Significance
Activating Janus kinase (JAK) mutations occur only in a minority of T-cell malignancies, which would appear to limit the clinical application of JAK inhibition for these diseases. Our study suggests that targeting JAK might be of value in treating diverse forms of anaplastic lymphoma kinase (ALK)− anaplastic large cell lymphoma (ALCL). Most exogenous cytokine-independent ALK− ALCL cells of diverse origins responded to JAK inhibition regardless of JAK mutation status. The JAK inhibitor sensitivity of these cells correlated with their positive signal transducer and activator of transcription 3 (STAT3) phosphorylation status. Using retroviral shRNA knockdown, we showed that all JAK inhibitor-sensitive cells were dependent on JAK1/STAT3 for survival. Cytokine receptor signaling and gain-of-function JAK1/STAT3 mutations contribute to JAK1/STAT3 dependency. Our data suggest that JAK inhibition maybe a rational therapy for patients with phosphorylated STAT3+ ALK− ALCL.
Keywords: ALK− ALCL, JAK, STAT, mutations, cytokine receptor
Abstract
Activating Janus kinase (JAK) and signal transducer and activator of transcription (STAT) mutations have been discovered in many T-cell malignancies, including anaplastic lymphoma kinase (ALK)− anaplastic large cell lymphomas (ALCLs). However, such mutations occur in a minority of patients. To investigate the clinical application of targeting JAK for ALK− ALCL, we treated ALK− cell lines of various histological origins with JAK inhibitors. Interestingly, most exogenous cytokine-independent cell lines responded to JAK inhibition regardless of JAK mutation status. JAK inhibitor sensitivity correlated with the STAT3 phosphorylation status of tumor cells. Using retroviral shRNA knockdown, we have demonstrated that these JAK inhibitor-sensitive cells are dependent on both JAK1 and STAT3 for survival. JAK1 and STAT3 gain-of-function mutations were found in some, but not all, JAK inhibitor-sensitive cells. Moreover, the mutations alone cannot explain the JAK1/STAT3 dependency, given that wild-type JAK1 or STAT3 was sufficient to promote cell survival in the cells that had either JAK1or STAT3 mutations. To investigate whether other mechanisms were involved, we knocked down upstream receptors GP130 or IL-2Rγ. Knockdown of GP130 or IL-2Rγ induced cell death in selected JAK inhibitor-sensitive cells. High expression levels of cytokines, including IL-6, were demonstrated in cell lines as well as in primary ALK− ALCL tumors. Finally, ruxolitinib, a JAK1/2 inhibitor, was effective in vivo in a xenograft ALK− ALCL model. Our data suggest that cytokine receptor signaling is required for tumor cell survival in diverse forms of ALK− ALCL, even in the presence of JAK1/STAT3 mutations. Therefore, JAK inhibitor therapy might benefit patients with ALK− ALCL who are phosphorylated STAT3+.
The term anaplastic large cell lymphoma (ALCL) is applied to a group of T-cell lymphomas sharing a high expression of CD30 and a cytotoxic immunophenotype (1). Several variants have been described based on genetic and clinical features, including anaplastic lymphoma kinase (ALK)+ systemic, ALK− systemic, and primary cutaneous ALCL (2, 3). ALK+ ALCL is characterized by recurrent chromosomal translocations involving the ALK gene (4, 5). ALK− ALCLs, a heterogeneous group of disorders, are defined as tumors morphologically and immunophenotypically similar to ALK+ ALCL, but lacking ALK expression.
Patients with newly diagnosed systemic ALCL are commonly treated with first-line chemotherapy regimens, such as CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone); nonetheless, the disease is more likely to relapse within 5 y in patients with ALK− ALCL than in those with ALK+ ALCL. The 5-y overall survival rate in patients with ALK− ALCL is 49%, compared with 70% in those with ALK+ ALCL and 90% in those with primary cutaneous (pc) ALCL (6). Recently, breast implant-associated ALCL was recognized as a distinct clinical and pathological entity that shares morphologic features with both primary ALK− ALCL and pc-ALCL (7). It arises in the seroma cavity surrounding breast implants, usually after many years, and in the absence of invasion of the breast tissue, the clinical outcome is usually excellent.
The JAK/STAT pathway is a major pathway that transmits signals from extracellular stimuli (e.g., cytokines) into cellular activities (8, 9). Gain-of-function JAK and STAT mutations have been reported in many mature T-cell malignancies, including ALK− ALCL (10–16). These mutations are usually associated with increased phosphorylation of protein and enhanced growth activity (17). However, most previous functional studies of JAK/STAT mutations were carried out in cells that do not represent the true nature of human T-cell malignancies, thus limiting our understanding of how the mutations contribute to specific tumor growth. Moreover, the JAK/STAT mutation rate in a specific T-cell tumor population is usually low (∼20% in ALK− ALCL), which would appear to make the clinical application of targeting the pathway more challenging.
In the present study, we investigated whether JAK inhibitors could be of value in treating diverse ALK− ALCL cell lines originating from systemic, cutaneous ALK− ALCL, and breast implant-associated ALK− ALCL. Interestingly, most exogenous cytokine-independent ALK− ALCL cell lines responded to JAK inhibition regardless of JAK mutation status. Their JAK inhibitor sensitivity was correlated with their STAT3 phosphorylation status. These JAK inhibitor-sensitive cell lines were dependent on JAK1 and STAT3 for survival, as demonstrated by retroviral shRNA knockdown. We found that cytokine receptors were required for cell survival even in the presence of gain-of-function JAK1/STAT3 mutations. Finally, ruxolitinib, a JAK1/2 inhibitor, demonstrated efficacy in vivo in a xenograft model of ALK− ALCL. In summary, our data suggest that ALK− ALCLs are dependent on JAK1/STAT3 and that JAK inhibitors may be of value in the treatment of ALK− ALCLs that are phospho-STAT3 (p-STAT3)+ regardless of the presence of JAK/STAT mutations.
Results
JAK Inhibitors Diminish Exogenous Cytokine-Independent ALK− ALCL Cell Growth in Vitro.
We studied ALK− ALCL cell lines that originated from diverse forms of ALCL, including systemic ALK− ALCL, cutaneous ALK− ALCL, and breast implant-associated ALK− ALCL. All ALK− cell lines grew independent of exogenous cytokines, with the exception of TLBR4, which needed exogenous IL-2 to grow and was excluded from the study.
