Abstract
Hepatitis B virus (HBV) infection is a major health problem worldwide. Recent evidence suggests that some viruses can manipulate the infection process by packing specific viral and cellular components into exosomes, small nanometer-sized (30–150 nm) vesicles secreted from various cells. However, the impact of HBV replication on the content of exosomes produced by hepatocytes has not been fully delineated. In this work, an HBV-inducible cell line HepAD38 was used to directly compare changes in the protein content of exosomes secreted from HepAD38 cells with or without HBV replication. Exosomes were isolated from supernantants of HepAD38 cells cultured with or without doxycycline (dox) and their purity was confirmed by transmission electron microscopy (TEM) and Western immunoblotting assays. Ion-intensity based label-free LC-MS/MS quantitation technologies were applied to analyze protein content of exosomes from HBV replicating cells [referred as HepAD38 (dox−)-exo] and from HBV nonreplicating cells [referred as HepAD38 (dox+)-exo]. A total of 1412 exosomal protein groups were identified, among which the abundance of 35 proteins was significantly changed following HBV replication. Strikingly, 5 subunit proteins from the 26S proteasome complex, including PSMC1, PSMC2, PSMD1, PSMD7 and PSMD14 were consistently enhanced in HepAD38 (dox−)-exo. Bioinformatic analysis of differential exosomal proteins confirmed the significant enrichment of components involved in the proteasomal catabolic process. Proteasome activity assays further suggested that HepAD38 (dox−)-exo had enhanced proteolytic activity compared with HepAD38 (dox+)-exo. Furthermore, human peripheral monocytes incubated with HepAD38 (dox−)-exo induced a significantly lower level of IL-6 secretion compared with IL-6 levels from HepAD38 (dox+)-exo. Irreversible inhibition of proteasomal activity within exosomes restored higher production of IL-6 by monocytes, suggesting that transmission of proteasome subunit proteins by HepAD38 (dox−)-exo might modulate the production of pro-inflammatory molecules in the recipient monocytes. These results revealed the composition and potential function of exosomes produced during HBV replication, thus providing a new perspective on the role of exosomes in HBV-host interaction.
Hepatitis B virus (HBV)1 infection is a major health care problem worldwide. It has been estimated that about 30% of the world's population shows serological evidence of current or past HBV infection with 248 million individuals suffering from chronic infection worldwide (1, 2). HBV infection may result in acute or chronic hepatitis that can ultimately lead to the development of liver cirrhosis and hepatocellular carcinoma (HCC). HBV is a partially double-stranded DNA virus, which belongs to the hepadnavividae family. In humans, HBV exclusively infects hepatocytes and is not considered cytopathic. The control of viral infection and extent of liver damage depend on the complex interplay between virus replication and host immune response (1). One of the possible mechanisms by which HBV-infected hepatocytes interact with other uninfected cells and host immune system is through exosome-mediated cell-to-cell communication pathways (3).
Exosomes represent a discrete population of vesicles of nanometer-sized (30–150 nm) that are formed in endocytic compartments and secreted from various cell types to the extracellular millieu. These nanoscale membrane-enclosed vesicles carry a variety of bio-macromolecules such as proteins, mRNA, microRNA (miRNA) as well as other noncoding RNAs (4, 5), and act as the coordinator of cell-cell information exchange between different cell types in the liver microenvironment (5). As the exosome biogenesis pathway has a considerable overlap with the assembly and egress of numerous viruses, it is suggested that some viruses can utilize the exosomal pathway for cell-to-cell spread to avoid the immune system surveillance (6). It is therefore reasonable to assume that the exosome content may be modulated by pathological conditions such as HBV infection of hepatocytes. The profile of proteins, which are packaged into the exosomes, may yield a molecular signature that is informative about physiological status and disease conditions induced by HBV infection. Therefore, the main goal of this study was to get insights into how the HBV gene replication modulates the protein content of exosomes secreted from hepatocytes.
Until now, only a few papers have reported the role of exosomes in liver microenvironment in response to HBV infection. Our previous work revealed that exosomes from IFN-α stimulated liver nonparenchymal cells (LNPCs) were rich in molecules with antiviral activity and could transfer the IFN-α- induced antiviral molecules from LNPCs, such as macrophage and liver sinusoidal endothelial cells (LSECs), to hepatocytes (7). Likewise, HBV-infected hepatocytes can secrete exosomes delivering functional bio-molecules and as such influencing the physiological activities of surrounding cells in the liver microenvironment. Recently, Yinli Yang's study demonstrated that exosomes circulating in the sera of chronic hepatitis B (CHB) patients contain both HBV nucleic acids and HBV proteins. These exosomes could be taken up by NK cells were shown to play a role in NK-cell dysfunction and in HBV transmission (8). Takahisa Kouwak 's group found that HBV infection of hepatocytes with HBV increased immunoregulatory microRNA levels in EVs and exosomes and these exosomes could regulate innate immune responses to HBV infection (9). HBV-mediated changes in exosomes' protein content were analyzed via comparative proteomic technology using stable isotope labeling by amino acids in cell culture (SILAC) in Huh-7 cells transfected with HBV plasmids (10). However, the low efficiency of the transfection of Huh-7 cells led to marginal differences, and only a limited number of exosomal proteins were identified and quantified. Therefore, further in-depth studies were needed to fully elucidate the roles of HBV replication on hepatocyte-secreted exosomes. In contrast to Huh-7 HBV-transient transfection system, HepAD38 cells permit high level of HBV DNA replication in an inducible manner strictly controlled by a tetracycline-responsive promoter. HepAD38 cell system thus allows a direct comparison of exosomes' content secreted by HepAD38 with or without HBV gene replication (11).
