Abstract
Cryptosporidium parvum causes severe diarrhea in infants in developing countries and in immunosuppressed persons, including those with AIDS. We are interested in the Asn-linked glycans (N-glycans) of C. parvum, because (1) the N-glycan precursor is predicted to contain five mannose and two glucose residues on a single long arm versus nine mannose and three glucose residues on the three-armed structure common in host N-glycans, (2) C. parvum is a rare eukaryote that lacks the machinery for N-glycan-dependent quality control of protein folding in the lumen of the Endoplasmic Reticulum (ER), and (3) ER and Golgi mannosidases, as well as glycosyltransferases that build complex N-glycans, are absent from the predicted proteome. The C. parvum N-glycans reported here, which were determined using a combination of collision-induced dissociation and electronic excitation dissociation, contain a single, unprocessed mannose arm ± terminal glucose on the trimannosyl chitobiose core. Upon nanoUPLC-MS/MS separation and analysis of the C. parvum tryptic peptides, the total ion and extracted oxonium ion chromatograms delineated 32 peptides with occupied N-glycan sites; these were derived from 16 glycoproteins. Although the number of potential N-glycan sites with Thr (NxT) is only about twice that with Ser (NxS), almost 90% of the occupied N-glycan sites contain NxT. The two most abundant C. parvum proteins modified with N-glycans were an immunodominant antigen on the surface of sporozoites (gp900) and the possible oocyst wall protein 1 (POWP1). Seven other glycoproteins with N-glycans were unique to C. parvum; five shared common ancestry with other apicomplexans; two glycoproteins shared common ancestry with many organisms. In summary, C. parvum N-glycans are remarkable for the absence of ER and Golgi modification and for the strong bias toward occupancy of N-glycan motifs containing Thr.
Cryptosporidium parvum and Cryptosporidium hominis are coccidian parasites (walled apicomplexans) that infect humans (both) and cows (C. parvum only) (1–3). C. parvum and C. hominis are leading causes of diarrhea and death in children in the developing world and cause chronic diarrhea in AIDS patients (4–7). Although the massive outbreak of C. parvum in Milwaukee in 1993 was associated with contamination of municipal water, Cryptosporidium in developing countries is likely spread by poor hygiene (8, 9). Although there are mouse vaccine models and veterinary vaccines for C. parvum, there are no human vaccines (10–14). Furthermore, Nitazoxanide, the drug used to treat C. parvum, is not effective in immune suppressed persons (15).
We are interested in the N-glycans of Cryptosporidium for numerous reasons. Like Toxoplasma gondii, Entamoeba histolytica, and Trichomonas vaginalis, there is secondary loss of ALG genes, so that the predicted N-glycan precursor of C. parvum has a single long arm rather than the three-arm structure common in the host (supplemental Table S1) (16–21). In contrast to most other eukaryotes, C. parvum has a paucity of predicted mannosidases and glycosyltransferases, which could modify N-glycans in the endoplasmic reticulum (ER)1 and Golgi (22). C. parvum sporozoites label with cyanovirin-N, an anti-retroviral lectin that binds to the high mannose N-glycans of gp120 in HIV (23, 24). C. parvum is a rare eukaryote that lacks the machinery for N-glycan-dependent quality control of protein folding, and there is no positive selection for N-glycan sites in secreted proteins of C. parvum (25–27). Antigenic proteins on the surface of Cryptosporidium sporozoites (e.g. gp900 and gp40/gp15), oocyst wall proteins (COWPs) and possible oocyst wall proteins (POWPs), are glycoproteins with numerous predicted N-glycan sites (24, 28–40). Finally, Concanavalin A, which binds some N-glycans and other mannose-containing structures, recognizes numerous C. parvum antigens, Whereas release of N-glycans reduces binding of immune sera to parasite proteins on Western blots (41).
Here we used tandem mass spectrometry to identify the proteins that contain N-glycans, and determine the structures of N-glycans released with PNGaseF. We found that the N-glycans of C. parvum contain a single long arm, are barely processed in the ER or Golgi, and show an extreme bias for sequons with threonine.
EXPERIMENTAL PROCEDURES
Parasites and Reagents
C. parvum oocysts were purchased from Bunch Grass Farm (Deary, ID) and handled under BSL-2 protocols approved by the Boston University Institutional Biosafety Committee. All chemicals and reagents, including proteomics grade trypsin, were obtained from Sigma-Aldrich (St. Louis, MO), unless otherwise stated. All solvents used for LC-MS were Fisher Scientific Optima™ grade (Thermo-Fisher Scientific, Waltham, MA). PNGase F was from New England Biolabs (Ipswich, MA).
Protein Extraction
Two distinct methods were utilized to extract proteins from whole C. parvum oocysts. The first method used a combination of mechanical disruption and detergent extraction. Briefly, 109 oocysts were concentrated by centrifugation at 1000 × g for 10 min at 4 °C. The oocysts were resuspended in phosphate buffered saline (PBS) with EDTA-free cOmpleteTM protease inhibitor (Roche, Basel, Switzerland). The oocysts were broken using 0.5-mm glass beads with 4 × 5 min cycles of vigorous bead beating at 4 °C. Samples were placed in an ice bath between cycles to mitigate any heating effect. Proteins were extracted using a buffer containing protease inhibitor (10 mm HEPES, 25 mm KCl, 1 mm CaCl2, 10 mm MgCl2, 2% CHAPS, 6 m guanidine HCl, 50 mm dithiothreitol (DTT), pH 7.4). Insoluble material was removed by centrifugation at 21,130 × g for 5 min at 4 °C in an Eppendorf (Hamburg, Germany) 5424R microcentrifuge. The supernatant was removed and added to a new microcentrifuge tube; proteins were precipitated by the addition of −20 °C acetone (acetone/sample v/v 8:1) and the tube was allowed to sit undisturbed for ≥18 h at −80 °C. The proteins were concentrated by centrifugation at 21,130 × g for 20 min at 4 °C. The supernatant was discarded, and the pellet was washed 3x with ice-cold acetone. Any remaining solvent was removed in an unheated SpeedVac Plus speed vacuum (Savant, Thermo-Fisher Scientific).
The second chemical method used hot phenol to kill and extract total proteins from 109 C. parvum oocysts (42, 43). C. parvum oocysts were pelleted by centrifugation, resuspended in 500 μl of distilled water, and added to a conical vial containing 1 ml of phenol, pre-heated to 68 °C in a heating block filled with sand. The vial was sealed, and the contents mixed by inversion every 2 min for 20 min. The vial was removed, placed on ice, and gently centrifuged to facilitate good phase separation. The aqueous layer was removed and discarded. The interphase and phenol layers were carefully separated and saved. The proteins were subsequently precipitated from the phenol and interphase layers by the addition of eight volumes of −20 °C MeOH containing 100 mm NH4OAc, and allowed to sit undisturbed for ≥18 h at −20 °C. The precipitated proteins were concentrated by centrifugation, and pellets were washed 3x with −20 °C MeOH/0.1 M NH4OAc prior to lyophilization.
Trypsin Digestions
Three sets of samples were prepared for proteomics experiments. The fraction obtained from the mechanical extraction is referred to as “CHAPS” in the analysis. Two fractions from the chemical extraction method came from the phenol layer (referred to as “phenol”) and the interphase layer (referred to as “interphase”). Precipitated proteins from these three samples were dissolved into 50 mm NH4HCO3, pH 8.0, reduced with 50 mm DTT for 20 min at 60 °C, cooled to RT, and then alkylated with iodoacetamide (IAA) for 20 min at RT, while protected from light. Excess IAA was quenched with DTT, and peptides were generated by digestion with proteomics grade trypsin, overnight at 37 °C (1:20, w/w). The resulting tryptic peptides were dried by speed vacuum and desalted with C18 ZipTip concentrators (EMD Millipore, Danvers, MA), according to the manufacturer's protocol.
Release and Processing of N-glycans
N-glycans were released from total protein isolated from oocysts (100 μg) by overnight treatment at 37 °C with ten units of glycerol-free PNGase F (New England Biololabs), according to the manufacturer's instructions, without the addition of Nonidet P-40. The product mixture was lyophilized, and the released N-glycans were separated from the proteins by addition of 0.1% trifluoroacetic acid (TFA) in LC-MS grade water. The aqueous phase was passed onto C-18 Sep-Pak cartridges (Waters Corporation, Milford, MA). The cartridges were washed with three bed volumes of 0.1% TFA/water, and the eluents were pooled and lyophilized. The N-glycan pool was reduced with 0.5 m NaBD4/2 M NH3 (aq) overnight at 55 °C. The reaction was quenched by dropwise addition of glacial acetic acid. The products were washed multiple times with 10% acetic acid/MeOH, dried with a gentle stream of nitrogen, and washed again multiple times with 100% MeOH.
