes of invertebrates and jawless vertebrates [from which jawed vertebrates diverged some 500 million years ago and which have developed an alternative means of adaptive immunity based on leucine-rich-repeats (5, 9, 10)]. Limited evidence has been obtained from the sequence analyses of the genomes of slowly evolving jawed vertebrates, such as the elephant shark (11): These analyses suggest a distant genetic link between B- and T-cell receptors, supporting the notion that these molecules diverged from a common primordial Ig-based receptor (12–14).
Although sequence-based phylogenetic reconstruction had failed to revealed detailed insights, more general efforts to understand mechanisms of primordial antigen receptors have been attempted: For instance, it has been suggested that such early receptors were encoded by a basic genetic ensemble consisting of V- and J-gene segments, as observed for modern-day antibody variable light (VL) domains and T-cell receptor variable (Vα) domains (5). Tethered to the surface of primordial lymphocytes, such receptor molecules may have recognized foreign antigens through homodimerization (5). The capacity of the receptors to dimerize would have allowed the formation of a large interaction site capable of forming high-affinity interactions and burying extensive antigen surface. However, in other respects, homodimerization may have limited the utility of primordial antigen receptors: For instance, such an arrangement would have restricted diversity, with the CDRs of both protomers displaying identical composition. Furthermore, it is not directly evident how a symmetrical receptor arrangement would have had the capability of binding the vast number of asymmetrical antigens in nature.
Such limitations do not apply to modern-day heterodimeric Ig receptors, which are believed to have evolved through processes of gene duplication and diversification, leading to distinct heavy and light chains, increased diversity, and a relaxation of symmetry constraints (Fig. 1 A and B). The diversification of duplicated components is an evolutionary common phenomenon, observed for many other protein families, resulting in the emergence of multimers of paralogous proteins (15, 16). This process is exemplified by structures of the photosystem I core, which is formed by a homodimer in bacteria and by a heterodimer in eukaryotes (17). In the case of rearranging Ig receptors, it has long been proposed that a primordial gene encoding a basic Ig-fold of ∼110 residues gave rise to a two-domain protein structure through an initial duplication event [analogous to the modern-day human antibody light chain (variable light [VL]-constant light [CL])] (18).
A second duplication and diversification event (including the addition of D-gene segments) would then have yielded the four-domain arrangement characteristic of the human antibody heavy chain [variable heavy (VH)-constant heavy (CH)1-CH2-CH3] (18) (Fig. 1B). Indeed, it has been suggested that this process may be correlated with two whole-genome duplications, which are believed to have occurred during early vertebrate evolution (9, 19).
We set out to investigate whether a homodimeric Ig receptor could be reconstructed in the laboratory and whether such a molecule would be capable of binding to an asymmetrical protein antigen with high affinity. As a genetic source for the reconstructive process, we used V- and J-segments of the human antibody VL family to assemble the basic Ig scaffold. This family was chosen because it displays relatively high thermodynamic and colloidal stabilities (20–22), and has long been known for its propensity to dimerize. Indeed, as early as 1848, the English physician Henry Bence-Jones described what were later identified as Ig light chain dimers in the urine of patients with multiple myeloma (23). Structures of such Bence-Jones proteins reveal a face-to-face homodimeric arrangement of the variable domains (24–27), similar to what has been observed for other homomers (15). This symmetrical structural pairing is in excellent agreement with the proposed evolutionary theory, because the homodimeric arrangement of variable domains in Bence-Jones proteins closely resembles the structure of the VH-VL heterodimer found in modern-day antibodies (5). Moreover, Bence-Jones proteins have long been known to be capable of providing hydrophobic pockets within their CDRs and interface regions, into which haptens (28) and small peptides (29) can be soaked. In contrast, the antibody heavy chain variable domain is less prone to the formation of homodimers, and is encoded by additional D-gene segments, which are believed to be of relatively recent evolutionary origin (30). Taken together, these attributes suggest that light chain more closely resembles the ancestral Ig molecule (18), rendering it a suitable starting point for the reconstruction of a primordial receptor.
Results
Sequence-Based Reconstruction.
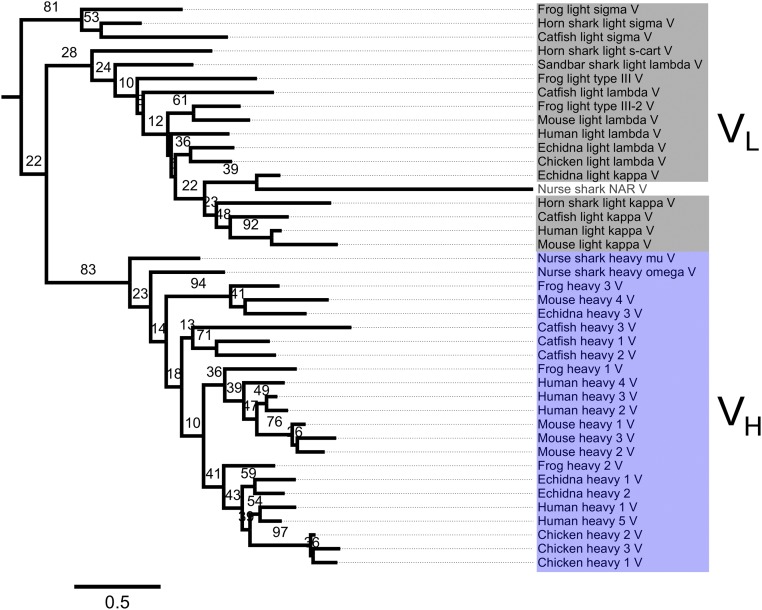
We first attempted to use ancestral protein reconstruction to gain insights into the evolutionary origins of Ig antigen receptors. Phylogenetic analysis broadly separated constant and variable domains into separate clades; however, the phylogenetic fine structure was not well resolved and was characterized by low bootstrap support (Fig. 1C and Fig. S1). This result is consistent with previous studies, which have illustrated the difficulties associated with sequence-based phylogenetic analysis of short and rapidly evolving Ig genes, including poor bootstrap support for major branches (6–8). The incapacity to produce robust phylogenies of antibody genes therefore precluded the direct reconstruction of an ancestral antigen receptor through statistical inference of sequences (31).
Fig. S1.
Maximum likelihood phylogeny of Ig heavy-chain and light-chain sequences. The number of substitutions per site is indicated. Bootstrap values were calculated from a total of 100 replicates.
Structural Reconstruction and Selection of Reconstructed Primordial Receptors.
For experimental reconstruction, human V-(O12/O2/DPK9) and J-(Jκ1) gene segments were used, which are among the most common in the human light chain repertoire (32). The segments were recombined, and amino acid diversity was introduced through site-directed mutagenesis of hypervariable CDRs [positions 28, 30–32, 50–51, 53, 91–94, and 96; numbering according to Kabat et al. (33)] to generate a large synthetic Ig repertoire (5 × 109 clones) displayed as protein fusions on the tip of filamentous bacteriophage (SI Text, Fig. S2, and Table S1). We used a phage display format that presents multiple copies of the receptor on the surface of each phage particle, thereby, in principle, allowing homodimerization (34). This repertoire was panned against the model antigen hen egg-white lysozyme (HEL) immobilized on a solid support; and, after four rounds of selection, binders were identified by antigen ELISA and sequenced (SI Text).
Fig. S2.
Library construction. Genes encoding O12/O2/DPK9 (V-) and Jκ1 (J-) segments were diversified at hypervariable CDR positions [CDR1: 28, 30–32; CDR2: 50–51, 53; CDR3: 91–94, 96; numbering according to Kabat (33)]. Combinatorial diversity was generated through recombination of the segments by splice overlap extension PCR.
Table S1.
