Significance
Higher plants have waxy surface layers that prevent uncontrolled water loss. Many wheat cultivars accumulate diketone epicuticular waxes in reproductive-age plants that produce a glaucous appearance. We identify INHIBITOR of WAX1 (Iw1), a dominant glaucous repressor, as a young miRNA gene (MIRNA) that produces an miRNA, miRW1, which targets the transcript of the biosynthetic gene WAX1-CARBOXYLESTERASE (W1-COE) for degradation. The high sequence similarity between the Iw1 hairpin sequence and W1-COE suggests that this MIRNA gene arose from an inverted duplication of its target. The cleavage specificity of miRW1 for its target gene defines the unique role of a young MIRNA gene in the regulation of an important agricultural trait related to stress tolerance.
Keywords: glaucous, inhibitor of wax, small RNA, long noncoding RNA, WAX1
Abstract
The cuticle of terrestrial plants functions as a protective barrier against many biotic and abiotic stresses. In wheat and other Triticeae, β-diketone waxes are major components of the epicuticular layer leading to the bluish-white glaucous trait in reproductive-age plants. Glaucousness in durum wheat is controlled by a metabolic gene cluster at the WAX1 (W1) locus and a dominant suppressor INHIBITOR of WAX1 (Iw1) on chromosome 2B. The wheat D subgenome from progenitor Aegilops tauschii contains W2 and Iw2 paralogs on chromosome 2D. Here we identify the Iw1 gene from durum wheat and demonstrate the unique regulatory mechanism by which Iw1 acts to suppress a carboxylesterase-like protein gene, W1-COE, within the W1 multigene locus. Iw1 is a long noncoding RNA (lncRNA) containing an inverted repeat (IR) with >80% identity to W1-COE. The Iw1 transcript forms a miRNA precursor-like long hairpin producing a 21-nt predominant miRNA, miRW1, and smaller numbers of related sRNAs associated with the nonglaucous phenotype. When Iw1 was introduced into glaucous bread wheat, miRW1 accumulated, W1-COE and its paralog W2-COE were down-regulated, and the phenotype was nonglaucous and β-diketone–depleted. The IR region of Iw1 has >94% identity to an IR region on chromosome 2 in Ae. tauschii that also produces miRW1 and lies within the marker-based location of Iw2. We propose the Iw loci arose from an inverted duplication of W1-COE and/or W2-COE in ancestral wheat to form evolutionarily young miRNA genes that act to repress the glaucous trait.
Plant epicuticular waxes deposited on the outer surface of the plant cuticle produce a water-resistant layer that serves to reduce nonstomatal water loss and mitigate the effects of heat and UV radiation as well as pathogen and insect attacks (1). Grasses in the Triticeae tribe, subfamily Pooideae, which include the cultivated species barley (Hordeum vulgare; 2n = 2x = 14), rye (Secale cereale, 2n = 2x = 14), durum wheat (Triticum durum; 2n = 4x = 28, AABB), and bread wheat (Triticum aestivum; 2n = 6x = 42, AABBDD), have two predominant pathways for wax production: (i) an alcohol- and alkane-rich wax pathway and (ii) a pathway leading to β-diketones and derivatives including hydroxy-β-diketones (2). The alcohol and alkane waxes are prevalent in earlier development and on leaves, whereas β-diketones dominate during the reproductive phase, particularly on leaf sheaths and flower heads (3, 4). β-Diketone wax is predominantly hentriacontane-14, 16-dione, which consists of a 31-carbon chain with carbonyl groups at C14 and C16. In durum wheat, about 20% of the β-diketone is hydroxylated to form 25-hydroxy-β-diketone, whereas in bread wheat hydroxylation is at C8 or C9 (5). β-Diketone wax deposition manifests visibly as glaucousness, a bluish-white coloration on stems, leaves, and flower heads. However, the relationship between a glaucous appearance and the total amount of cuticular wax can be inconsistent, especially during the later stages of wheat reproductive development (6, 7). Nonetheless, β-diketones are essential for the appearance of glaucousness and associated wax morphology (3). β-Diketone wax deposition and the development of glaucousness lead to a greater reflectance of incident light. Reduced light absorption can lower tissue temperatures, thereby reducing transpirational water loss, and also may reduce photosynthesis under nonsaturating illumination (1). In wheat, a glaucous appearance has been shown to be associated with stabilizing grain yield, particularly in growth environments that are water-limited and prone to heat stress (6, 8, 9). Thus, because of the protective nature of the waxy β-diketone layer, the glaucous appearance has generally been selected for during the breeding of cultivated durum and bread wheat varieties (3, 10). In contrast, a nonglaucous (NG) state is prevalent in the uncultivated relatives of wheat, including progenitor species wild emmer (Triticum dicoccoides, 2n = 4x = 28, AABB) and Aegilops tauschii (2n = 2x = 14, DD) (11, 12). As such, these species have been used in the development of NG wheat varieties and for studies on the characterization of the genes and genetic loci involved in wax deposition, in particular W1/W2 and Iw1/Iw2 (13, 14).
The complex evolution of durum and bread wheat as multilevel genome mosaics means each of the three subgenomes in wheat (A, B, and D) has the potential to contribute to the inheritance of glaucousness (15). However, only the B and D subgenomes contain major glaucousness loci, and the A genome progenitor Triticum urartu does not contain any appreciable β-diketones and is NG (5, 16). Within the B subgenome, WAX1 (W1) and INHIBITOR OF WAX1 (Iw1) have been mapped very close to each other on the distal end of 2BS. Conversely, in the D subgenome, W2 and Iw2 have been mapped far apart, with W2 at the proximal end and Iw2 at the distal end of 2DS (16). As the name suggests, the Iw loci provide epistatic dominant inhibition over the W loci. In recent years, several reports have furthered the characterization of these loci, including fine-mapping of Iw1 (11), demonstrating Iw1 suppression of β-diketone wax accumulation (17), fine mapping of Iw2 in Ae. tauschii (12), comparative mapping of Iw1 and Iw2 in hexaploid wheat (18), determining the impact of W and Iw loci on glaucousness and cuticle permeability (3), and fine mapping of W1 in hexaploid wheat (19). The synthesis and chemistry of diketone waxes has been studied extensively in barley, chiefly through the characterization of loss-of-function mutants of three tightly linked loci collectively referred to as “Cer-cqu” (2, 20, 21). The Cer-cqu operon, as it has been described, is associated with the cer-c, -q, and -u complementation groups and corresponding mutants glossy sheath 6 (gsh6), gsh1, and gsh8, respectively, and have been mapped close to the terminus of the short arm of chromosome 2H (22). A recent study identified the CER gene cluster including GSH1 (Cer-Q) as encoding a lipase/carboxylesterase, GSH6 (Cer-C), as a chalcone synthase-like polyketide synthase and GSH8 (Cer-U) as a cytochrome P450-type hydroxylase (23). More recently, the wheat W1 locus was identified as a gene cluster that is collinear to the barley CER gene cluster (24) and includes orthologs of Cer-Q, Cer-C, and Cer-U (23), which we define as W1-COE (carboxylesterase), W1-PKS (polyketide synthase), and W1-CYP (cytochrome P450 hydroxylase), respectively. However, it is not known which of the genes at the W1 locus are regulated by Iw1 and therefore are responsible for the presence or absence of diketone waxes and the glaucous phenotype. More importantly, the inhibitor genes Iw1 and Iw2 have not been identified, and their mechanism of action is unknown.
