Abstract
Progesterone is indispensable for differentiation of human endometrial stromal cells (HESCs) into decidual cells, a process that critically controls embryo implantation. We now show an important role for androgen receptor (AR) signaling in this differentiation process. Decreased posttranslational modification of the AR by small ubiquitin-like modifier (SUMO)-1 in decidualizing cells accounted for increased responsiveness to androgen. By combining small interfering RNA technology with genome-wide expression profiling, we found that AR and progesterone receptor (PR) regulate the expression of distinct decidual gene networks. Ingenuity pathway analysis implicated a preponderance of AR-induced genes in cytoskeletal organization and cell motility, whereas analysis of AR-repressed genes suggested involvement in cell cycle regulation. Functionally, AR depletion prevented differentiation-dependent stress fiber formation and promoted motility and proliferation of decidualizing cells. In comparison, PR depletion perturbed the expression of many more genes, underscoring the importance of this nuclear receptor in diverse cellular functions. However, several PR-dependent genes encode for signaling intermediates, and knockdown of PR, but not AR, compromised activation of WNT/β-catenin, TGFβ/SMAD, and signal transducer and activator of transcription (STAT) pathways in decidualizing cells. Thus, the nonredundant function of the AR in decidualizing HESCs, centered on cytoskeletal organization and cell cycle regulation, implies an important role for androgens in modulating fetal-maternal interactions. Moreover, we show that PR regulates HESC differentiation, at least in part, by reprogramming growth factor and cytokine signal transduction.
PROGESTERONE IS A pleiotropic hormone that regulates all aspects of female reproduction, from ovulation and embryo implantation to parturition. The actions of progesterone (P4) on reproductive target tissues are mediated predominantly by its cognate nuclear receptors, P4 receptor (PR)-A and PR-B, members of the superfamily of ligand-dependent transcription factors. In the uterus, the postovulatory rise in P4 levels induces differentiation of the endometrial mucosa in preparation for pregnancy (1). A cardinal event in this remodeling process is the transformation of endometrial stromal fibroblast into secretory, epithelioid-like decidual cells (2, 3, 4). In addition to the morphological changes, decidualization bestows some unique functional properties on human endometrial stromal cells (HESCs), including the ability to modulate local immune cells, to resist environmental stress signals, and to modulate trophoblast invasion (2, 3, 4). Mice deficient in PR fail to mount a decidual response and are sterile (1). However, activation of PR is in itself insufficient to induce decidualization in mice or humans. Initiation of HESC differentiation is strictly dependent upon elevated cAMP levels and sustained activation of the protein kinase A pathway, which in turn sensitize the cells to P4 (5, 6). Once decidualized, the endometrium becomes inextricably dependent upon continuous P4 signaling for homeostasis, and in the absence of pregnancy, falling P4 levels trigger a cascade of events that results in apoptosis, proteolytic breakdown of the superficial endometrium, focal bleeding, and menstrual shedding (7).
HESCs also abundantly express the androgen receptor (AR) (8, 9), yet little is known about the function of this nuclear receptor family member in the decidual process. AR and PR are phylogenetically closely related and share 54 and 80% sequence homology in their ligand- and DNA-binding domains, respectively (10). AR expression, which is confined to the stroma in cycling endometrium, decreases during the secretory phase, although the receptor remains detectable in the decidua of early pregnancy (11, 12). Serum androgen levels fluctuate throughout the menstrual cycle, with levels peaking around ovulation (13, 14). However, tissue androgen levels and conversion of androstenedione to testosterone are higher in secretory than proliferative endometrium (15). Moreover, a rise in circulating androgen levels in the late luteal phase is associated with a conception cycle and levels continue to rise in early pregnancy (16). Interestingly, both lack and excess of circulating androgens in premature ovarian failure and polycystic ovary syndrome, respectively, are associated with increased risk of early fetal loss and late obstetric complication due to impaired placental function, such as preeclampsia (17, 18, 19).
These observations provide compelling but circumstantial evidence that androgens play a role in decidual-trophoblast interactions in pregnancy. We now demonstrate that decidualization of HESCs is associated with increased responsiveness to androgen signaling. Compared with PR, AR controls a much smaller but focused network of genes essential for cytoskeletal organization and cell cycle regulation in decidualizing endometrium.
Materials and Methods
Primary endometrial cell culture
The Local Research and Ethics Committee at Hammersmith Hospitals NHS Trust approved the study, and patient consent was obtained before tissue collection. HESC cultures were established as previously described (2). Cultures were decidualized with 0.5 mm 8-Br-cAMP (Sigma Chemical Co., St. Louis, MO) and medroxyprogesterone acetate (MPA; Sigma), P4 (Sigma), dihydrotestosterone (DHT; Sigma), or bicalutamide (Casodex; AstraZeneca, London, UK), all at 1 μm bar for DHT, which was used at 0.1 μm concentration unless stated otherwise.
Transfections
Primary HESCs were transfected with DNA vectors or small interfering RNA (siRNA) by the calcium phosphate coprecipitation method using the Profection mammalian transfection kit (Promega, Madison, WI), as previously described (2). All transfections were performed in triplicate in 24-well plates and repeated at least three times. The expression plasmids for AR, PR-B, PIAS1, PIAS1(C351S, W372A), and EGFP-SUMO1 have been described (2, 6). The reporter constructs dPRL3000/Luc and PRE2/-32dPRL/Luc were a gift from B. Gellersen (Endokrinologikum Hamburg, Hamburg, Germany) The concentration of reporter constructs and expression vectors was 400 and 100 ng/well, respectively. The control vector pCH110 (50 ng/well), which leads to constitutive β-galactosidase expression, was used to compare transfection efficiency. For gene silencing studies, HESCs were cultured in six-well plates until confluency and transiently transfected with 100 nm of the following siRNA reagents (Dharmacon, Lafayette, CO): siCONTROL nontargeting (NT) siRNA Pool, AR siGENOME SMARTpool siRNA, PR siGENOME SMARTpool siRNA, and PIAS1 siGENOME SMARTpool siRNA.
Western blot analysis and prolactin (PRL) and insulin-like growth factor-binding protein-1 (IGFBP-1) assays
Whole-cell lysates and nuclear protein fractions were obtained as described elsewhere (4, 6). Proteins (30 μg) were separated on a 10% SDS-polyacrylamide gel before electrotransfer onto nitrocellulose membrane (Amersham, Little Chalfont, UK). The following primary antibodies were used: monoclonal AR (Biogenix, San Ramon, CA), mouse monoclonal PR (Novocastra Laboratories, Newcastle-Upon-Tyne, UK), mouse monoclonal β-actin (Abcam, Cambridge, UK), rabbit total and phosphorylated (Ser473) AKT (Cell Signaling, Hitchin, UK), rabbit total and phosphorylated (Thr202/204) ERK1/2 (Cell Signaling), mouse β-catenin (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit phosphorylated (Ser807/811) pRB (Cell Signaling), rabbit phosphorylated MLC (Ser19) (Cell Signaling), rabbit IL1R1 (Abcam), and rabbit signal transducer and activator of transcription 3 (STAT3) and STAT5b (Upstate Biotechnology, Lake Placid, NY). Primary antibodies were diluted to 1:1000 except β-actin, which was used at 1:100,000. Secondary antibodies were diluted at 1:2000 dilution and protein complexes visualized with a chemoluminescent detection kit (Amersham). PRL in the HESC culture media was measured by microparticle enzyme immunoassay (AxSYM system; Abbott Laboratories, North Chicago, IL). IGFBP-1 levels in culture media were determined using an amplified two-step sandwich-type immunoassay (R&D Systems, Minneapolis, MN).
Microarray and real-time quantitative (RTQ)-PCR
Gene expression profiling was performed on four independent primary cultures, established from proliferative-phase biopsies. Total RNA was isolated from cultured HESCs using Stat-60 (Tel-Test, Friendswood, TX). Genomic DNA was removed by deoxyribonuclease treatment, and the quality of the RNA was evaluated using a Bioanalyzer 2100 (Agilent Technologies Inc., Santa Clara, CA). All subsequent steps were carried out at the Finnish DNA-Microarray Centre using the Sentrix Human Illumina 6 V1 Expression BeadChips (Illumina, San Diego, CA), which contains over 47,000 known genes, gene candidates, and splice variants. Three hundred nanograms of each RNA sample, with 260/280 and 28S/18S ratio of greater than 1.8, was used to make double-stranded cDNA and then biotinylated cRNA using the Illumina RNA TotalPrep Amplification Kit (Ambion Inc., Austin, TX). Labeled cRNA was purified and hybridized to the BeadChip at 55 C, for 17 h after the Illumina Whole-Genome Gene Expression Protocol for BeadStation. Hybridized biotinylated cRNA was detected with cyanine3-streptavidin (Amersham). Arrays were scanned with the Illumina BeadArray Reader, which is a confocal-type imaging system with about 0.8-μm resolution and 532-nm laser illumination. The normalization and statistical analyses of the microarrays were performed using the statistical software R package limma (http://www.R-project.org). Genes of coefficient of variation values higher than 0.8 were filtered out from the analysis in preprocessing. The same software was used for single-gene analyses including fold-change calculations. The normalized data were analyzed by pair-wise comparisons to create a list of differentially expressed genes. Because we have shown that HESCs decidualized for 72 h are only modestly sensitive to androgen signaling, differentially expressed genes were defined by a lower boundary of a 99% confidence interval of fold change greater than 1.2 as validated by Student’s t test (P < 0.01). To interpret the biological significance of differentially expressed genes, a gene ontology analysis was conducted using Ingenuity Pathways Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com). RTQ-PCR analysis was performed as previously described (4). All measurements were performed in triplicate. Gene-specific primer pairs were designed using the ABI Primer Express software (supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http:// endo.endojournals.org).
