Abstract
Summary: The blood-brain barrier (BBB) is an impermeable cellular interface that physically separates the blood from the interstices of the brain. The endothelial cells lining the brain blood vessels form the principle barrier, and their unique phenotype is a consequence of dynamic interactions with several perivascular cell types present in the brain parenchyma. In addition, BBB dysfunction has been observed in the large majority of neurological diseases, but the causes of aberrant vascular behavior are generally unknown. Because of its barrier phenotype, drug delivery to the brain has also proven to be a very difficult task. Global genomics and proteomics analyses are currently being used to examine BBB function in healthy and diseased brain to better characterize this dynamic interface. It is becoming increasingly evident that these approaches have the potential to clarify the unique attributes of a healthy BBB, to identify therapeutic targets in diseased brain, and to identify novel conduits for noninvasive delivery of drugs against these targets. This review will discuss the application of genomics and proteomics to blood-brain barrier research and will offer views on the prospects of such approaches.
INTRODUCTION
The impermeable nature of the blood-brain barrier (BBB) requires it to act as a functional interface between the circulatory system and the parenchyma of the brain. Chemical, physical, cytokine, and cellular cues are transmitted across the blood-brain barrier during normal brain function to maintain homeostasis. In this way, the BBB plays an extremely important role in the regulation of trans-BBB information flow, and in essence functions as a molecular switchboard. In addition to the BBB contribution to normal brain function, BBB involvement has been implicated in a growing number of neurological disease states. This list includes stroke, human immunodeficiency virus, Alzheimer’s disease, brain cancer, and bacterial infections of the CNS, among many others. The BBB also participates in regular immune surveillance of the brain and responds to proinflammatory cytokines to help mediate recruitment and transmigration of immune cells. In pathological conditions, the anatomical attributes of the BBB are oftentimes altered with increases in permeability and restructuring of tight junctional proteins. Although the endothelium is the principle barrier and communication interface, the local microenvironment modulated by perivascular cells including astrocytes, neurons, pericytes, and smooth muscle contributes to BBB function. This collective composite of cells is often referred to as the neurovascular unit and intercellular communication is prevalent.
Although the existence of a BBB was confirmed in the early 20th century, the molecular origins of many of the unique properties of this interface remain elusive. This is partially a result of the inherent complexity of the BBB that results from its intimate interactions with several different cell types. Traditionally, blood-brain barrier studies have been constrained to evaluating the expression behavior and function of a few genes or proteins that are of interest in a particular functional pathway. However as discussed above, many different cells and factors interact synergistically in a time-dependent manner. Individual molecular interactions may eventually affect multiple pathways and BBB functions. In addition, the temporal and spatial progression of BBB involvement in disease is frequently controversial but of paramount importance when designing therapies for neurological diseases.
The relatively recent introduction of gene and protein expression profiling (genomics and proteomics) technologies affords researchers with an unsurpassed opportunity to address questions regarding the BBB. Unlike many biochemical methods that have been applied to the BBB, these techniques are particularly well suited for global molecular analyses of BBB function in health and disease. It is anticipated that these techniques will help elucidate the mechanistic underpinnings of BBB permeability regulation. In addition, these methods could shed light on the process of BBB maturation during development. Of clinical importance, genomics and proteomics approaches could also be used to direct drug development processes by unearthing pathways involved in disease pathogenesis where intervention may be most successful. Finally, genomics and proteomics techniques have the potential to identify candidate brain-specific transport systems that could be used to ferry drug cargo from the blood to the brain as a mode of noninvasive delivery. Ultimately, this last contribution may be very significant given that appropriate targeting and delivery strategies are critical for enabling the translation of basic neuroscience into successful clinical implementation.
In this review, different strategic approaches for genomics and proteomics of the blood-brain barrier will be discussed. To date, several functional genomics studies aimed at identifying the phenotypic determinants of the blood-brain barrier have been performed that have affirmed the rich functional diversity of the BBB. In addition, recent studies applying genomics to BBB response in disease have illuminated several potential therapeutic targets. On the other hand, proteomic studies have been more infrequent in BBB research and are complicated by the fact that membrane proteins are prominent contributors to BBB function. However, membrane protein profiling of the BBB is currently underway. A combination of both genomic and proteomic approaches is rapidly being embraced by the BBB field and promises to spawn a wealth of new views dealing with BBB involvement in both healthy and diseased brain.
BLOOD-BRAIN BARRIER GENOMICS
Source of gene transcripts
At first glance, it may seem that one could simply use mRNA extracted directly from whole brain tissue to analyze the differential expression of BBB transcripts under different physiological states. Such an approach would likely obscure many attributes if the system of interest is solely the BBB and not the entire brain. This is because the BBB comprises only 1/1000 of the total brain volume.1 Therefore, as an approximation, if mRNA were isolated from intact brain, only 1/1000 of the total transcript pool would have originated from the vasculature. In this case, only the most highly expressed BBB transcripts would be examined, and many potentially interesting gene transcript effects would be inadvertently overlooked. In addition, without secondary spatial confirmation of the brain region(s) where the transcript is being differentially expressed, it is difficult to attribute an expression profile differences as originating from the BBB component of total brain mRNA.
