Abstract
Plasmodium parasites contain several unique membrane compartments in which prenylated proteins may play important roles in pathogenesis. Protein prenylation has also been proposed as an antimalarial drug target because farnesyltransferase inhibitors cause potent growth inhibition of blood-stage Plasmodium. However, the specific prenylated proteins that mediate antimalarial activity have yet to be identified. Given the potential for new parasite biology and elucidating drug mechanism-of-action, we performed a large-scale identification of the prenylated proteome in blood-stage P. falciparum parasites using an alkyne-labeled prenyl analog to specifically enrich parasite prenylated proteins. Twenty high-confidence candidates were identified, including several examples of pathogen-specific prenylation activity. One unique parasite prenylated protein was FYVE-containing coiled-coil protein (FCP), which is only conserved in Plasmodium and related Apicomplexan parasites and localizes to the parasite food vacuole. Targeting of FCP to this parasite-specific compartment was dependent on prenylation of its CaaX motif, as mutation of the prenylation site caused cytosolic mislocalization. We also showed that PfRab5b, which lacks C-terminal cysteines that are the only known site of Rab GTPase modification, is prenylated. Finally, we show that the THQ class of farnesyltransferase inhibitors abolishes FCP prenylation and causes its mislocalization, providing the first demonstration of a specific prenylated protein disrupted by antimalarial farnesyl transferase inhibitors. Altogether, these findings identify prenylated proteins that reveal unique parasite biology and are useful for evaluating prenyltransferase inhibitors for antimalarial drug development.
Protein prenylation is the covalent attachment of hydrophobic prenyl chains to cysteine residues of protein substrates. This post-translational lipid modification often mediates membrane association important for the cellular function of prenylated proteins. During symptomatic infection, Plasmodium parasites reside inside mature red blood cells. Although the uninfected red blood cell lacks nucleus, mitochondria, and ER/Golgi and consists only of a cell membrane, Plasmodium-infected red blood cells contain numerous unique membranes both present within the parasite and induced by the parasite in the host cell. These include a specialized parasite organelle called the food vacuole in which host hemoglobin is digested, a parasitophorous vacuole that surrounds the parasite, and new membrane structures in the red cell cytoplasm. Plasmodium proteins modified with palmitoyl and/or myristoyl lipids have important functions at several of these new membranes (1, 2). It seems likely that prenylated Plasmodium proteins will also play critical roles within these unique parasite membranes.
Protein prenylation is catalyzed by three classes of protein prenyltransferases: farnesyltransferase (FT)1, geranylgeranyltransferase 1 (GGT1), and Rab geranylgeranyltransferase (RabGGT). These enzymes differ in the addition of either 15-carbon farnesyl or 20-carbon geranylgeranyl chains and their recognition of substrate proteins. Both FT and GGT1 recognize and modify a 4-amino acid Cysteine-aliphatic-aliphatic-terminal residue (CaaX) motif at the C-terminus of protein targets, whereas RabGGT exclusively recognizes a family of Rab GTPases and modifies two C-terminal cysteines. Plasmodium parasites encode at least a putative FT and RabGGT. In P. falciparum, two CaaX proteins which share homology with known prenylated mammalian proteins were shown to be prenylated, and 11 Rab GTPases are annotated in the genome (3–6). However, large-scale, unbiased approaches will be required to accelerate the identification of nonconserved prenylated parasite proteins that have unique functions in pathogenesis.
Given the diverse and often essential functions of prenylated proteins, protein prenylation is also a target for therapeutic intervention. Inhibitors of human FT and GGT1 have undergone extensive investigation for treatment of numerous cancers, the premature aging syndrome progeria, and hepatitis delta infection, resulting in multiple clinical trials (7). The mechanism-of-action of these inhibitors and their efficacy against specific diseases is highly dependent on the cellular functions of specific CaaX protein substrates. The prenylation of oncogenic Ras GTPases by FT and GGT1 is the desired target in cancer treatment, whereas disrupting the farnesylation of nuclear lamins underlies the efficacy of FT inhibitors in treatment of progeria.
To combat growing antimalarial drug resistance, FT inhibitors have been proposed as a new class of antimalarials based on several important attributes. First, these compounds potently inhibit the growth of blood-stage Plasmodium parasites (8, 9). Second, they show minimal to no effects on mammalian cell growth at the therapeutic doses required for parasite growth inhibition (8). Finally, the drug development process benefits enormously from the vast medicinal chemistry and biological knowledge accrued over decades of pharmaceutical research on FT inhibitors for cancer chemotherapy. The tetrahydroquinoline (THQ) series of FT inhibitors, which show low nm efficacy against blood-stage Plasmodium falciparum, have been investigated most extensively as antimalarials. Studies on their mechanism-of-action showed that THQ compounds block farnesylation activity in Plasmodium cell lysates and selectively inhibit farnesylation of unidentified cellular proteins (8). Notably, resistance to THQ compounds was associated with mutations in the parasite's putative FT β subunit (10, 11). These studies validated P. falciparum FT as the drug target but left unanswered questions about the specific CaaX protein(s) that mediate parasite growth inhibition.
To perform the first unbiased and large-scale identification of the prenylated proteome in P. falciparum, we took a chemical biology approach using metabolic labeling with an alkyne-modified farnesyl probe (AlkFOH) combined with mass spectrometry proteomics. Key questions we sought to answer were: What are the protein substrates of prenyltransferases in Plasmodium? How is protein prenylation in the parasite distinct from that in mammalian cells? Which prenylated proteins are important for mediating the antimalarial activity of FT inhibitors? Though the prenylated proteome is significantly reduced in P. falciparum compared with humans, we provide evidence for parasite-specific prenylation activity, including a novel prenylated protein and a novel site of modification. Furthermore, we use one of these newly identified prenylated proteins to investigate the mechanism-of-action of antimalarial farnesyltransferase inhibitors.
EXPERIMENTAL PROCEDURES
P. falciparum In Vitro Culture
All experiments described were performed in W2 (MRA-157) and Dd2attB (MRA-843) strain parasites, or with transgenic parasites generated in this study. Parasites were grown in human erythrocytes in RPMI 1640 media supplemented with 0.25% Albumax II (GIBCO Life Technologies; Carlsbad, CA), 2 g/L sodium bicarbonate, 0.1 mm hypoxanthine, 25 mm HEPES (pH 7.4), and 50 μg/L gentamycin, at 37 °C, 5% O2, and 5% CO2. Whole blood was obtained from the Stanford Blood Center. For comparison of growth between different treatment conditions, cultures were carried simultaneously and handled identically with respect to media changes and addition of blood cells.
Metabolic Labeling and Cu1+ Catalyzed Alkyne-azide Cycloaddition
Early trophozoite parasites were labeled for 6 h with 1 μm AlkFOH, unless otherwise noted. After labeling, parasites were washed and saponin pellets were prepared. Pellets were stored at −80°C. AlkFOH incorporation was monitored by in-gel fluorescence. Parasite pellets were lysed in 4% SDS, 50 mm TEA pH 7.4, and 150 mm NaCl. Lysate was immediately reacted with AzTAMRA (Life Technologies; Carlsbad, CA) under Cu1+ catalyzed alkyne-azide cycloaddition (CuAAC) conditions (12). Final concentrations were 1 mm TCEP (made fresh), 1 mm CuSO4, 100 μm TBTA, and 100 μm azide. The reaction was incubated for 1 h at room temperature. Protein was precipitated using methanol/chloroform/water (4/1.5/3 v/v/v) and washed three times with 1 ml cold methanol.