Permethylation was performed by published methods (44, 45). Briefly, a slurry of finely ground NaOH in dimethyl sulfoxide was added to the deutero-reduced sample; the suspension was mixed and methyl iodide was added. The solution was gently mixed at RT for one hr. To assure complete derivatization, the process was repeated three times. The product was isolated by extraction with water/chloroform, and the chloroform layer was dried in the SpeedVac.
MALDI-TOF MS
The purified deutero-reduced sample was dissolved in 20 μl of 1:1 MeOH/water, and 0.5 μl of this solution was spotted onto a stainless steel MALDI target with 2,5-dihydroxybenzoic acid as the matrix. The mass spectra were recorded with an ultrafleXtreme MALDI-TOF/TOF MS (Bruker Daltonics, Bremen, Germany) equipped with a smartbeam II Nd-YAG laser (355 nm, 3 nsec, 2 kHz). Each spectrum was acquired by summing the signals recorded after 500 shots from each of 10 locations within the sample spot.
Electron Excitation Dissociation (EED) Fourier Transform-Ion Cyclotron Resonance (FT-ICR) MS/MS
The released, deutero-reduced, and permethylated N-glycans were dried and re-suspended in 10 μl of 50% MeOH, 20 μm sodium acetate. The solution was loaded into a pulled glass capillary tube and directly infused into the ion source of a SolariX 12-T hybrid Qh-FT-ICR mass spectrometer (Bruker Daltonics), using a nano-ESI source. Each [M + Na]1+ parent ion was isolated by the quadrupole and accumulated in the collision cell for 8 s. The accumulated ions were then transferred into the ICR cell. Fragmentation by EED was achieved via 14-eV electrons generated from a cathode source heated with 1.5 A current. Electron density and energy were modulated using the following parameters: bias, 14 V; ECD lens, −13.85 V; pulse width 1.0 s. For each spectrum, 80 transients were averaged.
LC-MS/MS
The dried and desalted peptides were reconstituted in 2% ACN, 0.1% formic acid (FA) and separated on a NanoAcquity Ultra Performance Liquid Chromatography (UPLC) system (Waters), fitted with a nanoAcquity Symmetry C18 trap column (5-μm packing, 180 μm × 20 mm) and a BEH130C18 analytical column (1.7-μm packing, 150 μm × 10 cm). The mobile phase A was 99:1:0.1 HPLC grade water/ACN/FA), and mobile phase B was 99:1:0.1 ACN/HPLC grade water/FA. Each sample was loaded on the trapping column for 4 min at 4 μl/min flow rate and then separated on the analytical column using a 45 or 90 min 2–40% mobile phase B linear gradient at 0.5 μl/min flow rate. The column was washed between runs and equilibrated for 30 min.
The analytical column was coupled to a TriVersa NanoMate ion source (Advion, Ithaca, NY), and the ions were introduced into either an LTQ-Orbitrap-XL-ETD or a QE Plus mass spectrometer (both from Thermo-Fisher Scientific), which was operated in the positive-ion mode. MS spectra were obtained by scanning over the range m/z 350–2000. MS2 HCD spectra were acquired by isolating the top 5 (LTQ-Orbitrap) or top 20 (QE+) precursor ions with a 2-m/z window and fragmenting the selected precursor ions with 27, 35, or 45 V HCD energy. The MS2 HCD spectra were scanned from m/z 100 to a value that was dependent upon the parent ion.
Manual Interpretation of Glycopeptide MS/MS Spectra
Raw data files from LC-MS/MS experiments were manually interpreted using Qual Browser in the Xcalibur 2.2 software suite (Thermo-Fisher Scientific). HCD MS2 spectra containing oxonium ions were manually interpreted to determine the peptide sequence and the linear arrangement of the glycan. The y1 ion, corresponding to the residue Lys (K) or Arginine (R), was used as the starting point for most of the manually interpreted spectra. The resulting peptide tag was then searched using the online NCBI BLASTP algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the predicted C. parvum proteome, and the entire nr database (17–19). When a match was found, we determined the mass difference between the predicted trypsin generated peptide [M + H]1+ and that of the precursor, converted to [M + H]1+. The glycosidic bond fragment series, typically accounting for the most abundant peaks in the spectra, were sequenced in a similar manner, so far as each series could be followed. Missing residues were accounted for by calculating the difference between the highest member of the assigned series and the total observed molecular weight. Extracted ion chromatograms were generated to aid in the assignment of the numerous glycoconjugates.
LC-MS/MS Proteomics Database Search and Analysis
Once the possible N-glycoforms were discovered from the manual interpretation, these values could be utilized to search against the predicted C. parvum proteome as possible dynamic modifications. Database searches were performed using the PEAKS software suite version 7.5 (Bioinformatics Solutions Inc., Waterloo, ON, Canada). The following parameters were set for the search: the data refinement step corrected for the precursor m/z, for the PEAKSdenovo search stages, trypsin was specified as the enzyme, 8.0 ppm parent mass error tolerance, 0.05 Da fragment mass error tolerance, with carbamidomethyl cysteine set as a fixed modification, and possible dynamic modifications set to include methionine oxidation, HexNAc at serine/threonine; Hex6HexNAc2 and Hex5HexNAc2 on Asn. A maximum of five dynamic modifications was specified. The PEAKSDB search stage was identical to the PEAKSdenovo stage, with the exception that up to three missed trypsin cleavages were allowed, with the possibility of one nonspecific cleavage. Searches were performed against the C. parvum Iowa-II predicted proteome release-5.0 obtained from the Cryptosporidium Genome Resource (cryptodb.org), which contained 3803 entries (18, 19). False discovery rate (FDR) estimation was enabled. For the final PEAKSPTM stage, the de novo score average local confidence (ALC) threshold was 15 and the peptide hit threshold (-10 logP) was set to 30. All possible Unimod modifications were considered for this stage. The PEAKSPTM report was exported as a mzidentML with a FDR set to 5%, ALC 50% for de novo only, and proteins with a score of (-10 logP) ≥ 20 containing unique peptides ≥ 2. Each data file was analyzed individually for all samples and replicates.
Scaffold Analysis
The mzidentML files from the PEAKSPTM searches were imported into the computer program Scaffold version 4.6 for further analysis (Proteome Software, Inc., Portland, Oregon). Three “Biosamples” and two “categories” were specified for the samples. The two categories corresponded to the method of protein extraction, either “mechanical” or “chemical.” The sample names correspond to the subsample classification, the “CHAPS” was the mechanically broken 2% CHAPS extraction buffer soluble portion, and the “phenol” and “interphase” samples correspond to the phenol and interphase layers from the chemical extraction procedure. Each sample was analyzed independently, with experiment wide grouping and protein clustering. The probability model utilized was Peptide Prophet with delta mass correction. All spectra that were assigned by the software as possible N-glycosylated peptides were manually reviewed for quality and proper assignment to compile the final lists of glyopeptides and proteins that are available in supplemental table Excel S3.
Analysis of N-glycosylation Sites
For each protein observed to be N-glycosylated, the lists of occupied and total potential N-linked sites were compared. The “occupied” data set was created from the list of peptides modified with an N-glycan, taking for each a nine-amino acid window, centered on the modified asparagine. The same window was taken for all tryptic peptides that contained a canonical N-glycosylation sequon (Asn-Xxx-Ser/Thr, Xxx ≠ Pro) that could theoretically be generated from the group of observed glycoproteins. The program WebLogo v3.5.0 from the Department of Plant and Microbial Biology, University of California, Berkeley, was used to generate logos (46).
Bioinformatics
Predicted proteins of C. parvum with occupied N-glycan sites were analyzed for signal peptides and transmembrane helices using SignalP 4.0, TMHMM 2.0, and Phobius (47–51). Conserved domains were identified, and proteins were compared with those of other apicomplexans, eukaryotes, and bacteria (17–19, 50, 51). ALG enzymes, glucosidases, mannosidases, and OST peptides were predicted from whole genome sequences of C. parvum and T. gondii, using S. cerevisiae as a model (16, 20, 21, 25, 52, 53). Protein cartoon schematics were drawn using the program DOG 1.0 in combination with the software Inkscape 0.91 (54).
Analysis of Released N-glycans
To assist in the interpretation of the MS/MS spectra, we used the software GlycoWorkBench 2.1 (release 146) to generate theoretical fragmentation lists. Additional theoretical m/z values were generated using Microsoft Excel. Observed and theoretical peak lists were compared with obtain the best match. Assignments within 1-ppm error were considered to be a likely match. In the event that there were isobaric ion values, annotations were preferentially assigned to the ion that would be generated from a single fragmentation event. A single cross-ring fragment in combination with one or more glycosidic cleavages was considered only if the simple glycosidic bond fragment was also observed within the spectrum. All annotations were assigned only after a thorough manual review of the spectrum using Bruker DataAnalysis software suite version 4.0 SP5 build 283. Manual inspection helped to assign ions that didn't fit the list of theoretical values for expected cleavages.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the data set identifier PXD005503 and 10.6019/PXD005503. http://www.ebi.ac.uk/pride/archive/projects/PXD005503.
