Summary
RNA interference (RNAi) has enabled researchers to study the function of many genes. However, it is not understood why some RNAi experiments succeed, while others do not. Here, we show in C. elegans that pharyngeal muscle is resistant to RNAi when initially exposed to dsRNA by feeding, but sensitive to RNAi in the next generation. Investigating this observation, we find that pharyngeal muscle cells as well as vulval muscle cells require nuclear rather than cytoplasmic RNAi. Further, we find in these cell types that nuclear RNAi silencing is most efficiently triggered during early development, defining a critical period for initiating nuclear RNAi. Finally, using heat-shock induced dsRNA expression, we show that synMuv B class mutants act in part to extend this critical window. The synMuv B-dependent early development associated critical period for initiating nuclear RNAi suggests that mechanisms that restrict developmental plasticity may also restrict the initiation of nuclear RNAi.
Graphical abstract
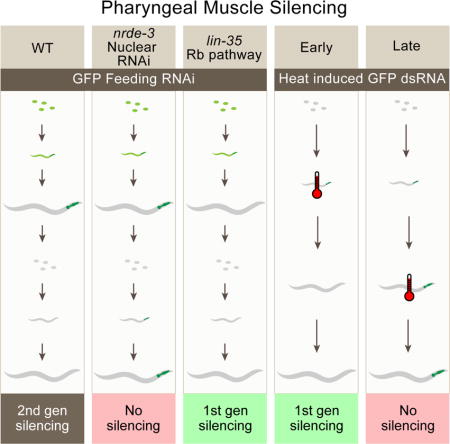
Introduction
RNA interference (RNAi) is a phenomenon in which double-stranded RNA (dsRNA) triggers silencing of cognate genes (Fire et al., 1998). RNAi is a particularly powerful research tool for studying the nematode C. elegans because simply feeding worms bacteria engineered to express gene-specific dsRNA can trigger RNAi, a process known as environmental RNAi (eRNAi) (Timmons and Fire, 1998). In C. elegans, RNAi silencing is associated with transcriptional gene silencing mechanisms (nuclear silencing) and post-transcriptional gene silencing mechanisms (cytoplasmic silencing) (Yigit et al., 2006, Guang et al., 2008).
Cytoplasmic and nuclear silencing processes share upstream dsRNA processing activities, but use divergent silencing effectors. In the common steps, cytoplasmic dsRNA is cleaved by the Dicer complex into short (~22 nucleotide) interfering RNA (siRNA) that bind to the Argonaute (Ago) protein RDE-1, which removes the passenger strand, resulting in a single-stranded primary siRNA that guides the RDE-1 complex to a complementary mRNA (Tabara et al., 1999, Tabara et al, 2002, Parrish and Fire, 2001, Steiner et al., 2009). The mRNA bound RDE-1 complex recruits an RNA-dependent RNA polymerase (e.g., RRF-1) which uses the mRNA as a template to synthesize abundant anti-sense secondary siRNAs (Sijen et al., 2001). In cytoplasmic RNAi silencing, these secondary siRNAs then bind to numerous secondary Argonautes that act redundantly to degrade siRNA targeted mRNA. Quadrupal mutation of the cytoplasmic argonautes sago-1, sago-2, ppw-1 and wago-4 causes strong defects in exogenous RNAi (Yigit et al., 2006). In nuclear RNAi silencing, secondary siRNAs bind the non-redundant Argonautes NRDE-3 and HRDE-1, which act, respectively in the soma and germline. NRDE-3 shuttles secondary siRNA into the nucleus where it complexes with other nuclear RNAi components, including NRDE-2 (Guang et al., 2008). This siRNA complex binds to nascent transcripts complementary to the siRNA to impede RNA polymerase elongation, and subsequently recruits histone methyltransferases to the silenced locus (Tabara et al., 1999). It is assumed that the cytoplasmic (PTGS) and nuclear (TGS) pathways function in parallel to efficiently repress gene expression.
Interestingly, nuclear RNAi is considerably more potent in the F1 progeny than in the P0 worms that were initially exposed to dsRNA (Burton et al., 2011, Zhuang et al., 2013). For example, the phenotype of dpy-11 RNAi is much stronger in the F1 progeny than in the P0 generation, and deposition of H3K9 methylation at the dpy-11 locus is also much higher in the F1 generation than the P0 generation (Burton et al., 2011). However, why nuclear RNAi is more potent in the second-generation remains unknown. One explanation is that germline transmission of silencing signals potentiates silencing ability. For example, it has been proposed that germline transmission might “mark” the locus to be silenced or that a particular siRNA silencing species may be created as a result of germline transmission (Burton et al., 2011). Consistent with the idea that the germline may be particularly efficient at RNAi, transformation of the soma into germline in Class B Synthetic Multivulva (synMuv B) mutants results in a significant increase in RNAi efficiency in the soma. This increase in efficiency accompanies somatic misexpression of germline RNAi genes (Wu et al., 2012).
In addition to the differences in RNAi efficiency between the germline and the soma, particular somatic tissues have different sensitivities to exogenous dsRNA. For example, neurons are generally resistant to RNAi, possibly because they lack the dsRNA uptake channel, SID-1 (Calixto et al., 2010). Indeed, overexpressing SID-1 in neurons enhances neuronal RNAi. Additional tissue-specific enhanced RNAi (Eri) phenotypes in response to particular Eri mutants (e.g., rrf-3 vs. eri-1) have been described, indicating that differing activity levels of endogenous small RNA metabolism genes compete to varying degrees with exogenous RNAi (Zhuang and Hunter, 2011). Finally, some groups have reported strikingly different silencing results for the same gene in the same tissue. For example, multiple groups have reported that the pharynx is resistant to RNAi (Kumsta and Hansen, 2012, Ashe et al., 2015), while others report success in knocking down gene expression in the pharynx (Horner et al., 1998, Winston et al., 2002). Understanding the differences in RNAi efficiency between different RNAi protocols and between different tissues in C. elegans may eventually shed light on the endogenous role of RNAi in C. elegans. Furthermore, understanding the differences in RNAi efficiency between tissues is essential for C. elegans researchers using RNAi as an experimental tool.
In this study, we examined the sensitivity of pharyngeal muscle cells to exogenous RNAi. We found that feeding RNAi fails to silence GFP expression in pharyngeal muscle of worms in the P0 generation but efficiently silences GFP expression in their progeny. The silencing in the progeny requires nuclear RNAi, but contrary to expectations, maternal RNAi activity is not required for silencing. Specifically, we used a heat-shock inducible promoter to express dsRNA at particular developmental time points, identifying a critical early developmental period for nuclear RNAi: earlier exposure to dsRNA results in stronger silencing in pharyngeal muscle cells. Furthermore, we find that vulval muscle cells are also dependent on nuclear RNAi for silencing and this silencing also has a critical period. Finally, we show that Eri mutants, including those in the synMuv B class, extend the critical period for pharyngeal muscle silencing. Together, our results suggest the possibility of an unexpected link between development and silencing by RNAi in C. elegans.