Mass Spectrometry Sample Preparation
Samples for mass spectrometry were generated by labeling 200 ml of 4% HCT, 10% early trophozoites (per condition) for 6 h with 1 μm AlkFOH or mock treated with DMSO. Parasites grown for each condition were mixed and separated immediately prior to labeling. Saponin pellets were prepared and stored at −80°C. Parasite pellets were lysed in 4% SDS, 50 mm TEA pH 7.4, 150 mm NaCl with protease inhibitor minitablets (Pierce) and Benzonase nuclease (Sigma-Aldrich; St. Louis, MO). At least 3 mg/ml protein underwent CuAAC with AzBiotin (Click chemistry tools) for 5 h at room temperature. CuAAC conditions were the same as described above. Cell lysate was diluted to 1 mg/ml for the reaction. In order to help prevent SDS contamination, after the 5 h incubation, reactions were diluted 10 times with 50 mm TEOA pH 7.4, 150 mm NaCl and precipitated using methanol and chloroform (methanol/chloroform/water at ratios of 4/1.5/3 v/v/v) overnight at −20 °C. Samples were centrifuged and washed 3 times with cold methanol. Protein pellets were dried for 1 h then solubilized in 1 ml 1% SDS, 50 mm TEOA pH 7.4, 150 mm NaCl using bath sonication. Samples were incubated with 100 μl high capacity streptavidin beads (Pierce) for 1 h, washed 3 times with 1% SDS, 50 mm TEOA pH 7.4, 150 mm NaCl. To help remove SDS, samples were washed 6 times with 6 m urea in 50 mm TEOA pH 7.4, 150 mm NaCl. Samples were then washed 3 times with 25 mm ammonium bicarbonate (ABC) then resuspended in 25 mm ABC. Samples were reduced using 10 mm DTT and alkylated using 500 mm iodoacetamide. Samples were digested using sequence-grade trypsin (Promega) overnight at 37 °C. Acetonitrile was added to the digestion buffer at a final concentration of 35%. The final solution containing the released peptides was removed from the beads and dried. Peptides were desalted using C18 stage tips and resuspended in 0.1% formic acid before being injected into the LC/MS-MS.
Mass Spectrometry Experimental Design and Statistical Rationale
Five biological replicates of each sample were analyzed by online capillary nanoLC-MS/MS. Samples were separated on an in-house made 20 cm reversed phase column (100 μm inner diameter, packed with ReproSil-Pur C18-AQ 3.0 μm resin (Dr. Maisch GmbH)) equipped with a laser-pulled nanoelectrospray emitter tip. Peptides were eluted at a flow rate of 400 nL/min using a two-step linear gradient including 2–25% buffer B in 70 min and 25–40% B in 20 min (buffer A: 0.2% formic acid and 5% DMSO in water; buffer B: 0.2% formic acid and 5% DMSO in acetonitrile) in an Eksigent ekspert nanoLC-425 system (AB Sciex). Peptides were then analyzed using a LTQ Orbitrap Elite mass spectrometer (Thermo Scientific). Data acquisition was executed in data dependent mode with full MS scans acquired in the Orbitrap mass analyzer with a resolution of 60,000 and m/z scan range of 340–1600. The top 20 most abundant ions with intensity threshold above 500 counts and charge states 2 and above were selected for fragmentation using collision- induced dissociation (CID) with isolation window of 2 m/z, collision energy of 35%, activation Q of 0.25 and activation time of 5 ms. The CID fragments were analyzed in the ion trap with rapid scan rate. Dynamic exclusion was enabled with repeat count of 1 and exclusion duration of 30 s. The AGC target was set to 1,000,000 and 5000 for full FTMS scans and ITMSn scans. The maximum injection time was set to 250 s and 100 s for full FTMS scans and ITMSn scans. The resulting spectra were searched against a “target-decoy” sequence database consisting of the PlasmoDB protein database (release 13.0, January 14, 2015), the Uniprot human database (released February 2, 2015) and the corresponding reversed sequences using the SEQUEST v. 28 rev. 12 algorithm (13). A total of 188,812 entries were in the combined dataset. The parent mass tolerance was set to 50 ppm and the fragment mass tolerance to 0.6 Da. Enzyme specificity was set to trypsin with up to three missed cleavages allowed. Oxidation of methionines was set as variable modification and carbamidomethylation of cysteines was set as static modification. Data was filtered to a 1% peptide and 5% protein false discovery rate using a linear discriminator analysis. MS1 peak area for each peptide were generated by extracting ion chromatograms (XICs) within 10 ppm of the observed m/z for the monoisoptopic peaks, as previously described (14). Areas were averaged between each biological replicate. The raw mass spectrometry data has been deposited at the Chorus Project (chorusproject.org) with the project identifier 1212.
Cloning and Transfections
E. coli codon optimized versions of FCP (PF3D7_1460100) and Rab5b (PF3D7_1310600) were designed and synthesized by GeneWiz. Quick change mutagenesis was used to mutate FCP cysteine 322 to a serine in the vector provided by GeneWiz. These constructs were then moved into the pLN transfection plasmid designed for Bxb1 mycobacteriophage integrase system (15). The In-fusion cloning kit (Clontech; Mountain View, CA) was used for all cloning. Restriction enzymes AvrII, BsiWI, and AflII were used to linearize the pLN vector. FCP and FCP(C322S) were designed to have an N-terminal GFP tag whereas Rab5b was C-terminally GFP tagged. All transgenes were driven with the ribosomal L2 protein (RL2) promoter (16).
Transfections were carried out as previously described (17). Briefly, 400 μl fresh red blood cells were preloaded with 50 μg of both pINT, which carries the bacteriophage integrase, and pRL2, which carries the gene of interest and the blasticidin resistance cassette, using a BioRad Gene-Pulser Xcell Electroporator. Electroporation conditions were infinite resistance, 310 V, and 950 μF using a 2-mm cuvette. Preloaded red blood cells were combined with 2.5 ml ∼20% schizont Dd2attB parasites and allowed to recover for 2 days before selection pressure was applied. Transfected parasites were selected with 2.5 μg/ml blasticidin and were detectable by thin smear within 20 days. Integration was confirmed by PCR.
SDS-PAGE and Western Blotting Analysis
Whole cell lysates were resolved on NuPage 4–12% Bis-Tris gels (Life Technologies). AlkFOH incorporation was monitored by in-gel fluorescence and measured using a Typhoon 9400 (General Electric; Piscataway, NJ) in the TAMRA channel. If further analysis by immunoblot was needed, protein was then transferred onto nitrocellulose membrane using Trans-Blot Turbo Transfer System (BioRad; Hercules, CA). The membrane was blocked for 1 h or longer at room temperature in 0.1% Casein (Casein, Hammersten Affymatrix) dissolved in 0.2× PBS. Membranes were probed overnight with 1:5000 Living Colors mouse monoclonal αGFP (Clonetech) or 1:20,000 rabbit polyclonal αAldolase (Abcam; Cambridge, UK) in 50% 0.1%Casein in 0.2×PBS and 50% 1×TBST. Secondary antibodies were probed for an hour or more at room temperature in the same buffer. Secondary antibodies used were either IRDye 680LT goat-αmouse or IRDye 800CW donkey-αrabbit when appropriate. Western blots were imaged using a LiCOR Odyssey Imaging System.
