Abstract
Herpes simplex virus (HSV-1) lytic infection results in global changes to the host cell proteome and the proteins associated with host chromatin. We present a system level characterization of proteome dynamics during infection by performing a multi-dimensional analysis during HSV-1 lytic infection of human foreskin fibroblast (HFF) cells. Our study includes identification and quantification of the host and viral proteomes, phosphoproteomes, chromatin bound proteomes and post-translational modifications (PTMs) on cellular histones during infection. We analyzed proteomes across six time points of virus infection (0, 3, 6, 9, 12 and 15 h post-infection) and clustered trends in abundance using fuzzy c-means. Globally, we accurately quantified more than 4000 proteins, 200 differently modified histone peptides and 9000 phosphorylation sites on cellular proteins. In addition, we identified 67 viral proteins and quantified 571 phosphorylation events (465 with high confidence site localization) on viral proteins, which is currently the most comprehensive map of HSV-1 phosphoproteome. We investigated chromatin bound proteins by proteomic analysis of the high-salt chromatin fraction and identified 510 proteins that were significantly different in abundance during infection. We found 53 histone marks significantly regulated during virus infection, including a steady increase of histone H3 acetylation (H3K9ac and H3K14ac). Our data provide a resource of unprecedented depth for human and viral proteome dynamics during infection. Collectively, our results indicate that the proteome composition of the chromatin of HFF cells is highly affected during HSV-1 infection, and that phosphorylation events are abundant on viral proteins. We propose that our epi-proteomics approach will prove to be important in the characterization of other model infectious systems that involve changes to chromatin composition.
Herpes simplex virus (HSV-1)1 leads to a contagious and persistent infection that affects about 95% of the human population. The HSV-1 genome is a double-stranded DNA molecule that replicates in the host cell nucleus (1). HSV-1 initially infects epithelial cells as a lytic infection, and then enters peripheral neurons where it establishes latency (2, 3). Processes such as viral transcription, viral DNA synthesis, virion assembly and DNA packaging take place in discrete virus-induced structures within the nucleus called replication compartments (1, 4). These processes are temporally regulated by the viral cascades of immediate-early (IE), early (E), and late (L) gene expression. The IE proteins are primarily responsible for counteracting intrinsic host defenses and for activating expression of early-phase genes (1). Early viral proteins include enzymes, transcriptional activators, and DNA-binding proteins necessary to initiate transcription and viral DNA replication. Early-late genes (E-L) are initially transcribed at low levels and up-regulated during viral DNA replication, whereas true-late genes are expressed exclusively after DNA replication and include proteins required for the assembly of the progeny HSV-1 virions. The efficacy of HSV-1 lytic infection depends on the ability to modulate the host cell proteome. Here we focus on proteomic changes during lytic infection of primary human foreskin fibroblast (HFF) cells, including protein post-translational modifications (PTMs) and proteins associated with the host chromatin.
Upon HSV-1 infection, viral genomes enter the nucleus and become “chromatinized” as the non-nucleosomal state is transformed into a chromatin structure that resembles the host genome (3, 5). The assembly and modulation of nucleosomes during infection is dependent on a plethora of viral and host chromatin components (6). Initially, viral genomes have been shown to carry repressive chromatin marks thought to represent a host silencing mechanism (5, 7). HSV-1 then overcomes this repression through histone H3 acetylation to promote viral gene expression (7). Many chromatin components are involved in the balance between heterochromatic suppression of the viral genome and the euchromatin transition that promotes the expression of viral genes (6). HSV-1 infection also causes extensive reorganization of cellular structures, with nuclear changes including margination of chromatin, enlargement of the nucleus, formation of replication compartments, disruption of the nuclear lamina and nucleoli, and cytoplasmic changes including disruption of the Golgi apparatus and microtubules and genetic damage to mitochondria (4, 8, 9). HSV-1 infection also impacts cellular metabolism, in part by diverting the central carbon metabolism toward production of pyrimidine nucleotide components (10).
HSV-1 proteins are extensively post-translationally processed by both cellular and viral enzymes. Several reports have demonstrated roles for viral kinases (US3 and UL13) and certain cellular kinases (PKA, PKC, CKI, JNK1 or CDC2) in modifications of HSV-1 IE proteins (11–15). Conversely, HSV-1 enzymes may also alter the regulation of PTMs on host proteins and histones. For example, HSV-1 UL13 directly phosphorylates host nuclear lamin proteins and this phosphorylation induces partial lamin disassembly or reorganization that allows virions to reach the inner nuclear membranes (16, 17). However, even though a prior study identified phosphorylation on HSV-1 proteins (18), a global picture of regulation of host phosphorylation upon HSV-1 infection has not been fully investigated. Moreover, virus infection globally affects the host epigenome (19), thus histone PTMs are of particular interest during HSV-1 infection. Histone PTM relative abundance and location is related to chromatin condensation, gene expression, or recruitment of other proteins to chromatin. Host and viral chromatin compete for cellular resources resulting in a battle between host defense mechanisms and viral neutralization of host proteins. For example, HSV-1 manipulates chromatin remodeling complexes such as the polycomb group (PcG) proteins (20), CoREST/REST (21, 22), and SWI/SNF (23) to facilitate viral gene expression.
In this study, we performed extensive proteomic analysis of host and viral proteomes during infection, including phosphorylation events and chromatin bound proteins. Results were clustered to provide an accurate representation of protein/PTM dynamics in a temporal manner, which can be correlated with the different stages of viral lytic infection. Using this approach, we show dynamic changes of virus and host protein levels and PTMs. We propose that changes in histone PTM levels are a direct result of virus infection, such as the accumulation of histone H3 acetylation marks that are indicators of open chromatin (H3K9ac and H3K14ac). By using label-free quantification we provide an estimation of relative protein abundance in infected cells. We also identified 571 phosphorylation sites on viral proteins, showing a relative enrichment of phosphorylated threonine as compared with the relative abundance in the host phosphoproteome. Selected candidate proteins and histone marks were validated by Western blot (WB) analysis and immunofluorescence (IF). In summary, we present a resource of host and viral proteomics that describes protein and PTM regulation during HSV-1 infection with unprecedented temporal resolution.
EXPERIMENTAL PROCEDURES
Cell Growth and Infection
We infected primary human foreskin fibroblast (HFF) and HeLa cells with HSV-1 strain 17 syn+ at a multiplicity of infection (MOI) of 3 according to standard methods (24). After virus adsorption, the HFF cells were harvested at 0 (mock-infected controls), 3, 6, 9, 12, and 15 h post-infection (hpi); and HeLa cells were harvested at 0, 4, and 8 hpi.
Antibodies
Primary antibodies were purchased from Abcam (H3K9me1 ab176880, H3K9me2 ab32521, H3K9ac ab177177, H3K14ac ab52946, H3K56me1 ab66857, Brd1 ab181060, SPOP ab137537, LSD1 ab17721, CKII ab76040), Millipore (Billerica, MA) (H3K27me3 07–449, HDAC1 06–720, HDAC2 05–814), Santa Cruz Biotechnology (Santa Cruz, CA) (ICP0 sc-53070), Sigma (actin A5441), Active Motif (Carlsbad, CA) (H3K9me3 39161), Cell Signaling Technology (Danvers, MA) (EZH2 3147), and Novus Biologicals, (Littleton, CO) (H3K4me3 NB21–1023). Secondary antibodies for immunoblotting were obtained from Jackson ImmunoResearch (West Grove, PA) and secondary antibodies for immunofluorescence were obtained from Life Technologies (Carlsbad, CA).
Immunofluorescence, Immunoblotting, and Chromatin Purification
Cells were grown on glass coverslips in 24-well plates and infected as described previously (24). Cells were harvested for immunofluorescence (IF) at the indicated time points, washed in PBS and fixed in 2% paraformaldehyde for 15 min. Coverslips were then blocked and stained as previously described (24) and mounted using ProLong Gold Antifade Reagent (Life Technologies). Immunofluorescence was visualized using a Zeiss LSM 710 Confocal microscope (Cell and Developmental Microscopy Core at UPenn) and ZEN 2011 software. Images were processed using ImageJ and assembled with Adobe CS6. Western blot (WB) analysis was carried out using standard methods. Salt fractionation of nuclei and preparation of salt fractions for nano-liquid chromatography tandem mass spectrometry (nLC-MS/MS) analysis was performed as described previously (25). nLC-MS/MS and data analysis settings were the same as the proteomic analysis described in the next paragraph.
Proteomics and Phosphoproteomics Analysis Using nLC-MS/MS
All chemicals used for preparation of nLC-MS/MS samples were of at least sequencing grade and purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise stated. Cells were lysed in 60 μl of cold lysis buffer (6 m urea/2 M thiourea, 50 mm ammonium bicarbonate, pH 8.2, Phosphatase and Protease Inhibitors Mix (Thermo Fisher Scientific, Waltham, MA)) by vortexing. Proteins were digested first with endopeptidase Lys-C (Wako, Cambridge, MA; MS grade) for 3 h, after which the solution was diluted 10 times with 20 mm ammonium bicarbonate. Subsequently, samples were reduced using 10 mm DTT for 1 h at room temperature and alkylated with 20 mm iodoacetamide (IAA) in the dark for 30 min at room temperature. Proteins were then digested with trypsin (Promega, Madison, WI) at an enzyme-to-substrate ratio of ∼1:50 for 12 h at room temperature. After digestion, the samples were concentrated to the volume of ∼100 μl by lyophilization. Phosphopeptide enrichment using titanium dioxide (TiO2) chromatographic resin was performed as previously described (26, 27). The lyophilized phosphorylated peptide samples were reconstituted in 0.1% trifluoroacetic acid (TFA) and desalted using Poros Oligo R3 RP (PerSeptive Biosystems, Framingham, MA) P200 columns. Unbound peptides from the TiO2 flow-through and subsequent TiO2 washes were combined and lyophilized to produce a non-modified peptide fraction. The non-modified peptide fraction was resuspended in 0.1% TFA and desalted using Sep-Pak tC18 Plus Light Cartridge (Waters, Milford, MA). The peptide samples were subsequently lyophilized and stored at −80 °C for further analysis.
