Significance
Cell response to physiological changes and oxidative stress involves the modulation of mitochondrial metabolism. Its dysfunction favors the development of hypoxia-dependent pathologies, including ischemia and cancer. A key modulator of mitochondrial activity is cytochrome c, whose cell function is regulated by tyrosine phosphorylation. However, how such modification affects cytochrome c structure and function is barely known. Here we report that a phosphomimetic mutant of cytochrome c exhibits enhanced dynamics, which could be responsible for the observed differences in cytochrome c functionality in oxidative stress and cell death. Thus, phosphorylation of cytochrome c becomes a target for further development of robust therapeutic approaches.
Keywords: cytochrome c, mitochondrial dysfunction, nuclear magnetic resonance, phosphorylation, respiratory supercomplexes
Abstract
Regulation of mitochondrial activity allows cells to adapt to changing conditions and to control oxidative stress, and its dysfunction can lead to hypoxia-dependent pathologies such as ischemia and cancer. Although cytochrome c phosphorylation—in particular, at tyrosine 48—is a key modulator of mitochondrial signaling, its action and molecular basis remain unknown. Here we mimic phosphorylation of cytochrome c by replacing tyrosine 48 with p-carboxy-methyl-l-phenylalanine (pCMF). The NMR structure of the resulting mutant reveals significant conformational shifts and enhanced dynamics around pCMF that could explain changes observed in its functionality: The phosphomimetic mutation impairs cytochrome c diffusion between respiratory complexes, enhances hemeprotein peroxidase and reactive oxygen species scavenging activities, and hinders caspase-dependent apoptosis. Our findings provide a framework to further investigate the modulation of mitochondrial activity by phosphorylated cytochrome c and to develop novel therapeutic approaches based on its prosurvival effects.
Oxidative phosphorylation (OxPhos) relies on the electron transport chain (ETC) to generate the membrane potential that drives ATP synthesis (1). Components of the ETC oxidize substrates to reduce molecular oxygen, thereby producing water. Nevertheless, around 2% of the electrons flowing across the ETC yield reactive oxygen species (ROS) (2). ROS are a source of oxidative stress and act as signaling molecules at low concentrations (3–5). Hindering redox reactions within the distinct ETC membrane complexes (complexes I to V) leads to enhanced ROS production (6). The activity of the ETC is tightly regulated by posttranslational modifications of its components, isoform swapping, and modulation of the equilibria for the association of the membrane protein complexes into supercomplexes (3, 7). Such associations allow substrate channeling while modulating ROS production (3, 7).
Oxidative stress response involves redox signals modulating protein phosphorylation (8). In the ETC, the major phosphorylation targets are NADH:UQ oxidoreductase, cytochrome c (Cc), and cytochrome c oxidase (CcO) (9). Besides being essential for oxidative respiration (Fig. 1A), Cc acts as a redox regulatory protein within the mitochondrial intermembrane space (10). In addition, Cc aids in controlling ROS levels by oxidizing superoxide anions (11) and exhibiting peroxidase activity (12); the latter, however, also yields lipid peroxidation (13, 14). Furthermore, Cc plays a crucial role in programmed cell death, a process that is only partially understood (15–21). In this context, a fraction of Cc binds and oxidizes cardiolipin (CL) at the internal mitochondrial membrane, thereby facilitating the release of unbound Cc to the cytoplasm (16, 17). In mammalian cells, extramitochondrial Cc interacts with apoptosis activating factor 1 (Apaf-1) in the cytoplasm to spark the caspase proteolytic cascade (15). It has recently been shown that Cc can interact with several other proteins outside the mitochondria in humans and plants (18–21). The similarities between the Cc signaling networks in both organisms suggest that key programmed cell-death pathways are conserved throughout evolution (22).
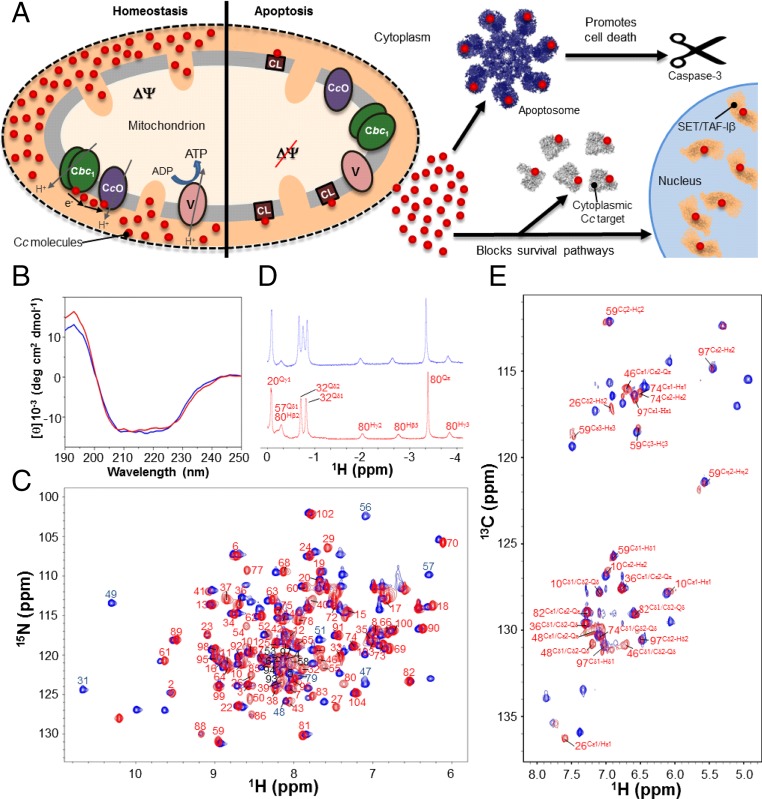
Fig. 1.
Control of human cell fate by the Cc-based signalosome, and biophysical and structural characterization of the Y48pCMF variant of Cc. (A) Diagram of the role of Cc in homeostasis and apoptosis. (A, Left) Under homeostatic conditions, Cc (red circles) transfers electrons from the cytochrome bc1 complex (Cbc1) to the cytochrome c oxidase complex (CcO). (A, Right) Upon apoptotic stimuli, Cc is released to the cytosol to induce apoptosome formation and block prosurvival pathways. A portion of Cc remains bound to cardiolipin. (B) Far-UV CD spectra of the reduced forms of WT (blue) and Y48pCMF Cc (red). The same color code is maintained in the following panels. (C) Superimposition of the 1H–15N HSQC spectra of uniformly 15N-labeled forms of WT and Y48pCMF Cc. Backbone amide resonances of Y48pCMF Cc are labeled in red and black. Particular amide resonances of WT Cc are labeled in blue. (D) Detailed view of the 1H NMR spectra of WT and Y48pCMF Cc at negative ppm values. Resonances for Met80 side-chain protons are shown for both Cc species. Assigned signals of all residues within this region are displayed for Y48pCMF Cc. The extra signal of WT Cc corresponds to the Qδ1 protons of Ile53. (E) Superimposition of the aromatic region of the 1H–13C HSQC spectra of WT and Y48pCMF Cc acquired in 13C natural abundance. Assigned aromatic resonances of Y48pCMF Cc are displayed in red.
Cc phosphorylation is an alleged modulator of the mitochondrial cell-death pathway (23, 24). Its deregulation is believed to be related to the onset of neurological disorders and cancer (24). Phosphorylation of Cc is easily reversed by phosphatases, hampering the isolation of the modified protein from tissues (25). Tyr-to-Glu substitutions of Cc, designed to emulate Tyr48 phosphorylation, impair both the electron donation to CcO and the activation of caspase-9 mediated by Apaf-1 in vitro (26, 27). Notably, nitration of this residue also hinders the ability of Cc to activate Apaf-1 (28–30). Thus, these posttranslational modifications alter both mitochondrial and cytoplasmic functions of Cc. Tyr48 phosphorylation also affects the physical-chemical properties of Cc, such as its alkaline transition and midpoint reduction potential (E0) (27, 31). Understanding the origin of these effects and how they affect Cc activities requires solving the 3D conformation of the phosphorylated species. However, deciphering the effects of Tyr48 phosphorylation on the structure and dynamics of Cc is highly challenging because of its reactivity toward phosphatases. Indeed, no atomic-resolution structure has been reported for either phosphorylated Cc or any reliable mimic mutant.
Here we have elucidated the structure of a phosphomimetic Cc variant in which Tyr48 is replaced by the synthetic, noncanonical amino acid p-carboxy-methyl-l-phenylalanine (pCMF). We show that such replacement induces local perturbations of the Cc structure and enhanced internal dynamics of the mutation’s surroundings. We used biochemical assays to show that the Y48pCMF mutation impairs Cc channeling between cytochrome bc1 (Cbc1) and CcO, enhances peroxidase activity, and induces an antiapoptotic function of Cc.
Results
Phosphorylation of Tyr48 Induces Local Structural Changes in Cytochrome c.
To understand how phosphorylation affects the structure of human Cc, we tackled the challenge of fully characterizing the phosphomimetic mutant Y48pCMF Cc in its reduced form, which is the redox state of Cc donating electrons to CcO in homeostasis and is essential for its apoptotic activity, because Cc becomes highly reduced upon its release from the mitochondria to the cytosol (32). Y48pCMF Cc maintains the overall secondary structure and global fold of wild-type (WT) Cc, as inferred from circular dichroism (CD) (Fig. 1B) and 1H–15N heteronuclear single-quantum correlation (HSQC) NMR spectra (Fig. 1C), respectively. Also, the native heme axial coordination was preserved, as indicated by 1D 1H NMR data (Fig. 1D). The NMR spectra of reduced Y48pCMF Cc were extensively assigned: Triple-resonance experiments (SI Appendix, Table S1) allowed us to detect and assign 91 backbone amide signals and the sequential connectivities for most residues of the protein. Aliphatic side-chain signals were assigned using 3D HBHA(CO)NH and 3D HCCH-total correlation spectroscopy (TOCSY) experiments, leading to the assignment of most (96.4%) of the side-chain proton resonances. The 15N–H resonance of pCMF48 was undetectable because the residue was not 15N-enriched. Four proline residues (Pro30, Pro44, Pro71, and Pro76) interrupt the sequential HN–HN connectivity. Additionally, in contrast to WT Cc, we could not detect the amide protons of the Thr49, Ala51, Gly56, and Ile57 residues. However, the WT spectra only had four amide protons that could not be detected (Gly1, Glu21, Thr28, and Gly84) under similar experimental conditions. Further, in contrast to WT Cc, the signals from Asn31, Gly45, and Ser47 in pCMF48 Cc were significantly weaker than the rest. The largest chemical-shift perturbations for backbone amides induced by the modified residue at position 48 are confined to nearby residues (SI Appendix, Fig. S1).
