Significance
The data presented address a fundamental mechanism controlling the expression of a master regulator of cellular differentiation, MyoD, by a member of the Deltex family of proteins. We show that MyoD expression is regulated by modulation of histone methylation in its promoter region by the histone demethylase, Jjmd1c. These data provide insight into the epigenetic control of gene expression in the regulation of myogenic differentiation.
Keywords: Deltex2, Jmjd1c, MyoD, myoblast, differentiation
Abstract
The myogenic regulatory factor MyoD has been implicated as a key regulator of myogenesis, and yet there is little information regarding its upstream regulators. We found that Deltex2 inhibits myogenic differentiation in vitro, and that skeletal muscle stem cells from Deltex2 knockout mice exhibit precocious myogenic differentiation and accelerated regeneration in response to injury. Intriguingly, Deltex2 inhibits myogenesis by suppressing MyoD transcription, and the Deltex2 knockout phenotype can be rescued by a loss-of-function allele for MyoD. In addition, we obtained evidence that Deltex2 regulates MyoD expression by promoting the enrichment of histone 3 modified by dimethylation at lysine 9 at a key regulatory region of the MyoD locus. The enrichment is attributed to a Deltex2 interacting protein, Jmjd1c, whose activity is directly inhibited by Deltex2 and whose expression is required for MyoD expression in vivo and in vitro. Finally, we find that Deltex2 causes Jmjd1c monoubiquitination and inhibits its demethylase activity. Mutation of the monoubiquitination site in Jmjd1c abolishes the inhibitory effect of Deltex2 on Jmjd1c demethylase activity. These results reveal a mechanism by which a member of the Deltex family of proteins can inhibit cellular differentiation, and demonstrate a role of Deltex in the epigenetic regulation of myogenesis.
During development, myogenic progenitors arise from a population of Pax3/7-expressing cells in the dermomyotome (1, 2) and begin to express the myogenic regulatory factor myogenic differentiation 1 (MyoD) in the somites and myotomes at embryonic day (E) 10.5 and in the limbs at E11.5 (3). During postnatal myogenesis, muscle stem cells (MuSCs, or “satellite cells”) give rise to MyoD-expressing cells on activation in response to stimuli such as injury or degenerative diseases (4–6). MyoD-expressing myoblasts ultimately withdraw from the cell cycle and fuse to form multinucleated myotubes, which then develop into myofibers, the mature cells of skeletal muscle. During the process of myoblast differentiation, MyoD expression first increases and then decreases (7, 8). Although MyoD knockout mice have only a modest phenotype (9), likely because Myf5 can compensate, subsequent studies have revealed a delayed differentiation during development (10) and impaired differentiation of MyoD−/− myoblasts despite the expression of Myf5 (11-13). Because of the critical role of MyoD in developmental and regenerative myogenesis, the regulation of its expression has been studied in detail.
Three regulatory elements have been identified in the MyoD promoter: a core enhancer region (CER) located ∼20 kb upstream of the transcriptional start site that is active in early embryonic myoblast development, a distal regulatory region (DRR) in the 5′ proximal 6 kb, and a proximal regulatory region (PRR). These three elements function together to drive MyoD transcription in adult muscle fibers and cultured muscle cells (14–18). Both serum response factor and MEF2 bind to the DRR to regulate MyoD transcription (19, 20). In terms of the complexity of the MyoD promoter and the expression profiles of MyoD during development and postnatal myogenesis, additional regulatory factors clearly play roles in the regulation of MyoD transcription.
Our previous studies revealed that the Notch signaling pathway plays a critical role in postnatal myogenesis (21, 22), consistent with previous in vitro observations of the inhibition of myogenic differentiation by activation of the Notch pathway (23). This may be attributed to its effects on down-regulation of MyoD. Indeed, ectopic expression of the intracellular domain of Notch (NICD) represses myogenesis by targeting the MyoD basic helix-loop-helix domain (24). In addition, canonical Notch signaling suppresses MyoD expression (25), and forced expression of the active form of the Notch coactivator, RBP-J, inhibits muscle differentiation by blocking the expression of MyoD (25, 26). Given the complexity of the regulation of myogenic differentiation by Notch signaling, it is obvious that Notch signaling needs to be tightly regulated during myogenesis. Therefore, regulators of the Notch pathway may be critical for regulating steps in the myogenic process by their effects on MyoD.
Deltex is a Notch-binding protein that acts as a positive regulator of Notch signaling in Drosophila (27–29). Although only one Deltex gene has been found in Drosophila (27), a Deltex gene family, including Deltex1, Deltex2, Deltex3, and Deltex4, has been found in both human and mouse (28). Deltex2 is further divided into a long form and a short form (Deltex2L and Deltex2S, respectively) owing to alternative splicing (30). The mammalian Deltex1 is the homolog most closely related to Drosophila Deltex (31). The N-terminal portion of the Deltex protein is necessary and sufficient to bind the ankyrin repeats of Notch (28). Deltex3, lacking key domains in the N-terminal region of Deltex1 and 2, does not bind to Notch (30), suggesting a Notch-independent function at least for this isoform. The potential role of Deltex in regulation of myogenic differentiation in mammals has not yet been investigated in any detail (30). Other than a decrease in myogenin mRNA levels by the overexpression of Deltex2 in C2C12 cells (30), the regulation of myogenic differentiation by Deltex family members has not been studied either in relationship to Notch signaling or via any Notch-independent mechanisms in mammalian cells.
In studies of the regulation of myogenesis by Notch signaling, we examined the effects of different regulators of Notch signaling, including the Deltex family members, on myogenic differentiation. We found that Deltex2, but not Deltex1, is expressed in murine myogenic progenitors, and that Deltex2 inhibits myogenic differentiation independent of the canonical Notch signaling pathway. Rather, the mechanism of this inhibition is under the control of MyoD expression involving the regulation of H3K9 methylation by the histone demethylase, Jmjd1c. Deltex2 binds to Jmjd1c and inhibits its demethylase activity by monoubiquitination to regulate the pattern of methylation of histones associated with key regulatory regions of the MyoD gene, and thus is a critical determinant of the inducibility of this essential early myogenic regulator. These results reveal a mechanism by which a member of the Deltex family of proteins inhibits myogenic differentiation by regulating MyoD expression, and demonstrate a role of Deltex in epigenetic regulation to control those processes.
Results
Enhanced Muscle Regeneration in Deltex2 KO Mice.
Notch signaling plays an important regulatory role in tissue morphogenesis both during development and during postnatal regeneration of skeletal muscle (32). That control is mediated by a series of regulatory proteins that enhance or inhibit Notch signaling by regulating protein processing, localization, activity, and stability (33). We have previously demonstrated a critical role of Notch signaling in postnatal regenerative myogenesis in mice (21). Given that Drosophila Deltex positively regulates Notch signaling (28), we were interested in exploring the potential regulation of myogenesis by members of the Deltex family of proteins in a mammalian system. We first examined which of the Notch-interacting Deltex family members (Deltex1, 2, and 4; Deltex3 does not have a Notch-interacting domain) are expressed in mouse myogenic progenitors. Published microarray data revealed the highest expression of Deltex2 in C2C12 myoblasts, which was decreased on differentiation, whereas Deltex1 was barely detectable and Deltex4 was expressed at low levels and increased with differentiation (Fig. S1A) (34).
Fig. S1.
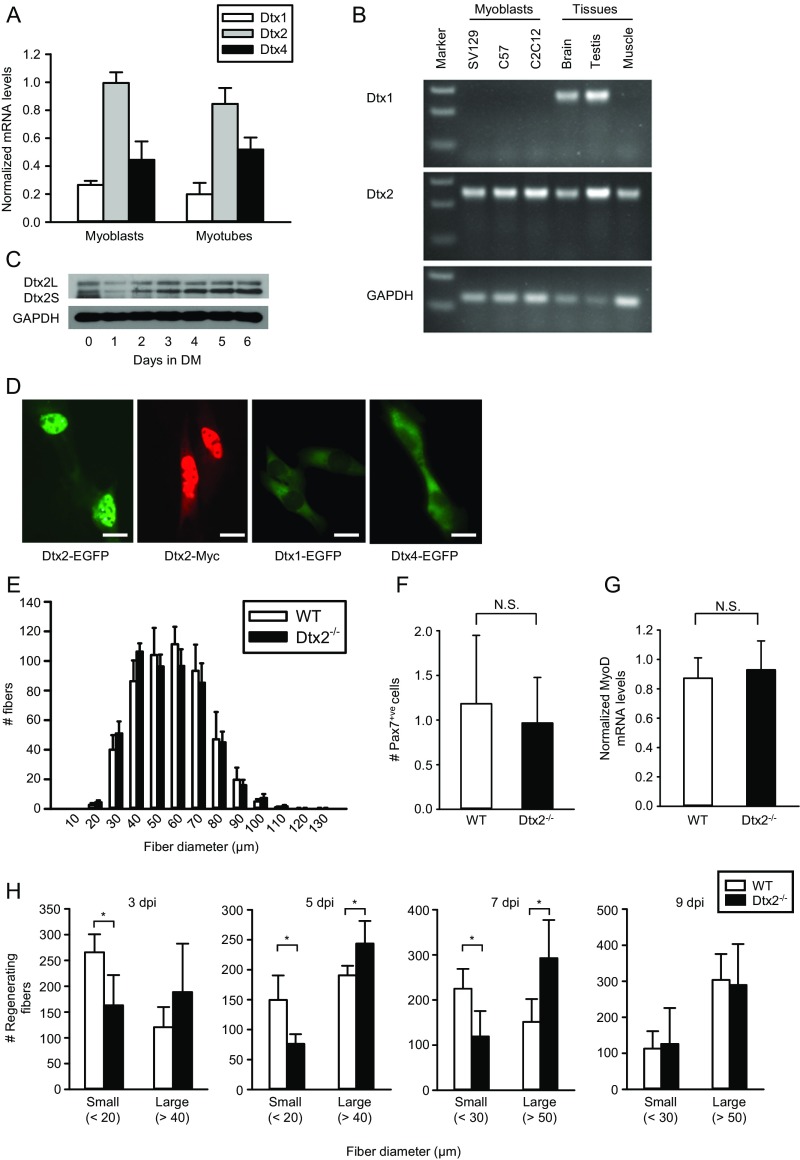
Expression patterns of Deltex1 and Deltex2. (A) Relative expression of Deltex family members as determined by microarray Geo Profiles. (B) Expression patterns of Deltex1 (Dtx1) and Deltex2 (Dtx2) mRNA were analyzed by RT-PCR in primary cultures of mouse myoblasts (SV129 and C57), C2C12 myoblasts, mouse brain and testis, and mouse skeletal TA muscle tissue. Marker represents the DNA 1-kb ladder. The primers used for detecting Deltex2 cannot distinguish the long and short forms. Primer sequences are listed in Table S1. (C) Expression patterns of Deltex2 (Dtx2L and Dtx2S) proteins were analyzed by Western blot analysis in C2C12 myoblast growth (day 0) and differentiation to myotubes (days 1–6 in DM). (D) C2C12 cells were transiently transfected with an EGFP-fused Deltex2L (Dtx2-EGFP), Myc-tagged Deltex2L (Dtx2-Myc), EGFP-fused Deltex1 (Dtx1-EGFP), or EGFP-fused Deltex4 expression vectors, and the cellular localization of either EGFP (green) fluorescence or Myc (red) immunostaining was analyzed microscopically. (Scale bar: 10 μ.) (E) TA muscles of control and Deltex2 KO mice were immunostained for Laminin to determine muscle fiber size (fiber diameter). (F) TA muscles of control and Deltex2 KO mice were immunostained for Pax7 to determine MuSC number per field. (G) Quiescent MuSCs were isolated from uninjured muscles and analyzed by qRT-PCR for the expression of the marker of activation, MyoD. (H) WT and Deltex2 knockout mice were injured and TA muscles fixed and stained for Laminin at 3, 5, 7, and 9 d after injury. Fiber diameters were quantified and plotted as the number of small and large fibers. Full histograms of the raw data are presented in Fig. 1A. *P < 0.05; N.S., not significant.
Using RT-PCR, Northern blot, Western blot, and immunofluorescence analyses, we confirmed robust expression of Deltex2 in proliferating C2C12 myoblasts and freshly isolated murine myogenic progenitors, which declined immediately on the onset of myogenic differentiation in C2C12 myoblasts (Fig. S1 B and C). In contrast, expression of Deltex1 was not detected, and Deltex4, although expressed, exhibited a distinct cytosolic localization when expressed as a GFP-fusion protein, indicating that it has a distinct function (Fig. S1D).
Based on these in vitro findings, we wanted to test for any functional role of Deltex2 in myogenesis by analyzing Deltex2 knockout (KO) mice (35). We observed no differences between KO and control mice in terms of muscle morphology, Pax7-positive cells, and transcript levels of the activation marker MyoD in MuSCs (Fig. S1 E–G), suggesting that developmental myogenesis does not depend on Deltex2 expression. As is the case with many important regulators of myogenesis, such as MyoD (9), the KO yields a negligible phenotype unless the tissue is stressed in some way (11); thus, we examined the regenerative response of the muscles of Deltex2 KO mice to injury. At 3 d postinjury, KO muscles exhibited on average significantly larger and more regenerating fibers compared with control muscles (Fig. 1 A and B and Fig. S1H). The differences in regenerating fiber sizes persisted through 7 d postinjury, but disappeared by 9 d postinjury (Fig. 1A and Fig. S1H). These data indicate that the processes of myogenic differentiation and muscle regeneration are accelerated in Deltex2 KO muscles, and suggest that Deltex2 may suppress the myogenic program in myogenic progenitors.
Fig. 1.
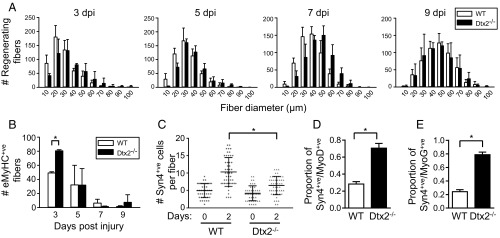
Knockout of Deltex2 accelerates muscle regeneration and enhances satellite cell differentiation capacity. (A) The kinetics of muscle regeneration are enhanced in the absence of Deltex2. Tibialis anterior (TA) muscles of control and Deltex2 KO mice were injured by injection of BaCl2. Cryosections of regenerating muscles were immunostained to detect early regenerating myofibers at 3, 5, 7, and 9 d postinjury, and the numbers of centrally nucleated regenerating fibers were quantified and graphed according to fiber diameter. P < 0.0001 for 3, 5, and 7 d postinjury, χ2 test. (B) The numbers of regenerating myofibers expressing embryonic myosin heavy chain, assessed in the studies described in A. *P < 0.05. (C) Number of Syn4+ve cells counted on single fiber explants from WT and KO (Dtx2−/−) extensor digitorum longus (EDL) muscles immediately after dissociation (day 0) or cultured for 2 d (day 2). The gray line represents the mean. (D and E) Quantitative analysis of the proportion of Syn4+ve SCs expressing MyoD (D) or MyoG (E) per fiber in single fiber explants from WT or Deltex2 KO mice, cultured for 2 d. n = 50. *P < 0.0001.