Dried samples were resuspended in buffer-A (0.1% formic acid) and loaded onto an Easy-nLC system (Thermo Fisher Scientific, San Jose, CA), coupled online with an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Peptides were loaded into a picofrit 18 cm long fused silica capillary column (75 μm inner diameter) packed in-house with reversed-phase Repro-Sil Pur C18-AQ 3 μm resin. The gradient length was 165 min for the (phospho)proteome and 105 min for the salt fractions. The gradient was from 2–26% buffer-B (100% ACN/0.1% formic acid) for phosphopeptides and from 4–28% B for unmodified peptides, including salt fractions, at a flow rate of 300 nl/min. The MS method was set up in a data-dependent acquisition (DDA) mode. For full MS scan, the mass range of 350–1200 m/z was analyzed in the Orbitrap at 120,000 FWHM (200 m/z) resolution and 5 × 10e5 AGC target value. HCD collision energy was set to 27, AGC target to 10e4 and maximum injection time to 200 msec. Detection of MS/MS fragment ions was performed in the ion trap in the rapid mode using the TopSpeed mode (3 s). MS raw files were analyzed by MaxQuant software (28) version 1.5.2.8 for the salt fractions and version 1.5.5.1 for proteome and phosphoproteome analysis. MS/MS spectra were searched by the Andromeda search engine (29) against the Human UniProt FASTA database [9606] (20,160 reviewed canonical entries, version December January 2015). Viral proteins were identified using Human Herpesvirus strain 17 (17syn+) [10299] (73 reviewed canonical entries, version December January 2015). Additionally, the database included 247 common contaminants, discarded during data analysis. The search for total proteome included variable modifications of methionine oxidation and N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Analysis of phosphoproteome included carbamidomethylation on cysteine residues as fixed modification, whereas phosphorylation on serine, threonine and tyrosine residues was set as variable modification. Trypsin (cleaves K/R) was specified as digestive enzyme with two missed cleaves allowed. Minimal peptide length was set to seven amino acids. Peptide identification was performed with an allowed precursor mass deviation up to 4.5 ppm after time-dependent mass calibration and an allowed fragment mass deviation of 20 ppm. Protein identification required at least one unique or razor peptide per protein group. iBAQ, intensity-based absolute quantification (30), was enabled for protein label-free quantification. For matching between runs, the retention time alignment window was set to 20 min and the match time window was 1 min. All other values were kept as default, including false discovery rate (FDR, <0.01). Protein tables were filtered to eliminate the identifications from the reverse database, only identified by site and common contaminants. iBAQ values were log2 transformed and normalized by subtracting the average of all protein values for each sample (in log2). Viral proteins were not normalized, as this type of normalization assumes that the total protein level is approximately the same in all samples, which is not applicable for viral proteins. Phosphorylation site abundance was retrieved using the columns “Intensity” from the table Phospho(STY) generated by MaxQuant. Normalization was performed using the same approach as the protein tables, and then the abundance of the phosphorylation was corrected for the protein abundance by subtracting the normalized protein iBAQ value (in log2). In cases where the respective protein abundance could not be assessed for the phosphorylation site in all samples/conditions, no correction by protein abundance was performed; i.e. missing values were not imputed. All result tables are given as supplemental Tables S1–S7. The mass spectrometry proteomics and phosphoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD005467.
Quantification of Histone PTMs
Histone purification and analysis was performed as previously described (31). Briefly, histones were acid extracted from nuclei with 0.2 m H2SO4 for 2 h and precipitated with 33% trichloroacetic acid (TCA) for 1 h. Purified histones were dissolved in 30 μl of 50 mm NH4HCO3, pH 8.0. Derivatization reagent was prepared by mixing propionic anhydride with acetonitrile in a ratio of 1:3 (v/v), and such reagent was mixed with the histone sample in the ratio of 1:4 (v/v) for 15 min at 37 °C. This reaction was performed twice. Histones were then digested with trypsin (enzyme/sample ratio 1:20, overnight, room temperature) in 50 mm NH4HCO3. After digestion, the derivatization reaction was performed again twice to derivatize peptide N termini. Samples were desalted prior nLC-MS/MS analysis by using C18 Stage-tips. Samples were analyzed by using a nLC-MS/MS setup. Chromatography was configured with the same type of column and HPLC as for the proteomics analysis. The HPLC gradient was as follows: 2% to 28% solvent B (A = 0.1% formic acid; B = 95% MeCN, 0.1% formic acid) over 45 min, from 28% to 80% solvent B in 5 min, 80% B for 10 min at a flow-rate of 300 nl/min. nanoLC was coupled to an LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). A full scan MS spectrum (m/z 300–1100) was acquired in the Orbitrap with a resolution of 120,000 (at 200 m/z) and an AGC target of 5 × 10e5. MS/MS was performed using a data-independent acquisition (DIA) mode; the entire mass range (300–1100 m/z) was fragmented at every cycle using windows of 50 m/z (16 MS/MS scans total). MS/MS AGC target was 3 × 10e4, the injection time limit was 50 msec and the CID collision energy was 35. MS/MS data were collected in centroid mode. EpiProfile (32) was used to retrieve the extracted ion chromatograms and estimate the relative abundance of each peptide as compared with the total respective histone. All MS-histone raw files have been deposited in the CHORUS database (https://chorusproject.org/) under project number 1254.
Experimental Design and Statistical Rationale
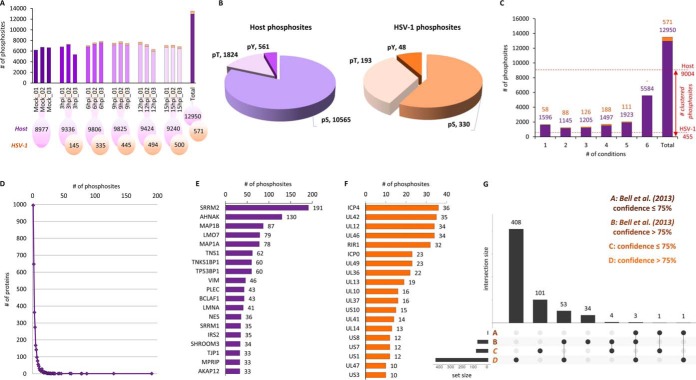
We performed three distinct proteomics studies on our model system (Fig. 1A). First, to investigate the temporal changes in cellular and viral proteome and phosphoproteome, the infected HFF cells with HSV-1 virus were harvested at 0, 3, 6, 9, 12, and 15 hpi in three biological replicates. The 0 hpi control sample represents the non-infected HFF cells. The time points were chosen based on the rate of synthesis immediate-early (IE), early (E), and late (L) HSV-1 genes (1). Secondly, we investigated the temporal changes in histone PTMs during HSV-1 infection using the same conditions and number of biological replicates. Finally, we examined the dynamics of the chromatin bound proteome during the HSV-1 infection by performing a high salt nuclei fractionation. To obtain larger quantities of material, the nuclei were isolated from HeLa cells in biological triplicate at 0, 4 and 8 hpi. Only the highest salt fraction (600 mm NaCl) was used for label-free nLC-MS/MS. Each biological replicate of the histone and chromatin bound proteome sample was performed in technical triplicates.
Fig. 1.
Proteome characterization during HSV-1 infection by mass spectrometry. A, Overview of the HSV-1 lytic lifecycle (top) across the six time points of viral infection in HFF cells. For both host and virus we identified and quantified proteome, phosphoproteome and chromatin-associated proteins. The host proteome was also investigated for histone PTMs. B, Total number of quantified proteins for host (blue) and virus (orange) for each time point and biological replicate. C, Histogram representing the number of host proteins quantified across all conditions. Most proteins were detected and quantified at all six time points. Proteins quantified in at least 5 out of 6 conditions were analyzed using fuzzy c-means clustering and functional enrichment using Gene Ontology (GO), whereas the others were only subjected to GO analysis. D, Top 50 host proteins with the largest coefficient of variation across the six time points, i.e. the proteins with the largest changes in intensity during virus infection. Data are sorted by ratio early/late time points. E, Log2 transformed absolute intensity (iBAQ) of all identified viral proteins. Data were clustered using a hierarchical tree (Euclidean).
ANOVA test was used in the time-course study of the proteome, phosphoproteome, and histone PTMs to identify proteins/PTMs that are significantly different between at least two time points. We performed paired sample t test to identified proteins with significant differential expression in mock in comparison with 4 hpi and 8 hpi in the chromatin bound proteome study. Every MS-analysis was performed with three biological replicates to provide enough statistical power to apply parametric tests (t test or ANOVA). No samples were excluded as outliers (this applies to all proteomics analyses described in this manuscript). Proteins with ANOVA and t test p value smaller than 0.05 were considered as significantly regulated. Data distribution was assumed to be normal but this was not formally tested.