Direct isolation of brain capillaries and generation of an mRNA pool from only this subset of brain tissue can overcome this problem. Because the BBB is ensheathed by a durable basement membrane, the vessels can be preferentially extracted from the surrounding brain tissue. This can be accomplished by following mechanical or enzymatic dissociation of brain tissue with size fractionation steps designed to enrich for the capillary fraction, while minimizing recovery of larger microvessel fragments and venules. Although multiple methods of capillary isolation have been demonstrated, it is important to consider mRNA quality when selecting an isolation procedure. If enzymatic isolation is applied to the generation of capillaries, one must take into account the multihour digestion at physiological temperature required to dissociate the capillaries from the surrounding brain tissue. During this time, the cellular transcription program is likely changing rapidly and the resultant mRNA isolate may not be very representative of the actual in vivo situation. Laser-capture microdissection can also be used to selectively remove capillaries and subsequently mRNA from tissue sections. Unfortunately, this method also suffers from drawbacks such as endothelial cell contamination with surrounding tissue, and the fact that gene expression is analyzed by using mRNA derived from localized sites. In contrast, when mechanical homogenization methods are used to dissociate the capillaries from surrounding brain tissue, the complete procedure can be performed at 4°C with little degradation or changes in the mRNA profiles.2
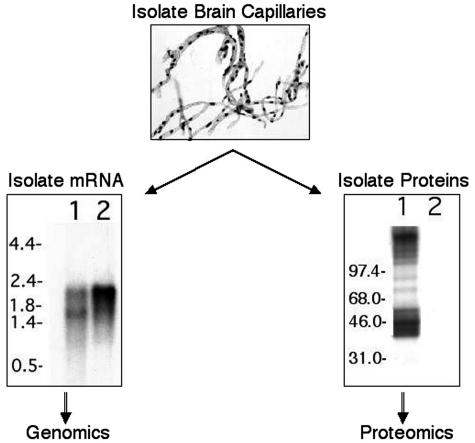
Isolated capillaries that are generated by mechanical homogenization are free of adjoining tissue and include predominantly endothelial cells (FIG. 1). During the isolation procedure, the capillary basement membrane remains intact, and as a consequence the isolates also contain pericytes, which share the basement membrane with the endothelial cells. In addition, smooth muscle cells present on small diameter, precapillary arterioles are inevitably present to some extent.3 This is not particularly problematic as interactions between these cell types clearly contribute to the coordinated function of the neurovascular unit. mRNA can then be readily isolated from these capillary preparations using standard techniques (FIG. 1).2 Although fine-tuning of enzymatic techniques can result in dissociation of pericytes and endothelial cells,4 the mRNA samples may not be representative of the in vivo situation for reasons discussed previously. Once capillary mRNA representative of the in vivo state has been generated by mechanical homogenization techniques, comprehensive gene expression profiling can be performed using one of the methods outlined below.
FIG. 1.
Schematic of BBB genomics and proteomics strategy. Top panel: Mechanically isolated bovine brain capillaries stained with o-toluidine blue. Left panel: Northern blot of bovine brain capillary mRNA (lane 1) and total brain mRNA (lane 2) with a radiolabeled actin probe. Transcript size is given in kilobases. Note the BBB actin signature seen in lane 1 compared with that of total brain seen in lane 2. The presence of an additional low molecular weight smooth muscle actin transcript is due to the presence of smooth muscle in the capillary preparation. Right panel: Western blot of bovine brain capillary proteins with polyclonal antiserum raised against isolated bovine brain capillary membranes. Blot probed with the specific antiserum (lane 1) or control preimmune antiserum (lane 2). Note the large number of putative membrane proteins recognized by the polyclonal antiserum. Molecular masses are indicated in kilodaltons.
Genomics methodologies
Several genomics methodologies are commonly used to profile gene expression. These include gene microarray analysis, suppression subtractive hybridization (SSH), and serial analysis of gene expression (SAGE). Although each technique has its associated advantages and disadvantages, a brief discussion of each method’s applicability to a BBB genomics program is warranted. The gene microarray is a widely used and successful technique that yields relatively quantitative measures of differential gene expression between two independent mRNA samples. One drawback is that most gene chips allow transcriptional profiling of on the order of 104 genes, whereas estimates of transcriptome size for humans are typically in the range of 30,000-40,000 genes.5,6 In addition, it can be difficult to analyze low abundance transcripts such as transcription factors using microarrays,7 and this is particularly important when profiling the BBB as it is a responsive and regulatory signaling interface. In contrast, SSH allows differential profiling of all the transcripts present in the independent mRNA samples without requiring a priori knowledge of gene identity or its presence on a prefabricated gene chip. SSH is also particularly effective in identifying variation in low abundance transcripts due to the careful design of the technique.8 Also, because a library of differentially expressed cDNA fragments is generated in the SSH process, the fragments can be used directly as probes for cloning of full-length cDNA products.9 This is critical for downstream functional analyses of novel clones that are identified in the differential screening process. Although SSH and microarrays are well suited for differential gene profiling between two tissues or tissues in different pathological states, they are generally incapable of generating a full blueprint of the BBB transcriptome. However, SAGE analysis results in a comprehensive catalog of the transcriptome and the method does not rely on a gene being differentially expressed (SSH) or the presence of a gene on a prefabricated microarray chip (gene microarray).10 The SAGE analysis has the power to reveal the identity and relative quantity of all genes expressed in a given tissue.10 Unlike microarray or SSH technologies, SAGE analysis is not sufficient to perform differential profiling on its own. It instead requires SAGE databases of transcript expression in other tissues to allow for quantitative differential comparison. However, SAGE data are being generated for a large number of tissues and disease states and are rapidly being compiled in publicly available databases. Thus, eventually one could imagine performing microarray experiments simply by comparing and contrasting complete transcriptomes generated using SAGE technology. To date, comprehensive gene expression profiling of the blood-brain barrier has been approached using SSH3,9,11 and SAGE12 technologies with the goal of unearthing the molecular origins of unique BBB attributes at the gene expression level. In addition, several studies focused on the molecular mechanisms of disease have used microarray profiling to examine differential gene expression under disease conditions.
SSH analysis of the BBB transcriptome
In a concerted effort to elucidate the phenotypic determinants of the BBB, differential expression analyses of rat and human brain capillaries have been conducted. In these studies, comparative profiling using SSH technology was performed to compare gene expression patterns of the BBB to those exhibited by the kidney and liver. Because the kidney and liver tissues are highly vascularized, these experiments were designed to meet two goals: 1. Identify BBB-specific genes that differentiate the brain vasculature from kidney and liver vasculature, 2. Identify potential BBB targets that are not present in kidney or liver tissue as a whole.