Fluorescence microscopy, motility, and proliferation assays
Immunofluorescence analysis was performed on cells seeded onto glass coverslips, fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton, and stained with tetramethylrhodamine isothiocyanate-labeled phalloidin (Chemicon, Temecula, CA) and counterstained with 4′,6-diamidino-2-phenylindole. The number of cells with actin stress fibers per 100 cells was determined by an independent assessor, blinded to the treatment, in three independent experiments. Cell motility was assessed by time-lapse microscopy using an inverted microscope with a motorized stage. Images were captured every 15 min over a 48-h period using a Hamamatsu C4742-95 CCD camera, and the distance each cell moved was analyzed using Image Pro Plus software (Media Cybernetics, Silver Spring, MD). Triplicate experiments were performed, and at least 15 cells per field of view, chosen randomly, were analyzed. Proliferation was ascertained using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega).
Results and Discussion
Increased androgen responses in decidualizing HESCs
Secretion of PRL and IGFBP-1 in response to cAMP and P4 signaling is the hallmark of decidual transformation of HESCs (2). To test whether androgens modify this differentiation process, primary HESC cultures were treated with 8-Br-cAMP, P4, and DHT, either alone or in combination over a time course lasting 8 d. Treatment with P4 alone was insufficient to trigger expression of either differentiation marker (Fig. 1A). In contrast, 8-Br-cAMP rapidly increased PRL and IGFBP-1 secretion, but expression declined after 4 d of treatment. As expected (2), enhanced and sustained expression of both IGFBP-1 and PRL in long-term cultures required both 8-Br-cAMP and P4 signaling. DHT also modestly enhanced the expression of both marker proteins in 8-Br-cAMP-treated cultures, but in contrast to P4, DHT was insufficient to sustain the decidual response in prolonged culture. Interestingly, DHT markedly enhanced PRL but not IGFBP-1 secretion in cultures treated with 8-Br-cAMP plus P4. This androgen response in decidualizing cultures increased in magnitude over time in a dose-dependent manner (Fig. 1B). RTQ-PCR analysis demonstrated that the changes in PRL transcript levels mirrored those at the protein level (Fig. 1C). In addition, transfection studies with dPRL3000/Luc, a luciferase reporter construct coupled to 3 kb of the decidua-specific PRL promoter region (20), revealed that the pattern of PRL expression in response to cAMP, P4, and DHT corresponded to promoter activation (Fig. 1D).
Fig. 1.
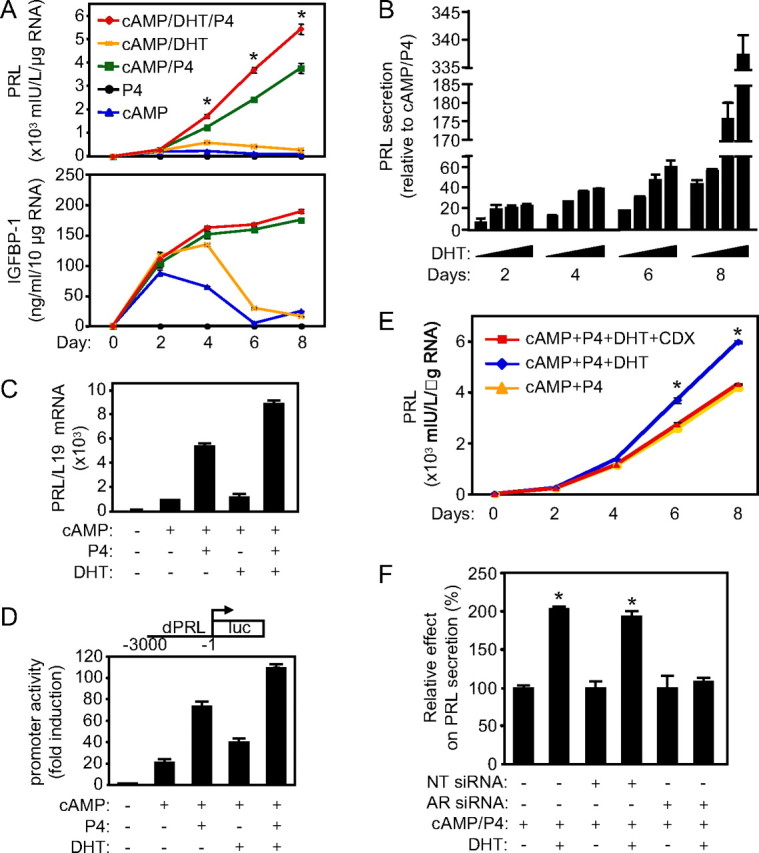
DHT selectively enhances decidual PRL expression in a time-dependent manner. A, Primary HESCs were cultured in the presence of 8-br-cAMP, P4, and DHT as indicated. The medium was collected and cells harvested every 48 h. The data represent the mean PRL (upper panel) and IGFBP-1 (lower panel) concentrations (±sd) in the supernatant, with normalization for RNA content at each time point, of triplicate cultures. Significant differences were found in terms of PRL secretion between cultures treated with cAMP/P4 and cAMP/P4/DHT at 4, 6, and 8 d (*, P < 0.05). B, Primary HESCs were decidualized with 8-Br-cAMP, P4, and increasing concentrations of DHT (0.001, 0.01, 0.1, and 1 μm). The medium was collected and assayed for PRL concentration. The data are expressed as percentage increase over cAMP+P4 alone (mean ±sd). C, RTQ-PCR analysis was carried out for PRL mRNA levels in cultures treated as in A. The results show mean PRL mRNA levels (±sd) normalized to L19 mRNA of three independent cultures. D, HESCs were treated as in A for 48 h followed by transfection with dPRL-3000/Luc. Subsequently, the cells were maintained in the same culture conditions for 24 h. Luciferase and galactosidase assays were performed, and the results represent the mean (±sd) of triplicate measurements of one representative experiment. E, Primary HESCs were cultured in the presence of 8-br-cAMP, P4, DHT, and bicalutamide (Casodex; CDX) as indicated and PRL secretion determined as described above. PRL secretion was significantly different between cAMP/P4/DHT- and cAMP/P4/DHT/CDX-treated cultures (*, P < 0.05). F, HESCs, decidualized with a combination of 8-Br-cAMP, P4, and DHT for 48 h, were either mock-transfected or transfected with NT or AR siRNAs. The treatments were continued for 72 h and the supernatants assayed for PRL. The data represent the relative effect of DHT (percent) on decidual PRL secretion (mean ± sd) of triplicate cultures normalized to total protein content of each well (*, P < 0.01).
To determine whether AR mediates the effect of DHT on PRL expression, primary cultures were first treated with 8-Br-cAMP plus P4 in the presence or absence of DHT and the nonsteroidal pure AR antagonist bicalutamide. This antiandrogen entirely negated the ability of DHT to enhance PRL secretion (Fig. 1E). Next, we transfected primary cultures, pretreated for 48 h with 8-Br-cAMP and P4 in the presence or absence of DHT, with either NT siRNA or siRNA targeting AR. Notably, AR depletion was very efficient (see Fig. 3A) and abolished the ability of DHT to enhance PRL secretion in cells differentiated with 8-Br-cAMP and P4 (Fig. 1F). Together, the results unequivocally demonstrate that androgen actions in decidualizing HESCs are dependent upon AR activation.
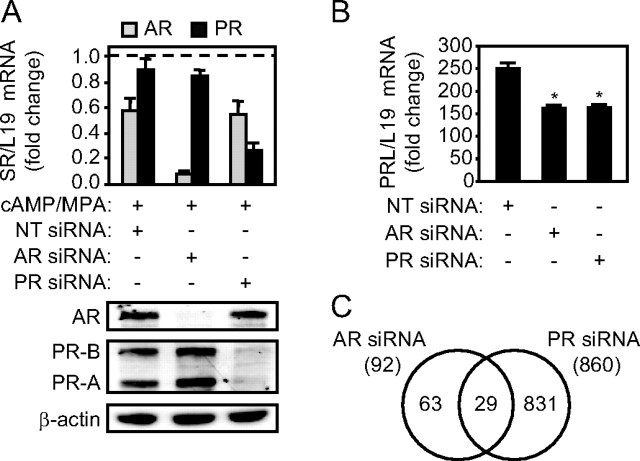
Fig. 3.
AR and PR knockdown perturbs the expression of distinct gene sets in decidualizing HESCs. A, Validation of AR and PR silencing. The upper panel shows RTQ-PCR analysis of AR and PR transcript levels in cells transfected with NT, AR, or PR siRNAs before treatment with 8-Br-cAMP and MPA for 72 h. AR and PR mRNA levels were normalized to that of L19 mRNA, and the results are the mean (±sem) of four separate cultures measured in triplicate. The results are fold change relative to transcript levels in undifferentiated cells transfected with NT siRNA (dotted line). The lower panel shows Western blot analysis of AR and PR expression in protein lysates from parallel cultures. β-Actin served as a loading control. B, RTQ-PCR analysis of PRL transcript levels, normalized to L19 mRNA, in cells first transfected with NT, AR, or PR siRNA followed by differentiation with 8-Br-cAMP and MPA for 72 h. The results are fold induction of PRL mRNA expression relative to the levels in undifferentiated cells transfected with NT siRNA. *, P < 0.01. C, Venn diagram showing the number of differentially expressed genes in decidualizing cells upon AR or PR knockdown.