Results
Pharyngeal muscle is resistant to exogenous dsRNA by feeding
It has been reported that the pharynx is strongly resistant to eRNAi (Kumsta and Hansen, 2012, Ashe et al., 2015, Devanapally et al., 2015). To confirm these observations, we exposed worms expressing GFP in the pharyngeal and body-wall muscle to feeding RNAi targeting GFP. We found that pharyngeal muscle cells are indeed resistant to eRNAi (Figure 1A, 1B). Embryos hatched and grown to adulthood on GFP RNAi food displayed dim body-wall muscle, indicating that eRNAi was effective (Figure 1A, middle panel), but bright pharyngeal muscle, confirming that pharyngeal muscle cells are not responsive to eRNAi.
Figure 1.
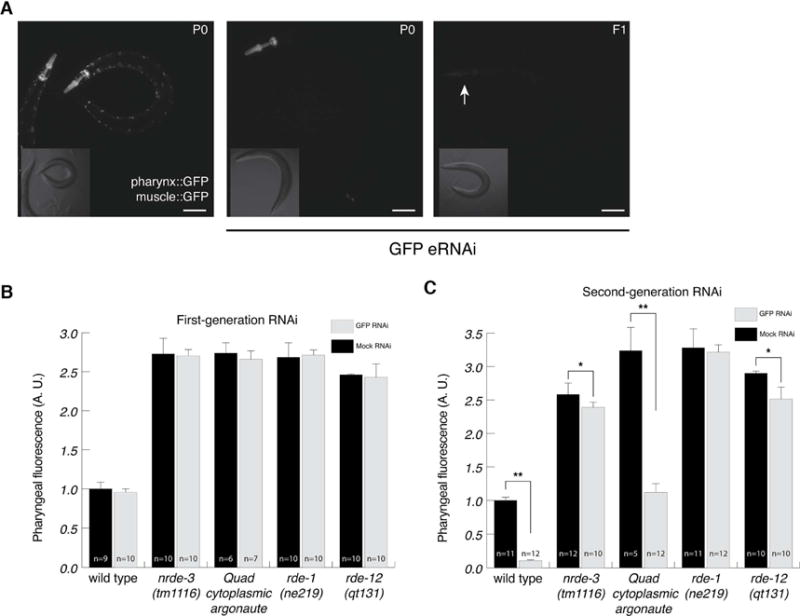
Pharyngeal muscle cells are sensitive to second-generation RNAi and require nuclear RNAi. (A) The pharyngeal muscle is defective for first-generation RNAi, but competent for second-generation RNAi. Representative photos of worms not exposed to dsRNA (left), an adult grown on GFP RNAi from hatching (first-generation RNAi, middle), and the F1 progeny of worms grown on GFP RNAi (second-generation RNAi, right). Scale bars represent 10 μm. (B and C) Quantified fluorescence intensity of pharyngeal muscle following first-generation (B) or second-generation (C) feeding RNAi. Wild type on mock RNAi is defined as 1 arbitrary unit (A. U.). Error bars show standard deviation, n= number of animals scored (*p<.05, ** p<.01).
Although our results are consistent with previous reports that pharyngeal muscle is resistant to RNAi, other researchers have reported effective RNAi in the pharynx (Horner et al., 1998, Winston et al., 2002). Interestingly, when we examined the F1 progeny of worms fed bacteria expressing GFP dsRNA, these worms had dim pharyngeal muscle (Figure 1A, 1C). To avoid confusion with the term “heritable RNAi”, which is increasingly associated with hrde-1-dependent silencing, in this paper we term this “second-generation RNAi.” Worms initially exposed to ingested dsRNA are worms exposed to “first-generation RNAi.” Thus, pharyngeal muscle cells are sensitive to second-generation RNAi, but not to first-generation RNAi. The pharyngeal muscle’s particular resistance to first-generation RNAi might arise if the pharyngeal muscle is competent at particular aspects of the RNAi pathway but not others.
Nuclear RNAi is required for second-generation pharyngeal silencing
Distinct genes act in diverse RNAi pathways. To test the role of specific RNAi components in first- and second-generation silencing of the pharyngeal muscle, we crossed pharyngeal and body-wall muscle GFP reporters into several RNAi mutants and then placed them onto bacteria expressing GFP dsRNA for one or two generations.
Consistent with the known role of these four genes in transgene silencing (Yigit et al., 2006, Grishok et al., 2005, Fischer et al., 2013, Yang et al., 2014, Shirayama et al., 2014, Shiu et al., 2014), expression of the GFP reporter was significantly enhanced in all four mutants, but in no mutant was significant pharyngeal silencing in the first-generation observed (Figure 1B). RDE-1, an Argonaute essential for primary siRNA maturation (Steiner et al., 2009, Sijen et al., 2001), is required for second-generation pharyngeal muscle silencing (Figure 1C). Likewise, RDE-12, which acts downstream of primary siRNA production, and is required for secondary siRNA amplification (Yang et al., 2014, Shirayama et al., 2014, Shiu et al., 2014), is required for strong second-generation pharyngeal muscle silencing. However, consistent with our previous report that rde-12 is dosage-sensitive (Yang et al., 2014), there was some statistically significant rde-12-independent silencing (Fig 1C).
Downstream of primary and secondary siRNA production are two parallel silencing pathways: nuclear and cytoplasmic RNAi. Cytoplasmic RNAi, which acts in parallel to nuclear RNAi, uses multiple redundant worm-specific Ago proteins to silence gene expression. Although a quadruple cytoplasmic Ago mutant [sago-1 (tm1195), sago-2 (tm894), ppw-1(tm914), wago-4(tm1019)] was, as expected, completely resistant to strong RNAi foods targeting act-5 (Figure S1) and unc-22 (Yigit et al., 2006), we found that it was sensitive to second-generation RNAi in the pharyngeal muscle (Figure 1C). However, as reported previously (Yigit et al., 2006), this strain is not fully resistant to eRNAi, as GFP is silenced in body-wall muscle cells (Table S1). This RNAi sensitivity likely represents either other muscle-expressed cytoplasmic Ago proteins and or compensating nuclear RNAi. Nuclear RNAi requires NRDE-3, an Ago protein that shuttles siRNA into the nucleus. We found that, although nrde-3 is not required for second-generation RNAi in body-wall muscle cells (Table S1), it is required for second-generation RNAi in the pharyngeal muscle (Figure 1C). Similar to rde-12, there was some statistically significant nrde-3 independent silencing (Fig 1C). These observations suggest that, in the absence of compensating cytoplasmic silencing, nuclear silencing is required for pharyngeal RNAi. Further, it is likely that the requirement for second-generation RNAi to effectively silence gene expression in pharyngeal muscle cells reflects this dependence on nuclear RNAi.
