Abstract
The rat placentation site is distinctly organized into interacting zones, the so-called labyrinth, junctional, and metrial gland compartments. These zones house unique cell populations equipped to undertake myriad prescribed functions including transport, hormonal responses, and immune interactions. Although much is known about the genesis of these cell types and specific markers that characterize each zone, a detailed global overview of gene expression in the three zones is absent. In this report, we used massively parallel sequencing (RNA-seq) to assess mRNA expression profiles and generated transcriptomic maps for each zone of the late-gestation rat placentation site (18.5 d postcoitum). Analysis of expression profiles revealed that each compartment expressed a unique signature, characterized by biological processes specific to the zone. Transport and vasculature-related processes predominated in the labyrinth, hormone secretion in the junctional, and immune interactions in the metrial gland. Furthermore, our analysis identified approximately 4000 differentially expressed genes within the zones. Using k-means clustering, we identified transcription factors with highest expression in either labyrinth, junctional, or metrial gland. Direct interaction (pathway) analysis revealed unique transcription factor networks operating in each compartment. The site-specific expression of 27 transcription factors in the three zones was ascertained via quantitative PCR and protein expression of six transcription factors was confirmed by immunohistochemistry. Finally, we elucidated the expression of key developmentally important families (Sox, GATA, Fox, Wnt, Tead, and IGF/IGFBP) in the placentation site to reveal novel expression of these several factors. The present dataset provides a novel resource to understand zonal gene expression and function in the placenta.
In all eutherian mammals, the placenta plays an indispensible role in supporting embryonic and fetal development throughout intrauterine life. Being one of the earliest fetal organs to develop, the placenta is entrusted with a number of critical functions requisite for a successful pregnancy. In early pregnancy, the placenta facilitates implantation and anchoring of the embryo to the uterine wall and through the production of luteotrophic hormones induces maternal recognition of pregnancy (1). After implantation, the placenta establishes the interface through which transport of nutrients, gases, and waste can occur between the mother and the developing fetus. In addition, the placenta also orchestrates a number of local and systemic changes in the mother via production of paracrine and endocrine mediators throughout the course of pregnancy, which profoundly influence maternal physiology. To accomplish these varied functions, the placenta is composed of multiple specialized cell types that originate either from the primordial trophectoderm (trophoblast lineage) or the extraembryonic mesenchyme (stromal and vascular lineages) (2–4). Broadly, although some of the trophoblast cells differentiate into the highly branched villous or labyrinth structures designed for nutritional exchange [syncytiotrophoblasts (SyT)], others differentiate into cell types specialized to secrete hormones and peptides [spongiotrophoblasts (SpT) and trophoblast giant cells (TGC)].
Structurally, the late-gestation rat utero-placental unit can be divided into three interacting but fairly distinct functional areas: 1) the labyrinth zone, 2) the junctional zone, and 3) metrial gland or the decidualized mesometrial triangle (5). The labyrinth zone is composed of both maternal and fetal vasculature and the specialized SyT. In both rodents and primates, the SyT of chorionic villi are directly bathed in maternal blood (referred to as hemochorial placentation), allowing for a highly efficient exchange surface. The materno-fetal interdigitation differs significantly between rodents and primates; rats and mice have a labyrinthine type of placenta compared with the villous type found in humans and other primates. In both rats and mice, maternal blood is separated from the fetal endothelial cells by two layers of SyT and a single layer of mononuclear cells (hemotrichorial) (1). The junctional zone is located at the maternal-placental interface and houses three cell types: SpT, TGC, and glycogen cells. Finally, akin to human placentation, in the rat, some trophoblast cells exit the placenta to invade the vasculature-rich mesometrial triangle and endovascularize uterine blood vessels (5, 6). This highly chimeric region of uterine stroma and invasive trophoblasts has been referred to as the metrial gland or the decidualized mesometrial triangle (7). Consistent with the specialized nature of these areas, several uniquely expressed marker genes for individual zones have been reported. Previous studies have also assessed global gene expression patterns using microarrays during placental development in the mouse and human placenta (8–10). However, information about the differential gene expression between the functional zones in the mature (late-gestation) placenta currently remains unavailable. In this report, we use ultra high-throughput sequencing of mRNA-derived cDNA libraries (RNA-seq) to assemble a genome-wide transcriptomic view of the rat placentation site. The present studies had three objectives. First, we examined whether the three utero-placental compartments of the placentation site have distinct gene expression signatures and catalogued the differentially expressed genes between the compartments. Second, we examined enrichment of key biological processes within the compartments based on gene expression patterns. Specifically, we examined whether enrichment of biological pathways reflected the functional specialization of the cell types constituting the compartments. Finally, we elucidated expression of key families of transcription factors known to influence placental development and identified novel expression of transcription factors in specific compartments. Our data provide an expression-level dissection of the rat placentation site and provide an important new resource in understanding key transcriptional networks in the maintenance of unique gene expression signatures.
Materials and Methods
Experimental design
All experimental treatments were conducted in accordance with the ethical guidelines established and approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences (Protocol 2971). Samples used in this study were collected as part of a separate unrelated ongoing analysis, examining the effect of overfeeding on placental gene expression. However, only tissues from the control group were employed herein to assess the transcriptomic differences within different utero-placental compartments. Virgin female Sprague Dawley rats (150–175 g; Charles River Laboratories, Wilmington, MA) were housed in an American Association for Accreditation of Laboratory Animal Care-approved animal facility. Animals were intragastrically cannulated and allowed to recover for 10 d as previously described (11–13). Rats (n = 10) were fed liquid diets (at 155 kcal/kg3/4·d) for 23 h/d using computer-controlled syringe pumps for 4 wk. Caloric intake was determined from previous studies and mimicked body weights and body composition of rats consuming standard commercial diets ad libitum (12–15). Diets met National Research Council nutrient recommendations, including essential fatty acids, and were 20% protein (casein), 75% carbohydrate (dextrose and maltodextrin), and 5% fat (corn oil) as percentage of total calories. After 4 wk of diets, each female rat was housed with one male, and successful mating was confirmed by the presence of sperm in the vaginal lavage the next morning [designated as day postcoitum (dpc) 0.5]. On dpc 18.5, pregnant dams were killed under Nembutal anesthesia. Placentas were collected from dams after dissecting the uterus from the antimesometrial side. From each placenta, the junctional and labyrinth-enriched zones were separated by dissection under a stereomicroscope (16). Metrial glands were dissected from the uterus. In this study, corresponding fetal livers were also collected and frozen in liquid nitrogen. Sex of the fetus was determined using DNA from the liver, and only placentas from male embryos were used in this analysis. Tissues were frozen in liquid nitrogen and stored at −70 C for RNA and protein analyses.
Preparation of RNA-seq libraries
Total RNA was isolated from each utero-placental compartment using a combination of TRI reagent and RNeasy-mini columns (QIAGEN, Valencia, CA), including on-column deoxyribonuclease digestion (15). Two biologically separate pools containing equal amounts of RNA from seven to eight individual placentas from at least five distinct litters were used for each utero-placental compartment. Thus, n = 15 utero-placental samples from n = 10 dams were represented over two biological replicate pools. Isolation of polyadenosine RNA and construction of RNA-seq libraries is described in Supplemental Materials (published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org). Quantification of the RNA-seq libraries was done via quantitative real-time PCR (qPCR) using SYBR green chemistry (Kapa Biosystems, Woburn, MA). Diluted libraries (1:10,000) were quantitated using standards ranging from 0.0002–20 pm.