Downstream Bioinformatics
Gene Ontology (GO) biological process (BP) information ranked by p value enrichment score was obtained from GeneGo's MetaCore pathways analysis package (Thomson Reuters) with false discovery rate (FDR) < 0.05. Perseus (33) software was used to visualize data from non-supervised hierarchical clustering, principal component analysis plot (PCA), and the matrix of correlation plots. PCA plot and matrix of correlation plots were performed on the normalized iBAQ protein intensity values (for proteome) or normalized phosphopeptide intensities (for phosphoproteome). Clustering analysis was performed using fuzzy c-means (34).
RESULTS AND DISCUSSION
To investigate the dynamics of protein abundance during HSV-1 infection, we evaluated the total proteome at different stages of viral infection. We infected primary HFF cells with HSV-1 strain 17 syn+ at an MOI of 3. After virus adsorption, the cells were harvested at 0 (mock-infected controls), 3, 6, 9, 12 and 15 h post-infection (hpi). The time points were chosen based on expression of immediate-early (IE), early (E) and late (L) HSV-1 genes (1) (Fig. 1A).
To evaluate the robustness of our method we performed PCA analysis of the host proteome results, which showed a high degree of reproducibility among biological replicates (supplemental Fig. S1A), with mock samples distinctly separated from infected conditions. We observed the highest correlation between 3 hpi/6 hpi and subsequently between 9 hpi/12 hpi samples, whereas the 15 hpi condition clustered separately. Interestingly, 9 hpi and 12 hpi were highly dispersed as compared with other HSV-1 infected conditions. This may reflect temporarily increasing protein dynamics because of the synthesis of IE, E, and L viral genes. Within 9 hpi and 12 hpi both viral DNA replication events and late gene expression are thought to reach their maximum efficiency, whereas at 15 hpi the virus triggers cell death of the infected cell (1). Although the rate of these processes may vary between different infected cells within the population tested, our PCA analysis shows similarities at time points consistent with the overall viral life cycle.
Host Proteome Depth and Quantification
Analysis of the full host cell proteome resulted in total identification of 4612 host protein groups, of which 4030 (∼87%) were quantified across all six time points of HSV-1 infection (Fig. 1B; supplemental Table S1A). We detected 4,326 proteins in at least five of the six different conditions, and those proteins were further subjected to fuzzy c-means clustering analysis (Fig. 1C; supplemental Table S1B). Proteins identified in fewer than four conditions (286 proteins) were considered time point specific and subjected to Gene ontology (GO) enrichment (GeneGo's MetaCore software, Thomson Reuters; FDR < 0.05) (Table S1C). The GO analysis showed that the majority regulate processes such as immune response, viral processes, cell death, structural components assembly, or mitochondrion organization (supplemental Fig. S2). We then sorted for proteins present only in the first three conditions (mock, 3 and 6 hpi). Interestingly, ∼50% of these were involved in immune response processes (supplemental Table S1C) known to be suppressed upon virus infection (35). Several of these proteins are reported to be degraded upon viral infection such as ITCH, which was found downregulated at 9–15 hpi (36). Alternatively, some have been shown to participate in antiviral responses early during HSV-1 infection as either positive regulation (e.g. PUM2) (37) or negative regulation (XIAP, SP1) (38). Other proteins that follow the same trend as known viral degradation targets over the time course may represent potential novel candidates for degradation by HSV-1. By sorting for proteins identified primarily at the last three time points (9, 12, and 15 hpi), we observed pro-apoptotic proteins relatively more enriched (e.g. CRADD, PPP2R1B, TRAF5, SH3RF1, SRA1, or LIG4). This may reflect an attempt by the host cell to induce apoptosis as a defense mechanism against infection. This brief overview of the data highlights potential host defense mechanisms and novel targets of HSV-1.
To obtain a comprehensive global picture of the dynamic host proteome during HSV-1 infection we performed unsupervised clustering of protein intensities using fuzzy c-means preceded by z-score normalization (Fig. 2A; supplemental Fig. S3) (34). We obtained the best minimum centroid distance by grouping our dataset into 16 different clusters. Each cluster represents the trend of protein abundance during HSV-1 infection with an average 270 cellular proteins per cluster. Proteins known to be degraded show a general downward trend (e.g. cluster 10) whereas proteins stimulated in response to infection are increased in abundance (e.g. cluster 6). The complex nature of the proteome warranted 14 other patterns depicted by each cluster for the best representation of global trends (supplemental Fig. S3).
Fig. 2.
Changes in host proteome over the time course of HSV-1 infection. A, Schematic of different phases during the HSV-1 infection cycle. Representative cluster trends showing changes in abundance of proteins involved in the different stages of virus infection are displayed in boxes. For example, proteins involved in viral transcription are activated at 3 hpi, whereas proteins responsible for viral transport or release of new virions increase in abundance at early (3–6 hpi) and late time points (15 hpi). B, Heatmap of GO biological process enrichment of host proteins detected in 16 independent clusters (all displayed in supplemental Fig. S3).
To define further the biological enrichment of the 16 clusters we used GeneGo's MetaCore software (FDR < 0.05) to determine the cell metabolic pathways and protein functions represented in each cluster. The nine major functions found to be enriched overall were RNA metabolic processes, DNA metabolic processes, gene expression, chromatin organization, cell cycle, intracellular transport, response to stress, cell death, and other (Fig. 2B). The global overview of cluster's GO-enrichment is described in supplementary information section 1. For the purpose of this study, we focused on chromatin remodeling complexes and histone modifiers (supplemental Table S1D). Proteins involved in chromatin and histone modifications were detected in all clusters, with highest enrichment in clusters 10 and 15 (supplemental Fig. S3), which show respectively monotonic (steady) or early (at 3 hpi) decrease in protein abundance in the cell after HSV-1 infection. Interestingly, within these two clusters, we detected a number of proteins involved in regulation of methylation of lysine at position 4 of histone H3, particularly tri-methylation (H3K4me3) (DPY30, WDR82 or OGT) (39–41); histone H2A mono-ubiquitylation (SKP1) (42); or proteins facilitating transcriptional repression (CTBP2, GATAD2B, LOXL2) (43–48). Furthermore, cluster 15 contained transcriptional regulator ATRX protein, which in complex with DAXX and histone H3.3 is important for heterochromatin silencing at multiple genomic regions (49). ATRX predominantly localizes to highly repetitive heterochromatic sequences via its recognition of 'repressive' histone modifications such as H3K9me3 (50). It has been reported that ATRX and DAXX act as a complex in their role in intrinsic resistance to HSV-1 infection (51). The next largest group of chromatin modifiers was detected in cluster 1 (supplemental Fig. S3), which peaks at 3–6 hpi with a decrease at late time points of infection (9–15 hpi). These proteins included histone-lysine N-methyltransferase SETD7 which is involved in mono-methylation of H3K4 (52), histone-lysine N-methyltransferase SMYD3 which has been suggested to methylate H3K4me inducing di- and tri-methylation but not monomethylation (53); transcription elongation factor SPT6H which is required for suppression of the repressive histone mark H3K27me3 (54); and RNA polymerase II-associated factor 1 homolog PAF1, which is involved in histone modifications such as ubiquitylation of histone H2B and methylation on histone H3K4me3 (55, 56). There was also a group of chromatin binding proteins present in cluster 3 (supplemental Fig. S3), which decreases after 6 hpi. Within this cluster, we identified several heterochromatin proteins such as CBX1, CBX3 and CBX5/HP1 which recognize and bind histone H3 tails methylated at lysine 9 (H3K9me), leading to epigenetic repression (57, 58). CBX proteins are also known to interact with lamin-B receptor (LBR). CBX dual binding function is involved in the association of heterochromatin with the inner nuclear membrane. Infection with HSV-1 results in changes in nuclear architecture consistent with disruption of the nuclear lamina, including enlargement of the nucleus. Furthermore, several studies support the hypothesis that phosphorylation of the lamina by protein kinase C (PRKCA, PRKCD) mediates lamina disruption during HSV-1 nuclear egress (59, 60). Interestingly, the trend of PRKCA (supplemental Fig. S3, cluster 1) suggests the recruitment of this kinase at early (3–6 hpi) and late (12 hpi) time points of infection. The correlation of CBX protein levels with those of lamin kinases points to specific proteins that may be manipulated by HSV-1 to facilitate entry and egress. The other intriguing protein present in cluster 3 was E3 ubiquitin-protein ligase HUWE1 which has been reported to ubiquitylate all four core histones in vitro (61) as well as control the levels or activities of important cellular regulators such as CDC6 (62), MCL1 (63), MYC (64) and p53 (65). Previous studies reported that p53 plays both, positive and negative roles in HSV-1 replication, but overall promotes HSV-1 infection (66). The observed decrease in abundance of HUWE1 protein at early time points of infection may be necessary for HSV-1 to control processes such as cell survival, apoptosis, DNA damage and DNA replication. All proteins described were significantly regulated according to calculated ANOVA p value (< 0.05). Together, this indicates that several epigenetic and structural factors are actively regulated during HSV-1 infection, and future studies will further examine the functional relevance of these factors.
