Significance
The transcription factor RelB has been thought to be required for dendritic cell (DC) development, although analysis of radiation bone marrow chimeras has raised some questions regarding this issue that have never been resolved. We have reevaluated the role of RelB in DC and myeloid development. We found that DC development was independent of a cell-intrinsic action of RelB in most tissues and that only the terminal maturation of Notch2-dependent splenic cDC2 cells was partially reduced in the absence of cell-intrinsic RelB expression. Moreover, the profound myeloid expansion seen in Relb−/− mice was due to an unrecognized action of RelB in nonhematopoietic cells, indicating that RelB is a critical component of the niche regulating the normal myeloid compartment.
Keywords: dendritic cells, hematopoiesis, transcription factors, hematopoietic niche
Abstract
RelB is an NF-κB family transcription factor activated in the noncanonical pathway downstream of NF-κB–inducing kinase (NIK) and TNF receptor family members including lymphotoxin-β receptor (LTβR) and CD40. Early analysis suggested that RelB is required for classical dendritic cell (cDC) development based on a severe reduction of cDCs in Relb−/− mice associated with profound myeloid expansion and perturbations in B and T cells. Subsequent analysis of radiation chimeras generated from wild-type and Relb−/− bone marrow showed that RelB exerts cell-extrinsic actions on some lineages, but it has remained unclear whether the impact of RelB on cDC development is cell-intrinsic or -extrinsic. Here, we reevaluated the role of RelB in cDC and myeloid development using a series of radiation chimeras. We found that there was no cell-intrinsic requirement for RelB for development of most cDC subsets, except for the Notch2- and LTβR-dependent subset of splenic CD4+ cDC2s. These results identify a relatively restricted role of RelB in DC development. Moreover, the myeloid expansion in Relb−/− mice resulted from hematopoietic-extrinsic actions of RelB. This result suggests that there is an unrecognized but critical role for RelB within the nonhematopoietic niche that controls normal myelopoiesis.
RelB was initially identified as a serum-induced factor in NIH 3T3 cells and recognized as a member of the NF-κB family of transcription factors (1). Subsequent analysis showed RelB to be highly expressed in thymic classical dendritic cells (cDCs), suggesting a possible role in their signaling or development (2). Germline inactivation of Relb in mice causes a complex phenotype of myeloid hyperplasia, extramedullary hematopoiesis, and multiorgan inflammation and disturbs the development of T cells, B cells, and cDCs in the spleen and thymus (3, 4). These abnormalities are not all due to cell-intrinsic requirements for RelB (5–7). First, in Relb−/− → wild-type (WT) BM chimeras, in which thymic epithelium is normal, T-cell development is normal, excluding a cell-intrinsic requirement for RelB in T-cell development (5). Likewise, the loss of natural killer T (NKT) cells in Relb−/− mice is normalized in Relb−/− → Rag2−/− chimeras, excluding a cell-intrinsic requirement for RelB for NKT cell development (6). By contrast, conditional deletion of RelB demonstrated a cell-intrinsic requirement for RelB in production of IL-17 by γδ T cells (7).
For other perturbations observed in Relb−/− mice, the precise cellular site of action for RelB has not been determined. For example, although marginal zone (MZ) B cells fail to develop in both Relb−/− mice and in Relb−/− → WT BM chimeras (8), it has not been shown whether this requirement is intrinsic to B cells or is due to an action of RelB in another hematopoietic cell controlling MZ B-cell development. Likewise, the impaired isotype switching of B cells in Relb−/− → WT chimeras could result from either a B-cell–intrinsic RelB requirement for switching or from the previously reported impaired immunogenicity of Relb−/− DCs (4) that might impair development of T follicular helper cells (9). Relb−/− B cells do show a cell-intrinsic impairment in proliferation in vitro in response to CD40 stimulation, but secretion of IgM is normal and in vitro switching to all non-IgM non-IgD isotypes is intact (10). These results imply that the observed in vivo requirement for RelB in class switching is B-cell–extrinsic.
The actions of RelB in DC development and function, also remain incompletely defined. An initial study claimed that the defects in cDC development seen in Relb−/− mice are present but less severe in Relb−/− → WT BM chimeras (4), but data supporting this statement were not shown. That study was cited in a subsequent publication (11) to support the claim that Relb−/− → WT chimeras lack cDCs derived from Relb−/− BM as well as to implicate a role for RelB in follicular DCs in regulation of class switching. However, this subsequent study (11) also lacked direct analysis of cDCs in BM chimeras. A later study stated that CD8α− cDCs do develop in Relb−/− → WT chimeras (12), but did not directly analyze cDC development and cited an earlier report (5), which also lacked direct analysis of cDCs in chimeras. However, a contemporary review from these authors referred to unpublished data that the impact of RelB on DC development is cell-extrinsic (13). Analysis by others showed that thymic CD8α+ cDC1s develop normally in Relb−/− → WT chimeras, yet argued for a cell-intrinsic action in CD8α− DEC-205− cDC development (14). Another report confirmed decreased cDC numbers in Relb−/− mice but did not examine BM chimeras to test for cell-intrinsic requirements for their development or function (15). Recently, a cell-intrinsic requirement for NF-κB–inducing kinase (NIK) in DCs for their ability to induce normal T-cell responses was reported (16), suggesting a role for noncanonical NF-κB signaling in cDC responses. However, that study did not address the role of RelB in cDCs or the specific cDC subset affected by loss of NIK. Finally, no studies using conditional RelB deletion in B cells or DCs have appeared as of yet.
Since the initial studies on RelB in DCs, knowledge of DC development has advanced substantially, allowing for the identification of distinct subsets of cDCs and related myeloid lineages (17). However, no studies have clarified the unsettled role of RelB in cDC development using either germline or conditional deletion. A study recently examined the expression of a RelB–Venus fusion protein, identifying populations of DCs expressing high levels of RelB in the spleen, but not in other tissues like the colon (18). However, this study did not examine the basis for the myeloid expansion and perturbations of DC development observed in Relb−/− mice.