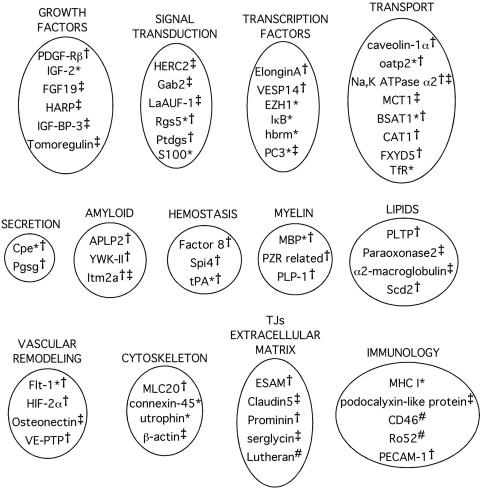
Freshly isolated human or rat capillaries were used to generate full-length mRNA to use in the subtraction process. Subsequently, transcripts commonly expressed in the BBB, liver, and kidney were subtracted using the SSH technique yielding a pool of transcripts with differentially elevated expression at the BBB. The differentially expressed genes resulting from one study regarding the human BBB3 and two studies regarding the rat BBB9,11 have been compiled in Figure 2. The transcripts that encode known proteins are clustered according to putative function to demonstrate the multiple functions orchestrated by the BBB in vivo. The clustering is somewhat arbitrary given that many proteins in each set have multiple functions, yet some themes resonate regardless of a protein placement in a particular functional grouping. The BBB is clearly enriched for tight junction and extracellular matrix proteins that contribute to the impermeable nature of the BBB. In particular, claudin5 has been shown to regulate paracellular transport and is present in tight junctions.13 Recently, claudin5 knockout mice were shown to have impaired BBB function with increased permeability to small molecule tracers, but not to molecules greater than 1.9 kDa.14 Endothelial cell-selective adhesion molecule (ESAM) is another protein that is localized to tight junctions,15 and the SSH study indicated enhanced expression of this protein at the BBB.11 The prevalence of growth factors, signal transduction molecules, and transcription factors extracted in the SSH screen support the hypothesis that the BBB aids in regulating brain plasticity and does so in an inductive manner through the release of growth factors [heparin affinity regulatory peptide (HARP), fibroblast growth factor 19, insulin-like growth factor (IGF)-2, IGF binding protein (IGFBP)-2]. The BBB can also act in an inhibitory mode by expression of osteonectin, a glycoprotein that has been shown to inhibit angiogenesis by binding vascular endothelial growth factor,16 and PC3, an antiproliferative protein induced by the p53 protein.17 The BBB is also rich in expression of molecular transporters and proteins involved in cellular trafficking. The differentially expressed transport systems that were identified in these studies facilitate organic anion transport (oatp2), sodium and potassium transport (Na+,K+ ATPase), energy substrate transport in the form of lactic acid and ketone bodies [monocarboxylate transporter (MCT1)], amino acid transport [cationic amino acid transporter (CAT1)], and protein transport [transferrin receptor (TfR)]. In addition, calveolin-1α is a main component of caveolae18 and was identified as BBB-enriched in the rat study. Ion transport regulator 5 (FXYD5) is also enriched at the BBB and is abundant in other transporting tissues.19 Although several dozen genes with enriched expression at the BBB compared with the liver or kidney were identified, many BBB-enriched clones still remain to be sequenced from the original pool. The estimated number of enriched cDNA clones generated by the SSH process for the human BBB study was 200,000.3 Thus far, only 71 clones have been sequenced and analyzed. Of these 71 clones, only 37 were found to be distinct as several clones were identified multiple times during the sequencing process. However, 25 of the 37 clones were single isolates, indicating from a statistical perspective that many genes remain to be identified from the pool, and that additional clone sequencing will continue to yield distinct BBB-enriched transcripts.
FIG. 2.
Functional clusters of BBB-enriched genes resulting from: rat SSH study 1 denoted by *,9 rat SSH study 2 denoted by †,11 human SSH study denoted by ‡,3 and bovine proteomic study denoted by #.27–29 PDGF-Rβ = PDGF receptor β subunit; Gab2 = Grb-2 associated binder-2; LaAUF-1 = AU-rich RNA binding factor; Rgs5 = G protein signaling regulator-5; Ptdgs = prostaglandin D synthase; VESP14 = vascular endothelial cell-specific protein 14; hbrm = human homolog of yeast SW12 transcription factor; PC3 = B-cell translocation gene-2; BSAT-1 = BBB-specific anion transporter type 1; FXYD5 = FXYD domain-containing ion transport regulator 5; Cpe = carboxypeptidase E; Pgsg = secretory granule proteoglycan core protein precursor; APLP2 = amyloid precursor-like protein 2; YWK-II = sperm membrane protein related to A4 amyloid protein; Itm2a = integral membrane protein 2a; Spi4 = serine protease inhibitor 4; tPA = tissue plasminogen activator; MBP = myelin basic protein; PZR related = protein zero-related protein 1; PLP-1 = proteolipid protein; PLTP = phospholipid transfer protein; Scd2 = stearoyl-CoA desaturase 2; Flt-1 = vascular endothelial growth factor receptor type 1; HIF-2α = hypoxia-inducible factor 2α; VE-PTP = vascular endothelial receptor-type protein tyrosine phosphatase; MLC20 = regulatory myosin light chain isoform C; Ro52 = 52-kDa ribonucleoprotein.
SAGE analysis of the BBB transcriptome
Although the SSH studies above have illuminated many of the differentially expressed classes of genes present at the BBB, the SAGE technology has recently been applied for expression profiling of all expressed BBB genes.12 This technique yields data that contrast with the differential expression data generated by SSH or microarray in that every transcribed product at the BBB can be identified if comprehensive sequencing is performed. Unlike the SSH methodology, the data set includes genes commonly expressed in other tissues, and yields quantitative data about relative expression levels of transcripts at the BBB. Similar to the SSH procedure above, mRNA was isolated from purified rat microvessels and used to prepare a microvessel SAGE library. Sequence analysis of ∼78,000 transcripts from the library identified nearly 11,000 different transcripts that are expressed at the BBB. The catalog of BBB transcripts was compared with compiled SAGE databases for the cerebral cortex and hippocampus, to identify BBB-enriched transcripts. Nearly 700 genes were found to be at least 10-fold enriched at the BBB compared with cortex and hippocampus and the genes clustered into functional categories of transporters (11%), receptors (10%), vesicular trafficking (7%), structural proteins (12%), and signal transduction (18%). The relative distributions of gene products into functional categories also agrees well with the three SSH studies detailed above, suggesting once again that the BBB functions as a molecular switchboard communicating between blood and brain. Although both of these major profiling studies examined BBB function under normal conditions, they are generally applicable to examining changes in gene expression in pathological states and could be used to help dissect molecular mechanisms of disease. Approaches with this goal will be presented in the final section of this review.