Long noncoding RNAs (lncRNAs) are a large and diverse class of RNA transcripts with a length of more than 200 nt that do not encode proteins. LncRNAs are emerging as important regulators in a wide range of essential biological processes. In humans, lncRNAs represent more than 68% of the transcriptome, and 79% of the lncRNAs were previously unannotated (25). Our current knowledge of their biological functions is limited, and lncRNA research in plants lags behind lncRNA research in animals. To date, very few lncRNAs have been characterized in detail (26). Some lncRNAs can be precursors of small RNAs (sRNAs) including miRNAs, which are a class of sRNA ranging from 20–24 nt in length that regulate numerous pathways and biological processes (27). miRNAs play significant roles in posttranscriptional gene regulation through base pairing with specific target sequences in their complementary mRNA targets, leading to transcript degradation (28, 29). The mechanism of action of miRNAs implies that they typically act as genetically dominant-negative regulators. Therefore, because the Iw loci are dominant repressors of the glaucous trait, we investigated the possible involvement of sRNAs in the regulation of wax accumulation. Using near-isogenic lines (NILS) from durum wheat that differed in glaucousness (30, 31), we compared the sRNAs in each of the isogenic pairs and identified a set of related miRNAs associated with repression of β-diketone deposition. We show that the Iw1 locus is a miRNA gene (MIRNA) that encodes a miRNA precursor and represses β-diketone deposition via miRNA-mediated cleavage of W1-COE transcripts.
Results
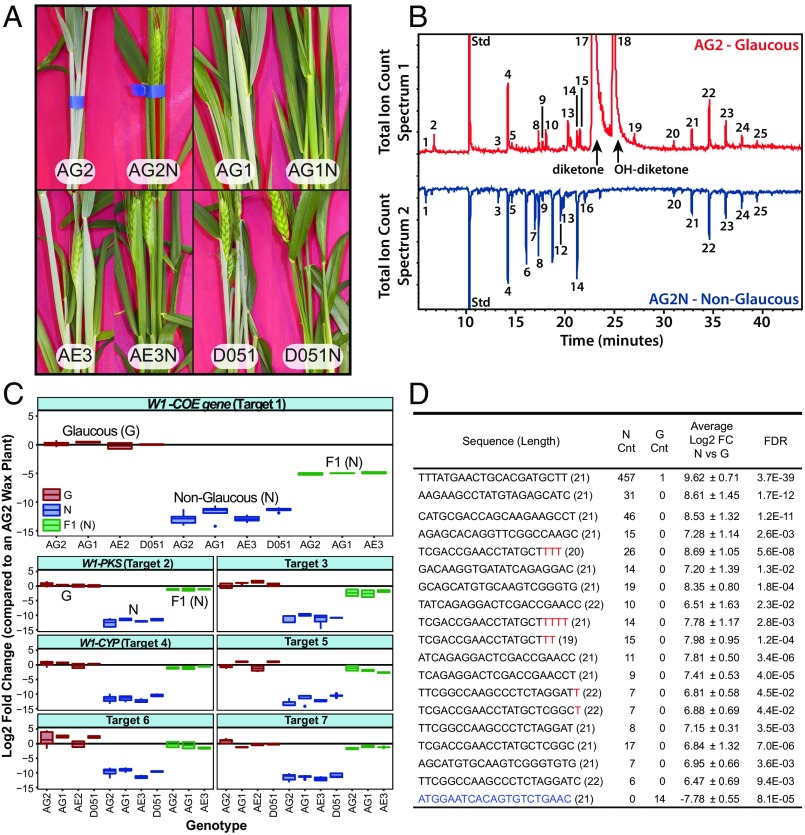
To investigate the genetic basis for glaucousness in wheat, we characterized four pairs of NILs of durum wheat, AG1, AG2, AE3, and D051, defined by the presence or absence of the glaucous trait (30, 31). These lines were produced by back-crossing a NG cultivar to a glaucous parent and then maintaining heterozygosity for glaucousness in the F4 and later generations (SI Appendix, Table S1A) (31). Glaucous lines showed bluish-white coloration from the booting stage in the stems, leaves, and floral tissues, whereas NG lines were green and glossy (Fig. 1A). The cuticular wax content and composition from leaf sheaths of all four NIL pairs was analyzed by GC-MS. Glaucous lines contained primarily β-diketone (hentriacontane-14, 16-dione) and 25-hydroxy-β-diketone (25-hydroxyhentriacontane-14, 16-dione) (5, 32), whereas the NG lines contained no detectable diketone waxes (Fig. 1B and SI Appendix, Table S1). Importantly, crosses between three pairs of NILs, AG1, AG2, and AE3 (glaucous × NG), resulted in F1 plants that were 100% NG (SI Appendix, Fig. S1), confirming the dominance of the NG trait, as observed previously (30, 31).
Fig. 1.
Characterization of durum wheat NILs differing in the presence or absence of the glaucous trait. (A) Phenotypes of the four NIL pairs, AG2/AG2N, AG1/AG1N, AE3/AE3N, and D051/D051N. Each pair consists of a glaucous line and a corresponding NG (N) isoline. (B) GC-MS analysis of the surface waxes confirms that the glaucous appearance is caused by the presence of β-diketones. (C) qPCR analysis of seven target genes present on chromosome 2BS including the W1 cluster genes W1-COE, W1-PKS, and W1-CYP. Target genes were identified through differential expression analysis using RNA-seq (Dataset S1). (D) Differentially expressed sRNAs common to all four NIL comparisons described in A. Black sequence color indicates a perfect match to the putative Iw1 sequence; red indicates probable sRNA U/A tailing modifications; blue-colored sRNA up-regulated in glaucous lines does not map to Iw1. Total read counts (Cnt) were normalized to 10 million total reads; FDR represents the highest significance of an isogenic pair; the significance threshold was at an adjusted P value (FDR) ≤ 0.05.
Transcripts Associated with Wax Production in the Durum NILs.
Transcripts in NIL pairs were compared to identify differences that were consistently associated with the loss of diketone wax production and glaucousness. Potential wax-related genes that were strongly down-regulated in NG lines were first identified by mapping reads to the National Center for Biotechnology Information (NCBI) unigene set and determining significant differences based on count data (edgeR, P ≤ 0.05). We found 16 unigenes that were commonly down-regulated in all four glaucous/NG NIL comparisons (Dataset S1). Consistent with previous mapping studies locating W1 and Iw1 on 2BS (16), blasting the 16 unigenes against the International Wheat Genome Sequencing Consortium (IWGSC) wheat survey sequences from AABB genomes revealed that most of the contigs were located on 2BS scaffolds (Dataset S1). Through further bioinformatics analyses, we defined these 16 unigenes into seven potential target genes (Dataset S1). Three of these target genes, targets 1, 2, and 4, were from the W1 locus gene cluster: W1-COE, W1-PKS, and W1-CYP. To confirm the significance of the differentially expressed genes from the NCBI unigene reference, we also used the IWGSC transcript set (v1 from EnsemblPlants) as a reference for analysis with the addition of the unannotated W1-COE sequence. Using this reference, the RNA sequencing (RNA-seq) data were reanalyzed using the pseudoalignment program kallisto and the Bioconductor package DESeq2 (adjusted P value ≤ 0.05) (33, 34). Similar differentially expressed target genes were obtained and included the W1 gene cluster: W1-COE, W1-CYP (Traes_2BS_163390FC4), target 6 (Traes_2BS_D6F1011EA), and W1-PKS (Traes_2BS_9E10D26DB) on 2BS and transcripts with high homology to W1-PKS: Traes_3B_FC275A64D, Traes_4BS_AB8E1AD32, and Traes_6BS_C400F1983 (Dataset S2). With respect to expression level, all seven of the potential target genes showed virtually no expression in NG NILs with an average down-regulation of more than 2,800-fold (Fig. 1C). In F1 heterozygous lines, W1-COE expression was still down-regulated by 31.3-fold on average, but the expression of all other targets, including W1-PKS and W1-CYP, recovered to a large extent, showing down-regulation between 1.7- and 4.7-fold (Fig. 1C), suggesting that W1-COE is most likely the gene controlling the glaucous trait in these durum NILs.