Sequencing, alignment, and data analysis
Single-read 36-bp sequencing of libraries was performed with a Genome Analyzer IIX (details in Supplementary Materials). Reads for each placental zone were collected by sequencing an entire lane, which were later de-multiplexed into the respective biological pool. Alignment to the rat Rn4 genome was carried out using ELAND (Efficient Large-Scale Alignment of Nucleotide; Illumina, San Diego, CA). All aligned reads were exported in SAM format, and subsequent data analysis was performed in Avadis NGS (Strand Scientific Intelligence Inc., San Francisco, CA) and SeqMonk software packages (http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk/; Babraham Bioinformatics, Cambridge, UK). Uniquely aligned reads were quantified in Avadis NGS, and both gene and exon-level reads per kilobase per million mapped reads (RPKM) values were calculated. RPKM values represent counts of reads mapping to a feature (gene, exon, etc.) normalized to both the overall sequencing coverage and the size of the feature. Similarly, we also used SeqMonk to quantify gene-level RPKM and to assess overall data distribution. Differentially expressed genes between the three zones were identified based on P value ≤ 0.05 using one-way ANOVA, followed by post hoc analysis using Tukey's honestly significant difference (HSD) and minimum fold change of ±5-fold (in pair-wise comparisons). Corrections for multiple testing were performed using the false discovery rate method (17). Venn diagrams were generated using Avadis NGS. Using the union of the differentially expressed genes, we performed k-means and correlation-based hierarchical clustering. Known biological functions of genes were queried using NetAffx (Affymetrix, Santa Clara, CA) and gene ontology (GO) analyses for biological/molecular function performed using Avadis NGS using false discovery rate-corrected P value < 0.01 (14, 15). Furthermore, the lists of differentially expressed genes were analyzed for GO biological and molecular function enrichment using Avadis NGS.
To examine temporal changes in expression of select transcription factors during placental development, we mined a previously reported microarray expression profiling dataset of the mouse placenta and decidua from dpc 8.5 to dpc 19.0 (GSE11224) (8, 18). Because our RNA-seq analysis was limited to only one time point (dpc 18.5), information about developmental changes in specific transcription factors might be additionally informative. Data analysis was carried out using GeneSpring version 11.5 (Agilent Technologies, Santa Clara, CA) (12, 14, 15). The .CEL files containing probe-level intensities were processed using robust multiarray analysis algorithm for background adjustment, normalization, and log2 transformation of perfect match values. Subsequently, for each probe, the baseline was normalized to the median of all samples. Normalized expression values for select transcripts (identified via RNA-seq analysis), were queried and expressed relative to the values at dpc 8.5.
Real-time RT-PCR
One microgram of total RNA was reverse transcribed (n = 15 per group) using the IScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-time PCR analysis was performed as described previously (12–15). Briefly, amplifications were performed using 2× SYBR green master mix (Applied Biosystems, Foster city, CA) with 25 ng cDNA and 0.25 μm forward and reverse primers in a final reaction volume of 10 μl. PCR were monitored using an ABI Prism 7500 Fast sequence detection system (Applied Biosystems, Foster City, CA). Gene-specific primers were designed using Primer Express Software (Supplemental Table 1). Relative amounts of mRNA were quantitated using the ΔCt method and normalized to the expression of SRP14 mRNA (19).
Immunohistochemical staining
Immunolocalization of proteins in placental sections was performed using Vectastain Elite reagents. Details of the procedure are described in Supplemental Materials.
Statistical analysis
Data are expressed as means ± sem. Statistical differences between different compartments of the placentation site were determined using one-way ANOVA, followed by post hoc analysis with Tukey's HSD. Statistical significance was set at P < 0.05. Statistical analyses were performed using SigmaStat versiona 3.3 software (Systat Software Inc., San Jose, CA).
Results
Utero-placental compartments of the rat placentation site exhibit distinct expression signatures
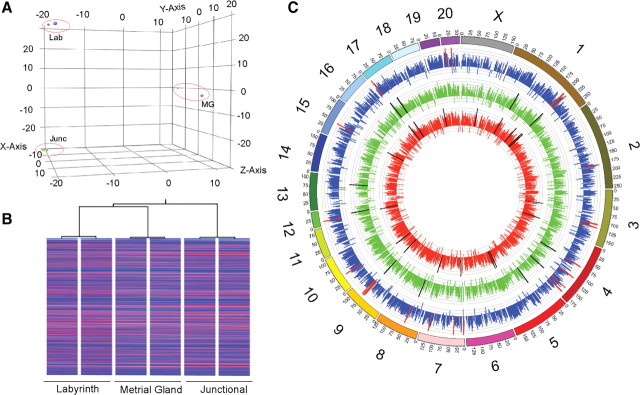
De-multiplexed reads for each biological replicate resulted in approximately 18–20 million reads (Table 1). Alignment results revealed that more than 94% of the uniquely mapped reads were localized to genic regions of the rat genome. The majority of reads (>70%) in all samples mapped to exonic regions, and approximately 11% mapped to intronic regions. Percentages of reads mapping to 3′-untranslated regions (UTR) was almost twice as that in 5′-UTR (20 vs. 10%), consistent with the twice longer average length of 3′-UTR in eukaryotes (20). These analyses indicated that overall read alignment was comparable between all samples. To assess global gene expression profiles, read counts were converted into RPKM. Percent cumulative distribution of RPKM indicated that approximately 45% of values were more than 1 (Supplemental Fig. 1A). Moreover, the range and distribution of RPKM (median and lower and upper quartiles) were similar for all samples, suggesting comparable transcriptomic coverage (Supplemental Fig. 1B). Both principal component analysis (Fig. 1A) and unsupervised condition clustering (Fig. 1B) of RPKM revealed clustering of expression profiles based on utero-placental compartment of origin, suggesting that each compartment expressed a unique gene expression signature. To visualize genome-wide RPKM abundance, we generated radial histograms using Circos (21) and identified the location of the 25 most abundant genes in each zone (Fig. 1C). A rank ordered list of these highly expressed transcripts in each zone is presented in Supplemental Fig. 2A. As expected, the majority of genes (>75%) were uniquely expressed in either of the compartments (Supplemental Fig. 2B). Only two transcripts, H19 and fibronectin 1, were expressed among the most abundant in all three compartments.
Table 1.