Immunoprecipitation. GFP-FCP, GFP-FCP(C322S) and Rab5b-GFP were purified from transgenic parasites using the GFP-TrapA immunoprecipitation kit (Chromotek; Planegg, Germany). Five ml cultures of mixed stage parasite between 2–5% parasitemia at 4%HCT were treated with BMS-386914 or lonafarnib for 1 h, then 1 μm AlkFOH was added to the media and parasites were labeled for 6 h. Saponin pellets were lysed in RIPA buffer with benzonase and protease inhibitor minitablets (Pierce) and incubated on ice for 20 min, with periodic vortexing. Whole cell lysates were diluted 5× with GFP-Trap dilution buffer and incubated with 25 μl of a 50% slurry of GFP-Trap agarose beads for 1 h at RT. Beads were washed three times in 500 μl wash buffer using a 27 G needle and syringe to remove any excess buffer. Protein was reacted with AzTAMRA on bead in 50 μl 1× PBS as described above. After a 1 h click reaction, beads and protein were precipitated using MeOH/chloroform/water and washed twice with 1 ml MeOH to remove any excess AzTAMRA. Pellets were dried and resuspended in 2× NuPage running buffer, then analyzed by SDS-PAGE and Western blotting as described above.
Live Epifluorescence Microscopy
Transgenic parasites were stained with 2 μg/ml Hoechst 33342. Epifluorescent images were collected using a Nikon Eclipse Ti-E using μManager software for acquisition. Images were processed using ImageJ. Single z-stack images were collected. Images were scored as showing either cytosolic localization, food vacuole localization, or a localization that had a weak cytosolic signal with brighter rings surrounding the food vacuole, which we termed “other.” Examples of scored images are shown in supplemental Fig. S2.
RESULTS
An Alkyne-modified Prenyl Lipid Analog Specifically Labels Parasite Proteins
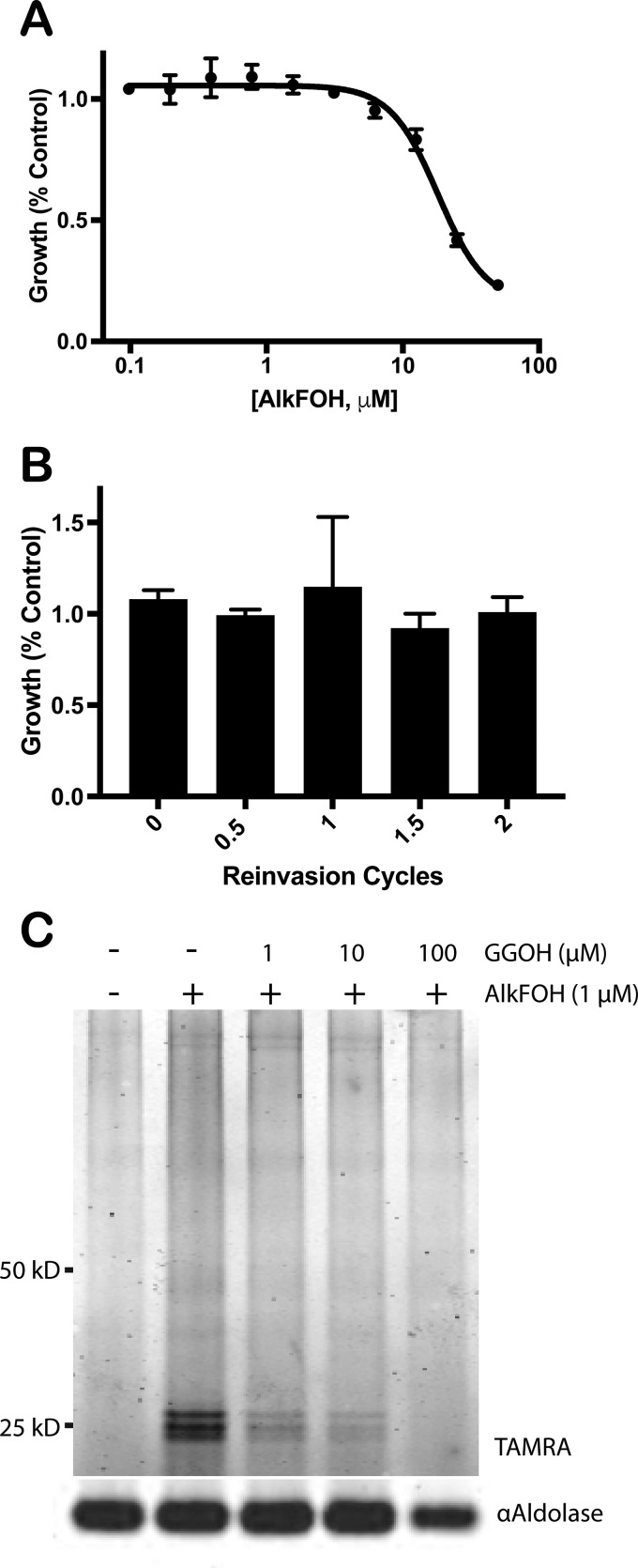
To enable large-scale identification of prenylated proteins, we performed metabolic labeling with an alkyne-modified farnesol analog, AlkFOH, in Plasmodium-infected red blood cells. Previously AlkFOH has been successfully used to identify prenylated proteins in mammalian cells (18). Mature red blood cells lack prenyltransferase activity, therefore incorporation of AlkFOH into proteins is due exclusively to the activity of protein prenyltransferases expressed by the infecting parasite (19). P. falciparum parasites were cultured with varying amounts of AlkFOH to determine its effect on parasite growth. Parasites could tolerate up to 10 μm AlkFOH with minimal growth inhibition (Fig. 1A). Parasites treated with 1 μm AlkFOH across the entire parasite replication cycle showed no growth defect nor any other observable phenotype, and 1 μm AlkFOH was sufficient to robustly label proteins in the parasite lysate (Fig. 1B and 1C). The most prominent bands incorporating the AlkFOH label were observed at ∼25 kDa and correspond to the expected size of Rab GTPases. Incorporation of AlkFOH into parasite proteins is specific, as protein labeling was competed away with increasing amounts of geranylgeraniol in vivo (Fig. 1C). Thus, this alkyne-modified prenyl lipid analog is a suitable tool for proteomic identification of prenylated proteins in blood-stage P. falciparum parasites.
Fig. 1.
AlkFOH specifically labels prenylated proteins in P. falciparum. A, P. falciparum growth was measured in the presence of varying concentrations of AlkFOH for 72 h. Growth relative to parasites treated with DMSO was plotted against AlkFOH concentration (μm, log scale, based on 3 biological replicates). B, Growth of parasites, treated with either 1 μm AlkFOH or DMSO, were monitored over the course of two replication cycles. Growth relative to parasites treated with DMSO was plotted against the number of reinvasion cycles (each cycle is 48 h; based on 3 biological replicates). C, Early trophozoites were labeled with 1 μm AlkFOH for 6 h with 0, 1, 10, or 100 μm GGOH or treated only with DMSO. Whole cell lysates were reacted with AzTAMRA under CuAAC conditions and resolved by SDS-PAGE, and in-gel fluorescence was measured.
The Prenylated Proteome Reveals a Limited Number of Prenylated Targets
Having successfully introduced an alkyne label into prenylated proteins, we used Cu1+ catalyzed cycloaddition (click chemistry) to append an azido-biotin tag, affinity purified the labeled proteins, and identified them by mass spectrometry. Labeling began during the trophozoite stage when the protein prenyltransferases are maximally expressed (20). Five biological replicates were performed, and protein abundance was quantified by the total MS1 area of all matched peptides. The average abundance of proteins in AlkFOH- versus mock-labeled samples was plotted to distinguish proteins that were enriched by AlkFOH labeling from nonspecifically bound proteins. As shown in Fig. 2, a distinct population of proteins, which included all known prenylated Plasmodium proteins, was only detected in the AlkFOH-labeled samples and absent from DMSO controls. Notably, several low-abundance prenylated proteins were highly enriched in the AlkFOH-labeled samples, suggesting that our method can specifically identify prenylated proteins.