Enhanced Numbers of MuSC Progeny with High Levels of MyoD and MyoG in Deltex2 KO ex Vivo Cultures.
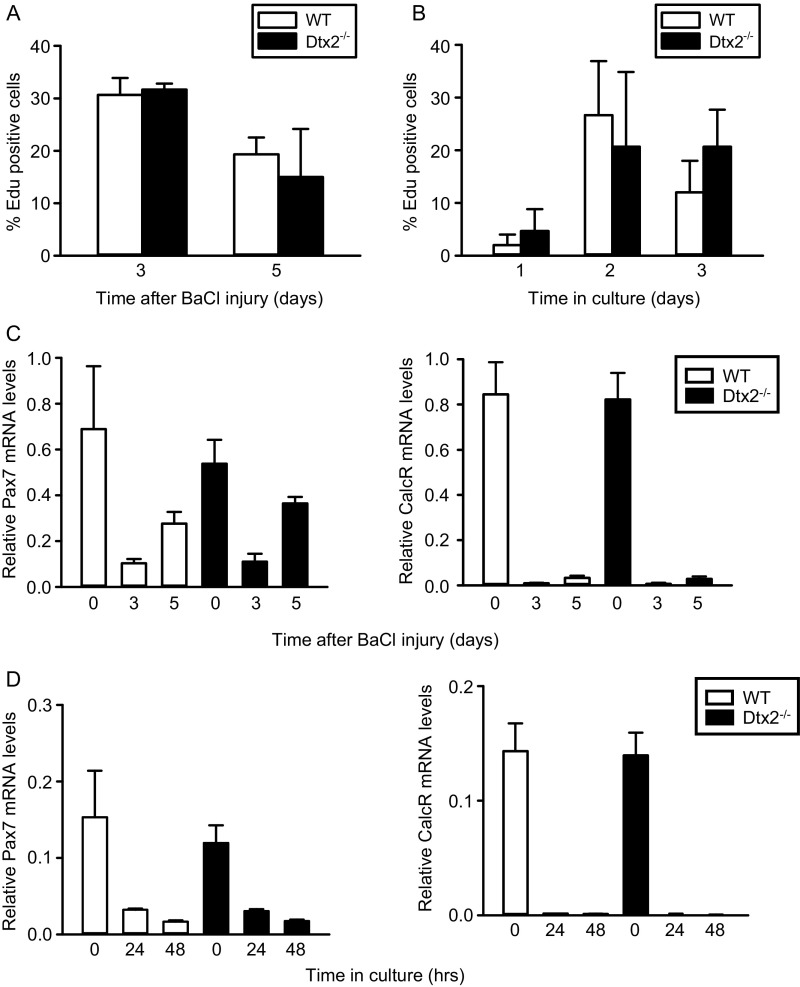
To test whether Deltex2 also might regulate myogenesis by mediating MuSC proliferation, we evaluated EdU incorporation in vivo and in vitro in MuSCs from wild-type (WT) and Deltex2 KO mice. We observed no differences (Fig. S2 A and B), suggesting that proliferation is unperturbed in Deltex2 KO MuSCs (Fig. S2B). Likewise, we found comparable levels of quiescence and activation markers during the early stages of MuSC activation in vivo and in vitro (Fig. S2 C and D). We next isolated single fiber explants from WT and Deltex2 KO mice and analyzed MuSCs and their progeny over time to study myogenesis ex vivo. Immediately on isolation, there was no difference in the number of MuSCs associated with individual myofibers from WT mice compared with those from Deltex2 KO mice (Fig. 1C); however, over the next 2 d, the number of MuSC progeny showed a significantly greater increase in the WT cultures compared with the Deltex2 KO cultures (Fig. 1C), suggesting that MuSCs from Deltex2 KO muscles may enter the myogenic differentiation program earlier. Analysis of cultures for markers of myogenic differentiation revealed a dramatic increase in the number of cells expressing high levels of MyoD (Fig. 1D) and MyoG (Fig. 1E) in Deltex2 KO explants. These results suggest that Deltex2 allows for the proliferative expansion of MuSCs by suppressing myogenic differentiation, consistent with the precocious myofiber formation after injury in Deltex2 KO mice.
Fig. S2.
MuSC proliferation and activation. (A) Mice were injected i.p. with a single dose of EdU at 12 h before sacrifice at and killed at 3 or 5 d postinjury. MuSCs were isolated by flow cytometry and stained for EdU. (B) MuSCs from uninjured mice were isolated, cultured, and pulsed with EdU 6 for h before fixation. Cells were fixed and stained at indicated time points. (C) MuSCs were isolated at indicated days postinjury and analyzed by qRT-PCR for Pax7 and calcitonin receptor expression. (D) MuSCs were isolated from uninjured mice, grown in culture for indicated times, and analyzed by qRT-PCR for Pax7 and calcitonin receptor expression.
Knockdown of Deltex2 Enhances the Differentiation of Myogenic Progenitors.
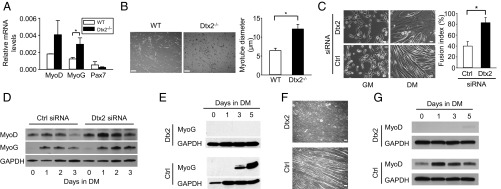
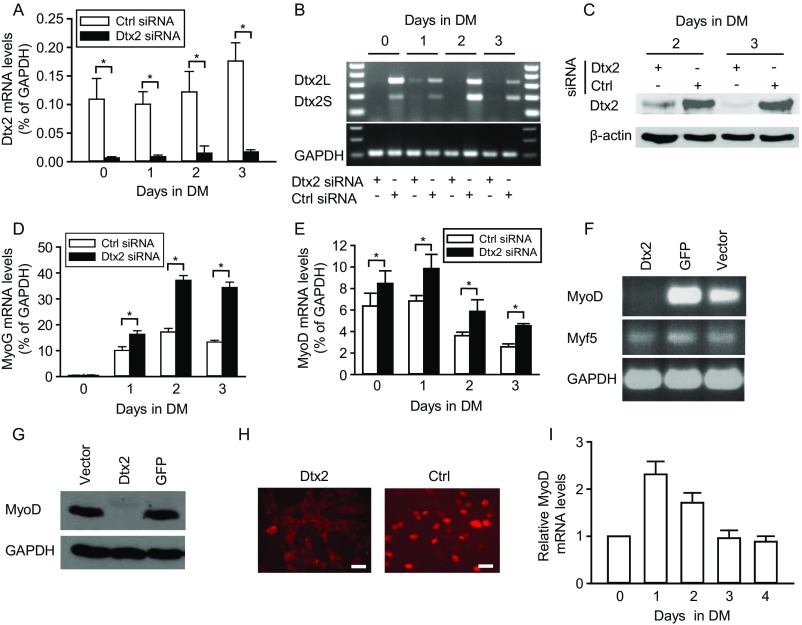
To further explore the role of Deltex2 in regulating myogenic differentiation, we isolated MuSCs by flow cytometry and differentiated them in culture. Differentiating Deltex2 KO cells expressed higher levels of MyoD and MyoG compared with control cells and gave rise to larger myotubes (Fig. 2 A and B). To confirm that this phenotype depends on Deltex2, we used siRNA duplex oligoribonucleotides to knock down endogenous Deltex2 in primary myoblasts. After 24 h of differentiation posttransfection, mRNA levels of Deltex2 (both Deltex2L and Deltex2S) were reduced by 90% (Fig. S3 A and B), and endogenous Deltex2 protein levels were markedly decreased (Fig. S3C). Cells with reduced levels of Deltex2 exhibited accelerated differentiation, as shown by the formation of larger myotubes compared with control cultures (Fig. 2C). In siRNA-treated cells, reduction of Deltex2 levels resulted in higher expression levels of MyoD and myogenin compared with control-transfected cultures (Fig. 2D and Fig. S3 D and E). These data clearly show that myoblasts with decreased Deltex2 expression levels have enhanced differentiation, indicating that Deltex2 plays an important inhibitory role in myogenic differentiation.
Fig. 2.
The effects of Deltex2 on differentiation of primary myoblasts. (A) Primary myoblasts were differentiated in vitro and analyzed for differentiation markers by qRT-PCR. (B, Left) Representative images of primary myoblasts from WT and Deltex2 KO mice differentiated for 6 d in vitro. (Scale bar: 100 μ.) (B, Right) Graph depicting quantitation of myotube diameters from studies illustrated in the representative images. n = 3. *P < 0.05. (C) Primary myoblasts were treated with Deltex2 (Dtx2) or control siRNA (Ctrl) and analyzed either in growth medium (GM) or after 3 d in differentiation medium (DM). (Left) Representative images of myoblasts and differentiated myotubes in response to siRNA treatment. (Scale bar: 40 μ.) (Right) Graph depicting fusion index of cells illustrated in representative images. n = 3. *P < 0.05. (D) Endogenous MyoD and Myogenin protein levels analyzed by Western blot analysis of primary myoblasts treated with Deltex2 or control siRNA. (E) Myogenin levels detected by Western blot analysis of C2C12 cells stably transfected with EGFP-Deltex2 (Dtx2; Upper) or a control EGFP vector (Ctrl; Lower). Myogenin levels were analyzed as a function of time in DM, as indicated. (F) C2C12 cells stably transfected with Deltex2 or control vectors were cultured in DM for up to 8 d and analyzed microscopically for the formation of multinucleated myotubes. (Scale bar: 100 μ.) (G) MyoD levels detected by Western blot analysis of differentiating C2C12 cells stably transfected with Deltex2 or control vectors. MyoD levels were analyzed as a function of time in DM, as indicated.
Fig. S3.
Knockdown of Deltex2 in primary myoblasts. (A) Deltex2 siRNA or negative control siRNA were transiently transfected into muscle primary culture cells. Growth medium was replaced with DM at 24 h after transfection. Real-time PCR was used to measure endogenous Deltex2 mRNA levels, which were reduced to <10% of control levels at all time points. n = 3. *P < 0.05. (B) Levels of both long and short forms of Deltex2 (Dtx2L and Dtx2S) mRNA were examined by RT-PCR with the primers of Dtx2L/S (Table S1) from muscle primary culture cells treated with siRNA as in A. GAPDH mRNA levels were detected as a loading control. (C) Endogenous Deltex2 protein levels were detected by Western blot analysis in siRNA-treated primary muscle cells. β-actin served as a loading control. (D) Endogenous Myogenin mRNA levels were detected by real-time PCR during differentiation of primary cultures treated with Deltex2 or control siRNA. Myogenin levels were enhanced in Deltex2 siRNA-treated cells. n = 4. *P < 0.05. (E) Endogenous MyoD mRNA levels were also enhanced during differentiation of muscle primary culture cells treated with siRNA against Deltex2. n = 3. *P < 0.05. (F) MyoD and Myf5 mRNA levels were examined by RT-PCR in Deltex2-transfected C2C12 cells (Dtx2) and compared with levels in control transfected cells (GFP and vector). MyoD mRNA could not be detected in Deltex2-transfected cells, whereas Myf5 RNA level was unchanged. The primer sequences are listed in Table S1. (G) MyoD protein levels were not detectable by Western blot analysis in Deltex2-transfected C2C12 cells (Dtx2), whereas levels were unaffected in control-transfected cells (GFP and vector). (Scale bar: 20 μ.) (H) Immunostaining showed the absence of MyoD (red) in the nuclei of Deltex2-transfected cells (Upper) and the presence of MyoD in the nuclei of control transfected cells (Lower). (I) Real-time PCR was used to measure relative MyoD mRNA levels during normal differentiation of C2C12 cells. n = 3.
Deltex2 Inhibits Myogenic Differentiation.
To directly test the sufficiency of Deltex2 for suppressing myogenic differentiation, we generated stable transfectants of Deltex2 fused with EGFP at its COOH terminus in C2C12 myoblasts. The Deltex2-expressing cells were induced to differentiate and analyzed for myogenin expression. At times when myogenin was clearly expressed in control cells, it was undetectable in Deltex2-expressing cells (Fig. 2E), suggesting an inhibition of myogenic differentiation by Deltex2. This is consistent with findings from previous studies of the role of Deltex2 in neurogenesis (30). Morphological analysis revealed that Deltex2-expressing cells did not form myotubes under differentiation-inducing conditions (Fig. 2F), even after maintenance of cultures in differentiation medium (DM) for 8 d. Thus, Deltex2 is a potent inhibitor of myogenic differentiation.
To study the mechanism(s) by which Deltex2 inhibits myoblast differentiation, we analyzed the effect of Deltex2 on the expression of the early myogenic regulatory factors (MRFs), Myf5 and MyoD. Myf5 transcript levels were negligibly affected (Fig. S3G), but both MyoD mRNA and protein levels were dramatically decreased in Deltex2-expressing cells compared with control cell populations (Figs. S3 G–I). We further analyzed MyoD protein levels in Deltex2-expressing cells under differentiation culture conditions, and found that MyoD protein was not detectable in cultures maintained for up to 5 d in DM, whereas the control cells showed a normal MyoD expression pattern (Fig. 2G and Fig. S3I). Therefore, from both loss-of-function and gain-of-function studies, our results suggest that Deltex2 might inhibit myogenic differentiation by suppressing MyoD expression.
Mechanism of Deltex2 Inhibition of Myogenesis: Inhibition of MyoD Expression.
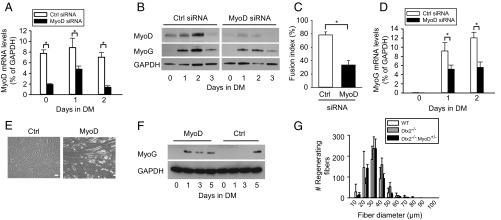
The regulation of myogenesis is clearly controlled by MRFs with overlapping activities, and MyoD is a critical factor in regulating myogenic differentiation (8). As such, knockdown of MyoD by siRNA (Fig. 3 A and B) led to a decrease in the formation of multinucleated myotubes and reduced myogenin expression in primary myoblasts induced to differentiate (Fig. 3 B–D), phenocopying the effects of Deltex2 expression. Indeed, the expression pattern of endogenous MyoD protein was inversely correlated with that of endogenous Deltex2 protein during normal myogenic differentiation (Fig. 2G and Figs. S1C and S3I). MyoD levels were maximal at the onset of differentiation, whereas Deltex2 levels were minimal at that same time.