To establish whether W1-COE is indeed involved in diketone wax production, we used virus-induced gene silencing (VIGS) to block its expression transiently in wheat. Fragments of W1-COE were integrated into a VIGS system using the barley stripe mosaic virus (35) and were applied to the leaves of glaucous AG2 plants at the tillering stage, before visible glaucousness was apparent. Then the development of glaucousness was monitored for 4–6 wk. W1-COE fragments all produced large reductions in visible glaucousness relative to waxy controls (SI Appendix, Fig. S2) and in total diketone wax accumulation in leaf sheaths (SI Appendix, Figs. S3A and S4). Control infections with a PHYTOENE DESATURASE (PDS) fragment produced a slight reduction in wax content which may be attributed both to the general effects of viral infection and to the reduction in pigment accumulation resulting from the inhibition of PDS. The levels of W1-COE expression in VIGS-treated plants were measured by quantitative PCR (qPCR), which showed that all four of the tested fragments reduced the expression of the gene (SI Appendix, Fig. S3B). In addition, there was a linear correlation between the amount of β-diketone wax and the expression of W1-COE (SI Appendix, Fig. S3C). The VIGS result further confirmed the hypothesis that W1-COE has a primary role in regulating diketone wax production in the durum NILs and is in agreement with recent reports identifying the barley Cer-cqu and wheat W1 gene clusters (23, 24).
sRNAs Associated with the NG State Show Targeting Specificity for W1-COE.
Differential expression analysis of sRNAs of 19–28 nt in length (edgeR adjusted P value ≤ 0.05) revealed a series of 19- to 22-nt sequences in NG lines that were almost completely absent in glaucous lines (Fig. 1D and Dataset S3A). These sRNA sequences up-regulated in NG lines could not be perfectly mapped to the IWGSC wheat genome survey sequences. However, the most abundant sRNA, 21 nt in length with 9,403 total reads and 457 average reads per 10 million (Fig. 1D), mapped to W1-COE with one mismatch (Dataset S3 A and B). From the sRNA reads, five other 19- to 22-nt sequences also mapped to W1-COE with one mismatch or less (Dataset S3B). Because the most abundant sRNA was complementary to a specific sequence in W1-COE, we designated the sRNA sequence as “microRNA specific to W1-COE” (miRW1) (Fig. 2). As mentioned, expression of miRW1 and other related sRNAs was almost absent in glaucous lines but was present in NG lines, including the F1 heterozygous progeny of crosses between the glaucous and NG NILs (Fig. 3A and Dataset S3 A and B). Because miRW1 had no sequence homolog other than W1-COE sequences in the wheat NCBI unigene set or coding sequences within the IWGSC survey sequence (Dataset S4A), we considered the possibility that miRW1 was derived from W1-COE. However, RNA structure-prediction software indicated that the W1-COE transcript could not fold to form a hairpin loop structure characteristic of miRNA precursors. Furthermore, the concept of W1-COE as a miRNA precursor is inconsistent with the inverse correlation between W1-COE and miRW1 expression (i.e., W1-COE being expressed in glaucous lines and miRW1 being preferentially expressed in NG lines). Therefore, the more likely explanation is that miRW1 is produced from an unknown precursor gene and targets W1-COE for suppression.
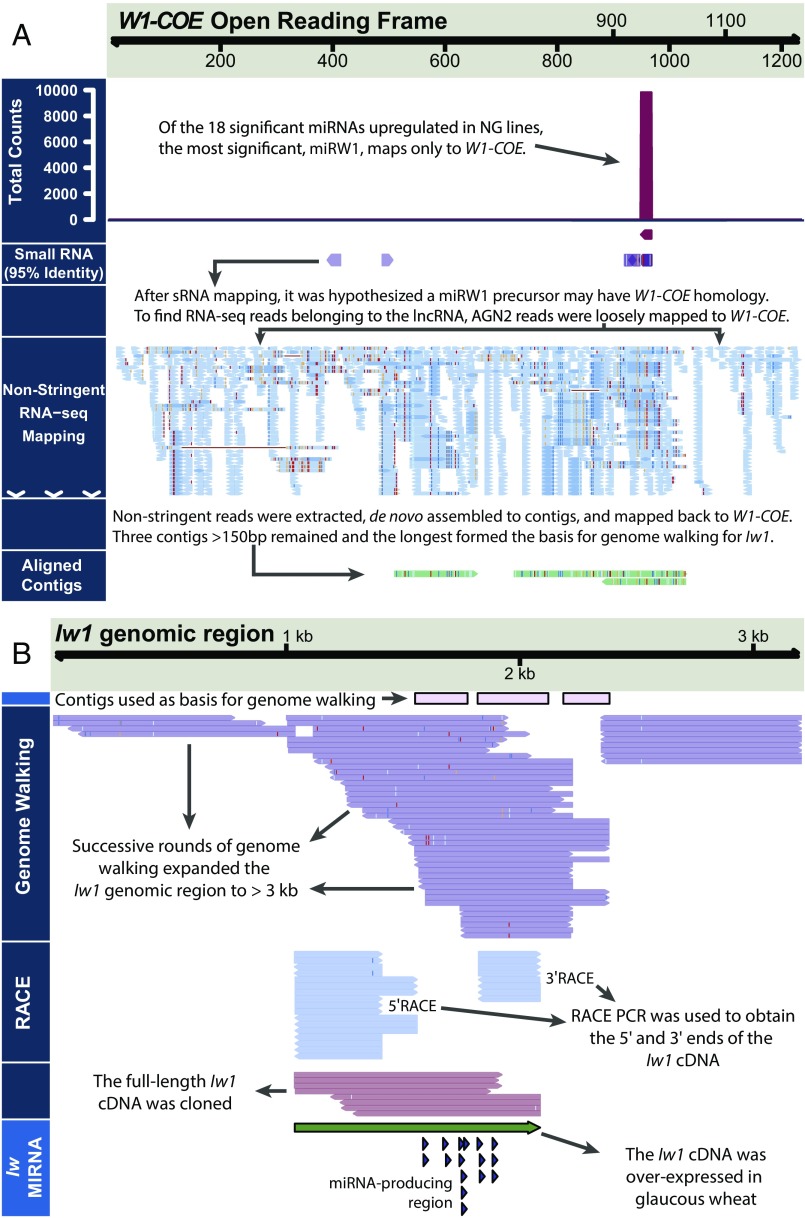
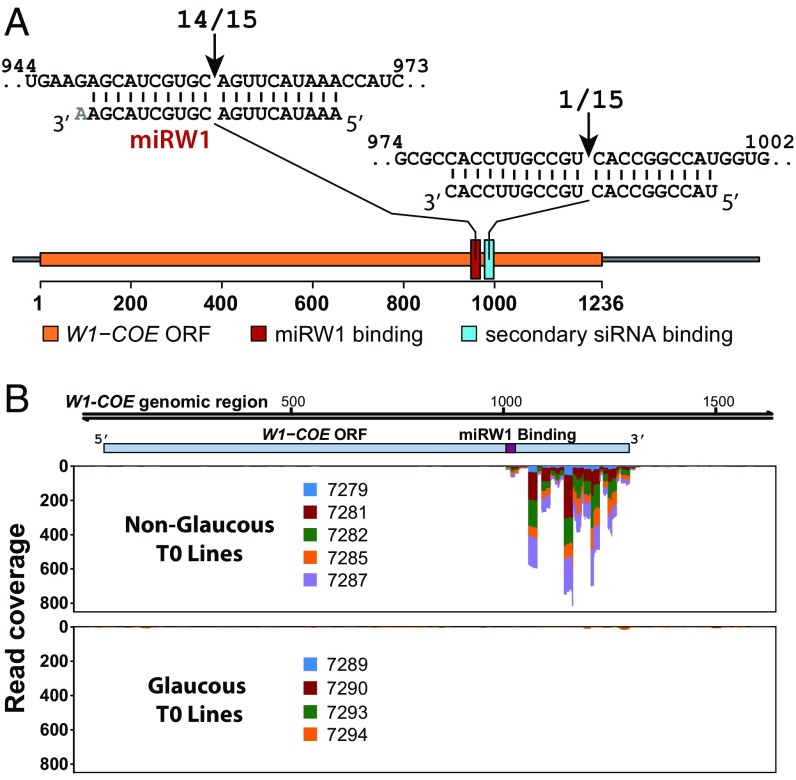
Fig. 2.