A critical period for nuclear RNAi
One difference between first-generation RNAi and second-generation RNAi is that in second-generation RNAi, dsRNA passes through the germline and into the progeny. It has been proposed that this germline transmission of silencing signals may promote nuclear RNAi (Burton et al., 2011), and may possibly even be required for silencing. Alternatively, it is possible that because we cannot feed embryos dsRNA before they hatch, that nuclear RNAi may simply require early, pre-hatching, exposure to dsRNA and/or siRNAs to establish silencing. To distinguish between these possibilities, we used a heat-shock inducible promoter, hsp-16.2, to induce GFP dsRNA at different developmental time-points. The hsp-16.2 promoter is active in a variety of tissues, including the pharynx (Stringham et al., 1992, Fire et al., 1990). To test the suitability of using the hsp-16.2 promoter to drive GFP-hairpin expression at successive developmental times, we first measured pharyngeal muscle GFP fluorescence intensity from a hsp-16.2∷GFP construct 24 hours after heat-shock. We found that the promoter is activated at all tested developmental time points, but more GFP fluorescence is detected in the pharynx and in non-pharyngeal tissues of late larvae and adults than in young larvae (Figure S2A; and data not shown). Although it is difficult to compare promoter activity between young larvae and adult worms, because of changes in volume and/or translation efficiency, we note that the significantly higher pharyngeal fluorescence intensity in older worms suggests that it is very unlikely that the hsp-16.2 promoter is more active in young worms than L4 larvae or adults.
We first obtained synchronized populations of hsp-16.2p∷gfp-hp; myo-2p∷gfp worms at successive developmental time points, then heat-shocked these worms. These worms were then returned to 20C for two days and then scored for GFP silencing by counting the fraction of their pharyngeal muscle that lacked GFP expression (Figure S2C).
If maternally produced and deposited silencing signals are strictly required for pharyngeal muscle silencing, then we expect no silencing, since we are examining heat-shocked worms themselves, rather than their progeny. In contrast, the critical period hypothesis posits that earlier exposure to dsRNA should lead to stronger silencing than later silencing. Our data is consistent with the hypothesis that there is an early critical period for nuclear RNAi. Heat-shock induced GFP dsRNA expression in embryos, directly after they were laid, generated the strongest silencing (Figure 2A). The response to heat-shock induced GFP dsRNA expression decreased as the worms aged; the older the worms were at the time of heat shock-induced GFP dsRNA expression, the less the pharyngeal muscle was silenced. Indeed, induction of GFP dsRNA expression after the L3 stage caused virtually no silencing (Figure 2A). Additionally, this data confirms that a maternal contribution is not required for pharyngeal silencing.
Figure 2.
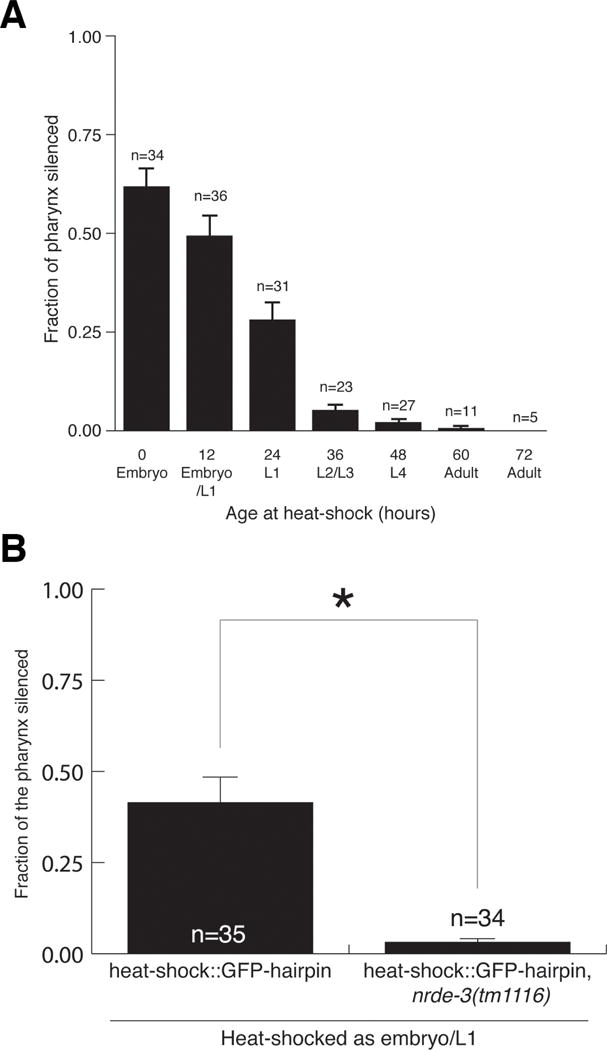
A critical period for nuclear RNAi. (A) Earlier dsRNA exposure results in stronger pharyngeal silencing. Bars show SEM, n = number of animals scored. (B) Pharyngeal RNAi by heat-shock-induced GFP dsRNA requires nrde-3-dependent nuclear RNAi. Bars show SEM from three trials (P<.05). (A and B) Silencing of individual sections of the pharyngeal muscle was scored.
An alternate explanation for the above result is that GFP protein is unusually stable in the adult pharyngeal muscle. Therefore, we used reverse transcription quantitative PCR to directly measure GFP mRNA levels following heat-shock induced GFP dsRNA expression. We compared GFP mRNA levels between heat-shocked worms with and without the hsp-16.2p∷GFP dsRNA array 48 hours post-heat shock. We note that presence of the GFP dsRNA array, similar to RNAi mutants, caused de-silencing of myo-2∷GFP, possibly due to titration of silencing factors. We found that GFP mRNA levels were strongly reduced by heat-shock induced GFP dsRNA expression in embryos (Figure S3). However, consistent with measured GFP fluorescent intensity, inducing GFP dsRNA expression in L4 worms did not result in a decrease in GFP mRNA (Figure S3). Thus, the observed critical window is the result of differences in stage-specific RNAi efficiency, not GFP stability. Finally, the reduced silencing observed at later developmental times is not due to poor hsp-16.2 promoter activity at these times; this promoter is actually more active within the pharynx and other tissues in older worms than young worms (Figure S2). Together, these results support the hypothesis that early exposure to dsRNA more readily initiates RNAi than later exposure to dsRNA.
Nuclear RNAi is required to silence pharyngeal GFP in response to second-generation feeding RNAi. To determine whether nuclear RNAi is also required for pharyngeal silencing from heat-shock induced GFP dsRNA, we tested nrde-3 (tm1116) worms in this assay. In contrast to wild type, induction of GFP dsRNA via heat-shock did not result in pharyngeal silencing in nrde-3(tm1116) embryos and L1 larvae (Figure 2B). Thus, we propose that for effective nuclear RNAi in the pharyngeal muscle, dsRNA must be delivered before or during a critical period, corresponding to embryonic and early larval development.