Here, employing unbiased label-free LC-MS/MS-based quantification analyses of exosomal proteins, we demonstrated that exosomes secreted from HepAD38 cell line, which supports HBV gene replication, are packed with proteins specifically associated with HBV that can modulate production of IL-6 by monocytes. Collectively, these findings shed new lights on interactions between HBV and the host and provide the foundation for future research into roles played by exosomes in HBV infection and pathogenesis.
EXPERIMENTAL PROCEDURES
Chemicals and Antibodies
Antibodies against Cox IV (4844) and EEA1 (3288S) were obtained from CST (Beverly, MA); anti-TSG101 rabbit polyAb (14497-1-AP) was purchased from Proteintech (Rosemont, IL); antibodies against LAMP-2 (sc-18822), CD63 (SC-15363), ALB (sc-51515), PSMC1 (sc-243890), PSMC2 (sc-166972), PSMD1 (sc-166038), PSMD7 (sc-390705, XRCC5 (sc-9034) were obtained from Santa Cruz (Dallas, TX); anti-Histone H4 (07–108) antibody was obtained from Millipore (Darmstadt, Germany); GM130 (ab52649) were purchased from Abcam (Cambridge, MA); abclonal HSPB1 (Grp94) (A6272) were purchased from ABclonal Technology (Wuhan, China). Doxycycline (dox), G418 and antibody against GAPDH (G8795) were purchased from Sigma (Darmstadt, Germany). PR-171 was purchased from Medchem Express (Monmouth Junction, NJ), human IL-6 ELISA Ready-SET-Go Kit was purchased from BD (Rosemont, IL), human CD14 Microbeads were purchased from MACS (Bergisch Gladbach, Germany).
Cell Culture
HepAD38 is a stably transfected hepatoblastoma cell line that replicates human HBV under the control of a tetracycline-regulated promoter (11). HepAD38 cells were maintained in Dulbecco's modified Eagle's/F12 medium (DMEM/F-12; GIBCO BRL/Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin G/streptomycin sulfate, and 200 μg/ml G418 at 37 °C and 5% CO2. HepAD38 cells in the doxycycline (dox) group were cultured in the presence of 1 μg/ml dox.
Production and Isolation of Exosomes
Exosome-depleted serum was prepared by ultracentrifugation of normal FBS at 120,000 × g for 18 h, followed by filtering the supernatant through 0.22 μm syringe filter (Millipore, Darmstadt, Germany). HepAD38 cells were cultured in exosomes free FBS for 48 h, and the collected cell culture supernatants were pooled and used for exosomes extraction. The collected supernatants of HepAD38 cultured with or without dox were processed in parallel in subsequent steps. Six pairs of independently prepared exosomes were analyzed as biological replicates.
Exosomes were isolated using a differential centrifugation protocol, as previously described, with minor modifications (7). Briefly, culture medium was collected and centrifuged at 300 × g for 10 min and 2000 × g for 20 min to eliminate cells and cell debris contamination. The cleared supernatants were further centrifuged at 10,000 × g for 30 min at 4 °C. Exosomes were pelleted from the supernatants by ultracentrifugation at 120,000 × g using an SW32 Ti swinging bucket rotor (Beckman Coulter, Fullerton, CA) for 100 min at 4 °C. The exosome pellets were resuspended in PBS, pooled, and ultracentrifuged again at 120,000 × g using a Type 100Ti rotor (Beckman Coulter) for 90 min at 4 °C. The final pellet of exosomes (referred as crude exosomes) was resuspended in PBS to 1/1000th of the original volume of the culture supernatant, aliquoted and immediately stored at −80 °C.
Crude exosomes were further purified by immunoprecipitation (IP) to remove contamination by HBsAg particles. 200 μl crude exosomes isolated by differential centrifugation were suspended in PBS and transferred into a 1.5 ml tube. 15 μg of anti-HBsAg antibody was added into the sample, and incubated at 4 °C for 2 h on a rotator. Then, 30 μl of Protein A or G Agarose (prewashed in the PBS for 3 times) were added and incubated overnight at 4 °C on a rotator followed by centrifugation at 1000 × g for 3 min at 4 °C to pellet the beads. The supernatant (referred as purified exosomes) were carefully drawn to a new 1.5 ml tube. A portion of purified exosomes were used for electron microscopy analysis and monocyte incubation experiment. The rest were vacuum dried and lysed in 50 μl lysis buffer (30 mm Tris, pH 8.5, 7 m Urea, 2 m Thiourea, 0.1% SDS, and proteases inhibitors) and the protein concentration was measured using the Bradford assay (Bio-Rad, Hercules, CA). The extracted proteins were stored at −80 °C and used for proteomic and Western immunoblotting analyses. Purified exosomes extracted from HBV replicating HepAD38 cells cultured without dox were referred as HepAD38 (dox−)-exo. Purified exosomes extracted from HepAD38 cultured in the presence of dox were referred as HepAD38 (dox+)-exo.
Electron Microscopy
For negative staining microscopy, 10 μl of purified exosomes were layered and absorbed on a formvar coated 300 mesh copper grid and stained with 2% phosphotungstic acid (PTA). Sample was imaged using a Philips CM120 transmission electron microscope (Eindhoven, The Netherlands) equipped with a tungsten filament and operated at an acceleration of 60 kV. Images were taken with a pixel size of 0.195 nm, and a direct magnification of 46,000× using a Gatan CCD camera.