Decidualization decreases small ubiquitin-like modifier (SUMO)-1 modification of the activated AR
We postulated that enhanced AR expression could account for the gradual increase in androgen sensitivity upon HESC differentiation. However, treatment of primary cultures with 8-Br-cAMP alone resulted in rapid but transient reduction in AR levels (Fig. 2A). Conversely, P4 had little effect on AR levels in short-term cultures but down-regulated receptor levels after 6–8 d of treatment. Combined 8-Br-cAMP plus P4 treatment resulted in a rapid and sustained decrease in cellular AR levels. DHT strongly increased AR levels in undifferentiated cells (Fig. 2A), as described in other cell systems (21), but only partially antagonized the down-regulation of the receptor in decidualizing cells. Thus, as reported for P4 (2), increased sensitivity to androgens in HESCs is paradoxically associated with decreasing receptor levels.
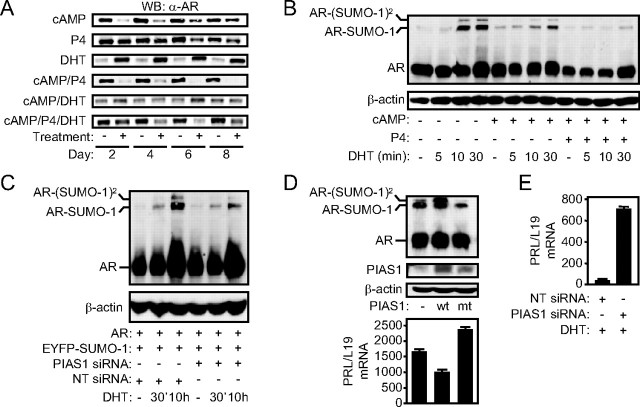
Fig. 2.
PIAS1 attenuates AR sumoylation and regulates endogenous androgen responses in differentiating HESCs. A, DHT antagonizes AR down-regulation in decidualizing HESCs. Primary cultures were treated with a combination of 8-Br-cAMP, P4, DHT, and MPA, as indicated. Whole-cell lysates, extracted every 48 h for 8 d, were immunoprobed for AR expression. B, Primary cultures transfected with AR and EGFP-SUMO-1 were treated with vehicle or decidualized with 8-Br-cAMP with or without P4 for 48 h and then pulsed with DHT, as indicated. Total protein lysates were probed for AR expression. Single and double EGFP-SUMO-1-modified AR species are indicated. β-Actin served as a loading control. C, HESCs were transfected with AR, EGFP-Sumo-1, and either NT siRNA or siRNA targeting PIAS1. Cells were left untreated for 2 d and then treated with DHT as indicated. D, Parallel cultures were treated with 8-Br-cAMP and DHT for 48 h and then subsequently transfected with pSG5 or pSG5-PIAS1 (wt or mt) and were analyzed for PRL mRNA expression by RTQ-PCR (lower panel) and immunoprobed for AR and PIAS1 (upper panel), as indicated. E, Undifferentiated HESCs were transfected with NT or PIAS1 siRNAs and treated 2 d later with vehicle or DHT for 48 h. The results show mean (±sd) PRL transcript levels normalized to L19 mRNA of three independent cultures.
In the case of P4, increased responsiveness has been linked to global changes in cellular sumoylation upon HESC differentiation (6). More specifically, decidualization is characterized by a gradual decline in the expression of the E3 SUMO ligase protein inhibitor of activated STAT1 (PIAS1), resulting in attenuated ligand-dependent sumoylation of PR, increased transcriptional activity, and enhanced receptor turnover. PIAS1 also serves as an E3 ligase for AR (22). This prompted an analysis of AR sumoylation in undifferentiated and decidualizing HESCs. Untreated cultures and cells first decidualized with 8-Br-cAMP or 8-Br-cAMP plus P4 for 48 h were transfected with expression vectors encoding AR and enhanced green fluorescent protein (EGFP)-tagged SUMO-1 and pulsed 24 h later with DHT. Immunoblotting of cell lysates with an anti-AR antibody demonstrated the presence of two slower migrating forms of AR in undifferentiated cells, first apparent after 10 min DHT stimulation, which represent SUMO-1 modification of the two known acceptor sites (K386 and K520) in AR (Fig. 2B) (22). Compared with undifferentiated HESCs, DHT-dependent sumoylation of AR was attenuated in cells treated with 8-br-cAMP and much more so in cultures decidualized with 8-Br-cAMP plus P4. Next, we confirmed in COS-1 cells that increasing expression of PIAS1 enhances ligand-dependent AR sumoylation (supplemental Fig. 1) and, conversely, that PIAS1 knockdown reduces AR SUMO-1 modification in HESCs treated with DHT (Fig. 2C). To explore whether PIAS1 regulates endogenous androgen responses in HESCs, we first overexpressed wild-type PIAS1 or a PIAS1 mutant (C351S, W372A) devoid of E3-ligase activity in cells decidualized with 8-Br-cAMP and DHT for 48 h. Parallel cultures were harvested after 24 h for protein and mRNA analyses. As shown in Fig. 2D, wild-type but not mutant PIAS1 enhanced SUMO-1 modification of AR, which corresponded to a 50% decrease in PRL mRNA expression. Notably, expression of the E3-deficient mutant antagonized endogenous PIAS1, resulting in decreased AR sumoylation and higher PRL mRNA levels. Furthermore, PIAS1 knockdown in undifferentiated HESCs was sufficient to induce PRL expression in response to DHT without the need of additional decidualizing stimuli (Fig. 2E). Together, the results demonstrate that down-regulation of PIAS1 upon decidualization sensitizes HESCs not only to P4 (6) but also to androgen signaling.
Identification of decidual AR and PR target genes
MPA, a 17-OH P4 derivative with known androgenic actions (23), is widely used in combination with 8-Br-cAMP to differentiate HESCs in vitro (2, 20). We confirmed that MPA, like DHT but not P4, enhances cellular AR levels in HESCs, induces its nuclear accumulation, and transactivates the receptor in a reporter assay (supplemental Fig. 2). We exploited the progestogenic and androgenic properties of MPA to search for specific AR- and PR-dependent genes in decidualizing HESCs. Four separate primary cultures were first transfected with either NT siRNA oligos or a siRNA pool targeting AR or PR and then treated with 8-Br-cAMP plus MPA for 72 h. Parallel cultures were harvested for mRNA and protein analysis. As shown in Fig. 3A, the siRNA knockdown approach for AR and PR was effective and selective at both the mRNA and protein level. Furthermore, knockdown of either receptor was equally efficient in attenuating PRL mRNA expression in differentiating HESCs (Fig. 3B). Total RNA was then processed for genome-wide expression profiling, and the data were interrogated using parametric statistical testing. Figure 3C represents the Venn diagram of regulated genes identified by the following pair-wise comparisons: AR siRNA-transfected cells (AR) vs. NT siRNA-transfected cells and PR siRNA-transfected cells (PR) vs. NT siRNA-transfected cells. AR knockdown affected the expression of a relatively small pool of genes. Of the 92 transcripts deregulated upon AR depletion, the expression of 42.4 and 57.6% of transcripts were up- and down-regulated, respectively (Tables 1 and 2). In contrast, PR knockdown perturbed the expression of 860 genes, 55.6% of which were up-regulated and 44.4% down-regulated. Tables 3 and 4 list the 50 most induced and repressed PR-dependent genes, respectively. We identified only 29 genes under control of both nuclear receptors in decidualizing cells, although 10 were regulated in an opposing manner (Table 5). Thus, the data confirm the major role of PR in regulating decidual gene expression and define, for the first time, a smaller but distinct set of genes under AR control.
TABLE 1.