Here, we reevaluated cDC development in Relb−/− mice in chimeras generated with WT and Relb−/− BM. Our results confirmed the dramatic myeloid and DC disturbances reported for germline Relb−/− mice. However, analysis of several types of BM chimeras indicated that most of these abnormalities were mediated by actions of RelB in cells extrinsic to the hematopoietic system. Specifically, neither the abnormal myeloid expansion nor the impaired DC development seen in germline Relb−/− mice was found in Relb−/− → WT chimeras. Moreover, both abnormalities found in germline Relb−/− mice were also found in WT → Relb−/− chimeras. These results indicate that both abnormalities arose as a result of the altered niche formed by Relb−/− cells in the radiation-resistant nonhematopoietic compartment of Relb−/− recipient mice. Furthermore, competitive mixed-BM chimeras showed that Relb−/− DCs had no competitive defect for plasmacytoid DCs (pDC) or any cDC subset in any tissue, with one exception. The splenic Relb−/− CD4+ ESAM+ cDC2 subset, which we recently showed to require Notch2 and lymphotoxin (LT) signaling for its terminal maturation (19, 20), was reduced approximately threefold relative to the WT counterpart. Finally, RelB was not required for antigen presentation by cDCs to CD4 or CD8 T cells. These results indicate that DC development does not have a cell-intrinsic requirement for RelB except for one subset of splenic CD4+ cDC2s that rely on noncanonical NF-κB signaling downstream of LT.
Results
DCs Accumulate in Relb−/− Mice Despite Myeloid Hyperplasia.
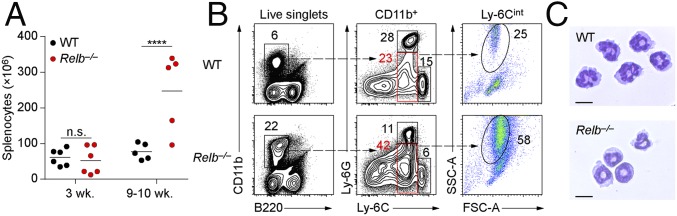
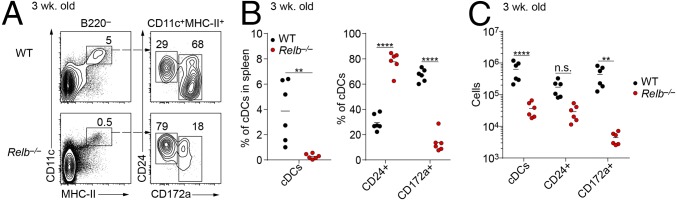
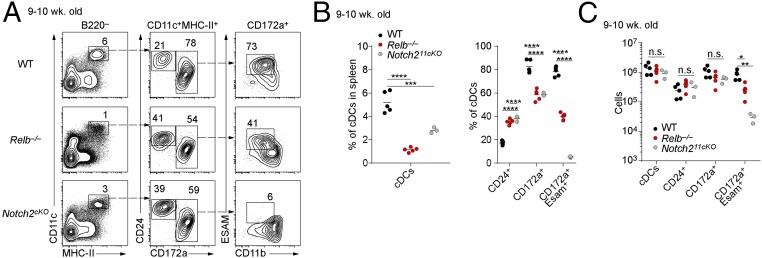
We recently showed that NIK is required for the development of the CD4+ ESAM+ subset of cDC2s that are also dependent on Notch2 signaling (20). LT, which signals through the NIK-dependent noncanonical NF-κB pathway (21), is also required for the development of CD4+ cDC2s (19). Thus, we wondered if RelB, a known transcription factor in the noncanonical NF-κB pathway, might also act downstream of LT and NIK to mediate development of CD4+ ESAM+ cDC2s, prompting us to examine Relb−/− mice (3). We observed that as Relb−/− mice aged, they developed splenomegaly (Fig. 1A) and accumulated a population of CD11b+ Ly-6Cint SSChi myeloid cells that resemble immature polymorphonuclear neutrophils (PMNs) (Fig. 1 B and C) as described in previous reports (3, 4). Because these hematopoietic abnormalities might influence DC development, we analyzed cDCs in 3-wk-old mice before these abnormalities developed. Relb−/− mice at 3 wk of age had normal numbers of splenocytes (Fig. 1A) but still showed a severe reduction in the numbers of cDCs compared with WT mice (Fig. 2). Splenic cDCs (B220−CD11c+MHC-II+) were reduced 10-fold in 3-wk-old Relb−/− mice compared to age-matched WT mice, both in terms of total cDC numbers and as a percentage of total splenocytes (Fig. 2 A–C). The cDC2 (CD172a+) subset demonstrated a greater reduction than the cDC1 (CD24+CD172a−) subset (Fig. 2B), with observed reductions of 20- and 6-fold, respectively (Fig. 2 A–C). This defect persisted in Relb−/− mice at 9 wk of age, with a fivefold reduction in the percentage of splenic cDCs compared with WT mice (Fig. 3A), but with a higher number of cDC2 cells than observed at 3 wk (Fig. 2A). At 9 wk, Relb−/− mice showed only a twofold reduction of CD4+ ESAM+ cDC2s compared with WT controls (Fig. 3 A and B), in contrast to Notch211cko mice, which lack this population entirely (20, 22). Due to the overall increase in spleen cellularity, total cDC numbers were almost equivalent at 9 wk of age between WT and Relb−/− mice (Fig. 3C). Thus, germline Relb−/− mice show a severe reduction in cDCs at 3 wk of age, but cDCs can accumulate even as these mice develop splenomegaly and myeloid expansion.
Fig. 1.