Oligonucleotides used for library construction
| Region | Nucleotide sequence |
| CDR1 | 5′-CATCACTTGCCGGGCAAGTCAGKMTATTKMTKMTKMTTTAAATTGGTATCAGCAGAAAC-3′ |
| CDR2 | 5′-CAGGGAAAGCCCCTAAGCTCCTGATCTATKMTKMTTCCKMTTTGCAAAGTGGGGTCCCATCAAG-3′ |
| CDR3 | 5′-ACTTACTACTGTCAACAGXxxXxxXxxXxxCCTXxxACGTTCGGCCAAGGGACCAAG-3′ |
Diversified positions are shown in boldface. CDR1 and CDR2 oligonucleotides were reverse-complemented and phosphorylated before use in Kunkel mutagenesis. KMT-encoded amino acids (codons): 25% Y(TAT), 25% S(TCT), 25% A(GCT), 25% D(GAT). Xxx-encoded amino acids (TRIM codons): 19.7% Y(TAC); 15.2% S(TCT); 6.6% A(GCT); 6.6% D(GAC); 16.7% G(GGT); 3.9% I(ATC), L(CTG), P(CCG), R(CGT), T(ACT), V(GTT); 1.6% E(GAA), F(TTC), H(CAT), K(AAA), M(ATG), N(AAC), Q(CAG), W(TGG).
Receptor Characterization.
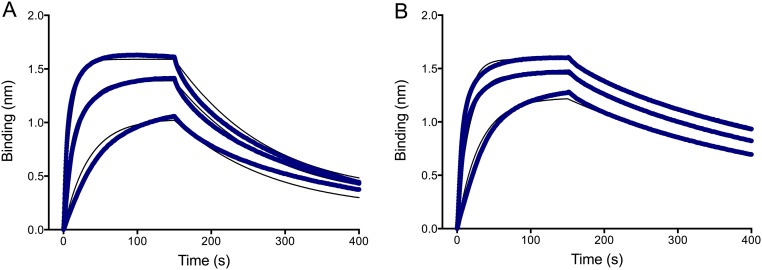
DNA sequencing revealed 12 unique clones, which clustered into two families. A representative of each family, designated Ig5 and Ig12 (amino acid sequences in Fig. S3C) was overexpressed in Escherichia coli, and the purified proteins were further investigated by a range of biophysical techniques. Size-exclusion chromatography coupled to multiangle laser light scattering (35) revealed the stoichiometry of both receptor–HEL complexes to be consistent with two Ig domains bound to a single HEL molecule (Fig. S3 A and B). Moreover, binding studies using biolayer interferometry revealed affinities in the nanomolar range, with equilibrium-binding constants for HEL binding of 130 nM and 30 nM for Ig5 and Ig12, respectively (Fig. S4).
Fig. S3.
SEC coupled to multiangle laser light scattering of the Ig–HEL complexes. (A) Elution profiles for Ig5 (black), HEL (gray), and their complex (red) reveal molecular weight estimates of the complex to be consistent with a 2:1 stoichiometry of Ig/HEL. (B) Elution profiles for Ig12 (black), HEL (gray), and their complex (red) similarly reveal a 2:1 stoichiometry for the Ig–HEL complex. (C) Amino acid sequences of Ig5 and Ig12 with CDRs underlined: CDR1 (purple), CDR2 (green), and CDR3 (blue); amino acids diversified in the library are highlighted in red. AU, arbitrary units.
Fig. S4.
Analyses of binding kinetics. HEL antigen was immobilized on streptavidin biosensors using biotinylated HyHEL-10 (Ig5) and HyHEL-5 (Ig12) capture antibodies in a Fab format using a Forte Bio biolayer interferometry system. (A) Ig5 (at 2,000 nM, 1,000 nM, and 500 nM concentrations) [ka = 4.3 × 104 M−1⋅s−1; Kd = 5.7 × 10−3 s−1; equilibrium-binding constant (KD) = 1.3 × 10−7 M]. (B) Ig12 (at 1,000 nM, 750 nM, and 375 nM concentrations) (Ka = 8.4 × 104 M−1⋅s−1; Kd = 2.5 × 10−3 s−1; KD = 3.0 × 10−8 M).
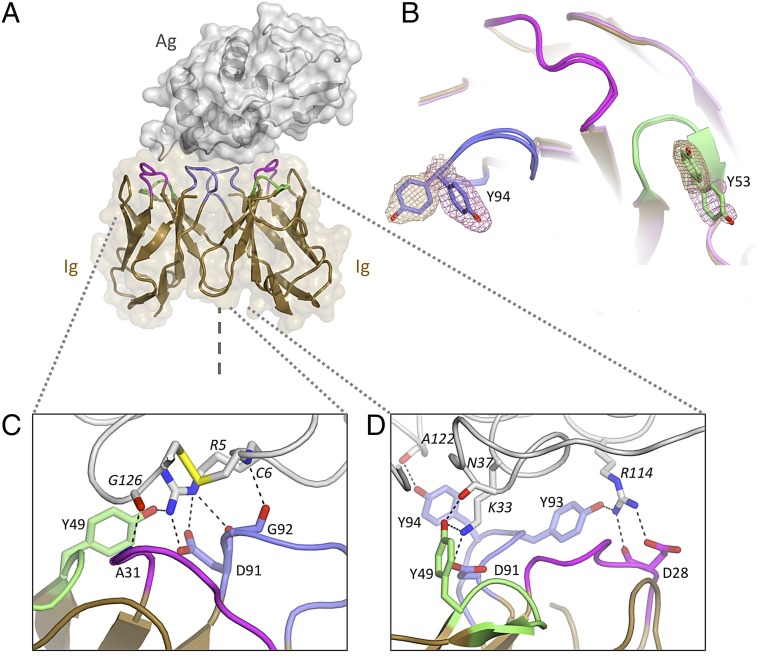
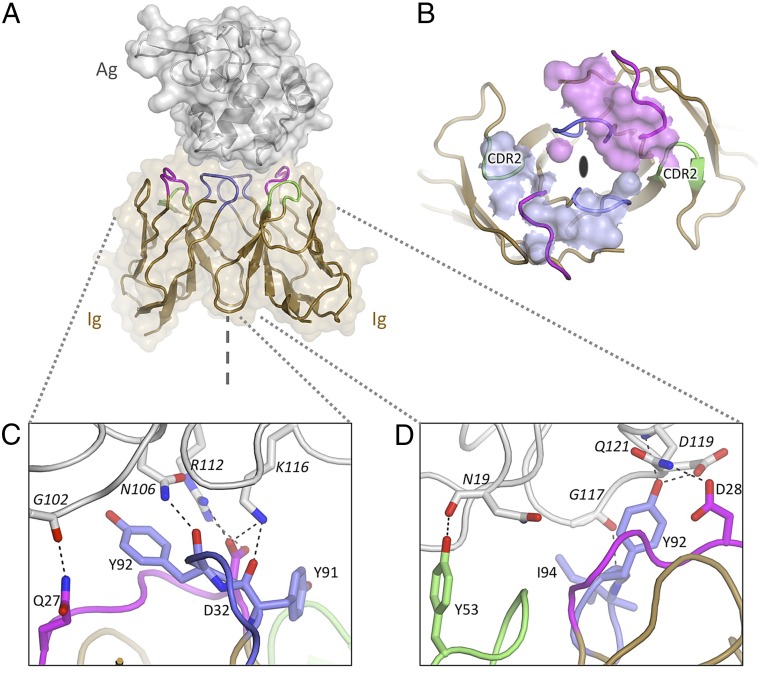
Crystals of Ig5 and Ig12 in complex with HEL were obtained, and their structures were solved by molecular replacement and refined to resolutions of 2.2 Å and 1.7 Å, respectively (Figs. 2A and 3A, and Table S2). In both structures, a receptor dimer is bound to a single HEL molecule, although the HEL epitopes targeted are completely different (Figs. 2A and 3A). Unlike previously reported Ig receptor structures in complex with antigen, in which VH and VL paralogs interact in a heterodimeric arrangement (36), both structures reveal a unique homodimeric nature of the receptor components. Strikingly, the CDRs of each receptor protomer, even though identical in amino acid sequence, form discrete and extensive contacts with different parts of the antigen surface (which lacks the twofold rotational symmetry of the receptor). In the case of the Ig52–HEL structure, the Ig domains individually bury ∼400 Å2 and 450 Å2 of the HEL surface (850 Å2 combined). Similar buried surface areas are observed for the Ig122–HEL structure (∼300 Å2, 400 Å2, and 700 Å2 combined). Indeed, the antigen contact surfaces are comparable in size to the antigen contact surfaces of the Ig-Ig homodimer interfaces (at 600 Å2 and 700 Å2 for Ig52 and Ig122, respectively). Both receptors form large and well-defined interfaces (Fig. S5), similar in extent to what has been observed for heterodimeric antibody–antigen complexes (560–855 Å2 combined) (37).