Identification and cloning of Iw1 from NG durum wheat NILs. (A) Schematic representation of the Iw1 cloning strategy. The premise of the experiment was to identify the Iw1 sequence based on its suspected loose homology to the suspected regulatory target W1-COE. (B) After identification of potential Iw1 contigs, genome walking was performed to isolate a 3-kb genomic DNA fragment from NG NILs. Using 5′ and 3′ RACE PCR, a 1-kb cDNA was obtained that included a region where differential sRNA mapped (Fig. 1D and Dataset S3). Nucleotide polymorphisms within sequencing reads and contigs are represented by colored bars.
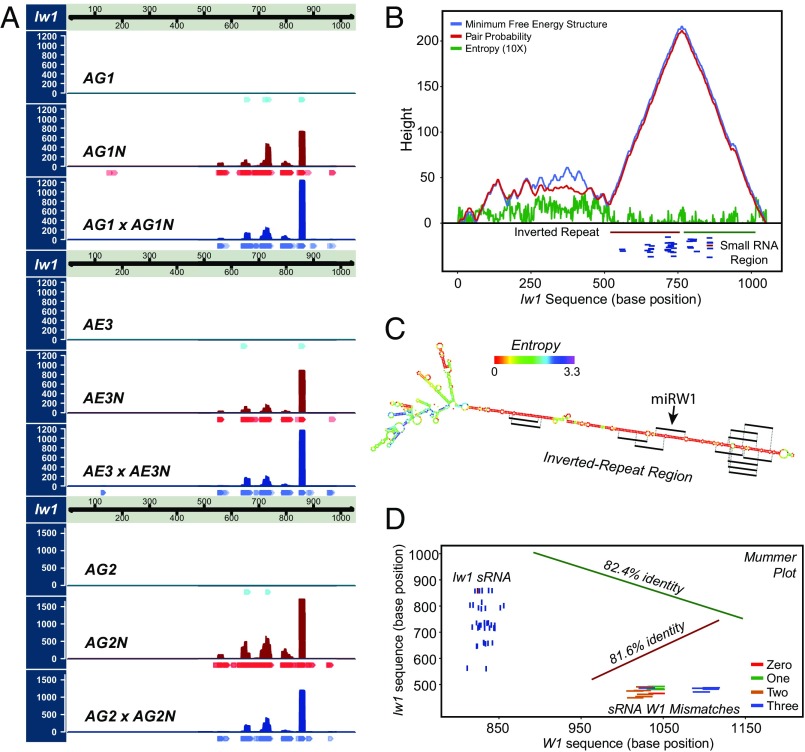
Fig. 3.
Characterization of the Iw1 lncRNA. (A) Expression of sRNA mapping to Iw1 in three pairs of NILs and in the F1 heterozygous generation resulting from genetic crosses between each pair of isolines. (B) Iw1 contains an IR with high base-paring probability. The mountain plot demonstrates that the 3′ end of Iw1 (∼500 bp) has a high probability of forming a hairpin structure in the region of an IR. An sRNA-producing region is identified within this hairpin IR. (C) The Iw1 hairpin structure. The IR region has the highest base-pairing probability consistent with low entropy values. Significant sRNAs with perfect homology are shown. (D) A mummer plot displays the relationship of Iw1 and its potential target gene W1-COE. The IR region shows greater than 80% identity with portions of the W1 gene. sRNA mapping to Iw1 (perfect match) and to W1-COE (up to three mismatches) are also displayed.
The miRW1 Precursor Contains a Hairpin-Forming Inverted Repeat with Homology to W1-COE.
We hypothesized that the putative miRW1 precursor could have weak homology to W1-COE based on evidence from the literature indicating that miRNAs and their targets can have sequence similarities that extend beyond the sequence of the miRNA itself (36, 37). Because 8 of the 18 differentially expressed miRNAs had homology to W1-COE (with three mismatches or fewer) (Dataset S3B), we surmised that the NG lines may contain RNA-seq reads from a precursor sequence with homology to W1-COE. Thus, all RNA-seq reads from NG lines, in which the putative miRNA precursor, but not W1-COE, would be expressed were pooled and aligned against W1-COE with low stringency (requiring a contiguous aligned read length of >20% to W1-COE with >80% homology). The mapped sequences were extracted and collected for de novo assembly. Three contigs of greater than 150 nt were obtained (Fig. 2A). Several of the differentially expressed sRNA sequences, including the most abundant miRW1 sequence, could be perfectly mapped to two of these contigs, suggesting that one or more of the contigs was part of the miRW1 precursor (Fig. 2B). These contigs were the starting point for a series of genome-walking experiments that allowed us to obtain a putative 3,207-nt genomic sequence fragment (Fig. 2B). Because many MIRNA genes have a 5′ cap structure and 3′ polyadenylation (38, 39), we performed 5′ and 3′ RACE-PCR from primers designed around the location where the sRNAs mapped within the precursor fragment obtained from genome walking (Fig. 2B and SI Appendix, Table S4). Through RACE, two primary miRNA sequences differing by three bases at the 5′ end were obtained, the longest of which was 1,051 nt. Sequence analysis revealed that the miRW1 precursor is a lncRNA that contains an inverted repeat (IR) from nucleotides 520–756 and from nucleotides 770–1014 with high base-paring probability (Fig. 3B). Structure prediction indicated with high confidence that the lncRNA could fold into a long hairpin loop structure with the IR forming the stem (Fig. 3C). Analysis of the similarity between the lncRNA and W1-COE showed that each repeat of the MIRW1 hairpin shares ∼82% identity with the W1-COE sequence (Fig. 3D), suggesting that the IR of the lncRNA originated from an inverted duplication of W1-COE, the mechanism proposed by Allen et al. (40). Of the 18 sRNAs significantly up-regulated in NG lines, 13, including the miRW1 sequence, mapped perfectly to the foldback region of the lncRNA, and all the sRNAs up-regulated in NG lines can be mapped to the lncRNA if sRNA tailing is considered (Fig. 3C and Dataset S4B). Tailing involves the nontemplated addition of bases to the 3′ end of sRNAs through adenylation or uridylation (41–43). Taking these findings together, we conclude that the miRW1 precursor forms a long hairpin structure that is processed to produce miRW1 and other sRNAs that specifically target W1-COE.
Expression of the miRW1 Precursor in Glaucous Wheat Creates an NG Phenotype Through Repression of W1-COE and W2-COE.
Introduction of the 1,051-nt miRW1 precursor driven by the maize ubiquitin1 promoter into the bread wheat cultivar Bobwhite resulted in an obvious NG appearance in 20 of 29 T0 plants (Fig. 4 and Dataset S5A). Analysis of diketone waxes in T0 transgenic lines also revealed that the NG trait was the result of the absence of β-diketones (SI Appendix, Fig. S5). The NG phenotype and the absence of diketone waxes was heritable and carried over to the T1 generation (SI Appendix, Figs. S6 and S7).