Western Blotting
Exosomes or cell lysate proteins (5 μg) were mixed with 2 × SDS sample buffer (4% SDS (m/v),100 mm DTT, 20% glycerol (v/v), 100 mm Tris-HCL pH 6.8), separated on 7.5%, 10% or 15% SDS-acrylamide gels, and transferred onto PVDF membrane. The membranes were blocked with 5% nonfat milk and incubated overnight at 4 °C with either anti-Cox IV (1/500 dilution), anti-EEA1 (1/300 dilution), anti-TSG101 (1/500 dilution), anti-LAMP2 1/300 dilution), anti-CD63 (1/500 dilution), anti-ALB (1/100 dilution), anti-Grp94 (1/400 dilution), anti-GM130 (1/1000 dilution), anti-Histone H4 (1/100 dilution), anti-PSMC1 (1/100 dilution), anti-PSMC2 (1/100 dilution), anti-PSMD1 (1/100 dilution), anti-PSMD7 (1/100 dilution), anti-XRCC5 (1/100 dilution) or anti-GAPDH (1/5000 dilution) antibodies, followed by incubation with horseradish-peroxidase-conjugated secondary antibody for 1 h at room temperature. Protein bands were detected using ImmobilonTM Western Chemiluminescent HRP Sustrate (Millipore, Bedford, MA).
Analyses of HBV Antigens and DNA
HBsAg and HBeAg level in the culture media of HepAD38 cells were quantified using the commercial HBsAg and HBeAg ELISA Kit (Kehua, Shanghai, China). Extraction of HBV DNA from HepAD38 culture media was performed using a HBV DNA Quantification Kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer.
MS Sample Preparation and Tryptic Digestion
A protein amount of 5 μg per sample was loaded on an 18% Tris-glycine polyacrylamide gel (Anamed Elektrophorese, Groβ-Bieberau, Germany) and allowed to run slightly into the gels (15 min, 100 V). The protein bands were stained with Coomassie dye and cut from the gels. Trypsin digestion of proteins was performed overnight at 37 °C (Serva Electrophoresis, Heidelberg, Germany) and the peptides were extracted from digested bands with 20 μl of 50% ACN in 0.1% TFA, vacuum dried and subsequently dissolved in 0.1% TFA. The peptide concentration was determined by amino acid analysis as previously described (12).
LC-MS/MS
The LC-MS/MS analysis was performed using an Ultimate 3000 RSLCnano system coupled online to a QExactive mass spectrometer (both from Thermo Scientific, Bremen, Germany). Three hundred nanograms of peptide sample were injected and preconcentrated on a trap column (Acclaim PepMap 100, 300 μm×5 mm, C18, 5 μm, 100 Å; flow rate 30 μl/min). Subsequently, the peptides were separated on the analytical column (Acclaim PepMap RSLC, 75 μm×50 cm, nano Viper, C18, 2 μm, 100 Å) by a gradient from 5% to 40% solvent B over 120 min (solvent A: 0.1% FA, solvent B: 0.1% FA, 84% ACN; flow rate 400 nL/min; column oven temperature 60 °C). The MS instrument was operated in a data-dependent mode. The 10 most abundant precursor ions detected in the full MS survey scan (m/z range of 350–1400, r = 70,000) were isolated with a 2 m/z mass window for further high-energy collisional dissociation (HCD) MS/MS analysis with a resolution of 35,000. Spectra were acquired under automatic gain control (AGC) for survey spectra (AGC: 3 × 106) and MS/MS spectra (AGC: 2 × 105). In all cases, one microscan was recorded using dynamic exclusion of 30 s.
Mass Spectrometric Data Analysis
Ion intensity based label-free quantification technique allows the determination of precursor ion abundances at MS level across samples. The peptide identification was performed using Proteome Discoverer Software (ver. 1.4.0.288, Thermo Fisher Scientific, Rockford, IL) (13). The mass spectra were searched against UniProtKB/Swiss-Prot database (2014_10; 546,790 entries) restricted to Homo sapiens (20,194 sequences) using Mascot (ver. 2.3.0.2, Matrix Science Ltd., London, UK). HBV genome type D protein sequences were downloaded from UniProtKB database (2014_10, 135 sequences) and a search against the generated fasta file was conducted using Sequest HT search algorithm integrated in Proteome Discoverer software (ver. 1.4.0.288). Respective identifications of HBV proteins were later used in relevant samples. For each of the database searches, the mass tolerance was set to 5 ppm for precursor ions and 0.02 Da for fragment ions. One tryptic miscleavage was considered as well as variable modification of methionine (oxidation) and cysteine (propionamide). The Target Decoy PSM Validator function implemented in Proteome Discoverer was used to estimate peptide confidence and only peptides that passed a false discovery rate < 1% were considered for analysis. Protein grouping function (strict maximum parsimony principle) was enabled in Proteome Discoverer. Protein groups identified with a minimum of two peptides (at least one unique peptide) were considered for further analysis (14). The peptide quantification was performed using Progenesis QI software (ver. 2.0, Nonlinear Dynamics Ltd., Newcastle upon Tyne, U.K.). Equal amount of peptides (25 ng) from the 12 samples (6 replicates×2 groups) were mixed before LC-MS/MS analysis and the resulted data were used as reference in Progenesis analysis. All runs were aligned to the reference run and a master list of features considering m/z values and retention times was generated. In Progenesis QI, peptide ions were filtered using the criteria as below: delete matching peptide ions with 2 isotopes or less, retain peptide ions with a charge state of 2 to 7. Then the peptide identifications exported from Proteome Discoverer were imported into Progenesis QI software where they were matched to the respective features. Those matching peptides with mass error (ppm) greater than 5 ppm were deleted. The remaining matching peptides were used for proteins quantification analysis.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data identifier PXD004724 (15) and the following submission details. Conversion of msf files (result files of Proteome Discoverer) into the mzIdentML standard format was conducted using ProCon - PROteomics CONversion tool (ver. 0.9.641) (16).