Fig. 2.
Structure of the Ig5 homodimeric antigen receptor in complex with antigen: Evidence for conformational diversity. (A) Homodimer of Ig domains (tan cartoon with CDR1, CDR2, and CDR3 loops colored in purple, green, and blue, respectively) bound to a single HEL molecule (gray cartoon and surface). The symmetry axis is indicated (dashed line). (B) Antigen-binding site (superposed Ig protomers, chains C/D): Multiple side-chain conformations are observed at positions Y53 and Y94. Electron density (mesh) is contoured at 1 SD above average (sigma-A weighted 2mFo-DFc map). (C) Antigen-binding site 1 (left-hand side). (D) Antigen-binding site 2 (right-hand side). Hydrogen bonds are highlighted; distinct contact networks are observed for each Ig subunit.
Fig. 3.
Structure of the Ig12 homodimeric antigen receptor in complex with antigen: Evidence for selective recruitment. (A) Homodimer of Ig domains (tan cartoon with CDR1, CDR2, and CDR3 colored in purple, green, and blue, respectively) bound to a single HEL molecule (gray cartoon and surface). The symmetry axis is indicated (dashed line). (B) HEL contact footprint mapped onto the surface of the Ig12 dimer (viewed down its twofold axis as indicated). The contact surfaces of the two Ig domains are colored in light blue and purple. The CDR2s are indicated and highlight the selective use of otherwise identical surface loops. (C) Antigen-binding site 1 (left-hand side). (D) Antigen-binding site 2 (right-hand side). Hydrogen bonds are highlighted; distinct contact networks are observed for each Ig subunit.
Table S2.
Diffraction Data: Data collection and refinement statistics (molecular replacement)
| Dataset | Ig52-HEL | Ig122-HEL |
| Space group | P21 | P63 |
| Unit cell dimensions: a, b, c (Å); β (°) | 114.9, 39.4, 178.3; 106.4 | 126.8, 126.8, 40.7; 90 |
| Wavelength, Å | 1.54179 | 0.95369 |
| Resolution range, Å | 45.7–2.23 | 41.5–1.7 |
| Observed reflections | 447,632 | 328,598 |
| Unique reflections* | 72,711 | 41,399 |
| Completeness,*,† % | 93.5 (99.6) | 99.8 (99.5) |
| Multiplicity, *,† % | 6.2 (4.2) | 7.9 (6.5) |
| Rmeas*,† | 0.087 (0.29) | 0.098 (0.62) |
| Mean,*,† (SD) | 14.3 (4.7) | 12.9 (2.8) |
| Wilson B, Å2 | 34.6 | 19.3 |
| Refinement | ||
| Protein molecules, asu | 8 Ig, 4 HEL | 2 Ig, 1 HEL |
| Amino acids modeled, asu | 1,351 | 330 |
| Waters modeled, asu | 130 | 154 |
| Ramachandran: favored,‡ % | 96.2 | 96.3 |
| Ramachandran: outliers,‡ % | 0.23 | 0.62 |
| R | 0.22 | 0.20 |
| Rfree (5% data) | 0.26 | 0.23 |
| RMSD bond lengths, Å | 0.014 | 0.013 |
| RMSD bond angles, ° | 1.45 | 1.38 |
| Buried surface§ | ||
| VL dimer AB; subunits-HEL, Å2 | 592; 363, 467 | 706; 285, 406 |
| VL dimer CD; subunits-HEL, Å2 | 640; 409, 512 | — |
| VL dimer EF; subunits-HEL, Å2 | 617; 406, 458 | — |
| VL dimer GH; subunits-HEL, Å2 | 556; 372, 419 | — |
| Alternate side-chain conformers contacting HEL | ||
| VL dimer AB | B-Tyr53 unresolved | — |
| B-Tyr94 alternative | ||
| VL dimer CD | d-Tyr53 alternative | — |
| d-Tyr94 alternative | ||
| VL dimer EF | — | — |
| VL dimer GH | H-Tyr-94 unresolved | — |
| PDB ID code | 4N1E | 4N1C |
asu, asymmetric unit; PEL, pheasant egg-white lysozyme.
As output by SCALA (55).
Values in parentheses are of the highest resolution shell.
As calculated by the MOLPROBITY validation server (59).
As calculated by PDBePISA (60).
Fig. S5.
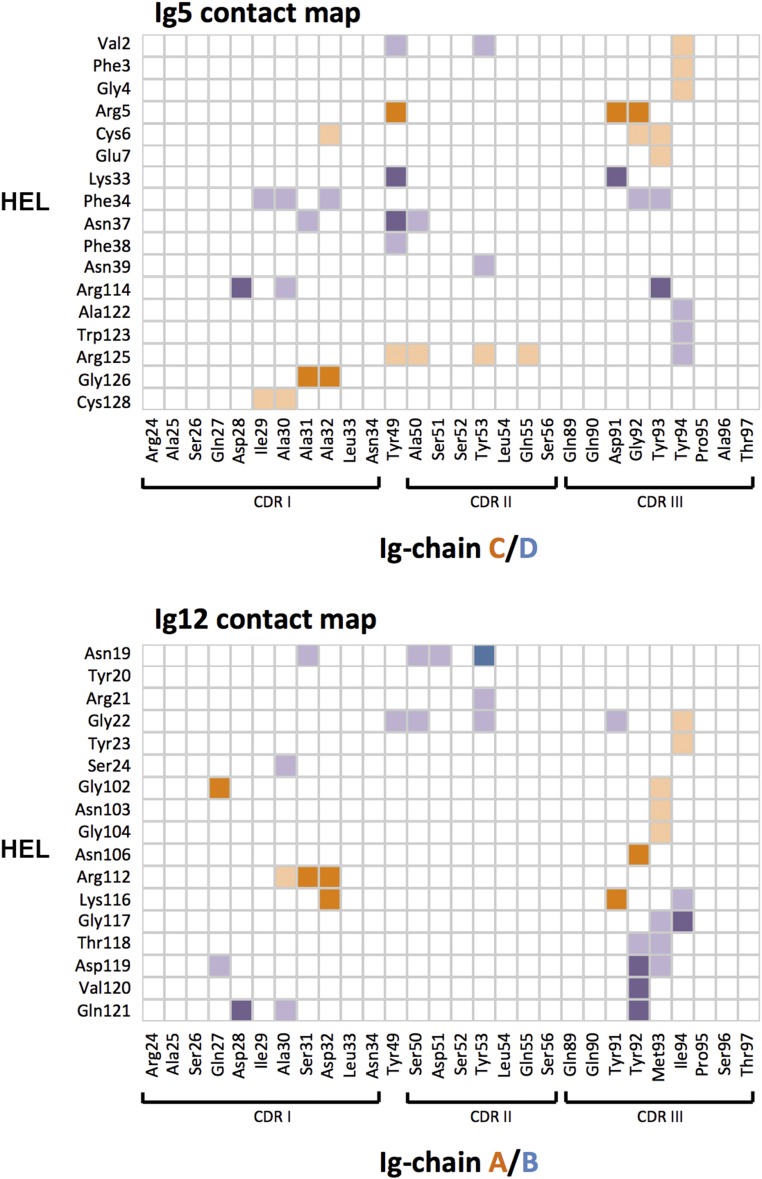
Contact map. Surface residues of the two protomers of Ig5 (Top) or Ig12 (Bottom) that contact the surface of HEL. Protomers are distinguished by color (shades of purple or orange, respectively). Light shades indicate hydrophobic interactions. Darker shades indicate hydrogen-bonding interactions or salt bridges. Contact information was derived from analysis output by PDBePISA.