Fig. 3.
The inhibition of differentiation by Deltex2 occurs through the down-regulation of MyoD. (A) MyoD or control siRNA were transiently transfected into primary muscle cell cultures. GM was replaced with DM at 24 h after transfection. Real-time PCR was used to measure endogenous MyoD mRNA levels. n = 3. *P < 0.05. (B) MyoD and Myogenin protein levels were analyzed by Western blot analysis of primary myoblasts treated with MyoD or control siRNA. (C) Primary myoblast cultures were treated with siRNA as in A. Morphological changes were analyzed by quantification of fusion indices after 3 d of differentiation. n = 3. *P < 0.05. (D) Endogenous myogenin mRNA levels were detected by real-time RT-PCR during differentiation of primary myoblasts treated with MyoD or control siRNA. n = 4, *P < 0.05. (E) C2C12 cells stably transfected with Deltex2 were cultured in GM and transiently transfected with either MyoD or control vector (Ctrl). The medium was changed to DM at 1 d after transfection. The cells were cultured in DM for 5 d and then analyzed microscopically for the formation of multinucleated myotubes. (Scale bar: 50 μ.) (F) Experiments as in E were analyzed for Myogenin levels by Western blot analysis as a function of time in DM. (G) TA muscles of control, Deltex2−/−, and Deltex2−/−MyoD+/− mice were injured by injection of BaCl2. Cryosections of regenerating muscles were immunostained to detect early-regenerating myofibers at 5 d postinjury, and the numbers of regenerating fibers was were quantified and graphed according to fiber diameter. (Scale bar: 20 μ.) P < 0.0001 comparing Deltext2−/− and Deltex2−/−MyoD+/−.
To test the hypothesis that the inhibition of differentiation by Deltex2 occurs through the down-regulation of MyoD, we performed rescue experiments by ectopic expression of MyoD in Deltex2-expressing myoblasts. Proliferating cells were transiently transfected with a MyoD expression vector and then induced to differentiate at 1 d after transfection. After incubation in DM for 5 d, Deltex2-expressing myoblasts with constitutive MyoD expression formed myotubes, whereas the cells transfected with a control vector did not (Fig. 3E). Myogenin expression could be detected in the MyoD-expressing cells 1 d after the cells were cultured in DM, whereas the control cells expressed myogenin only at day 5 after being cultured in DM (Fig. 3F), even though myotubes were still not detected at this time point (Fig. 3E). These data indicate that forced expression of MyoD can rescue the inhibition of myogenic differentiation by Deltex2.
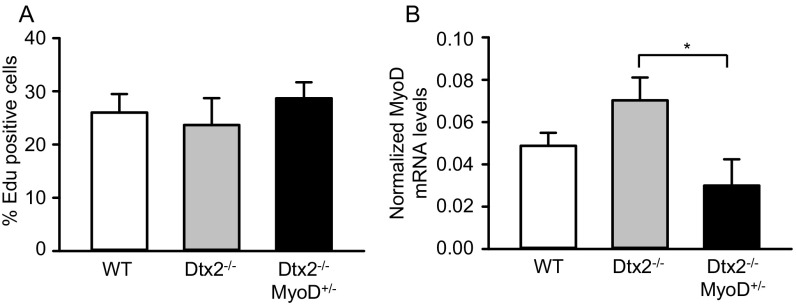
To test whether Deltx2-dependent regulation of MyoD underlies the regeneration phenotype observed in vivo in Deltex2 KO muscles, we bred Deltex2 KO mice with a MyoD KO model to obtain Deltex2−/−MyoD+/− mice. On muscle injury to these mice, we observed comparable proliferative activity but reduced levels of MyoD expression in activated MuSCs of Deltex2−/−MyoD+/− mice compared with Deltex2−/− mice (Fig. S4 A and B); however, there was a significant decrease in the size of regenerating fibers in the Deltex2−/−MyoD+/− mice, comparable to that in controls and significantly smaller than that in Deltex2 KO mice (Fig. 3G). Taken together, these data demonstrate that the reduction in MyoD expression can rescue the Deltex2 KO phenotype in vivo and suggest that Deltex2 inhibits myogenic differentiation in vivo by limiting MyoD expression.
Fig. S4.
Rescue of Deltex2 null phenotype. (A) Mice were injected i.p. with a single dose of EdU at 12 h before sacrifice and killed at 5 d postinjury. MuSCs were isolated by flow cytometry and stained for EdU. (B) MuSCs were isolated at 5 d postinjury and analyzed by qRT-PCR for MyoD.
Inhibition of Myogenic Differentiation by Deltex2 Is Independent of the Canonical Notch Signaling Pathway.
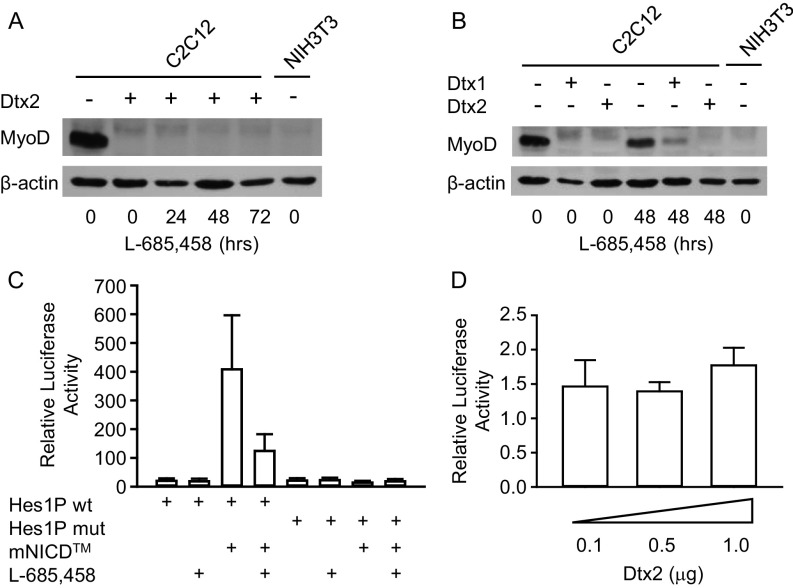
To test whether the inhibition of MyoD expression and myogenic differentiation by Deltex2 is Notch-dependent (28), we analyzed expression levels of MyoD in Deltex2-expressing cells with or without concurrent inhibition of Notch signaling by l-685,458 (36). Like untreated cells, the Deltex2-expressing cells treated with the Notch inhibitor for up to 3 d did not express MyoD protein (Fig. S5A), suggesting that Notch signaling is not required for the inhibition of MyoD expression by Deltex2. In contrast, after introducing Deltex1 into C2C12 cells, we observed similar inhibition of myogenic differentiation and MyoD expression (Fig. S5B), but this could be restored by treatment with l-685,458 (Fig. S5B). These results indicate that the inhibition of MyoD expression by Deltex2 is independent of canonical Notch signaling.
Fig. S5.
The inhibition of myogenic differentiation by Deltex2 is independent of the canonical Notch signaling pathway. (A) MyoD protein expression levels in cell lysates were detected by Western blot analysis in Deltex2 stably transfected C2C12 cells treated with the Notch inhibitor l-685,458 (5 μM) for up to 3 d. Untransfected C2C12 and NIH 3T3 cells served as positive and negative controls, respectively, for MyoD expression. (B) MyoD protein levels were detected by Western blot analysis of C2C12 cells expressing Deltex1 or Deltex2 and treated with l-685,458 (5 μM) for 2 d. Untransfected C2C12 and NIH 3T3 cells served as positive and negative controls as in C. The Notch inhibitor rescued MyoD expression in Deltex1-transfected, but not in Deltex2-transfected, C2C12 cells. (C) C2C12 cells were transiently cotransfected with a mouse NICD expression vector (mNICD) and either the pGL3-Basic vector carrying the mouse Hes1 promoter sequence from +46 to −197 relative to the transcription start site (Hes1P wt) or the same vector but with the mutation at the CSL-binding site (Hes1 P mut). The Renilla luciferase expression vector pRL-TK was cotransfected as a transfection efficiency control. The cells were maintained in the presence or absence of 5 μM of the γ-secretase inhibitor, l-685,458. The transcriptional activity was evaluated as relative light intensity, which is the ratio of light intensity produced by Photinus (firefly) luciferase from the pGL3-Basic vector to that produced by Renilla luciferase from the pRL-TK vector. n = 4. (D) C2C12 cells were transiently cotransfected with the Hes1P wt and a Deltex2 expression vector. The value of the luciferase activity in the absence of Deltex2 was arbitrarily assigned a value of 1, and the values of the activities in cells transfected with the Deltex2 construct were calculated relative to the control. n = 3. No significant difference was observed.
In a final test for an effect of Deltex2 on Notch signaling activity, we used a reporter construct (Hes1P wt) in which luciferase is driven by the mouse Hes1 promoter containing a CSL [named for CBF1 (also known as RBP-J), suppressor of hairless (SuH), and Lag-1] binding site. The luciferase activity of this reporter was enhanced by cotransfection with a Notch expression construct that expresses NICD and the associated transmembrane domain, thus requiring cleavage by γ-secretase to liberate the active form of the protein, NICD. This enhancement could be reduced by treating the cells with l-686,458 (Fig. S5C). A mutant reporter construct (Hes1P mut) in which the CSL binding site GTGGGAA was mutated to GTGAAAA did not show any activity, even in the presence of activated Notch (Fig. S5C). Therefore, luciferase activity from the WT construct is a reliable read-out of Notch signaling. Transfection of Deltex2 did not enhance the luciferase activity significantly (Fig. S5D). These data further suggest that Deltex2 acts in a Notch-independent fashion.
Deltex2 Binds to the MyoD Promoter and Enhances H3K9 Dimethylation.
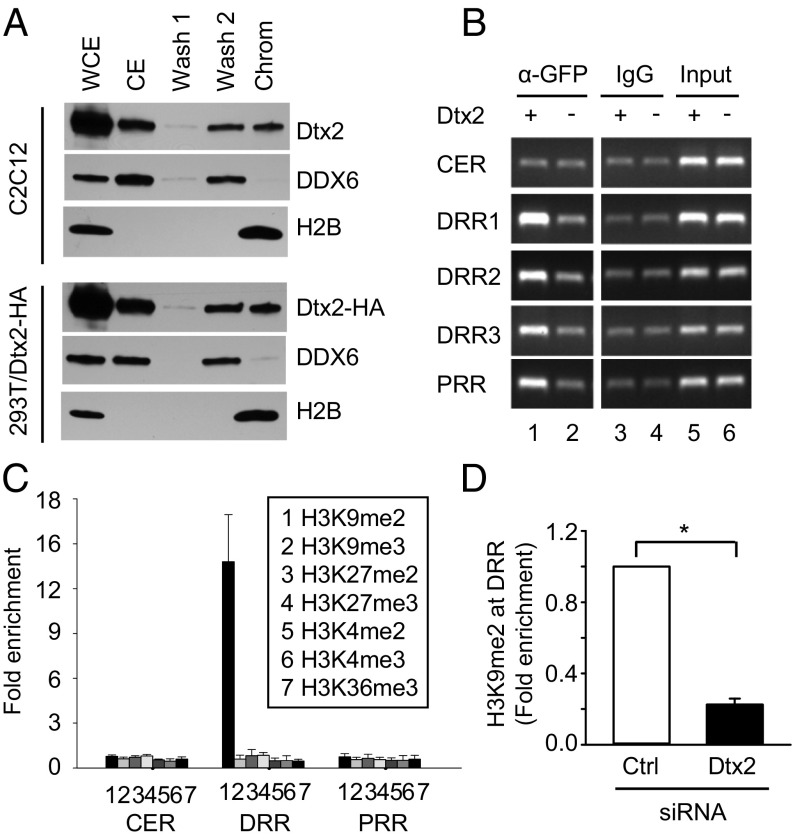
We sought to explore the mechanism by which Deltex2 may regulate MyoD expression. Because protein structure analysis did not reveal any potential DNA-binding domain within the Deltex2 protein (30, 37, 38), we tested whether Deltex2 binds to chromatin in myoblasts using a chromatin association assay (39). Both endogenous Deltex2 and exogenous Deltex2 transfected into 293T cells could be detected in the chromatin fraction together with histone H2B, as well as in the soluble nuclear wash (Fig. 4A). In contrast, a nuclear protein, DDX6, known to not be associated with chromatin (40), was detected in the soluble nuclear wash fraction but not in the chromatin fraction (Fig. 4A). These data support the idea that Deltex2 is a chromatin-associated protein.
Fig. 4.
Deltex2 is a MyoD promoter-binding protein that causes enrichment of dimethylated Histone H3K9 in the DRR of the MyoD promoter. (A) C2C12 and 293T cells transfected with HA-Deltex2 were fractionated. The indicated fractions were probed for endogenous Deltex2 or exogenous HA-Deltex2 by Western blot analysis. The non–chromatin-associated protein DDX6 was detected with an anti-DDX6 antibody as a control. An anti-H2B antibody was used to detect chromatin protein. (B) C2C12 cells were stably transfected with Deltex2-EGFP (lanes 1, 3, and 5) or EGFP (lanes 2, 4, and 6). Anti-GFP (lanes 1 and 2) or IgG control (lanes 3 and 4) antibodies were used to precipitate protein/DNA complexes. PCR products amplified from the CER, DRR, and PRR regions were separated in 1.5% agarose gels. Three consecutive primers—DRR1, DRR2, and DRR3 (primer sequences listed in Table S1)—were used to analyze the DRR region. (C) ChIP assays were performed with indicated antibodies on cell lysates from C2C12 cells stably transfected with Deltex2-EGFP or EGFP alone. ChIP-enriched DNA was quantified by real-time PCR (primer sequences listed in Table S1). H3 antibody served as a control. Values are expressed as fold-change of DNA in Deltex2-EGFP–transfected cells compared with EGFP-transfected control cells (set to 1). (D) Primary muscle cell cultures were treated with Deltex2 or control siRNA for 2 d cultured in GM. ChIP assays were performed to detect H3K9me2 enrichment at the DRR region of MyoD promoter in primary myoblasts. The primers and data analysis are the same as in C. n = 3. *P < 0.05.