Fig. 2.
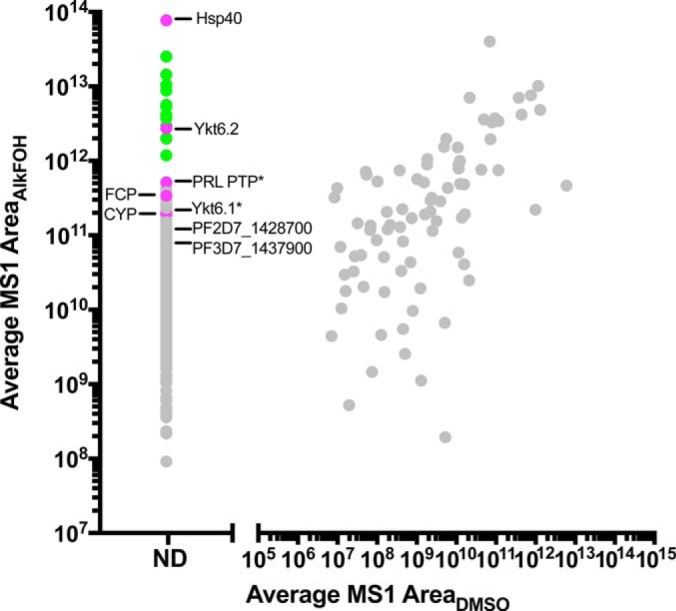
Proteomic identification of prenylated proteins in P. falciparum reveals a limited number of substrates. Average MS1 area measured in AlkFOH-labeled samples is plotted against average MS1 area measured for the same protein in DMSO mock-treated samples. Proteins which were detected only in AlkFOH labeled samples are shown on the left of the graph (ND = not detected). All data points are shown. Rab GTPases are marked in green, whereas proteins with a C-terminal CaaX box are shown in magenta. CaaX proteins which have previously been shown to be prenylated are designated with an asterisk. Results are based on five biological replicates.
We used both proteomic and biological criteria to segregate the list of identified proteins. Medium-confidence hits were selected if they satisfied criteria based on proteomic results: (1) identified by >1 unique peptide, (2) present in 3 out of 5 biological replicates, and (3) detected in AlkFOH-labeled samples and absent in DMSO controls. The full list of medium-confidence prenylated proteins are listed in supplemental Table S1. High-confidence prenylated proteins also fulfilled biological criteria, either containing cysteine residues within four amino acids of the C-terminus or had homology to known prenylated proteins. These criteria were based on established features of substrate recognition by the three classes of protein prenyltransferases.
A total of 20 high-confidence prenylated proteins were identified (Table I). We assigned the proteins to “prenylated protein clusters” identified in PRENbase, a database of known and predicted protein prenylation substrates (19). Each cluster is comprised of protein homologs with known or predicted prenylation motifs that are evolutionarily-conserved and likely functionally related. Thus, prenylated parasite proteins that cannot be assigned to a PRENbase cluster are more likely to have new, pathogen-specific functions that have not been described previously. A majority of the identified proteins clustered with homologs for which prenylation is both validated and highly conserved among eukaryotes, including 10 Rab GTPases and 4 CaaX proteins. Two candidates, a DUF544 protein of unknown function (PF3D7_1319100) and a peptidyl-prolyl cis-trans isomerase (PF3D7_0322000), are conserved eukaryotic proteins containing CaaX motifs whose prenylation are predicted but have not been previously validated in any organism (21). Significantly, two prenylated proteins could not be assigned to a PRENbase cluster: a FYVE and coiled-coil domain-containing protein, FCP (PF3D7_1460100) and a conserved protozoan protein of unknown function (PF3D7_1428700). Finally, we identified 2 unexpected Rab GTPase substrates: P. falciparum Rab5b (PF3D7_1310600) which lacks any C-terminal cysteines and human Rab35, a host red blood cell protein.
Table I. High-confidence prenylated proteins identified by proteomicsa.
| Gene ID | Annotation | Motif | PrePS prediction | PRENbase cluster |
|---|---|---|---|---|
| CaaX box proteins | ||||
| PF3D7_0322000 | peptidyl-prolyl cis-trans isomerase (CYP19A) | CGEL | FT– GGT1– | Predicted conserved |
| PF3D7_0910600 | SNARE protein, putative (YKT6.1) | CCSIM | FT+++ GGT1+++ | Known conserved |
| PF3D7_1113100 | protein tyrosine phosphatase (PRL) | CHFM | FT– GGT1– | Known conserved |
| PF3D7_1319100 | Conserved DUF544 protein, unknown function | CTIM | FT+++ GGT1+++ | Predicted conserved |
| PF3D7_1324700 | SNARE protein, putative (YKT6.2) | CCSLY | FT+ GGT1– | Known conserved |
| PF3D7_1428700 | Conserved protein, unknown function | CNFM | FT– GGT1– | Unknown Not conserved |
| PF3D7_1437900 | HSP40, subfamily A, putative (ERdj3) | CAQQ | FT++ GGT1– | Known conserved |
| PF3D7_1460100 | FYVE and coiled-coil domain-containing protein (FCP) | CNIM | FT+ GGT1++ | Unknown Not conserved |
| Rab GTPases | ||||
| PF3D7_0106800 | Ras related protein, Rab5c | KKCC | RabGGT+++ | Known conserved |
| PF3D7_0211200 | Ras related protein, Rab5a | KGCC | RabGGT+++ | |
| PF3D7_0512600 | Ras related protein, Rab1b | KKCC | RabGGT+++ | |
| PF3D7_0807300 | Ras related protein, Rab18 | NCAC | RabGGT+++ | |
| PF3D7_0903200 | Ras related protein, Rab7 | SRCC | RabGGT+++ | |
| PF3D7_1144900 | Ras related protein, Rab6 | KCLC | RabGGT+++ | |
| PF3D7_1231100 | Ras related protein, Rab2 | FSCC | RabGGT+++ | |
| PF3D7_0513800 | Ras related protein, Rab1a | FCSC | RabGGT+++ | |
| PF3D7_1320600 | Ras related protein, Rab11a | NKCC | RabGGT+++ | |
| PF3D7_1340700 | Ras related protein, Rab11b | VKCC | RabGGT++ | |
| PF3D7_1310600 | Ras related protein, Rab5b | Noneb | RabGGT – | |
| H. sapiens Rab GTPase | ||||
| Q15286 | Ras related protein, Rab35 | KRCC | GGT2+++ | Known conserved |
a High confidence proteins were defined by (1) homology to known prenylation substrates, (2) presence of a motif, (3) present in three or more biological replicates, and (4) detected only in AlkFOH labeled samples.
b No cysteine residues found in last 5 amino acids of predicted protein sequence.