Assigning 1H resonances to the heme substituents and 1H (and 13C) signals to the aromatic side chains was required for analyzing additional 2D maps (i.e., COSY and 1H–15N NOESY) (SI Appendix, Table S1). Assignment of the Cδ1, Cδ2, Cε1, and Cε2 aromatic signals from the pCMF48 side chain required the acquisition of an aromatic 1H–13C HSQC spectrum recorded in a natural abundance of 13C (Fig. 1E). The assignment of the pCMF48 side-chain signals involved the complete assignment of the 2D 1H–15N NOESY.
Numerous residues (Val20 to Asn31, Thr40 to Trp59, and Ile75 to Glu90) displayed signals attributable to a second, minor protein conformation, which had a 1:10 ratio in intensity. Hereafter, only the major form was considered for structure calculations.
We assigned 96% of all the 1H signals for the major form. Structural information derived from 2D and 3D NOESY maps supported the presence of five α-helical regions (labeled α1 to α5) with typically strong HN–HN (i, i+1), medium-range Hα–HN (i, i+3), and Hα–HN (i, i+4) interresidual NOEs. These regions spanned the sequence stretches Val3 to Lys13, Ala50 to Asn54, Glu61 to Glu69, Pro71 to Tyr74, and Lys88 to Thr102, resembling those in the NMR structure of reduced WT Cc (33). In total, we observed and assigned 2,176 meaningful NOEs, corresponding to 20.8 relevant restraints per residue on average (SI Appendix, Fig. S2). The 71 ϕ and 71 ψ dihedral-angle constraints were derived from 15N, 13C′, 13Cα, 13Cβ, and Hα chemical shifts, using TALOS+ (34). The heme moiety was included in the calculations as previously reported for WT cytochromes (35, 36), assuming an intact heme iron coordination as supported by X-ray absorption spectroscopy (XAS) data (next section).
Two hundred structures were calculated by CYANA (37), and the 20 structures with the lowest target-function (TF) value were selected to form a representative family. The range of TF values was 0.38 to 0.98 Å2, highlighting the high accuracy between calculated and experimental distances. Further refinement of the 20 lowest TF structures involved restrained energy minimization and restrained molecular dynamics (RMD) computations. A final restrained energy minimization was carried out on the structure with the lowest root-mean-square deviation (rmsd) from the average for each of the 20 trajectories. The overall quality of the 20 lowest TF ensembles was good, according to the PROCHECK G factor (38), MolProbity clashscores (39), and other structural quality indicators (SI Appendix, Table S2). Most residues were in the Ramachandran plot favored regions, whereas Cys14, Cys17, His18, Val20, Lys37, and Asn70 were in the generously allowed ones, as also observed for the solution structure of WT Cc (33, 40). Cys14, Cys17, and His18 are covalently bonded to the heme moiety, thereby straining their backbone conformation, as already described for c-type cytochromes (35, 36). As an additional control, we performed 20-ns unrestrained molecular dynamics (MD) simulations of the final minimized conformers without any geometrical restraint. The rmsd for the main-chain atoms was about 1.55 Å at the plateau, hardly drifting (0.122 pm⋅ns–1), as expected for a stable structure (SI Appendix, Fig. S3).
The overall fold of Y48pCMF Cc is very similar to that of the WT species (SI Appendix, Fig. S4), with rmsd values for the backbone nuclei of 1.67 ± 1.01 Å (Fig. 2 A and B). However, the two structures differ in the mutation-containing loop ΩNY (residues 40 to 57; Fig. 2B). This is consistent with the chemical-shift differences (SI Appendix, Fig. S1) and the rmsd between the NMR solution structure of WT Cc [Protein Data Bank (PDB) ID code 1J3S] (33) and the refined lowest-TF Y48pCMF Cc (SI Appendix, Fig. S5A). An increased dynamics in the ΩNY-loop has recently been observed in the G41S variant of Cc (42). The ΩNY-loop (also known as foldon V) and helix α2 constitute the less-stable folding unit of Cc (43). Unlike Tyr48 in the WT species, the pCMF48 residue presents a very low number of 1H–1H NOEs (SI Appendix, Fig. S2) and a high rmsd value within the family (Fig. 2C and SI Appendix, Fig. S5B). A decrease in the number of detectable NOEs can generally be attributed to either a partial assignment of the residue or an increased internal mobility. In our case, the highly dynamic behavior of the pCMF48 residue was confirmed from further analysis of internal motions within the nanosecond-to-picosecond timescale (see below).
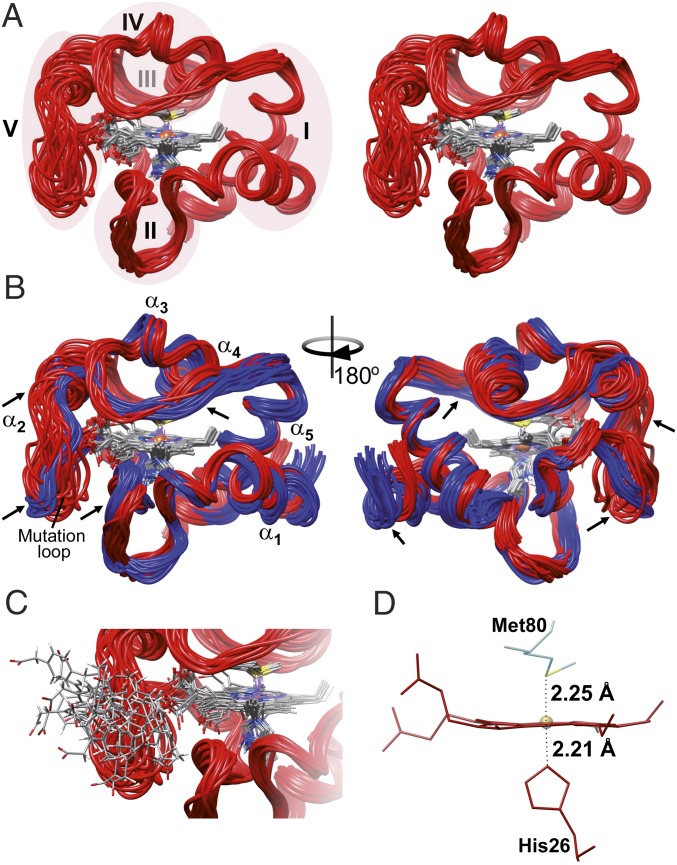
Fig. 2.
NMR solution structure of the Y48pCMF variant of Cc. (A) Stereoview ribbon representation of the 20 best conformers of Y48pCMF Cc. Heme group atoms are displayed for all conformers. Ribbons are colored in red, whereas atoms from the heme group are colored following the CPK (Robert Corey, Linus Pauling, and Walter Koltun) color scheme. Foldons of Y48pCMF Cc are shadowed and marked with roman numerals, except for foldon III, which is located behind foldon IV. (B) Comparison between the NMR solution structures of WT Cc (PDB ID code 1J3S) (33) and Y48pCMF Cc (this work). The ribbon for WT Cc is in blue. The five α-helices of both Cc species, as well as the mutation-containing loop of Y48pCMF Cc, are marked. Arrows point to the regions on the Y48pCMF Cc ribbon with substantial structural changes compared with the WT form. (C) Detailed view of the loop harboring the pCMF48 residue. pCMF48 atoms follow the CPK color scheme. Protein structures are presented by UCSF Chimera software (41). (D) Detail of the heme group and axial ligands. Labels display iron-to-axial ligand distances for the Y48pCMF mutant obtained from the EXAFS analysis (SI Appendix, Fig. S5).
Other regions of the Y48pCMF Cc structure that differed from those of WT Cc belong to the 19 to 36 ΩG-loop (part of foldon II) and the 71 to 85 ΩR-loop (foldon IV) comprising Met80 and helix α4. The rmsd values for backbone atoms of residues from Val20 to Gly29 of Y48pCMF Cc are higher (2.75 ± 1.50 Å) than those for the same residues of WT Cc (SI Appendix, Fig. S5A). In the second loop, the observed differences are mainly restricted to side chains. In fact, the rmsd values for backbone and heavy atoms are 1.18 ± 0.37 and 1.77 ± 0.71 Å, respectively. Residues included in the ΩR-loop indeed displayed a double conformation, suggesting the presence of conformational equilibria (SI Appendix, Fig. S5A).
The ensemble of structures for Y48pCMF Cc is very precise, except for residues surrounding the mutation. The backbone rmsd from the mean is 0.89 ± 0.01 Å for the whole protein and drops to 0.53 ± 0.12 Å when the mutation surroundings are excluded (residues 40 to 57) (SI Appendix, Fig. S5B). As expected, the highest rmsd values correspond to the 40 to 57 ΩNY-loop and the nearby residues Val20 to Gly29 in loop ΩG, whose global backbone rmsd values were 1.79 ± 0.53 and 0.78 ± 0.23 Å, respectively. These segments also exhibited a larger conformational variability in their secondary structure elements along the MD trajectory (SI Appendix, Fig. S3 B and C). High rmsd values along the Val20-to-Gly29 stretch are typical in Cc homologs (35, 36). They are also consistent with NMR signals revealing secondary conformations for the His26-to-Pro30 stretch. Moreover, the nearby Asn31 amide signal is weak, suggesting high mobility. Notably, all these residues contact the ΩNY-loop, comprising the Y48pCMF mutation.