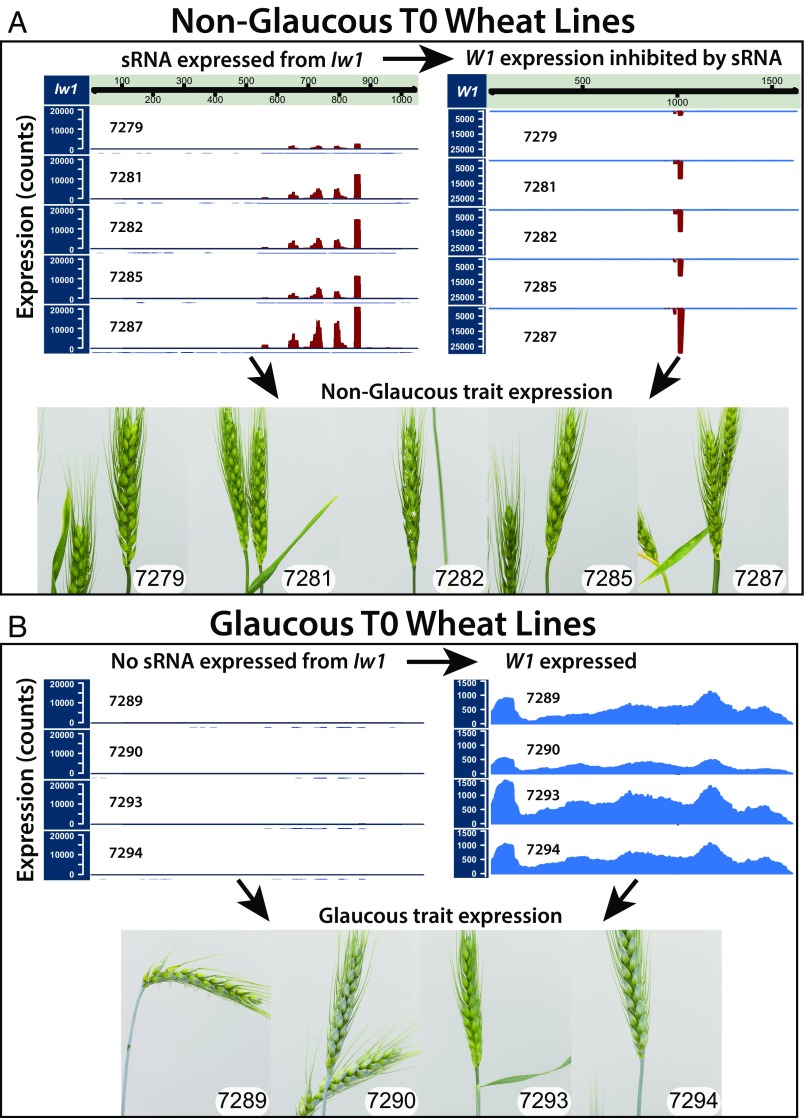
Fig. 4.
Characterization of transgenic lines overexpressing Iw1 through sRNA and RNA-seq analyses. (A) NG T0 overexpression lines displaying expression of sRNA derived from Iw1 and repression of W1-COE. (B) In contrast, glaucous T0 lines showed almost no expression of sRNA from Iw1 and showed expression of W1-COE. Analyses of sRNA and RNA-seq data demonstrated that the differences between the glaucous and NG lines were caused by the expression of sRNA from Iw1 and the resulting repression of W1-COE (SI Appendix, Tables S2 and S3 and Dataset S5). GC-MS experiments in the T0/T1 generation confirmed that the NG trait was caused by the absence of β-diketones (SI Appendix, Figs. S5–S7).
RNA-seq analyses of five NG and four glaucous T0 plants were carried out to determine both differentially expressed genes and sRNAs. Similar to the analysis of the wax NILs, we used two approaches: mapping to the NCBI unigene set and pseudoalignment to the IWGSC v1 transcript set. Four unigenes in the NG T0 lines were significantly down-regulated (edgeR adjusted P value ≤ 0.05); all were fragments of W1-COE or its paralog on chromosome 2DS, W2-COE (SI Appendix, Table S2). Following pseudoalignment with kallisto, the only differentially expressed transcript was W1-COE (DESeq2 adjusted P value ≤ 0.05) (SI Appendix, Table S3). Of the sRNAs, 222 were differentially up-regulated in the NG T0 lines (edgeR adjusted P value ≤ 0.05) (Dataset S5B), and 208 of the 222 could be mapped to the miRW1 precursor when tailing was considered (Dataset S4C). Other significantly up-regulated sRNAs mapped perfectly to W1-COE and its paralog on 2DS (W2-COE) (Dataset S4D). The numbers of sRNAs mapping to the miRW1 precursor and to the targets W1-COE and W2-COE are consistent with the observed reductions in W1-COE and W2-COE transcript levels in NG phenotypes (Fig. 4 and SI Appendix, Fig. S8).
To validate that W1-COE is the target of miRNA-guided cleavage in the transgenic lines, we performed a 5′ RACE assay to map possible cleavage sites within W1-COE (Fig. 5A). The principal position of cleavage within W1-COE was within the miRW1-binding site located between nucleotides 10 and 11 and was present in 14 of 15 cloned sequences. Furthermore, in NG transgenic lines with active cleavage of W1-COE, we detected the presence of secondary siRNA in the 3′ cleavage fragment (Fig. 5B). The majority of these siRNAs were 21 nt in length and were positively related to both the abundance of miRW1 and its monouridylated form (SI Appendix, Fig. S9 A and B). Secondary siRNAs also were apparent in the 3′ region adjacent to the miRW1-binding site in W2-COE (SI Appendix, Fig. S9C). The presence of secondary siRNAs also was detected in the heterozygous F1 generation in the isogenic lines but, interestingly, not in the NG homozygous lines (SI Appendix, Fig. S9D).
Fig. 5.
Validation of the cleavage of the W1-COE mRNA target by miRW1. (A) W1-COE cleavage sites identified by 5′ RACE. Sequences of 14 of 15 clones showed cleavage between nucleotides 10 and 11 of the miRW1-binding site. One cleavage site was located within a minor sRNA-binding site between the 10th and 11th nucleotide. (B) Mapping of secondary siRNAs to W1-COE in Iw1 T0 transgenic lines. (Upper) sRNAs with perfect homology to W1-COE mapped to a region 3′ of the miRW1-binding site in NG lines. (Lower) Almost none did so in glaucous lines.
These results from the introduction of the miRW1 precursor into glaucous wheat provide further validation that miRW1 acts as a repressor of wax production through miRNA-mediated suppression of W1-COE/W2-COE expression. The results from the overexpression experiments suggest that the miRW1 precursor is the wax inhibitor Iw1, the expression of which has the ability to silence specifically both W1-COE and W2-COE.
The IR Region of Iw1 Maps to the Location of Iw2.