Distribution of reads after mRNA-seq of rat placental zones
| Parameter | Labyrinth |
Junctional |
Metrial gland |
|||
|---|---|---|---|---|---|---|
| Pool 1 | Pool 2 | Pool 1 | Pool 2 | Pool 1 | Pool 2 | |
| Total reads sequenced | 18.81 | 20.03 | 19.51 | 21.31 | 21.03 | 18.39 |
| Uniquely aligned (%) | 72.37 | 71.57 | 74.31 | 74.11 | 75.68 | 75.78 |
| Reads (%) | ||||||
| Mapped to genic regions | 94.58 | 95.01 | 95.26 | 95.29 | 97.23 | 96.76 |
| Mapped to exons | 72.92 | 78.65 | 78.06 | 77.91 | 76.31 | 75.51 |
| Introns | 12.63 | 11.28 | 12.15 | 12.44 | 11.13 | 11.77 |
| 5′-UTR | 9.18 | 9.39 | 11.86 | 11.95 | 9.35 | 9.35 |
| 3′-UTR | 20.30 | 20.66 | 17.49 | 17.48 | 20.15 | 19.93 |
Single-read 36-bp sequencing, followed by a 7-bp index read of the clustered libraries was performed with a Genome Analyzer IIX using TruSeq version 5 reagents. Two biologically separate pools, each containing equal amounts of RNA from seven to eight individual placentas from at least five distinct litters were utilized for each placental zone. Thus, 15 placental samples were represented over two biological replicate pools. Cluster and base calling were performed by RTA version 2.6 (Illumina). Total number of reads passing Phred score higher than 30 is indicated in millions. The percentage of uniquely aligned reads was calculated after alignment with ELAND using CASAVA package (Illumina). The percentage of reads mapping to genomic features of the rat genome (Rn4) were calculated in SeqMonk package.
Fig. 1.
Overall gene expression patterns of utero-placental functional compartments. RPKM values for all Refseq genes in rat labyrinth zone, junctional zone, and metrial gland. A and B, Principal component analysis (A) and hierarchical clustering on conditions of global gene expression profiles from rat labyrinth zone, junctional zone, and metrial gland (B). Expression profiles of individual samples belonging to the same compartment are highly related and cluster together. C, Genome-wide visualization of mRNA abundance in utero-placental compartments using Circos. RPKM values are plotted over the whole rat genome. The outer ring represents chromosomes oriented pter-qter in a clockwise direction. Outside numbers indicate chromosome number. RPKM data are represented for the labyrinth zone in red (innermost track), junctional zone in green (center track), and metrial gland in blue (outer track). The inner scale for each track (in gray) is log transformed. Bars in black (for labyrinth and junctional zones) and red (for metrial gland) represent the genomic localizations of the top 25 most abundant transcripts in each respective compartment. A complete list is included in Supplemental Materials.
Utero-placental gene expression reveals distinct enrichment of GO terms
To characterize gene expression differences between the utero-placental compartments, we performed one-way ANOVA followed by pair-wise comparisons. A conservative threshold (±5-fold change) within each comparison was used to generate lists of differentially expressed genes. Comparisons between labyrinth and junctional zone, junctional zone and metrial gland, and labyrinth zone and metrial gland identified, respectively, 2102, 2204, and 2600 genes that were differentially expressed (Fig. 2B). GO biological process and molecular function for these transcripts were queried using Avadis NGS and NetAffx (Supplemental Table 2). Hierarchical clustering of the union of differentially expressed genes showed groups of transcripts that are distinctly expressed in one or more compartments (Fig. 2A). Using these lists, we assessed enrichment of biological processes in each functional compartment. In the labyrinth zone, anatomical structure, transporter, blood vessel/vasculature development, oxygen and substrate-specific transport, and adhesion were the top statistically enriched biological processes (Fig. 2C, P value < 0.0001). Genes highly expressed in junctional zone were those with hormone activity and response to hormones, female pregnancy, stress and hypoxia response, tissue regeneration, and cell proliferation (Fig. 2D). On the other hand, GO terms of immune and inflammatory response, T cell, leukocyte and lymphocyte activation, and response to stimulus were most evident in the metrial gland. Furthermore, the metrial gland also showed highly significant enrichment of extracellular matrix, vasculature remodeling, and hormone stimulus genes (Fig. 2E). These analyses underscore that RNA-seq-based transcriptomic analysis faithfully reflected the known functional specialization of utero-placental compartments as discerned by GO term analysis.
Fig. 2.
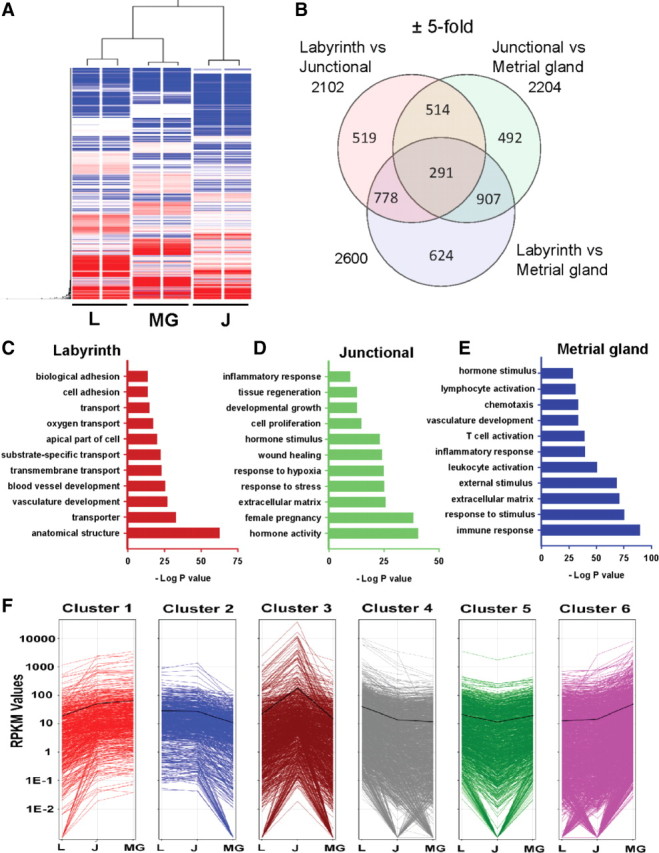
Identification of gene signatures within utero-placental functional compartment. A, Hierarchical clustering of the union of differentially altered transcripts between labyrinth zone, junctional zone, and metrial gland. Differentially expressed genes were generated using one-way ANOVA, followed by pair-wise comparisons and a minimum threshold of ±5-fold change within each comparison. Statistical analysis and clustering were carried out using Avadis NGS. B, Venn diagram representing overlap between differentially expressed genes in the labyrinth zone, junctional zone, and metrial gland (n = 2 biological replicates, each representing pools of seven to eight separate placenta from five separate rat dams). C–E, Functional enrichment analysis of GO biological processes in labyrinth zone (C), junctional zone (D), and metrial gland (E). The x-axis indicates −log10 corrected P values for enrichment with the indicated GO term. F, k- means clustering of union of differentially altered transcripts between labyrinth zone, junctional zone, and metrial gland. Differentially expressed genes were resolved into six clusters. The y-axis represents log10 RPKM, and L, J, and MG indicate means for labyrinth zone, junctional zone, and metrial gland.