Identification of transcripts encoding proteins with unknown function
One of the benefits of using SSH or SAGE technologies is that many of the genes identified as BBB-enriched are considered novel. That is, the function of the protein that the gene encodes is unknown, and even putative functional assignment based on gene homology is rarely possible. In the three SSH case studies above, 45% (rat),9 49% (rat),11 and 47% (human)3 of the genes that were identified to be BBB-enriched encoded proteins with unknown function. The fact that nearly 50% of the BBB-enriched genes encode proteins with novel function is quite exciting. However, elucidation of the molecular functions of these proteins is one of the biggest challenges in the functional genomics field. Even in the cases where one can identify a protein as a putative kinase or transcription factor, determining the substrates regulated by these proteins is not trivial. Although it may seem that nearly 50% of transcripts identified as BBB-enriched are of little use because their function is not known, this is clearly not the case. The following case study helps to illustrate this point.
Novel transporter discovery via genomics approaches
Because the BBB is quite impermeable, it helps regulate the molecular and cellular communication between the blood and the brain. Thus, one might expect the presence of an inordinately large number of molecular transporters that regulate bidirectional transport of nutrients between the luminal and abluminal membranes ofthe endothelium. This appears to be the case when SSH/SAGE results are compared with functional analyses generated by the human genome project. It has been estimated that ∼3% of the proteins encoded by the human genome function as molecular transporters.6 Thus, of the 30,000–40,000 predicted proteins in the human genome, around 1000 of these are transporters. In the SSH and SAGE studies, transporters are observed at a 10-15% frequency3,9,11,12 in the list of identified transcripts with known function. Therefore, it is likely that a significant portion of the genes encoding proteins with unknown functional roles are also molecular transporters.
The types of transporters identified in the genomics analyses are indicative of the types of transport proteins that might be expected to populate the collection of genes that encode proteins with unknown function. Ion, amino acid, energy substrate, small molecule, and protein transporters were all identified in the genomics studies and novel transporters are likely to be representative of these major categories. From the perspective of drug delivery, profiling the BBB transport system is crucial for two reasons. First, the polarized distribution of efflux transporters can be a significant hindrance in small molecule drug delivery to the brain. Also, some of the identified transport systems could be exploited as conduits for novel drug delivery strategies. Other reviews in this issue discuss these concepts in more detail.
The SSH identification of a novel transporter coined BSAT1 (BBB-specific anion transporter type 1) is an intriguing example of the way in which genomics can aid in transporter discovery. The BSAT1 clone has a distant similarity to a liver anion transporter and was demonstrated as BBB-enriched in both of the rat SSH studies.9,11 The BBB-enriched expression profile of BSAT1 was further confirmed by Northern blotting. In addition, the clone appeared at an extremely high frequency in both rat SSH studies with BSAT1 comprising 16%9 and 12%11 of the total clones sequenced. After the BBB-specific anion transporter was identified using genomics methods, future experiments confirmed that BSAT1 is the 14th member of the organic anion transporter family (Oatp14) and that it is expressed in brain endothelial cell membranes. It was also demonstrated that Oatp14 facilitates the transport of thyroxine and thus might help regulate the concentration of this hormone in the CNS.20 The BSAT1 example illustrates the potential of using genomics analyses as a means for new transporter identification. Without genomics studies, it is unlikely that novel clones like BSAT1 would be investigated by BBB scientists, although they may play critical functional roles at the BBB. The BSAT1 study also emphasizes the crucial contribution made by postgenomics investigations that focus on identifying the functional roles of novel proteins.
In silico genomics
Because the number of genomic databases continues to increase at a rapid pace, new opportunities for genomics studies have presented themselves. Because genomics strategies are often used to identify candidate genes involved in a given biological process, performing these screens on the computer or in silico would provide rapid access to potential factors involved in disease, development, and cellular differentiation. This method has recently been applied to clone novel endothelial specific genes through the use of expressed sequence tag (EST) databases. By comparing endothelial EST databases to a pool of nonendothelial ESTs, and by using database-assisted SAGE differential analysis, four novel genes were identified having endothelial-specific expression.21 Confirmation of the expression localization to endothelium was performed with RT-PCR analysis and illustrated upregulation of these transcripts in human umbilical vein cultures (HUVEC) and human microvascular endothelium cultures. Although these experiments were not performed with the BBB endothelium, the principle would be the same if BBB cDNA or SAGE databases were used for the comparative in silico profiling. Thus, an added importance of the BBB profiling programs mentioned above is the ability to use them for differential gene expression analysis in silico. However, it is important to be careful when using online resources for comparison of tissue expression profiles because ESTs, microarray data, and SAGE can yield conflicting results.22 Interestingly, differential gene expression analyses using brain and vascular endothelium databases correlated quite well,22 suggesting applicability of this method to BBB expression profiling.
A somewhat different in silico approach has been used to successfully clone an ATP binding cassette transporter (ABC transporter) that is highly expressed in human cerebral endothelial cells.23 In this study, a search of human brain EST databases using the ATP binding cassette domain revealed 15 ABC transporters that were expressed in brain tissue. Using the results of this in silico search, the clones were analyzed for BBB expression. One clone, ABCG2, was particularly highly expressed at the BBB, mediated efflux of small molecule substrates, and was shown to be upregulated in glioblastoma vessels.23 This approach can be successful if there is a conserved sequence that can be searched. However, many of the novel genes discovered in the SSH and SAGE BBB analyses do not contain readily identifiable conserved domains at the nucleotide level. Irregardless, in silico cloning of BBB-specific genes is a potentially powerful tool and will likely increase as generation of BBB genomics data continues.