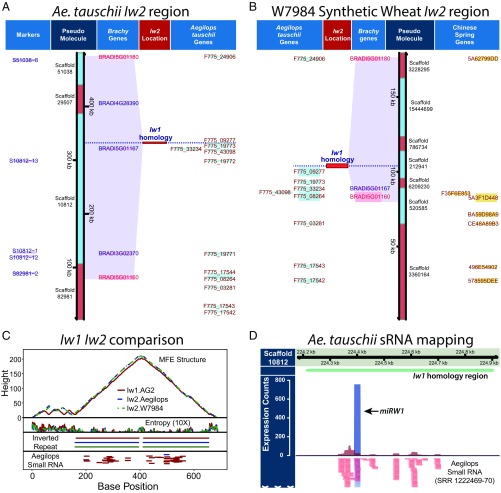
We were unable to map the Iw1 sequence to the IWGSC wheat genome survey sequences, indicating that Iw1 is not represented in the currently available reference genomes for the Chinese Spring cultivar. However, a 689-nt Iw1 fragment was mapped to scaffold 10812 of chromosome 2 of the D genome progenitor Ae. tauschii (44). Further, this Iw1 fragment maps to the precise location in Ae. tauschii to which Iw2 has been fine mapped previously (12) (Fig. 6A and SI Appendix, Fig. S10), and the location is consistent with genetic markers from syntenic blocks from species such as Brachypodium distachyon that include the genes BRADI5G01180 and BRADI5G01160 (17, 18, 44). The Iw1 homology region lies within the promoter region of the Ae. tauschii gene F775_09277 encoding cytochrome P450 84A1 (Fig. 6A). Additional mapping evidence comes from the synthetic hexaploid wheat W7984, for which a shotgun survey sequence assembly is available (45). Comparative analysis of the collinear regions of Ae. tauschii and W7984 revealed that the Iw1 fragment maps to W7984 scaffold 212941 in a similar context as in Ae. tauschii (Fig. 6B and SI Appendix, Fig. S10). Therefore, based on the dominant-negative effect of Iw1 and miRW1 on the glaucous trait and the colocalization of an Iw1 fragment with markers for Iw2, we propose that two miRW1 precursors, Iw1 and Iw2, are present on chromosomes 2BS and 2DS, respectively. The IR and sRNA-producing regions of Iw1 and Iw2 are highly homologous (94%), and both are energetically favored to form a hairpin structure (Fig. 6C). Additional evidence of an Iw2 hairpin-forming RNA comes from sRNA sequencing experiments downloaded at the NCBI short-read archive from Li et al. (46). Similar to Iw1, the Iw2 region also produces a series of sRNAs, including the most dominant sequence miRW1 (Fig. 6D).
Fig. 6.
Iw2 in the D subgenome is a paralog of Iw1 from the B subgenome. (A and B) The IR, sRNA-producing sequence of Iw1 maps to the Iw2 region in Ae. tauschii and the W7984 synthetic hexaploid wheat (SHW). The location of the Iw1 homology region is consistent with Ae. tauschii S10812 markers and the Brachypodium distachyon (Brachy) genes BRADI5G01180 and BRADI5G01160. (C) Comparison of Iw1 and the homologous Ae. tauschii and W7984 SHW sequences reveals a high similarity including ∼95% sequence identity between Iw1 and the D-genome sequences, the presence of IRs, and high base-pairing probabilities in the minimum free energy (MFE) structure. (D) sRNA libraries from spikes of Ae. tauschii revealed a similar pattern of sRNAs in the 689-bp Iw1 homology region on Ae. tauschii scaffold 10812 with the 21-bp sequence miRW1 predominating.
Discussion
We have identified and validated the regulatory function of Iw1 and confirmed the role of a key gene, W1-COE, within the W1 locus. Iw1 and its homolog Iw2 are young MIRNA genes with long hairpin precursors which ultimately suppress β-diketone wax production. Iw1 and Iw2 produce miRW1, which specifically targets and represses the expression of the putative carboxylesterase genes that are necessary for the production of β-diketone waxes in wheat (24). Identification of the Iw loci represents a major step forward in our regulatory understanding of the glaucous trait in wheat and related species, from both a functional and an evolutionary standpoint.
miRNAs have a significant regulatory role in plants and target a wide range of transcripts for degradation and therefore are inherently dominant-negative genetic factors (28, 41). Many evolutionarily conserved miRNA families play critical roles in plant development and adaptation to diverse environments. There also are many nonconserved, evolutionarily recent miRNAs and their corresponding targets that are present only within a few closely related species or appear to be unique to specific species (47–49). Wheat miRNA sequencing, identification, profiling, and characterization have been reported extensively (50–56). However, neither miRW1 nor its precursor has been reported, possibly because of the atypical characteristics of Iw1 and Iw2. In fact, the Iw1 sequence does not exist in any available wheat genome reference or sequencing database such as the NCBI. The identification of Iw1 and Iw2 as long noncoding, hairpin-forming, sRNA-producing RNAs with IRs similar to their target sequence places them among the few functional lncRNAs described in monocots, the most notable previous example being the maize Mu killer locus (57). Mu killer arose from an inverted duplication of a sequence similar to its target, the MuDR transposon; however Mu killer acts via the production of siRNAs and an epigenetic mechanism (58, 59) instead of the miRNA-based silencing mechanism of the Iw genes. lncRNA-mediated gene regulation is emerging as a common regulatory mechanism in plants. A variety of lncRNA-mediated regulation mechanisms have been unraveled, including target mimicry, transcription interference, PRC2-associated histone methylation, and DNA methylation (26). However, although the number of known plant lncRNAs is expanding, the great majority have no known function (60–65). In wheat, the lncRNA landscape has been profiled during fungal responses and heat stress, but the characterization of function is deficient (66, 67).
As described here, Iw1 and Iw2 serve as miRNA precursors and repress target gene expression through a miRNA-mediated mechanism. Several lines of evidence indicate that Iw1 and Iw2 are evolutionarily young MIRNA genes that arose by inverted duplication of their target gene (39, 49). First, the foldback region of Iw1 has extended similarity (>80%) with the target W1-COE beyond that of the miRW1 region. Second, the Iw1 primary transcript (1,051 nt) is much longer than typical miRNA primary sequences; 98% of miRNA precursor lengths are <336 nt with a mean of 146 nt (68). Third, the foldback regions of Iw1 and Iw2 are hairpin structures >200 nt that resemble a dsRNA and do not resemble the typical short structure of miRNA hairpins. In Arabidopsis, Ben Amor et al. (27) identified nine ncRNAs corresponding to miRNA, trans-acting siRNA, and 24-nt siRNA precursors, including a young MIRNA gene MIR869A. The transcript of MIR869A is processed by DCL4, because its secondary structure is closer to that of dsRNA than to that of a typical, short miRNA precursor processed by DCL1 (69). The example of MIR869a might indicate the evolutionary path of the Iw genes, with younger dsRNA-forming MIRNA genes evolving through the production of miRNA-like siRNA, because the Iw1 and Iw2 hairpin precursors also produce other sRNAs in addition to the predominant 21-nt miRW1. Fourth, we show evidence that miRW1, the predominant miRNA, is primarily responsible for the cleavage of the W1-COE transcript. Moreover, cleavage between nucleotides 10 and 11 of miRW1 is consistent with the principle hallmark of miRNA-guided degradation (40, 70, 71). The presence of secondary siRNAs mapping to the 3′ cleavage fragment in the NG Iw1 overexpression and F1 heterozygous crosses of NIL pairs provides additional evidence of miRNA-directed degradation of W1-COE and W2-COE. In plants there are two models, “one-hit” and “two-hit,” for secondary siRNA production. In the one-hit model, the trigger can be binding of 22-nt miRNAs, and in the two-hit model the trigger can be two neighboring miRNA target sites on the same mRNA (72, 73). Both models are possible triggers for the secondary siRNAs arising from the cleavage of W1-COE (and W2-COE). A 22-nt monouridylated form of miRW1 supports the one-hit model; alternatively, additional miRNAs arising from Iw1 with W1-COE as a potential binding target support the two-hit model. As mentioned above, one curious aspect of the discovery of secondary siRNA arising from W1-COE is the absence of secondary siRNA in the homozygous NILs, and this difference leads into a discussion of whether W1 genes are present in NG lines and cultivars.