We initially examined pharyngeal muscle expression from an integrated transgenic array, mIs11[myo-2p∷GFP + pes-10p∷GFP + F22B7.9p∷GFP]. Complex transgenic arrays are often silenced by RNAi (e.g., Figure 1B). To determine if our results held for non-array transgenes, we repeated our experiments with a single-copy integrated myo-2∷GFP transgene (Norris et al., 2015, and see experimental procedures). We found that, like the multi-copy array, the single-copy insertion is resistant to first-generation silencing, sensitive to second-generation silencing, and that this silencing depends, at least in part, on nrde-3 (Figure S4A). Furthermore, we found that early exposure to dsRNA by heat-shock generates stronger silencing than late exposure (Figure S4B and S4C). These results eliminate complications arising from the structure or nature of the complex multi-copy array as being responsible for the critical period and dependence on nrde-3.
A critical period in vulval muscle silencing
While examining the efficiency of GFP silencing in the body-wall muscle using a myo-3∷GFP transgene, we noticed that strong silencing in the vulval muscle was dependent on nrde-3 (Table S1, Figure 3A, B). When exposed to first-generation GFP feeding RNAi 90% of wild-type animals had silenced vulval muscle cells, while no nrde-3 (tm1116) animals had silenced vulval muscle cells (Figure 3B). To study this further, we examined a second GFP transgene, arg-1∷GFP, which expresses GFP in the vm1 and vm2 vulval muscle cells as well as the head mesodermal cell and four enteric muscles (Kostas and Fire, 2002). We found that silencing of this arg-1∷GFP transgene in vulval muscle cells also was dependent on nrde-3 (Figure 3A, B).
Figure 3.
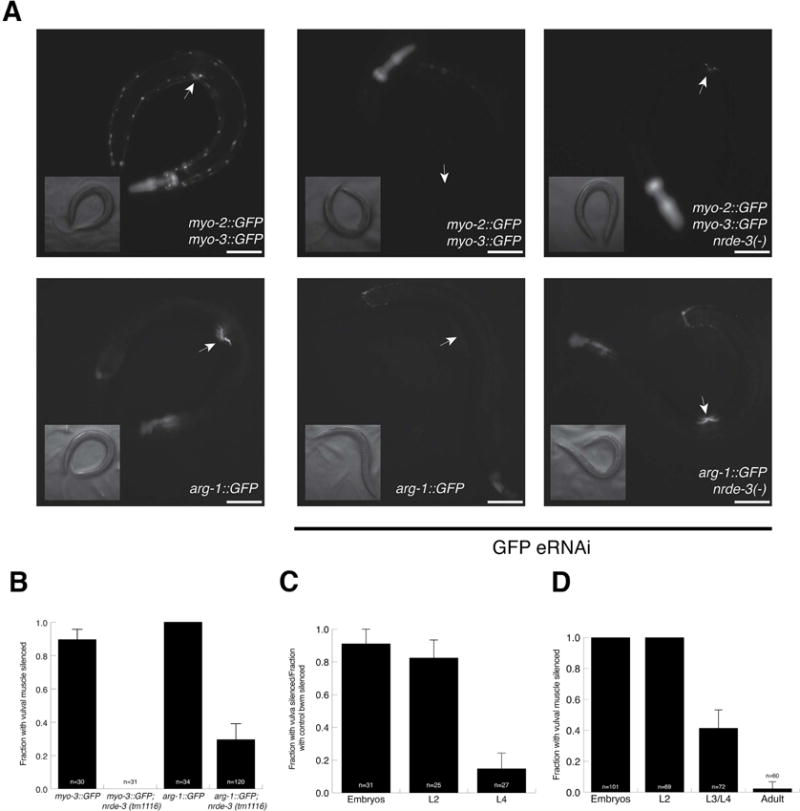
Vulval muscle cell silencing requires nuclear RNAi and has a critical period. (A) myo-3∷GFP (top) and arg-1∷GFP (bottom) are expressed in vulval muscle cells (left panels), are silenced by first-generation feeding RNAi (middle panels), which requires nuclear RNAi (right panels). Arrow points to the vulva. Scale bars represent 10 μm. (B) Vulval muscle cell silencing requires nuclear RNAi. (C and D) A critical period for vulval muscle cell silencing. In (C), two body-wall muscle cells directly anterior to the vulval muscle cells were used as an internal control for RNAi efficiency. (B–D) n = number of animals scored, bars show standard deviation.
Although these two transgenes could be silenced in the vulval muscle cells in the first-generation, because their silencing is dependent on nrde-3 we wondered whether vulval muscle silencing might also have a critical period. To test this, we fed early- and late-staged worms bacteria expressing GFP dsRNA for three days and examined silencing in the vulval muscle. Indeed, consistent with the critical period hypothesis, feeding RNAi initiated after the L4 larval stage resulted in little to no silencing (Figure 3B, C). In the myo-3∷GFP array, we used the two body-wall muscle cells directly anterior to the vulval muscle as a convenient internal control (Figure 3B), demonstrating that this defect in silencing is particular to the vulval muscle cells, which specifically require nrde-3 for silencing.
Enhanced RNAi mutants extend the pharyngeal critical period
Enhanced RNAi (Eri) mutants are thought to reflect greater sensitivity to low dsRNA concentrations. However, given the temporal constraints revealed by our discovery of a critical period to induce nuclear RNAi silencing in pharyngeal muscle cells, we hypothesized that Eri mutants may also act to broaden the critical period. To test this hypothesis, we crossed our hsp∷GFP dsRNA construct into eri-1 and lin-15ab mutants. ERI-1 is required to produce endogenous primary siRNAs that act in the Ergo and Ago-3/4 RNAi pathways (Duchaine et al., 2006, Gabel and Ruvkun 2008, Han et al., 2009). The resulting secondary siRNAs are thought to compete with exogenous secondary siRNAs for limiting RNAi machinery, including secondary cytoplasmic and nuclear Argonautes (Gabel and Ruvkun, 2008). In contrast, loss of lin-15b or lin-35, and other members of the synthetic Multivulva class B genes, results in a soma-to-germline transformation (Wu et al., 2012). This transformation results in a variety of misexpressed genes, including a number of germline RNAi factors, which is thought to cause an Eri phenotype (Wu et al., 2012).