Further, the highest rmsd values mapped to the ΩNY-loop. Consistently, signals from this region undergo a drastic reduction in their 1H–1H NOE cross-peaks. The intensities of the amide signals of Gly45 and Ser47 in Y48pCMF Cc were severely decreased, whereas those from Thr49, Ala51, Gly56, and Ile57 were undetectable. The ΩR-loop also contains some residues with high rmsd values (SI Appendix, Fig. S5B). The global rmsd value for backbone atoms of the 71 to 85 ΩR-loop is 0.76 ± 0.20 Å. Notably, the end of the ΩR-loop shows high rmsd values in WT cytochromes (35, 36).
Heme Iron Coordination Is Insensitive to Tyr48 Phosphorylation.
We tested the effects of the mutation on the heme iron coordination environment and the axial coordination restraints used in our structure computations by using X-ray absorption spectroscopy, studying both WT and Y48pCMF species (Fig. 2D and SI Appendix, Fig. S5 C–F). The absorption spectra of the two proteins are almost identical. The X-ray absorption near-edge structure regions of the absorption spectra for both proteins are superimposed (SI Appendix, Fig. S5C). The absence of any shift in the energy position of the absorption edge indicates that the mutation did not affect the electron density at the Fe center.
Likewise, the extracted extended X-ray absorption fine structure (EXAFS) signals of the WT and Y48pCMF Cc species were highly similar. However, small differences in high wave vector (k) values can be observed (SI Appendix, Fig. S5D), as well as in the slightly lower amplitude of the Y48pCMF protein. Whereas the corresponding Fourier transforms are also very similar (SI Appendix, Fig. S5E), the amplitude of the first peak is lower and broader in the Y48pCMF Cc species, suggesting a larger degree of disorder. The dynamic disorder for the two proteins should be similar, as the measurements were performed at cryogenic temperatures in both cases. Hence, the differences in disorder are due to a larger static disorder in Y48pCMF Cc. The main scattering paths contributing to this peak originate from the four nitrogen atoms of the porphyrin ring, although contributions of the nitrogen and sulfur axial ligands have also been included. The fit to the data requires the addition of the paths involving the eight porphyrin carbons closest to the iron atom, beyond the first coordination sphere. In addition, the multiple scattering paths involving these atoms were also included. Fits were performed in R space but also reproduced the spectra well in q space (SI Appendix, Fig. S5F). The parameters obtained from the best fit to the data revealed that the distances between the iron atom and its first coordinating ligands are insensitive to the Y48pCMF mutation (SI Appendix, Table S3). Specifically, the distances from the iron center to the axial S ligand are 2.26 ± 0.001 and 2.25 ± 0.001 Å in the WT and phosphomimetic mutant species, respectively (Fig. 2D). Data analyses also showed that the value for the Debye–Waller factor corresponding to the path involving the four porphyrin nitrogen atoms increased from 0.0012 ± 0.0006 Å2 in the WT species to 0.003 ± 0.001 Å2 in the Y48pCMF mutant. These data are consistent with the preserved chemical-shift pattern of the iron axial ligands and heme substituents observed in 1D 1H NMR (Fig. 1D), which have been reported to be sensitive indicators of the heme iron electronic structures (35, 36). The pattern of the observed NOEs for the heme substituents also supports an overall intact heme pocket, with the exception of the mutation site. Still, the ensemble of structures shows a small change in the orientation of Phe82 with respect to the porphyrin ring (SI Appendix, Fig. S6).
Phosphorylation of Tyr48 Enhances Internal Mobility in Cytochrome c.
NMR relaxation measurements were performed to evaluate the dynamics of WT and Y48pCMF Cc. The Y48pCMF substitution slightly affected both relaxation rate (R1 and R2) parameters (SI Appendix, Table S4). The rotational correlation time of the phosphomimetic mutant (6.96 ± 0.02 ns) was higher than that of the WT form (6.33 ± 0.02 ns), in agreement with the small increase (pnul values of ∼10−251, where pnul is the probability for the null hypothesis being the right one) in the average gyration radius, from 12.95 ± 0.07 to 13.04 ± 0.06 Å, as calculated by MD. Indeed, phosphorylation can alter protein dynamics at different timescales and cause conformational rearrangements, such as the formation of secondary conformations (44, 45).
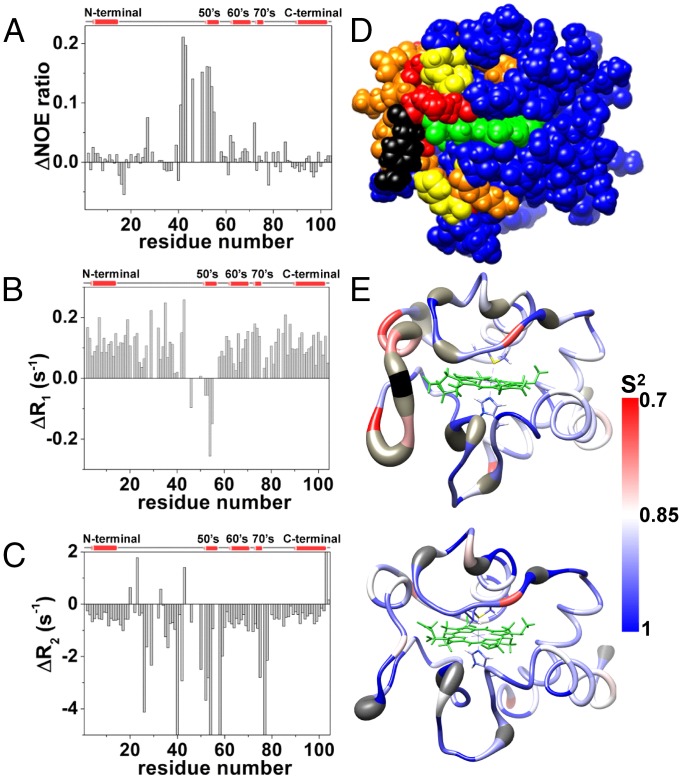
Comparing R1, R2, and heteronuclear NOE (HetNOE) relaxation measurements recorded on the two proteins revealed that the ΩNY-loop of Y48pCMF Cc exhibits a high mobility in the picosecond-to-nanosecond timescale (Fig. 3). Indeed, the Gly41-to-Lys55 segment showed a drastic drop of HetNOE values in the mutant species (Fig. 3A). Further, the amide R1 rates for the sequence stretch of Tyr46 to Lys55 in Y48pCMF Cc differ from those in WT Cc (Fig. 3B). In addition, R2 analysis reveals three regions undergoing conformational exchange in the microsecond-to-millisecond timescale: His26 to Thr28, Thr40 to Trp59, and Ile75 to Thr78 (Fig. 3C). This behavior agrees with the reduced intensity or lack of detection of the amide signals belonging to these stretches, compared with the WT form. Furthermore, some regions within the protein displayed signals corresponding to double conformations in the 2D 1H NOESY spectra—namely Val20 to Asn31, Thr40 to Trp59, and Ile75 to Glu90—indicating the presence of conformational equilibria between two different structures occurring on a slow timescale with respect to the NMR chemical shift. This dynamics involves residues located in a defined region surrounding the noncanonical amino acid (Fig. 3D). Altogether, the NMR relaxation measurements of Y48pCMF Cc agree with the per-residue S2-order parameter values computed with TENSOR (46) (Fig. 3E).
Fig. 3.
Relaxation NMR measurements and dynamic properties of WT and Y48pCMF Cc. (A–C) Differences in heteronuclear NOE (A), relaxation rate R1 (B), and relaxation rate R2 (C) between the experimental values for the reduced forms of WT and Y48pCMF Cc, plotted as a function of the residue number. Gaps in data result from overlapping resonances, broadened resonances beyond the detection limit, and unassigned resonances. A scheme of the secondary structure elements is included (Top). (D) Map of the Y48pCMF Cc residues colored according to their dynamic properties. Affected residues in the heteronuclear NOE and relaxation rate R2 parameters are colored in yellow and orange, respectively. Residues with backbone amide resonances that are undetectable in the 1H–15N HSQC spectrum of Y48pCMF Cc but detectable in the 1H–15N HSQC spectrum of WT Cc are colored in red. pCMF48 is shown in black, and the heme group is in green. Unaffected, unassigned, and proline residues are in blue. (E) Internal mobility comparison between Y48pCMF and WT Cc. S2-order parameter values per residue for Y48pCMF (Upper) and WT (Lower) Cc are represented on the respective NMR ribbon structures using a blue–red scale. Undetectable backbone resonances are in gray. Heme atoms are in green, with the axial ligands depicted as sticks.
Hydrogen–deuterium exchange experiments showed that a common core region is protected from solvent amide–hydrogen exchange in both Cc species. Nevertheless, a substantial number of amides become unprotected in pCMF48 Cc (SI Appendix, Fig. S7). The newly accessible amide protons in the phosphomimetic mutant (Gly29, Gly37, Arg38, Thr40, Gln42, and Trp59) are located in the surroundings of pCMF48 and the nearby ΩG-loop, in agreement with their high mobility in the microsecond-to-millisecond timescale.
Tyr48 Phosphorylation Modulates the Interaction of Cytochrome c with Its Mitochondrial Partners in the Electron Transfer Chain.
Cc carries electrons from Cbc1 to CcO within the ETC. To elucidate more details about the process, we analyzed the molecular recognition between the soluble N-terminal domain of plant cytochrome c1 (Cc1) or bovine CcO and the phosphomimetic human Y48pCMF Cc. The human and plant N-terminal domains of Cc1 have a 62% overall sequence identity and similar charge distributions on their molecular surfaces (47), whereas the bovine and human CcO are evolutionarily related proteins, with 91 and 96% sequence identity and homology, respectively.