One feature of the results leading to the identification of W1 and Iw1 is that in homozygous NG NILs other genes in addition to W1-COE are strongly down-regulated (Fig. 1C), notably W1-PKS and W1-CYP in the W1 gene cluster on chromosome 2BS, which also are involved in the β-diketone and OH-β-diketone synthesis pathways (24). However, Iw1 dominantly regulates glaucousness through miRW1-promoted cleavage and mRNA degradation of W1-COE, and overexpression of Iw1 in bread wheat down-regulated only W1-COE and its paralog W2-COE (Fig. 4 and SI Appendix, Tables S2 and S3). Moreover, in NG F1 heterozygous lines, which manifest Iw1 dominance, W1-COE was the key down-regulated gene related to diketone wax synthesis (Fig. 1C). These results show, first, that Iw1-mediated repression of W1-COE causes loss of the glaucous phenotype and, second, that there is another mechanism that down-regulates multiple genes at the W1 locus in NG homozygotes. Interestingly, Hen-Avivi et al. (24) provide evidence that W1-COE, W1-PKS, and W1-CYP are missing from the W1/Iw1 genomic interval in the glossy, Iw1-containing wild emmer accession TTD140 but are present in the W1 metabolic gene cluster found in the glaucous cultivar Zavitan. The idea that the W1 genes are missing or have moved to a transcriptionally inactive part of the genome in the NG genotype is interesting and is consistent with our observations of very strong down-regulation of the W1 cluster and the lack of secondary siRNAs from W1-COE in the homozygous NILS. However, the relationship between the W1 gene cluster and Iw needs to be explored further by sequencing more glaucous and NG cultivars.
The presence of Iw in selected species within the Triticeae tribe allows us to propose an approximate evolutionary origin of Iw. Barley, which contains diketone wax but in which there are no reports of a dominant wax inhibitor gene, diverged from wheat 8–12 Mya, suggesting that the inverted duplication event that created Iw occurred after this date (74–77). The inverted duplication may have been a single event in an ancestral wheat genome lineage or separate later events resulting in convergent evolution in B (Iw1) and D (Iw2) genome species. A single inverted duplication of W1-COE in an ancestral B genome is plausible, based on the evidence presented by Marcussen et al. (15), who suggest that a hybridization event between A and B lineages occurred ∼5.5 Mya and led to the origin of the D genome lineage. The time of Iw creation at <12 Mya is comparable to the creation of young MIRNA genes in the Arabidopsis genus at <20 Mya or in Arabidopsis thaliana itself at <10 Mya (49, 78–81). In contrast, more ancient conserved MIRNA genes (e.g., miR156) predate the separation of the monocots and dicots at ∼150 Mya (82).
In summary, the specific and unique interaction between Iw and miRW1 with W-COE represents a mechanism for dominant gene repression and provides a basis for genome-wide identification of other nonconserved lncRNA functions or atypical MIRNA genes. Furthermore, the identification of the Iw genes as a major regulatory mechanism governing W-COE expression and β-diketone deposition suggests the possibility of precise gene editing or marker-based manipulation of glaucousness for better adaptation to specific conditions and environments.
Materials and Methods
Details of sample preparation, experimental procedures, and data analysis with associated references can be found in SI Appendix, Materials and Methods.
The sRNA and RNA-seq data have been submitted to the Sequence Read Archive (SRA) at the NCBI with the accession numbers SAMN05725181–SAMN05725246 (mRNAs and sRNAs in Triticum durum and Triticum aestivum). The Iw1 full-length cDNA sequence was submitted to dbEST (NCBI). The accession number is KX823910.
Supplementary Material
Acknowledgments
We thank Drs. Jitao Zou of the National Research Council of Canada (NRC), Andrew Sharpe (Global Institute for Food Security, University of Saskatchewan), and Weiren Wu (Fujian Agriculture and Forestry University) for helpful comments during the course of the project and in the preparation of this paper; Dr. Ron Knox (Agriculture and Agri-Food Canada) for providing the NILs used in this research; Mr. Joe Hammerlindl and Mr. Allan Kolenovsky of the NRC-Saskatoon Plant Transformation Service Facility for wheat transformation and selection of transformants; Dr. Shawn Clark and Mr. Enwu Liu for suggestions regarding VIGS experiments; Mr. Darwin Reed for optimizing GC-MS conditions for wax analyses and for locating wax standards synthesized by the late Dr. Pat Tulloch and colleagues; the NRC-Saskatoon Genomics Service Facility for DNA, RNA, and sRNA sequencing; Mr. Dustin Cram for bioinformatics assistance; and Drs. Assaf Distelfeld (Tel-Aviv University) and Cristobal Uauy (John Innes Centre) for providing access to additional genome sequence data. Funding for this project was provided by the NRC through the Canadian Wheat Alliance. This paper is NRCC No. 56262.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The small RNA and RNA-sequencing data have been submitted to the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) [accession nos. SAMN05725181–SAMN05725246 (mRNAs and sRNA in Triticum durum and Triticum aestivum)]. The Iw1 full-length cDNA sequence was submitted to the NCBI expressed sequence tags database (dbEST) (accession no. KX823910).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617483114/-/DCSupplemental.
References
- 1.Shepherd T, Wynne Griffiths D. The effects of stress on plant cuticular waxes. New Phytol. 2006;171:469–499. doi: 10.1111/j.1469-8137.2006.01826.x. [DOI] [PubMed] [Google Scholar]
- 2.Von Wettstein-Knowles P. Plant Waxes. John Wiley & Sons, Ltd; Chichester, UK: 2012. [Google Scholar]
- 3.Zhang Z, Wang W, Li W. Genetic interactions underlying the biosynthesis and inhibition of β-diketones in wheat and their impact on glaucousness and cuticle permeability. PLoS One. 2013;8:e54129. doi: 10.1371/journal.pone.0054129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, et al. Developmental changes in composition and morphology of cuticular waxes on leaves and spikes of glossy and glaucous wheat (Triticum aestivum L.) PLoS One. 2015;10:e0141239. doi: 10.1371/journal.pone.0141239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tulloch AP, Baum BR, Hoffman LL. A survey of epicuticular waxes among genera of Triticeae. 2. Chemistry. Can J Bot. 1980;58:2602–2615. [Google Scholar]
- 6.Clarke JM, McCaig TN, DePauw RM. Relationship of glaucousness and epicuticular wax quantity of wheat. Can J Plant Sci. 1993;73:961–967. [Google Scholar]
- 7.Johnson DA, Richards RA, Turner NC. Yield, water relations, gas exchange, and surface reflectance of near-isogenic lines differing in glaucousness. Crop Sci. 1983;23:318–325. [Google Scholar]
- 8.Richards RA, Rawson HM, Johnson DA. Glaucousness in wheat: Its development and effect on water-use efficiency, gas exchange and photosynthetic tissue temperature. Aust J Plant Physiol. 1986;13:465–473. [Google Scholar]
- 9.Monneveux P, et al. Relationships between grain yield, flag leaf morphology, carbon isotope discrimination and ash content in irrigated wheat. J Agron Crop Sci. 2004;190:395–401. [Google Scholar]
- 10.Tsunewaki K. Comparative gene analysis of common wheat and its ancestral species. II. Waxiness, growth habit and awnedness. Jpn J Bot. 1966;19:175–254. doi: 10.1093/genetics/53.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, Yuan C, Wang J, Fu D, Wu J. Mapping the glaucousness suppressor Iw1 from wild emmer wheat ‘PI 481521’. Crop J. 2015;3:37–45. [Google Scholar]
- 12.Nishijima R, Iehisa JCM, Matsuoka Y, Takumi S. The cuticular wax inhibitor locus Iw2 in wild diploid wheat Aegilops tauschii: Phenotypic survey, genetic analysis, and implications for the evolution of common wheat. BMC Plant Biol. 2014;14:246. doi: 10.1186/s12870-014-0246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmonds JR, et al. Mapping of a gene (Vir) for a non-glaucous, viridescent phenotype in bread wheat derived from Triticum dicoccoides, and its association with yield variation. Euphytica. 2008;159:333–341. [Google Scholar]
- 14.Liu Q, et al. Molecular mapping of a dominant non-glaucousness gene from synthetic hexaploid wheat (Triticum aestivum L) Euphytica. 2007;155:71–78. [Google Scholar]
- 15.Marcussen T, et al. International Wheat Genome Sequencing Consortium Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345:1250092. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- 16.Tsunewaki K, Ebana K. Production of near-isogenic lines of common wheat for glaucousness and genetic basis of this trait clarified by their use. Genes Genet Syst. 1999;74:33–41. [Google Scholar]