SI Text
Library Design and Construction.
The genes encoding V-gene (O12/O2/DPK9) and J-gene (Jκ1) segments were amplified by PCR and cloned into the phage display vector pHEN1 (54). V-segment diversity was introduced at hypervariable CDR1 and CDR2 positions using Kunkel mutagenesis, whereas J-segment diversity was introduced by PCR using degenerate TRIM oligonucleotides (55, 56). Combinatorial diversity was generated through recombination of the segments using splice overlap extension PCR, followed by cloning into pHEN1 and transformation into Escherichia coli TG1 bacteria. This procedure resulted in a bacterial library encompassing 5 × 109 members, which was rescued using KM13 helper phage (57).
Selection of Antigen Binders.
The phage library was selected against HEL (Sigma–Aldrich), which had been biotinylated using NHS-PEG4-biotin (Thermo Fisher Scientific). For rounds 1 and 3 of the selection, the HEL antigen was captured using Neutavidin (Thermo Fisher Scientific)-coated wells of a MaxiSorp Immunoplate (Nunc). For rounds 2 and 4 of the selection, magnetic streptavidin beads (Invitrogen) were used as an alternative means of capture (to prevent the selection of binder against the capture reagents). After four rounds of selection, binders were identified by soluble-fragment ELISA (58).
Protein Expression and Purification.
Regions encoding Ig receptors were amplified by PCR from phagemid vectors and cloned into the periplasmic expression vector pET12a (Novagen). The resulting constructs were transformed into E. coli BL21-Gold (DE3) (Agilent). Expression was induced in midexponential cultures by addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (Gold Biotechnology), and Ig domains were purified from filtered supernatant using Protein L affinity resin (Genscript).
Determination of Binding Stoichiometry.
Size-exclusion chromatography (SEC) coupled with multiangle laser light scattering was used to determine molecular weight. Complexes were generated by incubating equimolar amounts of purified Ig domains and HEL antigen for 1 h at room temperature. Analyses were performed on a Superdex 75 10/300 GL SEC column (GE Healthcare) using an Äkta Purifier HPLC system (GE Healthcare) at a flow rate of 0.5 mL⋅min−1 in PBS. Column eluates were analyzed using a miniDAWN Tristar laser light scattering photometer and an Optilab DSP interferometric refractometer (Wyatt Technology Corporation). Molecular weights were calculated using Debye fitting of elution peaks.
Affinity Measurements.
Binding kinetics were analyzed by bio-layer interferometry (ForteBio). HEL antigen was immobilized on streptavidin biosensors using biotinylated HyHEL-10 (Ig5) and HyHEL-5 (Ig12) capture antibodies in a Fab format [expressed using the Expi-293 expression system (Thermo Fisher Scientific)]. Affinity of soluble purified Ig5 and Ig12 to HEL was analyzed using serial dilutions of the Ig component on a ForteBio BLItz instrument, utilizing global fitting and the BLItz Pro-1.1 software.
Crystal Growth, Structure Determination, and Refinement.
Complexes of Ig52–HEL and Ig122–HEL were purified by SEC on a Superdex 75 10/300 GL SEC column. Peak fractions were collected and concentrated using 10-kDa Amicon Ultra microconcentrators (Millipore). Initial crystallization conditions were determined using a JCSG- plus crystal screen (Molecular Dimensions), a TTP Labtech Mosquito crystallization robot, and 96-well MRC2 sitting-drop crystallization plates (Swissci).
Crystals of the Ig52–HEL complex were grown at room temperature in a hanging-drop format by combining 2 μL of protein solution (at 6.5 mg/mL in PBS) with an equivalent volume of well solution [200 mM sodium formate, 18% (wt/vol) PEG 3350]. Crystals were cryoprotected by brief soaking in well solution doped with ethylene glycol to a concentration of 20% (vol/vol). Crystals were snap-frozen in a nitrogen gas stream at 100 K, and diffraction data were recorded using a Rigaku Micromax-007HF generator and MAR345dtb imaging plate (kindly provided by the University of Sydney). Crystals of the Ig122–HEL complex were grown at room temperature in a sitting-drop format by combining 0.4 μL of protein solution (at 5 mg/mL in PBS) with an equivalent volume of well solution [200 mM ammonium acetate, 100 mM BisTris (pH 5.5), 25% (wt/vol) PEG 3350]. Crystals were snap-frozen without any further cryoprotection steps. Diffraction data were collected at 100 K on beamline MX2 at the Australian Synchrotron.
Reflections were indexed and integrated with iMOSFLM (59), analyzed for symmetry with POINTLESS (60), scaled with SCALA (60), and imported into the CCP4i software package (61). The structures were solved by molecular replacement using PHASER (62). The search models for the Ig and HEL components were PDB ID code 3UPA (κ-light chain) and PDB ID code 1ZVY (HEL), both stripped of side-chain moieties. In both cases, a 2:1 ratio of Ig/HEL was observed in the crystal; a pair of Ig monomers, arranged in Bence-Jones dimer configuration, is bound to each HEL molecule. Four essentially identical Ig2–HEL complexes were found in the asymmetrical unit of the Ig52–HEL structure, where Ig dimers are chained AB, CD, EF, and GH and corresponding HEL molecules are chained I, J, K, and L. Noncrystallographic restraints were used during early stages of refinement, although these restraints were completely removed before the introduction of solvent molecules, and for the duration of latter stages of refinement. A single Ig2–HEL complex was observed in the asymmetrical unit of the Ig122–HEL structure.
Initial rounds of rigid body refinement were followed by B-factor–restrained refinement using REFMAC5 (63), and used torsion-libration-screw parameterization (61). During later stages of refinement, water molecules were modeled into appropriate features of difference density. Models were compared with maps and manipulated in real space using Coot (62). Models were scrutinized using the MOLPROBITY validation server (64). Buried surfaces were calculated with PDBePISA (65).
Phylogenetic Analysis.
Heavy-chain and light-chain protein sequences from human (Homo sapiens), mouse (Mus musculus), echidna (Tachyglossus aculeatus), frog (Xenopus laevis/Xenopus tropicalis), catfish (Ictalurus punctatus), horn shark (Heterodontus francisci), nurse shark (Ginglymostoma cirratum) and sandbar shark (Carcharhinus plumbeus) were collected from the National Center for Biotechnology Information database; in total, 82 gene segments were used. CH1 domain sequences were extracted from heavy-chain sequences using ScanProsite (66). V and C domain sequences were both truncated at the last conserved cysteine residue. The multiple sequence alignment was constructed using the international ImMunoGeneTics information system (IMGT) unique numbering convention, which is based on structural alignments of both domains (67). A guide alignment of 11 V- and C-domain sequences was produced using IMGT/DomainDisplay (68). The full sequence set was then added to this alignment using MAFFT (69). Finally, the guide sequences and positions consisting mostly of gaps were removed. Phylogenetic trees were inferred using the maximum likelihood method in PhyML (70) and bootstrapped with 100 replicates. Model selection using PROTTEST (71) recommended use of the WAG substitution model for this analysis, with rate heterogeneity accounted for by the discrete-gamma model with five rate categories and a proportion of invariant sites. Figures were generated using FigTree (tree.bio.ed.ac.uk/software/).
Discussion
Ancestral Protein Reconstruction in the Absence of Sequence Conservation.