BLOOD-BRAIN BARRIER PROTEOMICS
Genomics versus proteomics
The use of genomics technologies is increasing at a rapid rate, and the techniques are moving into the scientific mainstream. A wealth of information can result from these types of studies, and due to constant refinement of genomic techniques, little prior experience is needed to generate meaningful results. However, genomics experiments do not tell the complete story as proteins are the predominant functional units of the cell. Because of this disconnect, genomics technologies have inherent shortcomings from a couple of standpoints. First, the amount of a given cellular protein does not necessarily correlate with the coordinate amount of mRNA present in the cell.24,25 This is a consequence of mRNA being dynamically regulated by factors that are distinct from those that govern protein turnover in the cell. Thus, it is possible for misinterpretation of data if solely genomic readouts are used. Second, the protein product can undergo modifications such as glycosylation, phosphorylation, and proteolysis that are critical for function. These alterations are not completely predicted by mRNA sequence and are extremely important to cellular function. Thus, it is important to have a combined genomics and proteomics program to gain the most comprehensive information about the cellular system in question. A few examples of applying standard proteomic methods to BBB research are included in the disease section of this review. The following section will focus instead on a new methodology that allows for the study of membrane proteins, a very difficult subset of proteins to analyze by standard methods.
Membrane proteomics and subtractive antibody expression cloning
The very nature of the BBB as an impermeable interface between the blood and brain indicates a significant reliance on membrane proteins for molecular/cellular communication and transport. Analysis of BBB membrane proteins is therefore required for full elucidation of brain disease mechanisms, and may help guide therapeutic intervention. Unfortunately, proteome-wide analysis of membrane protein expression has long been hampered by solubility problems inherent to the hydrophobic, lipid bilayer-spanning segments of membrane proteins. Also, the glycosylated nature of many membrane proteins results in diffuse banding in gel-based separations. These two factors have greatly limited the applicability of the current gold standard of proteomic technologies, two-dimensional electrophoresis coupled with mass spectrometry, for the analysis of membrane proteins.26
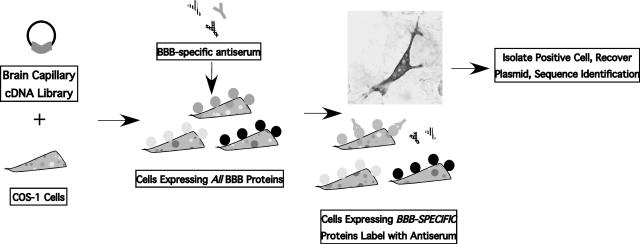
As an alternative to gel-based methods, an approach known as subtractive antibody expression cloning has been developed and was initially applied for differential membrane protein expression profiling of the BBB.27–29 In this method, a BBB cDNA library expressed in mammalian cells is used in conjunction with a BBB-specific antiserum for the cloning of membrane proteins that are differentially expressed at the BBB without a priori knowledge of the protein target. Because the BBB proteins are expressed in mammalian cells, they are probed in a native membrane environment and solubility or glycosylation constraints do not hinder the analysis. The method works as follows (FIG. 3). A polyclonal antiserum raised against isolated bovine BBB membranes was used as a specific probe against BBB membrane proteins. To identify membrane proteins that are specifically recognized at the BBB that likely contribute to its unique phenotype, the antiserum was depleted of antibodies recognizing common antigens that are also expressed in the liver or kidney. The subtracted antiserum specifically targets a large collection of BBB-specific proteins (FIG. 1). This antiserum was then used to probe a bovine BBB cDNA library generated from freshly isolated capillaries that was expressed in the COS-1 mammalian cell line. In principle, all BBB proteins were being expressed in the COS-1 cells in their native context, including membrane proteins. Finally, probing transfected monolayers with the BBB-specific probe for membrane proteins allowed for identification of full-length cDNA clones that are differentially expressed at the blood-brain barrier compared to liver or kidney tissues. In this way, it is possible to perform differential membrane protein expression profiling of the BBB as a complement to differential gene expression profiling.27
FIG. 3.
Cartoon of subtractive antibody expression cloning strategy.
Thus far, this method has been used to clone three differentially expressed BBB membrane proteins; Lutheran membrane glycoprotein,27 membrane cofactor protein CD46,29 and Ro52.28 Lutheran protein is involved in basal cell adhesion30 and functions as a laminin receptor.31 It is specifically expressed at the BBB compared with liver and kidney and can be used as a BBB marker in brain.32 CD46 is a regulator of complement activation, has been shown to actively bind the measles virus,33 and thus could potentially function in measles virus entry into the brain.34 Ro52 is a component of the SS-A ribonucleoprotein complex and has been implicated as an autoantigen in Sjogren’s syndrome and systemic lupus erythematosus, both of which can elicit CNS involvement.35,36 This finding suggests that Ro52 may be involved in the regulation of autoimmune responses at the brain microvasculature.
The localization of CD46 and Ro52 to the blood-brain barrier strengthens the link between the brain endothelium and the immune system. Because circulating immune components encounter the BBB directly, one could expect a large role for the BBB in immune survey and leukocyte recruitment to the brain. In addition, the immune response at the BBB can be upregulated during bouts of disease and inflammation. Combined with genomic data (FIG. 2) that indicates differential expression of major histocompatibility complex class I, PECAM (platelet/endothelial cell adhesion protein)-1, and podocalyxin-like protein, proteomic data confirms that immune molecules appear with high frequency in differential BBB expression analyses. The utility of these analyses in identifying immunologically relevant targets for disease intervention is becoming increasingly clear and could direct therapeutic design. With this added mechanistic knowledge, it may be possible to target the immune system to diseased regions of the brain parenchyma before the onset of pathological BBB disruption.