We found eri-1 and lin-15ab mutants extend the detected pharyngeal muscle critical period (Figure 4A). When a population of L2 and L3 worms were heat-shocked, the pharynxes of eri-1 and lin-15ab animals were significantly more silenced than those of wild type. lin-15ab mutants also enhanced the proportion of silenced pharyngeal muscle in the earliest time, suggesting that in addition to expanding the critical period, they increased RNAi sensitivity. However, when animals were heat-shocked as L4s and young adults eri-1 and wild-type animals showed a similar minimal silencing, while lin-15ab mutants showed significantly more silencing (Figure 4A). This indicates that the effect of eri-1 mutants to extend the critical period may be limited to enhanced sensitivity to dsRNA, while lin-15ab mutants also extend the boundary of the critical period into later development. Because eri-1 and lin-15ab mutants both extend the sensitivity and/or breadth of the critical period, we hypothesized that these mutants might be sensitive to first-generation feeding RNAi. Indeed, while first-generation feeding RNAi failed to silence pharyngeal GFP in wild-type animals both eri-1 and lin-15ab mutants displayed strong first-generation RNAi pharyngeal silencing (Figure 4B).
Figure 4.
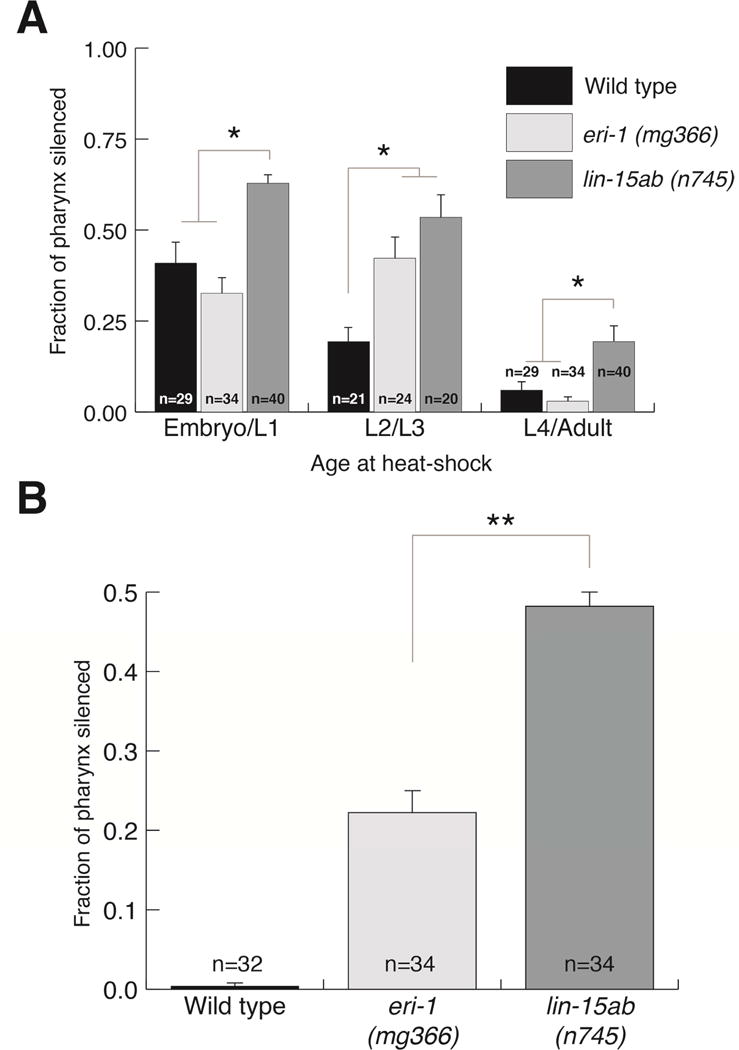
Enhanced RNAi mutants extend the critical period. (A) Pharyngeal silencing of the mIs11 integrated array in control and Eri mutants (B) Eri mutants are sensitive to feeding RNAi in the first generation. (A and B) Silencing of individual sections of the pharyngeal muscle was scored. Bars show SEM from three trials, n= number of animals scored (P<.01).
The enhanced RNAi phenotype of synMuv Class B mutants depend on nuclear RNAi
The Eri phenotypes of eri-1 and rrf-3 is known to depend on nuclear RNAi (Guang et al., 2008, Zhuang et al., 2013). Although pharyngeal silencing in wild type is dependent on nuclear RNAi, the synMuv B Eri phenotype could result from either or both enhanced nuclear RNAi or re-activated cytoplasmic RNAi. Therefore, we asked whether the Eri phenotype of a lin-35 mutant depends on nuclear RNAi. Nrde-3 is thought to function exclusively in the soma (Guang et al., 2008, Burton et al., 2011). To test both germline and somatic RNAi targets, we used nrde-2 mutants for these tests. We exposed wild-type or mutant worms to first-generation RNAi against a panel of targets known to require enhanced RNAi. Indeed, we found that the Eri phenotype of lin-35 was strongly dependent on nrde-2 (Figure 5 A–F). For example, first-generation cel-1 RNAi in lin-35 worms caused these worms to become sterile, but lin-35; nrde-2 worms had significantly lower levels of sterility (Figure 5A). Furthermore, first-generation his-14 RNAi caused lin-35 worms to arrest prior to the L3 stage, but lin-35; nrde-2 worms did not arrest (Figure 5B). Interestingly, his-14 RNAi exposed lin-35; nrde-2 worms were sterile (Figure 5C), suggesting that loss of synMuv B genes may also enhance cytoplasmic RNAi.
Figure 5.
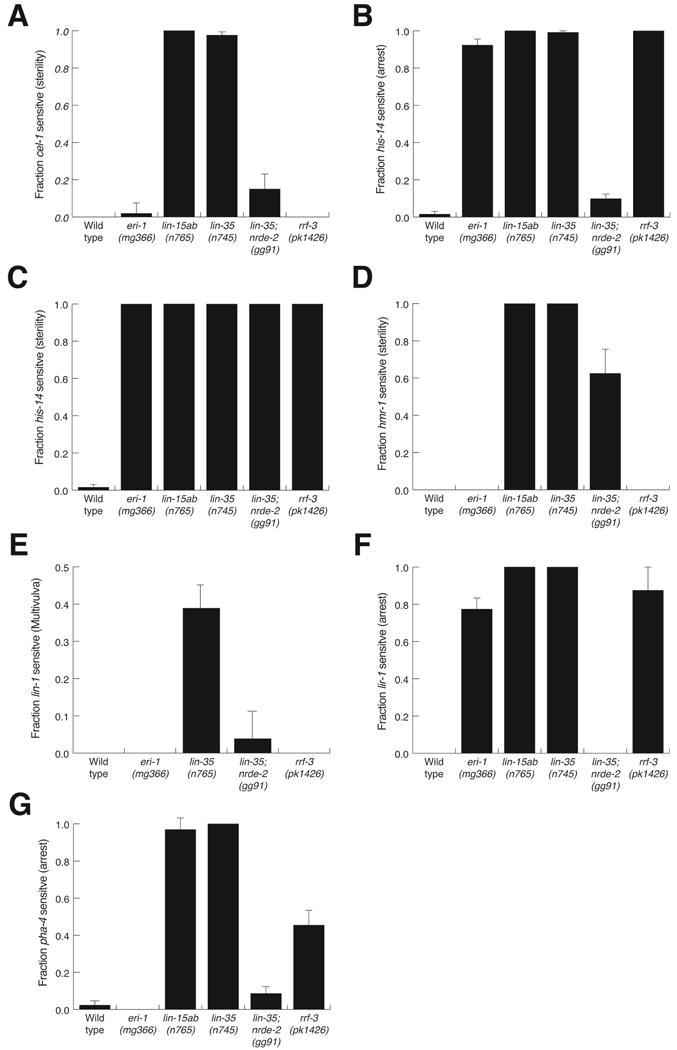
nrde-2 dependent single-generation RNAi in synMuv B class mutants. (A–G) The fraction of single and double-mutants that displayed the indicated phenotypes when placed as embryos on E. coli expressing the listed dsRNA. Bars show SEM from three trials, n>45 animals tested.