The high levels of sequence diversity within the Ig antigen receptor family had previously precluded detailed analyses of evolutionary origins (as confirmed by further sequence-based ancestral protein reconstruction attempts as outlined above). Here, we have a used different approach based on a combination of in vitro selection and crystallographic structure determination to investigate evolutionary ancestry. More specifically, we have reconstructed antigen receptors that resemble the Ig-fold and homodimeric state of ancestral proteins from which modern-day heterodimeric B- and T-cell receptors are believed to have originated through process of gene duplication and diversification.
Interaction of the Homodimeric Receptors with Antigen.
How is it possible for the reconstructed symmetrical Ig dimers to bind an asymmetrical target with high specificity and affinity? Closer inspection of the structures reveals two distinct mechanisms. In the Ig52–HEL structure, which contains four Ig52–HEL complexes in the asymmetrical unit, superposition of the Ig domains within each complex reveals that the side chains of key residues adopt a range of conformations (Fig. 2B). This conformational diversity is particularly pronounced for tyrosine residues at positions 53 and 94, which adopt strikingly different conformations (largely due to rotation about the chi1 dihedral angle, leaving the main chain fold unaffected) (Fig. 2B, Table S3, and Movie S1). Hence, in the Ig52–HEL structure, homodimer symmetry is relaxed through conformational side-chain diversity within CDR loops to complement the asymmetrical antigen.
Table S3.
Ig5–HEL contact interface
| Residue | Chain | HEL contact | CC | RMSD, Å |
| Asp28 | C | None | 0.93 | 0.42 |
| D | H-bonds side chain of Arg114 | 0.96 | ||
| Tyr49 | C | H-bonds side chain of Arg5 | 0.98 | 1.63 |
| D | H-bonds main chain of Asn37 | 0.98 | ||
| Tyr53 | C | Packs against side chain of Arg125 | 0.96 | 4.28 |
| D | None | 0.93 | ||
| Asp91 | C | H-bonds side chain of Arg5 | 0.96 | 1.18 |
| D | H-bonds side chain of Lys33 | 0.96 | ||
| Tyr93 | C | Packs against side chain of Cys6 | 0.96 | 1.99 |
| D | H-bonds side chain of Arg114 | 0.96 | ||
| Tyr94 | C | Packs against side chain of Val2 | 0.97 | 4.87 |
| D | Packs against side chain of Trp123 | 0.97 |
Real-space correlation coefficient (CC) for the 2Fo-Fc electron density map was calculated using Phenix (72), proving a measure of how well the model fits the electron density on a residue basis. For Ig5 chains C and D, the mean CC for all residues was 0.96 (ranging from 0.68 to 0.99). For interface CDR residues 28, 49, 91, 53, 93, and 94, the mean CC was also 0.96, strongly supporting the accuracy of the modeled side-chain conformations in these regions. The CC for these CDR residues in other Ig5 protomers was similarly high (ranging from 0.95 to 0.96). Root mean square deviation (RMSD) between protomers for residues in the Ig5 structure was calculated using CCP4i (73). For interface CDR residues 28, 49, 91, 53, 93, and 94, the RMSD equaled 2.40 Å (side-chain atoms), whereas “core” residues (defined as those residues with a solvent-accessible surface area less than 10%) displayed an RMSD of 0.28 Å using side-chain atoms of chains C and D. These results demonstrate large structural variation at CDR positions relative to a relatively rigid core.
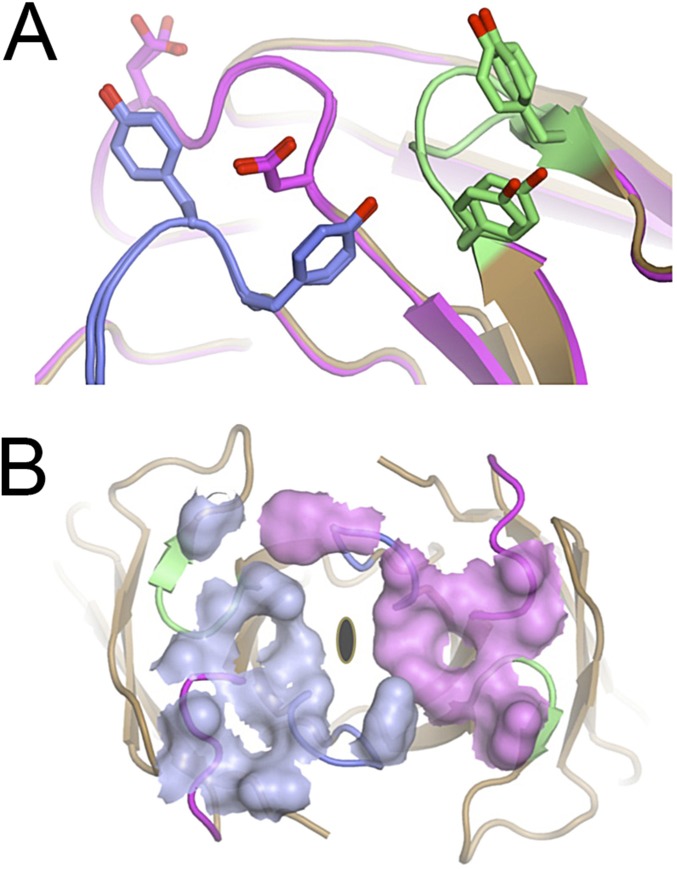
In contrast, such examples of structural plasticity are not observed in the Ig122–HEL structure, where superposition of the Ig protomers within the single complex in the asymmetrical unit reveals few detectable differences in side-chain conformations (Fig. S6A). Instead, a different mechanism based on the selective recruitment of CDRs is dominant in this second structure. Here, each Ig subunit uses different combinations of contact residues, within otherwise structurally similar CDR loops, to form unique hydrogen-bonding networks with the HEL surfaces (Fig. 3 C and D). The alternative utilization of otherwise identical CDR surfaces is particularly evident when mapping the contact footprint of the antigen onto the two Ig protomers (Fig. 3B). This projection reveals selective use of CDR2, with only one of the Ig12 subunits making extensive contacts through this CDR (detailed in Fig. S6). Such CDR contact selectivity is also observed in the Ig52–HEL complex (Fig. 2 C and D and Fig. S6B), complementing conformational CDR side-chain plasticity.
Fig. S6.
Additional perspectives of Ig52–HEL and Ig122–HEL interactions. (A) Superposition of Ig12 protomers. In contrast to what is observed for Ig5 (Fig. 2B), side-chain conformations are broadly conserved within the Ig12 antigen-binding site (despite different contacts with the asymmetrical HEL surface). (B) HEL contact footprint mapped onto the surface of the Ig5 dimer, as viewed down its twofold axis, reveals different contact networks of the Ig subunits, similar to what is observed for the Ig12 structure (Fig. 3B).
Comparisons with Previously Reported Antigen Receptor Structures.
Further comparison with structures from the Protein Data Bank reveal an intriguing similarity between Ig122–HEL and a heterodimeric antibody–antigen complex reported in the early 1990s: the 2.4-Å resolution structure of the D11.15 Fv fragment complexed with pheasant egg-white lysozyme (38). Comparison of the two structures (Fig. 4 A and B) reveals that, despite the absence of CDR sequence similarity, the receptors contact equivalent parts of the lysozyme surface and bury comparable surface areas (Ig12: ∼700 Å2 combined; D11.15: ∼650 Å2 combined). Similarities are further highlighted by structural superposition of the lysozyme components and examination of CDR3s (Fig. 4C), which contribute the majority of binding energy in antibody–antigen interactions (39). Although the CDR3 sequences and molecular details of the interactions are different, strong overall convergence is observed, with one of the Ig protomers resembling VH and the other resembling VL (Fig. 4C). The observation that Ig12 is capable of interacting with HEL antigen in a manner highly reminiscent of the modern-day D11.15 antibody raises questions relating to which evolutionary advantage duplication and divergence of an ancestral homodimer into a heterodimer may have been conferred. Although the asymmetrical nature of the HEL antigen used here prevents further experimental validation, it is likely that any homodimeric receptor would display an inherent preference for symmetrical antigens. The response would thereby be biased, diminishing neutralization potential. Moreover, pathogens could potentially exploit such epitope preference by expressing symmetrical decoy antigens. This behavior suggests that the need for broad epitope coverage and the avoidance of immune evasion may have shaped the evolution of modern-day heterodimeric antigen receptors.