Mice lacking RelB develop splenomegaly and myeloid hyperplasia. (A) Splenocyte numbers in 3- and 9-wk-old WT and Relb−/− mice. Each dot represents a biological replicate from four independent experiments. (B) Representative flow cytometry analysis of CD11b+ splenocytes from WT and Relb−/− mice. Data are representative of three independent experiments. (C) Microscopy of WT and Relb−/− splenic Ly-6G+ PMNs stained with Wright–Giemsa stain. (Scale bars: 10 µm.) n.s., not significant. ****P < 0.0001.
Fig. 2.
cDCs are diminished in 3-wk-old Relb−/− mice. (A) Flow cytometry analysis of B220− splenocytes from 3-wk-old WT and Relb−/− mice. Data are representative of three independent experiments. (B) Percentage and (C) cell counts of splenic cDCs (CD11c+ MHC-II+) in 3-wk-old WT and Relb−/− mice. Each dot represents a biological replicate from three independent experiments. n.s., not significant. **P < 0.01, ****P < 0.0001.
Fig. 3.
cDC2s develop in Relb−/−mice. (A) Flow cytometry analysis of splenic cDCs in 9-wk-old WT, Relb−/−, and Notch211cKO (Notch2f/f × Itgax-Cre) mice. Data are representative of three independent experiments. (B) Percentage and (C) cell counts of splenic cDCs in spleens of WT, Relb−/−, and Notch211cKO. Each dot represents a biological replicate from three independent experiments. n.s., not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Hematopoietic-Extrinsic Requirement for RelB in DC Development.
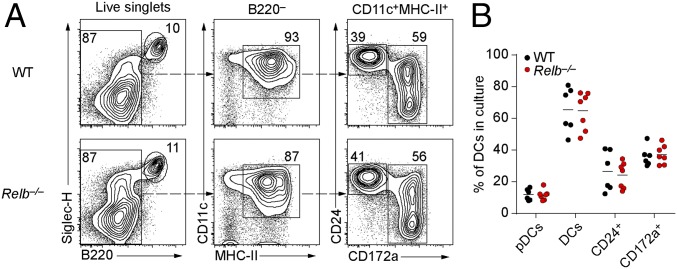
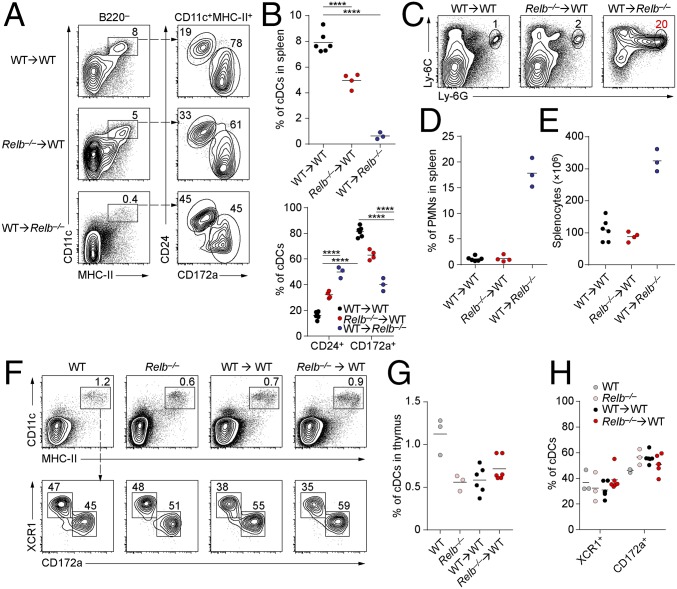
To examine the cell-autonomous actions of RelB in DC development, we first examined in vitro differentiation of Relb−/− BM in response to Flt3L, which supports the development of bona fide counterparts of in vivo DC subsets (23). In this setting, pDCs (SiglecH+B220+), cDCs1, and cDCs2 developed normally from Relb−/− BM compared with WT BM (Fig. 4 A and B), suggesting that DCs may not have a strict cell-intrinsic requirement for RelB for their development. To test this in vivo, we generated BM chimeras by transferring Relb−/− BM into congenically marked WT recipients and by transferring congenically marked WT BM into Relb−/− recipients. As a control, we transferred CD45.2+ WT BM into CD45.1+ WT recipients. Eight weeks after reconstitution, we analyzed the spleens and thymi of these chimeras for the development of cDCs and neutrophils (Fig. 5). Relb−/−→WT chimeras showed a partial reduction of cDCs as a percentage of total splenocytes compared with WT→WT chimeras (Fig. 5 A and B). There was a shift in the ratio of cDC1 to cDC2 cells, with cDC2 cells in Relb−/− → WT chimeras being slightly reduced by ∼20% as a percentage of all DCs compared with WT → WT chimeras, whereas cDC1s were increased (Fig. 5 A and B). In contrast to these mild changes in Relb−/−→WT chimeras, WT → Relb−/− chimeras showed a severe loss of total cDC development as a percentage of splenocytes (Fig. 5 A and B). Furthermore, WT → Relb−/− chimeras developed splenomegaly and myeloid hyperplasia similar to that observed in Relb−/− mice (Fig. 1B), whereas Relb−/− → WT chimeras had normal neutrophil development and splenocyte numbers compared with WT → WT controls (Fig. 5 C–E). Finally, previous studies have claimed a role for RelB in the development of DCs in the thymus (14). However, we observed normal development of both cDC subsets in thymi of either Relb−/− mice or Relb−/− → WT chimeras compared with controls (Fig. 5 F–H). Thus, the major action of RelB for cDC development is through a role in nonhematopoietic radiation-resistant cells that support cDC development in vivo.
Fig. 4.
cDCs lacking RelB develop in vitro. (A) Flow cytometry analysis of BM from WT or Relb−/− mice cultured for 9 d with Flt3 ligand. Data are representative of three independent experiments. (B) Percentage of pDCs (Siglec-H+B220+) and cDCs (B220−CD11c+MHC-II+) from WT and Relb−/− BM cultures generated as in A. Each dot represents a biological replicate from three independent experiments.