Pha-4 RNAi requires nuclear RNAi and is enhanced by synMuv B mutants
To confirm that the genetic requirements for silencing pharyngeal GFP extend to endogenous genes, we examined the genetics of pha-4 RNAi silencing. PHA-4 is a FoxA transcription factor that specifies pharyngeal organ identity (Horner et al., 1998, Mango et al., 1994). Although pha-4 is required embryonically and post-embryonically, wild-type worms exposed to pha-4 dsRNA from hatching develop to adulthood (Figure 5G). However, like pharyngeal GFP RNAi, second-generation pha-4 RNAi results in a strong phenotype: larvae do not develop past the L2 stage. Furthermore, this second-generation RNAi requires nuclear RNAi (Table S1). Additionally, like pharyngeal GFP RNAi, loss of synMuv B genes confers sensitivity to first-generation RNAi (Figure 5G, Table S1). In contrast to GFP silencing, pha-4 second-generation RNAi also requires cytoplasmic Argonautes (Table S1).
Discussion
In this study we examined the requirement for efficient pharyngeal and vulval muscle RNAi. We discovered that pharyngeal RNAi is highly dependent on nuclear RNAi silencing, and that this silencing mechanism imposes unique temporal requirements for dsRNA delivery. We also found that two continuous generations of ingested dsRNA exposure are required for detectable eRNAi in pharyngeal muscle cells. However, this requirement does not necessarily reflect two generations of silencing. Rather, the maternal delivery of dsRNA is required to initiate nuclear RNAi during a critical early developmental period. Additionally, vulval muscle cell silencing is also dependent on nuclear RNAi and also displays a critical period. Unexpectedly, some Eri mutants extend the boundaries of the pharyngeal critical period, enabling single-generation nuclear RNAi in the pharynx and other tissues.
Pharyngeal RNAi
We hypothesize that for most tissues, cytoplasmic and nuclear RNAi work in parallel and redundantly, enabling efficient RNAi. For example, rde-1-dependent body wall muscle cell silencing can occur in the absence of either the cytoplasmic or nuclear pathways (Figure S1). In contrast, we found that RNAi silencing in the pharyngeal muscle is dependent on nrde-3-dependent nuclear RNAi and may be independent of cytoplasmic Ago-dependent processes (Figure 1C). Therefore, one hypothesis to explain the nrde-3-dependence of pharyngeal silencing is a lack of redundant cytoplasmic silencing factors in the pharyngeal muscle. Blazie et al. recently performed tissue-specific RNA-Seq in mixed-staged worms comparing gene expression between the pharyngeal muscle, body-wall muscle and intestine (Blazie et al., 2015). When we examined their data, we found that cytoplasmic argonaute mRNAs (sago-1, sago-2, and ppw-1) were detected in the body-wall muscle and intestine samples, but were not detected in the pharyngeal muscle (Table S2) (Blazie et al., 2015). The lack of these three cytoplasmic argonautes is known to confer strong resistance to RNAi targeting the body wall muscle expressed unc-22 gene (Yigit et al., 2006). If the pharyngeal muscle is resistant to eRNAi because it lacks these three cytoplasmic argonautes, then restoring their expression in pharyngeal muscle cells should restore sensitivity to first-generation RNAi. However, co-expressing sago-1, sago-2 and ppw-1 under the pharyngeal-specific myo-2 promoter did not enable first generation eRNAi (data not shown), possibly because other cytoplasmic RNAi factors are also not expressed in the pharyngeal muscle.
A critical period for nuclear RNAi
We note that there is a graded temporal response to nuclear RNAi, with early exposure to dsRNA triggering stronger nuclear RNAi. Eventually, exposure to dsRNA is insufficient to trigger efficient silencing, defining a critical period for nuclear RNAi. The observation that nuclear RNAi must be initiated during an early critical period likely explains the lack of first-generation pharyngeal silencing. In first-generation eRNAi exposure of cells to ingested dsRNA is delayed, as the consumed dsRNA must be transported into the intestine, exported from the intestine, and finally imported into the recipient cells. By the time sufficient dsRNA accumulates in the pharyngeal muscle, the critical period has passed. The observation that second-generation eRNAi requires SID-1 activity in the mother (Table S1) suggest that maternally acquired dsRNA may be deposited in the embryo. Consequently, silencing can be initiated before the embryo hatches and begins consuming dsRNA itself. Similarly, heat-shock induced dsRNA expression in embryos and early larvae promotes immediate expression and accumulation of abundant dsRNA directly in the pharyngeal muscle cells. The abundance of heat-shocked induced dsRNA may explain the first generation post-embryonic silencing observed in some larvae. Consistent with this hypothesis, extending the critical window through loss of lin-15ab enables efficient first-generation silencing (Figure 4B).
It has been observed that progeny of worms exposed to dsRNA have greater quantities of H3K9 trimethylation marks within the silenced gene than the worms initially exposed to dsRNA (Burton et al., 2011). This suggested that there might be a requirement for germline transmission for efficient nuclear RNAi. Our heat-shock data demonstrates that there is no strict requirement for germline transmission of silencing signals for efficient nuclear RNAi, however, we cannot rule out the possibility that germline transmission might potentiate silencing. We propose that a critical period determines the strength of nuclear RNAi. By depositing dsRNA and perhaps abundant secondary siRNAs directly in the fertilized embryo, germline transmission of dsRNA allows for early initiation, and therefore, more potent nuclear RNAi.