Fig. 4.
Molecular mimicry at a conserved interface. Comparison of D11.15–pheasant egg-white lysozyme (PEL) and Ig122–HEL structures. (A) D11.15–PEL structure (PDB ID code 1JHL) comprises the classic heterodimer pairing of heavy chain (blue) and light chain (cyan) bound to lysozyme (gray cartoon and surface). The active site cleft is indicated (*). (B) Ig122–HEL structure, viewed from the same perspective as displayed in A. (C) Superposition of lysozyme molecules reveals the CDR3 loops of the D11.15 and Ig12 components (colored as in A and B) to be contacting equivalent surface areas of the lysozyme antigens.
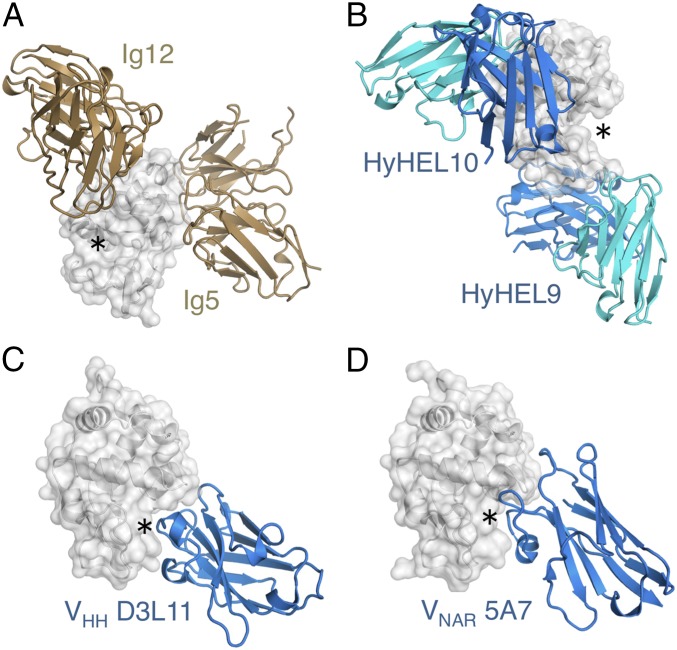
The epitope contacted by Ig122 and D11.15 corresponds to a flat part of the lysozyme surface, which is adjacent, but distinctly outside, the active site cleft (Fig. 4 A and B, the active site is indicated by asterisks); similarly, Ig52 targets a planar epitope distant from the catalytic site (Fig. 5A). Similar preferences for the binding of flat molecular surfaces are observed for other paired Ig receptors, including the model anti-HEL monoclonals HyHEL10 and HyHEL9, both of which bind away from the active site cleft (Fig. 5B). This behavior is in marked contrast to single-domain Ig formats, such as the heavy-chain-only receptors of camelids, which are naturally devoid of light chain partners (and are believed to be of relatively recent evolutionary origin) (40, 41). Crystal structures of these domains, as well as crystal structures of single domains from sharks [shark new antigen receptors (42)], have been reported in complex with HEL antigen. These previously reported structures reveal monomeric binding, with strict 1:1 stoichiometry and a strong preference for targeting the active site cleft of HEL (43, 44) (Fig. 5 C and D).
Fig. 5.
Epitope preferences of Ig receptor formats. (A) Ig5 and Ig12 homodimers (tan cartoons) target flat epitopes on the HEL surface (gray cartoon and surface), distant from the HEL active site cleft (indicated by *). (B) Flat epitope surfaces apart from the active site are also observed for the model anti-HEL monoclonals HyHEL10 (PDB ID code 1YQV) and HyHEL9 (PDB ID code 3D9A), which are paired receptors formed by VH-VL domain assembly (blue and cyan cartoons, respectively). Structures of nonpaired Ig antigen receptors of camels (C; VHH; PDB ID code 1ZVY) and sharks (D; shark new antigen receptor (VNAR); PDB ID code 1T6V) reveal a 1:1 stoichiometry with antigen, and binding in the HEL active site cleft.
The Ig52 and Ig122 complexes reported here clearly outline two distinct strategies by which symmetry mismatches can be tackled: relaxation of symmetry through structural side-chain plasticity and differential recruitment of CDR loops. Such mechanisms are not dissimilar to what has been observed in other cases of symmetrical receptors bound to asymmetrical protein ligands, as exemplified by the structures of the lectin-like homodimeric immunoreceptor NKG2D (natural-killer group 2, member D) bound to three different signaling proteins (45–47). Although these receptors are not Ig-based, they nonetheless structurally and functionally mimic the T-cell receptor Vα-Vβ pairing, recognizing ligands structurally related to MHC counterparts. In these structures, both receptor conformational plasticity and selective recruitment are used, and are supplemented, particularly in the case of one of the ligands, by significant conformational rearrangement of the ligand itself relative to its receptor-free state (ligand hyperplasticity) (45). This observation is in contrast to the two structures reported here, where the structure of the HEL ligand is strongly conserved, with only minor differences observed at surface loop positions. Also, it is also important to note that, unlike antibody–antigen interactions, the NKG2D–ligand complexes are not the products of adaptive immunity, but rather have coevolved over evolutionary time spans.
Differences to Naturally Occurring Bence-Jones Proteins.
The Ig5 and Ig12 receptors generated here were selected from phage display repertoires based on common human light-chain segments (DPK9 Vκ). Consequently, overall structures of the receptors and interdomain pairing closely resemble the homodimeric arrangement of VL domains observed in structures of Bence-Jones proteins derived from patients with multiple myeloma (25–27, 48). However, outside the VL domain–domain interface, the similarities between the in vitro selected receptors and naturally occurring Bence-Jones protein begin to fade. This behavior is exemplified by previously reported structures of the Bence-Jones protein MCG, which has been crystallized with a series of synthetic peptides (Tables S4 and S5). As had been observed for the NKG2D structure, but in contrast to the structures reported here, the capacity of MCG to bind to a range of peptides of increasing length is underpinned by both the plasticity within the antibody-binding site and the conformational malleability of the peptide antigens (29, 49) (Tables S4 and S5). The MCG/peptide interface is small, with buried surface areas ranging from 462–557 Å2 compared with 691–830 Å2 observed for the Ig12 and Ig5 complexes. Moreover, MCG interactions are strongly dominated by hydrophobic contacts, with limited hydrogen bonds and no salt bridges observed (Tables S4 and S5). Several of the hydrophobic interactions in the MCG peptide complex are mediated by antibody framework residues, which do not commonly interact with antigen in antibody–antigen complexes (Fig. S7). Overall, the MCG/peptide structures therefore outline a picture of a hydrophobic antibody pocket into which a wide range of nonpolar ligands can be soaked in a promiscuous fashion. Although details have not been reported, it is likely that affinities of the MCG/peptide are extremely low (micromolar), reflecting the noncognate nature of the ligands and the absence of adaptive selection (29, 49).
Table S4.