Fig. 5.
Radio-resistant cells require RelB to support normal hematopoiesis. (A) Flow cytometry analysis of live B220− splenocytes from chimeras generated with CD45.2+ WT or Relb−/− BM into CD45.1+ B6.SJL recipients or B6.SJL BM into Relb−/− mice, analyzed 8 wk after lethal irradiation and transplantation. Data are representative of two independent experiments. (B) Percentage of cDCs (CD11c+MHC-II+) in the indicated BM chimeras generated as in A. Each dot represents a biological replicate from two independent experiments. (C) Flow cytometry analysis of live granulocytes in spleens of BM chimeras generated as in A. (D) Frequency of Ly-6G+ PMNs in spleens of the indicated chimeras. Each dot represents a biological replicate from two independent experiments. (E) Splenocyte counts in the indicated BM chimeras 8 wk after irradiation and reconstitution. (F) Thymic DCs in the indicated mice were analyzed by flow cytometry. BM chimeras were generated as in A. Shown are two-color histograms representative of thymocyte samples pregated as CD45.2+B220−. (G) Frequency of DCs within CD45.2+B220− thymocytes in the indicated mice. (H) Frequency of thymic DC subsets in the indicated mice. Each dot represents an individual biological replicate from two independent experiments. ****P < 0.0001.
Cell-Intrinsic Actions of RelB Are Tissue-Specific and Restricted to Splenic ESAM+ CD4+ cDC2s.
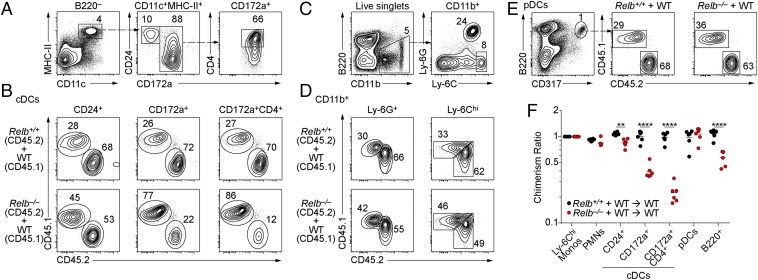
B cells can provide the LT required for normal cDC proliferation (19) and are reduced in Relb−/− mice (8). Conceivably, the slightly reduced number of cDCs developing in Relb−/− → WT chimeras (Fig. 4B) could result from the loss of RelB in a non-DC lineage, such as B cells, that supports cDC development. To test this, we generated mixed BM chimeras reconstituted with equal ratios of WT (CD45.1+) and Relb−/− (CD45.2+) BM (Fig. 6). In such chimeras, a cDC-intrinsic requirement for RelB could be observed by a selective reduction in Relb−/− CD45.2+ cDCs that develop in the RelB-sufficient cellular environment provided by the WT donor BM. We analyzed several tissues from these chimeras 8 wk after BM transplantation and measured the development of various cells, including cDC1s, CD4+, and CD4− cDC2s (Fig. 6 A and B); Ly-6Chi monocytes and PMNs (Fig. 6 C and D); and pDCs (Fig. 6E) and B cells (Fig. 6F). In the spleen, the generation of pDCs, cDC1s, and PMNs was similar for WT and Relb−/− BM cells (Fig. 6 A–F). There was a selective reduction in Relb−/− cDC2 compared with WT cDC2, specifically a threefold reduction in CD172a+ cDC2s and a fourfold reduction in CD4+ CD172a+ cDC2s (Fig. 6 A, B, and F). This selective effect on cDC2 is consistent with similar effects on cDC2 maturation in cDCs lacking LTβR (19) or NIK (20). Notably, the myeloid hyperplasia seen in germline RelB−/− mice was not present in mixed WT + Relb−/−→WT chimeras (Fig. 6 D and F), suggesting that this abnormal myeloid expansion was dependent on cell-extrinsic, nonhematopoietic actions of RelB. Finally, Relb−/− B cells (B220+) were slightly reduced in number by about twofold relative to WT B cells (Fig. 6F).
Fig. 6.
Cell-intrinsic actions of RelB in splenic cDC2s. (A–C) Flow cytometry analysis of live splenocytes from chimeras generated with equal mixes of CD45.1+ B6.SJL and CD45.2+ Relb+/+ or Relb−/− BM analyzed 8 wk after lethal radiation and transplant. Shown are representative two-color histograms for (A and B) cDCs, (C and D) monocytes (CD11b+ Ly-6Chi) and neutrophils (CD11b+ Ly-6G+), and (E) pDCs (B220+CD317+). (F) Contributions of Relb−/− BM to the indicated lineages in chimeras generated as in A shown as the ratio of monocyte contribution in the same mouse. Each dot represents a biological replicate from two independent experiments. **P < 0.01, ****P < 0.0001.