Protect Name: Label-free Proteomic Analysis of Exosomes Secreted by HBV-inducible HepAD38 Cell Line.
Project accession: PXD004724.
Reviewer account details:
Username: reviewer94189@ebi.ac.uk.
Password: Hk7M7Wls.
Experimental Design and Statistical Rationale
Six pairs of independently prepared HepAD38 (dox−/dox+) exosomes were analyzed as biological replicates to increase the reliability and achieve adequate predictive power for global label-free quantitative proteomics profiling. The peptide quantification was performed using Progenesis QI software. Relative ratio quantification was performed using quantities of unique peptides and required a minimum of two unique peptides. Proteins quantified with p value ≤ 0.05 and a fold change of ≥ 1.50 or ≤0.66 were considered to be significantly differential and used for further evaluation. The statistical analysis of label-free discovery study was analyzed by means of unpaired ANOVA implemented in the Progenesis QI software. For functional analyses of differential proteins, the statistical analysis was performed using two-tailed student's t test in Prism 5 (Graphpad) software.
Gene Ontology Analysis and Networks, Functional and Pathway Mapping
The functional annotations of the identified proteins were initially assigned using The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (https://david.ncifcrf.gov/home.jsp) (17). Three main types of annotations were obtained from the Gene Ontology (GO) Consortium website, including cellular components, molecular functions, and biological distribution. The KEGG pathway enrichment analysis was conducted by DAVID. The protein-protein interaction network was generated by STRING 10 (http://string-db.org/) (18).
Proteasome Activity Assays
The proteasome activity was measured based on detection of the fluorophore 7-Amino-4 Metholcoumarin (AMC) cleaved from the labeled substrate Leu-Leu-Val-Tyr-AMC (Enzo Life Sciences, Farmingdale, NY) by 20S proteasome as described previously (19). HepAD38 cells cultured with or without dox were washed 3 times with PBS and lysed with assay lysis buffer (50 mm Tris-HCl, pH 7.5; 150 mm NaCl; 1% Triton X-100; 2 mm ATP). Cell lysates were incubated on ice for 30 min, mixed by vortex every 5 min, and centrifuged at 12,000 × g for 10 min. The supernatants were collected and the protein concentrations were determined with BCA reagents (Thermo Scientific, Rockford, IL). 10 μg cell proteins were diluted in assay buffer (50 mm Tris-HCl, pH 7.5; 150 mm NaCl) to a total volume of 90 μl. For detection of proteasome activity of HepAD38 secreted exosomes, 2 μg exosomes suspended in PBS were lysed with equal volume of assay lysis buffer and diluted to 90 μl with proteasome assay buffer. Then 10 μl of the synthetic fluorogenic substrates Suc-Leu-Leu-Val-Tyr-AMC (500 μm, Enzo Life Sciences) was added to each assay reaction (final concentration of 50 μm in a total volume of 100 μl). The samples were incubated at 37 °C and the fluorescence was read in a PerkinElmer 1420 Multilabel Counter (Fremont, CA) plate reader at an excitation wavelength of 355 nm and an emission wavelength of 460 nm.
Coculture of HepAD38 or HepG2 Secreted Exosomes with Monocytes
Monocytes isolated from peripheral blood mononuclear cells (PBMCs) by positive immunomagnetic selection using MACS human CD14 Microbeads (Bergisch Gladbach, Germany), as described previously (20), were cultured in RPMI 1640 (HyClone, Utah) with 10% FBS and 1% penicillin/streptomycin (both from Life Technologies). PBMCs from anonymous healthy donors were provided by the Red Cross Blood Center of Shanghai. HepG2 were cultured in DMEM (HyClone, Utah, USA) with 10% FBS and 1% penicillin/streptomycin (both from Life Technologies). HepG2 derived exosomes were prepared using the same protocol as described for HepAD38 cells.
To monitor exosomes transfer to primary monocytes, purified HepAD38 (dox+/dox−) or HepG2 exosomes were labeled with a red fluorescent lipid dye (PKH26) that could be visible by confocal microscopy. Labeled exosomes were washed in 5 ml of PBS, collected by ultracentrifugation, and resuspended in PBS. Primary monocytes were incubated for 2 h at 37 °C with 10 μg/ml labeled exosomes. After cells were fixed and stained with DAPI, they were visualized with a confocal fluorescence microscope.