Two binding sites for human and plant Cc on plant Cc1 have been recently reported (47, 48). The proximal site is located near the heme moiety and is compatible with electron transfer, whereas the distal site lies far from the heme group and probably constitutes a local energy minimum of the encounter ensemble.
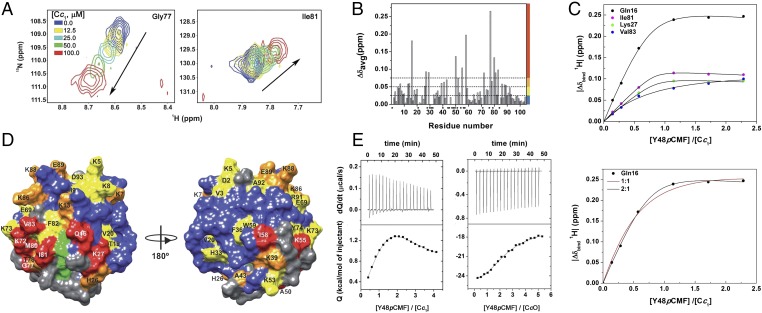
To test how Tyr48 phosphorylation affects the conformation of the Cc1–Cc complex, we recorded 1H–15N HSQC spectra upon titration of 15N-labeled reduced Cc with unlabeled reduced Cc1. Several amide signals exhibited significant chemical-shift perturbations (CSPs), thus indicating a fast exchange rate within the NMR timescale (Fig. 4 A and B). Average amide CSPs (Δδavg) were larger than 0.075 ppm for 11 residues: Gln16, Lys27, Gly29, Ala50, Lys55, Ile58, Lys72, Gly77, Met80, Ile81, and Val83 (Fig. 4B). The fits of CSPs along the titration suggested that Cc1 binds two Y48pCMF Cc molecules, as previously found with the WT Cc species (47, 48) (Fig. 4C). Using NMR spectroscopy, the dissociation constant (KD) values for the Cc1–Y48pCMF Cc complex were estimated to be 0.6 and 102 μM for the proximal and distal binding sites, respectively. All of the perturbed residues, except Lys55 and Ile58, surround the heme crevice, as previously described for the interaction between WT Cc and Cc1 (47, 48) (Fig. 4D). In fact, this region constitutes a very well conserved interaction surface in c-type cytochromes (49–52). Lys55 and Ile58, in turn, are located in the ΩNY-loop, which undergoes a conformational exchange in free Y48pCMF Cc (Fig. 4D). In addition, significant CSPs (Δδavg ≥ 0.05 ppm) were detected for Lys7, Lys13, His26, Lys39, Ala43, Thr78, Lys86, Lys88, and Glu89. Interestingly, Lys8, Lys13, Lys27, Lys72, and Lys86 also experienced large CSPs, as previously reported for the interaction between WT Cc and Cc1 (47, 48).
Fig. 4.
Binding assays between Y48pCMF Cc and its respiratory partners. (A) Overlay of selected residues of 1H–15N HSQC spectra of 15N-labeled Y48pCMF Cc along with titration with Cc1. Signals corresponding to different titration steps are colored according to the code indicated. (B) Plot of chemical-shift perturbations of 15N-labeled Y48pCMF Cc as a function of residue number. Proline and nonassigned residues are marked by asterisks. Color bars stand for the Δδavg categories: insignificant Δδavg < 0.025 ppm, blue; small 0.025 ≤ Δδavg < 0.050 ppm, yellow; medium 0.050 ≤ Δδavg < 0.075 ppm, orange; and large ≥ 0.075 ppm, red. (C, Upper) Curves representing the best global fit of several amide signals in the 1H dimension to a 2:1 ratio for the Y48pCMF Cc–Cc1 binding model with two different global KD values. (C, Lower) Binding curves of Gln16. Lines represent the best fit to 1:1 (red) and 2:1 (black) binding models. (D) CSP map of reduced Y48pCMF Cc upon addition of reduced Cc1 at a 1:1 ratio. Residues are colored according to Δδavg categories, as indicated in B. Proline and nonassigned residues are in gray. (E) ITC measurements of the Y48pCMF Cc–Cc1 and Y48pCMF Cc–CcO complexes in their reduced states. Experimental data were fitted to a 2:1 binding model. Thermograms (Upper); binding isotherms (Lower).
To obtain further data on the binding affinity and stoichiometry of the interaction between the two redox proteins, isothermal titration calorimetry (ITC) experiments were performed on both redox states. The isotherms obtained by titrating reduced Y48pCMF Cc with reduced Cc1 are displayed (Fig. 4E, Left). For the interaction between Cc and Cc1, the isotherm clearly shows two components with opposite signs in their respective enthalpy terms, thereby indicating the presence of two binding sites, in agreement with the NMR data herein presented and previous reports on the WT species (47, 48). Indeed, all data fit a model with two independent binding sites in the corresponding Cc partner (SI Appendix, Table S5). The interaction between Y48pCMF Cc and Cc1 was entropy-driven. At pH 7.4, the KD value at the proximal site was half that observed for WT Cc, whereas that at the distal site was four times higher (SI Appendix, Table S5). This could be ascribed to the extra negative charge at position 48, which alters the surface electrostatic potential of the hemeprotein (SI Appendix, Fig. S8). Both Cc1–Y48pCMF Cc and Cc1–Cc complexes in their oxidized states showed similar thermodynamic and equilibrium parameters at both acidic and basic pH values. The only exception was the KD value for the oxidized Cc1–Y48pCMF Cc complex, at the proximal site of Cc1, which was approximately four times lower than that for the oxidized Cc1–WT Cc adduct at pH 8.5. ITC measurements of the oxidized Cc1–Y48pCMF Cc complex were not run at pH 7.4 because the pKa for the alkaline transition, which is specific to the ferric state (53), shifts to neutral pH upon Tyr48 phosphorylation (31). Hence, the WT and phosphomimetic forms of oxidized Cc can differ in their sixth axial ligand at pH 7.4 (SI Appendix, Table S5).
In addition, we analyzed the binding affinity of the cross-complex between the reduced species of bovine CcO and human Y48pCMF Cc by ITC. The isotherm inflection point lies at a 2:1 Y48pCMF-Cc:CcO ratio, thereby indicating the presence of two binding sites on CcO, as previously found for WT Cc (47, 54, 55). The best fit was achieved when the model used distinct KD values for the two independent binding sites (SI Appendix, Fig. S9). The resulting isotherms likewise fit to such a model, yielding different micromolar-ranging KD values (Fig. 4E, Right and SI Appendix, Table S6) and enthalpy-driven interactions. Both the CcO proximal and distal sites had lower affinities for Y48pCMF Cc than the WT species, as previously reported (47) (SI Appendix, Table S6). Altogether, these data agree with previous steady-state kinetic analysis that demonstrated a binding model with more than one molecule of Cc per molecule of Cbc1 (56) and with kinetic-based models that proposed the existence of alternative, nonproductive binding sites for horse Cc in bovine CcO (55). In addition, direct binding studies performed by gel filtration between several mammalian Ccs and bovine CcOs evidenced a 2:1 stoichiometry, in which Cc bound to a first site with a dissociation constant within the nanomolar range and to a second site with less affinity (54).
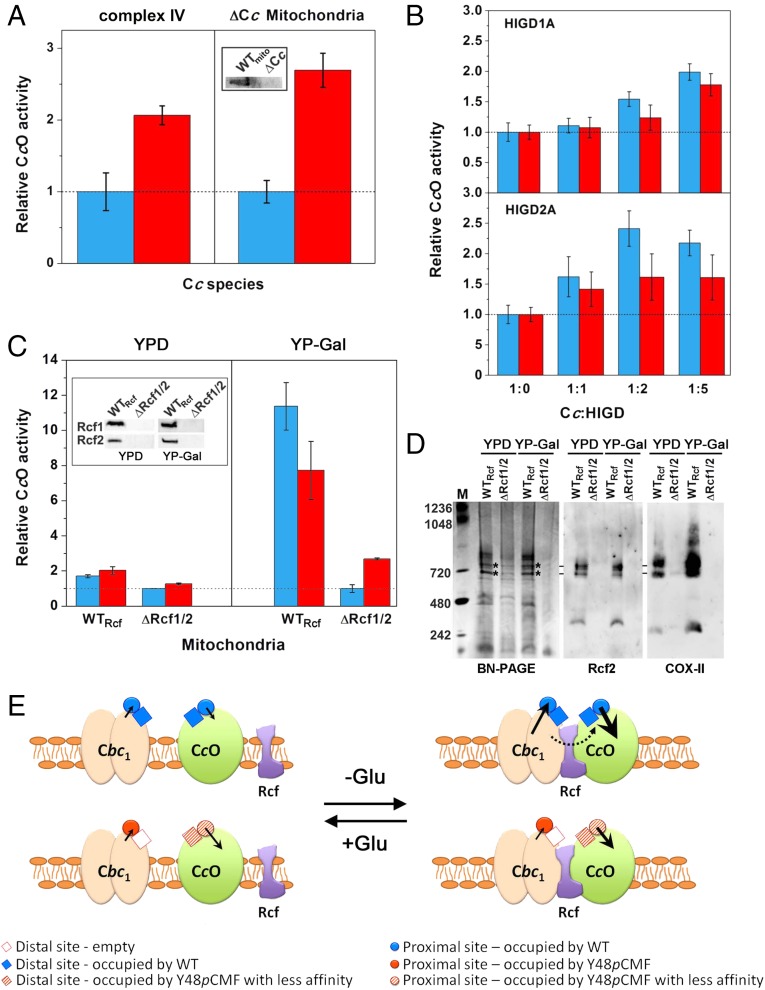
To assess the functional ability of Y48pCMF Cc to reduce respiratory complex IV, we tested the CcO activity of isolated complex IV or Cc-free mitochondria (ΔCc) from yeast cells grown with glucose as a carbon source (Fig. 5A and SI Appendix, Fig. S10A). In both cases, the Cc oxidation rate was at least twofold higher with Y48pCMF Cc than with WT Cc, thereby suggesting that Tyr48 phosphorylation enhances the ability of Cc to donate electrons to complex IV. Direct measurements of O2 consumption were consistent with such data (SI Appendix, Fig. S10B). Interestingly, the CcO activity was positively regulated by the human membrane proteins hypoxia-inducible domain family members 1A and 2A (HIGD1A and HIGD2A), which promote cell survival under hypoxia. HIGD2A was successfully expressed in cell-free expression systems combined with n-dodecyl-β-d-maltoside, as previously reported for HIGD1A (57) (SI Appendix, Fig. S11 A–C). HIGD1A significantly increased the rate of CcO-catalyzed oxidation of Y48pCMF Cc (Fig. 5B), as reported for WT Cc (58). Strikingly, HIGD2A induced an even stronger positive effect than HIGD1A (Fig. 5B). Nevertheless, the HIGD-dependent increase in CcO activity was slightly lower with Y48pCMF Cc (Fig. 5B). This may be due to HIGD-dependent changes in either the complex IV affinity toward Cc or the restraints of Cc diffusion (channeling) from Cc1 to complex IV. A direct Cc–HIGD interaction can also not be excluded.