Genes down-regulated upon AR knockdown
| Gene symbol | Gene name | Fold change |
|---|---|---|
| AR | AR (DHT receptor) | −1.81 |
| CUTL1 (37 ) | Cut-like 1 CCAAT displacement protein (Drosophila) | −1.49 |
| WASPIP (38 39 ) | Wiskott-Aldrich syndrome protein interacting protein | −1.48 |
| DUSP3 | Dual-specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) | −1.46 |
| LOC255065 | LOC255065 | −1.45 |
| IL1R1 (32 40 ) | IL-1 receptor type I | −1.43 |
| PFTK1 (41 ) | PFTAIRE protein kinase 1 | −1.42 |
| PLA2G4F | Phospholipase A2, group IVF | −1.37 |
| FAM135A | Family with sequence similarity 135, member A | −1.35 |
| ALEX2 (42 ) | Armadillo repeat protein ALEX2 | −1.35 |
| SDCCAG1 | Serologically defined colon cancer antigen 1 | −1.35 |
| GNPDA1 | Glucosamine-6-phosphate deaminase 1 | −1.33 |
| LPGAT1 | Lysophosphatidylglycerol acyltransferase 1 | −1.33 |
| WDSUB1 | WD repeat, sterile α-motif and U-box domain containing 1 | −1.32 |
| CAST (43 ) | Calpastatin | −1.32 |
| ZBTB1 | Zinc finger and BTB domain containing 10 | −1.28 |
| LOC286470 | LOC286470 | −1.27 |
| PLS3 (44 ) | Plastin 3 (T isoform) | −1.27 |
| ARHGEF7 (45 ) | ρ-Guanine nucleotide exchange factor (GEF) 7 | −1.27 |
| ACTR3 (46 ) | ARP3 actin-related protein 3 homolog (yeast) | −1.27 |
| AMBRA1 (47 ) | Autophagy/beclin-1 | −1.27 |
| PB1 | Polybromo 1 | −1.26 |
| FLJ00060 | Hypothetical gene FLJ00060 | −1.25 |
| KLHDC5 | Kelch domain containing 5 | −1.25 |
| NR2F2 | Nuclear receptor subfamily 2 group F member 2 | −1.25 |
| WDR40A | WD repeat domain 40A | −1.23 |
| GDI1 (48 ) | GDP dissociation inhibitor 1 | −1.22 |
| RPC8 | RNA polymerase III subunit RPC8 | −1.22 |
| C5orf24 | Chromosome 5 open reading frame 24 | −1.22 |
| WBSCR16 | Williams-Beuren syndrome chromosome region 16 | −1.22 |
| AHI1 | Abelson helper integration site | −1.21 |
| FBXW1B | F-box and WD-40 domain protein 1B | −1.21 |
| POMT2 (49 ) | Protein-O-mannosyltransferase 2 | −1.21 |
| SMTN (50 ) | Smoothelin | −1.21 |
| MADHIP | Mothers against decapentaplegic homolog (Drosophila) interacting protein | −1.20 |
| C19orf26 | Chromosome 19 open reading frame 26 | −1.20 |
| DYRK1A (51 52 ) | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | −1.20 |
Gene symbols in bold indicate genes known to be regulated in endometrium. Genes implicated in cytoskeletal organization are referenced.
TABLE 2.
Genes up-regulated upon AR knockdown
| Gene symbol | Gene name | Fold change |
|---|---|---|
| DIPA | Hepatitis δ-antigen-interacting protein A | 1.45 |
| CADPS2 | Ca2+-dependent activator protein for secretion 2 | 1.41 |
| UGP2 | UDP-glucose pyrophosphorylase 2 | 1.39 |
| MCM4 | MCM4 minichromosome maintenance deficient 4 (S. cerevisiae) | 1.37 |
| OIP5 | Opa-interacting protein 5 | 1.33 |
| CDT1 | DNA replication factor | 1.33 |
| POLR1D | Polymerase (RNA) I polypeptide D 16 kDa | 1.30 |
| CCDC99 | Coiled-coil domain containing 99 | 1.26 |
| FDXR | Ferredoxin reductase | 1.26 |
| TMEM160 | Transmembrane protein 160 | 1.25 |
| TBC1D13 | TBC1 domain family member 13 | 1.25 |
| C3orf26 | Chromosome 3 open reading frame 26 | 1.25 |
| NDFIP2 | Nedd4 family interacting protein 2 | 1.25 |
| ANKRD36 | Ankyrin repeat domain 36 | 1.25 |
| WDR51A | WD repeat domain 51A | 1.24 |
| IQCC | IQ motif containing C | 1.23 |
| MEIS2 | Meis1 myeloid ecotropic viral integration site 1 homolog 2 (mouse) | 1.22 |
| LOC121642 | Similar to prostate cancer antigen-1 | 1.22 |
| ACAS2 | Acetyl-coenzyme A synthetase 2 (AMP forming)-like | 1.22 |
| RGS10 | Regulator of G-protein signaling 10 | 1.21 |
| CHTF18 | CTF18 chromosome transmission fidelity factor 18 homolog (S. cerevisiae) | 1.21 |
| PDE7B | Phosphodiesterase 7B | 1.20 |
| RPL10A | Ribosomal protein L10a | 1.20 |
| HBLD1 | HESB like domain containing 1 | 1.20 |
| ECE2 | Endothelin converting enzyme 2 | 1.20 |
| PRO0386 | Hypothetical protein PRO0386 | 1.20 |
Gene symbols in bold indicate genes known to be regulated in endometrium.
TABLE 3.
Top 50 down-regulated genes upon PR knockdown
| Gene symbol | Gene name | Fold change |
|---|---|---|
| APCDD1 | Adenomatosis polyposis coli down-regulated 1 | −5.37 |
| CNR1 | Cannabinoid receptor 1 | −4.21 |
| LOC347348 | Similar to heat-shock 27-kDa protein (HSP27) | −3.34 |
| CHST7 | Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 | −3.19 |
| HSPB1 | Heat-shock 27-kDa protein 1 | −3.19 |
| RASD1 | RAS dexamethasone-induced | −3.12 |
| FKBP5 | FK506 binding protein 5 | −3.10 |
| IGF1 | IGF-I (somatomedin C) | −3.04 |
| RORB | RAR-related orphan receptor B | −3.01 |
| C13orf33 | Chromosome 13 open reading frame 33 | −3.00 |
| ACPL2 | Acid phosphatase-like 2 | −2.71 |
| SORBS1 | Sorbin and SH3 domain containing 1 | −2.59 |
| PCDH19 | Protocadherin 19 | −2.59 |
| HSD11B1 | Hydroxysteroid (11-β) dehydrogenase 1 | −2.56 |
| AOX1 | Aldehyde oxidase 1 | −2.51 |
| PDLIM1 | PDZ and LIM domain 1 (elfin) | −2.51 |
| RASL10B | RAS-like family 10 member B | −2.5 |
| BTBD3 | BTB (POZ) domain containing 3 | −2.46 |
| MDM1 | Nuclear protein double minute 1 | −2.46 |
| ZNF145 | Zinc finger protein 145 (Kruppel-like expressed in promyelocytic leukemia) | −2.41 |
| ADAMTS1 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif 1 | −2.41 |
| IMPA2 | Inositol(myo)-1(or 4)-monophosphatase 2 | −2.38 |
| NRXN3 | Neurexin 3 | −2.32 |
| HSPB6 | Heat-shock protein, α-crystallin-related, B6 | −2.32 |
| METTL7A | Methyltransferase like 7A | −2.28 |
| OSR2 | Odd-skipped-related 2A protein | −2.27 |
| SIPA1L2 | Signal-induced proliferation-associated 1 like 2 | −2.25 |
| DSIPI | δ-Sleep inducing peptide immunoreactor | −2.24 |
| PRPS2 | Phosphoribosyl pyrophosphate synthetase 2 | −2.21 |
| GATA6 | GATA binding protein 6 | −2.20 |
| PPAP2B | Phosphatidic acid phosphatase type 2B | −2.20 |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit polypeptide 1 (p85 α) | −2.16 |
| SERPINE1 | Serine (or cysteine) proteinase inhibitor clade E | −2.15 |
| XYLT1 | Xylotransferase 1 | −2.14 |
| ATAD2 | ATPase family, AAA domain containing 2 | −2.13 |
| TIPARP | TCDD-inducible poly(ADP-ribose) polymerase | −2.13 |
| ABLIM3 | Actin-binding LIM protein family member 3 | −2.12 |
| CARD9 | Caspase recruitment domain family member 9 | −2.12 |
| LARGE | Like-glycosyltransferas | −2.09 |
| NPR1 | Natriuretic peptide receptor A/guanylate cyclase A | −2.09 |
| PGR | Progesterone receptor | −2.08 |
| DKK1 | Dickkopf homolog 1 (Xenopus laevis) | −2.05 |
| RAB40A | RAB40A member RAS oncogene family | −2.03 |
| LMCD1 | LIM and cysteine-rich domains 1 | −2.02 |
| RACGAP1 | Rac GTPase activating protein 1 | −2.00 |
| SLC27A3 | Solute carrier family 27 (fatty acid transporter) member 3 | −1.96 |
| FLJ11539 | Hypothetical protein FLJ11539 | −1.95 |
| SLC7A8 | Solute carrier family 7 (cationic amino acid transporter y+ system) member 8 | −1.95 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | −1.94 |
| TNFRSF1B | TNF receptor superfamily member 1B | −1.92 |
Gene symbols in bold indicate genes known to be regulated in endometrium.
TABLE 4.