To test whether Deltex2 is a component of protein complexes binding to any one of the three known regulatory regions—the CER, DRR, and PRR (16, 17)—on the MyoD promoter, we performed a chromatin immunoprecipitation (ChIP) analysis to screen the MyoD promoter sequence. An antibody against GFP was used to pull down Deltex2-DNA complexes from cells expressing Deltex2-EGFP, with cells expressing EGFP alone serving as controls. Within the 24-kb MyoD promoter sequence, Deltex2-EGFP was enriched in the DRR and the PRR, but not in the CER (Fig. 4B).
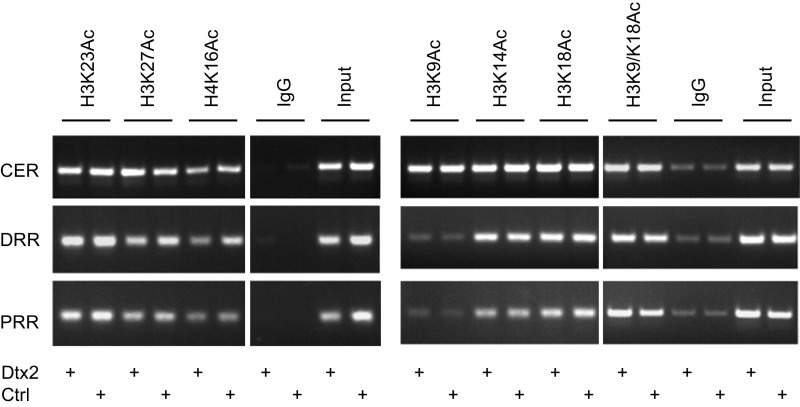
Because Deltex2 protein binds to both the DRR and PRR regions, we further explored whether this binding is correlated with modification of either Histone H3 or H4 associated with the MyoD promoter. The acetylation and methylation of Histones H3 and H4 play critical roles in regulating gene transcription (41–43). Although there are subtle changes in H3K27Ac and H4K16Ac, our ChIP assays on Deltex2-expressing and control cells did not reveal any significant changes in the acetylation status in histone H3 or H4 at the CER, DRR, and PRR (Fig. S6). Intriguingly, however, in Deltex2-expressing cells, histone H3 was dimethylated at lysine 9 (H3K9me2) and was highly enriched in the DRR region; all other modifications tested (histone H3 dimethylated at lysines 4 or 27 or trimethylated at lysines 4, 9, 27, or 36) remained unchanged in all three regions (Fig. 4C). Furthermore, the H3K9me2 enrichment in the DRR region was reduced in primary myoblasts in which the Deltex2 levels were knocked down compared with the control cells (Fig. 4D). Given that methylation of histone H3K9 is generally associated with gene repression (44), these data suggest that Deltex2 may inhibit myogenic differentiation by repressing MyoD expression by promoting a specific pattern of histone methylation.
Fig. S6.
Overexpression of Deltex2 does not change the histone acetylation status on MyoD promoter. ChIP assays were performed in C2C12 cells stably transfected with vectors expressing Dletex2-EGFP (Dtx2) or EGFP (Ctrl) with indicated antibodies. Acetylation status of lysines in histone H3 or H4 remained unchanged in the CER, DRR, and PRR.
Deltex2 Binds to Jmjd1c, a Histone H3K9 Demethylase, and Inhibits Its Activity.
Because Deltex2 has no known intrinsic histone-modifying activity, we performed a yeast two-hybrid assay to find proteins that bind to Deltex2, which could explain the enrichment of H3K9me2 at the DRR of the MyoD promoter by Deltex2. We used the second domain of Deltex2 protein as bait (amino acids 192–405; NP_001243025) (30). Among the first clones identified in the screen, a putative histone demethylase, Jmjd1c (also known as Jhdm2c), was identified. Jmjd1c belongs to a subfamily of Jumonji domain-containing protein members that includes Jmjd1a–c and Hairless (45), and both Jmjd1a and Jmjd1c have been shown to have demethylase activity specific for H3K9me2 (46, 47).
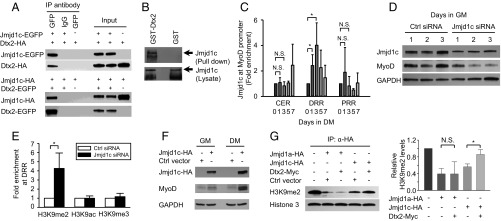
To confirm the interaction between Deltex2 and Jmjd1c, we performed coimmunoprecipitation (co-IP) and GST pull-down studies. Because all available antibodies are unsuitable for co-IP of endogenous Deltex2 and Jmjd1c, we used several independent approaches to test for direct interactions between Deltex2 and Jmjd1c. First, EGFP-fused Jmjd1c and HA-tagged Deltex2 or EGFP-fused Deltex2 and HA-tagged Jmjd1c were cotransfected in HEK293T cells and then subjected to co-IP using an anti-GFP antibody. HA-tagged Deltex2 and HA-tagged Jmjd1c were detected by an anti-HA antibody. Deltex2 or Jmjd1c protein was detected in the immunoprecipitated complexes from Jmjd1c and Deltex2 cotransfected cells, but not in control cells (Fig. 5A). Second, lysates of HEK293 cells expressing EGFP-Jmjd1c were run over agarose resins to which bacterially produced GST-Deltex2 was bound. Full-length Jmjd1c was pulled down by GST-Deltex2 agarose but not by control glutathione agarose alone (Fig. 5B). These results strongly suggest that Deltex2 binds to Jmjd1c, consistent with the results of the yeast two-hybrid assay.
Fig. 5.
The inhibition of myogenic differentiation by Deltex2 is mediated through Jmjd1c. (A) Jmjd1c-EGFP or EGFP vectors were cotransfected with HA-Deltex2 into HEK293T cells. After 24 h, proteins were immunoprecipitated with an anti-GFP or control antibody. Deltex2 and Jmjd1c were detected by Western blot analysis using antibodies against HA and GFP, respectively. The reciprocal IP is shown as well. (B) Lysates of HEK293T cells transfected with EGFP- Jmjd1c were incubated with glutathione-Sepharose 4B bound with GST-Deltex2 or GST only. Jmjd1c was detected by an anti-GFP antibody. Arrows indicate the position of Jmjd1c-EGFP. The Jmjd1c bands from the lysate blot indicate equal loading. (C) The enrichment of Jmjd1c at MyoD promoter regions was analyzed with ChIP assays in differentiating human primary myoblasts. During differentiation, the enrichment of Jmjd1c at the DRR was significantly greater than at the CER and PRR. n = 4. *P < 0.05. (D) Endogenous Jmjd1c and MyoD proteins were analyzed by Western blot analysis in primary myoblasts treated with control or Jmjd1c siRNA. (E) Primary myoblasts were treated with Jmjd1c or control siRNA for 2 d. Antibodies against H3K9me2, H3K9me3, and H3K9Ac were used for ChIP analysis. The DRR region of the MyoD promoter was analyzed by real-time PCR. The primers and data analysis are the same as in C. n = 3. *P < 0.05). (F) MyoD protein levels in Deltex2-expressing C2C12 cells transfected with either a Jmjd1c or control vector were analyzed by Western blot analysis. (G) HA-Jmjd1a, partial HA-Jmjd1c, Myc-Deltex2, and control vectors were transiently transfected into HEK293T cells. Jmjd1a and Jmjd1c proteins were immunoprecipitated with an anti-HA agarose and eluted with an HA peptide for in vitro histone demethylase activity assays. (Right) H3K9me2 and H3 were detected by Western blot analysis and quantified as shown. n = 3. *P < 0.05; N.S., not significant.
Given that Deltex2 interacts with Jmjd1c and is enriched at the MyoD promoter regulatory regions, we performed ChIP assays to test whether Jmjd1c protein is similarly enriched at MyoD regulatory regions. Among the three regulatory regions, Jmjd1c protein was most highly enriched at the DRR and even more enriched during differentiation (Fig. 5C), suggesting that Deltex2 and Jmjd1c act together at this MyoD promoter region to regulate MyoD transcription by modifying the methylation status of H3K9 there.
Knockdown of Jmjd1c Leads to Enrichment of H3K9me2 at the MyoD Promoter and Decreases MyoD Levels in Myogenic Progenitors.
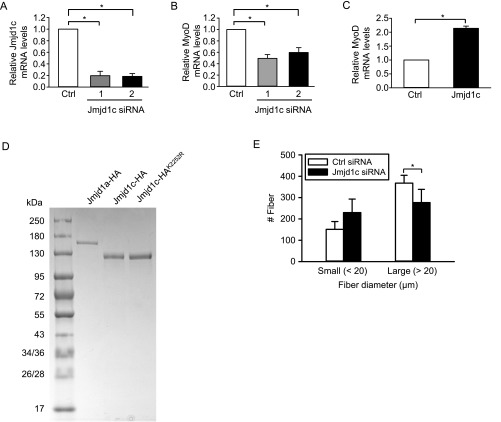
To test whether this Deltex2/Jmjd1c/H3K9me2 axis is a key regulator of MyoD expression and myogenic differentiation, we sought to directly test whether manipulating endogenous Jmjd1c levels would affect the enrichment of H3K9me2 at the MyoD promoter and the expression of MyoD. When Jmjd1c siRNAs were transfected into primary myoblasts, Jmjd1c mRNA and protein levels were reduced to 20% of control levels 1 d later (Fig. 5D and Fig. S7A). Jmjd1c knockdown led to enhanced H3K9me2 levels in the DRR region (Fig. 5E), whereas H3K9ac and H3K9me3 levels were unchanged. In these cells, the expression of MyoD was clearly repressed compared with control cells (Fig. 5D and Fig. S7B). In contrast, when a Jmjd1c cDNA expression vector was transfected into Deltex2-expressing myoblasts, MyoD mRNA levels and protein levels were significantly increased (Fig. 5F and Fig. S7C), suggesting that ectopic expression of Jmjd1c can rescue MyoD expression that was inhibited by the expression of Deltex2.
Fig. S7.
The effects of Jmjd1c on MyoD expression. (A) Two siRNA duplex oligoribonucleotides targeting Jmjd1c or negative control siRNA were transiently transfected into primary myoblasts. At 1 d after transfection, real-time PCR was used to measure endogenous Jmjd1c mRNA levels, which were reduced to approximately 20% of control levels. n = 3. *P < 0.05. (B) Jmjd1c and control siRNA oligonucleotides were transfected into primary myoblasts, and the levels of MyoD transcripts were measured by real-time PCR. The MyoD mRNA levels in siRNA-treated cells were reduced to approximately 50%. Values from negative control siRNA-treated samples are designated arbitrarily as 1. n = 3. *P < 0.05. (C) Deltex2-expressing C2C12 cells were transfected with Jmjd1c expression vector (Jmjd1c) or control empty vector (control). MyoD mRNA levels were analyzed by real-time PCR. n = 3. *P < 0.05. (D) Coomassie blue staining of purified recombinant Jmjd1a, partial Jmjd1c, and Jmjd1c mutant proteins separated on SDS/PAGE gel. (E) Muscles of Deltex2 KO mice were injured and treated with i.m. injections of siRNA after 30 and 54 h. Regenerating fiber diameters were quantified at 5 d postinjury.
As a further test of the Deltex2/Jmjd1c/H3K9me2 axis, and to test whether Deltex2 protein functionally inhibits the activity of Jmjd1c, we performed histone demethylase activity assays (48) using HA-tagged Jmjd1a and Jmjd1c proteins expressed in HEK293T cells, with or without cotransfection of Myc-tagged Deltex2 and incubated with a bulk preparation of histones from calf thymus as a substrate. Because the size of mouse Jmjd1c (285 kDa) precludes protein purification, we tested a partial Jmjd1c (amino acids 1560–2530) containing the Zn finger and Jumonji C domains, as described previously (47). When this partial Jmjd1c was added into the reaction, the H3K9me2 levels were decreased, albeit to a lesser extent than with the positive control Jmjd1a (Fig. 5D and Fig. S7D); however, when Deltex2 was present, the ability of Jmjd1c to demethylate H3K9me2 was inhibited (Fig. 5G). In contrast, Deltex2 had no effect on the demethylase activity of Jmjd1a (Fig. 5G). These results clearly demonstrate that Jmjd1c has H3K9me2 demethylase activity, and that Deltex2 can inhibit that activity specifically. In addition, because H3K9me2 is highly enriched only in the DRR region in Deltex2-expressing myoblasts (Fig. 4C), and because both Deltex2 and Jmjd1c proteins are enriched in the DRR region, these observations support the idea that Deltex2 inhibits the demethylase activity of Jmjd1c at the DRR region, leading to high H3K9me2 levels locally, repression of MyoD transcription, and inhibition of myogenic differentiation.
To test whether Jmjd1c is essential for the regenerative phenotype observed in Deltex2 KO muscles, we injured Deltex2 KO muscles and injected siRNA against Jmjd1c into the muscles after 30 and 54 h. We killed the animals at 5 d postinjury and measured the diameters of regenerating fibers. Injured Deltex2 KO muscles treated with siRNA against Jmjd1c exhibited a modest but significant reduction in large regenerating fibers compared with muscles treated with control siRNA, demonstrating that transient knockdown of Jmjd1c can at least partially rescue the regenerative phenotype in Deltex2 KO animals (Fig. S7E). These data are consistent with the model in which Deltex2 inhibits myogenic differentiation by inhibiting Jmjd1c and thereby suppressing MyoD expression.
Deltex2 Inhibits Jmjd1c Activity by Mediating Its Monoubiquitination.