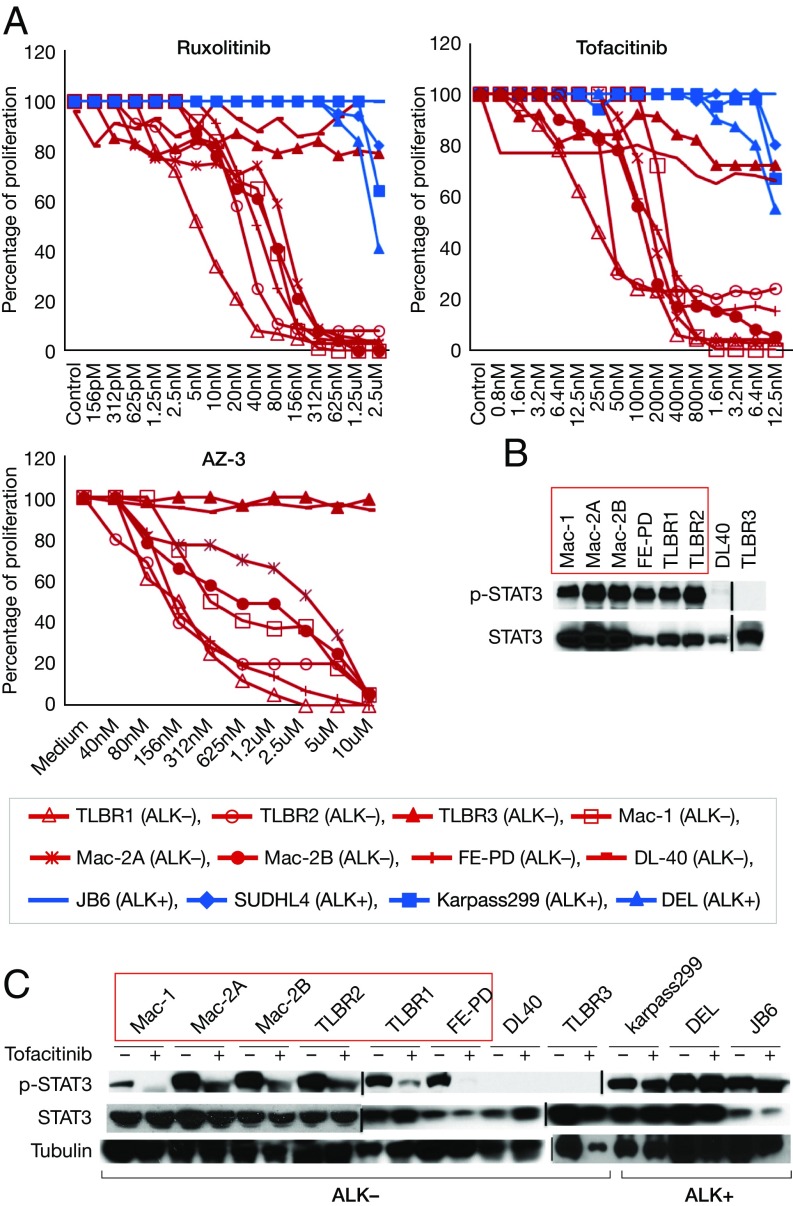
To investigate whether JAK inhibitors could be of value in treating ALK− ALCL, we treated eight exogenous cytokine-independent ALK− ALCL cell lines with the following JAK inhibitors: tofacitinib, a pan-JAK inhibitor; ruxolitinib, a JAK1/2 inhibitor; and AZ-3, a JAK1 inhibitor. All three JAK inhibitors are ATP-competitive inhibitors that bind to the ATP-binding site in the catalytic cleft of the kinase domain of JAK. Dose-dependent inhibition of proliferation was observed in six cell lines with all inhibitors (Mac-1/2A/2B, FE-PD, and TLBR1/2) (Fig. 1A). Interestingly, all JAK inhibitor-sensitive ALK− ALCL cells exhibited STAT3 activation (Fig. 1B), and tofacitinib treatment significantly decreased STAT3 phosphorylation in these cells (Fig. 1C). This suggests that STAT3 may be a major downstream target in the JAK inhibitor-sensitive cells. As a control, ALK+ ALCL cells did not respond to JAK inhibitors, and STAT3 phosphorylation was not affected by tofacitinib (Fig. 1 A and C), likely because the oncoprotein nucleophosmin-ALK activates STAT3 directly or indirectly without the involvement of JAKs in ALK+ ALCL cells (18).
Fig. 1.
JAK inhibitors diminished various p-STAT3+ ALK− ALCL cell line growth in vitro. (A) JAK inhibitors ruxolitinib, tofacitinib, and AZ-3 inhibited cell proliferation in most exogenous cytokine-independent ALK− ALCL cells. The cells were treated with a series of increasing concentrations of inhibitors. Blue, ALK+ ALCL cell lines; red, ALK− ALCL cell lines. The data are representative of three independent experiments. (B) Western blot analysis of STAT3 phosphorylation in ALK− ALCL cells. (C) Tofacitinib treatment decreased STAT3 phosphorylation in ALK− ALCL cells. The cells were treated with tofacitinib (1 μg/mL) for 4 h before harvesting.
JAK1 Is Required for the Survival of JAK Inhibitor-Sensitive Cells.
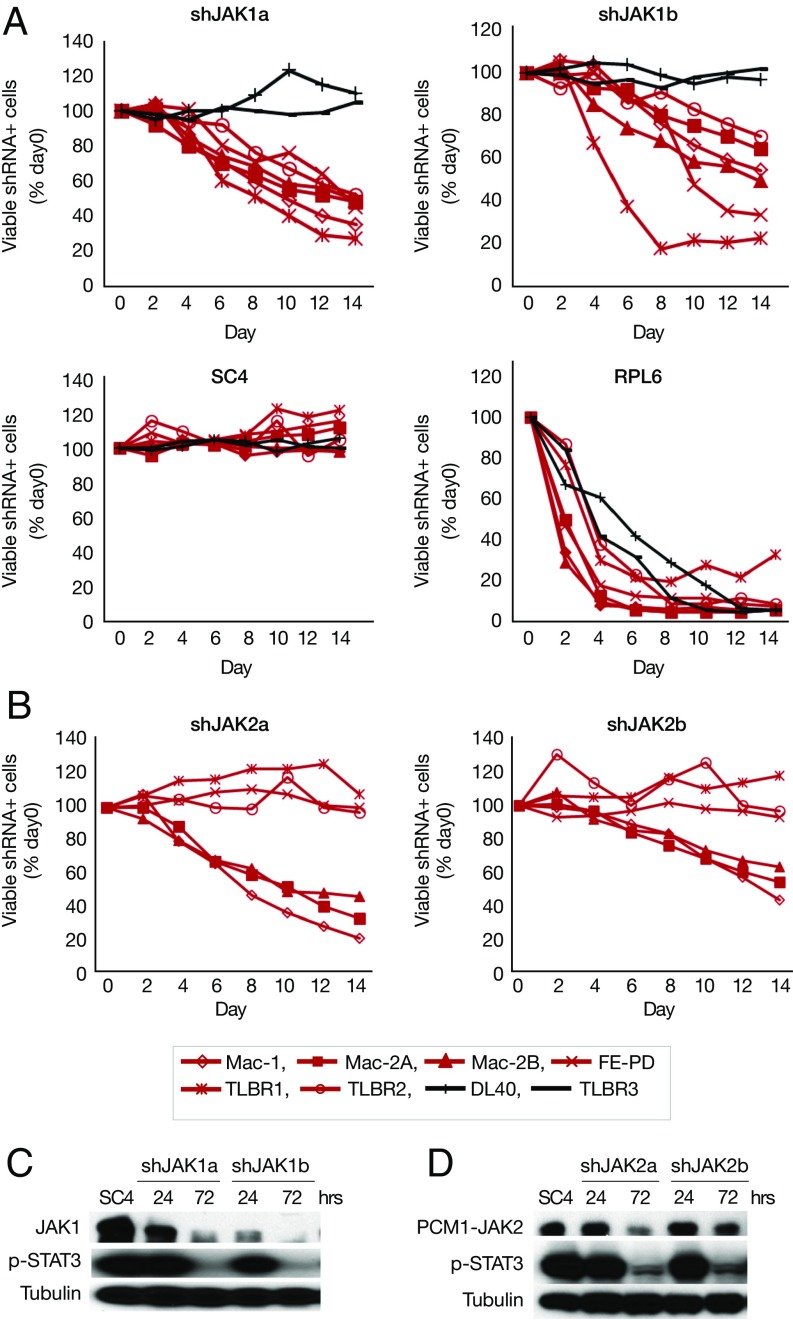
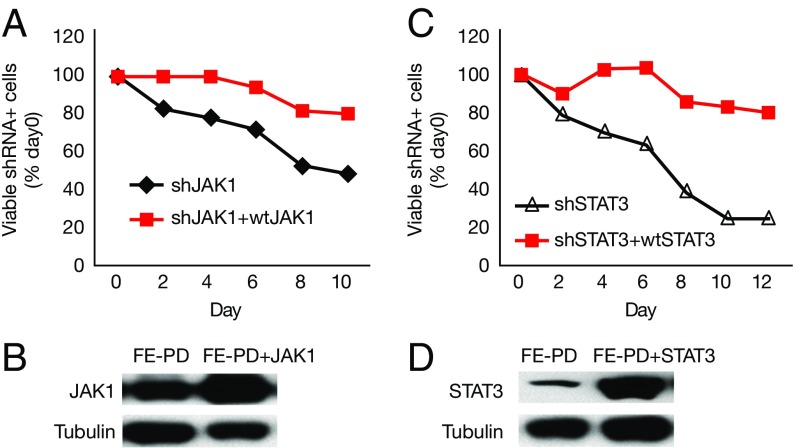
To investigate which JAK is responsible for JAK inhibitor sensitivity, we knocked down JAK1 and JAK2 with retroviral shRNA. Knockdown of JAK1 induced cell death in all six JAK inhibitor-sensitive lines (Fig. 2A), whereas knockdown of JAK2 induced cell death in three cell lines (Mac-1/2A/2B) that have JAK2 translocation (19) (Fig. 2B). For each knockdown experiment, two shRNAs were used, and similar effects were observed for both shJAK1 and shJAK2 (Fig. 2 A and B). JAK inhibitor-insensitive ALK− ALCL cell lines (DL-40 and TLBR3) served as negative cell controls (Fig. 2 A and B). Western blot analyses showed that JAK1/2 shRNA not only decreased JAK1 and JAK2 protein expression but also significantly decreased STAT3 phosphorylation (Fig. 2 C and D). This finding suggests that JAK1 (and JAK2) are required for cell survival in JAK inhibitor-sensitive cells and that STAT3 may be a major downstream target for JAK inhibition.