RESULTS
N-glycans of C. parvum Are Much Simpler Than Those of the Host and Most Other Parasites
The predicted N-glycan precursor of C. parvum, based upon its Alg enzymes, is Glc2Man5GlcNAc2 (supplemental table Excel S1) (16, 20, 21, 27). MALDI-TOF MS of N-glycans that had been deutero-reduced and permethylated after being released by PNGaseF from oocyst glycoproteins showed Hex6HexNAc2 ([M + Na]1+ m/z 1800.906) to be the most abundant form, whereas Hex5HexNAc2 ([M + Na]1+ m/z 1596.805) is less abundant (Fig. 1). The C. parvum N-glycans are much simpler than those of calf glycoproteins, suggesting oocysts, which are washed with PBS and purified on a CsCl gradient, are clean of host tissues (56). The released N-glycans also match those present on glycopeptides that were separated by reversed phase C18 nanoflow chromatography, and identified by manual interpretation of HCD MS/MS spectra (see below).
Fig. 1.
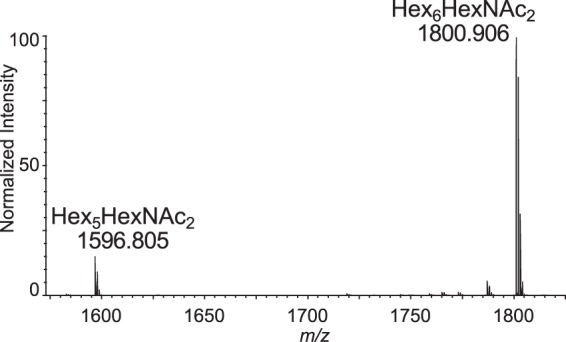
MALDI-TOF MS: released, deutero-reduced, permethylated N-glycans [M + Na]1+. Total N-glycans released from C. parvum glycoproteins with PNGase-F, reduced with sodium borodeuteride, and permethylated. Only two glycoforms are observed: Hex5HexNAc2 and Hex6HexNAc2.
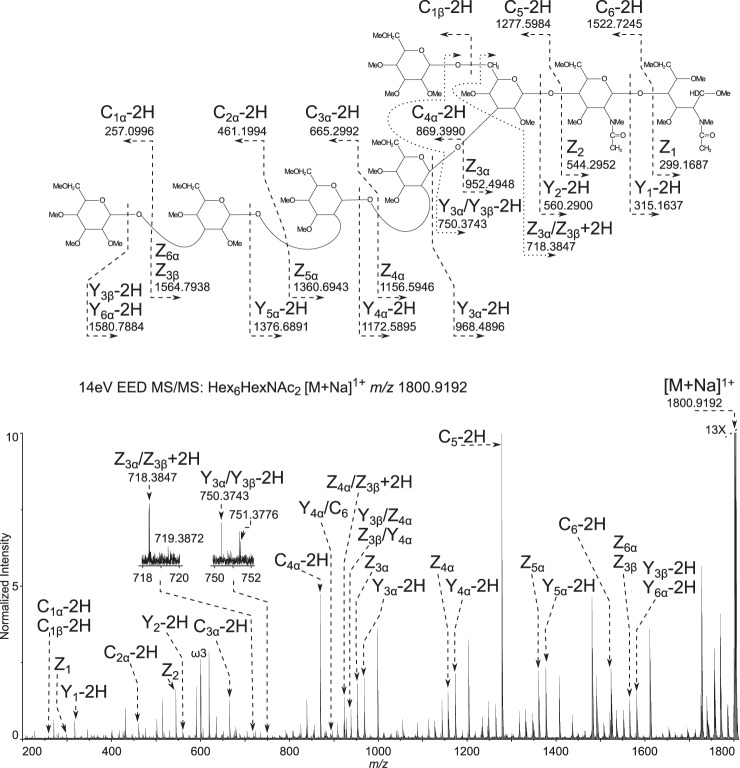
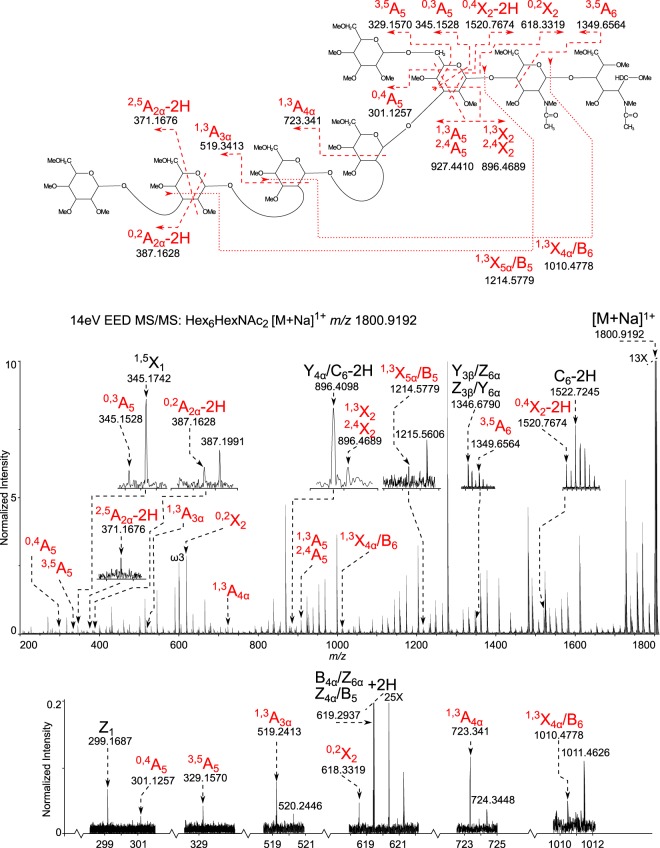
EED FT-ICR MS/MS was performed on the deutero-reduced and permethylated Hex6HexNAc2 in order to generate the glycosidic fragments that provide topographic information (Fig. 2) and the cross-ring fragments that provide linkage information (Fig. 3).
Fig. 2.
Topology of the most abundant glycoform Hex6HexNAc2 determined by EED FT-ICR MS/MS. 14-eV EED FT-ICR MS/MS: Hex6HexNAc2 [M + Na]1+ m/z 1800.9192. Glycosidic fragments provide topological information. The spectrum is labeled only with glycosidic fragments that indicate the topology of the glycoform, revealing a single long arm and an unmodified core. All assignments can be viewed in supplemental table Excel S1.
Fig. 3.
Glycosidic linkage determination of the most abundant glycoform Hex6HexNAc2 determined by EED FT-ICR MS/MS. 14-eV EED FT-ICR MS/MS: Hex6HexNAc2 [M+Na]1+ m/z 1800.9192. Cross-ring fragments provide linkage information. The spectrum is labeled with only the informative cross ring fragments that provide linkage information. All assignments can be viewed in supplemental table Excel S1.
The glycan topology is indicated by complete glycosidic bond fragmentation, shown dominated by the non-reducing end fragments (Cn-2H) series; in addition, reducing end glycosidic bond fragments of the (Yn-2H), and Zn series are prominent (Fig. 2). The single long arm topology is suggested by the sequential Z3α to Z6α ions, and the parallel sequential (Y3α-2H) to (Y6α-2H) series, where 1–4 hexoses are attached without branching. If branching were present in this tetrasaccharide moiety, there would be a gap in the linear series, and double glycosidic bond fragments might be observed along the chain. Instead, the only double glycosidic bond fragment series (Z3α/Z3β + 2H) through (Z6α/Z3β + 2H) correspond to cleavages involving the short arm (Fig. 3, supplemental table Excel S1). The (Z3α/Z3β + 2H) ion indicates there is no branching from the chitobiose core. The remaining double glycosidic fragments of the (Zn/Z3β) series, where Zn is Z4α to Z6α, show loss of the single hexose Z3β branch, with no branch points down the long arm (Fig. 3, supplemental table Excel S1).
The EED spectrum also allowed assignment of the linkage positions, as shown in Fig. 3. The key cross-ring fragment 0,4A5 and its paired reducing end fragment (0,4X2-2H), show that the short arm has a single hexose attached via a 1,6-linkage to the central hexose. The observation of 3,5A5 and 0,3A5 ions support this assignment. The ion pairs (1,3A5 and 1,3X2) indicate the longer arm, containing four hexose residues, is attached at the 2 or 3 position to this central hexose. Observation of the 0,2X2 ion eliminates the 2 position as the linkage site, thus, narrowing the possibility for the long arm linkage to position 3. Although the 1,3A5 and 1,3X2 ions are isobaric to the 2,4A5 and 2,4X2 ions and these might allow assignment of the linkage to the 3 or 4 position, the presence of the 3,5A5 ion rules out linkage at the 4 position. In sum, these cross-ring fragments indicate that the long arm containing four hexoses is attached by a 1,3-linkage to the central mannose.