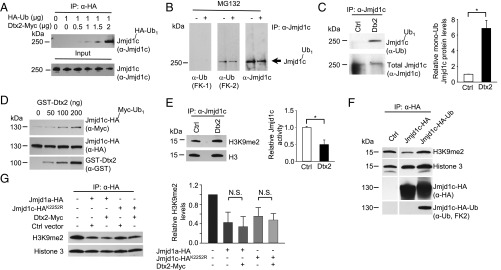
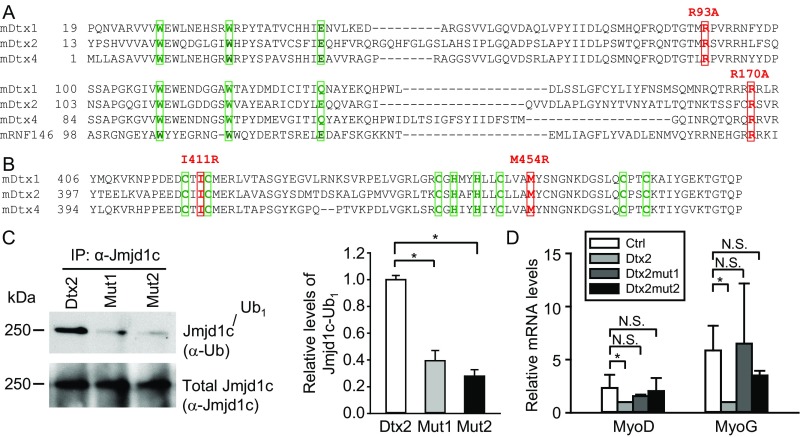
To examine the mechanism by which Deltex2 negatively regulates Jmjd1c, we focused on the reported enzymatic activity of Deltex2 as an E3 ubiquitin ligase (37), postulating that perhaps Deltex2 inhibits Jmjd1c by causing its ubiquitination and degradation. After ectopic expression of Deltex2 in 293T cells, we found that ubiquitinated Jmjd1c was undetectable in control cells but steadily increased as the amount of Deltex2 was increased (Fig. 6A). Interestingly, only monoubiquitination of Jmjd1c was observed. In myoblasts, as in 293T cells, we observed monoubiquitinated, but not polyubiquitinated, Jmjd1c (Fig. 6B), and the level of monoubiquitinated protein was greater in cells expressing Deltex2 (Fig. 6C). This enhancement was not observed when the cells were transfected with Deltex2 expression constructs mutated in either the WWE domain (Mut1, Dtx2R93A-R170A) or the RING domain (Mut2, Dtx2I411R-M454R) (Fig. S8 A–C), suggesting that both domains are important in Deltex2-mediated monoubiquitination of Jmjd1c. Of note, the catalytic Deltex2 mutants were unable to reduce MyoD and MyoG expression, demonstrating that Deltex2 inhibits differentiation through its enzymatic activity (Fig. S8D).
Fig. 6.
Deltex2 inhibits Jmjd1c activity by mediating its monoubiquitination. (A) Analysis of Jmjd1c ubiquitination. Lysates of 293T cells expressing HA-ubiquitin and Myc-Deltex2 were immunoprecipitated with an anti-HA antibody and analyzed by immunoblotting using an anti-Jmjd1c antibody. (B) C2C12 cells were treated with MG132 or DMSO for 6 h and then lysed. Proteins were immunoprecipitated with an anti-Jmjd1c antibody and then analyzed by immunoblotting for Jmjd1c, polyubiquitinated proteins (FK-1), and monoubiquitinated or polyubiquitinated proteins (FK-2). The arrow indicates the positions of nonubiquitinated and monoubiquitinated Jmjd1c, which are not resolved because of the small difference in molecular weights. No polyubiquitinated Jmjd1c protein was detected. (C) Analysis of of Jmjd1c monoubiquitination levels. C2C12 cells were transfected with Deltex2 or control vectors for 24 h and then lysed. (Left) Proteins were immunoprecipitated with an anti-Jmjd1c antibody and then analyzed by immunoblotting for anti-ubiquitin (FK-2) and Jmjd1c. (Right) The levels of monoubiquitinated Jmjd1c (shown quantitatively) were determined as the ratio of monoubiquitinated to total Jmjd1c protein, normalized to control. n = 3. *P < 0.05. (D) In vitro ubiquitination assay. Purified HA-Jmjd1c and GST-Deltex2 were incubated with Myc-ubiquitin and an E1 and E2 mixture and then analyzed by immunoblotting for Myc. The blot was then stripped and reprobed for HA and GST. (E) Nuclei of C2C12 cells transfected with Deltex2 (Dtx2) or empty (Ctrl) vectors were isolated and lysed under nondenaturing conditions. Total (native and monoubiquitinated) Jmjd1c was immunoprecipitated and used in demethylation activity assays. (Left) The assays were immunoblotted for H3K9me2. The blot was then stripped and reprobed for H3. (Right) The relative Jmjd1c activity was designated as the inverse value of H3K9me2 protein levels (shown quantitatively) standardized to total H3, and then normalized to control. n = 3. *P < 0.05. (F) Jmjd1c-HA or Jmjd1c-HA-Ub proteins were immunoprecipitated from lysates of transfected 293T cells and used in Histone demethylation assays in vitro. H3K9me2, H3, and HA were detected by Western blot analysis. The blot was then stripped and reprobed for anti-ubiquitin (FK2). (G) Purified HA-Jmjd1a or HA-Jmjd1cK2252R proteins from transfected 293T cells were tested for histone demethylase activity in vitro. H3K9me2 and H3 were detected by Western blot analysis and quantified as shown. n = 3. N.S., not significant.
Fig. S8.
Multiple sequence alignment of WWE and RING domains in mouse Deltex family members. (A) Mouse Deltex family proteins and RNF146 protein sequences in WWE domains were aligned by a protein sequence alignment program, Clustal Omega (86). The three most conserved amino acid residues in two WWE domains are shown in green boxes. Two amino acid residues, R93 and R170, which are key residues for the binding of the WWE domains to PAR, are highlighted in red boxes. In Mut1, each residue was mutated to disrupt the binding of Deltex2 to PAR. (B) Alignment of protein sequences in RING domains of mouse Deltex family proteins. The key residues to form Zn fingers in the Deltex family RING domains are shown in green boxes. Two amino acid residues, I411 and M454, which are key residues for the interaction of RING domains with E2 ubiquitin-conjugating enzymes, are shown in red boxes. In Mut2, each residue is mutated to R to disrupt the interaction of Deltex2 with its E2 enzyme. (C) Analysis of the levels of Jmjd1c monoubiquitination. (Left) C2C12 myoblasts were transfected with Deltex2 (Dtx2), mutant Deltex2 R93A-R170A (Mut1), or mutant Deltex2 I411R-M454R (Mut2) expression constructs and lysed. Monoubiquitinated Jmjd1c was detected with an anti-ubiquitin antibody (FK2), and total Jmjd1c protein (native and monoubiquitinated) was assessed with an anti-Jmjd1c antibody. (Right) Quantitative analysis of the monoubiquitinated Jmjd1c. n = 3. (D) WT recombinant Deltex2 or two independent activate site mutants were expressed in primary differentiating myoblasts, and MyoD and MyoG mRNA levels were determined by qRT-PCR. *P < 0.05; N.S., not significant.
We next combined purified HA-tagged partial Jmjd1c and GST-fused Deltex2 proteins with a commercial E1-E2 mixture in an in vitro ubiquitination assay, and again observed increasing levels of monoubiquitinated Jmjd1c as the amount of recombinant Deltex2 was increased (Fig. 6D), further confirming that Deltex2 acts as an E3 ligase leading to the monoubiquitination of Jmjd1c. Taken together, the data both in cells and with purified proteins demonstrate that Deltex2 can directly monoubiquitinate Jmjd1c.
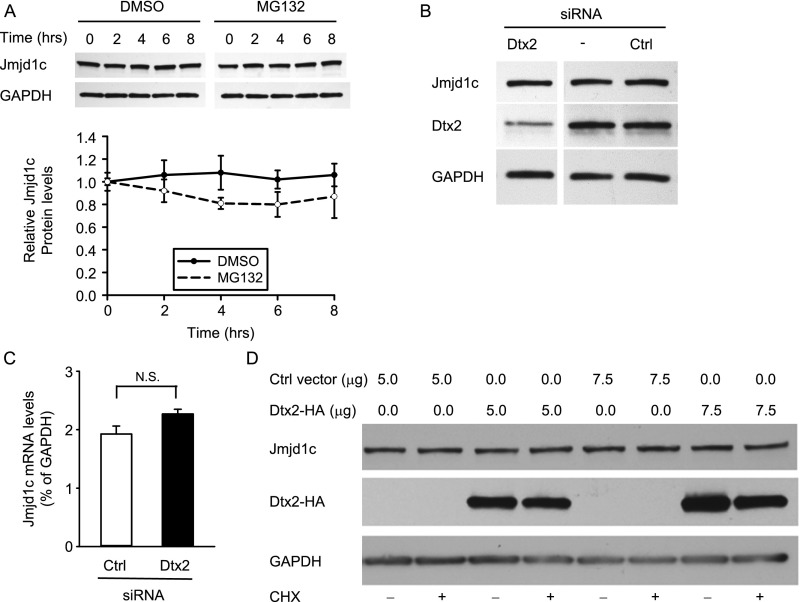
Although monoubiquitination is typically associated with altered protein function (49, 50), it also has been reported to lead to proteasomal degradation, similar to the effects of polyubiquitination (51). Thus, we asked whether the E3 ligase activity of Deltex2 could lead to the ubiquitination and proteasomal degradation of Jmjd1c. Treating C2C12 cells with the proteasome inhibitor MG132 did not lead to an accumulation of Jmjd1c protein (Fig. S9A), suggesting that Jmjd1c protein levels are not regulated by proteasomal degradation. In addition, levels of endogenous Jmjd1c were not affected by either overexpression or knockdown of Deltex2 in myoblasts, suggesting that Deltex2 does not regulate steady-state Jmjd1c protein levels (Fig. S9 B–D).
Fig. S9.
Deltex2 does not affect the expression levels of Jmjd1c. (A) Endogenous Jmjd1c protein levels were detected by immunoblotting in C2C12 cells treated with CHX and MG132 or control DMSO for the indicated times. (Upper) A representative example is shown above. (Lower) Quantitation of replicate experiments (n = 5). (B) Endogenous Jmjd1c and Deltex2 (Dtx2) protein levels were detected by Western blot analysis in primary muscle cell cultures transfected with Deltex2 siRNA, no siRNA (transfection reagent, X-TremeGene alone), or control siRNA. GAPDH served as a loading control. (C) Endogenous Jmjd1c mRNA levels were detected by real-time PCR in primary myoblasts treated with Deltex2 siRNA. (D) C2C12 cells were transfected with an HA-tagged Deltex2 expression vector or control empty vector for 1 d and then treated with CHX or DMSO for 8 h. Endogenous Jmjd1c protein levels and exogenous Deltex2 protein levels were detected by Western blot analysis with an anti-Jmjd1c antibody (Abcam, ab31215) and an anti-HA antibody (Roche, clone 3F10), respectively. Endogenous Deltex2 protein levels were analyzed by an anti-Deltex2 antibody provided by Masato Nakafuku, University of Tokyo. N.S., not significant.
Given that the ubiquitination of Jmjd1c by Deltex2 did not appear to inhibit demethylase activity due to Jmjd1c degradation, we asked whether the monoubiquitination of Jmjd1c might directly inhibit the enzymatic activity. We found that the histone demethylase activity of endogenous Jmjd1c present in C2C12 cells was reduced to <50% of control values in cells with elevated levels of monoubiquitinated Jmjd1c induced by the expression of Deltex2 (Fig. 6E). We tested for a causal relationship between Jmjd1c monoubiquitination and reduced demethylase activity through two independent approaches. The use of ubiquitin fusion proteins has proven to be a valuable approach to examining the effects of monoubiquitination on protein function (52). Along that line, we created a fusion protein to mimic the monoubiquitinated Jmjd1c protein and to test the effects on histone demethylase activity. Consistent with the finding that Jmjd1c histone demethylase activity is decreased when it is monoubiquitinated by Deltex2, enzyme activity was completely absent in the Jmjd1c-ubiquitin fusion protein (Fig. 6F).
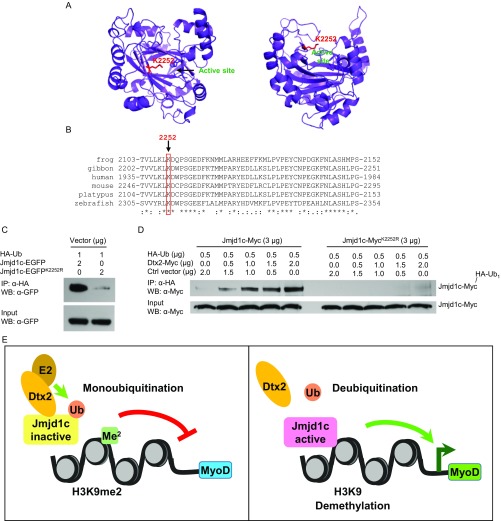
To further test whether Deltex2 negatively regulates Jmjd1c demethylase activity by monoubiquitination, we sought to identify and mutate the lysine residue targeted for ubiquitination by Deltex2 on Jmjd1c. The 285-kDa size of the Jmjd1c protein confounds purification for mass spectrometry analysis; thus, we turned to structural modeling to identify which of the 184 lysines in Jmjd1c might be targeted by Deltex2. Because the yeast two-hybrid experiments showed that Deltex2 binds to a part of the Jmjd1c protein that includes the demethylase domain, and because monoubiquitination affects demethylase activity, we generated and tested a structural model of the demethylase domain in Jmjd1c (Fig. S10A). In this structural model, a single lysine residue, lysine 2252 (NP_997104.2), protrudes from the rim of the enzymatic pocket with its side chain facing outwards. If ubiquitinated, it would sterically block substrates from entering the enzymatic pocket and thus inhibit demethylase activity. Moreover, multiple sequence alignments of vertebrate Jmjd1 demethylase domain sequences revealed that lysine 2252 is the only conserved lysine close to the enzymatic pocket (Fig. S10B). Thus, we mutated lysine 2252 to arginine to examine the potential role of lysine 2252 in the Deltex2-mediated inhibition of Jmjd1c function. The mutant protein is soluble (Fig. S7D) and retains its demethylase activity (Fig. 6G), indicating that it is properly folded. When transfected into 293T cells, the level of ubiquitinated mutant Jmjd1c was almost undetectable compared with that of WT Jmjd1c (Fig. S10C), indicating not only that lysine 2252 is a monoubiquitination target in Jmjd1c, but also that it appears to be the predominant site for ubiquitination. Furthermore, Deltex2 was not able to promote ubiquitination of the mutant Jmjd1c, confirming that lysine 2252 is the site monoubiquitinated by Deltex2 (Fig. S10D). Most importantly, the demethylase activity of the Jmjd1c mutant was not affected by Deltex2 (Fig. 6G), strongly suggesting that monoubiquitination at lysine 2252 is the mechanism by which Deltex2 negatively regulates the demethylase activity of Jmjd1c. Taken together, these date provide a mechanistic link between Deltex2 activity and enrichment of H3K9 dimethylation in the MyoD promoter by virtue of the ability of Deltex2 to inhibit the histone demethylase activity of Jmjd1c by monoubiquitination (Fig. S10E).
Fig. S10.