Fig. 2.
JAK1 (and JAK2) are required for cell survival in JAK inhibitor-sensitive cells. (A) shJAK1 induced cell death in all JAK inhibitor-sensitive cells. The percentage of viable shRNA+ cells at days 2, 4, 6, 8, 10, and 12 after addition of doxycycline was compared with that of day 0. shRNA-infected cells were GFP+; the shRNA expression was induced by doxycycline. shSC4 served as a negative control; shRPL6, as a positive control. (B) shJAK2 induced cell death in Mac-1/2A/2B cells. (C) Western blot analysis of JAK1 and p-STAT3 in Mac-1 cells stably infected with retroviral shJAK1. The shRNA expression was induced by doxycycline for 24 h or 72 h. (D) Western blot analysis of PCM1-JAK2 and p-STAT3 in Mac-1 cells stably infected with retroviral shJAK2.
STAT3 Is Required for the Survival of JAK Inhibitor-Sensitive Cells.
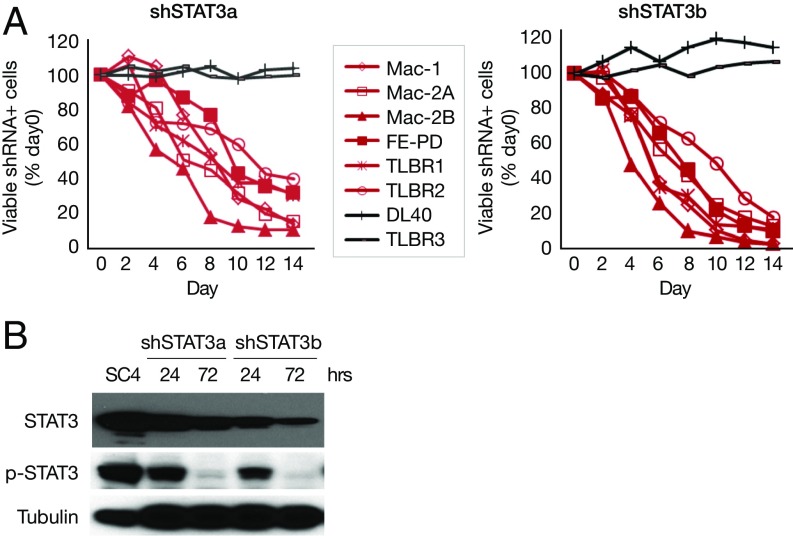
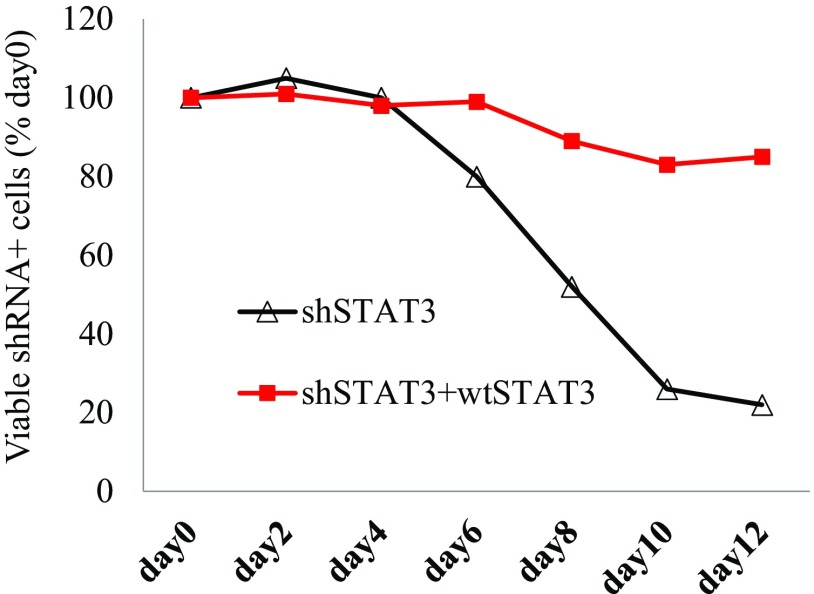
We next investigated whether STAT3 is required for cell survival in JAK inhibitor-sensitive cells. Knockdown of STAT3 induced cell death in all JAK inhibitor-sensitive cells (Fig. 3A). Two different shRNAs that target different regions of STAT3 were used, and similar effects were observed (Fig. 3A). p-STAT3− ALK− ALCL cell lines DL40 and TLBR3 (also JAK inhibitor-insensitive) served as negative cell controls (Fig. 3A). Western blot analysis showed decreased STAT3 protein expression and phosphorylation with shSTAT3 treatment (Fig. 3B).
Fig. 3.
STAT3 is required for cell survival in all JAK inhibitor-sensitive cells. (A) shSTAT3 induced cell death in all JAK inhibitor-sensitive cells. (B) Western blot analysis of STAT3 expression and p-STAT3 in Mac-1 cells stably infected with shSTAT3.
Activating JAK1 and STAT3 Mutations Are Identified in Some, But Not All, JAK Inhibitor-Sensitive Cells.
To investigate the underlying mechanism for JAK1/STAT3 dependency, we performed RNA-seq to examine whether JAK and STAT genes were mutated in these JAK inhibitor-sensitive cells. Interestingly, we found JAK1 and STAT3 gene mutations in some, but not all, JAK inhibitor-sensitive cells. The mutations were confirmed by Sanger sequencing (Table 1). Of these, all three STAT3 mutations—G618R, S614R, and D661Y—have been previously described in large granular lymphocytic (LGL), natural killer (NK)-cell, and γδ T-cell leukemias (11, 20). There was an additional JAK1 mutation, G1097V. The JAK2 translocation was previously reported in Mac-1/2A/2B cells (19). We confirmed the translocation by RNA-seq and Western blot analyses (Fig. S1).
Table 1.
JAK and STAT3 mutations in JAK inhibitor-sensitive cells
| Mutation | Mac-I | Mac-2A | Mac-2B | FE-PD | TLBR I | TLBR2 |
| JAK mutation | JAK2 (PCMI-JAK2) | JAK2 (PCMI-JAK2) | JAK2 (PCMl-JAK2) | JAK l (G1097V) | ||
| STAT3 mutation | STAT3 (G618R) | STAT3 (S614R) | STAT3 (D661Y) |
Fig. S1.
Western blot analysis of JAK2 and PCM1-JAK2.