An alternative model for synMuv B enhanced RNAi
An open question is why loss of synMuv B genes causes an Eri phenotype. SynMuv B genes, such as lin-15b and lin-35, are chromatin factors that function to repress gene expression, including germline gene expression. It has been postulated that transcriptional misregulation in lin-35 or lin-15b mutants result in their Eri phenotype (Wu et al., 2012). Four RNAi genes in particular, C04F12.1/vsra-1 (which encodes a cytoplasmic Argonatue), sago-2 (which encodes a cytoplasmic argonaute), rrf-2 (encoding a RNA-dependent RNA polymerase) and hrde-1 (encoding a nuclear RNAi argonaute expressed in the germline), are upregulated in the soma of synMuv B mutants. Curiously, hrde-1 mutants are weakly enhanced for RNAi, and this RNAi enhancement, like the eri-1 phenotype, is additive with synMuv B mutations (Wu et al., 2012). Thus, it is likely that, similar to eri-1 and rrf-3, hrde-1 dependent processes compete with exogenous RNAi pathways. Single loss of any of the other factors, C04F12.1/vsra-1, sago-2 and rrf-2, in conjunction with lin-35 or lin-15b caused at most a mild reduction in RNAi efficiency (Wu et al., 2012). Furthermore, overexpression of any of these four factors in the soma did not confer an Eri phenotype. Thus, transcriptional misregulation of these four factors likely does not fully explain the Eri phenotype of synMuv B mutants.
We speculate that the synMuv B Eri phenotype might be a direct result of soma-to-germline transformation, rather than any transcriptional misregulation. During development, chromatin and/or the factors that modify chromatin become less plastic. Because transcriptional gene silencing that accompanies nuclear RNAi may require malleable chromatin, the closing of the nuclear RNAi window may be coupled to developmental process that restrict developmental cell fate plasticity. Further, we hypothesize that in synMuv B mutants, the somatic chromatin and/or the factors that modify it are more germline-like and naïve. We speculate that the same mechanisms that result in closing of developmental plasticity may also play a role in restricting the initiation of nuclear RNAi. Thus, we hypothesize that synMuv B Eri phenotype represents delayed closing of the critical window for initiating nuclear RNAi.
Use of enhanced RNAi strains
In contrast to other reports (Lehner et al., 2006), we previously reported that synMuv B mutants had weaker Eri phenotypes than either eri-1 or rrf-3 (Zhuang et al., 2011). In contrast to this, in this paper we find that lin-35 and lin-15ab generally have stronger nuclear RNAi phenotypes than either eri-1 or rrf-3 (Figure 5). In our previous report, we exposed L3 staged worms to dsRNA and found that lin-15b or lin-35 did not have as robust of an Eri phenotype as either eri-1 or rrf-3. However, by the time worms are at the L3 stage, the critical window for nuclear RNAi may be nearly closed, leaving open only cytoplasmic RNAi silencing. Perhaps this explains why synMuv B worms did not have strong Eri phenotypes in the earlier report. This result highlights how understanding the fundamental aspects of C. elegans RNAi can guide its practical use. For experiments involving young worms, we now suggest using either lin-35 or, since loss of the two is additive, a lin-35; eri-1 double mutant. Furthermore, using a synMuv B Eri should allow for single-generation feeding RNAi screens.
Development, gene silencing and cancer
Why might nuclear RNAi have a critical period? Nuclear RNAi promotes histone H3K9 and histone H3K27 trimethylation, resulting in transcriptional silencing (Burton et al., 2011, Mao et al., 2015). One possibility is that cell division or development is coupled to nuclear RNAi. As embryos develop, developmental plasticity decreases (Mango, 2009, Meister et al., 2011). The critical period of nuclear RNAi may be related to this phenomenon; limiting histone methylation past a particular time may be useful in maintaining a particular developmental fate.
Consistent with the possibility that development and the critical period are linked is the fact that the critical period for vulval muscle extends later than the critical period for pharyngeal muscle. Although the pharyngeal muscle is resistant to first generation silencing, vulval muscle cells are not. However, by L4 and adulthood, feeding RNAi fails to silence GFP expressed in the vulval muscle cells. Interestingly, the vulval muscle cells are not generated until the L4 stage (Sternberg, 2005), thus, they and their precursors likely must remain developmentally plastic until at least this stage. By contrast, pharyngeal muscle cells are born during embryogenesis. Thus, we propose that the nuclear RNAi critical period and developmental plasticity may be functionally linked.
It is possible that this decreasing ability to silence gene expression by histone methylation might be a general rule for development in animals. If so, why might this be advantageous? Stabilizing transcription would prevent aberrant transcriptional profiles. One extreme example of this is cancer. Loss of Rb, the mammalian homolog of lin-35, results in high rates of cancer. We find that loss of lin-35 or lin-15ab results in an abnormal sensitivity to transcriptional silencing late in development. It is possible that Rb, in addition to regulating the cell cycle, may also play a role in regulating gene expression and a critical window for gene silencing. It may be possible that in cancers with loss of Rb, chromatin may be vulnerable to stochastic gene silencing; if tumor suppressor genes are silenced, then this could lead to further cancer progression.
Tissue-specificity in RNAi
One remaining question is why particular tissues are more or less sensitive to exogenous dsRNA. It is known that neurons are refractory to eRNAi unless sid-1 is experimentally overexpressed in them (Calixto et al., 2010). Thus far, the only known role for exogenous RNAi is for defense against viral infection (Schott et al., 2005, Felix et al., 2011). The only virus known to infect C. elegans is restricted to infecting the intestine (Franz et al., 2014). Therefore, it is possible that there is no selective pressure to maintain strong exogenous RNAi in the pharyngeal muscle, vulval muscle cells, or in neurons. There may be other roles for eRNAi (Sarkies and Miska, 2013). Identifying the natural substrates of eRNAi may inform an understanding about why particular tissues are differentially sensitive to dsRNA, while an understanding of these sensitivities may help guide that search.
Experimental Procedures
Feeding RNAi
For GFP RNAi, E. coli expressing either dsRNA targeting GFP or control dsRNA (L4440) was fed to L1 animals on agar plates containing 1 mM isopropyl β-D-1-thiogalactopyranoside (Timmons and Fire, 1998).
For act-5 RNAi, embryos were placed on E. coli expressing act-5 dsRNA, then 3 days later, the fraction of animals reaching adulthood was scored. For GFP RNAi, worms were placed on E. coli expressing GFP dsRNA, then 3 days later, animals were imaged then blindly scored for silencing. All feeding RNAi experiments were performed at 20° C. Bacteria engineered to express gfp dsRNA were prepared as described previously (Winston et al., 2003). All other bacteria expressing dsRNA were from the Ahringer library (Kamath and Ahringer, 2003).
Heat-shock
Adult worms were placed on seeded plates and allowed to lay embryos for approximately 6 or 12 hour time windows and then removed. The collected embryos developed for the specified time at 20C and were then heat-shocked on the growth plates at 34° C for two-hours in an air incubator. Two days post heat-shock, the pharynxes were scored as below for silencing.