Fig. 5.
CcO activity with WT or Y48pCMF Cc as the electron donor. (A) CcO activity of isolated complex IV and of mitochondria lacking Cc (ΔCc) upon addition of exogenous WT (blue bars) or Y48pCMF Cc (red bars). Western blot results confirmed the lack of endogenous Cc in ΔCc mitochondria (Inset). (B) In vitro modulation of CcO activity by HIGD1A and HIGD2A. WT Cc (blue bars) or Y48pCMF Cc (red bars), along with HIGD1A or HIGD2A at the indicated ratios, was added to isolated complex IV. (C) Effect of the modulators Rcf1 and Rcf2 on the CcO activity of mitochondria isolated from yeasts grown in YPD or YP-Gal media with either WT Cc (blue bars) or Y48pCMF Cc (red bars). All data represent the mean ± SD of three independent experiments. In all cases, CcO activity was detected only upon addition of exogenous Cc but not with endogenous Cc. (C, Inset) Western blots of WTRcf mitochondria (lane 1) and mitochondria lacking Rcf1 and Rcf2 (ΔRcf1/2) (lane 2). (D) BN/PAGE and Western blots of mitochondria from WTRcf and ΔRcf1/2 strains, using antibodies against Rcf2 and COX-II. Bands submitted to tryptic digestion (SI Appendix, Fig. S11D) are highlighted by asterisks. (E) Scheme of the interactions within the electron transport chain involving Cbc1, CcO, Rcf proteins, and WT or Y48pCMF Cc, as a function of glucose (Glu) availability. The Rcf proteins facilitate the interaction between Cbc1 and CcO to form OxPhos supercomplexes, mainly under glucose deprivation (Right). The thickness of solid arrows refers to the electron transfer rate at the Cc-binding proximal sites of Cbc1 and CcO by WT or Y48pCMF Cc—the longer and thicker the arrow, the more efficient the electron transfer. The dashed line highlights the channeling of WT Cc molecules.
To confirm the HIGD-mediated regulation of the CcO activity in a cellular context, we isolated mitochondria from different yeast strains grown either with glucose (YPD medium), which supports fermentation and respiration, or with the nonfermentable carbon sources lactate and galactose (YP-Gal) (59). Under both metabolic conditions, the isoformic respiratory supercomplex factors 1 (Rcf1, formerly Aim31) and 2 (Rcf2, formerly Aim38) are constitutively expressed (Fig. 5C, Inset), in agreement with their role in CcO activity and supercomplex stability (59–61). Rcf1 is a yeast ortholog of the human HIGD1A and HIGD2A proteins, whereas Rcf2 is specific to yeast. The external membranes of isolated mitochondria were then permeabilized to allow the entry of exogenous WT or Y48pCMF Cc. Under these conditions, mitochondria isolated from a yeast strain deficient in both Rcf1 and Rcf2 (ΔRcf1/2), as verified by Western blot, displayed an endogenous CcO activity lower than that isolated from WT yeast (WTRcf), no matter which type of exogenous hemeprotein—WT or Y48pCMF Cc—was used for supplementation (Fig. 5C). This indicates that Rcf1 and Rcf2 act as positive modulators of CcO activity, similar to human HIGD proteins. In mitochondria isolated from yeast grown with nonfermentable carbon sources, the Rcf-mediated increase in CcO activity was less prominent when Y48pCMF Cc rather than WT Cc was added as an exogenous electron donor, in agreement with the in vitro behavior of isolated proteins (Fig. 5B). This suggests that Tyr48 phosphorylation makes Cc less sensitive to the enhancer mechanism of the Rcf proteins and/or to the ability of Rcfs to stabilize the Cbc1–CcO supercomplexes (Fig. 5C). In fact, Rcf1 and Rcf2 promote the supercomplex assembly, preferably in mitochondria from yeasts grown in a respiratory-based medium (Fig. 5D and SI Appendix, Fig. S11D). This is especially remarkable when comparing the band intensities in an anti–COX-II immunoblot (Fig. 5D). Note that such OxPhos supercomplexes could show a certain degree of heterogeneity in their composition, because the gene transcription of COX-Va and COX-Vb—which encode for two CcO isoforms—is repressed and active, respectively, in the aerobic-to-anaerobic metabolic transition (62). Moreover, the faint band pattern of ΔRcf1/2 strains grown in YPD medium on blue native (BN)/PAGE corresponds to supercomplexes, thus suggesting that other factors may contribute to their assembly (Fig. 5D and SI Appendix, Fig. S11D).
Altogether, our data suggest that phosphorylation of Cc at Tyr48 modulates the mitochondrial ETC (Fig. 5E). Such a posttranslational modification allows a fast adaptation of the hemeprotein function to changing cell conditions. The population of Cbc1–CcO supercomplexes is less prominent in the presence than in the absence of glucose, although Rcf proteins are still expressed. In any case, Cc channeling from Cbc1 toward CcO is impaired. At a physiological pH, Tyr48 phosphorylation thus favors binding of Cc to the proximal rather than to the distal site of Cc1. Under respiration-based growth, the Rcf proteins preferably associate directly with the OxPhos supercomplex, bridging Cbc1 and CcO. However, the weaker Y48pCMF Cc–CcO binding, along with the loss of the distal site on Cc1 due to phosphorylated Cc, impairs the channeling of Cc molecules that functionally connects Cbc1 with CcO, which has been proposed to occur with WT Cc (47, 48). As a consequence, Y48pCMF Cc is less efficient than WT Cc as an electron carrier toward CcO in the context of OxPhos supercomplexes.
Tyr48-Phosphorylated Cytochrome c Acts as an Improved Peroxidase-Like Enzyme, in Particular When Bound to Cardiolipin-Containing Liposomes.
Assembly of the mitochondrial protein membrane complexes Cbc1 and CcO into OxPhos supercomplexes enables more efficient electron flow and decreases ROS levels generated by the ETC (63). The phospholipid CL, located in the inner mitochondrial membrane, also stabilizes the resulting supercomplexes (64) in a HIGD/Rcf-independent manner (59). In addition, CL interacts with Cc in a two-step binding reaction (65): In the first step, the so-called A site at the Cc surface makes transient electrostatic contacts with the membrane; in the second step, hydrophobic forces drive the formation of a tight and stable Cc–CL complex, with one of the CL acyl chains entering the hydrophobic groove of Cc (termed the C site) (66–68). C-site binding then triggers Cc-regulated CL peroxidation under oxidative stress and induces early apoptosis (40). Interestingly, several residues of Cc (Lys22, Lys27, and His33) become protonated at low pH to form an extra binding site—the so-called L site—that facilitates the electrostatic interaction of Cc with mitochondrial membranes (69).
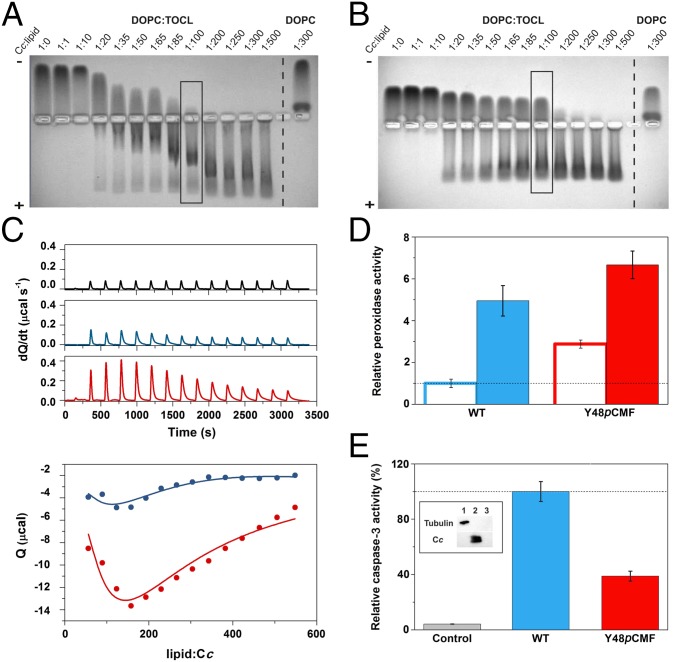
Within this framework, we analyzed how tyrosine phosphorylation can fine-tune the affinity of Cc toward CL-containing liposomes, analyzing binding of Cc species to liposomes in Hepes buffer at a physiological pH value by electrophoretic mobility shift assays (EMSAs) in native agarose gels (Fig. 6 A and B). To compare the binding properties of Cc species to the CL-containing liposomes of 4:1 DOPC:TOCL (1,2-dioleoyl-sn-glycero-3-phosphocholine:1,1′,2,2′-tetraoleoylcardiolipin) or to liposomes of DOPC alone, we measured the mobility profiles of Cc at different Cc:lipid ratios. Although both WT and Y48pCMF Cc bound to DOPC:TOCL and DOPC vesicles, their binding affinity for the CL-free DOPC liposomes seemed to be lower (Fig. 6 A and B). Notably, the presence of free hemeprotein at high lipid concentrations suggested that Y48pCMF Cc has a lower affinity than WT toward DOPC:TOCL liposomes (Fig. 6B), as recently observed for the phosphomimetic mutant S47D Cc (70). Another interesting finding is the unspecific interaction of Cc—either the WT or Y48pCMF species—with DOPC vesicles (Fig. 6 A and B).