Deltex2 does not monoubiquitinates a Jmjd1c mutant, Jmjd1cK2252R. (A) Structural model of the Jumonji C domain of Jmjd1c with lysine 2252 in red and active site residues highlighted in blue. (B) Multiple sequence alignment of the Jumonji C domain of Jmjd1c proteins from different species, with mouse lysine 2252 highlighted. (C) HA-tagged ubiquitin was transiently cotransfected with either Jmjd1c-EGFP or Jmjd1c-EGFPK2252R into 293T cells. Ubiquitinated Jmjd1c or its mutant proteins were immunoprecipitated with an anti-HA antibody and then detected with an anti-GFP antibody by Western blot analysis. (D) Myc-tagged Jmjd1c or its mutant and HA-tagged ubiquitin were transiently cotransfected with various amounts of Myc-tagged Deltex2 and control empty vector into 293T cells. Ubiquitinated Jmjd1c or its mutant proteins were detected as in C with an anti-Myc antibody. (E) Schematic model of Deltex2 regulation of MyoD transcription.
Discussion
In the present study, we have explored the role of the Deltex family of proteins in muscle cell differentiation. Surprisingly, we could detect strong expression of Deltex2, but not of Deltex1, in primary mouse muscle cells, C2C12 myoblasts, and skeletal muscle tissue. Interestingly, Deltex2 inhibited the differentiation of myogenic progenitors independent of Notch signaling; rather, it inhibited MyoD transcription. We found that Deltex2 is part of a chromatin complex that binds to the DRR and PRR regions of the MyoD promoter, leading to enrichment of H3K9me2 in the DRR region. Deltex2 binds to a member of the Jumonji family of histone demethylase proteins, Jmjd1c, and inhibits its demethylase activity. Manipulation of Jmjd1c levels leads to changes in MyoD expression levels associated with corresponding changes in the enrichment of H3K9me2 at the DDR region of the MyoD gene. Finally, we have demonstrated that monoubiquitinated Jmjd1c mediated by Deltex2 exhibits reduced its histone demethylase activity. Taken together, these results demonstrate a mechanism by which a member of the Deltex family of proteins regulates cellular differentiation epigenetically, a mechanism that is independent of the canonical Notch pathway and associated with H3K9me2 enrichment at the DRR of the MyoD promoter.
Although three key regulatory elements—the CER, DRR, and PRR—have been identified in the MyoD promoter, the mechanisms that control MyoD transcription are complex and remain incompletely defined (55). CpG demethylation in the CER might be required for enhancer activity in myogenic cells during development (56), but the CER appears not to be an essential regulatory region postnatally (18). Moreover, the CER of MyoD does not confer muscle specificity in cultured cells (17). Reporter assays using the DRR and PRR in C2C12 cells found that the DRR must be integrated to reflect endogenous gene activity (16). These observations suggest that MyoD may be regulated by epigenetic mechanisms, and that chromatin remodeling may be required. Because Deltex2 does not have any obvious DNA-binding domain, our finding that Deltex2 binds to histones associated with the MyoD promoter at the DRR and PRR suggests that Deltex2 associates with proteins that bind directly to DNA. Using ChIP, we found that in the presence of Deltex2, there was enrichment of H3K9me2 at the DRR, which demonstrates a direct link between histone methylation in the MyoD promotor and myogenic differentiation.
Histone methylation, a reversible process, is an important mechanism in the control of gene transcription (43, 44). In general, methylation of histone H3 at K9 and K27 and methylation of histone H4 at K20 are associated with repressed regions of chromatin, whereas methylation of histone H3 at K4, K36, and K79 is associated with active regions (44). Histone methylation is reportedly involved in the regulation of myogenic differentiation. The methylation of histone H3 lysine 9 by Suv39h occurs at several Rb/E2F target promoters and appears to be part of the control switch for C2C12 cells to exit the cell cycle and undergo differentiation (57). The removal of H3K27 methylation, regulated by dissociation of an active Ezh2-containing protein complex from the regulatory regions of myogenic genes, is required for muscle cell differentiation (58). The association of the histone methyltransferase Suv39h with MyoD, as well as methylation of H3K9 on the myogenin promoter by Suv39h, is essential for the inhibition of myogenic differentiation (59). Demethylation of H3K9me2 by the histone demethylase LSD1 at promoters of myogenic regulatory genes is associated with myogenic differentiation (60). Our results indicate that overexpression of Deltex2 changes the methylation status of H3K9 at the DRR of the MyoD promoter, a change that results in suppression of MyoD transcription and a resulting block of differentiation.
Recent observations indicate that Deltex acts as an E3 ligase, leading to the ubiquitination of its targets. Indeed, Deltex1 degrades MEKK1 through ubiquitination (61), Deltex4 targets a kinase TBK1 for degradation (62), Deltex forms a trimeric protein complex with Notch and Kurtz that mediates the degradation of the Notch receptor through a ubiquitination-dependent pathway (63), Deltex monoubiquitinates Notch in S2R+ cells, and Deltex is capable of self-ubiquitination (37, 64). However, no target of Deltex2 as an E3 ligase has been identified to date, nor has any study linked Deltex2 E3 ligase activity to myogenic differentiation.
Our data demonstrate that Deltex2 can inhibit Jmjd1c activity by monoubiquitination, suggesting that Deltex2 may function as an E3 ubiquitin ligase leading to either monoubiquitination or polyubiquitination. Similar functionalities have been found in other E3 ligases. Nedd4 regulates epidermal growth factor receptor pathway substrate clone 15 (Eps15) by monoubiquitination (65), but it also polyubiquitinates certain PPxY motif-containing proteins (66). Another well-studied example is Mdm2, which monoubiquitinates p53 but polyubiquitinates p53 when bound to MdmX (67). Recent studies suggest that specific E2 proteins are critical determinants that control protein monoubiquitination or polyubiquitination (68). Which E2 controls Deltex2 specificity of monoubiquitination remains to be determined. Polyubiquitination typically leads to proteasome degradation, whereas protein monoubiquitination plays important roles in regulating protein activity for such processes as protein trafficking, DNA repair, and gene transcription (69, 70). Our observation that monoubiquitination of Jmjd1c results in inhibition of enzymatic activity is consistent with this function.
Whereas Jmjd1a and Jmjd1b have been convincingly shown to have histone demethylation activity on H3K9me2 (46, 71), reports of the demethylase activity of Jmjd1c have been mixed. On the one hand, Jmjd1c overexpression was found to result in a global decrease in H3K9me2 (47), whereas knockdown of Jmjd1c led to an increase in H3K9me2 at specific genes (72), consistent with our findings at the MyoD promoter. On the other hand, recombinant Jmjd1c protein was shown to be ineffective at demethylating H3K9 in one study (71), but to be effective after supplementation with 293T cell nuclear extract in another study (72). This variable in vitro activity may reflect a dependency of the enzyme on specific cofactors, which is a topic for future studies.
In summary, we found that Deltex2 negatively regulates myogenic differentiation in a Notch-independent manner by inhibiting MyoD transcription. Deltex2 is a nuclear protein that causes enrichment of H3K9me2, associated with gene silencing, at a key regulatory region of the MyoD promoter. This effect is due to the binding of Deltex2 to Jmjd1c, a histone demethylase targeting H3K9me2, and to the negative regulation by Deltex2 of the demethylase activity of Jmjd1c via monoubiquitination. These data provide evidence for a mechanism of the regulation of myogenic differentiation by the epigenetic modulation of MyoD expression and present a paradigm for a novel role of the Deltex family of proteins in gene regulation.
Materials and Methods
All animal handling practices and procedures, including animal health monitoring, diet, primary enclosures, environmental control, and means of identification used in these studies, were in accordance with the current recommendations of the American Veterinary Medical Association. Animal protocols were approved by the Administrative Panel on Laboratory Animal Care of VA Palo Alto Health Care System. Detailed descriptions of the materials and methods used in this work are provided in SI Materials and Methods, including a descriptive list of chemicals and materials used, and details on the experimental protocols used to carry out in vitro and in vivo experiments.
SI Materials and Methods
Ethics Statement.
All animal handling practices and procedures, including animal health monitoring, diet, primary enclosures, environmental control, and means of identification used in these studies, were in accordance with current recommendations of the American Veterinary Medical Association.
Reagent and Plasmids.
The γ-secretase inhibitor l-685,458, cycloheximide (CHX), and MG132 were purchased from Sigma-Aldrich and dissolved in DMSO. Protease inhibitor mixture was obtained from Roche. The Myc-ubiquitin, ubiquitin aldehyde, and ubiquitin-conjugating enzyme fractions (K-960, fractions A and B) were purchased from Boston Biochem. Mouse Deltex1, Deltex2L, and Deltex2S cDNAs were isolated by RT-PCR from mouse brain tissue and C2C12 cells and then subcloned into pcDNA4/Myc-His (Invitrogen), pcDNA4/HA-His (with the Myc tag replaced with HA tag in the pcDNA4/Myc-His vector), and pEGFP-N3 vectors (Clontech), respectively, for Myc, HA-tagged, and EGFP fused expression. Mouse Deltex4 cDNA was isolated by RT-PCR from C2C12 cells and subcloned into pEGFP-N3 for EGFP-fused expression. EGFP-fused Deltex1, Deltex2L, and Deltex2S cDNAs were subcloned into pQCXIP, pQCXIH, and pQCIXN vectors (Clontech), respectively, for stable transfection driven by CMV promoter in C2C12 cells. The EGFP gene was cloned into pQCXIP, pQCXIH, and pQCIXN and used as an empty vector transfection control. The cells were first sorted by GFP expression and then cultured under selection by puromycin, hygromycin, or neomycin for stable transfection, with no more than 10 passages before analysis. Pools of transfectants were used in the experiments. The HA-tagged Deltex2L cDNA was also cloned into the pMXs vector (Cell Biolabs).
Using pMXs-Dtx2L-HA as a template, we created HA-tagged Deltex2L mutants, Dtx2LR93A-R170A and Dtx2LI411R-M454R, using the Agilent QuikChange II XL Site-Directed Mutagenesis Kit (Fig. S6 A and B). Mouse Jmjd1c cDNA was obtained by fusing the cDNA from AK173162 (mKIAA1380 protein, amino acids 218–2530, kindly provided by Kazusa DNA Research Institute) to the rest of the upstream region obtained from C2C12 cells using RT-PCR (amino acids 1–217; NM_207221, XM_907708, XM_975833), and then tagged with EGFP in pEGFP-N3 at its C-terminal. Even though sequence analysis did not indicate any error in the Jmjd1c cDNA, only the C-terminal half of the molecule (∼150 kDa) could be detected in co-IP studies as assayed by Western blot analysis using an anti-GFP antibody, whereas the full-length EGFP-fused Jmjd1c could be pulled down in GST pull-down assays. A partial Jmjd1c fragment (amino acids 1560–2530; NP_997104.2) was subcloned into pEGFP-N3 for EGFP-fused expression and into pcDNA4/HA-His or pcDNA4/Myc-His for HA- or Myc-tagged expression. EGFP-fused and HA- or Myc-tagged Jmjd1c (amino acids 1560–2530) mutants, Jmjd1cK2252R, were created using the Agilent QuikChange II XL Site-Directed Mutagenesis Kit. A ubiquitin cDNA sequence was inserted in-frame into the site between HA and His sequences of the pcDNA4/Jmjd1c-HA-His vector to create a partial Jmjd1c-ubiquitin fusion protein expression vector. Mouse Jmjd1a cDNA was kindly provided by Yi Zhang, Howard Hughes Medical Institute, and subcloned into pEF1/V5-His. Mouse MyoD cDNA was obtained by RT-PCR from C2C12 cells and cloned into pcDNA4/Myc-His. Mouse Notch intracellular domain (mNICD) cDNA was kindly provided by Raphael Kopan, Washington University, St. Louis, MO. The HA-tagged WT ubiquitin expression vector, pRK5-HA-Ub-WT, was obtained from Addgene. All cDNAs were sequenced to verify the correct sequences. Hes1 reporter vector (pHes1-luc; 2.5 kb) was kindly provided by Ryoichiro Kageyama, Kyoto University, Kyoto, Japan. The promoter sequence from +46 to −197 relative to the transcription start site was subcloned into the pGL3-basic luciferase reporter vector (Promega) and used in luciferase report assays. The CSL-binding site GTGGGAA in the Hes1 promoter was mutated to GTGAAAA using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega).
Mice.
Deltex2 KO mice were provided by Michael Bevan, University of Washington, Seattle, WA. MyoD1 KO mice were kindly provided by Michael Rudnicki, Ottawa Hospital Research Institute, Ottawa, ON, Canada. Animals were crossed with WT C57BL/6 mice and maintained as heterozygote breeding pairs to establish littermate controls for the experiments.
Muscle Regeneration Studies.
Analysis of muscle regeneration after BaCl2 injury was performed as described previously (73). In brief, the TA muscles were injured by injection of 50 μL of BaCl2, collected at different time points postinjury, and prepared for biochemical or histological analyses. The muscles were fixed in 0.5% PFA for 6 h, treated with 20% (wt/vol) sucrose overnight, and frozen in Tissue Tek (Sakura) in cooled isopentane. For staining, 7-μ sections were dried and fixed with 4% (wt/vol) PFA for 10 min, washed in PBS, permeabilized with 0.3% Triton, and blocked for 2 h in 20% (vol/vol) donkey serum. Primary antibodies (rat α-laminin; Abcam, ab11576, 1:500, to detect the basal lamina) and mouse α-eMyHC (Developmental Studies Hybridoma Bank, F1.652, 1:500, to detect early regenerating myofibers) were incubated for 2 h in 20% (vol/vol) donkey serum. After washing with PBS, secondary antibodies were added for 1 h. Sections were washed, counterstained with the nuclear stain DAPI, mounted, and immediately imaged. Pax7 staining was performed after antigen retrieval. The 7-μ sections were air-dried, fixed in 4% (wt/vol) PFA, washed in 0.2% Triton, and boiled for 30 min in HIER buffer (2.9 g sodium citrate in 1 L mQ, pH 6.0, plus 0.05% Tween-20). Then sections were blocked in multiblock solution (5% (vol/vol) donkey serum, 60 mg of nonfat dry milk, 60 mg of BSA, and 0.1% Triton in 50 mL of PBS) and stained overnight for Pax7 (Developmental Studies Hybridoma Bank, 1;100). Sections were rinsed with PBS, stained with secondary antibody in multiblock solution for 1 h, counterstained with DAPI, and then washed and mounted.
Proliferation Assays.
For in vivo EdU incorporation assays, animals were treated with a single i.p. injection of EdU at 12 h before sacrifice. MuSCs were isolated as described previously (74), seeded onto ECM-coated wells, and fixed. For in vitro EdU incorporation assays, freshly isolated MuSCs were seeded onto ECM-coated plates and pulsed for 6 h with EdU at the indicated time points before fixation. All cells were processed with Click-It chemistry. Images were obtained with a Zeiss Axiovision fluorescence microscope, and the numbers and diameters of centrally nucleated fibers were measured using Volocity software (Improvision).
