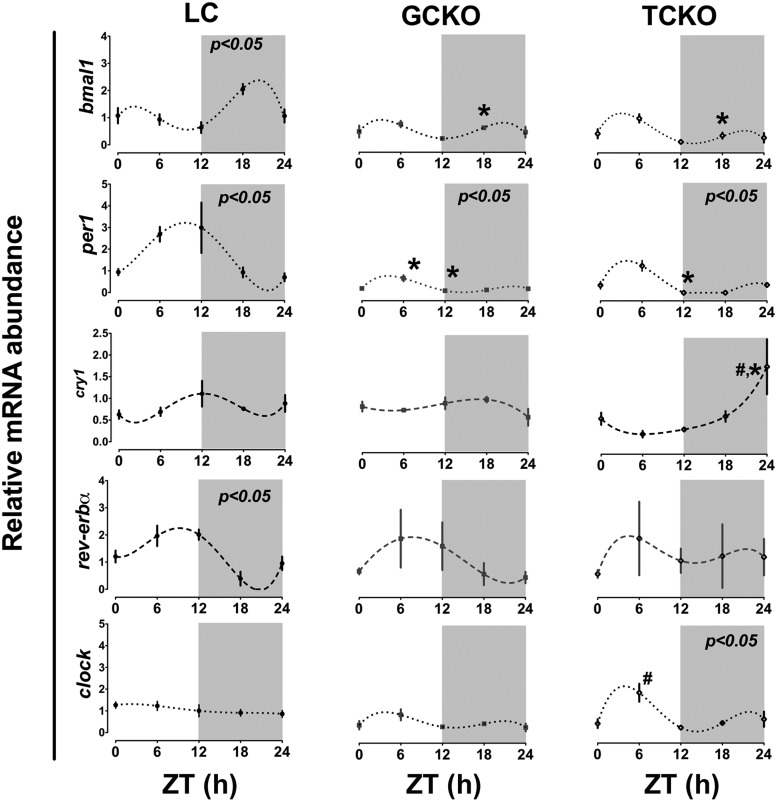
Figure 5. Rhythms of clock gene mRNA abundance in whole ovary are disrupted in both GCKO and TCKO transgenic mice.
Abundance of clock gene mRNA in ovaries recovered from LC (n = 3), GCKO (n = 3), and TCKO (n = 3) mice on proestrus. LC mice displayed diurnal rhythms of Bmal1, Per1, and Rev-erbα mRNA abundance (P < .05 indicated in top right of each panel). Rhythms of Bmal1 and Rev-erbα mRNA abundance were disrupted in both GCKO and TCKO mice. Although still rhythmic, the absolute level of Per1 mRNA was suppressed in both GCKO and TCKO mice. Although not significant by CircWave analysis, Cry1 mRNA abundance oscillated at low amplitude with a peak near midday in LC mice and was altered in both GCKO and TCKO mice. Clock mRNA displayed a low amplitude rhythm in TCKO mice that was not detected in LC or GCKO mice. Data are presented as mean ± SEM; n = 3 samples per time point except for ZT24 in TCKO mice (n = 2). (#, P < .05 GCKO vs TCKO; *, P < .05 LC vs GCKO or TCKO). Values for target mRNA abundance were calculated using the ΔΔCT method as described in Materials and Methods with β-actin as the reference gene. Data were fit to a nonlinear regression (see Materials and Methods).