Fig. 6.
Liposome-binding assays with caspase-3 activity induced by WT and Y48pCMF Cc. (A and B) EMSA of Cc in the presence of increasing concentrations of lipids. DOPC:TOCL (4:1) or DOPC liposomes were incubated with WT (A) or Y48pCMF (B) Cc. Note that free Cc species moved to the cathode, whereas liposome-bound Cc migrated to the anode. Lanes marked by rectangles correspond to the Cc:lipid ratio at which the peroxidase activity was determined (see below). (C) Calorimetric assays for lipid binding to Cc. (C, Upper) ITC thermograms, corresponding to titrations of DOPC:TOCL 4:1 liposomes (black), WT Cc (blue), or Y48pCMF Cc (red). (C, Lower) Binding isotherms with WT Cc (blue dots) or Y48pCMF Cc (red dots). Continuous lines represent the best fits to a sequential binding, as computed with NanoAnalyze software (TA Instruments) with a stoichiometry of 30 molecules of lipid per molecule of Cc. All data represent the mean ± SD of three independent experiments. (D) Relative peroxidase activities of WT Cc (blue) or Y48pCMF Cc (red) in the presence of liposomes containing DOPC (empty bars) or DOPC:TOCL (4:1) (filled bars). (E) Relative caspase-3 activity in HEK293 cell extracts devoid of endogenous Cc upon addition of exogenous WT Cc (blue) or Y48pCMF Cc (red). A lack of caspase autoactivation was verified in a run without the addition of Cc (gray). Western blots confirmed the lack of endogenous Cc in cytoplasmic cell extracts after immunoblotting with anti–α-tubulin (cytosolic marker) and anti-Cc antibodies (Inset). Lane 1, cytoplasmic cell extracts; lane 2, Cc; lane 3, BSA as a negative control. All data represent the mean ± SD of three independent experiments.
ITC measurements corroborated these EMSA data. ITC experiments were performed under aerobiosis to mimic physiological conditions, despite using degassed samples. This may result in partial oxidation of Cc upon binding to liposomes, as previously reported (71). ITC analysis yielded the apparent KD values for the first binding event of 427 μM (WT Cc) and 780 μM (Y48pCMF Cc) (Fig. 6C). Data for the interaction of WT Cc with liposomes are consistent with the KD values reported before (68, 72–74), but they diverge from the lower KD values reported in the literature (75) and the A/C two-site binding model described by Kinnunen and coworkers (66, 67). Such discrepancies are still unresolved, and further research is necessary to harmonize data from all these different binding assays in a unified and single model. The interaction with CL resulted in apparent ΔH values equal to 2.75 kcal⋅mol–1 for WT Cc and 38.97 kcal⋅mol–1 for Y48pCMF Cc. The differences in KD indicate a small change in binding energies (of ∼6 kcal⋅mol–1). Hence, the changes in ΔH are indicative of enthalpy–entropy compensation effects, which are compatible with the electrostatic change resulting from the extra carboxylate group of the Y48pCMF species. Further, an exothermic process can be observed, with longer peak equilibrium times upon successive lipid additions. In this case, the apparent ΔH values with the WT and Y48pCMF Cc species were equal to −6.97 and −47.84 kcal⋅mol–1, respectively, but the apparent KD value (of ∼1 mM) was practically the same with the two Cc species. Altogether, our EMSA and ITC assays indicate that Y48pCMF Cc binds to CL-containing liposomes with a slightly lower affinity than WT Cc.
WT Cc undergoes CL-dependent conformational changes that allow H2O2 to access the heme crevice (74). Hence, we addressed whether the affinity differences between WT and Y48pCMF Cc for CL could affect their peroxidase activity. In the absence of CL-containing liposomes, Y48pCMF Cc exhibited a threefold higher peroxidase activity than WT Cc (Fig. 6D). However, the presence of DOPC:TOCL vesicles, at a 1:100 Cc:lipid ratio, increased the enzymatic activity of both WT and Y48pCMF Cc, similar to that observed for other phosphomimetic Cc mutants (70). Note that the slightly lower peroxidase activity increment observed for Y48pCMF Cc is likely due to its higher population of free protein compared with WT Cc (Fig. 6 B and D).
Tyr48 Phosphorylation Is an Antiapoptotic Posttranslational Modification.
Phosphorylated Cc may be more easily released from mitochondria because of its lower affinity toward CL, as inferred from EMSA and ITC assays. Translocation to the cytosol could thus enable phosphorylated Cc to interact with Apaf-1 and to assemble the caspase-activating apoptosome. However, cytosolic caspase-3 activation was decreased by about 60% in the presence of the Y48pCMF Cc mutant (Fig. 6E). This is in agreement with the behavior previously reported for the Y48E Cc mutant, which exhibits a lower ability than WT Cc to activate not only caspase-3 (26) but also procaspase-9 by nonfunctional apoptosome assembly (27). These results could thus be indicative of an antiapoptotic function of Cc when phosphorylated at position 48.
Discussion
Here we tackle the structural and functional characterization of the Y48pCMF variant of human Cc. This mutation mimics protein phosphorylation at Tyr48 by adding a negative charge and slightly increasing the side-chain size while keeping the aromatic ring. A recent spectroscopic analysis of Y48pCMF Cc showed a singular shift of the typical alkaline transition pKa to physiological pH values (31), as is the case with the Y48E Cc mutant (26, 27). Here, we report NMR-based structure computations that indicate that Tyr48 phosphorylation maintains the core foldon of Cc but increases internal motions in the loops ΩNY, ΩR, and ΩG. Specifically, the ΩNY-loop, the most unstable folding unit of the hemeprotein, becomes looser and reaches conformational equilibria in Y48pCMF Cc. Enhanced motions at the ΩG- and ΩR-loops could be associated with the shift in the alkaline transition pKa. Indeed, these two loops hold the residues that provide the iron axial ligands, which are observed to at least partially lose their metal coordination in the alkaline form (76). This is in contrast to XAS data that suggest that axial coordination remains untouched. However, the cryogenic temperatures at which the XAS spectra were recorded may amplify the most stable structures and reduce contributions from minority species with a high disorder, which are undetectable.
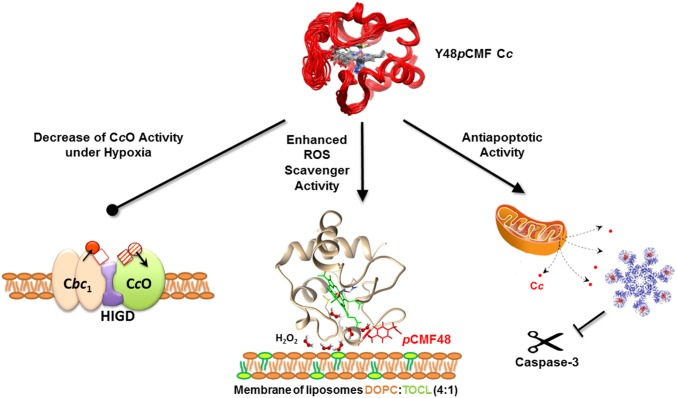
The local but substantial changes in conformation and dynamics of the regions surrounding the Y48pCMF mutation increased the solvent accessibility of the porphyrin ring, thereby enhancing cytochrome peroxidase activity. Thus, Y48pCMF Cc proficiently scavenges ROS and avoids damage of cellular components (Fig. 7). Our findings also indicate that Y48pCMF Cc binds to mitochondrial CL with a lower affinity than WT Cc but that Y48pCMF Cc shows a higher peroxidase activity upon binding to liposomes. According to the literature, both events may facilitate Cc translocation to the cytoplasm during apoptosis (77). However, phosphorylation at Tyr48 hinders the proapoptotic activity of extramitochondrial Cc, similar to the function of HIGD1A protein in ischemia and tumorigenesis (78). Indeed, both HIGD1A and Y48pCMF Cc act as prosurvival proteins by preventing apoptotic caspase activation (Fig. 7) (79).
Fig. 7.
Schematic diagram illustrating the changes induced in cell function by phosphomimetic Y48pCMF Cc. The negative charge at position 48 decreases CcO activity by disrupting Cc channeling in OxPhos supercomplexes (Left), enhances ROS scavenger activity by increasing the peroxidase activity of CL-bound Cc (Middle), and promotes the antiapoptotic function of Y48pCMF Cc by inhibiting its ability to activate the caspase-3 cascade (Right).
As expected, the Y48pCMF mutation affects the binding mode of Cc to its well-known respiratory partners Cbc1 and CcO. Not surprisingly, the enhanced internal motions of the loops ΩNY and ΩR and the electrostatic change at the interaction patch resulting from the additional carboxyl group affect the interaction of Y48pCMF Cc with the two membrane complexes. The mutation-induced changes in electrostatics and in solvation at the interface also concur for the interaction of reduced Y48pCMF Cc with the distal sites of CcO (and Cc1), as inferred from the high decrease in dissociation enthalpies and the strong (or moderate) enthalpy–entropy compensation herein reported. Indeed, the binding of Y48pCMF Cc on the Cc1 surface is weak and functionally irrelevant at the distal site but is favored at the proximal site (Fig. 5E). When the turnover of the redox carrier at the main site is considered, the high affinity of reduced Y48pCMF Cc inhibits its own replacement by oxidized Cc molecules, thereby impairing the electron transport.