The observation of 1,3A4α and 1,3A3α ions suggest that second and third hexoses on the long arm are either 2- or 3-linked. 0,2A3α or 0,2A4α fragments would be expected if these hexoses were linked at the 3 position; their absence suggests that the links are on the 2 position. The presence of an internal fragment 1,3X5α/B5 indicates that the terminal hexose is 1,3-linked and this assignment is confirmed by the observation of the 0,2A2α-2H ion that rules out the possibility that the terminal hexose is 2-linked (Fig. 3).
EED FT-ICR MS/MS was also performed on Hex5HexNAc2, which is the less abundant C. parvum N-glycan (supplemental Figs. S1 and S2, supplemental table Excel S1). The same topology as described for the aforementioned Hex6HexNAc2 (minus a terminal hexose on the long arm) is indicated by the complete (Cn - 2H), Z, (Yn - 2H), and (Znα/Z3β + 2H) series where n = 3, 4, or 5 (supplemental Fig. S1 and supplemental table Excel S1). A series of cross-ring fragments similar to the Hex6HexNAc2 glycan were observed for this glycoform. The observation of the ion pairs 0,4A4 and (0,4X2-2H) indicate that a single hexose is linked at the 6 position; this assignment is further supported by the ions 3,5A4 and 0,3A4 (supplemental Fig. S2). The longer trihexose arm is linked at the 3 position, as indicated by the ion pairs 1,3A4 and (1,3X2-2H), in conjunction with the 0,2X2 ion, ruling out the possibility of a 2 link for the isobaric pairs 2,4A4 and (2,4X2-2H). Therefore, it can be concluded that a single hexose is linked 1,6 to the first hexose on the core with the trisaccharide series linked 1,3 to the same residue. These results suggest the less abundant C. parvum N-glycan is likely Man5GlcNAc2 and has a structure identical to Hex6HexNAc2 without the terminal 1,3 linked hexose on the long arm. Although these methods cannot differentiate between isobaric monosaccharides, (e.g. mannose from glucose), these methods can accurately define the topology of the glycan and the linkages connecting each monosaccharide. The structures we have defined are consistent with the N-glycan structure that has been proposed on the basis of the presence or absence of the highly conserved N-glycosylation biosynthetic pathway enzymes identified in the C. parvum genome (supplemental table Excel S1 and supplemental Fig. S3). The topology of the Hex5HexNAc2 structure defined here, presumably Man5GlcNAc2, is different from the Man5GlcNAc2 glycoform produced when host Man9GlcNAc2 is processed by ER mannosidase 1 in higher organisms, which has 1,3 and 1,6 dimannosyl branches off the first 1,6-linked Man (16, 25).
Confident Assignment of Cryptosporidum parvum N-glycosylated Peptides
To rule out the possibility of host cell contamination that could occur because C. parvum is an obligate intracellular parasite, the MS/MS spectra of glycopeptides from a whole oocyst lysate were manually interpreted. Peptides generated from a trypsin digestion of proteins isolated from oocysts were separated on a reversed phase-C18 nanoflow column interfaced to a mass spectrometer, as described in the methods section. Extracted oxonium ion chromatograms for m/z 204.08 (HexNAc) and m/z 366.13 (HexNAc-Hex) were very abundant throughout the MS/MS spectra recorded across the HPLC separation (Fig. 4A). These XIC pointed out which spectra should be manually interpreted to obtain sequence information on both glycan and peptide. A representative HCD spectrum for [M + 2H]2+ m/z 1092.9426 eluting at 15 min, marked by the carat in Fig. 4A, is interpreted in Figs. 4B and 4C. Ions containing sequential glycosidic bond fragments dominate the spectrum and provide the linear sequence for Hex6HexNAc2 (Fig. 4B). The glycosidic fragments could be traced down to the aglycon, the peptide with [M + H]+ m/z 806.3992. Lower abundance peptide backbone fragments are observable in the magnified view of the spectrum (Fig. 4C). The complete y-series is interpretable, starting from y1 m/z 175.1189, to the full-length peptide, observed at m/z 806.3992, thus revealing the peptide sequence NSTTEVR, and indicating the Hex6HexNAc2 was linked to Asn (Fig. 4C). Checking the peptide sequence against the NCBI nr and C. parvum proteome databases, utilizing the blastp algorithm, revealed that the peptide belongs to the C. parvum protein POWP1 (Table I). In summary, the linear glycan sequence, conjugation site, and complete peptide sequence, can all be determined from a single spectrum.
Fig. 4.
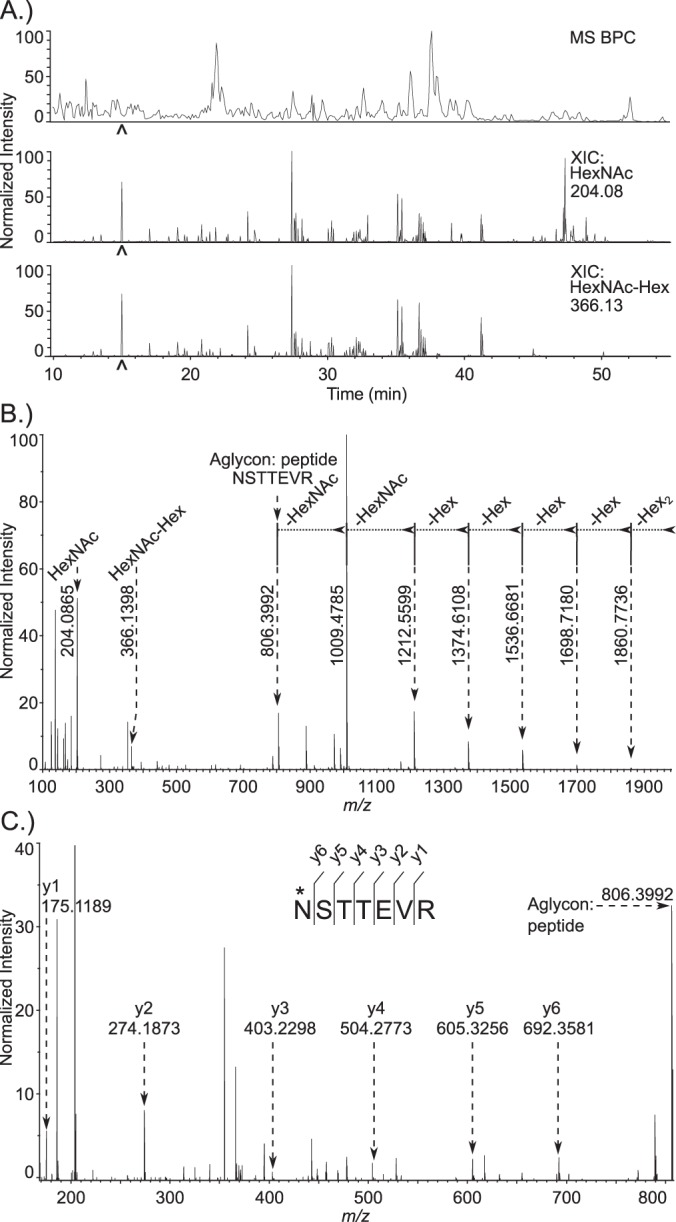
De novo identification of glycopeptides from a whole C. parvum oocyst lysate. A, Base peak and extracted oxonium ion chromatograms from a reversed phase C18 separation. The top trace shows the base peak chromatogram from the MS. The middle and bottom chromatograms are extracted oxonium ion chromatograms (XIC) from the 35-V HCD MS/MS spectra, corresponding to m/z 204.08 (HexNAc) and m/z 366.13 (HexNAc-Hex), respectively. The carat located at 15 min. indicates the time point for recording of the MS/MS spectrum shown in Fig. 4B and 4C. B, 35-V HCD MS/MS spectrum of an N-glycosylated peptide: NSTTEVR modified with Hex6HexNAc2, [M + 2H]2+ m/z 1092.9426. Prominent glycosidic bond fragmentation is observed, delineating the sequence of the glycan. C, Peptide sequence of the aglycon. Lower intensity y-ion peptide backbone fragments are observed in the same spectrum. Peptide fragment ion assignments are shown on this magnified view; these extend from y1 (Arg) at m/z 175.1189 to the complete aglycon, m/z 806.3992, defining the complete peptide sequence as NSTTEVR.