We then examined host proteins most dramatically regulated during virus infection, by selecting the top 50 host proteins according to the largest coefficient of variation between all stages, i.e. proteins with the highest dynamicity during the course of infection. Data were sorted for the ratio mock-3 hpi versus 12–15 hpi, in order to highlight proteins specifically enriched at early and late stages (Fig. 1D). The GO analysis revealed that extracellular matrix organization (SPARC, COL1A1/2, COL12A1, ATA2, ACTBL2), membrane bounded vesicle (AKTIP, NOSTRIN), immune response (IFITM3, CD63) and damaged DNA binding (PRKDC, CSE1L, HMGB2) among the GO-processes enriched at early time points. Proteins involved in RNA/mRNA processing, regulation of gene expression and cytokinesis were the most dynamic at late time points of infection. Microtubule and membrane organization related proteins were distributed evenly. Collectively, the global analysis of the host proteome revealed expected functional enrichments suggesting that our dataset accurately represents proteomic changes during HSV-1 infection.
Viral Proteome Depth and Quantification
We next analyzed the viral proteome during different phases of HSV-1 infection. We searched the MS/MS spectra using the reviewed Human herpesvirus 1 (strain 17; 17syn+) (HHV-1) database, which includes 73 proteins, although the total coding capacity of HSV-1 includes at least 84 transcripts that promote and regulate viral infection and spread (67). We identified and quantified 67 structural and nonstructural HSV-1 proteins (Fig. 1B, supplemental Table S2A). Most viral proteins were identified throughout infection but there was a wide dynamic range in abundance at different times (Fig. 1E). We detected 11 (∼16%) capsid proteins, 24 (∼36%) tegument proteins, 15 (∼22%) envelope proteins and 17 (∼25%) other proteins. We detected ∼90% each of the viral proteins expressed with IE (5 proteins), E (10 proteins) and L (52 proteins) expression kinetics, showing that all three classes of HSV-1 proteins were monitored during the selected infection period. The remaining six proteins not detected may be because of technical limitations such as insufficient solubility, very low abundance or masking by the signal of highly abundant host proteins. Three of these proteins belong to the viral tegument (UL11 and UL55) whereas the other three are in the viral envelope (UL20, UL49.5, US5, and US9) (supplemental Table S2B). Association with membranes is notoriously challenging for MS because of hydrophobicity and limited proteolytic peptides. For comparison, Bell et al. also reported the identification of 67 HSV-1 proteins with similar difficulties detecting UL11, UL20, UL49.5, and US5 proteins (18). Overall, our analysis detected ∼92% of all expected HSV-1 proteins, confirming accurate representation of the viral proteome in our dataset. We then investigated the dynamics of HSV-1 proteins during infection by applying the iBAQ protein label-free quantification to estimate absolute protein abundance. The iBAQ algorithm normalizes the summed peptide intensities by the number of theoretically observable peptides of the protein. The iBAQ values yield a good approximation of protein copy number (29), or at least relative abundance of a protein as compared with the others within the same MS run. Within the IE gene products present at 3 hpi, the ICP0 transactivator protein was the most abundant, followed by ICP47, ICP27, ICP4, and ICP22, respectively (Fig. 1E; supplemental Table S2A). The majority of these proteins peaked in abundance at 9 phi and 12 hpi, and then slightly decreased at 15 hpi. A similar pattern was observed for the 10 identified early proteins. Of these, UL9, UL8, and UL52 had about 8-fold lower abundance than the average of all early HSV-1 proteins. The data indicate that these proteins are expressed at lower abundance than other early proteins during HSV-1 infection. At 3 hpi we also identified 42 proteins expressed from genes characterized as late genes, even though their abundance on average was about 30-fold lower when compared with 15 hpi. Overall, most of the late proteins (38 of the 52 proteins) peaked at 9–12 hpi, whereas the remaining 14 proteins were characterized by monotonic increase across five time points (3–15 hpi) (supplemental Table S2A). These 14 proteins may be necessary for interaction with other cellular/viral targets at subsequent steps of the viral life cycle, or may be incorporated into mature extracellular virions. Indeed, Loret et al. confirmed the presence of 12 of these proteins in mature virions (68). Some reports have detected UL53 in mature virions (69, 70), whereas others did not (71). UL3 was identified in our data at the late time points of HSV-1 infection (9–15 hpi). Neurovirulence factor protein ICP34.5 showed an expression time-course similar to UL3. ICP34.5 contributes to HSV-1 resistance to the antiviral effects of α/β interferon (72, 73). Moreover, it recruits the serine/threonine-protein phosphatase PPP1CA to dephosphorylate the translation initiation factor EIF2A, thereby counteracting the host shutoff of protein synthesis involving double-stranded RNA-dependent protein kinase EIF2AK2 (74). Recently it has been shown that the interaction between ICP34.5 and C1QBP/p32 at late time points of infection leads to the disintegration of nuclear lamina and facilitates nuclear egress of HSV-1 particles (75). Interestingly, ICP34.5 targets peaked at 9–12 hpi except for EIF2AK2, which increased at 15 hpi (supplemental Fig. S3, cluster 13). Thus, the clustering analysis of ICP34.5 targets is consistent with its late translation. Two of the late genes, UL43 and US8.5, were detected only at 6 hpi and 12hpi. The function of both proteins is still unknown, although UL43 encodes a hydrophobic, myristoylated integral membrane protein (76, 77), whereas US8.5 encodes a nuclear protein that localizes to nucleoli and its mRNA is among the most abundant species packaged in virions (78). The protein intensities of US8.5 and UL43 were ∼40 and 270-fold (respectively) lower than the average intensity of all detected HSV-1 proteins. Thus, their lack of detection at 15 hpi may be because of masking by other highly abundant cell-associated virion proteins. In summary, our analysis of the viral proteome is consistent with previously studies and provides a basis for uncovering new viral targets (see supplementary Information section 2 for further discussion of comparisons with prior large-scale proteomics studies).
We next examined the abundance of viral proteins relative to host proteins by comparing intensities. For analysis of the time course-specific proteins in the total proteome, we ranked proteins according to their calculated raw iBAQ abundance within each analyzed condition (supplemental Fig. S4). As expected, we observed that viral proteins increased in relative abundance as compared with host proteins during all time points, however they remained below the absolute intensity of the most abundant host proteins by at least one order of magnitude. Based on this observation, we assume that viral proteins at late time points would not significantly mask or affect accurate quantification of host proteins.
Phosphoproteomics Analysis of Host and Viral Proteome During Infection
Phosphorylation is key to protein regulation and is extensively employed by virus and host (11–15). We therefore analyzed the phosphoproteome of both viral and cellular proteins over the time course of HSV-1 infection. Analysis of the phosphoproteome was highly reproducible in terms of number of phosphorylation sites identified across different replicates (Fig. 3A). Almost 13,000 phosphosites on host proteins were identified and quantified by combining all six conditions, highlighting the quality and sensitivity of our phosphoproteomic approach (supplemental Table S3A). Surprisingly, nearly all viral proteins were found to be phosphorylated (supplemental Table S4A). The relative ratio of host phosphorylation on serine, threonine and tyrosine amino acid residues (S/T/Y 80:20:1) matched that previously reported for the mammalian phosphoproteome (79, 80). Interestingly, this ratio was different (S/T/Y 58:34:8) within the viral phosphoproteome (Fig. 3B). In particular, phosphorylation on threonine residues showed a relative enrichment. However, it is important to note that the overall relative frequency of threonine residues in the virus proteome is about 1:1 with serine residues, whereas in humans the ratio S/T is about 1.6. This aspect partially compensates for the apparent enrichment of phosphothreonine in the viral phosphoproteome. Phosphorylation was also found to be more dynamic than total protein levels (supplemental Fig. S1A and S1B). From the 12,950 host phosphosites quantified, only 5584 were detected in all conditions. In contrast, 111 viral phosphosites out of 571 were identified across five conditions (3–15 hpi; Fig. 3C). In addition, the correlation between different time points was lower for phosphorylation compared with the protein level, both for host (supplemental Fig. S1C) and virus (supplemental Fig. S1D). This is particularly noticeable at the early time point of infection (3 hpi). This was not unexpected, because phosphorylation events are faster and more dynamic than protein expression. We therefore chose to cluster host phosphorylation events that were detected in at least four conditions across all time points, even if in some conditions they were detected in a single biological replicate. The ideal grouping of the ∼9000 phosphosites analyzed fell into 14 clusters (supplemental Fig. S6; supplemental Table S3B). For clustering of the phosphorylation sites on viral proteins we included only those which were identified in a minimum of two out of three biological replicates in at least one time point. Such filters were less stringent than the host phosphoproteome, because the viral phosphoproteome presented many phosphorylation events detected in a single time point. This allowed for the grouping of 455 phosphorylation sites into eight clusters (supplemental Fig. S7; Table S4B). To our knowledge, only 100 of the 571 identified viral potential phosphorylated residues have been previously described (18, 81) (see supplemental Table S4A and associated references).
Fig. 3.
Characterization of phosphoproteome during HSV-1 infection. A, Number of quantified phosphosites for host (purple) and viral (orange) proteins at each time point with biological replicates. B, Counts and relative frequency of amino acid residues modified by phosphorylation within host (purple) and viral (orange) proteins. Viral proteins have a ratio of S:T:Y significantly different from the standard ratio within the mammalian phosphoproteome (80:20:1). C, Histogram representing the number of phosphosites for host (purple) and viral (orange) proteins quantified in each condition. Host phosphosites quantified in at least 4 out of 6 conditions were analyzed using fuzzy c-means clustering. HSV-1 phosphosites were clustered if they were identified in a minimum of two out of three biological replicates in at least one time point. D, Histogram representing the number of phosphorylation sites as a function of the number of (host) proteins. Each mark reflects the number of proteins (y axis) that have the specific number of phosphorylation sites (x axis). Only a single protein has more than 150 phosphorylation sites (SRRM2). E, Top 20 host proteins with the highest number of phosphorylation sites. F, Top 20 viral proteins with the highest number of phosphorylation sites. G, Comparison of our identified viral protein phosphorylation sites (set C, D) to a prior large-scale HSV-1 phosphoproteomics study (set A, B) (18). Confidence - is the confidence score for site localization of the phosphorylation (100% means fully unambiguous).