Localization of a Novel Parasite Protein FCP is Dependent on Prenylation
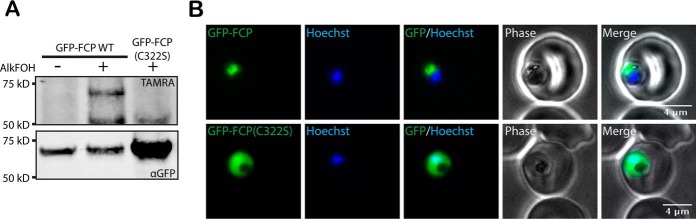
In our proteomic dataset, FCP stood out as a unique protein that is conserved in Plasmodium spp. and related Apicomplexan parasites that cause human diseases. In addition, there is evidence that FCP may be essential for blood-stage Plasmodium growth (22). Therefore, we further characterized the function of its CaaX prenylation. FCP was previously shown to localize to the Plasmodium food vacuole, site of host hemoglobin digestion (22). Unexpectedly, this localization did not require the binding of phosphatidylinositol 3-monophosphate (PI3P) by the FYVE domain but was dependent on the C-terminal 44 amino acids of FCP (22). Based on these previous findings, we hypothesized that prenylation of its CaaX motif may be responsible for membrane association and localization of FCP to the food vacuole. To test this hypothesis, we generated P. falciparum strains overexpressing N-terminal GFP-tagged wild-type or C322S mutant FCP. Cys322 is the modified cysteine residue in the CaaX motif, and its mutation to serine is expected to block prenylation of FCP. Indeed GFP-FCP purified from parasites treated with AlkFOH was covalently modified with this prenyl analog (visualized by reaction with fluorophore-azide), whereas labeling was not observed for GFP-FCP(C322S), consistent with loss of the prenyl modification when the CaaX motif is mutated (Fig. 3A). Live fluorescence microscopy also showed that GFP-FCP and GFP-FCP(C322S) differed in their cellular localization (Fig. 3B). GFP-FCP showed fluorescence overlapping or surrounding parasite hemozoin, consistent with its previously reported food vacuole localization. However, GFP-FCP(C322S) fluorescence appeared throughout the cell consistent with cytosolic mislocalization. Additional representative images are shown in supplemental Fig. S1. Taken together, these results suggest that the localization of FCP is dependent on prenylation of its C-terminal CaaX motif.
Fig. 3.
A CaaX motif is required for FCP localization. A, Parasites expressing Wt GFP-FCP or GFP-FCP(C322S) were labeled with AlkFOH and GFP-FCP was immunoprecipitated. AlkFOH labeling was visualized by a click reaction with AzTAMRA (top panel) which loading was visualized by an immunoblot against GFP (bottom panel). B, Live epifluorescent images of P. falciparum parasites expressing either Wt GFP-FCP (top panel) or GFP-FCP(C322S) (bottom panel). The nucleus was stained using Hoechst. Additional images are shown in the supplemental material (Supplemental Fig. S1).
Rab5b Prenylation Does Not Require C-terminal Cysteine Residues
The identification of Rab5b in the prenylated proteome was also not predicted as it lacks C-terminal cysteines that are the known site of modification by RabGGT in all other organisms. Instead Rab5b was previously shown to be N-myristoylated and palmitoylated, and these lipid modifications were believed to substitute for prenylation to mediate Rab5b membrane localization at the parasite food vacuole, parasite plasma membrane, and parasitophorous vacuole (1, 2, 6, 23, 24). However, Rab5b was strongly identified in our proteomic data set with 53% sequence coverage. Identified peptides mapped across the entirety of the full-length protein, making it unlikely that proteolytic processing exposes internal cysteines at a neo-C-terminus (Fig. 4A). To rule out the possibility that Rab5b was purified in the prenylated proteome because of nonspecific protein interactions, we constructed a P. falciparum strain expressing C-terminally tagged Rab5b-GFP and directly detected the prenyl modification on immunoprecipitated Rab5b-GFP from this strain. We observed labeling of Rab5b with AlkFOH, visualized by reaction with a fluorophore-azide tag (Fig. 4B). These results indicate that Rab5b is indeed prenylated at a nonconserved modification site.
Fig. 4.
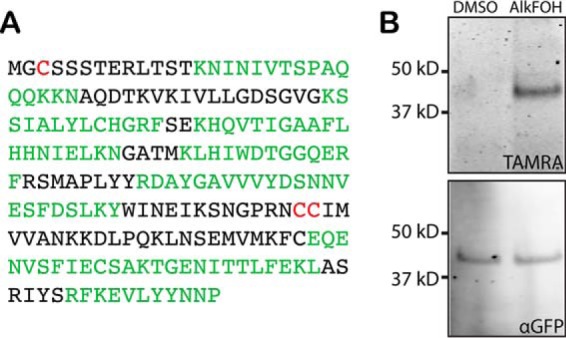
Rab5b is prenylated on internal cysteines. A, Peptides identified in AlkFOH samples are shown in green in the full-length sequence of Rab5b. B, Parasites expressing Rab5b-GFP were treated with either DMSO or AlkFOH and Rab5b-GFP was immunoprecipitated. AlkFOH incorporation was visualized by a click reaction with AzTAMRA (top panel) whereas Rab5b-GFP loading was visualized by an αGFP immunoblot (bottom panel).
The THQ Class Farnesyltransferase Inhibitor BMS-386914 Disrupts FCP Prenylation and Localization
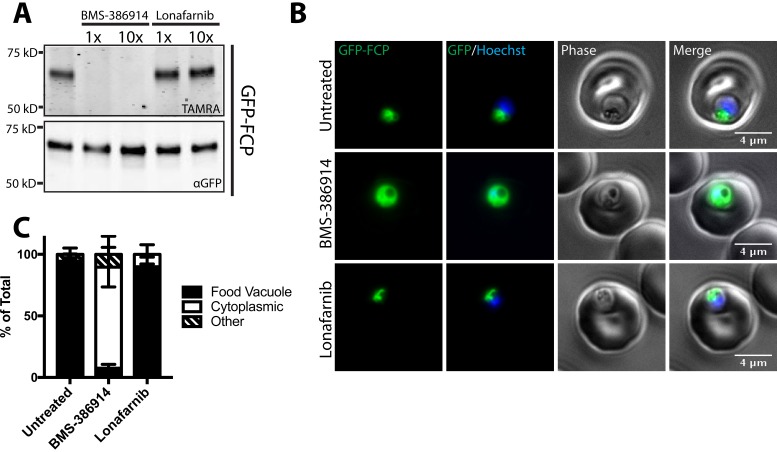
Although the mechanism-of-action of antimalarial FT inhibitors depends on the cellular functions of CaaX protein substrates whose prenylation is disrupted, no specific CaaX protein substrate has been linked to FT inhibition. In addition, it would be ideal to have an assay that correlates cellular prenylation activity with antimalarial growth inhibitory activity. Therefore, we determined the effect of FT inhibitors on cellular prenylation of FCP. The THQ compound BMS-386914 has been shown to inhibit the enzymatic activity of P. falciparum FT with an IC50 = 0.9 nm and blood-stage Plasmodium growth with an EC50 = 5 nm, whereas lonafarnib showed IC50> 250 nm and EC50 = 3 μm (8). Against P. falciparum W2 parasites, we determined the EC50 of BMS-386914 for growth inhibition was 25 nm and that of lonafarnib was 1.2 μm. We observed the amount of AlkFOH labeling of GFP-FCP in the presence of 1× and 10× EC50 concentrations of each drug. Consistent with its mechanism-of-action inhibiting PfFT, BMS-386914 blocked prenylation of FCP at similar concentrations in which parasite growth inhibition is observed (Fig. 5A). Furthermore, BMS-386914 inhibition of FCP prenylation also caused its cytosolic mislocalization, like that observed in the GFP-FCP(C332S) mutant (Fig. 5B and supplemental Fig. S2). Quantification of GFP-FCP localization from three independent biological replicates (two of which were blinded) showed that only 7.5% of parasites treated with BMS-386914 retained a food vacuole localization compared with 92.7% in untreated parasites (Fig. 5C). In contrast, lonafarnib showed no effect on FCP prenylation or localization even at 10x EC50 concentrations (Fig. 5B and 5C). These results show that BMS-386914 inhibits prenylation and proper localization of FCP as part of its antimalarial mechanism-of-action. In contrast, parasite growth inhibition by lonafarnib does not disrupt FCP prenylation, suggesting that lonafarnib's antimalarial activity is not because of FT inhibition.