Table I. Summary of identified N-glycosylated peptides.
| Name | Ac. Number | Unique N-glycosylated peptides | # Spectra |
|
|---|---|---|---|---|
| Hex5HexNAc2 | Hex6HexNAc2 | |||
| gp900 | cgd7_4020 | (107)-RMVDPVSLMLFDNSTGVMYDPNTNSILEGSIAGIR-(141) | 2 | 3 |
| (989)-SGNLVHPYTNQTMSGLSVSYLAAK-(1012) | 6 | 0 | ||
| (1224)-LINPTNNNTMDSSFAGAYK-(1242) | 4 | 0 | ||
| (1388)-DPVTNTQYSNTTGNIINPETGK-(1409) | 0 | 1 | ||
| (1460)-LPIPGSVAGDEILTEVLNITTDEVTGLPIDLETGLPR-(1496) | 20 | 20 | ||
| (1497)-DPVSGLPQLPNGTLVDPSNK-(1516) | 17 | 8 | ||
| (1524)-SGFINGTSGEQSHEK-(1538) | 11 | 0 | ||
| POWP1 | cgd2_490 | (31)-NSTTEVR-(37) | 1 | 3 |
| (195)-NQTSSSGNNPVNNLLNR-(211) | 7 | 44 | ||
| (261)-NNPLYNETSISSDGK-(275) | 6 | 47 | ||
| (277)-YNDTASPIK-(285) | 11 | 48 | ||
| (286)-TPEIVYYNNTSNLR-(299) | 3 | 58 | ||
| UCG1 | cgd5_1210 | (328)-APGSSNTMNQTTNLNNER-(345) | 5 | 2 |
| (362)-FALSPLNGTEVAPLFSK-(378) | 0 | 10 | ||
| (97)-VLLGNDSTVK-(106) | 1 | 4 | ||
| UCG2 | cgd6_710 | (130)-GTIYNITSVDDLIQNSR-(146) | 0 | 22 |
| UCG3 | cgd1_640 | (122)-EHVFNVTGQVPTLGEVK-(138) | 1 | 1 |
| UCG4 | cgd1_660 | (290)-EIDDIVPHNETIMK-(303) | 17 | 3 |
| UCG5 | cgd2_1290 | (37)-ATNQTNDSWFNLDLLR-(52) | 0 | 1 |
| UCG6 | cgd3_660 | (58)-ANVSTIFGDLLNSK-(71) | 0 | 1 |
| UCG7 | cgd8_4660 | (175)-ISFLESGSITETNFTMSTYR-(194) | 0 | 1 |
| (195)-NETGLLTNPK-(204) | 0 | 6 | ||
| GMCO | cgd2_2510 | (209)-VYNLFNVSDHGFR-(221) | 0 | 1 |
| (236)-NNTVIETSPVDILTNHLVTK-(255) | 0 | 4 | ||
| GAP50 | cgd2_640 | (109)-NNVTYDSNNDIFPR-(122) | 0 | 2 |
| (169)-VSEQAFQNLNATLHYGHK-(186) | 0 | 3 | ||
| COA | cgd3_3430 | (994)-VTLFVNK-(1000)-T | 0 | 1 |
| O-GNT4 | cgd7_1310 | (311)-SLNETQSGVDLEQR-(324) | 1 | 0 |
| CCP1 | cgd7_1730 | (339)-VSINLSANMTYQLK-(352) | 3 | 0 |
| CCP2 | cgd7_300 | (44)-SIIDTQDLGESNDTKK-(59) | 0 | 2 |
| (59)-KLNETQILSDAYEANINK-(76) | 0 | 1 | ||
| FNPA | cgd7_4810 | (144)-YNSTCGSQQSIVSSR-(158) | 0 | 10 |
This method of manual interpretation was continued systematically for the remaining spectra containing one or more oxonium ion(s). Many of the most abundant peptides mapped to the same protein, POWP1. Five N-glycosylation sites were mapped to this protein; all contain either Hex5HexNAc2 or Hex6HexNAc2 (Fig. 5 and Table I). Many of the MS/MS spectra assigned to glycopeptides contained Y1 and Y2 ions that arose via glycosidic cleavages adjacent to the HexNAc residues in the chitobiose core, with charge retention on the peptide fragment, but none of these MS/MS spectra contained (Y1 + 146) or (Y1 + 162) ions that would indicate the presence of a deoxyhexose or hexose branch on the inner HexNAc residue. No spectra indicated the presence of glycopeptides that did not originate from C. parvum. This result demonstrated that the preparation was clean from contaminating host material, and assured that the released N-glycans are of parasite origin, as the results had already suggested (Figs. 2, 3, supplemental Figs. S1, S2). The manual interpretations of the glycopeptide spectra are consistent with the results obtained by analysis of the released glycans and underscore the very limited repertoire N-glycans in this organism. This information could then be applied to perform semi-automated database searches to dig deeper in the spectra, allowing for faster processing of replicate samples.
Fig. 5.
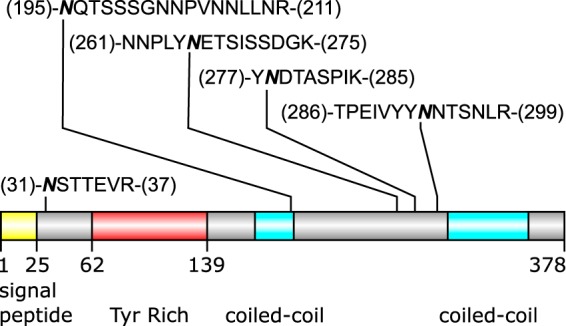
Occupied N-glycosylation sites of POWP1 (cgd2_490). The most densely N-glycosylated protein is represented as a cartoon schematic. The occupied peptides are shown; the bold and italicized asparagine residues indicate the site of attachment of the glycan.
Heavily Glycosylated Proteins Include an Immunodominant Protein (gp900) and a Putative Oocyst Wall Protein (POWP1)
Thirty-two tryptic peptides with occupied N-glycan sites were identified. These peptides derive from 16 glycoproteins, which include the vaccine candidate gp900 and a probable oocyst wall protein POWP1 (Fig. 5) (24, 30, 31, 35). Some proteins (e.g. gp900) but not others (e.g. POWP1) showed a higher relative abundance for Hex6HexNAc2 versus Hex5HexNAc2 (Table I), as did the released N-glycans (Fig. 1) (24). No other N-glycoforms were detected.
Like gp900, two Cryptosporidium N-linked glycoproteins that are also unique (UCG1 and UCG2) contain long runs of Thr, which are likely modified by O-linked GalNAc (30, 35, 57). Five other unique glycoproteins with occupied N-glycan sites (UCG3 to UCG7) remain uncharacterized. Other observed glycoproteins have analogs elsewhere in apicomplexa: the glideosome-associated protein (GAP50), three putative adhesion proteins with a Limulus coagulation factor C lectin (LCCL) domain (CCP1, CCP2, and FNPA), and a copper amine oxidase (CAO) are conserved throughout apicomplexa (17–19, 52, 51, 57–59). An O-GalNAc transferase 4 is present in apicomplexans and mammalian hosts, whereas GMC oxidoreductase (GMCO) is present in apicomplexans, metazoans, fungi, plants, and bacteria (56).
Nearly 90% of the Occupied N-glycan Sites Contain Thr Rather than Ser
N-glycans of C. parvum are not used for quality control of protein folding, and there is no positive selection for N-glycan sites in its secreted proteins (20, 25, 26). However, we observed a large difference in the rate of occupancy of potential N-glycosylation sequons. Despite the 5:3 ratio of Thr (100) and Ser (61) in the second position relative to Asn, the number of occupied N-glycan sites overwhelmingly (9:1) favors Thr (35) over Ser (4), as shown in the WebLogo in Fig. 6 and the data in Table I (46). Notably, only 11 total spectra were assigned to peptides with the asparagine modified in an Asn-Xxx-Ser sequon, compared with the 412 that correspond to N-glycosylation on the Asn-Xxx-Thr motif (Table I, supplemental table Excel S2).
Fig. 6.
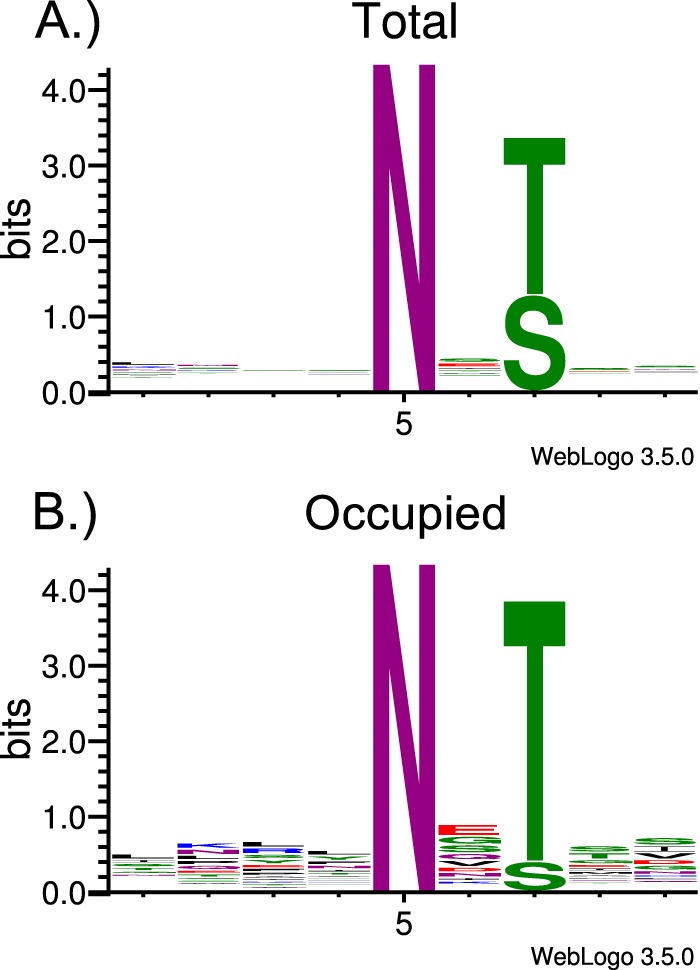
Total and occupied N-glycosylation sequons in the observed N-glycosylated proteins. A, Web-Logos, generated using WebLogo 3.5.0, were compiled for all the peptides containing the canonical N-glycosylation sequons from proteins observed to be glycosylated (Total). B, Peptides observed to be occupied are represented by the (Occupied) logo.