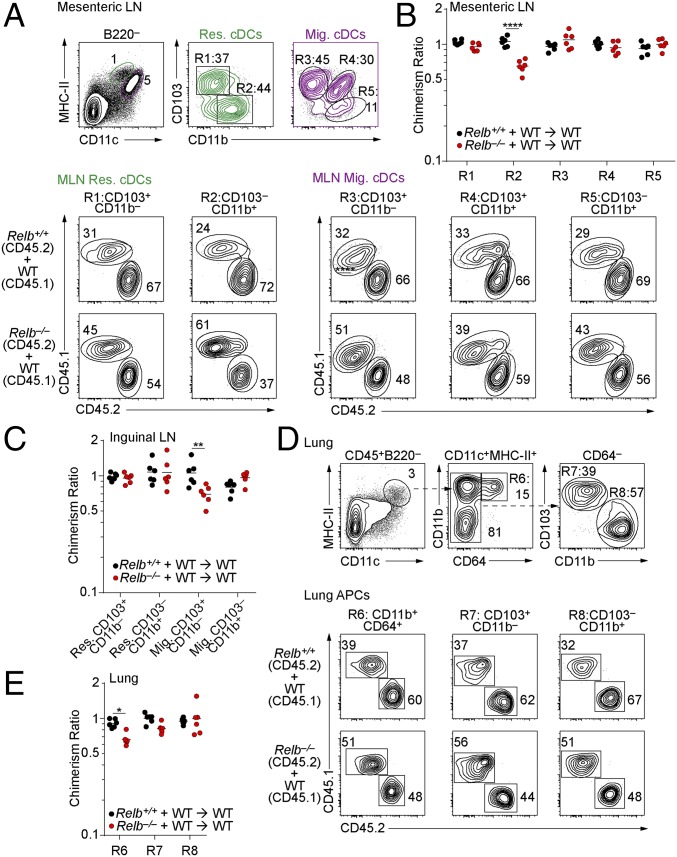
Using competitive chimeras, we previously showed that LtβR signaling is required for development of migratory CD103+ cDC2s in the mesenteric lymph node (MLN) (20). In contrast, the generation of migratory CD103+ and CD103− cDC2s in MLN was similar between WT and Relb−/− BM, with only resident Relb−/− cDC2s reduced about twofold compared with WT controls (Fig. 7 A and B). Furthermore, there was similar output of migratory and resident cDC1s from WT and Relb−/− BM (Fig. 7 A and B). To determine if this reduction of resident cDC2s in MLN was limited to this tissue, we also analyzed the inguinal lymph nodes of these chimeras (Fig. 7C). Here, the migratory Relb−/− cDC1s (CD103+ CD11b−) were reduced slightly, less than twofold, compared with WT cDC1s, with no other significant changes in migratory or resident cDC2s (Fig. 7C). Finally, no significant changes in output from Relb−/− BM were seen in lung resident cDC1s or cDC2s compared with WT BM (Fig. 7 D and E). Thus, the major cell-intrinsic role of RelB in cDC development appeared to be tissue-restricted to the spleen.
Fig. 7.
RelB is not necessary for the development of cDCs in peripheral tissues. (A–E) Mixed BM chimeras were generated as in Fig. 6. (A) Flow cytometry analysis of migratory and resident cDCs in the MLN of CD45.1+ B6.SJL and CD45.2+ WT or Relb−/− mixed BM chimeras. (B and C) Percentage of chimerism for the indicated populations in the MLN (B) or inguinal LN (C) shown as the ratio of splenic Ly-6Chi monocytes as in Fig. 6F. (D) Analysis of lung resident macrophages and cDCs in mixed BM chimeras generated as in Fig. 6. (E) Contributions of Relb−/− BM to the indicated populations from D shown as the ratio of splenic Ly-6Chi monocytes as in Fig. 6F. *P < 0.05, **P < 0.01, ****P < 0.0001.
Antigen Presentation by DCs to T Cells Does Not Require RelB.
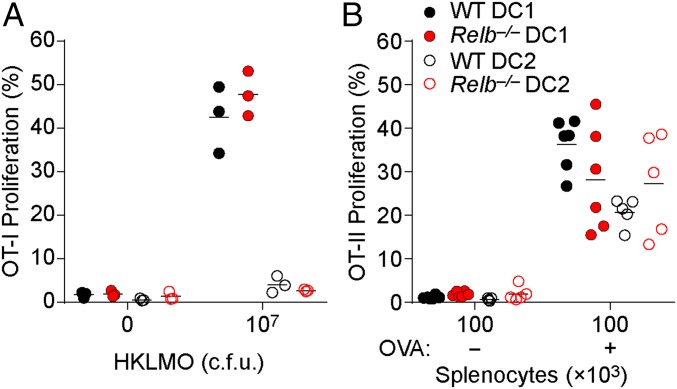
The actions of RelB in DC development so far have been limited to CD4+ ESAM+ cDC2s in the spleen, but DCs may require RelB for their function as well. In DCs, RelB may be activated by CD40 ligation, which signals through the noncanonical NF-κB pathway and enables optimal priming by DCs of CD8 T cells (24–26). In addition, DCs require NIK for their ability to deliver normal costimulation to CD4 T-cell responses (16), and this concievably could require RelB activity. To address this, we tested the ability of splenic cDC1s and cDC2s from Relb−/−→WT chimeras to present cell-associated antigens to OT-I and OT-II cells ex vivo. We find that both WT and Relb−/− cDC1s induced robust OT-I proliferation in response to heat-killed OVA-expressing Listeria monocytogenes (HKLMO) (Fig. 8A). Neither WT or Relb−/− cDC2s were able to cross-present HKLMO (Fig. 8A), as expected for this subset. Furthermore, Relb−/− cDC1 and cDC2 cells cultured with OVA-loaded splenocytes induced OT-II proliferation similar to their WT counterparts (Fig. 8B). In summary, we did not find a defect in the antigen processing or presentation in Relb−/−DCs, although other functions, such as cytokine production, induction of specific effector modalities in T cells, and tolerogenicitiy will still need to be addressed.
Fig. 8.
DCs lacking RelB are capable of priming T cells against cell-associated antigen. (A) Sorted DC1 (CD24+) or DC2 (CD172a+) cells were cocultured with CFSE-labeled OT-I cells and heat-killed OVA expressing L. monocytogenes (HKLMO) for 3 d. OT-I proliferation was determined as the frequency of CD44+ OT-I cells that had undergone at least one CFSE dilution. Each dot represents one biological replicate. (B) CFSE-labeled OT-II cells were cocultured with sorted DC1 or DC2 and OVA-loaded γ-irradiated Ciita−/− splenocytes for 3 d and assayed for proliferation as in A. Each dot represents one biological replicate from two independent experiments.