Single Myofiber Isolation, Immunofluorescence Staining, and Quantitative Microscopy.
Single myofibers from EDL muscles were isolated as described previously (75). In brief, EDL muscles were collected, digested in 2 mg/mL collagenase II, and then triturated to isolate single myofibers. Intact myofibers were washed three times to discard floating cells and debris, and then cultured for 1 or 2 d in Ham’s F-10 medium containing 10% (vol/vol) horse serum and 0.05% chicken embryo extract. Single fibers were fixed with 4% (wt/vol) paraformaldehyde and then immunostained. After fixation, single fibers were permeabilized and blocked for 2 h in PBS containing 5% (vol/vol) normal donkey serum and 0.25% Triton X-100. MyoD, Myogenin, and Syndecan4 were detected using a mouse monoclonal anti-MyoD antibody (1:1,000; Dako), a mouse monoclonal anti-MyoG antibody (1:250; BD Pharmingen), and a chicken anti-Syndecan4 antibody (1:500), a gift from Dr. Brad Olwin, University of Colorado, for 24 h at 4 °C, followed by 2 h with donkey anti-mouse Alexa Fluor 594 (Invitrogen) and donkey anti-chicken Alexa Fluor 488 (Invitrogen) secondary antibodies. Cell nuclei were stained with DAPI. After washing in blocking buffer, slides were mounted with Fluoromount-G (Southern Biotech). Immunofluorescence for all conditions was performed concomitantly and in parallel. Quantitative microscopy was performed as described previously (76).
Fusion Index and Myonuclear Number Analyses.
Analysis of myoblast fusion was performed as described previously (73). Myoblasts were cultured in low-serum medium to induce myogenic terminal differentiation. Phase-contrast images were obtained in random fields using a 20× objective and then analyzed using Volocity software. The fusion index was determined as the percentage of myonuclei in myotubes (defined as cells with three or more nuclei) compared with the total number of nuclei in the field.
Cell Culture.
Primary cultures of myoblasts derived from mouse skeletal muscle were established as described previously (73). Human skeletal muscle stem cells were isolated according to our previously published protocol (77). Myoblasts of the murine C2C12 cell line (American Type Culture Collection; CRL 1772) were maintained in growth medium consisting of DMEM supplemented with 10% (vol/vol) FBS, 10% (vol/vol) horse serum, penicillin, and streptomycin at 37 °C and 5% CO2. To induce the differentiation of myoblasts into multinucleated myotubes, myoblast cultures were maintained in DM consisting of 2% (vol/vol) horse serum in DMEM. Lipofectamine 2000 (Invitrogen), GENE6 (Roche), or X-tremeGENE HP DNA (Roche) transfection reagent was used in transfections according to the manufacturers’ protocols. The Promega Dual-Luciferase Reporter Assay System was used in the luciferase reporter assay. Standard immunostaining procedures were followed for cultured cells. An antiembryonic myosin heavy chain (eMyHC) antibody (clone F1.652) was obtained from the Developmental Studies Hybridoma Bank. Anti-Laminin antibody was obtained from Sigma-Aldrich. Anti-MyoD antibody (EMD Millipore; clone MoAb5.8A) and anti-Myc antibody (Invitrogen; clone R950) were used in the immunostaining protocol. The Alexa Fluor 594 donkey anti-mouse IgG and Alexa Fluor 488 donkey anti-rabbit IgG antibodies used as secondary antibodies were obtained from Invitrogen. A Cy3-conjugated secondary antibody against mouse IgG was obtained from GE Healthcare.
Western Blot Analysis.
Standard Western blot analysis procedures were followed (78). The endogenous Deltex2 protein was detected by a polyclonal anti-Deltex2 antibody that was kindly provided by Masato Nakafuku, University of Tokyo (79). The endogenous mouse Jmjd1c was detected by an anti-Jmjd1c antibody from either Abcam (ab31215) or EMD Millipore (09-817). Anti-Myogenin antibody (clone F5D; 556358) and anti-MyoD antibody (clone 5.8A; 554130) were obtained from BD Pharmingen. Anti-GFP antibody (clone B2) and anti-Myc antibody (clone 9E10) were obtained from Santa Cruz Biotechnology. Another anti-GFP antibody (A11122) was obtained from Thermo Fisher Scientific. Anti-HA tag antibody (clone 3F10) was obtained from Roche. Anti-GAPDH antibody (clone 6C5) was obtained from Invitrogen/Ambion, and anti–β-actin antibody (clone AC-15; A3854) was obtained from Sigma-Aldrich. Anti-histone H3K9me2 antibody (ab1220) and anti-Histone H3 antibody (ab1791) were obtained from Abcam. Anti-ubiquitinated protein antibodies, FK1 and FK2, were obtained from Enzo Life Sciences. Anti-DDX6 antibody (A300-460A) was obtained from Bethyl Laboratories. Anti-H2B antibody (07–371) was obtained from EMD Millipore. Anti-GST antibody (27–4577-01) was obtained from GE Healthcare.
RT-PCR and Real-Time PCR.
Reverse transcription was carried out using the SuperScript III First-Strand Kit (Invitrogen) or the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Taqman probes, SYBR Green PCR Master Mix, and MyoD, MyoG, Jmjd1c, GAPDH, and Deltex2 Taqman primers and probes for real-time PCR were purchased from Applied Biosystems. Primers used for regular RT-PCR or ChIP analysis, along with primers used for real-time PCR analysis in ChIP, are listed in Table S1.
Table S1.
Oligonucleotide sequences of primers
| Gene or region | Forward primer | Reverse primer |
| Primers for RT-PCR analysis | ||
| MyoD | CTGCCTTCTACGCACCTGGACCGCT | TGCCATCAGAGCAGTTGGAGCGC |
| Myf5 | CTCCAGGAGCTGCTGAGGGAACA | TGCCGTCAGAGCAGTTGGAGGTG |
| Dtx1 | GGCCACCGTGTGCCACCACATCGAGAA | TGTCCATGTCGTAGGCCGTCCACGC |
| Dtx2 | AGCACCCCAGCCCTGGGAAGCCATTCA | TAGTTGGGGTCCGGATAGCCGTGGCCT |
| Dtx2L/S | ACCACTGTGCCCGTGCAGATGCCAAA | CCTTGCTGTCAGTCATGTCACTGTA |
| GAPDH | CCGTGGGGCTGCCCAGAACATCATC | CGGAAGGCCATGCCAGTGAGCTT |
| Primers for regular PCR analysis in ChIP | ||
| mCER | ACAACATGAGCCCCACAG | AGAGTGCTGACCTCTCATGC |
| mDRR1 | GATCTACACTTGGTGGCAGG | AGCCTCTGGAAACCGGAT |
| mDRR2 | AAGACTAGCCAAGGGAGCTG | TCTCTTATCTCAAACCCGAGC |
| mDRR3 | AGCCCGCAGTAGCAAAGTAA | ATAGGTGGCCCCTTTGATTT |
| mPRR | TAGACACTGGAGAGGCTTGG | GCCTGTCAAGTCTATGTCC |
| Primers for real-time PCR analysis in ChIP | ||
| mCER | GCTTCTTTCGGCCAAGTATCCTCCTCC | CTGGCTGTGTTGTGAGTCACGGGTT |
| mDRR | AGTCCTTCAGCCCCCTAGACCCAAG | AACTAGCACCTGCCCCAAGCCTCAG |
| mPRR | GGACCCCAAGCTCCGCCCTACTACA | TAAAAGCCCGGGAGCTGGAAAGG |
| hCER | AGGAGCAGGGGGAATGAGCACA | TGCTGTGGGGCCCATGTTCT |
| hDRR | ACCCAGGAACAGGGAGGAAGGGAA | AAAGCATGAGTGGGCCTGGTGG |
| hPRR | GGTTTGGAAAGGGCGTGCCG | CAGGGAGGGAGGGAGCGACC |
m, mouse; h, human.
RNA Interference.
Stealth siRNA duplex oligoribonucleotides to mouse Deltex2 (MSS232443), mouse Jmjd1c (CMSS201048, 201049, 272663), and negative control siRNA were purchased from Invitrogen. SMART pool of ON-TARGET plus Deltex2, MyoD, Jmjd1c siRNA, and negative control siRNA (Dharmacon/GE Healthcare) were also used in RNA interference assays. X-tremeGENE siRNA transfection reagent (Roche) was used according to the manufacturer’s protocol. For in vivo knockdown experiments, 600 pmol siRNA was diluted with 30 μL Lipofectamine 2000 into 70 μL of Optimem I and injected into the injured muscles at 30 h and 54 h after BaCl2 injections.
Yeast Two-Hybrid Assay.
The Pretransformed Matchmaker mouse 17-d embryo cDNA library in yeast (638864; Clontech) was screened with a bait of Deltex2L fused to the GAL4 DNA-binding domain of pGBKT7 using the Matchmaker GAL-4 Two-Hybrid System 3 (Clontech). More than 100 positive clones were obtained and sequenced.
ChIP.
ChIP was conducted using the manufacturer’s protocol (Upstate/Millipore) with minor modifications. Protein G agarose (Upstate/Millipore) or Staphen A cells (Calbiochem) (80) were used to precipitate the antibody and protein/DNA complex. The precipitate was washed six times with low-salt solution, three times with high-salt solution, twice with LiCl wash solution, and three times with Tris-EDTA buffer. After reverse cross-linking and protease K treatment, DNA was purified with the Qiagen PCR purification kit or Agencourt AMPure XP magnetic beads (Beckman Coulter). The elution was used directly for PCR. The PCR primers for ChIP are listed in Table S1. Anti-GFP (ab290), anti-histone H3K4Me2 (ab7766), anti-histone H3K9Me2 (ab1220), anti-histone H3K27Me2 (ab24684), anti-histone H3K27Me3 (ab6002), anti-histone H3K36Me3 (ab9050), and anti-H3 (ab1791) antibodies were obtained from Abcam; anti-histone H3K4Me3 (07-473), anti-histone H3K9Me3 (07-442) and all anti–acetyl-histone H3 and H4 lysine antibodies were obtained from Upstate/Millipore. Anti-Jmjd1c antibody was obtained from Santa Cruz Biotechnology (sc-101073). SYBR green was used in real-time PCR to quantify ChIP samples. The quantitative real-time PCR results shown are the averages of at least three independent experiments, with error bars indicating SDs. In Figs. 4 C and D and 5E, the data analysis followed the “standard curve method” from Applied Biosystems, represented as fold enrichment. Each PCR result was normalized to input DNA first, and then each value obtained from each specific antibody was normalized to the value obtained from an anti-H3 antibody. The values from control groups were arbitrarily designated as 1. In Fig. 5C, all data were normalized to input first and then normalized to 0 h in DM in each group.
Immunoprecipitation.
EGFP-fused mouse Jmjd1c and HA-tagged Deltex2, or EGFP-fused Deltex2 and HA-tagged Jmjd1c, were cotransfected into 293T17 cells. Immunoprecipitation was performed from nuclear extracts following a previously reported protocol (81). The protein complexes were precipitated with an antibody against GFP (Roche; clones 7.1 and 13.1), and Deltex2 and Jmjd1c were detected in Western blots with an anti-HA antibody (Roche; clone 3F10). An anti-HA affinity matrix (Roche) or agarose conjugated with an anti-HA antibody (Sigma-Aldrich; A2095) was used to precipitate HA-tagged proteins. An anti-Jmjd1c antibody (Santa Cruz Biotechnology; S-17, sc-83421 or Millipore; 09–817) was used to precipitate endogenous mouse Jmjd1c proteins with a denaturing or nondenaturing protocol.
Preparation of Recombinant Proteins and in Vitro GST Pull-Down.
Glutathione Sepharose 4B (GE Healthcare) was used to purify GST-fused recombinant proteins. A modified pGEX-4T-1 was used to construct Jmjd1c or Deltex2 expression vectors, in which a DNA sequence (GAGAATCTTTATTTTCAGGGC) that codes a tobacco etch virus (TEV) protease specifically recognizing a 7-aa sequence (Glu-Asn-Leu-Tyr-Phe-Gln-Gly) was inserted into the site between BamHI and EcoRI. The partial Jmjd1c-HA proteins were eluted after on-column digestion of GST-Jmjd1c-HA with an AcTEV protease (Life Technologies) following the manufacturer’s instructions.
Glutathione resins to which GST-Deltex2 or control GST was bound were incubated with equal amounts of lysates of HEK293 cells that were transfected with EGFP-fused Jmjd1c, then washed and analyzed by Western blot analysis.
Chromatin Association Assay.
Chromatin fractions were prepared following a previously published method (39). In brief, C2C12 cells or 293T cells transfected with HA-tagged Deltex2 were resuspended in nuclei isolation buffer (NIB) without IGEPAL CA-630 (Sigma-Aldrich). Some of the resuspension was saved as whole-cell extract. An equal volume of NIB plus 0.6% IGEPAL CA-630 was added into the cell suspension to release nuclei. After centrifugation of nuclei at 600 × g, supernatant was saved as cytoplasmic extract and nuclei were washed with NIB plus 0.6% IGEPAL CA-630 buffer, and supernatant was saved as wash 1. The nuclei were washed with NIB once, resuspended in NIB, and then treated with micrococcal nuclease and spun at 10,000 × g. The supernatant was saved as wash 2, and the pellet was resuspended in ice-cold 2 mM EDTA, pH 8.0. After centrifugation at 10,000 × g, the supernatant containing solubilized nucleosomes was collected as a chromatin fraction. All fractions were analyzed by Western blot analysis with anti-Deltex2, anti-HA, anti-DDX6, and anti-H2B antibodies.
In Vitro Histone Demethylase Assay.