DISCUSSION
ALG enzymes, which are required for the synthesis of N-glycan precursors, are reliable predictors of the types of N-glycans transferred to the nascent peptides, because these glycosyltransferases are constitutively expressed in the ER (16, 21). Two peculiarities present themselves with regards to the ALG enzymes of C. parvum. First, the ALG13 peptide of the glycosyltransferase that adds the second GlcNAc to the pyrophosphate-linked precursor can easily be identified in Cryptosporidium muris and in all other organisms that make N-glycans, but ALG 13 is absent from the predicted proteins of C. parvum and C. hominis (supplemental table Excel S1) (17–19, 52). Second, although the C. parvum N-glycan precursor is predicted to be Glc2Man5GlcNAc2, Hex7HexNAc2 was absent from the N-glycans released with PNGaseF and from tryptic glycopeptides (Figs. 1 to 4 and Table I). This result suggests that ALG8, which adds the second glucose to the N-glycan precursor, is not active, or a glucose residue is rapidly removed by glucosidase 2 from Glc2Man5GlcNAc2 after it is transferred to the nascent peptide. In contrast, T. gondii, which has a predicted N-glycan precursor composed of Glc3Man5GlcNAc2, has been shown to have glycoproteins containing Hex8HexNAc2 and Hex7HexNAc2 (57).
Although it is well-known that the oligosaccharyltransferase (OST) that adds N-glycans to the nascent peptide prefers N-glycan sites with Thr over those with Ser, such a strong bias for Thr as that observed here for occupied N-glycan sites of C. parvum has not previously been described, to our knowledge (26, 60). The composition of the C. parvum OST, which includes the catalytic STT3 subunit and three non-catalytic subunits (supplemental table Excel S1), is similar to that found in other apicomplexans, whereas the OSTs of some parasites (e.g. Giardia and Trypanosoma) only contain STT3 (53).
The binding of the anti-retroviral lectin cyanovirin-N to C. parvum strongly suggested that the parasite contains a high mannose N-glycan (23, 24). Cyanovirin-N also binds to Entamoeba and Trichomonas, each of which builds its N-glycans from a precursor composed of Man5GlcNAc2 (61–63). The N-glycan profile of C. parvum differs from those of the other parasites in the relative abundance of GlcMan5GlcNAc2 and the absence of mannosidase products (Man4GlcNAc2 and Man3GlcNAc2) and/or hybrid and complex N-glycans, which contain LacNAc arms (Trichomonas) or galactose capped with Glc (Entamoeba) (63, 64). Other parasites (Trypanosoma, Leishmania, and Acanthamoeba) and Dictyostelium have N-glycan precursors with three mannose arms and make numerous complex N-glycans that contain LacNAc, fucose, and xylose (65–68). Finally, the N-glycans of the mammalian hosts (mice, humans, cats, etc.) are much more complex than those of C. parvum, which are remarkable for their simplicity (56). Whether the high mannose N-glycans of C. parvum are involved in antigen masking and/or pathogenesis, as has been shown for high mannose N-glycans on gp120 of HIV and on HA of influenza virus, remains to be determined (69–71). It has been established that many of the C. parvum proteins that elicit a strong immune response are N-linked glycoproteins (35, 41). Attempts have been made to develop vaccines from several of these glycoproteins; however, the critical details such as which amino acids are modified and with what glycan structure(s) were left unanswered (72, 73). The results we have presented here fill in the missing details regarding the N-glycosylation of the immunodominant antigen gp900, and we also expand upon the number of N-glycosylated proteins previously described in the literature. Of particular interest is the abundant and densely glycosylated protein POWP1. The function of POWP1 remains to be determined. These details may be crucial in providing a means to finally developing an effective, synthetic glycoprotein or glycopeptide-based vaccine against cryptosporidiosis.
The protein names referred to in this publication map to the following UniProtKB accession numbers: UCG3, UniProtKB = UniProt # Q5CT01; UCG4, UniProtKB = UniProt # Q5CSZ9; POWP1, UniProtKB = UniProt # Q5CU33; GAP50, UniProtKB = UniProt # Q5CU19; UCG5, UniProtKB = UniProt # Q5CTW5; GMCO, UniProtKB = UniProt # Q5CTL6; UCG6, UniProtKB = UniProt # Q5CV13; COA, UniProtKB = UniProt # Q5CUC0; UCG1, UniProtKB = UniProt # Q5CRW6; UCG2, UniProtKB = UniProt # Q5CXK6; CCP2, UniProtKB = UniProt # Q5CZ08; O-GAT4, UniProtKB = UniProt # Q5CYR4; CCP1, UniProtKB = UniProt # Q5CYM9; gp900, UniProtKB = UniProt # Q5CY21; FNPA, UniProtKB = UniProt # Q5CXV2; UCG7, UniProtKB = UniProt # Q5CVB8.
Supplementary material is freely available for downloading from the MCP website. It contains the EED MS/MS spectrum of the Hex5HexNAc2 glycan, glycosidic and cross-ring fragment assignments, a list of the C. parvum glycosyltransferases and a figure showing the predicted glycans of C. parvum. Excel files S1 and S2 contain the lists of fragment ion assignments. Excel file S3 contains the complete list of glycopeptides, proteins, and related bioinformatics data.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the dataset identifier PXD005503 and 10.6019/PXD005503 (http://www.ebi.ac.uk/pride/archive/projects/PXD005503).
Supplementary Material
Acknowledgments
We thank Yi Pu and Cheng Lin for their assistance setting up the EED FT-ICR MS/MS experiment. Furthermore, Cheng Lin provided valuable feedback and questions pertaining to the interpretation of the EED spectra. Additionally, we thank Giulia Bandini for her insight on glycosyltransferases and knowledge of T. gondii.
Footnotes
Author contributions: J.R.H., J.S., and C.E.C. designed research; J.R.H. performed research; D.R.L. contributed new reagents or analytic tools; J.R.H., D.R.L., J.S., and C.E.C. analyzed data; J.R.H., J.S., and C.E.C. wrote the paper.
* Support for this study came from NIH grants R01 AI110638 and R01 GM031318 (J.S.), and P41 GM104603, S10 RR025082 and S10 OD010724 (C.E.C.), and NIH-NHLBI contract HHSN268201000031C (C.E.C.). We thank Thermo-Fisher Scientific for loan of the Q Exactive Plus system. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- AIDS
- Acquired Immune Deficiency Syndrome
- ALG
- Asparagine-Linked Glycosylation
- ALC
- average local confidence
- EED
- Electronic Excitation Dissociation
- FA
- formic acid
- FDR
- false discovery rate
- GAP50
- glideosome-associated protein
- HA
- Hemagglutinin
- HCD
- higher-energy collision-induced dissociation
- IAA
- iodoacetamide
- MeOH
- methanol
- AcOH
- acetic acid
- HIV
- human immunodeficiency virus
- IAA
- iodoacetamide
- OST
- oligosaccharyltransferase
- PNGaseF
- peptide:N-glycanaseF
- POWP1
- possible oocyst wall protein 1
- ppm
- parts-per-million
- UCG
- unique Cryptosporidium glycoprotein
- UPLC
- Ultra Performance Liquid Chromatography.