Discussion
Recent work has established that several DC lineages develop from a common dendritic cell progenitor through various transcriptional programs (17). The transcription factors that are known to influence DC development are IRF8, IRF4, PU.1, Id2, Batf3, E2-2, Nfil3, Notch2, Bcl6, and Zeb2 (17, 27, 28). An intrinsic requirement for each of these factors has been established through the examination of lineage-specific conditional deletion in vivo, analysis of mixed BM chimeras, or defective development in Flt3L-treated BM cultures. RelB was one of the earliest transcription factors to be associated with DC development (2). Initial analysis found that germline Relb−/− mice exhibit decreased cDC numbers, but also have a marked myeloid expansion of immature PMNs leading to splenomegaly, impaired lymph node development, and T-cell–dependent autoimmunity (3, 4, 29). Several subsequent studies examined BM chimeras produced from Relb−/− BM to evaluate the cell-intrinsic requirements for these defects (5, 6, 11, 12). Although cell-extrinsic actions for RelB have been clearly established for some cell types, such as B cells, the nature of the RelB requirement in cDC development has not been entirely resolved.
We confirmed that germline loss of RelB caused decreased numbers of both cDC1 and cDC2 populations in vivo that was apparent at 3 wk of age before the onset of splenomegaly and myeloid expansion. However, cDC1 and cDC2 cells could develop from Relb−/− BM in Flt3L-treated cell cultures (Fig. 4), suggesting that there may not be a strict cell-intrinsic developmental requirement for RelB for either cDC subset. Consistent with this result, we found that Relb−/−→WT chimeras produced donor-derived DCs of all types in the setting of a WT host. In contrast, WT→Relb−/− chimeras produced a phenotype very similar to that of germline Relb−/− mice, with a severe decrease in DCs and a myeloid expansion, suggesting that RelB plays a major role in regulating hematopoiesis through its expression in radio-resistant host cells. So far, no studies on gene regulation by RelB in BM stromal cells have been carried out that might reveal how RelB controls hematopoiesis.
We did observe one setting of mixed bone marrow chimeras in which RelB appeared to exert a cell-intrinsic action in DCs. In most locations, there was no competitive advantage of WT over Relb−/− BM for the generation of any DC subset. However, in the spleen, the Notch2-dependent CD4+ ESAM+ cDC2s cells derived from Relb−/− BM were reduced by two- to threefold relative to those derived from WT BM, in agreement with the previously reported reduction of CD8α−DEC-205− cDCs in spleens of Relb−/− → WT chimeras (14). Because this occurred in the setting of a mixed chimera, this likely indicates a DC-intrinsic role for RelB, and not an effect mediated by another hematopoietic lineage. Conceivably, B cells may require RelB to generate LTα1β2, the ligand for LtβR, that is intrinsically required for DC homeostasis in the spleen (19). Furthermore, others have described the necessary interactions between DCs and stroma for DC development. For example, deletion of the Notch ligand Delta-like 1 in splenic fibroblasts ablates the differentiation of ESAM+ cDC2s (30). Stroma cells also require LtβR for their development (31); however, it has not been determined if RelB is the requisite factor downstream of LT signaling. The exact cell-intrinsic and -extrinsic functions of RelB in DC development could be determined using lineage-specific conditional deletion of RelB (32).
Both Notch2 and LtβR signaling are known to regulate development of CD4+ ESAM+ DCs in the spleen and CD103+CD11b+ cDC2s in the small intestine (20). In the setting of a mixed BM chimera, WT BM outcompetes Ltbr−/− BM for the generation of both splenic CD4+ ESAM+ DCs and for intestinal CD103+CD11b+ cDC2s (20). By contrast, the Notch2-deficient BM and Ltbr−/− BM compete equally for the generation of both of these types of cDCs (20), suggesting that Notch2 and LtβR act in the same development pathway of terminal maturation of these DCs in these tissues. In the current study, loss of RelB affected only the development of CD4+ ESAM+ cDC2s in the spleen, but not that of CD103+CD11b+ cDC2s in the MLN. The basis for this more limited phenotype of RelB could be due to selective expression of RelB in splenic CD8α− cDCs in the spleen but not in cDC populations in other tissues (18).
We did not observe a deficiency in the ability of Relb−/− cDCs to present cell-associated antigens to either CD4 or CD8 T cells ex vivo; however, RelB may act on other immune models in DCs. The recent availability of a conditional Relb allele (32), combined with deletor strains that direct specific Cre expression in DC subsets (33), will facilitate future studies to determine what functional role, if any, RelB has in DCs for immune responses. For example, LtβR signaling in DCs is required for optimal IL-23 production by cDC2s in response to infection by Citrobacter rodentium infection (34) and for induction of IgA class-switching (35). However, the role for RelB in mediating either of these responses has not been examined.
In summary, we have documented a dramatic action of RelB in radio-resistant host cells that was required to support normal hematopoiesis. Loss of RelB in nonhematopoietic cells led to most of the developmental disturbances reported initially for Relb−/− mice. The cell-intrinsic actions of RelB that we found for DC development apply to the development of the CD4+ ESAM+ cDC2 subset the terminal maturation of which involves Notch2 and LTβR signaling in the spleen and is consistent with RelB acting downstream of LTβR. Future studies will need to address the likely role of RelB activation in the functioning of mature DCs independently of their development. For example, RelB activation in cDCs downstream of CD40 stimulation may take place in settings of CD4 help for CD8 T-cell responses (36), but analysis of these actions may require conditional deletion within specific DC subsets. Finally, the more severe hematopoietic defects, such as myeloid expansion, found in Relb−/− mice and in WT → Relb−/− chimeras, appear to result from loss of RelB activity in radio-resistant cells, such as BM-resident stromal cells. It will be important to determine the identity of such cells and to determine the RelB-dependent mechanisms that normally operate to control myelopoiesis.
Materials and Methods
Mice.