Fig. 5.
BMS-386914 inhibits FCP prenylation and causes mislocalization to the cytosol. A, GFP-FCP was immunoprecipitated from AlkFOH labeled parasites treated with either 1x EC50 or 10x EC50 BMS-386914 and lonafarnib. AlkFOH labeling was detected by reaction with AzTAMRA fluorescent tag (top panel) whereas GFP loading was visualized through an αGFP immunoblot (bottom panel). B, Live microscopy of GFP-FCP expressing parasites treated with 10× EC50 BMS-386914 or lonafarnib. The nucleus was stained using Hoechst. C, Quantification of GFP-FCP localization in untreated, BMS-386914 (10x EC50) and lonafarnib (10× EC50) images. Based on three biological replicates, n>50 for each condition. Additional scored images are shown in the supplemental material (supplemental Fig. S2).
DISCUSSION
Our findings indicate that Plasmodium parasites contain markedly few prenylated proteins, only 20 high-confidence identifications out of 5398 protein-coding genes representing 0.3% of the proteome. Other protein lipid modifications, such as S-palmitoylation and N-myristoylation, were reported to be more abundant in P. falciparum parasites using similar metabolic labeling and chemoproteomic approaches (1, 2). Though the low number of identifications is surprising, we believe our data set is an accurate estimate of prenylated proteins, and biological factors likely account for the low abundance of cellular prenylation in blood-stage Plasmodium. Plasmodium lacks homologs to several major families of prenylated proteins found in mammalian cells, including Rho/Rac GTPases, nuclear lamins, and Gγ subunits of heterotrimeric G-proteins (25). Other protein families have been significantly reduced. For example, Plasmodium encodes just 11 Rab GTPases compared with 66 in humans (6, 26). There are also fewer numbers of potential CaaX substrates with only 95 Plasmodium proteins containing the required cysteine residue at position −4 from the C-terminus, and only a small subset of these are expected to be recognized as a CaaX motif and prenylated. Even the prenylation site of the most well-known prenylated protein, Ras GTPase, is not conserved in thePlasmodium homolog. The single P. falciparum Ras GTPase (PF3D7_0616700) lacks C-terminal cysteines and was not identified among medium- or high-confidence candidates, though its transcript expression level is similar to PF3D7_1428700, the lowest expressing gene among the high-confidence candidates. Thus PfRas prenylation may no longer be required despite being highly conserved in other eukaryotes. Finally, the limited number of Plasmodium prenylated proteins is consistent with lower estimates of prenylated protein abundance in lower eukaryotes compared with mammalian cells (27). These biological factors predict that protein prenylation is drastically reduced in Plasmodium spp. parasites, consistent with our results.
The efficiency and broad incorporation of modified prenyl analogs into parasite proteins also suggests that our ability to detect prenylated proteins was not limited by this approach. AlkFOH was previously shown to be recognized by all three types of protein prenyltransferases and incorporated into >100 known and novel prenylated proteins in mammalian cells (12, 18). In fact, labeling in mammalian cells was significantly less efficient than in Plasmodium parasites, requiring 50-fold higher concentration of the analog and depletion of endogenous prenyl lipids by inhibitor treatment. In our experiments, 1 μm AlkFOH was able to compete with endogenous prenyl lipids under conditions that maintained parasite viability. Under these conditions, both Rab GTPases and CaaX substrates, which are recognized by different protein prenyltransferase classes, were enriched. Labeling with an alkyne-modified geraniol (10-carbon) yielded similar identifications as those in AlkFOH-labeled samples, suggesting that the specificity of Plasmodium prenyltransferases for substrate proteins is not altered by moderate modifications of the prenyl lipid (supplemental Table S2). Independent experiments performed using alternative alkyne- and azide-modified farnesol analogs also did not identify additional prenylated targets (R.A. Serwa, M. Jones, P. Bowyer, E.W. Tate, and J.C. Rayner; personal communication). Thus, this chemoproteomic approach was robust and likely quite sensitive.
In fact, a significant advantage of our unbiased approach was the ability to identify novel prenylated proteins such as FCP and PF3D7_1428700, because these would not have been recognized as high-priority candidates based on sequence homology with previously known prenylated proteins. Bioinformatic prediction of CaaX motifs recognized by FT and GGT1 is also not sufficiently sensitive, because the Prenylation Prediction Suite (PrePS) software recognized only five of the eight high-confidence CaaX protein candidates (Table I and supplemental Table S3) (28). Our high-confidence list assumes that Plasmodium FT/GGT1 enzymes recognize and prenylate CaaX-containing proteins, similar to other organisms. However, numerous medium-confidence candidates are neither homologs of known prenylated proteins nor have a recognizable prenylation site. One strategy to identify true novel prenylated proteins among these candidates would be to select lower abundance proteins (based on transcript expression levels) for validation, because these are less likely to be nonspecific contaminants. Importantly, though protein farnesylation and geranylgeranylation on cysteines are by far the most common prenyl modification, we cannot rule out the possibility that the AlkFOH probe is further metabolized to longer chain prenyl groups or other isoprenoid derivatives before covalent attachment to protein. Indeed protein dolicholyation (C55) has been reported in Plasmodium, based on the incorporation of radioactive label derived from farnesyl diphosphate into dolichols released from unidentified proteins (29). These alternative prenyl modifications were detected at low abundance and were labile to methyl iodide consistent with thioether attachment via cysteine residues. Lastly, our data set only includes proteins expressed and prenylated during trophozoite/schizont stages when AlkFOH labeling was performed and may miss very low abundance proteins (<10 RPKM transcript expression).
Despite the limited number of prenylated proteins in Plasmodium, we detected several examples of prenylation activity unique to the parasite that may target proteins to novel membranes in the Plasmodium-infected red cell. FCP in particular stood out as a novel parasite-specific protein, as it was the only protein in our data set that is only conserved among Apicomplexan parasites. FCP localizes to a parasite-specific compartment, the food vacuole, and we demonstrate that prenylation at its CaaX motif is necessary for this localization through both mutagenesis and FT inhibitor treatment. The Toxoplasma FCP homolog has also been localized to its vacuolar compartment, VAC, which is functionally analogous to the food vacuole (30). Furthermore, FCP function may be essential because, in a previous study, a truncated form of FCP (lacking the FYVE domain) still localized to the food vacuole but had a dominant-negative effect on parasite growth and abnormal food vacuole morphology (22). Finally, FCP is one of only 3 PI3P-binding proteins present in P. falciparum, binding PI3P via its FYVE domain (22). PI3P is normally found on early endosomes, however, in P. falciparum it is present on two parasite-specific compartments, the food vacuole and the apicoplast (31). Normally, PI3P-binding proteins are not themselves post-translationally lipidated but often interact with prenylated and palmitoylated proteins. For example, the mammalian protein early endosomal antigen (EEA1) binds PI3P and interacts with Rab GTPases (32). By contrast, FCP was previously identified in the palmitoylated proteome and is now shown to be prenylated, making it the first example, to our knowledge, of a lipidated PI3P-binding protein (2). Notably Plasmodium does not encode a recognizable EEA-1 homolog, and a lipidated, PI3P-binding, coiled-coil protein like FCP may serve a similar function as EEA-1. Considering all the evidence, FCP is likely to have an important and pathogen-specific membrane function dependent on its prenylation. Future studies will be necessary to determine whether it is essential for growth and identify its protein interaction partners.