REFERENCES
- 1. Checkley W., White A. C. Jr, Jaganath D., Arrowood M. J., Chalmers R. M., Chen X. M., Fayer R., Griffiths J. K., Guerrant R. L., Hedstrom L., Huston C. D., Kotloff K. L., Kang G., Mead J. R., Miller M., Petri W. A. Jr, Priest J. W., Roos D. S., Striepen B., Thompson R. C., Ward H. D., Van Voorhis W. A., Xiao L., Zhu G., and Houpt E. R. (2015) A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 15, 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Esch K. J., and Petersen C. A. (2013) Transmission and epidemiology of zoonotic protozoal diseases of companion animals. Clin. Microbiol. Rev. 26, 58–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fayer R., and Xiao L. (2007) Cryptosporidium and cryptosporidiosis, CRC press [Google Scholar]
- 4. Baldursson S., and Karanis P. (2011) Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2004–2010. Water Res. 45, 6603–6614 [DOI] [PubMed] [Google Scholar]
- 5. Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., Wu Y., Sow S. O., Sur D., Breiman R. F., Faruque A. S., Zaidi A. K., Saha D., Alonso P. L., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ochieng J. B., Omore R., Oundo J. O., Hossain A., Das S. K., Ahmed S., Qureshi S., Quadri F., Adegbola R. A., Antonio M., Hossain M. J., Akinsola A., Mandomando I., Nhampossa T., Acacio S., Biswas K., O'Reilly C. E., Mintz E. D., Berkeley L. Y., Muhsen K., Sommerfelt H., Robins-Browne R. M., and Levine M. M. (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222 [DOI] [PubMed] [Google Scholar]
- 6. Platts-Mills J. A., Babji S., Bodhidatta L., Gratz J., Haque R., Havt A., McCormick B. J., McGrath M., Olortegui M. P., Samie A., Shakoor S., Mondal D., Lima I. F., Hariraju D., Rayamajhi B. B., Qureshi S., Kabir F., Yori P. P., Mufamadi B., Amour C., Carreon J. D., Richard S. A., Lang D., Bessong P., Mduma E., Ahmed T., Lima A. A., Mason C. J., Zaidi A. K., Bhutta Z. A., Kosek M., Guerrant R. L., Gottlieb M., Miller M., Kang G., Houpt E. R., and Investigators M.-E. N. (2015) Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob. Health 3, Se564–Se575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wanyiri J. W., Kanyi H., Maina S., Wang D. E., Steen A., Ngugi P., Kamau T., Waithera T., O'Connor R., Gachuhi K., Wamae C. N., Mwamburi M., and Ward H. D. (2014) Cryptosporidiosis in HIV/AIDS patients in Kenya: clinical features, epidemiology, molecular characterization and antibody responses. Am. J. Trop. Med. Hyg. 91, 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mac Kenzie W. R., Schell W. L., Blair K. A., Addiss D. G., Peterson D. E., Hoxie N. J., Kazmierczak J. J., and Davis J. P. (1995) Massive outbreak of waterborne Cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin. Infect. Dis. 21, 57–62 [DOI] [PubMed] [Google Scholar]
- 9. Sarkar R., Kattula D., Francis M. R., Ajjampur S. S., Prabakaran A. D., Jayavelu N., Muliyil J., Balraj V., Naumova E. N., Ward H. D., and Kang G. (2014) Risk factors for cryptosporidiosis among children in a semi urban slum in southern India: a nested case-control study. Am. J. Trop. Med. Hyg. 91, 1128–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benitez A. J., McNair N., and Mead J. R. (2009) Oral immunization with attenuated Salmonella enterica serovar Typhimurium encoding Cryptosporidium parvum Cp23 and Cp40 antigens induces a specific immune response in mice. Clin. Vaccine Immunol. 16, 1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ehigiator H. N., Romagnoli P., Priest J. W., Secor W. E., and Mead J. R. (2007) Induction of murine immune responses by DNA encoding a 23-kDa antigen of Cryptosporidium parvum. Parasitol. Res. 101, 943–950 [DOI] [PubMed] [Google Scholar]
- 12. Jenkins M., Higgins J., Kniel K., Trout J., and Fayer R. (2004) Protection of calves against cryptosporiosis by oral inoculation with gamma-irradiated Cryptosporidium parvum oocysts. J. Parasitol. 90, 1178–1180 [DOI] [PubMed] [Google Scholar]
- 13. Ludington J. G., and Ward H. D. (2015) Systemic and mucosal immune responses to cryptosporidium-vaccine development. Curr. Trop. Med. Rep. 2, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mead J. R. (2014) Prospects for immunotherapy and vaccines against Cryptosporidium. Hum. Vaccin. Immunother. 10, 1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cabada M. M., and White AC. Jr (2010) Treatment of cryptosporidiosis: do we know what we think we know? Curr. Op. Infect. Dis. 23, 494–499 [DOI] [PubMed] [Google Scholar]
- 16. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 17. Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aurrecoechea C., Barreto A., Brestelli J., Brunk B. P., Cade S., Doherty R., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Hu S., Iodice J., Kissinger J. C., Kraemer E. T., Li W., Pinney D. F., Pitts B., Roos D. S., Srinivasamoorthy G., Stoeckert C. J. Jr, Wang H., and Warrenfeltz S. (2013) EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res. 41, SD684–SD691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heiges M., Wang H., Robinson E., Aurrecoechea C., Gao X., Kaluskar N., Rhodes P., Wang S., He C. Z., Su Y., Miller J., Kraemer E., and Kissinger J. C. (2006) CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 34, SD419–SD422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bushkin G. G., Ratner D. M., Cui J., Banerjee S., Duraisingh M. T., Jennings C. V., Dvorin J. D., Gubbels M. J., Robertson S. D., Steffen M., O'Keefe B. R., Robbins P. W., and Samuelson J. (2010) Suggestive evidence for Darwinian Selection against asparagine-linked glycans of Plasmodium falciparum and Toxoplasma gondii. Eukaryot. Cell 9, 228–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., and Robbins P. W. (2005) The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. U.S.A. 102, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Satoh T., Yamaguchi T., and Kato K. (2015) Emerging structural insights into glycoprotein quality control coupled with N-glycan processing in the endoplasmic reticulum. Molecules 20, 2475–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams E. W., Ratner D. M., Bokesch H. R., McMahon J. B., O'Keefe B. R., and Seeberger P. H. (2004) Oligosaccharide and glycoprotein microarrays as tools in HIV glycobiology; glycan-dependent gp120/protein interactions. Chem. Biol. 11, 875–881 [DOI] [PubMed] [Google Scholar]
- 24. Chatterjee A., Banerjee S., Steffen M., O'Connor R. M., Ward H. D., Robbins P. W., and Samuelson J. (2010) Evidence for mucin-like glycoproteins that tether sporozoites of Cryptosporidium parvum to the inner surface of the oocyst wall. Eukaryot. Cell 9, 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banerjee S., Vishwanath P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W., and Samuelson J. (2007) The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc. Natl. Acad. Sci. U.S.A. 104, 11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cui J., Smith T., Robbins P. W., and Samuelson J. (2009) Darwinian selection for sites of Asn-linked glycosylation in phylogenetically disparate eukaryotes and viruses. Proc. Natl. Acad. Sci. U.S.A. 106, 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuelson J., and Robbins P. W. (2015) Effects of N-glycan precursor length diversity on quality control of protein folding and on protein glycosylation. Semin. Cell Develop. Biol. 121–128, Elsevier; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ajjampur S. S., Sarkar R., Allison G., Banda K., Kane A., Muliyil J., Naumova E., Ward H., and Kang G. (2011) Serum IgG response to Cryptosporidium immunodominant antigen gp15 and polymorphic antigen gp40 in children with cryptosporidiosis in South India. Clin. Vaccine Immunol. 18, 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allison G. M., Rogers K. A., Borad A., Ahmed S., Karim M. M., Kane A. V., Hibberd P. L., Naumova E. N., Calderwood S. B., Ryan E. T., Khan W. A., and Ward H. D. (2011) Antibody responses to the immunodominant Cryptosporidium gp15 antigen and gp15 polymorphisms in a case-control study of cryptosporidiosis in children in Bangladesh. Am. J. Trop. Med. Hyg. 85, 97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barnes D. A., Bonnin A., Huang J. X., Gousset L., Wu J., Gut J., Doyle P., Dubremetz J. F., Ward H., and Petersen C. (1998) A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol. Biochem. Parasitol. 96, 93–110 [DOI] [PubMed] [Google Scholar]
- 31. Cevallos A. M., Zhang X., Waldor M. K., Jaison S., Zhou X., Tzipori S., Neutra M. R., and Ward H. D. (2000) Molecular Cloning and Expression of a Gene EncodingCryptosporidium parvum Glycoproteins gp40 and gp15. Infect. Immun. 68, 4108–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lazarus R. P., Ajjampur S. S., Sarkar R., Geetha J. C., Prabakaran A. D., Velusamy V., Naumova E. N., Ward H. D., and Kang G. (2015) Serum Anti-Cryptosporidial gp15 Antibodies in Mothers and Children Less than 2 Years of Age in India. Am. J. Trop. Med. Hyg. 93, 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonald A. C., Mac Kenzie W. R., Addiss D. G., Gradus M. S., Linke G., Zembrowski E., Hurd M. R., Arrowood M. J., Lammie P. J., and Priest J. W. (2001) Cryptosporidium parvum-specific antibody responses among children residing in Milwaukee during the 1993 waterborne outbreak. J. Infect. Dis. 183, 1373–1379 [DOI] [PubMed] [Google Scholar]