Top 50 up-regulated genes upon PR knockdown
| Gene symbol | Gene name | Fold change |
|---|---|---|
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 6.69 |
| MMP10 | Matrix metalloproteinase 10 (stromelysin 2) | 5.29 |
| STC1 | Stanniocalcin 1 | 5.20 |
| KIAA1199 | KIAA1199 protein | 5.01 |
| FJX1 | Four jointed box 1 (Drosophila) | 4.81 |
| TNFRSF11B | TNF receptor superfamily member 11b (osteoprotegerin) | 3.99 |
| ABCG1 | ATP-binding cassette sub-family G (WHITE) member 1 | 3.76 |
| MMP11 | Matrix metalloproteinase 11 (stromelysin 3) | 3.56 |
| IER3 | Immediate-early response 3 | 3.35 |
| SLC16A6 | Solute carrier family 16 (monocarboxylic acid transporters) member 6 | 3.14 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15 inhibits CDK4) | 2.86 |
| GDF15 | Growth differentiation factor 15 | 2.76 |
| IL13RA2 | IL-13 receptor α2 | 2.75 |
| ANGPTL2 | Angiopoietin-like 2 | 2.74 |
| THBS2 | Thrombospondin 2 | 2.72 |
| NR4A2 | Nuclear receptor subfamily 4 group A member 2 | 2.63 |
| C10orf10 | Chromosome 10 open reading frame 10 | 2.63 |
| SOX4 | SRY (sex determining region Y)-box 4 | 2.56 |
| FAM43A | Family with sequence similarity 43, member A | 2.53 |
| TEK | TEK tyrosine kinase endothelial | 2.50 |
| DPYSL4 | Dihydropyrimidinase-like 4 | 2.48 |
| NNAT | Neuronatin | 2.45 |
| MEX3A | Mex-3 homolog A | 2.45 |
| PDE4B | Phosphodiesterase 4B cAMP-specific | 2.41 |
| FOXP1 | Forkhead box P1 | 2.32 |
| AMSH-LP | Associated molecule with the SH3 domain of STAM (AMSH) like protein | 2.32 |
| FRMD4 | FERM domain containing 4 | 2.29 |
| TWIST1 | Twist homolog 1) | 2.25 |
| ARNT2 | Aryl-hydrocarbon receptor nuclear translocator 2 | 2.24 |
| CNIH3 | Cornichon homolg 3 | 2.23 |
| PPFIBP2 | PTPRF interacting protein binding protein 2 (liprin β 2) | 2.22 |
| LOH11CR2A | Loss of heterozygosity11 chromosomal region 2 | 2.22 |
| EHZF | Early hematopoietic zinc finger | 2.19 |
| DIO2 | Deiodinase iodothyronine type II | 2.19 |
| ABCA6 | ATP-binding cassette sub-family A (ABC1) member 6 | 2.18 |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 2.18 |
| TNFRSF21 | TNF receptor member 21 | 2.16 |
| FLJ22536 | Hypothetical locus LOC401237 | 2.16 |
| FHOD3 | Formin homology 2 domain containing 3 | 2.15 |
| WNT2 | Wingless-type MMTV integration site family member 2 | 2.15 |
| F13A1 | Coagulation factor XIII A1 polypeptide | 2.14 |
| KIAA1370 | Hypothetical protein LOC56204 | 2.14 |
| CSRP2 | Cysteine- and glycine-rich protein 2 | 2.07 |
| CHAC1 | ChaC, cation transport regulator homolog 1 | 2.06 |
| CREB5 | cAMP responsive element binding protein 5 | 2.06 |
| PBEF1 | Pre-B-cell colony enhancing factor 1 | 2.04 |
| FGF9 | Fibroblast growth factor 9 | 2.04 |
| MGC13057 | Hypothetical protein MGC13057 | 2.04 |
| FAP | Fibroblast activation protein α | 2.02 |
| LOC221091 | Hypothetical protein LOC221091 | 2.01 |
Gene symbols in bold indicate genes known to be regulated in endometrium.
TABLE 5.
Genes regulated by AR and PR
| Gene symbol | Gene name | AR siRNA fold change | PR siRNA fold change |
|---|---|---|---|
| INHBA | Inhibin β A | −1.56 | 2.38 |
| KCNK3 | Potassium channel subfamily K member 3 | −1.55 | −1.36 |
| FADS2 | Fatty acid desaturase 2 | −1.45 | −1.55 |
| LMOD1 (53 ) | Leiomodin 1 (smooth muscle) | −1.43 | −1.67 |
| WNT4 | Wingless-type MMTV integration site family member 4 | −1.39 | −1.60 |
| ELOVL4 | Elongation of very long chain fatty acids (FEN1/Elo2 SUR4/Elo3 yeast)-like 4 | −1.36 | 1.40 |
| GPR125 (54 ) | G protein-coupled receptor 125 | −1.30 | 1.37 |
| KIAA1377 | KIAA1377 protein | −1.27 | −1.28 |
| LRRK1 (55 ) | Leucine-rich repeat kinase 1 | −1.24 | −1.35 |
| LRCH2 | Leucine-rich repeats and calponin homology (CH) domain containing 2 | −1.23 | 1.49 |
| CTSO (56 ) | Cathepsin O | −1.21 | −1.23 |
| CKAP4 (57 ) | Cytoskeleton-associated protein 4 | −1.21 | −1.22 |
| TEAD3 (58 59 ) | TEA domain family member 3 | −1.20 | −1.26 |
| LOC401627 | Similar to hypothetical protein FLJ33610 | −1.20 | −1.21 |
| TCEAL7 | Transcription elongation factor A (SII)-like 7 | −1.20 | 1.59 |
| MRAS (60 ) | Muscle RAS oncogene homolog | −1.20 | 1.34 |
| MCM2 | MCM2 minichromosome maintenance deficient 2 mitotin (S. cerevisiae) | 1.39 | −1.25 |
| PCDH7 | BH-protocadherin (brain-heart) | 1.38 | 1.74 |
| PCDH7 | BH-protocadherin (brain-heart) | 1.28 | 1.63 |
| HNRPH2 | Heterogeneous nuclear ribonucleoprotein H2 (H’) | 1.27 | 1.23 |
| WEE1 | WEE1 homolog (S. pombe) | 1.26 | 1.32 |
| IMPDH2 | IMP (inosine monophosphate) dehydrogenase 2 | 1.25 | −1.24 |
| ACSL1 | Acyl-CoA synthetase long-chain family member 1 | 1.23 | −1.43 |
| DPP4 | Dipeptidylpeptidase 4 (CD26 adenosine deaminase complexing protein 2) | 1.23 | 1.51 |
| XTP3TPA | XTP3-transactivated protein A | 1.23 | 1.21 |
| XAB1 | XPA binding protein 1 | 1.22 | 1.43 |
| PAK1 | PAK1 interacting protein 1 | 1.22 | 1.35 |
| BM88 | BM88 antigen | 1.20 | 1.28 |
| NAP1L1 | Nucleosome assembly protein 1-like 1 | 1.20 | −1.29 |
Gene symbols in bold indicate genes known to be regulated in endometrium. Genes implicated in cytoskeletal organization are referenced.
Because all cultures were decidualized before array analysis, we cross-referenced our genes lists with the Endometrium Database Resource (http://endometrium.bcm.tmc. edu/edr/) and annotated the tables to indicate genes already reported to be regulated upon endometrial differentiation. RTQ-PCR validation of genes under the putative control of AR (IL1R1, DUSP3, and OIP5), PR (MMP10, TWIST1, and RASD1), or both AR and PR (KCNK3, PCDH7, and WNT4) yielded no false-positive results (Fig. 4). Interestingly, not all identified target genes were dependent upon ligand activation of the receptor. For instance, DUSP3 (dual-specificity phosphatase 3) mRNA levels remained unchanged upon HESC differentiation, yet transfection of AR siRNA resulted in a 3-fold decrease in the transcript levels of this negative regulator of the MAPK pathway. Similarly, MMP10 (matrix metalloproteinase 10) expression was strongly repressed in differentiating HESCs, and PR knockdown not only reversed this repression but also elicited a 4-fold increase in MMP10 mRNA levels when compared with undifferentiated cells, suggesting that even the unliganded PR plays a role in repressing MMP expression in human endometrium.
Fig. 4.
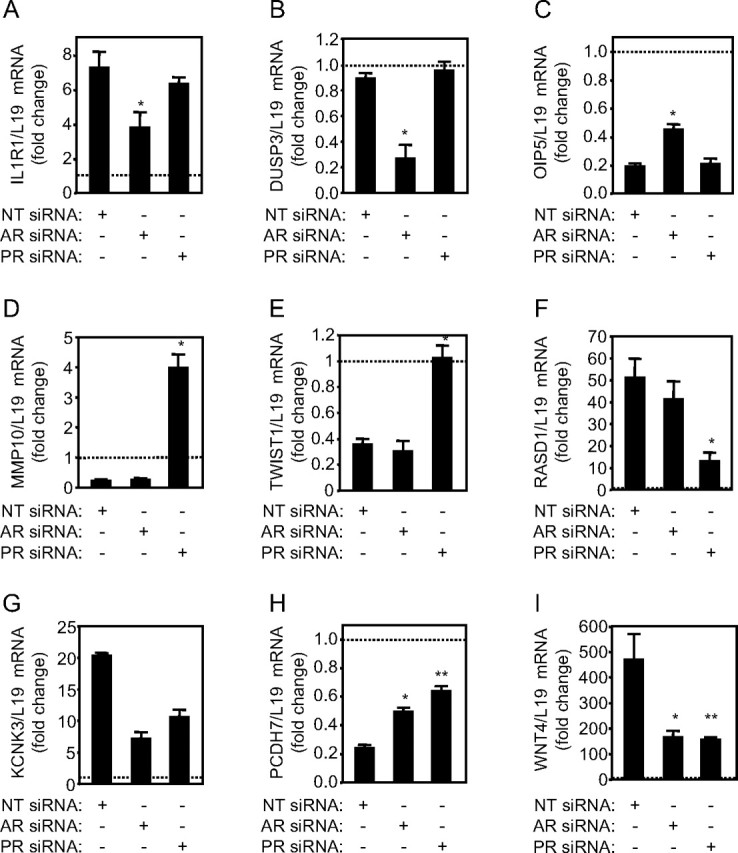
Validation of putative AR- and PR-dependent genes. For microarray validation, three separate cultures were first transfected with NT, AR, or PR siRNA and then treated with 8-Br-cAMP and MPA for 72 h, and mRNA levels of the indicated putative target genes were measured in triplicate for each sample by RTQ-PCR. The data normalized to L19 are expressed as fold change (±sem) relative to expression levels in undifferentiated HESCs transfected with NT siRNA (dotted lines). *, P < 0.05; **, P < 0.001.