Statistics
P-values were calculated using the Student’s t-test.
Live Microscopy
Worms were immobilized for imaging by placing plates on ice for 15–30 minutes. Images being compared in each figure were taken using the same nonsaturating exposure conditions and processed identically using Adobe Illustrator for display. 8-bit images were taken at 10× magnification at 8-bit using an Olympus SZX2 microscope, a Hamamatsu C8484 camera and HCI Imaging Software.
Quantification of pharyngeal fluorescence
The pharyngeal fluorescence was quantified in one of two complementary methods, whole pharynx fluorescence quantification or scoring of individual pharynx sections. In both methods, worms were imaged as above and mIs11 or the single-copy myo-2∷GFP transgene was the only GFP transgene. In the first method, the fluorescence of the entire pharynx was quantified. Worm images were analyzed with Fiji (an ImageJ distribution) by first tracing the pharynx, measuring the average fluorescence intensity and subtracting the background as performed in Gavet and Pines, 2010. We report whole pharynx fluorescence quantification in Figures 1B, 1C, S2A, S4A and S4C
Because silencing in the pharyngeal muscle is often partial and incremental, we also analyzed silencing of individual sections of the pharyngeal muscle, in which we divided the pharynx into eight sections (Fig S2C). Although the pharynx has three-fold rotational symmetry, for convenience of scoring, we divided the pharynx in half corresponding to half of each of the procorpus, metacarpus, isthmus and terminal bulb of the pharynx. The number of each of these sections were scored for strong silencing (Figure S2). The genotype of each worm was blinded from the scorer. We report scoring of individual pharynx sections in Figures 2A, 2B, 4A, 4B, and S4B. We note that these two methods result in similar results (Figures S4B, S4C), while qPCR and scoring of individual sections also are consistent (Figures 2A and S3).
Single-copy myo-2∷GFP transgene
This single-copy insertion myo-2∷GFP transgene (Norris et al., 2015) was generated by CRISPR/Cas9 genome editing, and is inserted at, and disrupts the function of, F32B4.4, which encodes a putative RNA binding protein. There is no evidence that F32B4.4 plays a role in RNAi (Figure S4A and data not shown).
Supplementary Material
Acknowledgments
We thank members of the Hunter Lab for useful discussions and suggestions, including Eddie Wang, Sonya Bhatia, Olga Minkina, Taliesin Lenhart, Alix Weisman, and Xi Ge. We appreciate critical reading of the manuscript by Susan Mango and Stephen Von Stetina. We thank Adam Norris and John Calarco for their generous gift of the myo-2∷GFP single copy insertion. We thank Renate Hellmiss for assistance with figure preparation. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIGMS grant GM089795 to C.P.H. and a NSF Graduate Research Fellowship (Grant DGE1144152) to P.K.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, P.K.S. and C.P.H.; Investigation, P.K.S.; Writing - Original Draft, P.K.S.; Writing-Review & Editing, P.K.S. and C.P.H.
References
- Ashe A, Sarkies P, Le Pen J, Tanguy M, Miska EA. Antiviral RNAi against Orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans. J Virol. 2015 doi: 10.1128/JVI.03664-14. JVI.03664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie SM, Babb C, Wilky H, Rawls A, Park JG, Mangone M. Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol. 2015;13 doi: 10.1186/s12915-015-0116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci. 2011;108:19683–8. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–9. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanapally S, Ravikumar S, Jose AM. Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc Natl Acad Sci U S A. 2015;112:2133–8. doi: 10.1073/pnas.1423333112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–54. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–98. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischer SEJ, Pan Q, Breen PC, Qi Y, Shi Z, Zhang C, et al. Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Genes Dev. 2013;27:2678–95. doi: 10.1101/gad.233254.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz CJ, Renshaw H, Frezal L, Jiang Y, Félix M-A, Wang D. Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology. 2014;448:255–64. doi: 10.1016/j.virol.2013.09.024. [DOI] [PubMed] [Google Scholar]
- Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat Struct Mol Biol. 2008;15:531–3. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing by RNAi in the soma of of a transgene C. elegans. Genes Dev. 2005;19:683–96. doi: 10.1101/gad.1247705.genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–41. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, et al. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–9. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–52. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–21. doi: 10.1016/S1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kostas SA, Fire A. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev. 2002;16:257–69. doi: 10.1101/gad.923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. C. elegans rrf-1 Mutations Maintain RNAi Efficiency in the Soma in Addition to the Germline. PLoS One. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, et al. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–31. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Mango SE. The molecular basis of organ formation: insights from the C. elegans foregut. Annu Rev Cell Dev Biol. 2009;25:597–628. doi: 10.1146/annurev.cellbio.24.110707.175411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Zhu C, Zong D, Weng C, Yang X, Huang H, et al. The Nrde Pathway Mediates Small-RNA-Directed Histone H3 Lysine 27 Trimethylation in Caenorhabditis elegans. Curr Biol. 2015;25:2398–403. doi: 10.1016/j.cub.2015.07.051. [DOI] [PubMed] [Google Scholar]
- Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Curr Opin Genet Dev. 2011;21:167–74. doi: 10.1016/j.gde.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Kim HM, Colaiácovo MP, Calarco JA. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics. 2015;201:449–58. doi: 10.1534/genetics.115.180679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–402. [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Miska EA. Is there social RNA? Science. 2013;341:467–8. doi: 10.1126/science.1243175. [DOI] [PubMed] [Google Scholar]
- Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:18420–4. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Stanney W, Gu W, Seth M, Mello CC. The Vasa homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr Biol. 2014;24:845–51. doi: 10.1016/j.cub.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PK, Zhuang JJ, Hunter CP. Animal Endo-SiRNAs. 2014;1173:71–87. doi: 10.1007/978-1-4939-0931-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–76. doi: 10.1016/S0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–11. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- Sternberg PW. Vulval development. WormBook. 2005:1–28. doi: 10.1895/wormbook.1.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham EG, Dixon DK, Jones D, Candido EP. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–33. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–71. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–9. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Wu X, Shi Z, Cui M, Han M, Ruvkun G. Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 2012;8:e1002542. doi: 10.1371/journal.pgen.1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Vallandingham J, Shiu P, Li H, Hunter CP, Mak HY. The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. Elegans. Curr Biol. 2014;24:832–8. doi: 10.1016/j.cub.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CCG, Tolia NH, et al. Analysis of the C. elegans Argonaute Family Reveals that Distinct Argonautes Act Sequentially during RNAi. Cell. 2006;127:747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zhuang JJ, Banse SA, Hunter CP. The nuclear Argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics. 2013;194:117–31. doi: 10.1534/genetics.113.149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, Hunter CP. Tissue Specificity of Caenorhabditis elegans Enhanced RNA Interference Mutants. Genetics. 2011;188:235–7. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