- 34. O'Connor R. M., Wanyiri J. W., Cevallos A. M., Priest J. W., and Ward H. D. (2007) Cryptosporidium parvum glycoprotein gp40 localizes to the sporozoite surface by association with gp15. Mol. Biochem. Parasitol. 156, 80–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petersen C., Gut J., Doyle P., Crabb J., Nelson R., and Leech J. (1992) Characterization of a> 900,000-M (r) Cryptosporidium parvum sporozoite glycoprotein recognized by protective hyperimmune bovine colostral immunoglobulin. Infect. Immun. 60, 5132–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Priest J. W., Kwon J. P., Arrowood M. J., and Lammie P. J. (2000) Cloning of the immunodominant 17-kDa antigen from Cryptosporidium parvum. Mol. Biochem. Parasitol. 106, 261–271 [DOI] [PubMed] [Google Scholar]
- 37. Sarkar R., Ajjampur S. S., Muliyil J., Ward H., Naumova E. N., and Kang G. (2012) Serum IgG responses and seroconversion patterns to Cryptosporidium gp15 among children in a birth cohort in south India. Clin. Vaccine Immunol. 19, 849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spano F., Puri C., Ranucci L., Putignani L., and Crisanti A. (1997) Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology 114, 427–437 [DOI] [PubMed] [Google Scholar]
- 39. Strong W. B., Gut J., and Nelson R. G. (2000) Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68, 4117–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wanyiri J. W., O'Connor R., Allison G., Kim K., Kane A., Qiu J., Plaut A. G., and Ward H. D. (2007) Proteolytic processing of the Cryptosporidium glycoprotein gp40/15 by human furin and by a parasite-derived furin-like protease activity. Infect. Immun. 75, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luft B. J., Payne D., Woodmansee D., and Kim C. W. (1987) Characterization of the Cryptosporidium antigens from sporulated oocysts of Cryptosporidium parvum. Infect. Immun. 55, 2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Howe C., Lloyd K. O., and Lee L. T. (1972) [20] Isolation of glycoproteins from red cell membranes using phenol. Methods Enzymol. 28, 236–245 [Google Scholar]
- 43. Hurkman W. J., and Tanaka C. K. (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81, 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ciucanu I., and Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydrate Res. 131, 209–217 [Google Scholar]
- 45. Ciucanu I., and Costello C. E. (2003) Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J. Am. Chem. Soc. 125, 16213–16219 [DOI] [PubMed] [Google Scholar]
- 46. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kall L., Krogh A., and Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, SW429–SW432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 49. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 50. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., Wang Z., Yamashita R. A., Zhang D., Zheng C., and Bryant S. H. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, SD222–SD226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., Sangrador-Vegas A., Scheremetjew M., Rato C., Yong S. Y., Bateman A., Punta M., Attwood T. K., Sigrist C. J., Redaschi N., Rivoire C., Xenarios I., Kahn D., Guyot D., Bork P., Letunic I., Gough J., Oates M., Haft D., Huang H., Natale D. A., Wu C. H., Orengo C., Sillitoe I., Mi H., Thomas P. D., and Finn R. D. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, SD213–SD221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bickel T., Lehle L., Schwarz M., Aebi M., and Jakob C. A. (2005) Biosynthesis of Lipid-linked Oligosaccharides in Saccharomyces cerevisiae Alg13p AND Alg14p form a complex required for the formation of GlcNAc2-PP-Dolichol. J. Biol. Chem., 280, 34500–34506 [DOI] [PubMed] [Google Scholar]
- 53. Kelleher D. J., and Gilmore R. (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R-62R [DOI] [PubMed] [Google Scholar]
- 54. Ren J., Wen L., Gao X., Jin C., Xue Y., and Yao X. (2009) DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273 [DOI] [PubMed] [Google Scholar]
- 55. Morelle W., Faid V., Chirat F., and Michalski J.-C. (2009) Analysis of N-and O-linked glycans from glycoproteins using MALDI-TOF mass spectrometry. Methods Mol. Biol., 534, 5–21 [DOI] [PubMed] [Google Scholar]
- 56. Bhat N., Wojczyk B. S., DeCicco M., Castrodad C., Spitalnik S. L., and Ward H. D. (2013) Identification of a family of four UDP-polypeptide N-acetylgalactosaminyl transferases in Cryptosporidium species. Mol. Biochem. Parasitol. 191, 24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fauquenoy S., Hovasse A., Sloves P.-J., Morelle W., Alayi T. D., Slomianny C., Werkmeister E., Schaeffer C., Van Dorsselaer A., and Tomavo S. (2011) Unusual N-glycan structures required for trafficking Toxoplasma gondii GAP50 to the inner membrane complex regulate host cell entry through parasite motility. Mol. Cell. Proteomics 10, M111.008953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baum J., Richard D., Healer J., Rug M., Krnajski Z., Gilberger T. W., Green J. L., Holder A. A., and Cowman A. F. (2006) A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J. Biol. Chem. 281, 5197–5208 [DOI] [PubMed] [Google Scholar]
- 59. Pradel G., Hayton K., Aravind L., Iyer L. M., Abrahamsen M. S., Bonawitz A., Mejia C., and Templeton T. J. (2004) A multidomain adhesion protein family expressed in Plasmodium falciparum is essential for transmission to the mosquito. J. Exp. Med. 199, 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kasturi L., Eshleman J. R., Wunner W. H., and Shakineshleman S. H. (1995) The Hydroxy Amino-Acid in an Asn-X-Ser/Thr Sequon Can Influence N-Linked Core Glycosylation Efficiency and the Level of Expression of a Cell-Surface Glycoprotein. J. Biol. Chem. 270, 14756–14761 [DOI] [PubMed] [Google Scholar]
- 61. Carpentieri A., Ratner D. M., Ghosh S. K., Banerjee S., Bushkin G. G., Cui J., Lubrano M., Steffen M., Costello C. E., O'Keefe B., Robbins P. W., and Samuelson J. (2010) The antiretroviral lectin cyanovirin-N targets well-known and novel targets on the surface of Entamoeba histolytica trophozoites. Eukaryot. Cell 9, 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chatterjee A., Ratner D. M., Ryan C. M., Johnson P. J., O'Keefe B. R., Secor W. E., Anderson D. J., Robbins P. W., and Samuelson J. (2015) Anti-retroviral lectins have modest effects on adherence of Trichomonas vaginalis to epithelial cells in vitro and on recovery of Tritrichomonas foetus in a mouse vaginal model. PLoS ONE 10, e0135340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Magnelli P., Cipollo J. F., Ratner D. M., Cui J., Kelleher D., Gilmore R., Costello C. E., Robbins P. W., and Samuelson J. (2008) Unique Asn-linked oligosaccharides of the human pathogen Entamoeba histolytica. J. Biol. Chem. 283, 18355–18364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Paschinger K., Hykollari A., Razzazi-Fazeli E., Greenwell P., Leitsch D., Walochnik J., and Wilson I. B. (2012) The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology 22, 300–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Manthri S., Guther M. L., Izquierdo L., Acosta-Serrano A., and Ferguson M. A. (2008) Deletion of the TbALG3 gene demonstrates site-specific N-glycosylation and N-glycan processing in Trypanosoma brucei. Glycobiology 18, 367–383 [DOI] [PubMed] [Google Scholar]
- 66. Hykollari A., Balog C. I., Rendic D., Braulke T., Wilson I. B., and Paschinger K. (2013) Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J. Proteome Res. 12, 1173–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schiller B., Hykollari A., Yan S., Paschinger K., and Wilson I. B. (2012) Complicated N-linked glycans in simple organisms. Biol. Chem. 393, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schiller B., Makrypidi G., Razzazi-Fazeli E., Paschinger K., Walochnik J., and Wilson I. B. (2012) Exploring the unique N-glycome of the opportunistic human pathogen Acanthamoeba. J. Biol. Chem. 287, 43191–43204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kobayashi Y., and Suzuki Y. (2012) Evidence for N-glycan shielding of antigenic sites during evolution of human influenza A virus hemagglutinin. J. Virol. 86, 3446–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pentiah K., Lees W. D., Moss D. S., and Shepherd A. J. (2015) N-linked glycans on influenza A H3N2 hemagglutinin constrain binding of host antibodies, but shielding is limited. Glycobiology 25, 124–132 [DOI] [PubMed] [Google Scholar]
- 71. van Montfort T., Eggink D., Boot M., Tuen M., Hioe C. E., Berkhout B., and Sanders R. W. (2011) HIV-1 N-glycan composition governs a balance between dendritic cell-mediated viral transmission and antigen presentation. J. Immunol. 187, 4676–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Petersen C. (2000). U.S. Patent No. 6,071,518 Washington, DC: U.S. Patent and Trademark Office.
- 73. Ward H., Hamer D., Keusch G., & Cevallos A. M. (2003). U.S. Patent No. 6,657,045 Washington, DC: U.S. Patent and Trademark Office.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the dataset identifier PXD005503 and 10.6019/PXD005503 (http://www.ebi.ac.uk/pride/archive/projects/PXD005503).