The generation of Relb−/− mice has been described (3). Notch211cKO mice were generated by crossing Notch2f/f (B6.129S-Notch2tm3Grid/J) to CD11c-Cre (B6.Cg-Tg(Itgax-cre)1–1Reiz/J) mice. WT B6.SJL (B6-Ly5.2/CR) mice were purchased from Charles River Laboratories. Ciita−/− (B6.129S2-Ciitatm1Ccum/J), OT-I [C57BL/6-Tg(TcraTcrb)1100Mjb/J], and OT-II [B6.Cg-Tg(TcraTcrb)425Cbn/J] were purchased from Jackson Laboratories. OT-I and OT-II mice were bred to B6.SJL mice and maintained as CD45.1+. All mice were maintained on the C57BL/6 background in a specific pathogen-free animal facility following institutional guidelines and with protocols approved by the Animal Studies Committee at Washington University in St. Louis. Experiments were performed with mice 8–12 wk of age using sex-matched littermates as controls.
Antibodies and Flow Cytometry.
Cells were kept at 4 °C while staining in PBS with 0.5% BSA and 2 mM EDTA in the presence of CD16/32 Fc block (BD clone 2.4G2).
The following antibodies were purchased from Becton Dickinson (BD): CD11b (M1/70), CD45.2 (104), MHC-II (M5/114.15.2), Ly-6C (AL-21), CD103 (M290), CD45R/B220 (RA3-6B2), and CD64 (X54-5/71); from eBioscience: CD4 (GK1.5), CD8α (53-6.7), CD11b (M1/70), CD45.1 (A20), CD44 (IM7), MHC II (M5/114/15/2), CD24 (M1/69), CD172a (P84), Ly-6G PE (IA8), Siglec-H (eBio440C), CD317/BST2 (eBio927), and ESAM (1G8); from Tonbo Biosciences: CD45.1 (A20) and CD11c (N418); from ThermoFisher Scientific: TCR Vα2 (B20.1); from BioLegend XCR-1 (ZET). Cells were analyzed or sorted with a FACS Canto II or FACS Aria Fusion flow cytometer (BD), respectively. Data were analyzed with FlowJo software (Tree Star).
Cell Isolation and Preparation.
Spleens, thymi, and lymph nodes were minced and digested for 60 min at 37 °C with stirring in 5 mL Iscove’s complete media (cIMDM) with 250 μg/mL collagenase B (Roche) and 30 U/mL DNase I (Sigma-Aldrich). Lungs were perfused by intracardiac injection with 10 mL ice-cold PBS into the right ventricle, minced, and digested for 90 min with stirring in cIMDM with 1 mg/mL collagenase D (Roche) and 30 U/mL DNase I. Red blood cells (RBCs) were lysed with ammonium chloride–potassium bicarbonate lysis buffer, and single-cell suspensions were passed through a 70-µm strainer. Cells were counted in a Vi-Cell XR (Beckman Counter), and 2 × 106 cells were stained as described above.
Bone Marrow Cultures.
Bone marrow from femurs and tibias was flushed with 10 mL of cIMDM using a 25-gauge syringe (BD). Single-cell suspensions of RBCs were lysed and passed through a 70-µm strainer as described above. Cells were counted in a Vi-Cell XR and 2 × 106 cells/mL were cultured at 37 °C in cIMDM supplemented with 100 ng/mL FLT3 ligand (Peprotech) for 9 d. Afterward, loosely adherent cells were recovered with gentle pipetting and analyzed by flow cytometry.
Bone Marrow Chimeras.
BM cells were isolated from femurs, tibias, and iliums. Briefly, bones were crushed using a mortar and pestle, and cells were isolated from debris by gradient centrifugation with Histopaque 1119 (Sigma-Aldrich). Cells were counted, and 0.5–1 × 107 total BM cells were transferred by retro-orbital injection into recipients 24 h after whole-body irradiation (10.5 Gy). For mixed BM chimeras, whole BM cells from two different genotypes were counted, mixed at an equal ratio, and transplanted into lethally irradiated recipients. Cells were distinguished on the basis of CD45.1 and CD45.2 staining. To determine chimerism, donor contributions were normalized to the ratio of Ly-6Chi monocyte output in the spleen.
Antigen Presentation Assays.
For class I antigen presentation assays, OVA expressing L. monocytogenes (LM-OVA), a gift from H. Shen (University of Pennsylvania, Philadelphia), was cultured in brain–heart infusion broth at 37 °C for 6 h and frozen for 24 h after dilution plating for titer enumeration. Bacteria were thawed, washed three times with Dulbecco’s PBS, inactivated at 80 °C for 1 h, and stored at −80 °C until used as described before (37). Splenic OT-I cells were sorted as B220−CD11c−CD45.1+TCR-Vα2+CD8α+CD4−, labeled with carboxyfluorescein succinimidyl ester (CFSE), and cultured at a density of 1.25 × 105 cells/mL with sorted DCs (5 × 104 cells/mL) and 1 × 107 cfu ∆LM-OVA for 3 d. Afterward, OT-I cells were assayed for CFSE dilution and expression of CD44 by flow cytometry. For class II antigen presentation assays, Ciita−/− splenocytes were osmotically loaded with OVA and irradiated (13.5 Gy) as described before (38, 39). Splenic OT-II cells were sorted as B220−CD11c−CD45.1+TCR-Vα2+CD8α−CD4+ and CFSE-labeled. Labeled OT-II cells (1.25 × 105 cells/mL) were cocultured with sorted DCs (1.25 × 105 cells/mL) and OVA-loaded irradiated Ciita−/− splenocytes (5 × 105 cells/mL) for 3 d; irradiated Ciita−/− splenocytes (5 × 105 cells/mL) without OVA were used as controls. After culture the frequency of CD44+ OT-II cells that had undergone one CFSE dilution was determined by flow cytometry.
Microscopy.