To evaluate the effect of exosomal proteasome proteins on cytokine secretion by monocytes the proteasome inhibitor PR-171 was employed. PR-171 powder was dissolved at 10 mm in DMSO and the aliquots of it diluted in PBS were used for experiments. For experiments, 25 μg of exosomes preparation was incubated with either 50 nm PR-171 or DMSO at 37 °C for 1 h following which, excess of PR-171 was washed with PBS and removed by ultra-filtration. In parallel, HepG2 derived exosomes were used as a control. PR-171-treated or DMSO-treated exosomes (10 μg/ml) from HepAD38 (dox+), HepAD38 (dox−) or HepG2 were then cocultured with monocytes for 48 h. Cell culture supernatants were analyzed for secretion of human IL-6 by ELISA Ready-SET-Go Kit (BD) according to the manufacturer's instructions.
RESULTS
General Experiment Design
To investigate differences in the protein content of exosomes produced by hepatocytes that replicate HBV, an ion intensity based label-free quantitative proteomic strategy was designed, as summarized in supplemental Fig. S1. We employed the HBV-inducible HepAD38 cell line, which allows a direct comparison of changes in cellular characteristics induced by HBV replication (11). The HBV pgRNA, mandatory for HBV replication in HepAD38, is under the control of a tetracycline-regulated promoter that is suppressed by doxycycline (dox). Upon the removal of dox from the culture medium, the cells initiate the expression of viral pgRNA, followed by the production of HBeAg and the secretion of virus-like particles into the supernatant. Exosomes were prepared from HepAD38 culture supernatants by ultracentrifugation followed by immunoprecipitation to remove contamination by HBsAg containing particles. Ion-intensity based label-free quantification strategy was used to analyze six pairs of independently prepared exosomes to get unbiased quantification results.
Characterization of HBV Replication in HepAD38 Cells
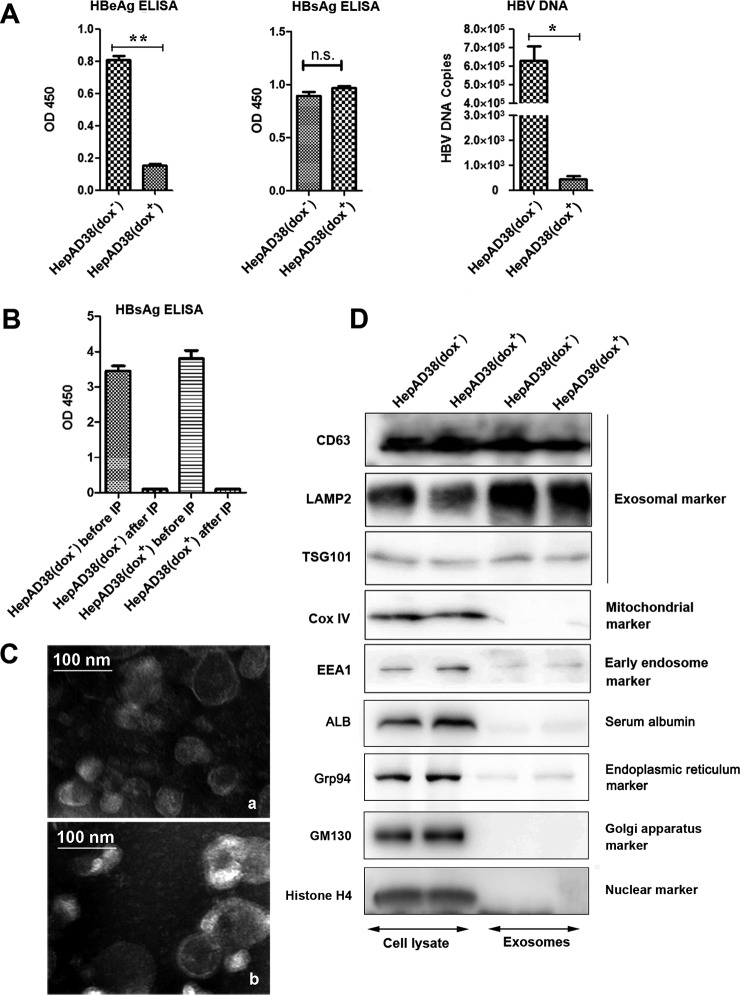
Prior to exosomes extraction, expression of HBV proteins and HBV replication in HepAD38 cultured in the presence or absence of dox were monitored by quantitative immunoassays of HBsAg and HBeAg proteins and by RT-PCR of HBV DNA in culture media. The results showed that secretion of HBeAg and HBV DNA to the supernatant of HepAD38 cells cultured without dox was significantly higher than in dox (+) controls (Fig. 1A). Treatment HepAD38 cells with dox resulted in very low expression levels of both HBeAg and HBV DNA, thus confirming the efficiency of dox in controlling the expression pgRNA and hence the replication of HBV in HepAD38. In contrast, the HBsAg level was not affected by dox treatment and remained very high because of the fact that the transcription of HBsAg was not controlled by the tetracycline-regulated promotor (21) (Fig. 1A). High level of HBsAg protein secreted from HepAD38 in the form of either HBsAg particles or viral particles, which have similar size as exosomes could potentially contaminate the exosomes purification during the ultracentrifugation process. Therefore, a special care was taken to get rid of contamination by HBsAg containing particles secreted from HepAD38 cells.
Fig. 1.
Characterization of exosomes secreted by HepAD38 cells. A, Monitoring HBV replication in HepAD38. HBV expression and replication in HepAD38 with or without dox were examined by quantitative immunoassay of HBeAg and HBsAg and RT-PCR of HBV DNA in culture media. B, Removal of secreted HBV contaminating particles during exosome preparations. Crude exosomes from ultracentrifugation were further purified by immunoprecipitation to remove contamination of exosome preparations by HBsAg particles. C, Electron microscopy of purified exosomes secreted by HepAD38 grown in the absence (a) or presence (b) of dox inhibitor. Scale bar, 100 nm. D, Validation of exosome purification by Western immunoblotting, as described in the Experimental Procedures section, using anti-exosome specific antibodies and antibodies to commonly found contaminations in exosomal preparations. Note: (* p < 0.05, ** p < 0.01).