We next investigated the density of phosphorylation sites across the phosphoproteome and found large differences. We detected 996 proteins with only a single phosphorylation site (Fig. 3D), whereas proteins such as SRRM2 and AHNAK carried respectively 191 and 130 distinct phosphorylation events (Fig. 3E). Viral proteins also displayed variations in the number of phosphorylation sites but the dynamic range was more narrow that that observed in host proteins (Fig. 3F). The viral proteins most enriched for phosphorylation were ICP4 (36 sites) and UL42 (35 sites). A prior report identified 96 unique phosphorylation sites on 37 different HSV-1 proteins (18), of which 62 were covered by our analysis (Fig. 3G). This prior report also found ICP4 (15 sites) as the most heavily phosphorylated viral protein (18). Interestingly, the overall increase in abundance of phosphorylation events on viral proteins appeared delayed as compared with the respective proteome level (supplemental Fig. S5A and S5B). The viral proteome has its highest distribution (expressed in quartiles of iBAQ values) at 12 hpi, whereas the intensities of phosphorylation events were highest at 15 hpi. Together, these data highlight the dynamics of host and viral phosphorylation and potentially dephosphorylation events, and point toward a role for this modification in regulation of processes during viral infection.
Changes to the Chromatin Bound Proteome During HSV-1 Infection
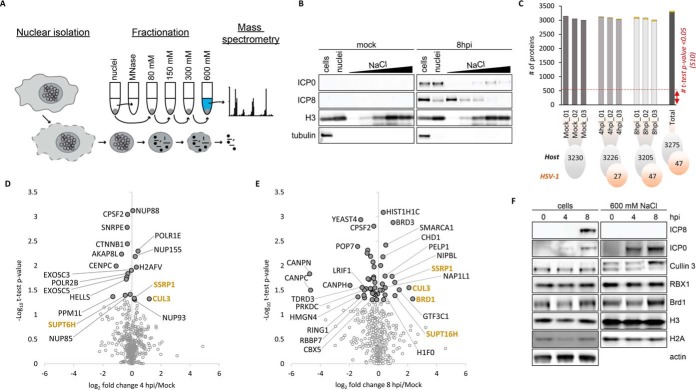
We hypothesized that nuclear replication of HSV-1 may impact the composition of host chromatin and histone PTMs during infection. To investigate chromatin composition during infection, we performed fractionation of nuclei using increasing concentrations of salt to separate proteins tightly bound to DNA (e.g. histones, histone-binding proteins) from those loosely associated with chromatin (e.g. transcription factors) (Fig. 4A) (82, 83). We found that proteins associated with viral DNA (represented by the viral single-stranded DNA binding protein ICP8) elute primarily in low salt fractions compared with proteins such as histones which are tightly bound to host DNA and elute in high salt fractions (Fig. 4B). We did find a subset of viral proteins accumulated in the high salt fraction (e.g. ICP0), suggesting association with host chromatin. We therefore examined whether specific chromatin proteins are differentially associated with host chromatin during infection independently from regulation of their total proteome abundance. To obtain a sufficient amount of material for purification and to test this idea in a second cell model, we performed this study in HeLa cells. We analyzed high salt fractions in uninfected cells (mock) and compared with HSV-1 infections at 4 hpi and 8 hpi. In total we identified and quantified 3275 host proteins, of which 510 were significantly different as compared with mock (t test p value < 0.05 for 4 hpi/mock and 8 hpi/mock) (Fig. 4C; supplemental Table S5A). MS data were also searched against the Uniprot Herpes Simples Virus (Strain 17; 17syn+) database and revealed 47 viral proteins (supplemental Fig. S8A; supplemental Table S6). HSV-1 transcription factor ICP4 and transcriptional regulator ICP22 have been shown to interact with components of the RNA polymerase II complex (84, 85), which are present in our chromatin-enriched fraction. We also detected tegument protein VP22 enriched in the chromatin fraction, consistent with the finding that it binds mitotic chromatin (86, 87). Enrichment may be because of a direct association with host chromatin or accumulation in other subnuclear structures such as the nucleolus that are also high salt soluble. This is also likely the case for viral mRNA export factor ICP27, which interacts with the nuclear pore complex (88). Interestingly, little is known about any chromatin association of viral protein ICP47, which was also detected in our high salt fraction. Future studies will investigate specific roles of these viral proteins on host chromatin and subnuclear structures.
Fig. 4.
Analysis of proteins accumulated in chromatin fraction during HSV-1 infection. A, Workflow of high salt fractionation of chromatin-associated proteins. Purified nuclei were digested with MNase enzyme to separate chromatin into soluble and insoluble fractions. Subsequently, the insoluble fractions were washed with increasing concentrations of salt (80–600 mm NaCl). The high salt fraction, which solubilized the majority of nucleosomes was subjected to MS-analysis. B, Western blots of selected viral (ICP0, ICP8), and host (H3, tubulin) proteins showing their abundance at mock and 8 hpi in the whole HeLa cell extract, whole nuclei extract, MNase treated fraction, and individual fractions over the salt gradient (80, 150, 300, and 600 mm NaCl). C, Number of quantified proteins for host (gray) and virus (orange) at each time point and biological replicate analyzed in the high salt fraction (600 mm NaCl). D, Volcano plot representing the fold change and significance of host protein abundance at 4 hpi versus mock and (E) 8 hpi versus mock. The standard significant threshold (t test p value < 0.05) transformed in -Log10 is 1.30. 'Light gray dots' highlight t test p value > 0.05. Only proteins involved in chromatin organization processes were highlighted (annotation retrieved from MetaCore database; False Discovery Rate (FDR < 0.05). F, Western blots of selected host proteins representing their abundance at 0, 4 and 8 hpi in the whole cell extract and the high salt fraction (600 mm NaCl). BRD1 and Cullin 3 accumulated in the high salt fraction at 8 hpi.
The number of host proteins within the high salt chromatin fraction became progressively more differentially regulated during infection (supplemental Fig. S8B and S8C). We filtered the dataset for proteins involved in chromatin organization based on the MetaCore and UniProt knowledge database (Fig. 4D and 4E). The majority of host chromatin and histone modifying proteins had relatively small differences in abundance across the analyzed time points, and only 14 significant proteins (t test p value < 0.05) had a fold change larger than 2 (supplemental Table S5B). Between 4 and 8 hpi we observed a gradual increase in abundance of some cellular proteins in the high salt fraction, such as cullin-3 (CUL3), FACT complex subunit SSRP1, and transcription elongation factor SPT6 (SUPT16H) (Fig. 4D and 4E). Cullin-3 is the component of cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex involved in ubiquitylation of histone macroH2A variant (89, 90). SSRP1 and SUPT16H are the components of the FACT (FAcilitates Chromatin Transcription) histone chaperone complex which is thought to reorganize nucleosomes through the destabilization of multiple intranucleosome contacts (91–93). The FACT complex activity has been linked to methylation of lysine 4 of histone H3, which is a mark enriched on gene promoters undergoing active transcription. This up-regulation of FACT in the high salt fraction is consistent with higher levels of H3K4me3 at early times (3–4 hpi) compared with late points in infection (9–12 hpi) (see Quantification of Histone Post-translational Modifications During Infection). H3K4me3 is associated with active transcription and the global decrease in this mark seen during HSV-1 infection is consistent with shut down of host transcription. Conversely, specifically at 8 hpi we observed a 5-fold increase for BRD1 as compared with mock (Fig. 4E). To validate our MS results, we used WB to examine the differential accumulation of CUL3, RBX1, and BRD1 proteins within both the total cell lysate and the high salt fractions (Fig. 4F). We confirmed enrichment of CUL3 and BRD1 proteins in the high salt fraction at 8 hpi, whereas RBX1 abundance was similar across analyzed time points for both total cellular extract and high salt fractions. BRD1 is a component of histone acetyltransferase (HAT) complexes of the MYST family together with Tip60 (KAT5), Hbo1 (KAT7), Moz (KAT6A)/Morf (KAT6B) and Mof (KAT8). BRD1 forms a novel HAT complex with Hbo1 and is responsible for the global acetylation of histone H3 at lysine 14 (H3K14ac) (94). Furthermore, BRD1 was found to bind regions that highly overlap with histone H3K9ac (94). We find a steady increase in H3K9ac throughout infection (see below and Fig. 6), consistent with an increase in BRD1 enriched in the high salt fraction. This may correlate with active transcription in response to infection which would reflect a different subset of genes than those with decreasing H3K4me3. Further studies using chromatin immunoprecipitation experiments to complement our proteomic approach will uncover which genes are marked by these dynamic methylated and acetylated histones.
Fig. 6.