We also detected atypical RabGGT activity in Plasmodium. Our results demonstrate that Rab5b, which has been localized to the parasite food vacuole, parasite plasma membrane, and parasitophorous vacuole, is prenylated even though it lacks C-terminal cysteines that are the only known modification site of Rab GTPases in other organisms. Instead we propose that Rab5b is prenylated on one or more internal cysteine residues. In our proteomic sample preparation, the peptide containing the modified residue remains attached to the agarose beads following on-bead trypsin digestion. Based on the sequence coverage of Rab5b, only three cysteines were not represented by identified peptides and are potential candidates for the modification sites: one at residue three near the N terminus (likely palmitolyation site) and two tandem cysteines at residues 141–142 (Fig. 4A). The dicysteine site has previously been suggested to serve as an internal prenylation site (6). Normally unprenylated Rab GTPases are bound by the Rab escort protein (REP), which presents their C-terminal cysteine residues to the RabGGT for prenylation. Given this mode of recognition, it is possible that a putative Plasmodium REP (PF3D7_1038100) may present internal cysteines for modification. Another alternative is that the prenyl modification occurs on a noncysteine residue but this seems unlikely as it would require a change in the chemical reaction catalyzed by the RabGGT. There is no known biosynthetic route that converts isoprenoids to other common protein modifications. Thus, the modification of Rab5b by AlkFOH cannot be explained by the presence of palmitoylation, myristolyation, or other known post-translational modifications. As this is the first report of Rab prenylation on a non-conserved site, it may be unique to Plasmodium but may also have gone undetected in other organisms. Interestingly Ara6, a plant Rab5 homolog, also lacks C-terminal cysteines and is both N-myristoylated and palmitoylated, thus it may also be prenylated on a non-conserved site (23, 33). Both N-myristoylation and palmitoylation of Rab5b are necessary for localization to two parasite-specific compartments, the food vacuole and the parasitophorous vacuole; it remains to be determined whether prenylation is also important for its localization (2, 23, 24). In particular, mutation of the internal dicysteine Cys141 and Cys142 may reveal the nonconserved prenylation site and show whether prenylation is important for Rab5b localization.
The most surprising prenylated protein identified in our high-confidence list was a human Rab GTPase, HsRab35. First, Rab GTPases in mature red blood cells should already be prenylated, therefore the C-terminal cysteines of HsRab35 should not be available for AlkFOH labeling. Second, in Plasmodium-infected red cells, the only PPTs are expressed by the parasite and present in the parasite cytosol, whereas HsRab35 is expected to localize to the red blood cell compartment (19). Despite these apparent discrepancies, HsRab35 was not identified in AlkFOH-treated, uninfected red blood cells, implying that AlkFOH labeling of HsRab35 is dependent on parasite infection. Only three unique peptides were detected for HsRab35, resulting in 6% protein coverage. Indeed, six other human Rab GTPases were also detected by single unique peptides in our prenylated proteome, representing seven of the nine HsRab GTPases identified in the human red blood cell proteome (19). The sparse peptide identifications likely reflect the low abundance of human Rab GTPases, which is not surprising because the proteomic samples were prepared from saponin-lysed parasites in which host cell proteins in the red blood cell membrane and cytoplasm were specifically depleted. The fact that these human Rab GTPases were detected in the prenylated proteome at all suggests they were specifically enriched. Human Rab GTPases may be more robustly detected in proteomic samples prepared from whole Plasmodium-infected red cells. Unfortunately, we were unsuccessful in purifying a tagged HsRab35 introduced into resealed red cells to directly detect its parasite-dependent prenyl modification. Although the detection of a human Rab GTPase in our prenylated proteome is intriguing and hints at a potential role for parasite-dependent prenylation at novel membranes in the host cell compartment, at the moment we are unable to provide direct evidence for prenylation of host cell proteins.
Finally, in addition to identifying unique parasite prenylation activity, the Plasmodium prenylated proteome will bolster the investigation of FT inhibitors as antimalarials. We identified eight CaaX proteins as candidate FT substrates, some or all of which may be responsible for parasite growth inhibition caused by FT inhibitors (Table I). Further validation of these candidates will require that (1) cellular prenylation of its CaaX motif is inhibited by FT inhibitors and (2) prenylation is required for parasite growth. As seen previously, we observed the loss of several ∼50 kDa AlkFOH-labeled proteins upon treatment with THQ BMS-386914 (supplemental Fig. S3) (8). A high-confidence candidate in our dataset, PF3D7_1437900 (Hsp40), may represent one of these abundant prenylated proteins that are disrupted. We also demonstrated that BMS-386914 disrupts prenylation and localization of Apicomplexan-specific protein FCP. As expected, it did not disrupt Rab5b prenylation which is most likely catalyzed by the Rab geranylgeranyltransferase (data not shown). In contrast, lonafarnib which potently inhibits human FT activity has no effect on FCP prenylation and likely causes parasite growth inhibition via an alternative non-FT Plasmodium target (8, 9, 34). The difference between the mechanism-of-action of FT inhibitors in mammalian cells and Plasmodium parasites reinforces the fact that Plasmodium protein prenyltransferases are divergent from human protein prenyltransferases and can be targeted selectively as a strategy toward novel anti-malarial development (8, 9).
Supplementary Material
Acknowledgments
We thank Dr. Nathan Westcott and Professor Howard Hang (Rockefeller University) for providing AlkFOH and significant experimental guidance. We also thank Drs. Remigiusz Serwa (Imperial College), Matthew Jones (Wellcome Trust Sanger Institute), Paul Bowyer (London School of Hygiene & Tropical Medicine), and Professors Ed Tate (Imperial College London) and Julian Rayner (Wellcome Trust Sanger Institute) for sharing their unpublished data. THQ compounds were provided by Professor Michael Gelb (University of Washington). Finally, we thank the Bioinformatics Group at Research Institute of Molecular Pathology (IMP) for generating PrePS predictions for the Plasmodium falciparum 3D7 genome.
Footnotes
Author contributions: J.E.G. and E.Y. designed research; J.E.G., L.Z., and E.Y. performed research; L.Z. and J.E.E. contributed new reagents or analytic tools; J.E.G., J.E.E., and E.Y. analyzed data; J.E.G. and E.Y. wrote the paper.
* Funding support for this project was provided by National Institutes of Health Grants 1K08AI097239 and 1DP5OD012119 (to E.Y.), the Burroughs-Wellcome Fund (to E.Y.), and the Dean's Postdoctoral Fellowship of the Stanford School of Medicine (to J.E.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- FT
- farnesyltransferase
- GGT1
- geranylgeranyltransferase
- RabGGT
- Rab geranylgeranyltransferase
- CaaX
- cysteine-aliphatic-aliphatic-terminal residue residue
- THQ
- tetrahydroquinoline
- AlkFOH
- alkyne-modified farnesyl probe
- CuAAC or click chemistry
- copper-catalyzed alkyne-azide cycloaddition
- PI3P
- phosphatidylinositol 3-monophosphate.
DATA AVAILABILITY
The raw mass spectrometry data has been deposited at the Chorus Project (chorusproject.org) with the project identifier 1212.