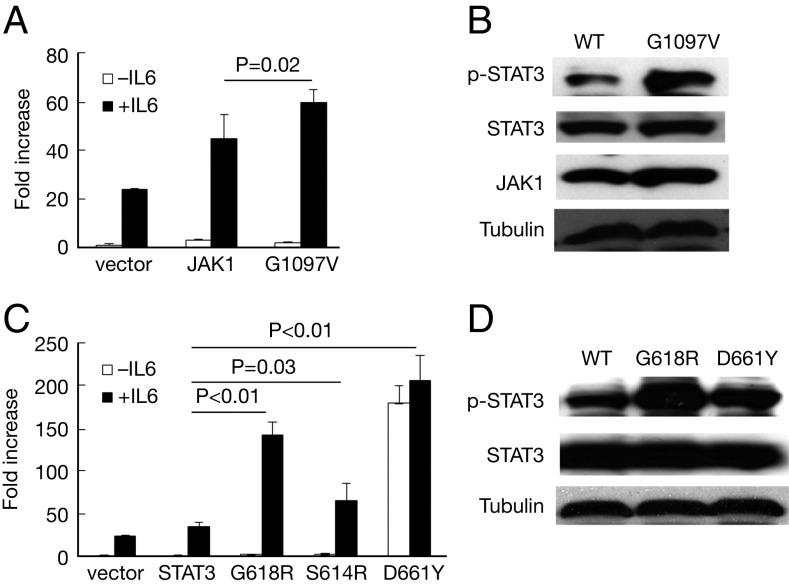
We next analyzed the activity of JAK1 and STAT3 mutants using a Cignal STAT3 reporter assay (Qiagen). JAK1G1097V showed a modest increase in STAT3 reporter activation and STAT3 phosphorylation in response to IL-6 (JAK1 vs. JAK1G1097V, P = 0.02) (Fig. 4 A and B). STAT3 mutations G618R, S614R, and D661Y demonstrated increased STAT3 reporter activation and increased phosphorylation in response to IL-6 (STAT3 vs. G618R, P < 0.01; STAT3 vs. S614R, P = 0.03; STAT3 vs. D661Y, P < 0.01) (Fig. 4 C and D). Of note, D661Y demonstrated high activity even in the absence of IL-6.
Fig. 4.
JAK1 and STAT3 mutations induce increased STAT3 activity in response to IL-6. (A) JAK1G1097V induced increased STAT3 reporter activity compared with WT JAK1 in response to IL-6 (JAK1 vs. JAK1G1097V, P = 0.02). (B) JAK1G1097V induced greater STAT3 phosphorylation in response to IL-6. (C) STAT3 mutants induced increased STAT3 reporter activity in response to IL-6 compared with WT STAT3 (STAT3 vs. G618R, P < 0.01; STAT3 vs. S614R, P = 0.03; STAT3 vs. D661Y, P < 0.01). (D) STAT3 mutants induced increased STAT3 phosphorylation in response to IL-6. IL-6 (10 ng/mL) was added (or not) to the cell culture at 30 min before harvesting.
Wild-Type JAK1 or STAT3 Is Sufficient to Promote Cell Survival in JAK Inhibitor-Sensitive Cells with Either JAK1 or STAT3 Mutations.
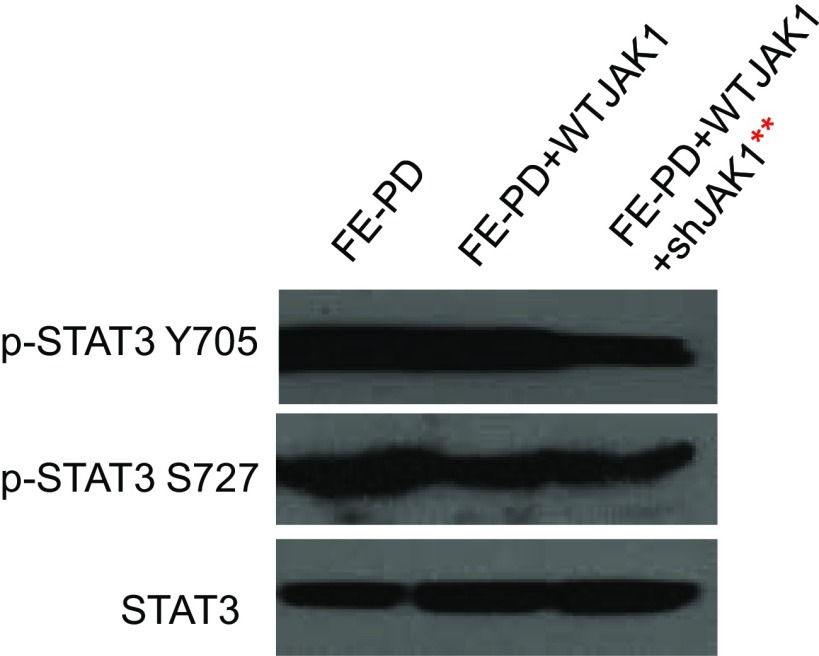
Activating JAK1 and STAT3 mutations have proven capable of promoting tumor growth and oncogenesis (10, 11). To investigate whether the JAK1/STAT3 dependency in the mutant-containing cells is due solely to the mutations, we knocked down the endogenous JAK1 (or STAT3) expression and overexpressed wild-type (WT) JAK1 (or STAT3) in FE-PD cells that had both JAK1 and STAT3 mutations. The expression of WT JAK1 abrogated the cell death induced by shJAK1 (Fig. 5 A and B), and p-STAT3 expression was maintained in both Y705 and S727 (Fig. S2). Similarly, expression of WT STAT3 also abrogated the cell death induced by shSTAT3 (Fig. 5 C and D). Moreover, expression of WT STAT3 abrogated the cell death induced by shSTAT3 in TLBR2, a cell line with a STAT3 D661Y mutation (Fig. S3). These data suggest that WT JAK1 and STAT3 are sufficient to promote cell growth in JAK1 or STAT3 mutant-containing cells, and thus the JAK1/STAT3 mutations alone cannot explain the JAK1/STAT3 dependency in these leukemic cell lines.
Fig. 5.
WT JAK1 or STAT3 is capable of promoting cell growth in JAK1/STAT3 mutant-containing FE-PD cells. (A) WT JAK1 abrogated cell death induced by shJAK1. FE-PD cells that stably expressed WT were infected with shJAK1 to knock down endogenous JAK1 expression. Cell growth was monitored in infected cells (GFP+). shJAK1 expression was induced by doxycycline. (B) Western blot analysis of JAK1 expression in WT and JAK1-overexpressing cells. (C) WT STAT3 abrogated cell death induced by shSTAT3. (D) Western blot analysis of STAT3 expression in WT and STAT3-overexpressing cells.
Fig. S2.
Replacing endogenous JAK1 with WT JAK 1 did not affect pSTAT3 Y705 or pSTAT3 S727 expression. **Endogenous JAK1 was knocked down by shJAK1, whereas WT JAK1 was not knocked down.
Fig. S3.
WT STAT3 abrogated cell death induced by shSTAT3 in TLBR2 cells.
Cytokine Receptors GP130 and IL-2Rγ Are Needed for Cell Survival in Select JAK Inhibitor-Sensitive Cells.
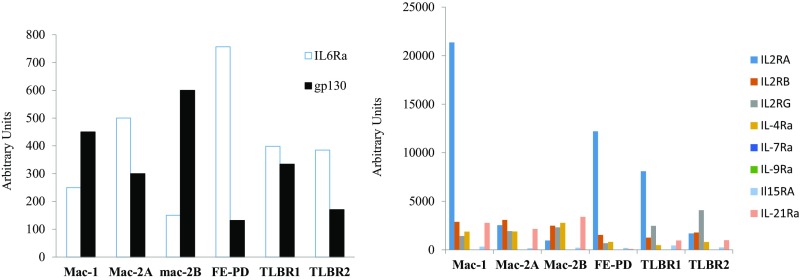
To investigate whether other mechanisms are involved in JAK1/STAT3 dependency, we examined cytokine/cytokine receptor expression in the JAK inhibitor-sensitive cells. Interestingly, these cells produced large amounts of cytokines, including IL-6, IL-10, and IFNγ (Table S1). They also expressed IL-6 receptors and γ cytokine receptors (Fig. S4).
Table S1.
Cytokine secretion by JAK inhibitor-sensitive ALK− ALCL cell lines
| Cell line | IL-6, pg/mL | IL-10, pg/mL | IL-13, pg/mL | IFN-γ, pg/mL | TNF-α, pg/mL | GM-CSF, pg/mL | IL-7, pg/mL |
| Mac-1 | 183.5 | 2.2 | 46.6 | >1,210 | 3.6 | 898.2 | 9.2 |
| Mac-2A | >774 | 2.9 | >492 | 100.2 | >321 | >911 | 10.7 |
| Mac-2B | 104.1 | 0.3 | >492 | 2.1 | 52.7 | >911 | 5.4 |
| TLBRl | 379.3 | >321 | >492 | >1,210 | 182.5 | >911 | 26.4 |
| TLBR2 | >774 | >321 | >492 | 5.3 | 1.2 | 2.6 | 27.1 |
| FE-PD | <0.76 | >321 | 2.1 | 2.8 | 0.4 | 1.8 | 37 |
Fig. S4.