- 17.Adamski NM, et al. The inhibitor of wax 1 locus (Iw1) prevents formation of β- and OH-β-diketones in wheat cuticular waxes and maps to a sub-cM interval on chromosome arm 2BS. Plant J. 2013;74:989–1002. doi: 10.1111/tpj.12185. [DOI] [PubMed] [Google Scholar]
- 18.Wu H, et al. Comparative high-resolution mapping of the wax inhibitors Iw1 and Iw2 in hexaploid wheat. PLoS One. 2013;8:e84691. doi: 10.1371/journal.pone.0084691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu P, et al. Comparative fine mapping of the Wax 1 (W1) locus in hexaploid wheat. Theor Appl Genet. 2015;128:1595–1603. doi: 10.1007/s00122-015-2534-9. [DOI] [PubMed] [Google Scholar]
- 20.von Wettstein-Knowles P, Søgaard B. The cer-cqu region in barley: Gene cluster or multifunctional gene. Carlsberg Res Commun. 1980;45:125–141. [Google Scholar]
- 21.von Wettstein-Knowles P. Biosynthesis and genetics of waxes. In: Hamilton RJ, editor. Waxes: Chemistry, Molecular Biology and Functions. Oily, Allowry, Ayr; Scotland: 1995. pp. 91–130. [Google Scholar]
- 22.Anonymous Description of glossy sheath 1. Barley Genet Newsl. 1996;26:292. [Google Scholar]
- 23.Schneider LM, et al. The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. J Exp Bot. 2016;67:2715–2730. doi: 10.1093/jxb/erw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hen-Avivi S, et al. A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell. 2016;28:1440–1460. doi: 10.1105/tpc.16.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Mujahid H, Hou Y, Nallamilli BR, Peng Z. Plant long ncRNAs: A new frontier for gene regulatory control. Am J Plant Sci. 2013;4:1038–1045. [Google Scholar]
- 27.Ben Amor B, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke JM, McCaig TN, DePauw RM. Inheritance of glaucousness and epicuticular wax in durum wheat. Crop Sci. 1994;34:327–330. [Google Scholar]
- 31.Clarke JM, et al. Registration of seven pairs of durum wheat genetic stocks near-isogenic for glaucousness. Crop Sci. 1995;35:1241. [Google Scholar]
- 32.Bianchi G, Figini ML. Epicuticular waxes of glaucous and nonglaucous durum wheat lines. J Agric Food Chem. 1986;34:429–433. [Google Scholar]
- 33.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan C, et al. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One. 2011;6:e26468. doi: 10.1371/journal.pone.0026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Nozawa M, Miura S, Nei M. Origins and evolution of microRNA genes in plant species. Genome Biol Evol. 2012;4:230–239. doi: 10.1093/gbe/evs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Z, et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bologna NG, et al. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013;23:1675–1689. doi: 10.1101/gr.153387.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 41.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou MT, et al. Tailor: A computational framework for detecting non-templated tailing of small silencing RNAs. Nucleic Acids Res. 2015;43:e109. doi: 10.1093/nar/gkv537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai J, et al. Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell. 2013;25:2417–2428. doi: 10.1105/tpc.113.114603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia J, et al. International Wheat Genome Sequencing Consortium Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91–95. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- 45.Chapman JA, et al. A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol. 2015;16:26. doi: 10.1186/s13059-015-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li A, et al. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell. 2014;26:1878–1900. doi: 10.1105/tpc.114.124388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23:431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor RS, Tarver JE, Hiscock SJ, Donoghue PC. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014;19:175–182. doi: 10.1016/j.tplants.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Fahlgren N, et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun F, et al. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.) BMC Plant Biol. 2014;14:142. doi: 10.1186/1471-2229-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandey R, Joshi G, Bhardwaj AR, Agarwal M, Katiyar-Agarwal S. A comprehensive genome-wide study on tissue-specific and abiotic stress-specific miRNAs in Triticum aestivum. PLoS One. 2014;9:e95800. doi: 10.1371/journal.pone.0095800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han R, et al. Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.) BMC Genomics. 2014;15:289. doi: 10.1186/1471-2164-15-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Budak H, Khan Z, Kantar M. History and current status of wheat miRNAs using next-generation sequencing and their roles in development and stress. Brief Funct Genomics. 2015;14:189–98. doi: 10.1093/bfgp/elu021. [DOI] [PubMed] [Google Scholar]
- 54.Yao Y, Sun Q. Exploration of small non coding RNAs in wheat (Triticum aestivum L.) Plant Mol Biol. 2012;80:67–73. doi: 10.1007/s11103-011-9835-4. [DOI] [PubMed] [Google Scholar]
- 55.Meng F, et al. Development-associated microRNAs in grains of wheat (Triticum aestivum L.) BMC Plant Biol. 2013;13:140. doi: 10.1186/1471-2229-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agharbaoui Z, et al. An integrative approach to identify hexaploid wheat miRNAome associated with development and tolerance to abiotic stress. BMC Genomics. 2015;16:339. doi: 10.1186/s12864-015-1490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slotkin RK, Freeling M, Lisch D. Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics. 2003;165:781–797. doi: 10.1093/genetics/165.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slotkin RK, Freeling M, Lisch D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet. 2005;37:641–644. doi: 10.1038/ng1576. [DOI] [PubMed] [Google Scholar]
- 59.Tricker PJ. Transgenerational inheritance or resetting of stress-induced epigenetic modifications: Two sides of the same coin. Front Plant Sci. 2015;6:699. doi: 10.3389/fpls.2015.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bazin J, Bailey-Serres J. Emerging roles of long non-coding RNA in root developmental plasticity and regulation of phosphate homeostasis. Front Plant Sci. 2015;6:400. doi: 10.3389/fpls.2015.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heo JB, Lee YS, Sung S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013;21:685–693. doi: 10.1007/s10577-013-9392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, et al. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015;84:404–416. doi: 10.1111/tpj.13018. [DOI] [PubMed] [Google Scholar]
- 63.Zhang YC, et al. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014;15:512. doi: 10.1186/s13059-014-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014;15:R40. doi: 10.1186/gb-2014-15-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, et al. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014;24:444–453. doi: 10.1101/gr.165555.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xin M, et al. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H, et al. Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genomics. 2016;17:238. doi: 10.1186/s12864-016-2570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thakur V, et al. Characterization of statistical features for plant microRNA prediction. BMC Genomics. 2011;12:108. doi: 10.1186/1471-2164-12-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasschau KD, et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 71.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 72.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 73.Chen HM, et al. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang S, et al. Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol Biol. 2002;48:805–820. doi: 10.1023/a:1014868320552. [DOI] [PubMed] [Google Scholar]
- 75.Dvorak J, Akhunov ED. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics. 2005;171:323–332. doi: 10.1534/genetics.105.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chalupska D, et al. Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA. 2008;105:9691–9696. doi: 10.1073/pnas.0803981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Middleton CP, et al. Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS One. 2014;9:e85761. doi: 10.1371/journal.pone.0085761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 79.Wright SI, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol Biol Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- 80.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith LM, et al. Rapid divergence and high diversity of miRNAs and miRNA targets in the Camelineae. Plant J. 2015;81:597–610. doi: 10.1111/tpj.12754. [DOI] [PubMed] [Google Scholar]
- 82.Floyd SK, Bowman JL. Gene regulation: Ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.