Buried surface area and contact details for Ig5–HEL, Ig12–HEL, D11.15–PEL, and MCG–peptide complexes
| Protein | VL12 | VL5 | D11.15 | MCG | MCG | |||||
| Name | Human VL | Human VL | Mouse Fab | Human LC | Human LC | |||||
| Framework | Vκ1 | Vκ1 | VH1 | Vκ16 | Vλ2 | Vλ2 | ||||
| PDB code | 4N1C | 4N1E | 1JHL | 1MCB | 1MCC | |||||
| Binding partner | HEL | HEL | PEL | Ac-QfHp-OH | Ac-QfHp-NH2 | |||||
| Affinity, nM) | 64 | 130 | 15 | |||||||
| Shape complementarity* | 0.667 | 0.725 | 0.635 | 0.633 | 0.703 | |||||
| Chain | A | B | A | B | H | L | A | B | A | B |
| Hydrogen bonds† | 6 | 6 | 8 | 6 | 7 | 2 | — | 1 | 1 | 1 |
| Salt bridges† | 1 | — | 1 | 2 | 2 | — | — | — | — | — |
| BSA antibody† | 691 | 830 | 638 | 462 | 385 | |||||
| Total per chain | 285 | 406 | 363 | 467 | 439 | 200 | 283 | 179 | 208 | 176 |
| CDR1 | 67 | 129 | 68 | 109 | 117 | 37 | 67 | 29 | 75 | 56 |
| CDR2 | 82 | 56 | 31 | 93 | 18 | 21 | 3 | 6 | ||
| CDR3 | 218 | 184 | 190 | 242 | 173 | 158 | 139 | 96 | 97 | 75 |
| Framework | 11 | 49 | 86 | 56 | 5 | 59 | 33 | 33 | 39 | |
Table S5.
Buried surface area and contact details for Ig5–HEL, Ig12–HEL, D11.15–PEL, and MCG–peptide complexes
| Protein | MCG | MCG | MCG | MCG | ||||
| Name | Human LC | Human LC | Human LC | Human LC | ||||
| Framework | Vλ2 | Vλ2 | Vλ2 | Vλ2 | ||||
| PDB ID code | 1MCD | 1MCE | 1MCF* | 1MCH* | ||||
| Binding partner | Ac-fβHp-NH2 | Ac-QfHpβ-OH | Ac-QfHpββ-OH | Ac-QfHpββ-OH | ||||
| Shape complementarity† | 0.537 | 0.499 | 0.671 | 0.637 | ||||
| Chain | A | B | A | B | A | B | A | B |
| Hydrogen bonds‡ | 1 | — | 3 | — | 3 | 2 | 4 | 2 |
| Salt bridges‡ | — | — | — | — | — | — | — | — |
| BSA antibody‡ | 353 | 499 | 530 | 557 | ||||
| Total per chain | 157 | 196 | 297 | 202 | 306 | 224 | 284 | 273 |
| CDR1 | 36 | 72 | 93 | 81 | 88 | 94 | 64 | 63 |
| CDR2 | 13 | 6 | 3 | 6 | 2 | 20 | 26 | |
| CDR3 | 111 | 77 | 119 | 51 | 153 | 53 | 124 | 121 |
| Framework | 10 | 35 | 52 | 66 | 58 | 75 | 76 | 64 |
Fig. S7.
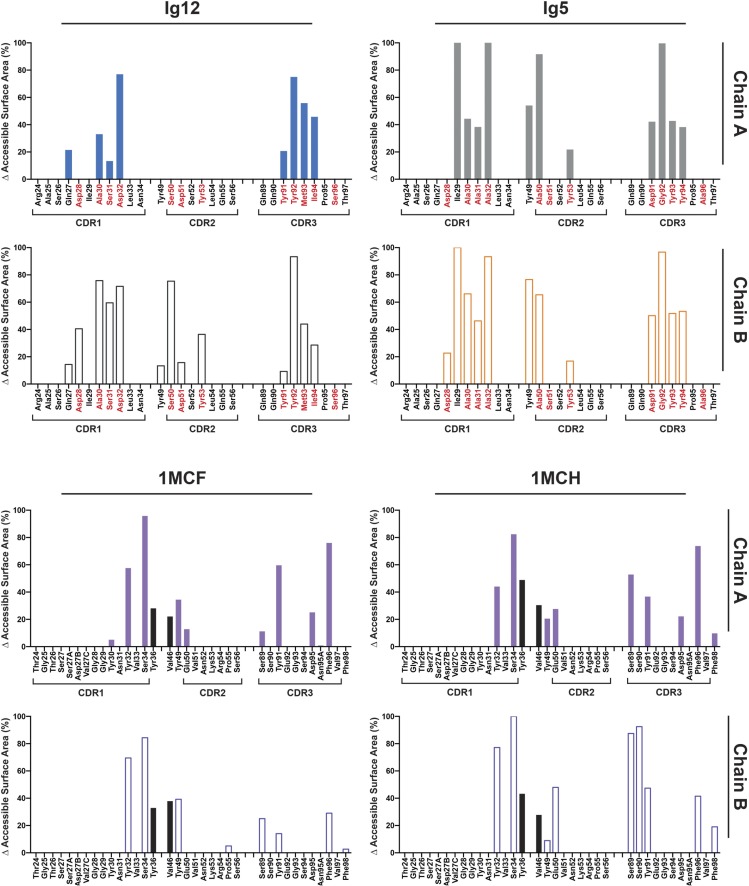
Accessible surface areas buried by contact with ligand. Percentages of accessible surface areas buried by ligand contact are displayed for residues involved in ligand binding. These area percentages are plotted for Ig12 and Ig5 (Top) and for two MCG structures (Bottom) containing the same ligand either soaked (PDB ID code 1MCF) or cocrystallized (PDB ID code 1MCH). The MCG structures use some contacts (black bars) in positions outside those contact positions normally observed in VH–VL ligand complexes.
In marked contrast, the Ig5 and Ig12 structures outlined here are characterized by affinities that are orders of magnitude higher (nanomolar) and an extensive interface encompassing numerous hydrogen bond and charge interactions (Tables S3–S5), reflecting selective pressure and their origins from combinatorial immune repertoires.
Conclusion
For many protein families, evolutionary ancestry can be reconstructed through analyses of modern-day sequences from a range of organisms. However, in other cases, evolutionary traces have been obscured through mutation and divergence, thereby hindering sequence-based reconstruction. Nevertheless, it is evident that, even in such cases, structural features of proteins are often highly conserved throughout evolution, despite the absence of detectable sequence identity (50, 51). Here, we have used this characteristic, by selecting antigen receptors resembling the fold and homodimeric nature of ancestral proteins from a repertoire of structural Ig building blocks, followed by biophysical confirmation and structure determination. Much as experimental archeology has allowed the validation of our perceptions of ancient materials and machinery (52, 53), the approach outlined here provides insights into molecular mechanisms that are believed to have shaped antigen receptor evolution. More specifically, the structures reported here provide definite validation that a symmetrical receptor is capable of interacting with asymmetrical ligands in an adaptive fashion, not only in principle but also in a specific and high-affinity manner typical of bona fide antibody–antigen interactions.
Methods
Library design and construction, selection of antigen binders, determination of binding stoichiometry, affinity measurements and crystal growth, structure determination and refinement, and phylogenetic analysis are described in detail in SI Text.
Supplementary Material
Acknowledgments
We thank the staff at the Australian Synchrotron MX2 beamline. This work was supported by the Australian National Health and Medical Research Council and the Australian Research Council (Grants DP140103465, DE160100608, APP1050146, and APP1022688).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4N1E and 4N1C).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613477114/-/DCSupplemental.