This assay was performed following a previously published protocol with slight modifications (48). Immunoaffinity-purified Jmjd1a, partial Jmjd1c, partial Jmjd1c mutant, or endogenous Jmjd1c proteins expressed in mammalian cells were used in this assay. Jmjd1a-HA, partial Jmjd1c-HA (amino acids 1560–2530), partial Jmjd1c-HAK2252R (amino acids 1560–2530), and Dtx2L-Myc cDNA expression vectors and control empty vectors were transfected into 293T17 cells using standard methods with X-tremeGENE HP DNA Transfection Reagent or Lipofectamine 2000. Nuclei isolation and micrococcal nuclease (MNase; Roche; 10107921001) digestion were performed following a previously published protocol (81). After MNase digestion, the nuclei were lysed with a lysis buffer (50 mM Tris⋅HCl pH 7.5, 137 mM NaCl, 0.5% Triton X-100, 0.5% CHAPS, 10% (vol/vol) glycerol, and Roche Protease Inhibitor mixture). For HA-tagged proteins, the nuclear lysates were incubated with agarose conjugated with an anti-HA antibody (Sigma-Aldrich; A2095). The agarose was washed three times with the lysis buffer and then with a wash buffer (20 mM Tris⋅HCl pH 7.5, 100 mM NaCl, and 10% (vol/vol) glycerol). The HA-tagged proteins were eluted with an HA peptide solution (1 mg/mL in the wash buffer without glycerol). The purity of eluted proteins was assessed by SDS/PAGE separation and Coomassie blue staining (Fig. S6S). Endogenous Jmjd1c protein (nonubiquitinated and monoubiquitinated) was immunoprecipitated with an anti-Jmjd1c antibody (Millipore; 09–817). Protein G agarose was used to pull down antibody/protein complexes. The agarose was washed five times with wash buffer (20 mM Tris⋅HCl pH 7.5, 0.1M NaCl, and 0.05% Tween-20), and then three times with freshly prepared reaction buffer [20 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2 + 6(H2O), 1 mM α-ketoglutarate, and 2 mM ascorbic acid]. The reaction was performed at 37 °C for 3 h in 30 μL of reaction buffer containing 1.5 μg of bulk histone preparation from calf thymus (Sigma-Aldrich; H9250) dissolved in the reaction buffer with either purified enzymes or protein G agarose to which the endogenous Jmjd1c proteins were bound. Western blot analysis was performed using an anti-H3K9me2 antibody (Abcam; ab1220), and the blot was reprobed with an anti-H3 antibody (Abcam; ab1791).
Ubiquitination Assays.
To study the effects of Deltex2 on endogenous Jmjd1c proteins in 293T cells, the lysates of cells that had been cotransfected with HA-tagged ubiquitin and Myc-tagged Deltex2 expression vectors were immunoprecipitated with anti-HA antibody. The ubiquitinated endogenous Jmjd1c protein was then detected with anti-Jmjd1c antibody (Santa Cruz Biotechnology; BA-09, sc-101073), and the ubiquitinated transfected Deltex2 protein was detected with anti-Myc antibody.
To detect the ubiquitination of Jmjd1c, the endogenous Jmjd1c proteins were immunoprecipitated from C2C12 cell lysates with an anti-Jmjd1c antibody (Santa Cruz Biotechnology; S-17, sc-83421) with a denaturing method and detected on Western blot analysis with an anti-ubiquitin antibody (Enzo Life Sciences; clone FK1 or FK2) for ubiquitination, and then redetected with anti-Jmjd1c antibody (Santa Cruz Biotechnology; BA-09, sc-101073) for total Jmjd1c protein levels.
The in vitro ubiquitination assay was performed with purified HA-tagged Jmjd1c (amino acids 1560–2530) and GST-fused Deltex2 proteins. The Jmjd1c-HA (200 ng) and GST-Deltex2 (0–250 ng) proteins were mixed with Myc-ubiquitin (2.5 µg), ubiquitin aldehyde (3 µM), and an E1-E2 mixture from the conjugation fraction A of ubiquitin-protein conjugation kit in a 25-µL volume of the ubiquitination assay buffer containing 50 mM Tris⋅HCl pH 7.5, 2.5 mM MgCl2, 0.5 mM DTT, and 1× ATP energy regeneration solution and then incubated at 30 °C for 2 h. The ubiquitinated proteins were detected with anti-Myc antibody.
Homology Modeling of Jmjd1c.
The 3D structure of the Jumonji C domain of mouse Jmjd1c protein was obtained using homology modeling of SWISS MODEL Server version 8.05 (82). A template search was performed using the Jumonji C domain sequence from the protein sequence of Jmjd1C (NP_997104.2), and a single hit (2YPD.pdb) with >90% confidence was retrieved: the Jumonji C domain of the human JMJD1C protein (98% confidence). To confirm the specificity, we blasted the sequence of that human Jumonji C domain against the mouse protein database, retrieving Jmjdc1 as the top hit with 89% identity. We then structurally modeled the mouse Jumonji C domain sequence to the structure of 2YPD, centered on the predicted active site residues histidine 2326, glutamate 2328, and histidine 2456 (83). Based on crystallography analyses of human Jmjd2A, Jumonji C domains fold into eight β-sheets, thereby forming an enzymatically active pocket that coordinates Fe2+ and α-ketoglutarate (83). Three amino acid residues within the Jumonji C domain bind to Fe2+. For catalyzing histone demethylation, the cofactor-bound Jumonji C domain is thought to produce a highly reactive oxoferryl species that hydroxylates the methylated substrate, allowing spontaneous loss of the methyl group as formaldehyde (45). Based on sequence homology, we predict that the Jumonji C domain of mouse Jmjd1c binds Fe2+ in the same manner. We note that the overall amino acid conservation between the Jumonji C domains of the two proteins is 23%, but that there is 100% conservation of Fe2+-coordinating residues between the two proteins. We confirmed the alignment with T-Coffee (84). We assessed the quality of the model using Procheck (85). More than 99% of amino acids fall with the allowable regions in a Ramachandran plot, and >98% of amino acid bond lengths and bond angles fall within limits (85), indicating that the model is of high stereochemical quality.
We performed multiple sequence alignments in T-Coffee with the Jumonji C domains of vertebrate orthologs of mouse Jmjd1c. The domains showed >55% sequence identity conservation.
Statistical Analysis.
Data from quantification are presented as mean ± SD. Statistical analyses were performed using Student’s t test (unpaired or paired) or two-way ANOVA (for the data shown in Fig. 5C) using SigmaPlot 12.5. P < 0.05 was considered to indicate statistical significance. All histograms were tested with the χ2 test in GraphPad Prism 5.
Acknowledgments
We thank Dr. Michael Bevan for providing the Deltex2 KO mice, Dr. Michael Rudnicki for providing the MyoD KO mice, Dr. Brad Olwin for the chicken anti-syndecan4 antibody, Dr. Masato Nakafuku for the anti-Deltex2 antibody, and the members of the T.A.R. laboratory for helpful comments and discussion. This work was supported by the Glenn Foundation for Medical Research and by grants from the Muscular Dystrophy Association (Development Grant 313960) (to A.d.M.) and from the National Institutes of Health (Grants R01 AG23806, R01 AR056849, and P01 AG036695) and the Department of Veterans Affairs (Merit Reviews) (to T.A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613592114/-/DCSupplemental.
References
- 1.Relaix F, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172(1):91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minetti GC, et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med. 2006;12(10):1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 3.Sassoon D, et al. Expression of two myogenic regulatory factors Myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341(6240):303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- 4.Cooper RN, et al. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112(Pt 17):2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- 5.Smith CK, 2nd, Janney MJ, Allen RE. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J Cell Physiol. 1994;159(2):379–385. doi: 10.1002/jcp.1041590222. [DOI] [PubMed] [Google Scholar]
- 6.Yablonka-Reuveni Z, Rivera AJ. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev Biol. 1994;164(2):588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16(4-5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Tapscott SJ, Davis RL, Lassar AB, Weintraub H. MyoD: A regulatory gene of skeletal myogenesis. Adv Exp Med Biol. 1990;280:3–5, discussion 5–6. doi: 10.1007/978-1-4684-5865-7_1. [DOI] [PubMed] [Google Scholar]
- 9.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71(3):383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 10.Kablar B, et al. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124(23):4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- 11.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10(10):1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 12.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144(4):631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yablonka-Reuveni Z, et al. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol. 1999;210(2):440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: Conserved elements mediate expression in embryonic axial muscle. Dev Biol. 1995;171(2):386–398. doi: 10.1006/dbio.1995.1290. [DOI] [PubMed] [Google Scholar]
- 15.Chen JC, Love CM, Goldhamer DJ. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn. 2001;221(3):274–288. doi: 10.1002/dvdy.1138. [DOI] [PubMed] [Google Scholar]
- 16.Tapscott SJ, Lassar AB, Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol. 1992;12(11):4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldhamer DJ, Faerman A, Shani M, Emerson CP., Jr Regulatory elements that control the lineage-specific expression of myoD. Science. 1992;256(5056):538–542. doi: 10.1126/science.1315077. [DOI] [PubMed] [Google Scholar]
- 18.Goldhamer DJ, et al. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121(3):637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- 19.L’honore A, et al. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol Biol Cell. 2003;14(5):2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L’honore A, et al. Identification of a new hybrid serum response factor and myocyte enhancer factor 2-binding element in MyoD enhancer required for MyoD expression during myogenesis. Mol Biol Cell. 2007;18(6):1992–2001. doi: 10.1091/mbc.E06-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3(3):397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 22.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302(5650):1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 23.Nofziger D, Miyamoto A, Lyons KM, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126(8):1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 24.Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: A constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120(9):2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda K, et al. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274(11):7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124(20):4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 27.Xu T, Artavanis-Tsakonas S. deltex, a locus interacting with the neurogenic genes Notch, Delta, and Mastermind in Drosophila melanogaster. Genetics. 1990;126(3):665–677. doi: 10.1093/genetics/126.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121(8):2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 29.Gorman MJ, Girton JR. A genetic analysis of deltex and its interaction with the Notch locus in Drosophila melanogaster. Genetics. 1992;131(1):99–112. doi: 10.1093/genetics/131.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishi N, et al. Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int J Dev Neurosci. 2001;19(1):21–35. doi: 10.1016/s0736-5748(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 31.Matsuno K, et al. Human deltex is a conserved regulator of Notch signalling. Nat Genet. 1998;19(1):74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- 32.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 33.Luo D, Renault VM, Rando TA. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin Cell Dev Biol. 2005;16(4-5):612–622. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen IH, Huber M, Guan T, Bubeck A, Gerace L. Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis. BMC Cell Biol. 2006;7:38. doi: 10.1186/1471-2121-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehar SM, Bevan MJ. T cells develop normally in the absence of both Deltex1 and Deltex2. Mol Cell Biol. 2006;26(20):7358–7371. doi: 10.1128/MCB.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martys-Zage JL, et al. Requirement for presenilin 1 in facilitating lagged 2-mediated endoproteolysis and signaling of Notch 1. J Mol Neurosci. 2000;15(3):189–204. doi: 10.1385/jmn:15:3:189. [DOI] [PubMed] [Google Scholar]
- 37.Takeyama K, et al. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J Biol Chem. 2003;278(24):21930–21937. doi: 10.1074/jbc.M301157200. [DOI] [PubMed] [Google Scholar]
- 38.Aravind L. The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26(5):273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 39.Macfarlan T, et al. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J Biol Chem. 2005;280(8):7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 40.Smillie DA, Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J Cell Sci. 2002;115(Pt 2):395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- 41.Forcales SV, Puri PL. Signaling to the chromatin during skeletal myogenesis: Novel targets for pharmacological modulation of gene expression. Semin Cell Dev Biol. 2005;16(4-5):596–611. doi: 10.1016/j.semcdb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 42.McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14(6):763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 43.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 44.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 45.Klose RJ, Kallin EM, Zhang Y. JmjC domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7(9):715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 46.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 47.Kim SM, et al. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res. 2010;38(19):6389–6403. doi: 10.1093/nar/gkq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 49.Sadowski M, Sarcevic B. Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 2010;5:19. doi: 10.1186/1747-1028-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr Top Microbiol Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 51.Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2010;40(5):749–761. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130(2):349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 53.Busseau I, Diederich RJ, Xu T, Artavanis-Tsakonas S. A member of the Notch group of interacting loci, deltex encodes a cytoplasmic basic protein. Genetics. 1994;136(2):585–596. doi: 10.1093/genetics/136.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diederich RJ, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 1994;120(3):473–481. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- 55.Chen JC, Goldhamer DJ. Transcriptional mechanisms regulating MyoD expression in the mouse. Cell Tissue Res. 1999;296(1):213–219. doi: 10.1007/s004410051282. [DOI] [PubMed] [Google Scholar]
- 56.Brunk BP, Goldhamer DJ, Emerson CP., Jr Regulated demethylation of the myoD distal enhancer during skeletal myogenesis. Dev Biol. 1996;177(2):490–503. doi: 10.1006/dbio.1996.0180. [DOI] [PubMed] [Google Scholar]
- 57.Ait-Si-Ali S, et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23(3):605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18(21):2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25(14):3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi J, et al. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun. 2010;401(3):327–332. doi: 10.1016/j.bbrc.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Liu WH, Lai MZ. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol Cell Biol. 2005;25(4):1367–1378. doi: 10.1128/MCB.25.4.1367-1378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui J, et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13(4):387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukherjee A, et al. Regulation of Notch signalling by nonvisual beta-arrestin. Nat Cell Biol. 2005;7(12):1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 64.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol. 2011;195(6):1005–1015. doi: 10.1083/jcb.201104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woelk T, et al. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8(11):1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 66.Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J Biol Chem. 2002;277(11):9307–9317. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Jiang X. Mdm2 and MdmX partner to regulate p53. FEBS Lett. 2012;586(10):1390–1396. doi: 10.1016/j.febslet.2012.02.049. [DOI] [PubMed] [Google Scholar]
- 68.Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 2012;64(2):136–142. doi: 10.1002/iub.589. [DOI] [PubMed] [Google Scholar]
- 69.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2(3):195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 70.Hammond-Martel I, Yu H, Affar B. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012;24(2):410–421. doi: 10.1016/j.cellsig.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 71.Brauchle M, et al. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One. 2013;8(4):e60549. doi: 10.1371/journal.pone.0060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen M, et al. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors. Genes Dev. 2015;29(20):2123–2139. doi: 10.1101/gad.267278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quach NL, Rando TA. Focal adhesion kinase is essential for costamerogenesis in cultured skeletal muscle cells. Dev Biol. 2006;293(1):38–52. doi: 10.1016/j.ydbio.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 74.Liu L, Cheung TH, Charville GW, Rando TA. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc. 2015;10(10):1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zammit PS, et al. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp Cell Res. 2002;281(1):39–49. doi: 10.1006/excr.2002.5653. [DOI] [PubMed] [Google Scholar]
- 76.Boutet SC, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10(3):327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Charville GW, et al. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Rep. 2015;5(4):621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 79.Yamamoto N, et al. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem. 2001;276(48):45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- 80.Oberley MJ, Farnham PJ. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- 81.Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3(3):398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
- 82.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 83.Chen Z, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125(4):691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 84.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 85.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 86.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]