Unique transcription factor expression profiles in utero-placental compartments
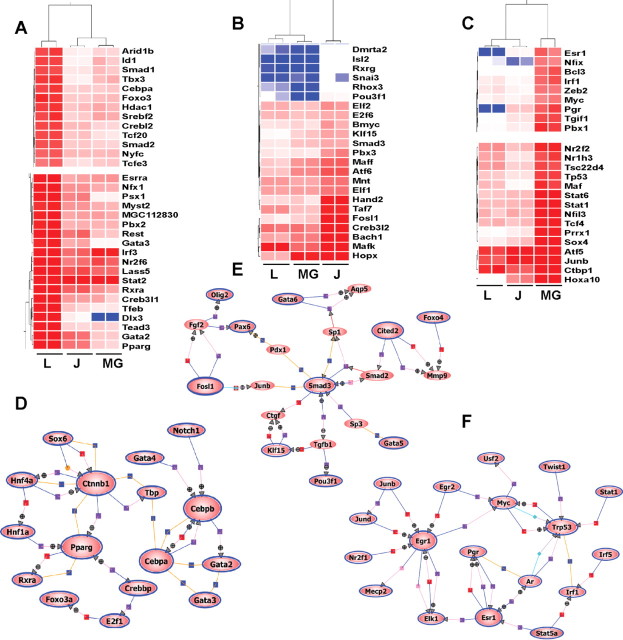
To identify uniquely expressed genes in each utero-placental compartment, we performed k-means clustering of the one-way ANOVA filtered list of genes. Using this approach, genes were classified into six clusters based on their group means and change with respect to other compartments (Fig. 2F). Detailed lists of genes in each cluster are included in Supplemental Table 3. Clusters 3, 4, and 6 consisted of genes highly expressed in junctional zone, labyrinth zone, and metrial gland, respectively. Among these clusters, we further queried for genes with functions as transcription factors or transcriptional regulators, because these are likely to be important in maintaining distinct gene expression patterns in each compartment. Correlation-based hierarchical clusters of transcription factors expressed in each compartment are presented in Fig. 3, A–C (Supplemental Fig. 3). A complete list of factors along with fold changes is included in Supplemental Table 4. Using these unique transcription factors, we performed direct interaction (pathway) analysis for each compartment. The labyrinth showed β-catenin, peroxisome proliferator activated receptor-gamma (PPAR-γ), CCAAT/enhancer binding protein (C/EBP)-α, and C/EBP-β to be key transcriptional factors functioning in this zone (Fig. 3D). Expression of Cited-2, Smad3, Klf15, and Fosl1 characterized the junctional zone transcriptional network (Fig. 3E). Nuclear hormone receptors estrogen receptor (ER)-α (ESR1) and progesterone receptor (PGR) and inflammation regulatory factors (IRF)-1 and -5 and early growth response 1, were highly expressed in the metrial gland (Fig. 3F).
Fig. 3.
Identification of unique transcription factors within utero-placental functional compartments. A–C, Representative subclusters of correlation-based hierarchical clustering of transcription factors highly expressed in labyrinth zone (A), junctional zone (B), and metrial gland (C). Transcription factors were queried from clusters 3, 4, and 6 for junctional zone, labyrinth zone, and metrial gland, respectively. Heat-map colors red, white, and blue represent up-regulation, no relative effect, and down-regulation of transcripts, respectively. Complete list of transcription factors and hierarchical clusters are included in Supplemental Materials. Gene symbols can be queried at http://www.ncbi.nlm.nih.gov/gene. D–F, Direct Interaction (pathway) analysis of transcription factors enriched in labyrinth zone (D), junctional zone (E), and metrial gland (F).
Next we used real-time RT-PCR to assess mRNA levels of nine transcription factors from each utero-placental compartment (n = 15 per compartment) to validate RNA-seq data. mRNA expression of developmental genes Tfeb, Tead3, β-catenin, and BMP-2 were 469-, 11-, 7.5-, and 3.5-fold greater in the labyrinth zone relative to the metrial gland (Fig. 4A, P < 0.0001). Expression of Tfeb showed a graded increase, with expression in junctional zone approximately 20-fold higher compared with metrial gland. However, expression of Tead3, BMP-2, and β-catenin was restricted to only the labyrinth zone. We also assessed expression of nuclear receptors PPAR-γ and RXR-α in the three compartments. PPAR-γ and RXR-α mRNA expression was highest in the labyrinth zone (14- and 8.5-fold higher expression than metrial gland), with modestly greater expression (4.5- and 2.1-fold) compared with the junctional zone. Lastly, we also assessed expression of factors involved in controlling cell proliferation and survival, C/EBP-β, FoxO3, and HDAC-1. Expression of both HDAC-1 and FoxO3 was selectively higher (∼3.5-fold) in the labyrinth zone relative to the metrial gland. On the other hand, C/EBP-β mRNA was approximately 2.5-fold higher in both junctional and labyrinth zones (relative to metrial gland).
Fig. 4.
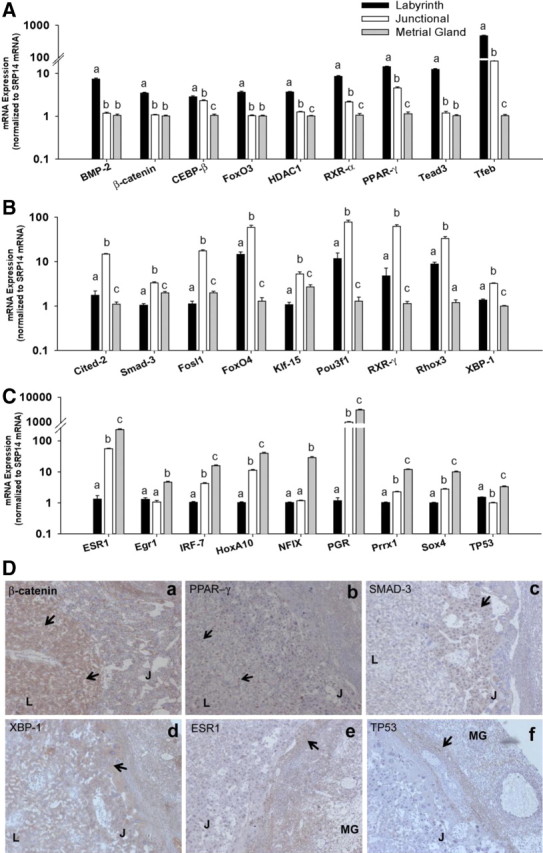
mRNA expression of transcription factors in labyrinth zone, junctional zone, and metrial gland. A–C, Placental expression of transcription factor mRNA at dpc 18.5 in labyrinth zone (A), junctional zone (B), and metrial gland (C) (n = 15 per group). Gene expression was assessed via real-time RT-PCR and normalized to abundance of SRP14 mRNA. Data are expressed relative to lowest group. Statistical differences were determined using one-way ANOVA followed by Tukey's HSD. Different letters indicate P < 0.05. The y-axis is set to log10 scale. D, Immunohistochemical staining for β-catenin (a), PPAR-γ (b), Smad-3 (c), XBP-1 (d), ER-α (e), and p53 (f) in placental sections. L, J, and MG indicate labyrinth, junctional, and metrial gland areas. Arrows show areas of positive staining. Magnification, ×50.