PR is indispensable for activation of secondary signaling pathways upon decidualization
Microarray analyses have been extensively used to examine endometrial responses to P4 in humans and various animal models (24, 25, 26). Our gene profiling complements these studies and confirms that PR controls the expression of a network of at least 860 genes in decidualizing HESCs. Ingenuity Pathway Analysis clustered PR-dependent genes into 28 different functional molecular and cellular categories (supplemental Fig. 3), and many can be functionally linked to known P4 actions in the endometrium. For instance, P4 critically ensures tissue integrity of the decidualizing endometrium before menstruation and during pregnancy (3, 7), and not unexpectedly, several genes repressed in a PR-dependent manner encode for matrix metalloproteinases (MMP2, -8, -10, -11, and -27), death receptors of the tumor necrosis factor receptor superfamily (TNFRSF10B, -10D, -11B, -19, and -21), apoptosis mediators (e.g. MCL1, NR4A2, BCL2L10, TRAF4, TP53INP, MOAP1, SEMA3F, DAD1, CIAPN1, DEDD, and MAP3K5), and oxidative stress defenses and DNA repair (e.g. TXNRD1, HNOX1, PPP1R15A, and XRCC5).
Although insufficient to trigger HESC differentiation, P4 is essential for maintaining the decidual phenotype both in vivo and in vitro. Compelling evidence has emerged to indicate that sustained expression of the decidual phenotype is also dependent on autocrine or paracrine signals, resulting in activation of various secondary signaling pathways (5, 27, 28). Strikingly, a significant number of PR-dependent genes encode for ligands, membrane-bound receptors, and intermediates in various signal transduction pathways (supplemental Fig. 4). This prompted us to examine the expression and/or activation status of critical signal intermediates in the STAT, MAPK (ERK1/2), PI3K, TGFβ/SMAD, and WNT/β-catenin pathways in undifferentiated HESCs and decidualized cells transfected with AR or PR siRNAs. In agreement with the gene profile, PR knockdown selectively abolished the induction of STAT3 and STAT5b upon HESC differentiation (Fig. 5). Total ERK1/2, AKT, SMAD2, and β-catenin levels remained unchanged upon differentiation of HESCs. However, decidualization was accompanied by a down-regulation in activated (phosphorylated) AKT levels and a reciprocal increase in ERK1/2 phosphorylation, yet the activation status of either pathway was unaffected by AR or PR depletion. In contrast, PR knockdown eliminated nuclear accumulation of activated SMAD2 and β-catenin in differentiating HESCs. The data imply that a substantial proportion of PR-dependent decidual genes are regulated indirectly, via autocrine or paracrine activation of the WNT/β-catenin, TGFβ/SMAD, and STAT pathways.
Fig. 5.
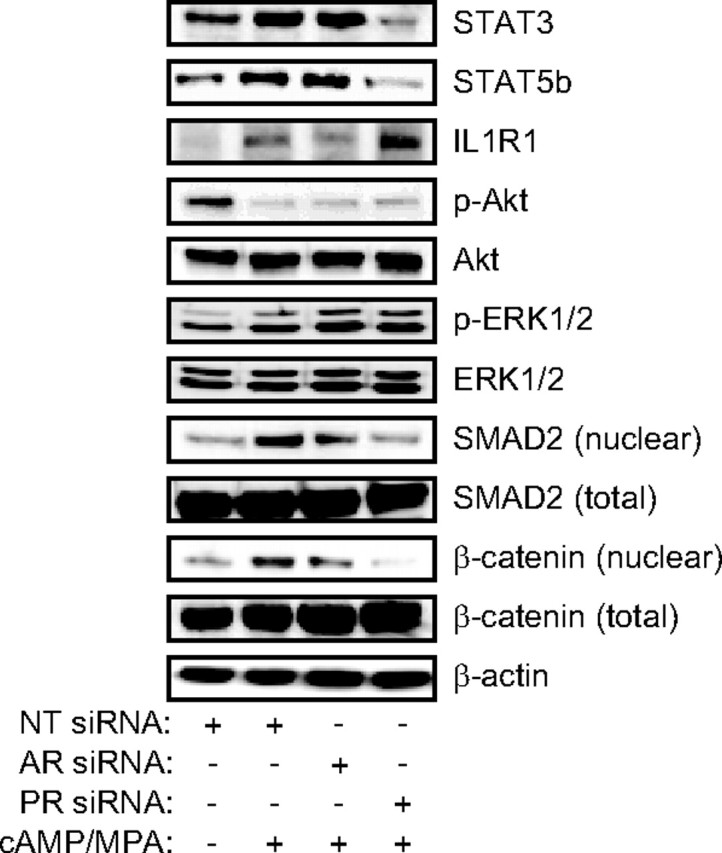
PR regulates STAT, TGFβ/SMAD, and WNT/β-catenin signaling in decidualizing cells. Whole-cell lysates or nuclear protein fractions from HESCs, transfected first with NT, AR, or PR siRNA and then treated with 8-Br-cAMP and MPA for 72 h, were immunoprobed for various signal intermediates, as indicated. β-Actin served as a loading control.
AR regulates cytoskeletal organization and cell cycle inhibition
Ingenuity pathway analysis complemented by manual mining of available literature implicated 40% (21 of 53) of genes down-regulated upon AR silencing in the regulation of cell morphology, cytoskeletal organization, and cell motility (Tables 1 and 5). This prompted us to examine more closely the differentiation-associated changes in the actin cytoskeleton by phalloidin staining of filamentous actin (F-actin). As shown in Fig. 6A, decidualization is characterized by a dramatic increase in F-actin polymerization and stress fiber formation. However, the proportion of cells that express elongated stress fibers was reduced by approximately 50% upon AR knockdown, whereas PR silencing had no apparent effect (Fig. 6A and supplemental Fig. 5). Time-lapse microscopy demonstrated that decidualization is also associated with a dramatic decrease in basal cell motility, which was partially reversed upon AR knockdown (Fig. 6B). Actin-myosin interactions are essential for cell motility and promoted by phosphorylation of the regulatory light chain of myosin 2 (MLC2) (29). In agreement with the motility studies, AR knockdown was sufficient to reverse loss of MLC2 phosphorylation upon decidualization of HESCs (Fig. 6C).
Fig. 6.
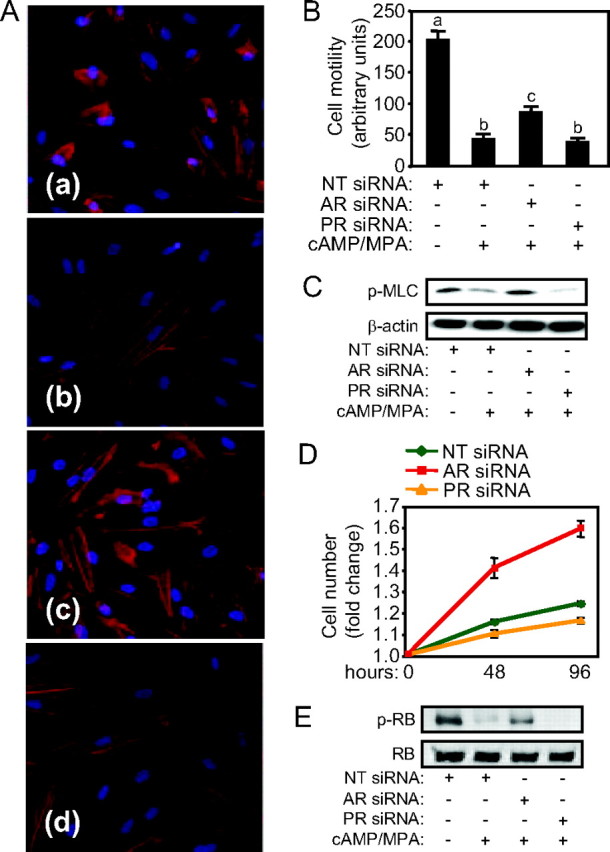
AR controls cytoskeletal organization, cell motility, and proliferation in differentiating HESCs. A, Phalloidin staining of F-actin in undifferentiated HESCs transfected with NT siRNA (a) and cells decidualized with cAMP and MPA for 72 h after transfection with NT siRNA (b), AR siRNA (c), or PR siRNA (d). B, Motility of HESCs transfected and treated as in A was analyzed by time-lapse microscopy, quantified, and expressed in arbitrary units. The results are the mean (±sd) of triplicate analyses. Different letters above the error bars indicate that those groups are significantly different from each other at P < 0.01. C, Protein lysates obtained from parallel cultures were immunoprobed for phosphorylated MLC2. β-Actin served as a loading control. D, Primary cultures were first transfected in six-well plates with NT, AR, or PR siRNAs, replated in 96-well plates, and treated with 8-Br-cAMP and MPA, and cell viability was measured at the indicated time points. The results show the relative fold change in cell number, and the data are the mean (±sd) of triplicate measurements. E, Protein lysates from HESCs transfected with NT, AR, or PR siRNA, then treated with 8-Br-cAMP and MPA for 72 h, were subjected to Western blot analysis for total and phosphorylated RB (p-RB) expression.
In addition to cell motility, the actin cytoskeleton is involved in many other biological functions, including endo- and exocytosis, cytokinesis, and signal transduction (30, 31), underscoring the importance of AR in regulating decidual cell function. Importantly, induction of the IL-1 receptor (IL1R1) in decidualizing cells is under AR control (Fig. 4). Embryonic signals, and in particular IL-1β, have been shown to activate focal adhesion kinase (FAK) and to further promote cytoskeletal reorganization in decidual cells (32). Together, these observations suggest AR plays a major role in coordinating decidual-trophoblast interactions during early pregnancy. This conjecture is further supported by the observation that inactivation of decidual RhoA, a Rho GTPase family member essential for cytoskeletal organization, blocks outgrowth but not attachment of blastocysts in a coculture model (33).