References
- 1.Zinkernagel RM, Hengartner H. Regulation of the immune response by antigen. Science. 2001;293(5528):251–253. doi: 10.1126/science.1063005. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242(4):309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18(1):1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 4.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124(4):815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Eason DD, et al. Mechanisms of antigen receptor evolution. Semin Immunol. 2004;16(4):215–226. doi: 10.1016/j.smim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Das S, Nikolaidis N, Klein J, Nei M. Evolutionary redefinition of immunoglobulin light chain isotypes in tetrapods using molecular markers. Proc Natl Acad Sci USA. 2008;105(43):16647–16652. doi: 10.1073/pnas.0808800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Criscitiello MF, Flajnik MF. Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol. 2007;37(10):2683–2694. doi: 10.1002/eji.200737263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards MH, Nelson JL. The evolution of vertebrate antigen receptors: A phylogenetic approach. Mol Biol Evol. 2000;17(1):146–155. doi: 10.1093/oxfordjournals.molbev.a026227. [DOI] [PubMed] [Google Scholar]
- 9.Cooper MD, Herrin BR. How did our complex immune system evolve? Nat Rev Immunol. 2010;10(1):2–3. doi: 10.1038/nri2686. [DOI] [PubMed] [Google Scholar]
- 10.Herrin BR, Cooper MD. Alternative adaptive immunity in jawless vertebrates. J Immunol. 2010;185(3):1367–1374. doi: 10.4049/jimmunol.0903128. [DOI] [PubMed] [Google Scholar]
- 11.Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505(7482):174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–157. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 13.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat Rev Genet. 2010;11(1):47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litman GW, et al. Phylogenetic diversification of immunoglobulin genes and the antibody repertoire. Mol Biol Evol. 1993;10(1):60–72. doi: 10.1093/oxfordjournals.molbev.a040000. [DOI] [PubMed] [Google Scholar]
- 15.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453(7199):1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8(4):R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schubert WD, et al. A common ancestor for oxygenic and anoxygenic photosynthetic systems: A comparison based on the structural model of photosystem I. J Mol Biol. 1998;280(2):297–314. doi: 10.1006/jmbi.1998.1824. [DOI] [PubMed] [Google Scholar]
- 18.Hill RL, Delaney R, Fellows RE, Lebovitz HE. The evolutionary origins of the immunoglobulins. Proc Natl Acad Sci USA. 1966;56(6):1762–1769. doi: 10.1073/pnas.56.6.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3(10):e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewert S, Huber T, Honegger A, Plückthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325(3):531–553. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 21.Dudgeon K, et al. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc Natl Acad Sci USA. 2012;109(27):10879–10884. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouet R, Lowe D, Christ D. Stability engineering of the human antibody repertoire. FEBS Lett. 2014;588(2):269–277. doi: 10.1016/j.febslet.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 23.Bernier GM, Putnam FW. Monomer–dimer forms of Bence Jones proteins. Nature. 1963;200:223–225. doi: 10.1038/200223b0. [DOI] [PubMed] [Google Scholar]
- 24.Novotný J, Haber E. Structural invariants of antigen binding: Comparison of immunoglobulin VL-VH and VL-VL domain dimers. Proc Natl Acad Sci USA. 1985;82(14):4592–4596. doi: 10.1073/pnas.82.14.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colman PM, Schramm HJ, Guss JM. Crystal and molecular structure of the dimer of variable domains of the Bence-Jones protein ROY. J Mol Biol. 1977;116(1):73–79. doi: 10.1016/0022-2836(77)90119-x. [DOI] [PubMed] [Google Scholar]
- 26.Fehlhammer H, et al. The structure determination of the variable portion of the Bence-Jones protein Au. Biophys Struct Mech. 1975;1(2):139–146. doi: 10.1007/BF00539775. [DOI] [PubMed] [Google Scholar]
- 27.Epp O, et al. Crystal and molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI. Eur J Biochem. 1974;45(2):513–524. doi: 10.1111/j.1432-1033.1974.tb03576.x. [DOI] [PubMed] [Google Scholar]
- 28.Edmundson AB, et al. Binding of 2,4-dinitrophenyl compounds and other small molecules to a crystalline lambda-type Bence-Jones dimer. Biochemistry. 1974;13(18):3816–3827. doi: 10.1021/bi00715a031. [DOI] [PubMed] [Google Scholar]
- 29.Edmundson AB, et al. Principles and pitfalls in designing site-directed peptide ligands. Proteins. 1993;16(3):246–267. doi: 10.1002/prot.340160304. [DOI] [PubMed] [Google Scholar]
- 30.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 31.Thornton JW. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat Rev Genet. 2004;5(5):366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 32.Tomlinson IM, Cox JP, Gherardi E, Lesk AM, Chothia C. The structural repertoire of the human V kappa domain. EMBO J. 1995;14(18):4628–4638. doi: 10.1002/j.1460-2075.1995.tb00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabat E, Wu TT, Perry HM, Kay S, Gottesman CF. 1992. Sequences of Proteins of Immunological Interest (DIANE Publishing, Collingdale, PA), 5th Ed.
- 34.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature. 1990;348(6301):552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 35.Ye H. Simultaneous determination of protein aggregation, degradation, and absolute molecular weight by size exclusion chromatography-multiangle laser light scattering. Anal Biochem. 2006;356(1):76–85. doi: 10.1016/j.ab.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 36.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: Contact analysis and binding site topography. J Mol Biol. 1996;262(5):732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 37.Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proc Natl Acad Sci USA. 1996;93(1):7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chitarra V, et al. Three-dimensional structure of a heteroclitic antigen-antibody cross-reaction complex. Proc Natl Acad Sci USA. 1993;90(16):7711–7715. doi: 10.1073/pnas.90.16.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217(1):133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 40.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 41.Rouet R, Dudgeon K, Christie M, Langley D, Christ D. Fully human VH single domains that rival the stability and cleft recognition of camelid antibodies. J Biol Chem. 2015;290(19):11905–11917. doi: 10.1074/jbc.M114.614842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenberg AS, et al. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374(6518):168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 43.De Genst E, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA. 2006;103(12):4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science. 2004;305(5691):1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 45.Li P, et al. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2(5):443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 46.Li P, McDermott G, Strong RK. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity. 2002;16(1):77–86. doi: 10.1016/s1074-7613(02)00258-3. [DOI] [PubMed] [Google Scholar]
- 47.Radaev S, Rostro B, Brooks AG, Colonna M, Sun PD. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity. 2001;15(6):1039–1049. doi: 10.1016/s1074-7613(01)00241-2. [DOI] [PubMed] [Google Scholar]
- 48.Edmundson AB, Ely KR, Abola EE, Schiffer M, Panagiotopoulos N. Rotational allomerism and divergent evolution of domains in immunoglobulin light-chains. Biochemistry. 1975;14:3953–3961. [PubMed] [Google Scholar]
- 49.Edmundson AB, Manion CV. Treatment of osteoarthritis with aspartame. Clin Pharmacol Ther. 1998;63(5):580–593. doi: 10.1016/S0009-9236(98)90109-6. [DOI] [PubMed] [Google Scholar]
- 50.Christ D, Winter G. Identification of functional similarities between proteins using directed evolution. Proc Natl Acad Sci USA. 2003;100(23):13202–13206. doi: 10.1073/pnas.2134365100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391(6663):203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 52.Reibold M, et al. Materials: Carbon nanotubes in an ancient Damascus sabre. Nature. 2006;444(7117):286. doi: 10.1038/444286a. [DOI] [PubMed] [Google Scholar]
- 53.Juleff G. An ancient wind powered iron smelting technology in Sri Lanka. Nature. 1996;379(6550):60. [Google Scholar]
- 54.Hoogenboom HR, et al. Multi-subunit proteins on the surface of filamentous phage: Methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19(15):4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouet R, Dudgeon K, Christ D. Generation of human single domain antibody repertoires by Kunkel mutagenesis. Methods Mol Biol. 2012;907:195–209. doi: 10.1007/978-1-61779-974-7_10. [DOI] [PubMed] [Google Scholar]
- 57.Lee CM, Iorno N, Sierro F, Christ D. Selection of human antibody fragments by phage display. Nat Protoc. 2007;2(11):3001–3008. doi: 10.1038/nprot.2007.448. [DOI] [PubMed] [Google Scholar]
- 58.Rouet R, et al. Expression of high-affinity human antibody fragments in bacteria. Nat Protoc. 2012;7(2):364–373. doi: 10.1038/nprot.2011.448. [DOI] [PubMed] [Google Scholar]
- 59.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 61.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 66.de Castro E, et al. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lefranc MP, et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol. 2005;29(3):185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 71.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 72.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 7):1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.