As assessed via qPCR, expression of Cited-2, Smad-3, Klf-15, and Fosl-1 mRNA was approximately 8-, 3-, 5-, and 17-fold higher in the junctional zone compared with the labyrinth zone (Fig. 4B). In contrast to FoxO3 expression, which was higher in the labyrinth zone, the junctional zone showed approximately 4-fold higher expression of FoxO4. Similarly, RXR-γ expression was approximately 12-fold higher in the junctional zone (relative to labyrinth zone). However, expression of both RXR-γ and FoxO4, was remarkably greater (>60-fold) in both labyrinth and junctional zones, relative to the metrial gland. Furthermore, we also found greater expression of Pou3f1/Oct-6 (∼7-fold) and the reproductive homeobox gene, Rhox3 (∼4-fold) in the junctional zone (relative to the labyrinth zone) (Fig. 4B). The metrial gland was characterized by high expression of ER-α (ESR1), progesterone receptor, and HoxA10 (∼230-, 3000-, and 40-fold, relative to the labyrinth zone), which were also expressed in the junctional zone, albeit at much lower levels (Fig. 4B). Expression of Sox4, Prrx1, and IRF-7 in the metrial gland was approximately 10-fold higher relative to labyrinth zone (Fig. 4C). Moreover, expression of both Egr-1 and Nfix was higher in metrial gland relative to labyrinth zone (Fig. 4C). Finally, we confirmed expression of several transcription factors at the protein levels via immunohistochemical staining. Protein expression of β-catenin and PPAR-γ was localized predominantly in the labyrinth zone (Fig. 4D). Expression of Smad-3 and XBP-1 was localized in the TGC and SpT of the junctional zone, and staining for ER-α and p53 was strongest in the metrial gland (Fig. 4D). Finally, we examined the temporal changes in expression of select transcription factors using the microarray dataset of murine placental development published by Knox and Baker (8). Expression of C/EBP-β, FoxO3, PPAR-γ, Cited-2, and Smad-3 showed significant increases in the placenta over embryonic development. Similarly, expression of transcription factors identified in the metrial gland, IRF-7, Sox-4, and Prrx-1, showed increases between dpc 12 and dpc 19.0 (Supplemental Fig. 4).
Expression profiles of developmental gene families in utero-placental compartments
Many developmentally regulated genes have been shown to be critical in placental physiology. Hence, we mined our RNA-seq data to map the relative expression of key gene families in different compartments within the placentation site. Heat maps depicting mRNA expression of Sox, Gata, and Tead families of transcription factors are presented in Fig. 5A. Sox family members Sox4, -12, -13, -17, and -18 were expressed in all three zones of the placenta to an appreciable degree. Of these, Sox13, -17, and -18 were predominantly expressed in the labyrinth zone and metrial gland, relative to the junctional zone. Furthermore, expression of Sox7, known to be important in vascular development, was restricted to the labyrinth zone. Conversely, expression of Sox4 was highest in the metrial gland, with diminishing expression in the junctional and labyrinth zones. Akin to expression of Gata2, expression of Gata3 was highest in the labyrinth zone. Interestingly, the GATA-domain-containing transcriptional repressors Gatad2a and -2b (p66-α, and p66-β) were also highly expressed in the placenta, especially in the labyrinth zone. Finally, mRNA of the Yap1/Tead signaling pathway were highly expressed in most compartments of the placentation site. Expression of Tead3 was highest in the labyrinth zone, similar to Yap1 and the upstream kinase Lats2. These results corroborate earlier evidence that the Yap1 signaling pathway is highly functional in the placenta.
Fig. 5.
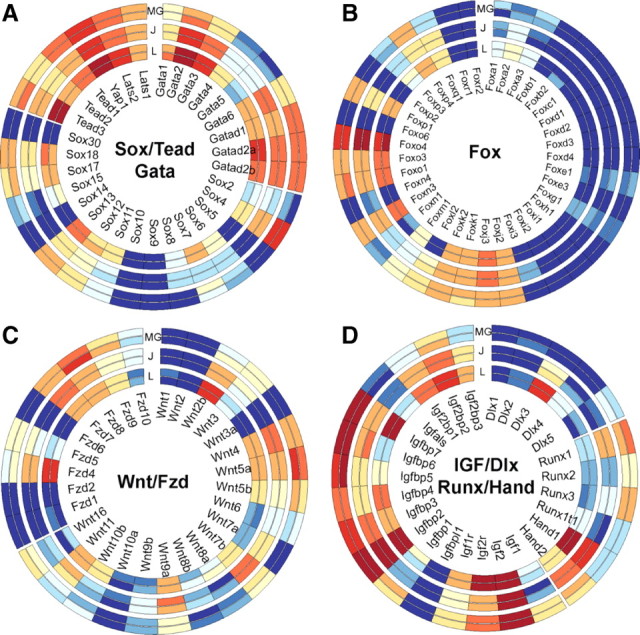
mRNA expression of select transcription factor families in the rat placentation site. Heat maps depicting expression levels of Sox, Tead, and Gata (A); Fox (B); Wnt/Fzd (C); and Igf, Dlx, Runx, and Hand (D) families of transcription factors and developmental regulators in rat labyrinth zone, junctional zone, and metrial gland. Six tracks represent log-transformed RPKM data from individual biological pools and are separated (by white space) into sets of two, of labyrinth zone (innermost), junctional zone (middle), and metrial gland (outer). Gene symbols are indicated in the middle. For RPKM data, red, white, and blue represent high, medium, and low absolute levels of abundance, respectively. Images were created using Circos.
mRNA expression of FoxA, -B, -C, -D, -E, -G, -H, and -I families was limited in all parts of the placentation site. Highest expression among all Fox family members was that of FoxO1, -3, and -4, with the greatest levels found in the labyrinth zone (Fig. 5B). We also found high levels of FoxJ and -K families, which were expressed similarly in all compartments of the placentation site. Expression of Wnt transcripts in the placentation site was restricted to specific ligands. mRNA for Wnt2b, -9a, and -10b showed greatest expression in the labyrinth zone, with intermediate expression in the junctional zone (Fig. 5C). Conversely, Wnt3 and -5a showed highest expression in the metrial gland, and Wnt4 and -7b showed junctional zone-specific expression (Fig. 5C). We also profiled expression of IGF-1 and -2 and IGFBP family members. As anticipated, expression of IGF-2 and IGFBP-2 was high throughout the placentation site (Fig. 5D). In addition, expression of IGF2-binding proteins (IGF2BP-1, -2, and -3) was also appreciable, especially in the labyrinth zone. We also indentified high expression of several other IGF-binding proteins (IGFBP-3, -4, -5, -6, and -7) with highest expression in the metrial gland. Expression of distal-less homeobox genes (Dlx) showed selective expression of Dlx3 that was restricted to the labyrinth zone. Runt-related transcription factor expression (Runx-1, -2, and -3) was highest in the metrial gland. Heart and neural crest derivatives-expressed proteins (Hand1 and -2), known to be important in placental development, showed highest expression in labyrinth and junctional zone, respectively. Overall, these data provide a comprehensive description of the abundance of many recognized developmental regulators in the rat placentation site.