Validation of changes in histone marks during HSV-1 infection of HFF cells. A, Western blot analysis of selected histone marks quantified by MS over the time course (3–15 hpi) of HSV-1 infection in HFFs. The different marks are indicated and compared with uninfected mock (M) extracts. B, Ponceau stain of lysates used in (A) showing even loading of histone proteins in all samples. C, Immunofluorescence imaging of H3K9ac (green) at 3, 6, and 9 hpi. ICP8 (red) marks viral replication compartments and DNA is stained with DAPI (blue). Scale bar is 10 μm. D, Same as (C) for H3K14ac.
Quantification of Histone Post-translational Modifications During Infection
Finally, we used our optimized in-house workflow to analyze cellular histone PTM changes during HSV-1 infection. To isolate histones we performed purification via acidic extraction, and followed this by derivatization with propionic anhydride (31). After digestion, we performed data-independent acquisition in MS to quantify accurately the canonical and isobaric peptides (95, 96). Chromatographic peak area extraction was performed using the software EpiProfile (32). We quantified 140 different histone peptides (Fig. 5A; supplemental Table S7A), including peptides from H3, H4, H2A, H2B, and H1. Because histone tails can contain several PTMs it is impossible to determine which modifications occur on the same protein. There are some exceptions such as H3.3K27 and K36 modifications, because the peptide aa 27-40 carries both modifications and the sequence variant. In our study, we found a high degree of correlation between replicates and conditions, with most comparisons having a Pearson correlation > 0.9 (supplemental Fig. S9A). Despite the minimal changes between time points, we could perform a clear grouping by PCA where replicates of the same condition clustered together (supplemental Fig. S9B). At 15 hpi we observed more scattered distribution of replicates, which may reflect the asynchronous nature of infection. To show the effect of HSV-1 infection on global histone PTMs, we plotted fold change and significance of peptide relative abundance. We compared 6 hpi and 15 hpi to show examples of the variation in changes. The volcano plot of 6 hpi versus mock showed smaller significant changes as compared with 15 hpi versus mock (Fig. 5B). Peptide abundance was then organized in a hierarchical clustering (supplemental Fig. S10) such that trends were grouped for all quantified forms.
Fig. 5.
Quantification of modified histone peptides during HSV-1 infection. A, Number of quantified histone peptides in each replicate and condition. B, Volcano plot representing fold-change and significance of histone peptide abundance at 6 hpi versus mock (top) and at 15 hpi versus mock (bottom). The standard significant threshold (t test p value < 0.05) transformed in -log2 is 4.32. C, Relative abundance of selected modified peptides. Error bars represent standard deviation (n = 3). D, Heatmap of deconvoluted histone H3 and H4 marks calculated using the sum of all peptides carrying a given mark. For instance, the abundance of H3K27me2 was calculated by summing all modified forms of the peptide aa 27–40 containing the given modification (with and without modified K36).
To analyze global histone modifications we calculated the abundance of single PTMs using all peptides and compared the trends of single marks (supplemental Table S7B). For example, to estimate globally the relative abundance of H3K14ac, we summed the relative abundances of all peptides carrying this modification (aa 9–17) (i.e. K9unmodK14ac + K9me1K14ac + K9me2K14ac + K9me3K14ac + K9acK14ac). One example of increased abundance over time was H1K25me2 (Fig. 5C). In addition, the adjacent phosphorylation event on histone H1S26ph was found to be significantly reduced after infection (Fig. 5C). We generated a simplified heatmap including the overall abundance of each single mark (Fig. 5D). H3K9ac and H3K14ac were found to increase steadily during the course of infection, and this was confirmed by WB (Fig. 6A). The marks H3K4me1 and H3K4me3 are enriched on active enhancers and promoters, respectively. H3K4me3 was not detected by MS because of its low abundance, however, WB showed a monotonic decrease in abundance during infection (Fig. 6A). A similar trend was observed for H3K4me1 from MS data (Fig. 5D). We also performed IF to examine histone marks during HSV-1 infection (Fig. 6). We observed a decrease in H3K4me3, as well as increases in immunostaining for H3K9ac and H3K14ac. Intriguingly, the increased abundance of H3K9ac and H3K14ac did not appear to accumulate at viral replication compartments (Fig. 6C and 6D), suggesting that these two modifications are predominantly altered on histones that are in host chromatin and not on viral genomes. A further interesting observation was the PTM H3S10ph, which was found to increase after infection and peak at 9 hpi (Fig. 5D and 6A). This mark is usually enriched on chromatin during mitosis (97). Interestingly, the most abundant PTM combination of this phosphorylation was on the peptide modified as H3K9me3S10ph (supplemental Table S7A). This mark is usually associated with interruption of gene repression as an example of a methyl/phospho switch, because the H3K9me3 reader CBX5/HP1 cannot bind the histone PTM in the presence of S10 phosphorylation (98). HSV-1 was recently shown to take advantage of the methyl/phospho switch to reactivate from latency (99), and this might also benefit the lytic phase of infection. The additionally tested histone PTMs such as H3K9me1, H3K27me3 and H3K56me1 have not shown significant difference across six time points of HSV-1 infection (Fig. 6A). Together, our data highlight that histone PTMs are dynamically affected during viral infection and provide a resource for future studies into chromatin control during HSV-1 infection.
Summary and Future Directions
Our study demonstrates the potential for combining multiple proteomic datasets with clustering analysis to define pathways that regulate the global proteome of human cells during viral infection. Collectively, these proteomic approaches provide unprecedented resolution and the largest and most comprehensive proteomic analysis of HSV-1 infection. The work includes host and virus proteome, phosphoproteome, chromatin enriched proteome and comprehensive analysis of histone PTMs. A comparison with a recent publication investigating the same model system (100) and GO analysis confirmed the robustness of the dataset, making it an important repository for future biological studies. In addition, the chromatin-related data we provide, including enrichment of chromatin bound proteins and histone marks, will pave the way for a deeper understanding of epigenetic alterations to host chromatin during HSV-1 infection. Our experiments were performed using multiple time points to provide temporal resolution and allow correlation analysis across datasets, e.g. proteome and histone PTMs. Here we provide examples of how our data can be used to guide investigation into epigenetic changes on host and/or viral chromatin (see supplemental Table S1-S7 for a layout of navigating our data set). First, our histone PTM data revealed that methylation on H3K4 (H3K4me) was downregulated (Fig. 5D and 6A). Proteomics data highlighted that several proteins involved in the catalysis of these marks, including DPY30, WDR82 or OGT, were also found downregulated at early times (3 hpi) or throughout the infection process (supplemental Fig. S3, clusters 10 and 15). Even more striking was the regulation of SETD7 and PAF1 (supplemental Fig. S3, cluster 1), up-regulated until 6 hpi and then downregulated, matching the trend of H3K4me1 (Fig. 5D) and H3K4me3 (Fig. 6A). Together, these data suggest that H3K4 methylation and proteins involved in depositing and reading this mark are affected by HSV-1 infection. Heterochromatin binding proteins CBX1, CBX3 and CBX5/HP1 were all also found downregulated from 9 hpi (supplemental Fig. S3, cluster 3), providing another example of chromatin factors affected by HSV-1 infection. These proteins bind H3K9me3, and to a lesser degree H1K25me2, which are both marks commonly associated with heterochromatin. As mentioned above, H3S10ph, which can prevent CBX5/HP1 binding to H3K9me3 (98), peaked at 9 hpi (Fig. 5D; Fig. 6A). The increase in abundance of H3S10ph is therefore consistent with the reduced abundance of CBX5/HP1 proteins within the chromatin-enriched proteome (Fig. 4E; supplemental Table S5B). The interaction of CBX proteins with lamin B receptor (LBR) contributes to the association of heterochromatin with the inner nuclear membrane. HSV-1 has been shown to induce CBX5/HP1 dissociation from LBR resulting in destabilization of the nuclear envelope. Our study thus presents a combinatorial approach that confirms the existing data that HSV-1 disrupts the nuclear envelope likely through interrupting H3K9me3-based heterochromatin and lamin association of CBX proteins. Phosphorylation of lamin proteins and associated disruption of the lamina is a conserved feature of herpesvirus infection (16, 101–106). Phosphorylation of lamina components during infection is mediated by both viral and cellular kinases. Previous studies suggested that viral US3 kinase may affect the lamin A/C (LMNA) disrupting function of the envelopment apparatus by UL31 phosphorylation (59). The phosphorylated form of UL31 (at S26, S27, and S43) was suggested to be associated with the inner nuclear membrane, thereby preventing UL31/UL34 aggregation and premature budding of virus (107). Interestingly, the trends of US3 and UL31 expression patterns and identified UL31 phosphosites (S24, S26, S27) (supplemental Fig. S11A and S11B; supplemental Fig. S7, cluster 4) followed the trend of LMNA protein (supplemental Fig. S11C; supplemental Fig. S3, cluster 13). This may support the significance of S24, S26, S27 sites in the regulation of UL31 localization and function. Overall, our data provide specific temporal information of protein and PTM abundance during HSV-1 infection, which will guide future studies into their significance for the virus lifecycle.
DATA AVAILABILITY
The mass spectrometry proteomics and phosphoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD005467. All MS-histone raw files have been deposited in the Chorus database (https://chorusproject.org/) under project number 1254.
Acknowledgments
We thank members of the Weitzman and Garcia labs for scientific input and G. Cohen for generously providing antibodies to ICP4.
Footnotes
Author contributions: K.K., S.S., B.A.G., and M.D.W. designed research; K.K., D.C.A., C.H., A.N.D., and E.K. performed research; K.K. and S.S. analyzed data; K.K., D.C.A., S.S., B.A.G., and M.D.W. wrote the paper.