Exosomes Enrichment and Characterization
Crude exosomes were prepared from culture supernatants of HepAD38 cells following a differential centrifugation. To remove contaminating HBsAg particles, anti-HBsAg antibody was used to immunoprecipitate (IP) HBsAg. IP significantly decreased the inclusion of HBsAg containing particles in purified exosome preparation in both dox+ and dox− groups (Fig. 1B). To further ensure the purity of exosomal preparation, an aliquot of the purified exosomes from HepAD38 (dox−/dox+) cells was also analyzed by electron microscopy, which demonstrated a presence of small membrane vesicles of about 30–150 nm in diameter with the cup-shaped structure typical of exosomes (Fig. 1C, a and b, respectively). Successful purification of exosomes lacking possible contamination by cellular membranous structures was also confirmed by Western immunoblotting using antibodies to specific protein markers, including CD63 antigen (CD63), lysosome-associated membrane protein 2 (LAMP2), tumor susceptibility gene 101 (TSG101), cytochrome C oxidase IV (Cox IV) and early endosome antigen 1 (EEA1) (Fig. 1D). As expected, the recognized exosomal marker LAMP2 (3) was highly enriched in the exosome preparations compared with cell lysates from the corresponding parental cells (Fig. 1D, second panel from the top, last two lanes). CD63 and TSG101, two commonly used protein markers for exosomes (3, 22) were also present in exosome samples from HepAD38 cell lines (Fig. 1D). To assess the presence of contaminating cellular debris, blots were stained for the early endosome antigen EEA1, the mitochondrial marker Cox IV, serum albumin (ALB), the endoplasmic reticulum marker (Grp94), the Golgi apparatus marker (GM130) and the nuclear marker (histones H4). The results shown that these contamination markers were present in the cell lysates, whereas their presence in the exosome samples were very low. Moreover, it should be noted that the expression of none of these tested proteins was affected by the dox treatment, which as expected only regulates expression of HBV pgRNA in HepAD38 cells. Together, these findings indicate that the preparation of exosomes secreted from HepAD38 cells was characterized by expression of common exosomal marker proteins and showed minimal contamination by cellular debris and by HBV-derived viral and subviral particles.
Global Profiling of HepAD38 Exosome Proteins
Global profiling of protein content in exosomes secreted by rat hepatocytes and human HCC cell line has been reported (10, 23, 24). These studies led to only limited identification of exosomal protein content. We therefore wished to extend these earlier studies by employing highly sensitive LC-MS/MS approaches. Six pairs of independently prepared HepAD38 (dox−/dox+) exosomes were analyzed by a QExactive mass spectrometer. The 12 MS raw data were processed by Proteome Discoverer and Progenesis QI software for global identification and quantification of HepAD38 derived exosomes, respectively. In total, 1412 protein groups were identified in HepAD38 (dox−/dox+) exosomes (Fig. 2A). The identified proteins and peptide information are listed in Supplementary data supplemental Table S1. Compared with earlier reports, overall our analyses led to identification of a much larger number of proteins in exosomes secreted by hepatocytes (10, 23, 24).
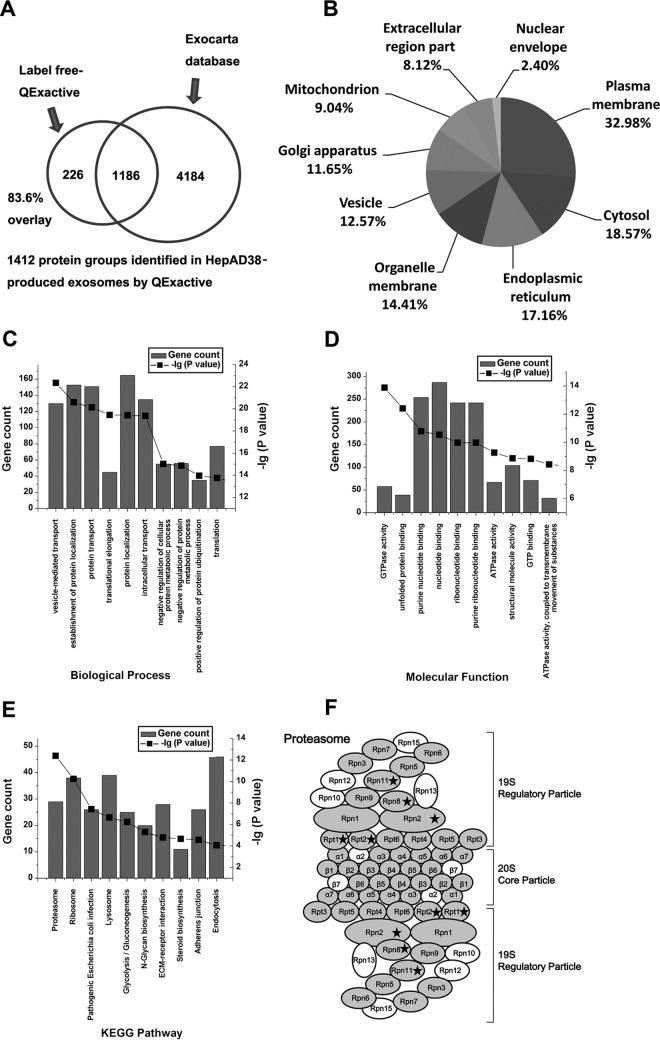
Fig. 2.