Discussion
By virtue of its role as the conduit between mother and offspring, the placenta requires mechanisms that integrate a number of physiological processes. To this end, a complex interaction of different cell types and molecular processes are integral to maintaining the multiple roles required of the placenta. In the current study, we leveraged mRNA-seq technology to identify the major transcriptomic differences between interacting compartments of the late-gestation placentation site. Sequencing-based mRNA profiling affords a number of distinct advantages. These include being open-ended (i.e. independent of a predefined set of probes), having a large dynamic range for quantification (>105), and providing a digital readout (counts) for gene expression (22). To our knowledge, this is the first report to use RNA-seq to examine utero-placental gene expression patterns in the rat. Several novel findings are evident from the present study: 1) individual compartments of the placentation site that are characterized by distinct expression profiles with biological processes reflecting compartment-specific function; 2) a description of unique transcriptional factor networks in each functional compartment within the placentation site; and 3) compartment-specific expression maps of key developmentally critical transcriptional regulators.
In this study, our goal was to understand key gene expression signatures that characterize each compartment of the mature placentation site. Because the nature of differentiated trophoblasts that populate each compartment is relatively distinct, we reasoned that elucidating differential gene expression might help uncover molecular components associated with maintaining specialization. The innermost part of the placenta, the labyrinth zone, is composed of multinucleated SyT resulting from cell fusion of postmitotic labyrinth trophoblast cells arising from the chorion. SyT separate the maternal blood spaces and fetal vasculature and carry out the essential function of exchange of nutrients, gases, and waste (4). Consistent with this, our analysis of GO biological processes for genes enriched in the labyrinth zone clearly indicated vasculature/blood vessel development and transport as most significant. Juxtaposed to the labyrinth zone are SpT that along with TGC and glycogen cells make up the junctional zone. One subtype of the TGC, the parietal TGC, form the boundary of the implantation site and are in direct contact with the uterine decidua. During midgestation, a population of trophoblasts (invasive trophoblasts) that arise in the junctional zone invade the mesometrial compartment, remodel uterine blood vessels, and replace uterine endothelial cells to facilitate greater blood flow to the labyrinth zone (5, 16). Invasive trophoblasts that endovascularize maternal blood vessels or exist in interstitial or perivascular locations are the only trophoblast-origin cells in the metrial gland. Immune cells, specifically natural killer cells, endothelial cells, and uterine decidual cells are the other major constituents of the mesometrial compartment. SpT and TGC in the junctional zone and invasive trophoblasts in the metrial gland are endocrine in nature, secreting a variety of hormones, angiogenic proteins, and tissue remodeling factors. GO analysis of transcripts expressed highly in the junctional zone again clearly indicated hormone activity, female pregnancy, and hormone stimulus to be the most enriched terms. Furthermore, GO analysis of metrial gland genes showed unequivocal enrichment not only for immune function genes but also for vasculature development, highlighting the presence of genes involved in vascular remodeling. Due to the large number of differentially regulated transcripts (>4000 genes) between compartments, we used analysis of GO terms (biological processes), which provides a birds-eye view of the most significant processes. Overall, our results suggest that gene signatures after seq-based expression profiling precisely confirm the widely ascribed functions to placental compartments.
Gene expression to a large extent is controlled by cis-acting transcriptional activators and repressors. Previous reports have examined gene expression profiles in the murine and human placenta using microarrays (8–10, 18, 23). Sood et al. (9) examined gene expression signatures in different areas of the term human placenta, amnion, chorion, and umbilical cord. Each of these was readily distinguished by unique expression patterns. Similarly, monitoring changes in gene expression at the materno-fetal interface (basal plate) over gestation showed only few changes from 14–24 wk of human gestation (10). However, several hundred genes involved in immunity, angiogenesis, extracellular matrix remodeling were altered between midgestation and term. More recently, Knox and colleagues (8, 18) carried out a comprehensive analysis of the gene expression changes associated with placentation in the mouse from dpc 8.5 to postnatal d 0. Their analysis revealed a major molecular transition occurring in midgestation in both the maternal and fetal compartments of the placentation site (8). In the present report, we examined transcription factors based on their expression in different compartments of the placentation site and carried out pathway analysis to identify key networks. Most importantly, using qPCR, we established the relative expression of 27 transcription factors, many of which have not been previously described in the rat placenta. Moreover our analysis clearly identified expression of several factors known to be critical in placental development. Previous studies have shown the requirement of bone morphogenetic protein 2 (BMP-2), C/EBP-β, Tfeb, and PPAR-γ in placental development, especially in SyT differentiation and/or labyrinth zone formation (24–27). Consistent with these reports, our results indicate highest expression of all four transcription factors in the labyrinth zone. In addition, we found several other factors with high expression in the labyrinth zone including FoxO3, β-catenin, Sox7, HDAC-1, and Tead3. Lappas and colleagues (28) recently reported expression of FoxO3 limited to SyT in human placenta. Although forkhead box protein O1 (FoxO1) is essential for vasculature development in the embryo, FoxO3a−/− mice are viable, suggesting that this isoform is not necessary for embryonic vascular development (29). Nonetheless, it is plausible that FoxO3 plays redundant roles in vascular development in the labyrinth zone. BMP and Wnt signaling play crucial evolutionarily conserved roles in embryogenesis. Both BMP-2 and its downstream target Smad-1 are required for extraembryonic development, and recent evidence implicates BMP-2 in promoting angiogenesis via Wnt/β-catenin signaling (27, 30). Canonical Wnt signaling, which requires β-catenin and its DNA-binding partners (members of the TCF family), plays critical roles in blastocyst activation and implantation (31, 32). Mice with Tcf-1/Lef-1 deletions fail to develop a functional placenta due to impaired chorio-allantoic fusion (33). Similarly in mice, Wnt2b and -7b and frizzled family receptor 5 (Fzd5) are critical for either labyrinth zone development or vascularization (34–38). Indeed, our analysis of Wnt and Fzd expression clearly showed high expression of Wnt2b and Wnt11 in the labyrinth zone. However, our results also suggest that in the labyrinth zone of the rat, Fzd4 is expressed at much greater levels than Fzd5. Interestingly, Fzd4 has also been shown to promote angiogenesis and vascularization in the retina (39). Additional studies are needed to address the functional role of Fzd4 in the rat labyrinth zone. The labyrinth zone also showed high expression of SoxF family members. SoxF genes that include, Sox7, -17, and -18 perform pivotal roles in cardiovascular development, orchestrating endothelial cell fate and differentiation in developing heart, blood vessels, and lymphatic vessels (40, 41). Although previously not described in the rat placenta, presumably SoxF members are involved in angiogenesis in the labyrinth zone. Another novel finding from our studies is the identification of higher levels of histone deacetylase-1 (HDAC-1) in the labyrinth zone. Under hypoxic conditions, HDAC-1 has been shown to promote angiogenesis via negative regulation of tumor suppressor genes (42). Furthermore, recent studies implicate inhibition of HDAC-1 in the differentiation of trophoblast stem cells into chorionic and SyT (43). Hence, our RNA-seq-based analysis of labyrinth zone transcription factors revealed several potential regulators with known roles in angiogenesis and vascular development. Further analysis of specific genes enriched in the labyrinth zone is warranted.