The Cc–CcO complex keeps its interface solvated, according to the recent X-ray diffraction structure (80), which displays a protruding lysine side chain in CcO pointing to Tyr48 in Cc. A salt bridge involving phosphorylated Tyr48 would restrain the ΩNY-loop. The increase in the KD of the complex between Y48pCMF Cc and CcO (this work) and the decrease in the Michaelis constant (KM) of phosphorylated Cc with CcO (81) indicate that KM is mainly governed by the catalytic step. This agrees with the larger CcO-driven oxidation rate of Y48pCMF Cc compared with that of WT Cc at the limiting protein concentrations in the enzymatic activity assays used here. In this context, the ca. 60-mV decrease in the E0 of Y48pCMF Cc (31) may facilitate the electron flow from Cc to CcO, although it may impair Cc reduction by Cbc1. In other words, Y48pCMF Cc exhibits a reduced ability as oxidizing agent (it is less capable than WT Cc of receiving electrons from Cc1) but displays an increased capacity as reducing agent (it is a better electron donor to CcO than WT Cc). It is well-known that the catalytic step can be tuned by conformational changes of Cc upon binding to CcO (52). Strikingly, these changes resemble the prominent internal dynamics of Y48pCMF Cc, which could also explain not only the change in E0 but also the shift in the alkaline transition pKa of oxidized Y48pCMF Cc (31). Therefore, the transition from reduced to oxidized Y48pCMF Cc, which is accompanied by a change in the iron axial ligand at physiological pH, is expected to have a high-energy barrier (82). Hence, such conformational changes might be the limiting step for Y48pCMF Cc oxidation inside the complex.
Under nutrient and oxygen depletion, a dysfunctional mitochondrial ETC and OxPhos can lead to many human diseases, including pathologies such as ischemia and cancer (24). The nexus in both disorders could be the control of mitochondrial ET rate by Cc phosphorylation (23), which has indeed been reported to efficiently fine-tune respiratory rates (81). Even though HIGD-mediated assembly of Cbc1 and CcO into OxPhos supercomplexes preferably occurs under low-glucose conditions, the decrease in both the CcO activity and binding affinity toward Cc1 and CcO resulting from the enhanced dynamics of Y48pCMF Cc could explain the disruption of Cc channeling inside the Cbc1–CcO supercomplex, thus slowing down the ETC flow (Fig. 7).
Intriguingly, not only respiratory supercomplex formation (60, 61)—whose structure has recently been solved (83, 84)—but also Cc phosphorylation, and the resulting decrease in the ETC rate, could help to keep ROS levels low and guarantee cell survival.
Tyr48-phosphorylated Cc could be targeted as a biomarker of mitochondrial dysfunction for associated pathological states such as ischemia/reperfusion and cancer. Deciphering the details of the phosphorylated Cc-controlled complex network requires accurate structural and dynamic analyses to eventually develop robust therapeutic approaches to foster or silence—as required—the prosurvival action of phosphorylated Cc reported here.
Methods
Methods and associated references are available in SI Appendix, Methods. All biological samples were obtained from bacteria, yeast, or human cell extracts, in full compliance with University of Seville Ethical Committee bylaws.
Supplementary Material
Acknowledgments
We acknowledge the NMR services at the Centro di Ricerca di Risonanze Magnetiche (CERM; Florence), Centro de Investigación, Tecnología e Investigación (CITIUS; Seville), and Biointeractomics Platform (BIP-cicCartuja; Seville), as well as TA Instruments. We also thank Diamond Light Source for access to beamline I20-scanning (Proposal SP-6011). Experimental work was performed in part at the Grenoble INSTRUCT Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL), with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. Financial support was provided by the Spanish Ministry of Economy and Competitiveness (Grants BFU2015-71017-P/BMC and BFU2015-19451/BMC, cofounded by FEDER EU), European Union (Bio-NMR-00130 and CALIPSO-312284), Ramon Areces Foundation, and Andalusian Government (BIO198). B.M.-B. was awarded a PhD fellowship from the Spanish Ministry of Education (AP2009-4092) and a short-term traveling fellowship from the European Bio-NMR Project. A.G.-C. was awarded a PhD fellowship from the CSIC (JaePre-2011-01248).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.L.B. is a Guest Editor invited by the Editorial Board.
Data deposition: The NMR assignments and coordinates for Y48pCMF cytochrome c reported in this paper have been deposited in the Biological Magnetic Resonance Data Bank (BMRB entry code 25660) and Protein Data Bank (PDB ID code 2N3Y) databases, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618008114/-/DCSupplemental.
References
- 1.Papa S. Molecular mechanism of proton translocation by the cytochrome system and the ATPase of mitochondria. Role of proteins. J Bioenerg Biomembr. 1982;14:69–86. doi: 10.1007/BF00745021. [DOI] [PubMed] [Google Scholar]
- 2.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenaz G, Genova ML. Structure and organization of mitochondrial respiratory complexes: A new understanding of an old subject. Antioxid Redox Signal. 2010;12:961–1008. doi: 10.1089/ars.2009.2704. [DOI] [PubMed] [Google Scholar]
- 4.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou T, Wang X, Ma Q, Cheng H. Mitochondrial flashes: New insights into mitochondrial ROS signalling and beyond. J Physiol. 2014;592:3703–3713. doi: 10.1113/jphysiol.2014.275735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solaini G, Baracca A, Lenaz G, Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochim Biophys Acta. 2010;1797:1171–1177. doi: 10.1016/j.bbabio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Lenaz G, et al. Mitochondrial respiratory chain super-complex I–III in physiology and pathology. Biochim Biophys Acta. 2010;1797:633–640. doi: 10.1016/j.bbabio.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Corcoran A, Cotter TG. Redox regulation of protein kinases. FEBS J. 2013;280:1944–1965. doi: 10.1111/febs.12224. [DOI] [PubMed] [Google Scholar]
- 9.Helling S, et al. Multiple phosphorylations of cytochrome c oxidase and their functions. Proteomics. 2012;12:950–959. doi: 10.1002/pmic.201100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Díaz-Moreno I, García-Heredia JM, Díaz-Quintana A, De la Rosa MA. Cytochrome c signalosome in mitochondria. Eur Biophys J. 2011;40:1301–1315. doi: 10.1007/s00249-011-0774-4. [DOI] [PubMed] [Google Scholar]
- 11.Wegerich F, Turano P, Allegrozzi M, Möhwald H, Lisdat F. Cytochrome c mutants for superoxide biosensors. Anal Chem. 2009;81:2976–2984. doi: 10.1021/ac802571h. [DOI] [PubMed] [Google Scholar]
- 12.Florence TM. The degradation of cytochrome c by hydrogen peroxide. J Inorg Biochem. 1985;23:131–141. doi: 10.1016/0162-0134(85)83017-8. [DOI] [PubMed] [Google Scholar]
- 13.Radi R, Thomson L, Rubbo H, Prodanov E. Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide. Arch Biochem Biophys. 1991;288:112–117. doi: 10.1016/0003-9861(91)90171-e. [DOI] [PubMed] [Google Scholar]
- 14.Radi R, Turrens JF, Freeman BA. Cytochrome c-catalyzed membrane lipid peroxidation by hydrogen peroxide. Arch Biochem Biophys. 1991;288:118–125. doi: 10.1016/0003-9861(91)90172-f. [DOI] [PubMed] [Google Scholar]
- 15.Martinou JC, Desagher S, Antonsson B. Cytochrome c release from mitochondria: All or nothing. Nat Cell Biol. 2000;2:E41–E43. doi: 10.1038/35004069. [DOI] [PubMed] [Google Scholar]
- 16.Basova LV, et al. Cardiolipin switch in mitochondria: Shutting off the reduction of cytochrome c and turning on the peroxidase activity. Biochemistry. 2007;46:3423–3434. doi: 10.1021/bi061854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagan VE, et al. Cytochrome c/cardiolipin relations in mitochondria: A kiss of death. Free Radic Biol Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertini I, Chevance S, Del Conte R, Lalli D, Turano P. The anti-apoptotic Bcl-x(L) protein, a new piece in the puzzle of cytochrome c interactome. PLoS One. 2011;6:e18329. doi: 10.1371/journal.pone.0018329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Fábregas J, et al. New Arabidopsis thaliana cytochrome c partners: A look into the elusive role of cytochrome c in programmed cell death in plants. Mol Cell Proteomics. 2013;12:3666–3676. doi: 10.1074/mcp.M113.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Fábregas J, et al. Structural and functional analysis of novel human cytochrome c targets in apoptosis. Mol Cell Proteomics. 2014;13:1439–1456. doi: 10.1074/mcp.M113.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Arzola K, et al. Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc Natl Acad Sci USA. 2015;112:9908–9913. doi: 10.1073/pnas.1508040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Fábregas J, Díaz-Moreno I, González-Arzola K, Díaz-Quintana A, De la Rosa MA. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 2014;5:e1314. doi: 10.1038/cddis.2014.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hüttemann M, et al. Regulation of oxidative phosphorylation, the mitochondrial membrane potential, and their role in human disease. J Bioenerg Biomembr. 2008;40:445–456. doi: 10.1007/s10863-008-9169-3. [DOI] [PubMed] [Google Scholar]
- 24.Hüttemann M, Lee I, Grossman LI, Doan JW, Sanderson TH. Phosphorylation of mammalian cytochrome c and cytochrome c oxidase in the regulation of cell destiny: Respiration, apoptosis, and human disease. Adv Exp Med Biol. 2012;748:237–264. doi: 10.1007/978-1-4614-3573-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadenbach B. Incorporation of 32P-phosphate into phosphatides of rat liver mitochondria in vivo and in vitro. FEBS Lett. 1968;2:118–120. doi: 10.1016/0014-5793(68)80118-8. [DOI] [PubMed] [Google Scholar]
- 26.Pecina P, et al. Phosphomimetic substitution of cytochrome c tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry. 2010;49:6705–6714. doi: 10.1021/bi100486s. [DOI] [PubMed] [Google Scholar]