REFERENCES
- 1. Wright M. H., Clough B., Rackham M. D., Rangachari K., Brannigan J. A., Grainger M., Moss D. K., Bottrill A. R., Heal W. P., Broncel M., Serwa R. A., Brady D., Mann D. J., Leatherbarrow R. J., Tewari R., Wilkinson A. J., Holder A. A., and Tate E. W. (2014) Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat. Chem. 6, 112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones M. L., Collins M. O., Goulding D., Choudhary J. S., and Rayner J. C. (2012) Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pendyala P. R., Ayong L., Eatrides J., Schreiber M., Pham C., Chakrabarti R., Fidock D. A., Allen C. M., and Chakrabarti D. (2008) Characterization of a PRL protein tyrosine phosphatase from Plasmodium falciparum. Mol. Biochem. Parasitol. 158, 1–10 [DOI] [PubMed] [Google Scholar]
- 4. Ayong L., DaSilva T., Mauser J., Allen C. M., and Chakrabarti D. (2011) Evidence for prenylation-dependent targeting of a Ykt6 SNARE in Plasmodium falciparum. Mol. Biochem. Parasitol. 175, 162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howe R., Kelly M., Jimah J., Hodge D., and Odom A. R. (2013) Isoprenoid biosynthesis inhibition disrupts Rab5 localization and food vacuolar integrity in Plasmodium falciparum. Eukaryotic Cell 12, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quevillon E., Spielmann T., Brahimi K., Chattopadhyay D., Yeramian E., and Langsley G. (2003) The Plasmodium falciparum family of Rab GTPases. Gene 306, 13–25 [DOI] [PubMed] [Google Scholar]
- 7. Ochocki J. D., and Distefano M. D. (2013) Prenyltransferase Inhibitors: Treating Human Ailments from Cancer to Parasitic Infections. Medchemcomm 4, 476–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nallan L., Bauer K. D., Bendale P., Rivas K., Yokoyama K., Hornéy C. P., Pendyala P. R., Floyd D., Lombardo L. J., Williams D. K., Hamilton A., Sebti S., Windsor W. T., Weber P. C., Buckner F. S., Chakrabarti D., Gelb M. H., and Van Voorhis W. C. (2005) Protein farnesyltransferase inhibitors exhibit potent antimalarial activity. J. Med. Chem. 48, 3704–3713 [DOI] [PubMed] [Google Scholar]
- 9. Buckner F. S., Eastman R. T., Yokoyama K., Gelb M. H., and Van Voorhis W. C. (2005) Protein farnesyl transferase inhibitors for the treatment of malaria and African trypanosomiasis. Curr. Opin. Investig. Drugs 6, 791–797 [PubMed] [Google Scholar]
- 10. Eastman R. T., White J., Hucke O., Yokoyama K., Verlinde C. L. M. J., Hast M. A., Beese L. S., Gelb M. H., Rathod P. K., and Van Voorhis W. C. (2007) Resistance mutations at the lipid substrate binding site of Plasmodium falciparum protein farnesyltransferase. Mol. Biochem. Parasitol. 152, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eastman R. T., White J., Hucke O., Bauer K., Yokoyama K., Nallan L., Chakrabarti D., Verlinde C. L. M. J., Gelb M. H., Rathod P. K., and Van Voorhis W. C. (2005) Resistance to a protein farnesyltransferase inhibitor in Plasmodium falciparum. J. Biol. Chem. 280, 13554–13559 [DOI] [PubMed] [Google Scholar]
- 12. Charron G., Tsou L. K., Maguire W., Yount J. S., and Hang H. C. (2011) Alkynyl-farnesol reporters for detection of protein S-prenylation in cells. Mol. Biosyst. 7, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 14. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adjalley S. H., Lee M. C. S., and Fidock D. A. (2010) A method for rapid genetic integration into Plasmodium falciparum utilizing mycobacteriophage Bxb1 integrase. Methods Mol. Biol. 634, 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Balabaskaran Nina P., Morrisey J. M., Ganesan S. M., Ke H., Pershing A. M., Mather M. W., and Vaidya A. B. (2011) ATP synthase complex of Plasmodium falciparum: dimeric assembly in mitochondrial membranes and resistance to genetic disruption. J. Biol. Chem. 286, 41312–41322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spalding M. D., Allary M., Gallagher J. R., and Prigge S. T. (2010) Validation of a modified method for Bxb1 mycobacteriophage integrase-mediated recombination in Plasmodium falciparum by localization of the H-protein of the glycine cleavage complex to the mitochondrion. Mol. Biochem. Parasitol. 172, 156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Charron G., Li M. M. H., MacDonald M. R., and Hang H. C. (2013) Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc. Natl. Acad. Sci. U.S.A. 110, 11085–11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. D'Alessandro A., Righetti P. G., and Zolla L. (2010) The red blood cell proteome and interactome: an update. J. Proteome Res. 9, 144–163 [DOI] [PubMed] [Google Scholar]
- 20. Otto T. D., Wilinski D., Assefa S., Keane T. M., Sarry L. R., Böhme U., Lemieux J., Barrell B., Pain A., Berriman M., Newbold C., and Llinás M. (2010) New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 76, 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F. L., and Eisenhaber F. (2007) Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 3, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McIntosh M. T., Vaid A., Hosgood H. D., Vijay J., Bhattacharya A., Sahani M. H., Baevova P., Joiner K. A., and Sharma P. (2007) Traffic to the malaria parasite food vacuole: a novel pathway involving a phosphatidylinositol 3-phosphate-binding protein. J. Biol. Chem. 282, 11499–11508 [DOI] [PubMed] [Google Scholar]
- 23. Ezougou C. N., Ben-Rached F., Moss D. K., Lin J.-W., Black S., Knuepfer E., Green J. L., Khan S. M., Mukhopadhyay A., Janse C. J., Coppens I., Yera H., Holder A. A., and Langsley G. (2014) Plasmodium falciparum Rab5B is an N-terminally myristoylated Rab GTPase that is targeted to the parasite's plasma and food vacuole membranes. PLoS ONE 9, e87695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ebine K., Hirai M., Sakaguchi M., Yahata K., Kaneko O., and Saito-Nakano Y. (2016) Plasmodium Rab5b is secreted to the cytoplasmic face of the tubovesicular network in infected red blood cells together with N-acylated adenylate kinase 2. Malar. J. 15, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boureux A., Vignal E., Faure S., and Fort P. (2007) Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 24, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diekmann Y., Seixas E., Gouw M., Tavares-Cadete F., Seabra M. C., and Pereira-Leal J. B. (2011) Thousands of rab GTPases for the cell biologist. PLoS Comput. Biol. 7, e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Epstein W. W., Lever D., Leining L. M., Bruenger E., and Rilling H. C. (1991) Quantitation of prenylcysteines by a selective cleavage reaction. Proc. Natl. Acad. Sci. U.S.A. 88, 9668–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maurer-Stroh S., and Eisenhaber F. (2005) Refinement and prediction of protein prenylation motifs. Genome Biol. 6, eR55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. D'Alexandri F. L., Kimura E. A., Peres V. J., and Katzin A. M. (2006) Protein dolichylation in Plasmodium falciparum. FEBS Lett. 580, 6343–6348 [DOI] [PubMed] [Google Scholar]
- 30. Daher W., Morlon-Guyot J., Sheiner L., Lentini G., Berry L., Tawk L., Dubremetz J.-F., Wengelnik K., Striepen B., and Lebrun M. (2015) Lipid kinases are essential for apicoplast homeostasis in Toxoplasma gondii. Cell. Microbiol. 17, 559–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tawk L., Chicanne G., Dubremetz J.-F., Richard V., Payrastre B., Vial H. J., Roy C., and Wengelnik K. (2010) Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryotic Cell 9, 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simonsen A., Lippé R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., and Stenmark H. (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, 494–498 [DOI] [PubMed] [Google Scholar]
- 33. Ueda T., Yamaguchi M., Uchimiya H., and Nakano A. (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ha Y. R., Hwang B.-G., Hong Y., Yang H.-W., and Lee S. J. (2015) Effect of farnesyltransferase inhibitor R115777 on mitochondria of Plasmodium falciparum. Korean J. Parasitol. 53, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry data has been deposited at the Chorus Project (chorusproject.org) with the project identifier 1212.