IL-6 and γ cytokine receptor expression in JAK inhibitor-sensitive ALK− ALCL cells.
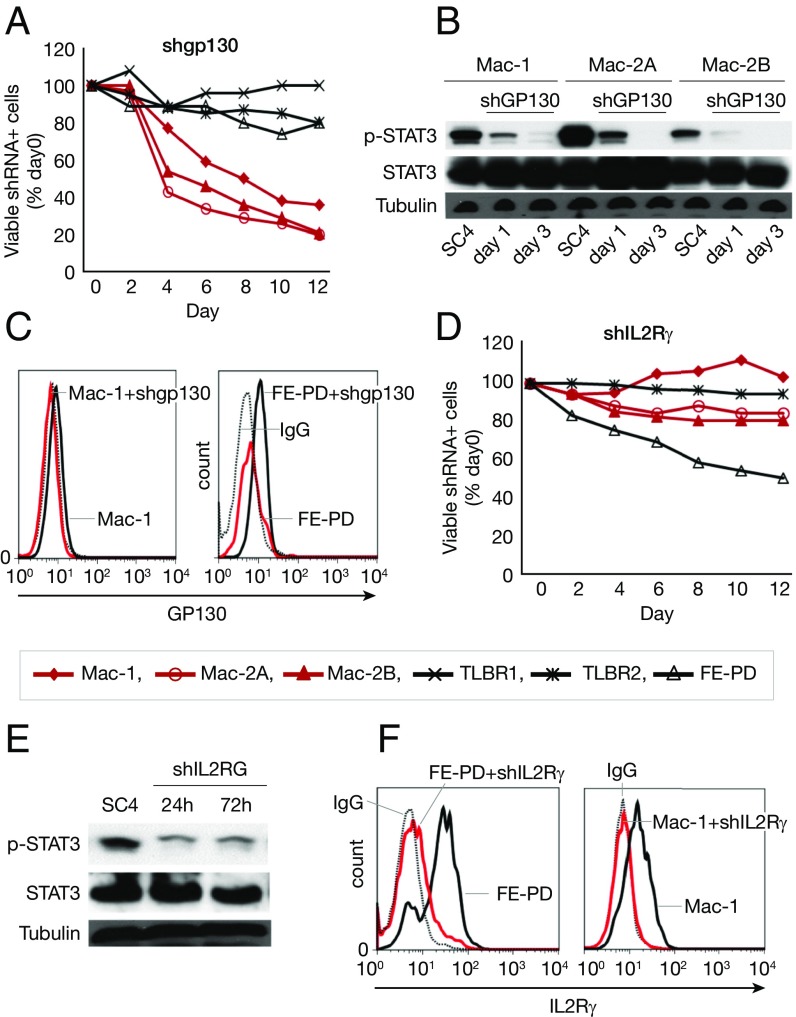
To investigate whether cytokine receptors are involved in the cell survival in JAK inhibitor-sensitive cells, we knocked down GP130 and IL-2Rγ. Knockdown of GP130 led to cell death in Mac-1/2A/2B cells (Fig. 6A) and also induced a dramatic decrease in STAT3 phosphorylation in these cells (Fig. 6B). Depletion of surface GP130 expression was demonstrated by flow cytometry (Fig. 6C). Knockdown of IL-2Rγ led to cell death in FE-PD cells (Fig. 6D), as well as decreased STAT3 phosphorylation (Fig. 6E). Depletion of surface IL-2Rγ expression was demonstrated by flow cytometry (Fig. 6F). These data suggest that cytokine receptors GP130 and IL-2Rγ are involved in the cell survival of select JAK inhibitor-sensitive cells.
Fig. 6.
Cytokine receptor GP130 and IL-2Rγ are needed for cell survival in select JAK inhibitor-sensitive cells. (A) shGP130 induced cell death in Mac-1/2A/2B cells. (B) shGP130 induced decreased STAT3 phosphorylation in Mac-1/2A/2B cells. (C) shGP130 depleted surface expression of GP130. The black dotted line represents IgG control; the black solid line, before shGP130 induction; red solid line, 72 h after shGP130 induction. (D) shIL-2Rγ induced cell death in FE-PD cells. (E) shIL-2Rγ induced decreased STAT3 phosphorylation in FE-PD cells. (F) shIL-2Rγ depleted surface expression of IL-2Rγ. The black dotted line represents IgG control; the black solid line, before shGP130 induction; the red solid line, 72 h after shGP130 induction.
High Expression of Cytokine Genes in ALK− ALCL Primary Tumors.
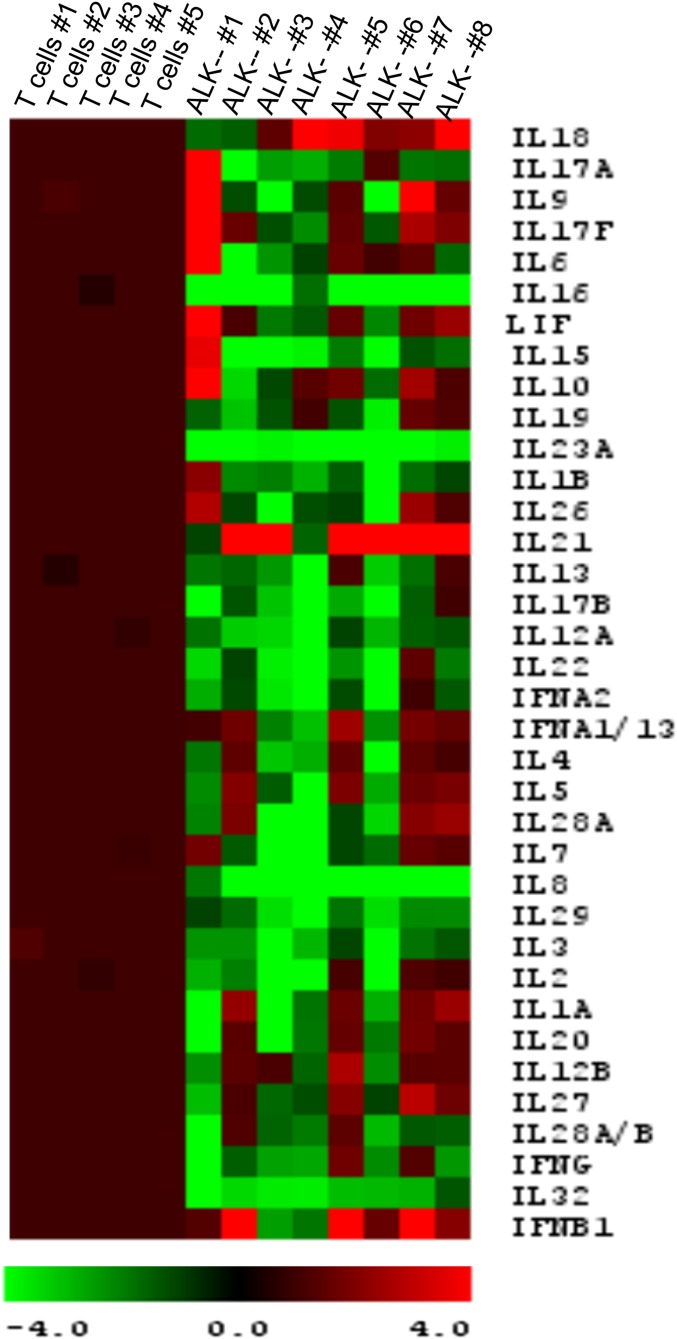
Chronic autocrine paracrine cytokine stimulation is common in T-cell disorders (21). To investigate whether there is cytokine stimulation in ALK− ALCL primary tumors, we analyzed cytokine gene expression with NanoString. The primary tumor tissues were obtained from eight systemic ALK− ALCLs, and the diagnosis was confirmed by pathology. NanoString demonstrated increased expression of cytokine genes, including IL-21, IL-18, IFNβ, IL-6, LIF, IL-17F, and IL-10, in the ALK− primary tumors (Fig. S5).
Fig. S5.
Cytokine gene expression is increased in primary ALK− ALCL tumors. Cytokine gene expression was measured by Nanostring. The expression levels were normalized to levels in 15 housekeeping genes and were compared with those of normal T cells.