BLOOD-BRAIN BARRIER GENOMICS AND PROTEOMICS APPLIED TO NEUROLOGICAL DISEASE
Genomics and proteomics methods have also been applied to mechanistic studies of BBB involvement in neurological disease with the goal of identifying cellular components that contribute to disease pathogenesis. To date, genomics techniques have been used more frequently than proteomics techniques, and in many cases, whole brain tissue is used rather than isolated brain capillaries. In many situations, highly expressed BBB transcripts appear in these whole brain tissue analyses and such studies could readily be expanded by further investigation of BBB involvement. Unlike the BBB phenotypic profiling genomics studies mentioned above where a normally functioning BBB blueprint was desired, these studies typically compare gene or protein expression profiles in diseased tissue to those found in normal tissue. In this way, single molecules and cellular pathways that exhibit aberrant expression behavior are identified and can provide the basis for additional, targeted biochemical analysis.
The integrity of the BBB is affected under stroke conditions of ischemia and hypoxia, resulting in BBB permeability increases and edema formation. SSH was used to investigate the molecular determinants of stroke-prone spontaneously hypertensive rats by comparing cerebral capillary gene expression profiles with those found in capillaries of stroke-resistant spontaneously hypertensive rats.37 The study indicated that the rat sulfonylurea receptor 2B was upregulated, whereas the G protein signaling 5 regulator was downregulated in stroke-prone rats. Two cDNA clones were also isolated that had no known function. In a related study, mRNA isolated from whole brain tissue of stroke-prone rats was compared with that derived from stroke-resistant rats by gene microarray.38 Differential expression was observed for kinases involved in the MAP and AKT signaling pathways and tyrosine kinase B (TrkB) receptor isoforms that function in neuronal survival and proliferation mechanisms. Two-dimensional gel electrophoresis was then used to identify altered kinase substrates in stroke-prone rats, and it was suggested that altered signaling and phosphorylation states were indicators of stroke susceptibility. Using an alternative stroke model, the cold-induced hypertensive rat, SSH was performed with total brain tissue and 76 genes were found to be overexpressed in stroke tissue.39 The differentially expressed genes again included those involved in signal transduction but also included genes associated with energy metabolism and transcription/translation. In an attempt to determine new strategies for treatment of stroke, Dhodda and co-workers40 used genomic and proteomic techniques to determine how ischemic preconditioning can induce protection against future ischemic events. Gene microarray studies using whole brain tissue indicated the induction of 40 neuroprotective transcripts after a 10-min transient middle cerebral artery occlusion (MCAO). Fourteen of these 40 transcripts were heat shock proteins. Two-dimensional gel electrophoresis also confirmed the upregulation of various heat shock proteins and stress response genes. Given the success of these studies including those that used whole brain tissue as a source of transcripts, BBB participation in stroke could likely be further discerned by using the same techniques with isolated capillaries as the source of transcripts.
Aberrant proliferative behavior of the brain vasculature has been associated with many neurological diseases. This observation has been reinforced by gene expression profiling of various disease states. In an effort to classify tumors based on gene expression profiles, microarray analysis was performed with tissue samples of low-grade astrocytoma, primary glioblastoma, and secondary glioblastoma. It was discovered that expression levels of angiogenic gene clusters could be used to differentiate tumor types. Genes such as vascular endothelial growth factor receptor (flt-1), and IGFBP2 helped serve as predictors of tumor status.41 In a similar investigation, it was determined that angiogenesis-related genes were reliable markers in the classification of benign versus malignant astrocytomas.42 Many differentially regulated transcripts involved in the hypoxia-inducible transcription factor (HIF-2α) pathway are upregulated in the high-grade tumor, including the epidermal growth factor receptor and the IGFBP2 gene. Human brain arteriovenous malformations can also lead to stroke, and active angiogenesis has been implicated in the recurrence/regression nature of the disease. Gene microarray analysis of brain tissue having arteriovenous malformations was performed and increases in angiogenesis-related genes, such as VEGF, VEGF receptors flt-1 and flk-1, and angiopoietin-1 and -2 were signatures of the malformations.43 Gene microarray experiments profiling mice deficient in the Alzheimer’s disease-associated presenilin-1 protein also indicated significant expression difference in genes involved in vascular development, in addition to signaling and neural differentiation.44
Given that many of the genes identified in the above studies implicate vascular involvement and that many of these genes have been shown to be upregulated at the BBB (compare FIG. 2 with genes mentioned in the previous paragraph), it may be particularly informative to perform analogous analyses with isolated capillaries rather than whole tissue. This approach has thus far been limited, but two independent studies using endothelial cell lines illustrate the potential benefits. The effects of the tumor suppressor protein, p53, on tumor cell expression profile were investigated using cDNA microarray.45 Array data demonstrated that tumor cells with wild-type p53 exhibited increased VEGF production after radiation treatment, whereas tumor cells with mutant p53 had unaltered VEGF expression. Subsequently, based in part on the VEGF finding, glioblastoma cells with wild-type p53 and mutated p53 were cocultured with HUVEC cells. It was discovered that endothelial cells had increased radiation sensitivity when the p53 function was blocked via mutation. Taken together, these data suggested that altered intercellular communication was the cause of different endothelial cell reactions to radiation, and that cytotoxic therapies could be influenced by the p53 status of tumor cells. Another study using HUVEC cells has been performed to determine candidate angiogenesis regulators. HUVEC cells grown in a three-dimensional collagen gel were stimulated by VEGF treatment, and differential transcript profiling was performed using SSH.46 Several dozen cDNA fragments that exhibited differential expression on VEGF exposure were identified, including 11 VEGF-responsive genes not previously known to participate in the angiogenic response. Although these two studies used HUVEC cells rather than brain endothelial cells as a model to investigate global gene response in disease, they illustrate the additional impact that studies can have when genomics studies are principally focused on the endothelial cells.