Cytospins of sorted splenic B220−CD11b+Ly-6G+ PMNs from WT and Relb−/− mice were stained with Wright–Giemsa stain using the Hema 3 kit (Fisher Scientific). Images were acquired at room temperature with an Axioskop microscope (objective: 40×, 1.25, oil) using an Axiocam ICc3 camera (Zeiss).
Statistical Analysis.
Statistical analyses were performed using two-way analysis of variance with Sidak’s multiple comparison test. Error bars indicate SE of mean. All statistical analyses were performed using Prism version 7 (GraphPad Software).
Acknowledgments
We thank Dr. Ansuman T. Satpathy and Xiaodi Wu for helpful discussions. V.D. is supported by a Ruth L. Kirschstein National Research Service Award F30 DK108498. K.M.M. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ryseck RP, et al. RelB, a new Rel family transcription activator that can interact with p50-NF-kappa B. Mol Cell Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrasco D, Ryseck RP, Bravo R. Expression of relB transcripts during lymphoid organ development: Specific expression in dendritic antigen-presenting cells. Development. 1993;118:1221–1231. doi: 10.1242/dev.118.4.1221. [DOI] [PubMed] [Google Scholar]
- 3.Weih F, et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 4.Burkly L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 5.DeKoning J, et al. Thymic cortical epithelium is sufficient for the development of mature T cells in relB-deficient mice. J Immunol. 1997;158:2558–2566. [PubMed] [Google Scholar]
- 6.Elewaut D, et al. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J Exp Med. 2003;197:1623–1633. doi: 10.1084/jem.20030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powolny-Budnicka I, et al. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in γδ T cells. Immunity. 2011;34:364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Weih DS, Yilmaz ZB, Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J Immunol. 2001;167:1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- 9.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snapper CM, et al. B cells lacking RelB are defective in proliferative responses, but undergo normal B cell maturation to Ig secretion and Ig class switching. J Exp Med. 1996;184:1537–1541. doi: 10.1084/jem.184.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerloni M, Lo D, Zanetti M. DNA immunization in relB-deficient mice discloses a role for dendritic cells in IgM→IgG1 switch in vivo. Eur J Immunol. 1998;28:516–524. doi: 10.1002/(SICI)1521-4141(199802)28:02<516::AID-IMMU516>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Gerloni M, Lo D, Ballou WR, Zanetti M. Immunological memory after somatic transgene immunization is positively affected by priming with GM-CSF and does not require bone marrow-derived dendritic cells. Eur J Immunol. 1998;28:1832–1838. doi: 10.1002/(SICI)1521-4141(199806)28:06<1832::AID-IMMU1832>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.Crowley MT, Lo D. Targeted Gene Knockouts: Insights into Dendritic Cell Biology. Academic; San Diego: 1999. pp. 579–593. [Google Scholar]
- 14.Wu L, et al. RelB is essential for the development of myeloid-related CD8alpha- dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 2003;19:353–363. doi: 10.1016/s1074-7613(03)00230-9. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann J, Mair F, Greter M, Schmidt-Supprian M, Becher B. NIK signaling in dendritic cells but not in T cells is required for the development of effector T cells and cell-mediated immune responses. J Exp Med. 2011;208:1917–1929. doi: 10.1084/jem.20110128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy TL, et al. Transcriptional control of dendritic cell development. Annu Rev Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki T, et al. Visualization of RelB expression and activation at the single-cell level during dendritic cell maturation in Relb-Venus knock-in mice. J Biochem. 2015;158:485–495. doi: 10.1093/jb/mvv064. [DOI] [PubMed] [Google Scholar]
- 19.Kabashima K, et al. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 2005;22:439–450. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Satpathy AT, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin L, et al. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 22.Lewis KL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naik SH, et al. Cutting edge: Generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174:6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 24.Ridge JP, Di Rosa F, Matzinger P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature. 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 25.Bennett SR, et al. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 26.Schoenberger SP, Toes RE, van der Voort EI, Offringa R, Melief CJ. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 27.Ohtsuka H, et al. Bcl6 is required for the development of mouse CD4+ and CD8α+ dendritic cells. J Immunol. 2011;186:255–263. doi: 10.4049/jimmunol.0903714. [DOI] [PubMed] [Google Scholar]
- 28.Scott CL, et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med. 2016;213:897–911. doi: 10.1084/jem.20151715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weih F, et al. Both multiorgan inflammation and myeloid hyperplasia in RelB-deficient mice are T cell dependent. J Immunol. 1996;157:3974–3979. [PubMed] [Google Scholar]
- 30.Fasnacht N, et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med. 2014;211:2265–2279. doi: 10.1084/jem.20132528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar V, et al. A dendritic-cell-stromal axis maintains immune responses in lymph nodes. Immunity. 2015;42:719–730. doi: 10.1016/j.immuni.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Silva NS, Silva K, Anderson MM, Bhagat G, Klein U. Impairment of mature B cell maintenance upon combined deletion of the alternative NF-κB transcription factors RELB and NF-κB2 in B cells. J Immunol. 2016;196:2591–2601. doi: 10.4049/jimmunol.1501120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durai V, Murphy KM. Functions of murine dendritic cells. Immunity. 2016;45:719–736. doi: 10.1016/j.immuni.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tumanov AV, et al. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reboldi A, et al. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 2016;352:aaf4822. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laidlaw BJ, Craft JE, Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol. 2016;16:102–111. doi: 10.1038/nri.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kretzer NM, et al. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. J Exp Med. 2016;213:2871–2883. doi: 10.1084/jem.20160597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carbone FR, Bevan MJ. Class I-restricted processing and presentation of exogenous cell-associated antigen in vivo. J Exp Med. 1990;171:377–387. doi: 10.1084/jem.171.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Briseño CG, et al. Distinct transcriptional programs control cross-priming in classical and monocyte-derived dendritic cells. Cell Reports. 2016;15:2462–2474. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]