Ruxolitinib Inhibits Tumor Growth in Vivo in a Xenograft ALK− ALCL Model.
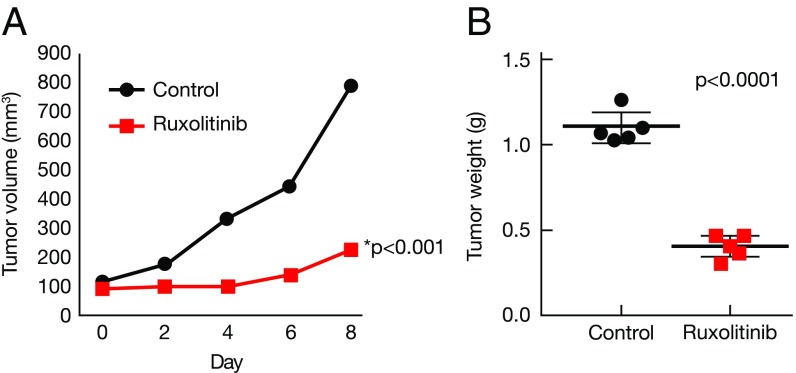
To investigate the possibility of using JAK inhibitor in the clinic, we tested the efficacy of the JAK1/2 inhibitor ruxolitinib in vivo in a xenograft ALK− ALCL mouse model. The mouse model was established by s.c. injection of FE-PD cells. Treatment started when the tumor volume reached 100 mm3. One week of continuous infusion of ruxolitinib (50 mg/kg/d) significantly inhibited tumor growth (n = 5; P < 0.01) (Fig. 7A). The ruxolitinib-treated mice also had lower tumor weights compared with the control group when measured at the end of the study (day 9; P < 0.0001) (Fig. 7B). Our data suggest that ALK− ALCL cells are dependent on JAK1 and STAT3 and that targeting JAK1 could be a rational approach to the clinical treatment of ALK− ALCL tumors.
Fig. 7.
Ruxolitinib inhibits tumor growth in a xenograft ALK− ALCL mouse model. (A) Ruxolitinib inhibited ALK− ALCL tumor growth in vivo. Ruxolitinib was given at 50 mg/kg/d continuously via a pump (Alzet) for 7 d. Treatment started when the tumor volume reached 100 mm3. Control vs. ruxolitinib, P < 0.01. (B) Ruxolitinib treatment decreased tumor weight when measured at day 9. Control vs. ruxolitinib, P < 0.001.
Discussion
Mature T-cell lymphomas are a rare, heterogeneous group of non-Hodgkin lymphomas with an aggressive disease course and poor overall survival. The advent of novel technologies, such as next-generation sequencing, not only has helped delineate the molecular pathogenesis of T-cell lymphomas, but also has led to the discovery of many actionable genetic alterations, which can be targeted either by specific therapeutic compounds or by monoclonal antibodies. The JAK/STAT pathway has emerged as one of these targets (11–14). JAK mutations have been identified in patients with adult T-cell leukemia, ALK− ALCL, early T-cell precursor acute lymphoblastic leukemia, T-cell prolymphocytic leukemia, and Sézary syndrome. STAT mutations have been identified in LGL, nasal type NK/T-cell lymphoma, γδ hepatosplenic T-cell lymphoma, and ALK− ALCL. Although the JAK/STAT mutations are quite common among T-cell malignancies in general, the mutation rate in any specific T-cell malignancy is quite low (e.g., ∼20% in ALK− ALCL). This would appear to limit the clinical application of targeting this pathway for a broader patient population.
In this study, we investigated the targeting of JAK for the treatment of diverse forms of ALK− ALCL using ALK− ALCL tumor cell lines originated from systemic, cutaneous ALK− ALCLs as well as breast implant-associated ALK− ALCLs. We tested three JAK inhibitors: tofacitinib, a pan-JAK inhibitor; ruxolitinib, a JAK1/2 inhibitor; and AZ-3, a JAK1-selective inhibitor. Surprisingly, most exogenous cytokine-independent ALK− ALCL cells (six of eight) responded to JAK inhibition (Fig. 1). The JAK inhibitor sensitivity correlated with the positive STAT3 phosphorylation status of the cells. Moreover, JAK inhibitor treatment significantly decreased STAT3 phosphorylation, suggesting that STAT3 might be a major downstream target for JAK inhibition (Fig. 1).
Janus kinase has four family members: JAK1, JAK2, JAK3, and TYK2. To further characterize the nature of JAK inhibitor sensitivity in ALK− ALCL cells, we knocked down JAK1 and JAK2 with shRNA. Knockdown of JAK1 led to cell death in all JAK inhibitor-sensitive cell lines (Fig. 2), whereas knockdown of JAK2 led to cell death only in PCM1-JAK2–containing Mac-1/2A/2B cell lines. Interestingly, knockdown of JAK1 and JAK2 led not only to decreased expression of JAK1 (or PCM1-JAK2) but also to significantly decreased p-STAT3 expression. This finding again suggests that STAT3 may be a major downstream target for JAK inhibition. This hypothesis was further confirmed by our demonstration that knockdown of STAT3 led to cell death in all JAK inhibitor-sensitive cells (Fig. 3).
To investigate the underlying mechanisms of JAK1/STAT3 dependency in ALK− ALCL cells, we considered two possibilities: gain-of-function JAK1/STAT3 mutations and activation of the pathway through cytokine receptors. Using RNA-seq followed by Sanger sequencing, we demonstrated gain-of-function mutations in JAK1 (G1097V) and STAT3 (S614R, G618R, and D661Y) in some, but not all, JAK inhibitor-sensitive cell lines (Table 1). We also confirmed PCM1-JAK2 translocation in Mac-1/2A/2B cells (Fig. S1). These mutations demonstrated greater STAT3 activity in response to IL-6 when transfected into 293T cells (Fig. 4). Only D661Y demonstrated STAT3 activity in the absence of IL-6, suggesting that D661Y may be a constitutive active mutation, or that it requires less cytokine stimulation, which may be achieved endogenously in 293T cells. Nevertheless, these data suggest that the mutations may facilitate and augment signals from upstream in the pathway, but alone cannot fully explain the JAK1/STAT3 dependency in JAK inhibitor-sensitive cells, given that most of the JAK1-dependent cells had no JAK1 mutation (Table 1). Similarly, Kücük et al. (17) demonstrated that activating STAT5b mutations were insufficient to initiate leukemic cell proliferation and only facilitated and prolonged signals from above by IL-2 stimulation.
We next investigated whether the JAK1/STAT3 mutations were responsible for the JAK1/STAT3 dependency in JAK1/STAT3 mutant-containing FE-PD cells. Surprisingly, we found that WT JAK1 or STAT3 was sufficient to promote cell growth in FE-PD cells (Fig. 5). Similarly, WT STAT3 was sufficient to promote cell growth in TLBR2, a cell line with a STAT3 D661Y mutation (Fig. S3). These data suggest that, even in JAK1/STAT3 mutant-containing cells, other mechanisms are involved in activating the JAK/STAT pathway.
Cytokine receptors are major components in the signaling pathway for transducing extracellular stimuli into cellular functions. They also act as scaffold/docking sites for the transactivation of JAKs and the recruitment of the STAT factors (22–24). In particular, Lu et al. (22) demonstrated that the expression of a homodimeric type 1 cytokine receptor is required for JAK2V617F-mediated transformation. In addition, Hornakova et al. (25) showed that IL-9Rα and IL-2Rβ homodimers efficiently mediate constitutive activation of an ALL-associated JAK1 mutant, again indicating that a cytokine receptor is required as a JAK activation scaffold and is a STAT docking site. We investigated whether cytokine receptors are involved in activating the JAK1/STAT3 pathway in JAK inhibitor-sensitive cells. Knockdown of GP130 and IL-2Rγ induced cell death in Mac1/2A/2B and FE-PD cells, respectively (Fig. 6). Mac-1/2A/2 B (25) cells produced large amounts of IL-6 (Table S1) and expressed IL-6 receptors (Fig. S4). The GP130 knockdown experiment suggests the possibility of an IL-6/IL-6R autocrine stimulation in these cells. This idea is in agreement with our finding that Mac-1/2A/2B cells are dependent not only on JAK1 but also on JAK2 for survival (Fig. 2), because IL-6 signals through JAK1, JAK2, and TYK2. However, neutralizing antibodies targeted to IL-6 or gp130 did not affect cell growth (Fig. S6). This may due to an intracellular interaction of IL-6 with its receptor or the inability of the commercial antibodies to neutralize large amounts of IL-6. The shJAK2 used in this study targeted both WT and PCM1-JAK2; thus, whether its effect is due to WT JAK2 or PCM1-JAK2 is unknown. Nevertheless, this observation supports a possible IL-6 autocrine loop in Mac-1/2A/2B cells, although we cannot not rule out the possibility that gp130 acts as a scaffold/docking site for PCM-JAK2–mediated transformation that involves JAK1 and STAT3.