A number of developmental signals including Cited-2 and Hand1 are involved in the formation of SpG and TGC of the junctional zone. These have been recently reviewed by Hu and Cross (3). Mice lacking Cited-2 die by GD14.5 and show impaired development of TGC and SpT. Consistent with this, we found high expression of Cited-2 in the junctional zone. Hand1 expression was highest in the labyrinth zone. Hand1 has been implicated in placental development and is essential for placentation and cardiac morphogenesis (44). Mice that lack Hand2 die at embryonic d 10.5 from right ventricular hypoplasia and vascular defects, whereas those lacking Hand1 have lower TGC numbers and a smaller ectoplacental cone (45, 46). Hand2 is expressed in the decidua and is potentially important in decidual development. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2 (47). Interestingly, the junctional zone also showed greater expression of previously unrecognized factors KLF-15, RXR-γ, Pou3f1/Oct-6, and FoxO4. At present, the precise function of these factors in the junctional zone is not clear. Studies in mice suggest that RXR-γ is not essential for placental development. RXR-γ is highly expressed in the thyroid gland and thyrotrope cells in the pituitary, and it is plausible that it is involved in regulating hormone synthesis in the junctional zone (48, 49). KLF-15 is expressed in multiple tissues, including liver, white and brown adipose tissues, kidney, heart, and skeletal muscle. KLF-15 is a glucocorticoid-responsive gene involved in regulation of vascular smooth muscle cells and amino acid catabolism and adipogenesis. Recent studies also suggest that KLF-15 regulates basal levels of low density lipoprotein receptor-related protein 5, which functions as a Frizzled coreceptor in the canonical Wnt/β-catenin pathway (50). The metrial gland showed prominent expression of estrogen and progesterone receptors. In addition, the steroid-responsive homeobox factor HoxA10, which is critical for implantation, was higher in metrial gland (51). We also found higher expression of transcription factors regulating inflammation: IRF-7, Egr-1, and Sox4. IRF constitute a family of transcription factors that possess a helix-turn-helix DNA-binding motif. IRF-7 is ubiquitously expressed and essential for the induction of IFN-α/β genes (52, 53). SOX4 is highly expressed in the thymus and in T lymphocyte and pro-B-lymphocyte lineages. Mice with embryonic deletion of SOX4 show impaired lymphocyte development (54). Although the metrial gland is clearly rich in lymphocytes and other immune cells, the precise role of Sox4 remains to be investigated.
In conclusion, we present a genome-wide transcriptomic view of the three compartments of the rat placentation site. RNA-seq analysis identified that global gene expression is unique in each of the utero-placental compartments. Biological processes enriched in these gene signatures follow the primary functions ascribed to the utero-placental compartments: transport and vasculature in the labyrinth zone, hormone secretion in the junctional zone, and immune interactions in the metrial gland. Most importantly, we identify numerous transcription factors expressed in individual compartments not previously described in the placentation site. These results provide an important new reference to understand the molecular pathways that orchestrate gene expression in the specialized cell types of the rat placentation site.
Acknowledgments
We gratefully acknowledge the assistance of Matt Ferguson and the members of the Arkansas Children's Nutrition Center Animal Research Core Facility for their assistance with total enteral nutrition. We are grateful to Dr. Damir Herman for his suggestions and discussions regarding RNA-seq data analysis. We also thank Ms. Crystal Combs and Michèle Perry for their technical assistance.
These studies were supported in part by the U.S. Department of Agriculture Research Service CRIS 6251-51000-007-04S.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- C/EBP
- CCAAT/enhancer binding protein
- dpc
- day postcoitum
- ER
- estrogen receptor
- FoxO1
- forkhead box protein O1
- Fzd5
- frizzled family receptor 5
- GO
- gene ontology
- HDAC-1
- histone deacetylase-1
- HSD
- honestly significant difference
- IGFBP
- IGF-binding protein
- IRF
- inflammation regulatory factor
- PPAR-γ
- peroxisome proliferator activated receptor-gamma
- qPCR
- quantitative real-time PCR
- RPKM
- reads per kilobase per million mapped reads
- SpT
- spongiotrophoblast
- SyT
- syncytiotrophoblast
- TGC
- trophoblast giant cell
- UTR
- untranslated region.
References
- 1. Cross JC. 2006. Placental function in development and disease. Reprod Fertil Dev 18:71–76 [DOI] [PubMed] [Google Scholar]
- 2. Maltepe E , Bakardjiev AI , Fisher SJ. 2010. The placenta: transcriptional, epigenetic, and physiological integration during development. J Clin Invest 120:1016–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu D , Cross JC. 2010. Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol 54:341–354 [DOI] [PubMed] [Google Scholar]
- 4. Rossant J , Cross JC. 2001. Placental development: lessons from mouse mutants. Nat Rev Genet 2:538–548 [DOI] [PubMed] [Google Scholar]
- 5. Ain R , Canham LN , Soares MJ. 2003. Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: novel endocrine phenotype and regulation. Dev Biol 260:176–190 [DOI] [PubMed] [Google Scholar]
- 6. Pijnenborg R , Robertson WB , Brosens I , Dixon G. 1981. Review article: trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta 2:71–91 [DOI] [PubMed] [Google Scholar]
- 7. Pijnenborg R. 2000. The metrial gland is more than a mesometrial lymphoid aggregate of pregnancy. J Reprod Immunol 46:17–19 [DOI] [PubMed] [Google Scholar]
- 8. Knox K , Baker JC. 2008. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res 18:695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sood R , Zehnder JL , Druzin ML , Brown PO. 2006. Gene expression patterns in human placenta. Proc Natl Acad Sci USA 103:5478–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winn VD , Haimov-Kochman R , Paquet AC , Yang YJ , Madhusudhan MS , Gormley M , Feng KT , Bernlohr DA , McDonagh S , Pereira L , Sali A , Fisher SJ. 2007. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 148:1059–1079 [DOI] [PubMed] [Google Scholar]
- 11. Badger TM , Ronis MJ , Lumpkin CK , Valentine CR , Shahare M , Irby D , Huang J , Mercado C , Thomas P , Ingelman-Sundberg M. 1993. Effects of chronic ethanol on growth hormone secretion and hepatic cytochrome P450 isozymes of the rat. J Pharmacol Exp Ther 264:438–447 [PubMed] [Google Scholar]
- 12. Shankar K , Harrell A , Kang P , Singhal R , Ronis MJ , Badger TM. 2010. Carbohydrate-responsive gene expression in the adipose tissue of rats. Endocrinology 151:153–164 [DOI] [PubMed] [Google Scholar]
- 13. Shankar K , Harrell A , Liu X , Gilchrist JM , Ronis MJ , Badger TM. 2008. Maternal obesity at conception programs obesity in the offspring. Am J Physiol Regul Integr Comp Physiol 294:R528–R538 [DOI] [PubMed] [Google Scholar]
- 14. Shankar K , Kang P , Harrell A , Zhong Y , Marecki JC , Ronis MJ , Badger TM. 2010. Maternal overweight programs insulin and adiponectin signaling in the offspring. Endocrinology 151:2577–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shankar K , Zhong Y , Kang P , Lau F , Blackburn ML , Chen JR , Borengasser SJ , Ronis MJ , Badger TM. 2011. Maternal obesity promotes a proinflammatory signature in rat uterus and blastocyst. Endocrinology 152:4158–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ain R , Konno T , Canham LN , Soares MJ. 2006. Phenotypic analysis of the rat placenta. Methods Mol Med 121:295–313 [DOI] [PubMed] [Google Scholar]