In silico analysis further revealed that several genes up-regulated in decidualizing cells upon AR depletion, and thus normally repressed in an AR-dependent manner, are involved in various aspects of cell cycle regulation (e.g. NAP1L1, WEE1, BM88, XTP3TA, and IMPDH2) including DNA replication licensing (e.g. CDT1, MCM4, and MCM2) and chromatid separation (e.g. CHTF18, DIPA) (Tables 2 and 5). This expression profile points toward a role for AR in safeguarding the genetic stability of the endometrium during rapid cyclic remodeling. Functionally, AR knockdown enhanced proliferation of HESCs decidualized with 8-Br-cAMP and MPA (Fig. 6D). In contrast, proliferation was modestly but consistently reduced upon PR knockdown. Inactivation of retinoblastoma protein (RB) by hyperphosphorylation enables the expression of E2F-target genes essential for coordinating entry into S phase of the cell cycle (34). As shown in Fig. 6E, differentiation of HESCs is strongly associated with loss of RB phosphorylation but much less so upon AR knockdown.
In summary, we have shown that HESCs become increasingly responsive to androgen signaling upon differentiation. This increased sensitivity to sex steroids, whether androgens or P4, is directly linked to global changes in cellular sumoylation and, more specifically, to differentiation-dependent down-regulation of PIAS1, the E3 SUMO-1 ligase of AR and PR. In comparison to PR, AR governs the expression of a limited decidual gene pool, responsible for cytoskeletal organization and inhibition of cell motility and proliferation. These cell functions under AR control may be critical for coordinated trophoblast invasion and placental development. This notion is supported by the observations in female AR-deficient mice, demonstrating that uterine responses to exogenous gonadotropins are impaired before developing premature ovarian failure (35). Furthermore, the earliest reproductive defect in these mice is a dramatic reduction in the number of pups per litter, and pregnancy is further characterized by placentamegaly, which suggests a compensatory response to defective uterine remodeling (35, 36). However, additional experiments are required to exclude the possibility that these early uterine defects in AR-deficient mice are a consequence of impaired ovarian steroidogenesis.
The identification of human AR signature genes could be exploited to assess the decidual responses before pregnancy, especially in patients with relative androgen deficiency, including older women and patients with premature ovarian failure receiving fertility treatment with donor oocytes. Such translational studies may provide the in vivo rational for the targeted use of selective AR modulators for the prevention of associated pregnancy complications.
Acknowledgments
We thank Ms. Ruba Kalaji and Drs. Osamu Ishihara, Steven Franks, Nick Macklon, Guy Whitley, Jenny Higham, Nicholas Panay, Charles Swanton, Vania Braga, and Malcolm Parker.
Footnotes
This work was supported by a Wellcome Trust Research Training Fellowship (to B.C.), by the IOG Trust, and Grant-in-Aid 19591909 from the Ministry of Education, Science, and Culture, Japan.
Disclosure Statement: The authors have nothing to disclose.
First Published Online May 29, 2008
Abbreviations: AR, Androgen receptor; DHT, dihydrotestosterone; EGFP, enhanced green fluorescent protein; F-actin, filamentous actin; HESC, human endometrial stromal cell; IGFBP-1, insulin-like growth factor-binding protein-1; MLC2, light chain of myosin 2; MPA, medroxyprogesterone acetate; NT, nontargeting; P4, progesterone; PIAS1, protein inhibitor of activated STAT1; PR, P4 receptor; PRL, prolactin; RB, retinoblastoma protein; RTQ, real-time quantitative; siRNA, small interfering RNA; STAT3, signal transducer and activator of transcription 3; SUMO, small ubiquitin-like modifier.
References
- 1.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery Jr CA, Shyamala G, Conneely OM, O'Malley BW 1995. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev 9:2266–2278 [DOI] [PubMed] [Google Scholar]
- 2.Brosens JJ, Hayashi N, White JO 1999. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology 140:4809–4820 [DOI] [PubMed] [Google Scholar]
- 3.Gellersen B, Brosens IA, Brosens JJ 2007. Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med 25:445–453 [DOI] [PubMed] [Google Scholar]
- 4.Kajihara T, Jones M, Fusi L, Takano M, Feroze-Zaidi F, Pirianov G, Mehmet H, Ishihara O, Higham JM, Lam EW, Brosens JJ 2006. Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol Endocrinol 20:2444–2455 [DOI] [PubMed] [Google Scholar]
- 5.Gellersen B, Brosens J 2003. Cyclic AMP and progesterone receptor cross-talk in human endometrium: a decidualizing affair. J Endocrinol 178:357–372 [DOI] [PubMed] [Google Scholar]
- 6.Jones MC, Fusi L, Higham JH, Abdel-Hafiz H, Horwitz KB, Lam EW, Brosens JJ 2006. Regulation of the SUMO pathway sensitizes differentiating human endometrial stromal cells to progesterone. Proc Natl Acad Sci USA 103:16272–16277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosens JJ, Gellersen B 2006. Death or survival: progesterone-dependent cell fate decisions in the human endometrial stroma. J Mol Endocrinol 36:389–398 [DOI] [PubMed] [Google Scholar]
- 8.Mertens HJ, Heineman MJ, Koudstaal J, Theunissen P, Evers JL 1996. Androgen receptor content in human endometrium. Eur J Obstet Gynecol Reprod Biol 70:11–13 [DOI] [PubMed] [Google Scholar]
- 9.Slayden OD, Nayak NR, Burton KA, Chwalisz K, Cameron ST, Critchley HO, Baird DT, Brenner RM 2001. Progesterone antagonists increase androgen receptor expression in the rhesus macaque and human endometrium. J Clin Endocrinol Metab 86:2668–2679 [DOI] [PubMed] [Google Scholar]
- 10.Hu X, Funder JW 2006. The evolution of mineralocorticoid receptors. Mol Endocrinol 20:1471–1478 [DOI] [PubMed] [Google Scholar]
- 11.Horie K, Takakura K, Imai K, Liao S, Mori T 1992. Immunohistochemical localization of androgen receptor in the human endometrium, decidua, placenta and pathological conditions of the endometrium. Hum Reprod 7:1461–1466 [DOI] [PubMed] [Google Scholar]
- 12.Milne SA, Henderson TA, Kelly RW, Saunders PT, Baird DT, Critchley HO 2005. Leukocyte populations and steroid receptor expression in human first-trimester decidua: regulation by antiprogestin and prostaglandin E analog. J Clin Endocrinol Metab 90:4315–4321 [DOI] [PubMed] [Google Scholar]
- 13.Dawood MY, Saxena BB 1976. Plasma testosterone and dihydrotestosterone in ovulatory and anovulatory cycles. Am J Obstet Gynecol 126:430–435 [DOI] [PubMed] [Google Scholar]
- 14.Massafra C, De Felice C, Agnusdei DP, Gioia D, Bagnoli F 1999. Androgens and osteocalcin during the menstrual cycle. J Clin Endocrinol Metab 84:971–974 [DOI] [PubMed] [Google Scholar]
- 15.Bonney RC, Scanlon MJ, Jones DL, Reed MJ, James VH 1984. Adrenal androgen concentrations in endometrium and plasma during the menstrual cycle. J Endocrinol 101:181–188 [DOI] [PubMed] [Google Scholar]
- 16.Castracane VD, Stewart DR, Gimpel T, Overstreet JW, Lasley BL 1998. Maternal serum androgens in human pregnancy: early increases within the cycle of conception. Hum Reprod 13:460–464 [DOI] [PubMed] [Google Scholar]
- 17.Abdalla HI, Billett A, Kan AK, Baig S, Wren M, Korea L, Studd JW 1998. Obstetric outcome in 232 ovum donation pregnancies. Br J Obstet Gynaecol 105:332–337 [DOI] [PubMed] [Google Scholar]
- 18.Castracane VD, Asch RH 1995. Testosterone and androstenedione in premature ovarian failure pregnancies: evidence for an ovarian source of androgens in early pregnancy. Hum Reprod 10:677–680 [DOI] [PubMed] [Google Scholar]
- 19.de Vries MJ, Dekker GA, Schoemaker J 1998. Higher risk of preeclampsia in the polycystic ovary syndrome. A case control study. Eur J Obstet Gynecol Reprod Biol 76:91–95 [DOI] [PubMed] [Google Scholar]
- 20.Gellersen B, Kempf R, Telgmann R, DiMattia GE 1994. Nonpituitary human prolactin gene transcription is independent of Pit-1 and differentially controlled in lymphocytes and in endometrial stroma. Mol Endocrinol 8:356–373 [DOI] [PubMed] [Google Scholar]
- 21.Yeap BB, Krueger RG, Leedman PJ 1999. Differential posttranscriptional regulation of androgen receptor gene expression by androgen in prostate and breast cancer cells. Endocrinology 140:3282–3291 [DOI] [PubMed] [Google Scholar]