Fig. S6.
Neutralizing antibodies to the γc cytokine/receptor, IL-6 family cytokine/receptor does not affect cell growth in JAK inhibitor-sensitive ALK− ALCL cells.
Interestingly, the breast implant-associated tumor cell lines TLBR1/2 were not dependent on GP130 for survival, even though they produced high levels of IL-6 (Table S1) and expressed IL-6 receptors (Fig. S4). Further study is needed to characterize the upstream signaling that activates JAK1 (and STAT3) in TLBR1/2. In agreement with the cell line data, expression of cytokines, including IL-6, IL18, IL21, LIF, and others, was demonstrated in primary ALK− ALCL tumors (Fig. S5).
Our demonstration of JAK1/STAT3 dependency in ALK− ALCL cell lines suggests that the JAK/STAT pathway could be an attractive target for therapy in diverse forms of ALK− ALCL that are p-STAT3+. This significantly broadens the scope of clinical application of targeting the JAK/STAT pathway, given that roughly 50% of ALK− ALCL primary tumors have p-STAT3 expressed in the nucleus (26). The STAT3 activation in ALK− ALCL may or may not be associated with JAK/STAT mutations; regardless, the prognosis is much worse in patients with p-STAT3+ tumors (26). Similarly, constitutive activation of the JAK/STAT pathway is more pervasive than JAK/STAT mutations among T-cell malignancies, suggesting that the JAK/STAT pathway may be an attractive target for therapy of T-cell malignancies in general (27).
STAT3 has been recognized as a potentially good target for therapy in many other cancers; however, very few of the STAT3 inhibitors that have been developed have undergone in vivo evaluation (28). On the other hand, several JAK inhibitors have successfully transitioned into clinical use. Tofacitinib, a pan-JAK inhibitor, has been approved by the Food and Drug Administration (FDA) for rheumatoid arthritis (29). Ruxolitinib, a JAK1/2 inhibitor, was recently approved by the FDA for the treatment of polycythemia vera and intermediate-risk and high-risk myelofibrosis (30, 31). To investigate whether a JAK inhibitor could be useful in patients with ALK− ALCL, we developed an in vivo xenograft ALK− ALCL mouse model using FE-PD cells. One week of continuous ruxolitinib infusions significantly inhibited tumor growth and decreased tumor weight (Fig. 7 A and B).
In summary, our data suggest that even in the presence of JAK1 or STAT3 mutations, signaling through cytokine receptors is required for cell survival through activation of JAK1 and STAT3 in ALK− tumor cells that originated from diverse forms of ALK− ALCL. JAK inhibitors demonstrated antitumor efficacy both in vitro and in vivo; therefore, JAK inhibitor therapy might benefit patients with an ALK− ALCL that is p-STAT3+.
Materials and Methods
Cell Lines.
Mac-1/2A/2B cell lines were established from different stages and sites of ALK− cutaneous ALCLs (32). FE-PD and DL-40 were established from aggressive leukemic ALK− ALCLs (33, 34). TLBR1/2/3/4 cell lines were established from four patients with breast implant-associated ALCLs (35, 36). JB6, SUDHL4, Karpass299, and DEL were established from ALK+ ALCLs. TLBR4 and NK-92 cells were cultured with 100 U of rhIL-2, and BaF3 cells were cultured with 10% (vol/vol) WEHI-3 conditioned medium. Cells were engineered to express ecotropic retroviral receptors and the TET repressor for infection and inducible expression of retroviral shRNA vectors as described previously (37). All human studies were approved by the Institutional Review Board of National Cancer Institute (NCI), and patients provided consent. Animal studies were approved by the Animal Care and Use Committee (ACUC) of the NCI.
Proliferation Assay.
Here 100-μL aliquots of cell culture suspensions (1 × 106/mL) were seeded into 96-well microtiter plates in triplicate and cultured for 3 d at 37 °C in 5% CO2. JAK inhibitors were added at the beginning of the assay. Cultures were pulsed with 1 μCi [3H]thymidine at 6 h before harvesting.
shRNA Gene Knockdown.
shRNAs were cloned into a doxycycline-inducible pRSMX-puroGFP vector. Retroviruses containing the shRNA were produced by cotransfection of 293T cells with helper plasmids expressing gag-pol genes, an ecotropic pseudotyping env gene, and a retroviral vector using FuGENE6 transfection reagent as described previously (37). Target cells were infected with the retroviruses in the presence of 8 µg/mL polybrene.
Retroviral Gene Expression.
cDNAs were cloned into pCMV/TO/puro, mscv-puro, or mscv-hyg. Retroviruses were produced in 293T cells, and the target cells were infected as described above. Stable cell lines were obtained by selection with puromycin or hygromycin for 3 wk.
STAT3 Reporter Assay.
JAK1/STAT3 expression constructs were cotransfected with STAT3 reporter (Qiagen) using the Fugene6 transfection reagent. Cells were lysed following the protocol of the Promega Dual-Luciferase Reporter Assay system and then analyzed for luciferase activity.
SI Materials and Methods
Reagents and Antibodies.
Anti-human IL-2Rγ and GP130 antibodies conjugated to PE were purchased from BD Biosciences. The following antibodies were purchased from Cell Signaling Technology: JAK1, JAK2 (3230), STAT3, and p-STAT3 (9145).
Flow Cytometry Analysis.
Cells were washed with PBS containing 2% FBS twice and incubated for 30 min on ice with anti-human GP130, IL-2Rγ, or control isotype IgG. After washing with FACS buffer (PBS/2% FBS) twice, the cells were analyzed on a FACSCalibur cell analyzer (BD Biosciences).
Immunoblot Analysis.
Cells were lysed at 4 °C with RIPA buffer. Cell lysates (50 μg) were resolved by electrophoresis on SDS/PAGE (4–12%) gels and then transferred to PVDF membranes. Proteins were detected with the following primary antibodies: p-STAT3, STAT3, JAK1, JAK2, and α-tubulin.
RNA-seq.
Total RNA was extracted using the Qiagen RNeasy Kit, and sequencing libraries were prepared using the Illumina TruSeq RNA Sample Prep Kit v2 according to the manufacturer’s instructions. Paired-end 108-bp read sequencing was performed on an Illumina HiSeq 2000 system. The mapping of paired-end reads and the extraction of putative single nucleotide variants (SNVs) were performed as described previously (1). In brief, paired-end reads were mapped to the RNA sequences in the RefSeq database [National Center for Biotechnology Information (NCBI) build 37] using Burrows–Wheeler Aligner software with default parameters. Reads that failed to map to RefSeq were mapped to RNA sequences in the Ensemble database. The remaining unmapped reads were mapped to the human genome assembly (NCBI build 37). Mutant SNV calls were declared if more than two reads were mutated and the ratio of mutant reads vs. total coverage was >20%. SNVs that corresponded to single nucleotide polymorphisms in the dbSNP database (build 132), the 1000 Genomes database (May 2011 release), and the National Heart, Lung, and Blood Institute’s Grand Opportunity Exome Sequencing Project database (ESP5400; December 2011 release) were excluded. RNA-seq data were submitted to the NCBI Gene Expression Omnibus (GEO) database (accession no. GSE96048).
Sanger DNA/RNA Resequencing.