- 17. Hochberg Y , Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med 9:811–818 [DOI] [PubMed] [Google Scholar]
- 18. Knox K , Leuenberger D , Penn AA , Baker JC. 2011. Global hormone profiling of murine placenta reveals Secretin expression. Placenta 32:811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Jonge HJ , Fehrmann RS , de Bont ES , Hofstra RM , Gerbens F , Kamps WA , de Vries EG , van der Zee AG , te Meerman GJ , ter Elst A. 2007. Evidence based selection of housekeeping genes. PloS one 2:e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cenik C , Derti A , Mellor JC , Berriz GF , Roth FP. 2010. Genome-wide functional analysis of human 5′ untranslated region introns. Genome Biol 11:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krzywinski M , Schein J , Birol I , Connors J , Gascoyne R , Horsman D , Jones SJ , Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z , Gerstein M , Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mao J , Zhang X , Sieli PT , Falduto MT , Torres KE , Rosenfeld CS. 2010. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci USA 107:5557–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steingrímsson E , Tessarollo L , Reid SW , Jenkins NA , Copeland NG. 1998. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 125:4607–4616 [DOI] [PubMed] [Google Scholar]
- 25. Barak Y , Nelson MC , Ong ES , Jones YZ , Ruiz-Lozano P , Chien KR , Koder A , Evans RM. 1999. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 4:585–595 [DOI] [PubMed] [Google Scholar]
- 26. Bégay V , Smink J , Leutz A. 2004. Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol Cell Biol 24:9744–9751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H , Bradley A. 1996. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 122:2977–2986 [DOI] [PubMed] [Google Scholar]
- 28. Lappas M , Lim R , Riley C , Menon R , Permezel M. 2010. Expression and localisation of FoxO3 and FoxO4 in human placenta and fetal membranes. Placenta 31:1043–1050 [DOI] [PubMed] [Google Scholar]
- 29. Potente M , Urbich C , Sasaki K , Hofmann WK , Heeschen C , Aicher A , Kollipara R , DePinho RA , Zeiher AM , Dimmeler S. 2005. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest 115:2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tremblay KD , Dunn NR , Robertson EJ. 2001. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128:3609–3621 [DOI] [PubMed] [Google Scholar]
- 31. Xie H , Tranguch S , Jia X , Zhang H , Das SK , Dey SK , Kuo CJ , Wang H. 2008. Inactivation of nuclear Wnt-β-catenin signaling limits blastocyst competency for implantation. Development 135:717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohamed OA , Jonnaert M , Labelle-Dumais C , Kuroda K , Clarke HJ , Dufort D. 2005. Uterine Wnt/β-catenin signaling is required for implantation. Proc Natl Acad Sci USA 102:8579–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galceran J , Farinas I , Depew MJ , Clevers H , Grosschedl R. 1999. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/−Tcf1−/− mice. Genes Dev 13:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ishikawa T , Tamai Y , Zorn AM , Yoshida H , Seldin MF , Nishikawa S , Taketo MM. 2001. Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development 128:25–33 [DOI] [PubMed] [Google Scholar]
- 35. Parr BA , Cornish VA , Cybulsky MI , McMahon AP. 2001. Wnt7b regulates placental development in mice. Dev Biol 237:324–332 [DOI] [PubMed] [Google Scholar]
- 36. Monkley SJ , Delaney SJ , Pennisi DJ , Christiansen JH , Wainwright BJ. 1996. Targeted disruption of the Wnt2 gene results in placentation defects. Development 122:3343–3353 [DOI] [PubMed] [Google Scholar]
- 37. Sonderegger S , Husslein H , Leisser C , Knofler M. 2007. Complex expression pattern of Wnt ligands and frizzled receptors in human placenta and its trophoblast subtypes. Placenta 28(Suppl A):S97–S102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sonderegger S , Pollheimer J , Knöfler M. 2010. Wnt signalling in implantation, decidualisation and placental differentiation: review. Placenta 31:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye X , Wang Y , Cahill H , Yu M , Badea TC , Smallwood PM , Peachey NS , Nathans J. 2009. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell 139:285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Francois M , Koopman P , Beltrame M. 2010. SoxF genes: Key players in the development of the cardio-vascular system. Int J Biochem Cell Biol 42:445–448 [DOI] [PubMed] [Google Scholar]
- 41. Matsui T , Kanai-Azuma M , Hara K , Matoba S , Hiramatsu R , Kawakami H , Kurohmaru M , Koopman P , Kanai Y. 2006. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J Cell Sci 119:3513–3526 [DOI] [PubMed] [Google Scholar]
- 42. Kim MS , Kwon HJ , Lee YM , Baek JH , Jang JE , Lee SW , Moon EJ , Kim HS , Lee SK , Chung HY , Kim CW , Kim KW. 2001. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med 7:437–443 [DOI] [PubMed] [Google Scholar]
- 43. Maltepe E , Krampitz GW , Okazaki KM , Red-Horse K , Mak W , Simon MC , Fisher SJ. 2005. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development 132:3393–3403 [DOI] [PubMed] [Google Scholar]
- 44. Riley P , Anson-Cartwright L , Cross JC. 1998. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet 18:271–275 [DOI] [PubMed] [Google Scholar]
- 45. Firulli AB , McFadden DG , Lin Q , Srivastava D , Olson EN. 1998. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet 18:266–270 [DOI] [PubMed] [Google Scholar]
- 46. Srivastava D , Thomas T , Lin Q , Kirby ML , Brown D , Olson EN. 1997. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16:154–160 [DOI] [PubMed] [Google Scholar]
- 47. Li Q , Kannan A , DeMayo FJ , Lydon JP , Cooke PS , Yamagishi H , Srivastava D , Bagchi MK , Bagchi IC. 2011. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 331:912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Q , Fujii H , Knipp GT. 2002. Expression of PPAR and RXR isoforms in the developing rat and human term placentas. Placenta 23:661–671 [DOI] [PubMed] [Google Scholar]
- 49. Krezel W , Dupe V , Mark M , Dierich A , Kastner P , Chambon P. 1996. RXRγ null mice are apparently normal and compound RXRα+/−/RXRβ−/−/RXRγ−/− mutant mice are viable. Proc Natl Acad Sci USA 93:9010–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J , Yang Y , Jiang B , Zhang X , Zou Y , Gong Y. 2010. Sp1 and KLF15 regulate basal transcription of the human LRP5 gene. BMC Genet 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Satokata I , Benson G , Maas R. 1995. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature 374:460–463 [DOI] [PubMed] [Google Scholar]
- 52. Honda K , Yanai H , Negishi H , Asagiri M , Sato M , Mizutani T , Shimada N , Ohba Y , Takaoka A , Yoshida N , Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 53. Taniguchi T , Ogasawara K , Takaoka A , Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19:623–655 [DOI] [PubMed] [Google Scholar]
- 54. van de Wetering M , Oosterwegel M , van Norren K , Clevers H. 1993. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J 12:3847–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]