* This work was supported by grants from the National Institutes of Health (R01-NS082240 to MDW, R01-GM110174 and R01-AI118891 to BAG). DCA was supported in part by National Institutes of Health Training grants (T32-CA115299 and F32-GM112414). We gratefully acknowledge additional funding from a Leukemia & Lymphoma Society Dr Robert Arceci Scholar Award (BAG), the Institute for Immunology of the University of Pennsylvania (BAG and MDW), and funds from the Children's Hospital of Philadelphia (MDW). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HSV-1
- herpes simplex virus type-1
- BP
- biological process
- CID
- collision-induced dissociation
- DDA
- data-dependent acquisition-based mass spectrometry
- DIA
- data-independent acquisition-based mass spectrometry
- DTT
- dithiothreitol
- E
- herpes simplex virus type 1 early gene expression
- E-L
- herpes simplex virus type 1 early-late gene expression
- FDR
- false discovery rate
- GO
- Gene Ontology
- HeLa
- epithelial adenocarcinoma cells
- HFF
- human foreskin fibroblast cells
- hpi
- hours post-infection
- IAA
- iodoacetamide
- iBAQ
- intensity-based absolute quantification
- IE
- herpes simplex virus type 1 immediate-early gene expression
- IF
- immunofluorescence
- L
- herpes simplex virus type 1 late gene expression
- MOI
- multiplicity of infection
- nLC-MS/MS
- nano-liquid chromatography tandem mass spectrometry
- PCA
- principal component analysis
- PTMs
- post-translational modifications
- SILAC
- stable isotope labeling by amino acids in cell culture
- TCA
- trichloroacetic acid
- TFA
- trifluoroacetic acid
- TiO2
- titanium dioxide
- WB
- Western blot.
REFERENCES
- 1. Knipe D. M., and Howley P. (2013) Fields Virology (Lippincott Williams & Wilkins; ) [Google Scholar]
- 2. Nicoll M. P., Proença J. T., and Efstathiou S. (2012) The molecular basis of herpes simplex virus latency. FEMS Microbiol. Rev. 36, 684–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knipe D. M., and Cliffe A. (2008) Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6, 211–221 [DOI] [PubMed] [Google Scholar]
- 4. Weller S. K., and Coen D. M. (2012) Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb Perspect Biol 4, ea013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oh J., and Fraser N. W. (2008) Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82, 3530–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kristie T. M. (2015) Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology 479- 480, 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cliffe A. R., and Knipe D. M. (2008) Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82, 12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saffran H. A., Pare J. M., Corcoran J. A., Weller S. K., and Smiley J. R. (2007) Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 8, 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cibulka J., Fraiberk M., and Forstova J. (2012) Nuclear Actin and Lamins in Viral Infections. Viruses 4, 325–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vastag L., Koyuncu E., Grady S. L., Shenk T. E., and Rabinowitz J. D. (2011) Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 7, e1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogle W. O., Ng T. I., Carter K. L., and Roizman B. (1997) The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology 235, 406–413 [DOI] [PubMed] [Google Scholar]
- 12. Wilcox K. W., Kohn A., Sklyanskaya E., and Roizman B. (1980) Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J. Virol. 33, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xia K., Knipe D. M., and DeLuca N. A. (1996) Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J. Virol. 70, 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xia K., DeLuca N. A., and Knipe D. M. (1996) Analysis of phosphorylation sites of herpes simplex virus type 1 ICP4. J. Virol. 70, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhi Y., and Sandri-Goldin R. M. (1999) Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J. Virol. 73, 3246–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cano-Monreal G. L., Wylie K. M., Cao F., Tavis J. E., and Morrison L. A. (2009) Herpes simplex virus 2 UL13 protein kinase disrupts nuclear lamins. Virology 392, 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu S., Pan S., Zhang L., Baines J., Roller R., Ames J., Yang M., Wang J., Chen D., Liu Y., Zhang C., Cao Y., and He B. (2016) Herpes simplex virus 1 induces phosphorylation and reorganization of lamin A/C through the γ134.5 protein that facilitates nuclear egress. J. Virol. 90, 10414–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bell C., Desjardins M., Thibault P., and Radtke K. (2013) Proteomics analysis of herpes simplex virus type 1-infected cells reveals dynamic changes of viral protein expression, ubiquitylation, and phosphorylation. J. Proteome Res. 12, 1820–1829 [DOI] [PubMed] [Google Scholar]
- 19. Knipe D. M., Lieberman P. M., Jung J. U., McBride A. A., Morris K. V., Ott M., Margolis D., Nieto A., Nevels M., Parks R. J., and Kristie T. M. (2013) Snapshots: chromatin control of viral infection. Virology 435, 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watson Z., Dhummakupt A., Messer H., Phelan D., and Bloom D. (2013) Role of polycomb proteins in regulating HSV-1 latency. Viruses 5, 1740–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou G., Te D., and Roizman B. (2010) The CoREST/REST repressor is both necessary and inimical for expression of herpes simplex virus genes. MBio 2, e00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor T. J., and Knipe D. M. (2004) Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78, 5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bryant K. F., Colgrove R. C., and Knipe D. M. (2011) Cellular SNF2H chromatin-remodeling factor promotes herpes simplex virus 1 immediate-early gene expression and replication. MBio 2, e00330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., and Weitzman M. D. (2005) DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U.S.A. 102, 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Avgousti D. C., Herrmann C., Kulej K., Pancholi N. J., Sekulic N., Petrescu J., Molden R. C., Blumenthal D., Paris A. J., Reyes E. D., Ostapchuk P., Hearing P., Seeholzer S. H., Worthen G. S., Black B. E., Garcia B. A., and Weitzman M. D. (2016) A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 535, 173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thingholm T. E., and Larsen M. R. (2016) The Use of Titanium Dioxide for Selective Enrichment of Phosphorylated Peptides. Methods Mol. Biol. 1355, 135–146 [DOI] [PubMed] [Google Scholar]
- 27. Engholm-Keller K., Birck P., Størling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 30. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 31. Sidoli S., Bhanu N. V., Karch K. R., Wang X., and Garcia B. A. (2016) Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J. Vis. Exp. e54112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan Z.-F., Lin S., Molden R. C., Cao X.-J., Bhanu N. V., Wang X., Sidoli S., Liu S., and Garcia B. A. (2015) EpiProfile quantifies histone modifications by extracting retention time and intensity in high-resolution mass spectra. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 34. Schwämmle V., and Jensen O. N. (2010) A simple and fast method to determine the parameters for fuzzy c-means cluster analysis. Bioinformatics 26, 2841–2848 [DOI] [PubMed] [Google Scholar]
- 35. Conwell S. E., White A. E., Harper J. W., and Knipe D. M. (2015) Identification of TRIM27 as a novel degradation target of herpes simplex virus 1 ICP0. J. Virol. 89, 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ushijima Y., Luo C., Kamakura M., Goshima F., Kimura H., and Nishiyama Y. (2010) Herpes simplex virus UL56 interacts with and regulates the Nedd4-family ubiquitin ligase Itch. Virol. J. 7, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Narita R., Takahasi K., Murakami E., Hirano E., Yamamoto S. P., Yoneyama M., Kato H., and Fujita T. (2014) A novel function of human Pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog. 10, e1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Angelova A., Tenev T., and Varadinova T. (2004) Expression of cellular proteins Bcl-X(L), XIAP and Bax involved in apoptosis in cells infected with herpes simplex virus 1 and effect of pavine alkaloid (-)-thalimonine on virus-induced suppression of apoptosis. Acta Virol. 48, 193–196 [PubMed] [Google Scholar]
- 39. Patel A., Dharmarajan V., Vought V. E., and Cosgrove M. S. (2009) On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 284, 24242–24256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu M., Wang P. F., Lee J.-S., Martin-Brown S., Florens L., Washburn M., and Shilatifard A. (2008) Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28, 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., Urh M., Daniels D. L., and Fuks F. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gearhart M. D., Corcoran C. M., Wamstad J. A., and Bardwell V. J. (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brackertz M., Boeke J., Zhang R., and Renkawitz R. (2002) Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 277, 40958–40966 [DOI] [PubMed] [Google Scholar]
- 44. Brackertz M., Gong Z., Leers J., and Renkawitz R. (2006) p66alpha and p66beta of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 34, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iturbide A., García de Herreros A., and Peiró S. (2015) A new role for LOX and LOXL2 proteins in transcription regulation. FEBS J. 282, 1768–1773 [DOI] [PubMed] [Google Scholar]
- 46. Shi Y., Sawada J.-I., Sui G., Affar E. B., Whetstine J. R., Lan F., Ogawa H., Luke M. P.-S., Nakatani Y., and Shi Y. (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738 [DOI] [PubMed] [Google Scholar]
- 47. Ray S. K., Li H. J., Metzger E., Schüle R., and Leiter A. B. (2014) CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells. Mol. Cell. Biol. 34, 2308–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuppuswamy M., Vijayalingam S., Zhao L.-J., Zhou Y., Subramanian T., Ryerse J., and Chinnadurai G. (2008) Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol. Cell. Biol. 28, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Voon H. P. J., and Wong L. H. (2016) New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res. 44, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Noh K.-M., Maze I., Zhao D., Xiang B., Wenderski W., Lewis P. W., Shen L., Li H., and Allis C. D. (2015) ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons. Proc. Natl. Acad. Sci. U. S. A. 112, 6820–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lukashchuk V., and Everett R. D. (2010) Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84, 4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Couture J.-F., Collazo E., Hauk G., and Trievel R. C. (2006) Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 13, 140–146 [DOI] [PubMed] [Google Scholar]