Total RNA was DNase-digested and reversed-transcribed using the Qiagen QuantiTect Reverse Transcription Kit. JAK1 and STAT3 coding regions flanking the mutation site (300–400 bp) were amplified by PCR. The PCR products were purified and sequenced using the Applied Biosystems Big Dye Terminator V1.1 Cycle Sequencing Kit and analyzed on an Applied Biosystems 3730 Genetic Analyzer.
NanoString Gene Expression Assay.
Three or four 10-μm tissue curls were extracted from formalin-fixed, paraffin-embedded (FFPE) blocks from ALK− ALCL tumors. Total RNA was extracted from the curls using the RNeasy FFPE Kit (Qiagen). The quality of the RNA was analyzed with an Agilent Bioanalyzer. The gene expression assay was conducted using a predesigned code set containing 581 human immunology genes (Human Immunology v2; NanoString Technologies). In brief, 100 ng of total RNA was hybridized with reporter and capture probes overnight. After washing, the signals were analyzed on an nCounter Digital Analyzer (NanoString Technologies). Analysis and normalization of the raw NanoString data were conducted using nSolver Analysis Software v1.1 (NanoString Technologies). Raw counts were normalized to internal levels of 15 reference genes: ABCF1, ALAS1, EEF1G, G6PD, GAPDH, GUSB, HPRT1, OAZ1, POLR1B, POLR2A, PP1A, SDHA, TBP, TUBB, and RPL19.
Acknowledgments
This study was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute at the NIH. M.E.K. receives grant support from the Aesthetic Surgery Education and Research Foundation. The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE96048).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700682114/-/DCSupplemental.
References
- 1.Campo E, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood. 2011;117(19):5019–5032. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kempf W. CD30+ lymphoproliferative disorders: Histopathology, differential diagnosis, new variants, and simulators. J Cutan Pathol. 2006;33(Suppl 1):58–70. doi: 10.1111/j.0303-6987.2006.00548.x. [DOI] [PubMed] [Google Scholar]
- 3.Falini B, Martelli MP. Anaplastic large cell lymphoma: Changes in the World Health Organization classification and perspectives for targeted therapy. Haematologica. 2009;94(7):897–900. doi: 10.3324/haematol.2009.008250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris SW, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263(5151):1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 5.Chiarle R, et al. The anaplastic lymphoma kinase is an effective oncoantigen for lymphoma vaccination. Nat Med. 2008;14(6):676–680. doi: 10.1038/nm1769. [DOI] [PubMed] [Google Scholar]
- 6.Vose JM, Neumann M, Harris ME. International T-Cell Lymphoma Project International peripheral T-cell and natural killer/T-cell lymphoma study: Pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- 7.Lazzeri D, et al. ALK-1–negative anaplastic large cell lymphoma associated with breast implants: A new clinical entity. Clin Breast Cancer. 2011;11(5):283–296. doi: 10.1016/j.clbc.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Jove R. The STATs of cancer: New molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 9.Jatiani SS, Baker SJ, Silverman LR, Reddy EP. Jak/STAT pathways in cytokine signaling and myeloproliferative disorders: Approaches for targeted therapies. Genes Cancer. 2010;1(10):979–993. doi: 10.1177/1947601910397187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crescenzo R, et al. European T-Cell Lymphoma Study Group, T-Cell Project: Prospective Collection of Data in Patients with Peripheral T-Cell Lymphoma and the AIRC 5xMille Consortium “Genetics-Driven Targeted Management of Lymphoid Malignancies” Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516–532. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koskela HL, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajala HL, Porkka K, Maciejewski JP, Loughran TP, Jr, Mustjoki S. Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations. Ann Med. 2014;46(3):114–122. doi: 10.3109/07853890.2014.882105. [DOI] [PubMed] [Google Scholar]
- 13.Koganti S, de la Paz A, Freeman AF, Bhaduri-McIntosh S. B lymphocytes from patients with a hypomorphic mutation in STAT3 resist Epstein-Barr virus-driven cell proliferation. J Virol. 2014;88(1):516–524. doi: 10.1128/JVI.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milner JD, et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125(4):591–599. doi: 10.1182/blood-2014-09-602763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo GC, et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. 2012;2(7):591–597. doi: 10.1158/2159-8290.CD-12-0028. [DOI] [PubMed] [Google Scholar]
- 16.Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012;36(4):529–541. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Küçük C, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK cells. Nat Commun. 2015;6:6025. doi: 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamo A, et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002;21(7):1038–1047. doi: 10.1038/sj.onc.1205152. [DOI] [PubMed] [Google Scholar]
- 19.Ehrentraut S, et al. t(8;9)(p22;p24)/PCM1-JAK2 activates SOCS2 and SOCS3 via STAT5. PLoS One. 2013;8(1):e53767. doi: 10.1371/journal.pone.0053767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jerez A, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–3057. doi: 10.1182/blood-2012-06-435297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, et al. Autocrine/paracrine cytokine stimulation of leukemic cell proliferation in smoldering and chronic adult T-cell leukemia. Blood. 2010;116(26):5948–5956. doi: 10.1182/blood-2010-04-277418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X, et al. Expression of a homodimeric type I cytokine receptor is required for JAK2V617F-mediated transformation. Proc Natl Acad Sci USA. 2005;102(52):18962–18967. doi: 10.1073/pnas.0509714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornakova T, et al. Acute lymphoblastic leukemia-associated JAK1 mutants activate the janus kinase/STAT pathway via interleukin-9 receptor alpha homodimers. J Biol Chem. 2009;284(11):6773–6781. doi: 10.1074/jbc.M807531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Huang LJ, Lodish HF. Dimerization by a cytokine receptor is necessary for constitutive activation of JAK2V617F. J Biol Chem. 2008;283(9):5258–5266. doi: 10.1074/jbc.M707125200. [DOI] [PubMed] [Google Scholar]
- 25.Hornakova T, et al. ALL-associated JAK1 mutations confer hypersensitivity to the antiproliferative effect of type I interferon. Blood. 2010;115(16):3287–3295. doi: 10.1182/blood-2009-09-245498. [DOI] [PubMed] [Google Scholar]
- 26.Khoury JD, et al. Differential expression and clinical significance of tyrosine-phosphorylated STAT3 in ALK+ and ALK− anaplastic large cell lymphoma. Clin Cancer Res. 2003;9(10 Pt 1):3692–3699. [PubMed] [Google Scholar]
- 27.Zenatti PP, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43(10):932–939. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue P, Turkson J. Targeting STAT3 in cancer: How successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson B, Krishnaswami S, van Vollenhoven RF. Tofacitinib versus methotrexate in rheumatoid arthritis. N Engl J Med. 2014;371(12):1164–1165. doi: 10.1056/NEJMc1408607. [DOI] [PubMed] [Google Scholar]
- 30.Vannucchi AM, et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N Engl J Med. 2015;372(5):426–435. doi: 10.1056/NEJMoa1409002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganetsky A. Ruxolitinib: A new treatment option for myelofibrosis. Pharmacotherapy. 2013;33(1):84–92. doi: 10.1002/phar.1165. [DOI] [PubMed] [Google Scholar]
- 32.Kadin ME, et al. Loss of receptors for transforming growth factor beta in human T-cell malignancies. Proc Natl Acad Sci USA. 1994;91(13):6002–6006. doi: 10.1073/pnas.91.13.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Mistro A, et al. A CD30-positive T cell line established from an aggressive anaplastic large cell lymphoma, originally diagnosed as Hodgkin’s disease. Leukemia. 1994;8(7):1214–1219. [PubMed] [Google Scholar]
- 34.Kubonishi I, et al. A Ki-1 (CD30)-positive T (E+, CD4+, Ia+)-cell line, DL-40, established from aggressive large cell lymphoma. Cancer Res. 1990;50(23):7682–7685. [PubMed] [Google Scholar]
- 35.Lechner MG, et al. Breast implant-associated, ALK-negative, T-cell, anaplastic, large-cell lymphoma: Establishment and characterization of a model cell line (TLBR-1) for this newly emerging clinical entity. Cancer. 2011;117(7):1478–1489. doi: 10.1002/cncr.25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lechner MG, et al. Survival signals and targets for therapy in breast implant-associated ALK− anaplastic large cell lymphoma. Clin Cancer Res. 2012;18(17):4549–4559. doi: 10.1158/1078-0432.CCR-12-0101. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]