Global profilling of exosomal proteins secreted by HepAD38 using Mass spectrometry.
A, Venn diagrams of proteins identified in HepAD38 exosomes by mass spectrometry. The total number of proteins identified was compared with results from the Exocarta database of published exosomal proteins. B, Intracellular protein locations assigned by Gene Ontology annotations. C, Top 10 significantly enriched GO biological processes in exosomes secreted from HepAD38 (dox−/dox+) cells; D, Top 10 significantly enriched GO molecular function in exosomes secreted from HepAD38 (dox−/dox+) cells; E, Top 10 significantly enriched KEGG pathways in exosomes secreted from HepAD38 (dox−/dox+) cells; F, structure of proteasome complex showing the subunit proteins labeled by gray were identified in this study. Proteins differentially packed into exosomes from HBV replicating are depicted by black ☆. Please note that the proteasome complex has symmetrical structure. The proteins were labeled according to the nomenclature adopted by the KEGG map (supplemental Table S3). The graphs  show the total number of genes and
show the total number of genes and  show the -lg (p value) that fall into the designated Ingenuity classification.
show the -lg (p value) that fall into the designated Ingenuity classification.
Of 1412 protein groups detected in all preparations of HepAD38 exosomes, 1186 were present in the Exocarta database of exosome proteome (Fig. 2A), including 29 of 30 the most frequently identified proteins (25, 26). A list of commonly described exosomal proteins identified in exosomes secreted by HepAD38 is presented in supplemental Table S2. These data confirm that the exosomes from our preparations contain abundant levels of exosomal proteins. The mass spectrometry analysis also identified 226 new exosome components not present in Exocarta as indicated by Venn diagram (Fig. 2A). Moreover, MS analysis of purified exosomes confirmed the presence of common exosomal markers, (23) such as tumor susceptibility gene 101, CD63, and CD9 as well as a specific marker for exosomes of hepatic origin ASGR receptor (23) (supplemental Table S2).
Exosomal proteins identified by MS were subsequently classified by bioinformatics analysis. Gene Ontology annotation was applied to classify identified proteins in terms of their subcellular localizations with each protein assigned at least one term. In total, 1412 protein groups were used in this analysis. The result shows that exosomes secreted by HepAD38 contain proteins residing in the major intracellular organelles. 32.98% proteins were annotated as belonging to plasma membrane-associated proteins, whereas the other two main categories were from cytosol (18.57%) and endoplasmic reticulum (17.16%) (Fig. 2B). 12.57% identified exosomal proteins were vesicle-associated proteins. To gain an insight into the functional roles of the proteins from hepatocyte-derived exosomes, we employed GO-based categories clustering on molecular function and biological processes. A significant enrichment in proteins involved in vesicle-mediated transport, establishment of protein location, protein transport, and translational elongation processes were identified (Fig. 2C) supporting the notion that exosomes play a central role in the transportation of cellular materials among cells. We also observed enrichment in molecules with GTPase activity, unfolded protein binding, purine nucleotide binding, ribonucleotide binding, and ATPase activity, suggesting that exosomes influence the extracellular protein homeostasis (Fig. 2D). The KEGG pathway enrichment analysis revealed high enrichment of the proteasome, ribosome pathways (Fig. 2E). Several proteasome subunits have been identified in HepAD38 exosomes. A list of 26S proteasome subunits identified in this study is shown in supplemental Table S3 and the identified proteins were depicted in by the gray color within the structure of proteasome complex in Fig. 2F. A total number of 27 subunit proteins from the 26S proteasome complex (33 subunit proteins in total) were identified raising the possibility that entire proteasome is carried by these exosomes.
Liver is considered as an immune organ (27), and exosomes secreted by hepatocytes have been proposed to play an important role in immune modulation (28). In this study, among the proteins identified, we found several immune response-related proteins included in exosomes. HepAD38-exosomal proteins involved in immune effector process were also analyzed by GO enrichment and shown in supplemental Table S4. These immune effector process-related proteins included several proteins that participate in the regulation of cytokine production or cytokine-mediated signaling pathway, such as Hsp60 (29–31), cell adhesion molecule 1 (32), clusterin (33), tyrosine-protein kinase Lyn (34) and receptor-type tyrosine-protein phosphatase C (35). The identified immune regulation-related proteins carried by HepAD38 exosomes may contribute to the modulation of immune response during HBV infection. Hence, hepatocyte-derived exosome population provides useful information to enhance our understanding of hepatic environment function.
To examine if viral proteins were also packed into exosomes secreted from HepAD38 cells sustaining HBV replication, MS data were searched against the HBV database. The results showed that several proteins encoded by the HBV genome were identified in exosomes (supplemental Table S1). These included large S, core and P proteins, suggesting that HBV-encoded proteins may be specifically packed into exosomes secreted by hepatocytes sustaining HBV replication. The label-free quantification results also showed that the core protein was significantly, by 69.9-fold, enriched in exosomes from HBV-replicating HepAD38 cells compared with exosomes from non-HBV-replicating HepAD38. Western blot analysis of exosomes confirmed specific enrichment of the core protein in exosomes secreted from HBV-replicating HepAD38 (supplemental Fig. S2), suggesting that HBV replication in hepatocytes enhances the core protein loading into exosomes. However, further research is required to conclusively demonstrate the involvement of virus -encoded proteins in exosomes.