Genomics and proteomics approaches have also been applied to analyze other functions of the BBB including immunological response, bacterial invasion, and transporter expression in epilepsy. The BBB responds to tumor necrosis factor α (TNFα), and TNFα has been shown to regulate BBB permeability. The effects of TNFα on human cerebral endothelial cells were profiled using microarray and two-dimensional gel electrophoresis.47 It was discovered that cell adhesion, apoptosis, and chemotaxis genes were differentially expressed, and these findings were corroborated by proteomic analysis. Interestingly, HUVEC cells were profiled in parallel with the brain endothelial cells and indicated that although many responses were similar, there were clearly responses to TNFα that were brain specific.47 This again emphasizes the importance of using appropriate cell sources for genomic/proteomic studies. In a similar vein, Doran and co-workers48 performed microarray experiments to understand the BBB response to infection in bacterial meningitis. To enter the brain, the bacteria must interact intimately with the BBB and this study illustrated the endothelial cell response. Group B streptococcus was found to elicit expression of interleukins (IL-6 and IL-8), chemokines (Groα and Groβ), and adhesion molecules (intercellular adhesion molecule-1), all of which can be attributed to the inflammatory response. The bacterial upregulation of these genes was found to be dependent on the B-hemolysin/cytolysin toxin expressed by the B streptococcus. Finally, antiepileptic drug resistance was investigated by microarray profiling of cerebral endothelial cells in epileptic versus nonepileptic tissue, with a special emphasis on transporter expression. It was discovered that MDR1 (multidrug resistance 1), MRP1 (multidrug resistance-associated protein), MRP2, MRP5, and cisplatin-resistance protein were overexpressed in epileptic tissue helping to explain the origin of drug resistance.49
The results detailed above are indicative of the types of information that can be gleaned from genomics/proteomics studies of disease states. The data can direct researchers to further investigation of pertinent signaling pathways, structural rearrangements, and chemokine/cytokine messengers. However, these techniques only identify targets for future research and do not unequivocally prove hypotheses or identify disease cures. Therefore, traditional reductionist approaches will continue to retain their importance in validation of clues that genomics and proteomics approaches may unearth.
CONCLUSIONS AND PERSPECTIVES
Study of the BBB using genomics and proteomics techniques is admittedly in its early stages. However, it is clear that such global analyses of the BBB have the potential to identify interacting pathways and mechanisms that may not be accessible to traditional techniques. Thus, these new tools should be integrated into existing BBB research programs to help clarify the unique aspects of the brain endothelium and its interactions with the surrounding cellular microenvironment. In addition, by identifying brain disease targets at the BBB, the potential exists to uncover new modes of therapeutic intervention. Without proper targeting and delivery strategies, however, the success of new therapeutic strategies will be limited. Therefore, genomics and proteomics should also be applied to brain drug delivery to characterize active efflux systems that can prevent drug access to the brain, and to identify new transporters that could be used as noninvasive drug delivery conduits.
REFERENCES
- 1.Pardridge WM. CNS drug design based on principles of blood-brain barrier transport. J Neurochem 70: 1781–1792, 1998. [DOI] [PubMed] [Google Scholar]
- 2.Boado RJ, Pardridge MM. A one-step procedure for isolation of poly(A)+ mRNA from isolated brain capillaries and endothelial cells in culture. J Neurochem 57: 2136–2139, 1991. [DOI] [PubMed] [Google Scholar]
- 3.Shusta EV, Boado RJ, Mathern GW, Pardridge WM. Vascular genomics of the human brain. J Cereb Blood Flow Metab 22: 245–252, 2002. [DOI] [PubMed] [Google Scholar]
- 4.Song L, Pachter JS. Culture of murine brain microvascular endothelial cells that maintain expression and cytoskeletal association of tight junction-associated proteins. In Vitro Cell Dev Biol Anim 39: 313–320, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science 291: 1304–1351, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature 409: 860–921, 2001. [DOI] [PubMed] [Google Scholar]
- 7.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470, 1995. [DOI] [PubMed] [Google Scholar]
- 8.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93: 6025–6030, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JY, Boado RJ, Pardridge WM. Blood-brain barrier genomics. J Cereb Blood Flow Metab 21: 61–68, 2001. [DOI] [PubMed] [Google Scholar]
- 10.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science 270: 484–487, 1995. [DOI] [PubMed] [Google Scholar]
- 11.Li JY, Boado RJ, Pardridge WM. Rat blood-brain barrier genomics. II. J Cereb Blood Flow Metab 22: 1319–1326, 2002. [DOI] [PubMed] [Google Scholar]
- 12.Enerson BE, Drewes LR. The rat blood-brain barrier transcriptome. Brain03, XXIst International Symposium on Cerebral Blood Flow, Metabolism, and Function, 2003. [DOI] [PubMed]
- 13.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 147: 185–194, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, et al. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol 161: 653–660, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasdala I, Wolburg-Buchholz K, Wolburg H, Kuhn A, Ebnet K, Brachtendorf G, et al. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J Biol Chem 277: 16294–16303, 2002. [DOI] [PubMed] [Google Scholar]
- 16.Kupprion C, Motamed K, Sage EH. SPARC (BM-40, osteonectin) inhibits the mitogenic effect of vascular endothelial growth factor on microvascular endothelial cells. J Biol Chem 273: 29635–29640, 1998. [DOI] [PubMed] [Google Scholar]
- 17.Guardavaccaro D, Corrente G, Covone F, Micheli L, D’Agnano I, Starace G, et al. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol 20: 1797–1815, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couet J, Belanger MM, Roussel E, Drolet MC. Cell biology of caveolae and caveolin. Adv Drug Deliv Rev 49: 223–235, 2001. [DOI] [PubMed] [Google Scholar]