- 27.García-Heredia JM, et al. Tyrosine phosphorylation turns alkaline transition into a biologically relevant process and makes human cytochrome c behave as an anti-apoptotic switch. J Biol Inorg Chem. 2011;16:1155–1168. doi: 10.1007/s00775-011-0804-9. [DOI] [PubMed] [Google Scholar]
- 28.Díaz-Moreno I, García-Heredia JM, Díaz-Quintana A, Teixeira M, De la Rosa MA. Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. Biochim Biophys Acta. 2011;1807:1616–1623. doi: 10.1016/j.bbabio.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 29.García-Heredia JM, et al. Specific nitration of tyrosines 46 and 48 makes cytochrome c assemble a non-functional apoptosome. FEBS Lett. 2012;586:154–158. doi: 10.1016/j.febslet.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Ly HK, et al. Perturbation of the redox site structure of cytochrome c variants upon tyrosine nitration. J Phys Chem B. 2012;116:5694–5702. doi: 10.1021/jp302301m. [DOI] [PubMed] [Google Scholar]
- 31.Guerra-Castellano A, et al. Mimicking tyrosine phosphorylation in human cytochrome c by the evolved tRNA synthetase technique. Chemistry. 2015;21:15004–15012. doi: 10.1002/chem.201502019. [DOI] [PubMed] [Google Scholar]
- 32.Ripple MO, Abajian M, Springett R. Cytochrome c is rapidly reduced in the cytosol after mitochondrial outer membrane permeabilization. Apoptosis. 2010;15:563–573. doi: 10.1007/s10495-010-0455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeng WY, Chen CY, Chang HC, Chuang WJ. Expression and characterization of recombinant human cytochrome c in E. coli. J Bioenerg Biomembr. 2002;34:423–431. doi: 10.1023/a:1022561924392. [DOI] [PubMed] [Google Scholar]
- 34.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 35.Baistrocchi P, et al. Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c. Biochemistry. 1996;35:13788–13796. doi: 10.1021/bi961110e. [DOI] [PubMed] [Google Scholar]
- 36.Banci L, Bertini I, Huber JG, Spyroulias GA, Turano P. Solution structure of reduced horse heart cytochrome c. J Biol Inorg Chem. 1999;4:21–31. doi: 10.1007/s007750050285. [DOI] [PubMed] [Google Scholar]
- 37.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 39.Word JM, et al. Visualizing and quantifying molecular goodness-of-fit: Small-probe contact dots with explicit hydrogen atoms. J Mol Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 40.Rajagopal BS, et al. The hydrogen-peroxide-induced radical behaviour in human cytochrome c-phospholipid complexes: Implications for the enhanced pro-apoptotic activity of the G41S mutant. Biochem J. 2013;456:441–452. doi: 10.1042/BJ20130758. [DOI] [PubMed] [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Karsisiotis AI, et al. Increased dynamics in the 40–57 Ω-loop of the G41S variant of human cytochrome c promote its pro-apoptotic conformation. Sci Rep. 2016;6:30447. doi: 10.1038/srep30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maity H, Maity M, Englander SW. How cytochrome c folds, and why: Submolecular foldon units and their stepwise sequential stabilization. J Mol Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Xiao Y, et al. Phosphorylation releases constraints to domain motion in ERK2. Proc Natl Acad Sci USA. 2014;111:2506–2511. doi: 10.1073/pnas.1318899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wauer T, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 47.Moreno-Beltrán B, et al. Respiratory complexes III and IV can each bind two molecules of cytochrome c at low ionic strength. FEBS Lett. 2015;589:476–483. doi: 10.1016/j.febslet.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Moreno-Beltrán B, et al. Cytochrome c1 exhibits two binding sites for cytochrome c in plants. Biochim Biophys Acta. 2014;1837:1717–1729. doi: 10.1016/j.bbabio.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 49.Díaz-Moreno I, et al. NMR analysis of the transient complex between membrane photosystem I and soluble cytochrome c6. J Biol Chem. 2005;280:7925–7931. doi: 10.1074/jbc.M412422200. [DOI] [PubMed] [Google Scholar]
- 50.Díaz-Moreno I, Díaz-Quintana A, Ubbink M, De la Rosa MA. An NMR-based docking model for the physiological transient complex between cytochrome f and cytochrome c6. FEBS Lett. 2005;579:2891–2896. doi: 10.1016/j.febslet.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 51.Volkov AN, Worrall JA, Holtzmann E, Ubbink M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci USA. 2006;103:18945–18950. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakamoto K, et al. NMR basis for interprotein electron transfer gating between cytochrome c and cytochrome c oxidase. Proc Natl Acad Sci USA. 2011;108:12271–12276. doi: 10.1073/pnas.1108320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boffi F, et al. pH-dependent local structure of ferricytochrome c studied by X-ray absorption spectroscopy. Biophys J. 2001;80:1473–1479. doi: 10.1016/S0006-3495(01)76119-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osheroff N, et al. The reaction of primate cytochromes c with cytochrome c oxidase. Analysis of the polarographic assay. J Biol Chem. 1983;258:5731–5738. [PubMed] [Google Scholar]
- 55.Garber EA, Margoliash E. Interaction of cytochrome c with cytochrome c oxidase: An understanding of the high- to low-affinity transition. Biochim Biophys Acta. 1990;1015:279–287. doi: 10.1016/0005-2728(90)90032-y. [DOI] [PubMed] [Google Scholar]
- 56.Speck SH, Margoliash E. Characterization of the interaction of cytochrome c and mitochondrial ubiquinol-cytochrome c reductase. J Biol Chem. 1984;259:1064–1072. [PubMed] [Google Scholar]
- 57.Klammt C, et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9:834–839. doi: 10.1038/nmeth.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi T, et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci USA. 2015;112:1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen YC, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 62.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 63.Louro RO, Díaz-Moreno I, editors. Redox Proteins in Supercomplexes and Signalosomes. CRC Press; Oxfordshire, UK: 2015. [Google Scholar]
- 64.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 65.Belikova NA, et al. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rytömaa M, Mustonen P, Kinnunen PK. Reversible, nonionic, and pH-dependent association of cytochrome c with cardiolipin-phosphatidylcholine liposomes. J Biol Chem. 1992;267:22243–22248. [PubMed] [Google Scholar]
- 67.Rytömaa M, Kinnunen PK. Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J Biol Chem. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 68.Sinibaldi F, et al. Insights into cytochrome c-cardiolipin interaction. Role played by ionic strength. Biochemistry. 2008;47:6928–6935. doi: 10.1021/bi800048v. [DOI] [PubMed] [Google Scholar]
- 69.Kawai C, et al. pH-dependent interaction of cytochrome c with mitochondrial mimetic membranes: The role of an array of positively charged amino acids. J Biol Chem. 2005;280:34709–34717. doi: 10.1074/jbc.M412532200. [DOI] [PubMed] [Google Scholar]
- 70.Guerra-Castellano A, Díaz-Moreno I, Velázquez-Campoy A, De la Rosa MA, Díaz-Quintana A. Structural and functional characterization of phosphomimetic mutants of cytochrome c at threonine 28 and serine 47. Biochim Biophys Acta. 2016;1857:387–395. doi: 10.1016/j.bbabio.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 71.Serpas L, Milorey B, Pandiscia LA, Addison AW, Schweitzer-Stenner R. Autoxidation of reduced horse heart cytochrome c catalyzed by cardiolipin-containing membranes. J Phys Chem B. 2016;120:12219–12231. doi: 10.1021/acs.jpcb.6b05620. [DOI] [PubMed] [Google Scholar]
- 72.Hanske J, et al. Conformational properties of cardiolipin-bound cytochrome c. Proc Natl Acad Sci USA. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pandiscia LA, Schweitzer-Stenner R. Coexistence of native-like and non-native partially unfolded ferricytochrome c on the surface of cardiolipin-containing liposomes. J Phys Chem B. 2015;119:1334–1349. doi: 10.1021/jp5104752. [DOI] [PubMed] [Google Scholar]
- 74.Pandiscia LA, Schweitzer-Stenner R. Coexistence of native-like and non-native cytochrome c on anionic liposomes with different cardiolipin content. J Phys Chem B. 2015;119:12846–12859. doi: 10.1021/acs.jpcb.5b07328. [DOI] [PubMed] [Google Scholar]
- 75.Domanov YA, Molotkovsky JG, Gorbenko GP. Coverage-dependent changes of cytochrome c transverse location in phospholipid membranes revealed by FRET. Biochim Biophys Acta. 2005;1716:49–58. doi: 10.1016/j.bbamem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 76.Assfalg M, et al. Structural model for an alkaline form of ferricytochrome c. J Am Chem Soc. 2003;125:2913–2922. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 77.Kagan VE, et al. The “pro-apoptotic genies” get out of mitochondria: Oxidative lipidomics and redox activity of cytochrome c/cardiolipin complexes. Chem Biol Interact. 2006;163:15–28. doi: 10.1016/j.cbi.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 78.Ameri K, et al. HIGD1A regulates oxygen consumption, ROS production, and AMPK activity during glucose deprivation to modulate cell survival and tumor growth. Cell Reports. 2015;10:891–899. doi: 10.1016/j.celrep.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.An HJ, et al. The survival effect of mitochondrial Higd-1a is associated with suppression of cytochrome c release and prevention of caspase activation. Biochim Biophys Acta. 2011;1813:2088–2098. doi: 10.1016/j.bbamcr.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 80.Shimada S, et al. Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode. EMBO J. 2017;36:291–300. doi: 10.15252/embj.201695021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu H, Lee I, Salomon AR, Yu K, Hüttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta. 2008;1777:1066–1071. doi: 10.1016/j.bbabio.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blouin C, Guillemette JG, Wallace CJA. Resolving the individual components of a pH-induced conformational change. Biophys J. 2001;81:2331–2338. doi: 10.1016/S0006-3495(01)75879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 84.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.