- 53. Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F. P., Li M., Yagyu R., and Nakamura Y. (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nature Cell Biol. 6, 731–740 [DOI] [PubMed] [Google Scholar]
- 54. Wang A. H., Zare H., Mousavi K., Wang C., Moravec C. E., Sirotkin H. I., Ge K., Gutierrez-Cruz G., and Sartorelli V. (2013) The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J. 32, 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wood A., Schneider J., Dover J., Johnston M., and Shilatifard A. (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 56. Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., and Shilatifard A. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 57. Kaustov L., Ouyang H., Amaya M., Lemak A., Nady N., Duan S., Wasney G. A., Li Z., Vedadi M., Schapira M., Min J., and Arrowsmith C. H. (2011) Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286, 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eissenberg J. C., and Elgin S. C. R. (2014) HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 30, 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Park R., and Baines J. D. (2006) Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Leach N. R., and Roller R. J. (2010) Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology 406, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Z., Oughtred R., and Wing S. S. (2005) Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell. Biol. 25, 2819–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hall J. R., Kow E., Nevis K. R., Lu C. K., Luce K. S., Zhong Q., and Cook J. G. (2007) Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell 18, 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhong Q., Gao W., Du F., and Wang X. (2005) Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 64. Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Kogel U., Scheffner M., Helin K., and Eilers M. (2005) The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123, 409–421 [DOI] [PubMed] [Google Scholar]
- 65. Chen D., Kon N., Li M., Zhang W., Qin J., and Gu W. (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 66. Maruzuru Y., Fujii H., Oyama M., Kozuka-Hata H., Kato A., and Kawaguchi Y. (2013) Roles of p53 in herpes simplex virus 1 replication. J. Virol. 87, 9323–9332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Honess R. W., and Roizman B. (1975) Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. U.S.A. 72, 1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Loret S., Guay G., and Lippé R. (2008) Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82, 8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Klupp B. G., Baumeister J., Dietz P., Granzow H., and Mettenleiter T. C. (1998) Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72, 1949–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Foster T. P., Rybachuk G. V., and Kousoulas K. G. (2001) Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75, 12431–12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hutchinson L., Roop-Beauchamp C., and Johnson D. C. (1995) Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J. Virol. 69, 4556–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cheng G., Brett M. E., and He B. (2001) Val193 and Phe195 of the gamma 1 34.5 protein of herpes simplex virus 1 are required for viral resistance to interferon-alpha/beta. Virology 290, 115–120 [DOI] [PubMed] [Google Scholar]
- 73. Leib D. A., Alexander D. E., Cox D., Yin J., and Ferguson T. A. (2009) Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J. Virol. 83, 12164–12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li Y., Zhang C., Chen X., Yu J., Wang Y., Yang Y., Du M., Jin H., Ma Y., He B., and Cao Y. (2011) ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2alpha (eIF2alpha) and protein phosphatase 1. J. Biol. Chem. 286, 24785–24792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y., Yang Y., Wu S., Pan S., Zhou C., Ma Y., Ru Y., Dong S., He B., Zhang C., and Cao Y. (2014) p32 is a novel target for viral protein ICP34.5 of herpes simplex virus type 1 and facilitates viral nuclear egress. J. Biol. Chem. 289, 35795–35805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Carter K. L., Ward P. L., and Roizman B. (1996) Characterization of the products of the U(L)43 gene of herpes simplex virus 1: potential implications for regulation of gene expression by antisense transcription. J. Virol. 70, 7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. MacLean C. A., Efstathiou S., Elliott M. L., Jamieson F. E., and McGeoch D. J. (1991) Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72 (Pt 4), 897–906 [DOI] [PubMed] [Google Scholar]
- 78. Georgopoulou U., Kakkanas A., Miriagou V., Michaelidou A., and Mavromara P. (1995) Characterization of the US8.5 protein of herpes simplex virus. Arch. Virol 140, 2227–2241 [DOI] [PubMed] [Google Scholar]
- 79. Sharma K., D'Souza R. C. J., Tyanova S., Schaab C., Wiśniewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 80. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 81. Antrobus R., Grant K., Gangadharan B., Chittenden D., Everett R. D., Zitzmann N., and Boutell C. (2009) Proteomic analysis of cells in the early stages of herpes simplex virus type-1 infection reveals widespread changes in the host cell proteome. Proteomics 9, 3913–3927 [DOI] [PubMed] [Google Scholar]
- 82. Teves S. S., and Henikoff S. (2012) Salt fractionation of nucleosomes for genome-wide profiling. Methods Mol. Biol. 833, 421–432 [DOI] [PubMed] [Google Scholar]
- 83. Zaret K. (2005) Micrococcal nuclease analysis of chromatin structure. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21.1 [DOI] [PubMed] [Google Scholar]
- 84. Lester J. T., and DeLuca N. A. (2011) Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediator in virus-infected cells. J. Virol. 85, 5733–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rice S. A., and Davido D. J. (2013) HSV-1 ICP22: hijacking host nuclear functions to enhance viral infection. Future Microbiol. 8, 311–321 [DOI] [PubMed] [Google Scholar]
- 86. Martin A., O'Hare P., McLauchlan J., and Elliott G. (2002) Herpes simplex virus tegument protein VP22 contains overlapping domains for cytoplasmic localization, microtubule interaction, and chromatin binding. J. Virol. 76, 4961–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ingvarsdottir K., and Blaho J. A. (2010) Association of the herpes simplex virus major tegument structural protein VP22 with chromatin. Biochim. Biophys. Acta 1799, 200–206 [DOI] [PubMed] [Google Scholar]
- 88. Malik P., Tabarraei A., Kehlenbach R. H., Korfali N., Iwasawa R., Graham S. V., and Schirmer E. C. (2012) Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J. Biol. Chem. 287, 12277–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhuang M., Calabrese M. F., Liu J., Waddell M. B., Nourse A., Hammel M., Miller D. J., Walden H., Duda D. M., Seyedin S. N., Hoggard T., Harper J. W., White K. P., and Schulman B. A. (2009) Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hernández-Muñoz I., Lund A. H., van der Stoop P., Boutsma E., Muijrers I., Verhoeven E., Nusinow D. A., Panning B., Marahrens Y., and van Lohuizen M. (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 102, 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., and Formosa T. (2009) yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., and Reinberg D. (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 [DOI] [PubMed] [Google Scholar]
- 93. Orphanides G., Wu W. H., Lane W. S., Hampsey M., and Reinberg D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 [DOI] [PubMed] [Google Scholar]
- 94. Fryland T., Christensen J. H., Pallesen J., Mattheisen M., Palmfeldt J., Bak M., Grove J., Demontis D., Blechingberg J., Ooi H. S., Nyegaard M., Hauberg M. E., Tommerup N., Gregersen N., Mors O., Corydon T. J., Nielsen A. L., and Børglum A. D. (2016) Identification of the BRD1 interaction network and its impact on mental disorder risk. Genome Med. 8, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sidoli S., Simithy J., Karch K. R., Kulej K., and Garcia B. A. (2015) Low Resolution Data-Independent Acquisition in an LTQ-Orbitrap Allows for Simplified and Fully Untargeted Analysis of Histone Modifications. Anal. Chem. 87, 11448–11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sidoli S., Lin S., Xiong L., Bhanu N. V., Karch K. R., Johansen E., Hunter C., Mollah S., and Garcia B. A. (2015) SWATH Analysis for Characterization and Quantification of Histone Post-translational Modifications. Mol. Cell. Proteomics 14, 2420–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., and Allis C. D. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 [DOI] [PubMed] [Google Scholar]
- 98. Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., and Allis C. D. (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 99. Cliffe A. R., Arbuckle J. H., Vogel J. L., Geden M. J., Rothbart S. B., Cusack C. L., Strahl B. D., Kristie T. M., and Deshmukh M. (2015) Neuronal stress pathway mediating a histone methyl/phospho switch is required for Herpes Simplex virus reactivation. Cell Host Microbe 18, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Berard A. R., Coombs K. M., and Severini A. (2015) Quantification of the host response proteome after herpes simplex virus type 1 infection. J. Proteome Res. 14, 2121–2142 [DOI] [PubMed] [Google Scholar]
- 101. Camozzi D., Pignatelli S., Valvo C., Lattanzi G., Capanni C., Dal Monte P., and Landini M. P. (2008) Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89, 731–740 [DOI] [PubMed] [Google Scholar]
- 102. Bjerke S. L., and Roller R. J. (2006) Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hamirally S., Kamil J. P., Ndassa-Colday Y. M., Lin A. J., Jahng W. J., Baek M.-C., Noton S., Silva L. A., Simpson-Holley M., Knipe D. M., Golan D. E., Marto J. A., and Coen D. M. (2009) Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5, e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Reynolds A. E., Liang L., and Baines J. D. (2004) Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78, 5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Simpson-Holley M., Baines J., Roller R., and Knipe D. M. (2004) Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78, 5591–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Simpson-Holley M., Colgrove R. C., Nalepa G., Harper J. W., and Knipe D. M. (2005) Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79, 12840–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mou F., Wills E., and Baines J. D. (2009) Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83, 5181–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics and phosphoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD005467. All MS-histone raw files have been deposited in the Chorus database (https://chorusproject.org/) under project number 1254.