- 19.Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics 68: 41–56, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama D, Kusuhara H, Taniguchi H, Ishikawa S, Nozaki Y, Aburatani H, et al. Functional characterization of rat brain-specific organic anion transporter (Oatp14) at the blood-brain barrier: high affinity transporter for thyroxine. J Biol Chem 278: 43489–43495, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Huminiecki L, Bicknell R. In silico cloning of novel endothelial-specific genes. Genome Res 10: 1796–1806, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huminiecki L, Lloyd AT, Wolfe KH. Congruence of tissue expression profiles from Gene Expression Atlas, SAGEmap, and TissueInfo databases. BMC Genomics 4: 31, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Mojsilovic-Petrovic J, Andrade MF, Zhang H, Ball M, Stanimirovic DB. The expression and functional characterization of ABCG2 in brain endothelial cells and vessels. FASEB J 17: 2085–2087, 2003. [DOI] [PubMed] [Google Scholar]
- 24.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18: 533–537, 1997. [DOI] [PubMed] [Google Scholar]
- 26.Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis 21: 1054–1070, 2000. [DOI] [PubMed] [Google Scholar]
- 27.Shusta EV, Boado RJ, Pardridge WM. Vascular proteomics and subtractive antibody expression cloning. Mol Cell Proteom 1: 75–82, 2002. [DOI] [PubMed] [Google Scholar]
- 28.Shusta EV, Li JY, Boado RJ, Pardridge WM. The Ro52/SS-A autoantigen has elevated expression at the brain microvasculature. Neuroreport 14: 1861–1865, 2003. [DOI] [PubMed] [Google Scholar]
- 29.Shusta EV, Zhu C, Boado RJ, Pardridge WM. Subtractive expression cloning reveals high expression of CD46 at the blood-brain barrier. J Neuropathol Exp Neurol 61: 597–604, 2002. [DOI] [PubMed] [Google Scholar]
- 30.Udani M, Zen Q, Cottman M, Leonard N, Jefferson S, Daymont C, et al. Basal cell adhesion molecule/lutheran protein. The receptor critical for sickle cell adhesion to laminin. J Clin Invest 101: 2550–2558, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zen Q, Cottman M, Truskey G, Fraser R, Telen MJ. Critical factors in basal cell adhesion molecule/Lutheran-mediated adhesion to laminin. J Biol Chem 274: 728–734, 1999. [DOI] [PubMed] [Google Scholar]
- 32.Boado RJ, Li JY, Pardridge WM. Selective Lutheran glycoprotein gene expression at the blood-brain barrier in normal brain and in human brain tumors. J Cereb Blood Flow Metab 20: 1096–1102, 2000. [DOI] [PubMed] [Google Scholar]
- 33.Hsu EC, Sarangi F, Iorio C, Sidhu MS, Udem SA, Dillehay DL, et al. A single amino acid change in the hemagglutinin protein of measles virus determines its ability to bind CD46 and reveals another receptor on marmoset B cells. J Virol 72: 2905–2916, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cattaneo R, Rebmann G, Baczko K, ter Meulen V, Billeter MA. Altered ratios of measles virus transcripts in diseased human brains. Virology 160: 523–526, 1987. [DOI] [PubMed] [Google Scholar]
- 35.Govoni M, Padovan M, Rizzo N, Trotta F. CNS involvement in primary Sjogren’s syndrome: prevalence, clinical aspects, diagnostic assessment and therapeutic approach. CNS Drugs 15: 597–607, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Jennekens FG, Kater L. The central nervous system in systemic lupus erythematosus. Part 1. Clinical syndromes: a literature investigation. Rheumatology (Oxford) 41: 605–618, 2002. [DOI] [PubMed] [Google Scholar]
- 37.Kirsch T, Wellner M, Luft FC, Haller H, Lippoldt A. Altered gene expression in cerebral capillaries of stroke-prone spontaneously hypertensive rats. Brain Res 910: 106–115, 2001. [DOI] [PubMed] [Google Scholar]
- 38.Fornage M, Swank MW, Boerwinkle E, Doris PA. Gene expression profiling and functional proteomic analysis reveal perturbed kinase-mediated signaling in genetic stroke susceptibility. Physiol Genom 15: 75–83, 2003. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Z, Zhao B, Wang X, Zhu S, Zhang Q, Xu Y, et al. Differentially expressed genes in hypertensive rats developing cerebral ischemia. Life Sci 74: 1899–1909, 2004. [DOI] [PubMed] [Google Scholar]
- 40.Dhodda VK, Sailor KA, Bowen KK, Vemuganti R. Putative endogenous mediators of preconditioning-induced ischemic tolerance in rat brain identified by genomic and proteomic analysis. J Neurochem 89: 73–89, 2004. [DOI] [PubMed] [Google Scholar]
- 41.Godard S, Getz G, Delorenzi M, Farmer P, Kobayashi H, Desbaillets I, et al. Classification of human astrocytic gliomas on the basis of gene expression: a correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res 63: 6613–6625, 2003. [PubMed] [Google Scholar]
- 42.Khatua S, Peterson KM, Brown KM, Lawlor C, Santi MR, LaFleur B, et al. Overexpression of the EGFR/FKBP12/HIF-2α pathway identified in childhood astrocytomas by angiogenesis gene profiling. Cancer Res 63: 1865–1870, 2003. [PubMed] [Google Scholar]
- 43.Hashimoto T, Lawton MT, Wen G, Yang GY, Chaly T Jr, Stewart CL, et al. Gene microarray analysis of human brain arteriovenous malformations. Neurosurgery 54: 410–423; discussion 423-435, 2004. [DOI] [PubMed] [Google Scholar]
- 44.Mirnics ZK, Mirnics K, Terrano D, Lewis DA, Sisodia SS, Schor NF. DNA microarray profiling of developing PS1-deficient mouse brain reveals complex and coregulated expression changes. Mol Psychiatry 8: 863–878, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Khodarev NN, Labay E, Darga T, Yu J, Mauceri H, Gupta N, et al. Endothelial cells co-cultured with wild-type and dominant/negative p53-transfected glioblastoma cells exhibit differential sensitivity to radiation-induced apoptosis. Int J Cancer 109: 214–219, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Wary KK, Thakker GD, Humtsoe JO, Yang J. Analysis of VEGF-responsive genes involved in the activation of endothelial cells. Mol Cancer 2: 25, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franzen B, Duvefelt K, Jonsson C, Engelhardt B, Ottervald J, Wickman M, et al. Gene and protein expression profiling of human cerebral endothelial cells activated with tumor necrosis factor-α. Brain Res Mol Brain Res 115: 130–146, 2003. [DOI] [PubMed] [Google Scholar]
- 48.Doran KS, Liu GY, Nizet V. Group B streptococcal β-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J Clin Invest 112: 736–744, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dombrowski SM, Desai SY, Marroni M, Cucullo L, Goodrich K, Bingaman W, et al. Overexpression of multiple drug resistance genes in endothelial cells from patients with refractory epilepsy. Epilepsia 42: 1501–1506, 2001. [DOI] [PubMed] [Google Scholar]