- 22.Poukka H, Karvonen U, Janne OA, Palvimo JJ 2000. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc Natl Acad Sci USA 97:14145–14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghatge RP, Jacobsen BM, Schittone SA, Horwitz KB 2005. The progestational and androgenic properties of medroxyprogesterone acetate: gene regulatory overlap with dihydrotestosterone in breast cancer cells. Breast Cancer Res 7:R1036–R1050 [DOI] [PMC free article] [PubMed]
- 24.Cheon YP, Li Q, Xu X, DeMayo FJ, Bagchi IC, Bagchi MK 2002. A genomic approach to identify novel progesterone receptor regulated pathways in the uterus during implantation. Mol Endocrinol 16:2853–2871 [DOI] [PubMed] [Google Scholar]
- 25.Jeong JW, Lee KY, Kwak I, White LD, Hilsenbeck SG, Lydon JP, DeMayo FJ 2005. Identification of murine uterine genes regulated in a ligand-dependent manner by the progesterone receptor. Endocrinology 146: 3490–3505 [DOI] [PubMed]
- 26.Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC 2002. Global gene profiling in human endometrium during the window of implantation. Endocrinology 143:2119–2138 [DOI] [PubMed] [Google Scholar]
- 27.Dimitriadis E, Stoikos C, Tan YL, Salamonsen LA 2006. Interleukin 11 signaling components signal transducer and activator of transcription 3 (STAT3) and suppressor of cytokine signaling 3 (SOCS3) regulate human endometrial stromal cell differentiation. Endocrinology 147:3809–3817 [DOI] [PubMed] [Google Scholar]
- 28.Mohamed OA, Jonnaert M, Labelle-Dumais C, Kuroda K, Clarke HJ, Dufort D 2005. Uterine Wnt/β-catenin signaling is required for implantation. Proc Natl Acad Sci USA 102:8579–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fumoto K, Uchimura T, Iwasaki T, Ueda K, Hosoya H 2003. Phosphorylation of myosin II regulatory light chain is necessary for migration of HeLa cells but not for localization of myosin II at the leading edge. Biochem J 370:551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Disanza A, Steffen A, Hertzog M, Frittoli E, Rottner K, Scita G 2005. Actin polymerization machinery: the finish line of signaling networks, the starting point of cellular movement. Cell Mol Life Sci 62:955–970 [DOI] [PubMed] [Google Scholar]
- 31.Lanzetti L 2007. Actin in membrane trafficking. Curr Opin Cell Biol 19:453–458 [DOI] [PubMed] [Google Scholar]
- 32.Ihnatovych I, Hu W, Martin JL, Fazleabas AT, de Lanerolle P, Strakova Z 2007. Increased phosphorylation of myosin light chain prevents in vitro decidualization. Endocrinology 148:3176–3184 [DOI] [PubMed] [Google Scholar]
- 33.Shiokawa S, Sakai K, Akimoto Y, Suzuki N, Hanashi H, Nagamatsu S, Iwashita M, Nakamura Y, Hirano H, Yoshimura Y 2000. Function of the small guanosine triphosphate-binding protein RhoA in the process of implantation. J Clin Endocrinol Metab 85:4742–4749 [DOI] [PubMed] [Google Scholar]
- 34.Cobrinik D 2005. Pocket proteins and cell cycle control. Oncogene 24:2796–2809 [DOI] [PubMed] [Google Scholar]
- 35.Hu YC, Wang PH, Yeh S, Wang RS, Xie C, Xu Q, Zhou X, Chao HT, Tsai MY, Chang C 2004. Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proc Natl Acad Sci USA 101:11209–11214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiina H, Matsumoto T, Sato T, Igarashi K, Miyamoto J, Takemasa S, Sakari M, Takada I, Nakamura T, Metzger D, Chambon P, Kanno J, Yoshikawa H, Kato S 2006. Premature ovarian failure in androgen receptor-deficient mice. Proc Natl Acad Sci USA 103:224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, D'Arrigo C, Ryder K, Menke A, Gress T, Downward J 2005. CUTL1 is a target of TGFβ signaling that enhances cancer cell motility and invasiveness. Cancer Cell 7:521–532 [DOI] [PubMed] [Google Scholar]
- 38.Anton IM, Jones GE 2006. WIP: a multifunctional protein involved in actin cytoskeleton regulation. Eur J Cell Biol 85:295–304 [DOI] [PubMed] [Google Scholar]
- 39.Lanzardo S, Curcio C, Forni G, Anton IM 2007. A role for WASP interacting protein, WIP, in fibroblast adhesion, spreading and migration. Int J Biochem Cell Biol 39:262–274 [DOI] [PubMed] [Google Scholar]
- 40.Qwarnstrom EE, MacFarlane SA, Page RC, Dower SK 1991. Interleukin 1β induces rapid phosphorylation and redistribution of talin: a possible mechanism for modulation of fibroblast focal adhesion. Proc Natl Acad Sci USA 88:1232–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang EY, Bai AH, To KF, Sy SM, Wong NL, Lai PB, Squire JA, Wong N 2007. Identification of PFTAIRE protein kinase 1, a novel cell division cycle-2 related gene, in the motile phenotype of hepatocellular carcinoma cells. Hepatology 46:436–445 [DOI] [PubMed] [Google Scholar]
- 42.Kurochkin IV, Yonemitsu N, Funahashi SI, Nomura H 2001. ALEX1, a novel human armadillo repeat protein that is expressed differentially in normal tissues and carcinomas. Biochem Biophys Res Commun 280:340–347 [DOI] [PubMed] [Google Scholar]
- 43.Franco SJ, Huttenlocher A 2005. Regulating cell migration: calpains make the cut. J Cell Sci 118:3829–3838 [DOI] [PubMed] [Google Scholar]
- 44.Giganti A, Plastino J, Janji B, Van Troys M, Lentz D, Ampe C, Sykes C, Friederich E 2005. Actin-filament cross-linking protein T-plastin increases Arp2/3-mediated actin-based movement. J Cell Sci 118:1255–1265 [DOI] [PubMed] [Google Scholar]
- 45.Rosenberger G, Jantke I, Gal A, Kutsche K 2003. Interaction of αPIX (ARHGEF6) with β-parvin (PARVB) suggests an involvement of αPIX in integrin-mediated signaling. Hum Mol Genet 12:155–167 [DOI] [PubMed] [Google Scholar]
- 46.Goley ED, Welch MD 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev 7:713–726 [DOI] [PubMed] [Google Scholar]
- 47.Le Bot N 2007. Autophagy: a new regulator of development. Nat Cell Biol 9:741. [DOI] [PubMed] [Google Scholar]
- 48.Rivero F, Illenberger D, Somesh BP, Dislich H, Adam N, Meyer AK 2002. Defects in cytokinesis, actin reorganization and the contractile vacuole in cells deficient in RhoGDI. EMBO J 21:4539–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akasaka-Manya K, Manya H, Nakajima A, Kawakita M, Endo T 2006. Physical and functional association of human protein O-mannosyltransferases 1 and 2. J Biol Chem 281:19339–19345 [DOI] [PubMed] [Google Scholar]
- 50.van Eys GJ, Niessen PM, Rensen SS 2007. Smoothelin in vascular smooth muscle cells. Trends Cardiovasc Med 17:26–30 [DOI] [PubMed] [Google Scholar]
- 51.Funakoshi E, Hori T, Haraguchi T, Hiraoka Y, Kudoh J, Shimizu N, Ito F 2003. Overexpression of the human MNB/DYRK1A gene induces formation of multinucleate cells through overduplication of the centrosome. BMC Cell Biol 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou Y, Lim S, Lee K, Deng X, Friedman E 2003. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J Biol Chem 278: 49573–49581 [DOI] [PubMed]
- 53.Conley CA 2001. Leiomodin and tropomodulin in smooth muscle. Am J Physiol Cell Physiol 280:C1645–C1656 [DOI] [PubMed]
- 54.Fredriksson R, Gloriam DE, Hoglund PJ, Lagerstrom MC, Schioth HB 2003. There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem Biophys Res Commun 301:725–734 [DOI] [PubMed] [Google Scholar]
- 55.Korr D, Toschi L, Donner P, Pohlenz HD, Kreft B, Weiss B 2006. LRRK1 protein kinase activity is stimulated upon binding of GTP to its Roc domain. Cell Signal 18:910–920 [DOI] [PubMed] [Google Scholar]
- 56.Bromme D, Okamoto K, Wang BB, Biroc S 1996. Human cathepsin O2, a matrix protein-degrading cysteine protease expressed in osteoclasts. Functional expression of human cathepsin O2 in Spodoptera frugiperda and characterization of the enzyme. J Biol Chem 271:2126–2132 [DOI] [PubMed] [Google Scholar]
- 57.Wesierska-Gadek J, Gueorguieva M, Kramer MP, Ranftler C, Sarg B, Lindner H 2007. A new, unexpected action of olomoucine, a CDK inhibitor, on normal human cells: up-regulation of CLIMP-63, a cytoskeleton-linking membrane protein. J Cell Biochem 102:1405–1419 [DOI] [PubMed] [Google Scholar]
- 58.Azakie A, Larkin SB, Farrance IK, Grenningloh G, Ordahl CP 1996. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J Biol Chem 271:8260–8265 [DOI] [PubMed] [Google Scholar]
- 59.Maeda T, Mazzulli JR, Farrance IK, Stewart AF 2002. Mouse DTEF-1 (ETFR-1, TEF-5) is a transcriptional activator in α1-adrenergic agonist-stimulated cardiac myocytes. J Biol Chem 277:24346–24352 [DOI] [PubMed] [Google Scholar]
- 60.Heo WD, Meyer T 2003. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell 113:315–328 [DOI] [PubMed] [Google Scholar]