HBV Replication Affects HepAD38 Exosomal Protein Contents
To get insight into the impact of HBV replication on inducing the changes in the content of exosomal proteins and to identify and quantify those proteins we employed an ion-intensity based label-free LC-MS/MS analyses using HepAD38 cell line in which HBV replication is tightly regulated. Six pairs of independent preparations of exosomes were analyzed. Among the 1412 protein groups identified (Fig. 2A), proteins quantified by Progenesis QI software with at least two unique peptides that were significantly increasing or decreasing in their abundance with a fold-change ≥ 1.50 or ≤0.66 were regarded as differentially abundant in exosomes secreted from HBV+ and HBV− HepAD38 cells. We identified 35 proteins that adhered to these critieria (Table I). Twenty-nine of these proteins showed higher presence in exosomes from HepAD38 cells permitting HBV replication, whereas 6 showed lower expression in this group. 85.7% (30 of 35) of the differentially included exosomal proteins have been identified in the ExoCarta database. For example, proteasome complex proteins protease regulatory subunits 4 and 7 and non-ATPase regulatory subunits 1, 7, and 14 of the 26S proteasome complex, were found to be 1.55- to 2.61-fold enriched in the exosomes secreted from HepAD38 (dox−)-exo (Table I). The results also showed that proteins related to HBV-induced pathogenic conditions (9, 36–39) were differentially included in the exosomes from HepAD38 (dox−/dox+) cells. For example, the levels of alpha-2-macroglobulin, versican core protein and X-ray repair cross-complementing protein 5, were enhanced, whereas cell division control protein 42 homolog and pigment epithelium-derived factor were reduced in the HepAD38 (dox−)-exo (Table I). In addition, our results demonstrated that the levels of cellular proteins belonging to cellular proteases, transporters, ion channels, cytosolic and nuclear proteins as well as proteins from the cell membrane were generally enhanced in the HepAD38 (dox−)-exo (Table I).
Table I. Proteins found to be significantly altered between HepAD38 exosomes with or without HBV replication in the label-free discovery study.
| UniProt accession | Gene symbol | Protein name | Fold change (HepAD38 (dox−)-exo/HepAD38 (dox+)-exo) | p-Value | Exocarta |
|---|---|---|---|---|---|
| 26S proteasome complex | |||||
| P62191 | PSMC1 | 26S protease regulatory subunit 4 | 1.63 | 0.011 | Yes |
| P35998 | PSMC2 | 26S protease regulatory subunit 7 | 1.56 | 0.004 | Yes |
| Q99460 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 1.55 | 0.013 | Yes |
| O00487 | PSMD14 | 26S proteasome non-ATPase regulatory subunit 14 | 1.72 | 0.006 | Yes |
| P51665 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7 | 2.61 | 0.037 | Yes |
| HBV induced pathogenetic condition | |||||
| P01023 | A2M | Alpha-2-macroglobulin | 1.55 | 0.031 | Yes |
| P60953 | CDC42 | Cell division control protein 42 homolog | 0.53 | 0.022 | Yes |
| P36955 | SERPINF1 | Pigment epithelium-derived factor | 0.64 | 0.043 | Yes |
| P13611 | VCAN | Versican core protein | 3.46 | 0.038 | Yes |
| P13010 | XRCC5 | X-ray repair cross-complementing protein 5 | 1.50 | 0.039 | Yes |
| Proteases | |||||
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | 1.87 | 0.049 | Yes |
| P41252 | IARS | Isoleucine–tRNA ligase, cytoplasmic | 1.50 | 0.043 | Yes |
| Q3LXA3 | DAK | Bifunctional ATP-dependent dihydroxyacetone kinase/FAD-AMP lyase (cyclizing) | 1.52 | 0.029 | No |
| P49591 | SARS | Serine–tRNA ligase, cytoplasmic | 1.57 | 0.013 | Yes |
| P47897 | QARS | Glutamine–tRNA ligase | 1.58 | 0.003 | Yes |
| O00754 | MAN2B1 | Lysosomal alpha-mannosidase | 0.34 | 0.003 | Yes |
| Q32P28 | LEPRE1 | Prolyl 3-hydroxylase 1 | 0.58 | 0.043 | No |
| Q9Y265 | RUVBL1 | RuvB-like 1 | 1.56 | 0.008 | Yes |
| Transporters & channels | |||||
| Q9UGQ3 | SLC2A6 | Solute carrier family 2, facilitated glucose transporter member 6 | 1.68 | 0.050 | No |
| P30825 | SLC7A1 | High affinity cationic amino acid transporter 1 | 1.54 | 0.023 | Yes |
| P53618 | COPB1 | Coatomer subunit beta | 1.65 | 0.002 | Yes |
| Q9Y678 | COPG1 | Coatomer subunit gamma-1 | 1.88 | 0.035 | Yes |
| Q96RQ1 | ERGIC2 | Endoplasmic reticulum-Golgi intermediate compartment protein 2 | 1.68 | 0.038 | Yes |
| O15431 | SLC31A1 | High affinity copper uptake protein 1 | 1.81 | 0.028 | No |
| O14980 | XPO1 | Exportin-1 | 1.59 | 0.004 | Yes |
| Cytosolic proteins | |||||
| O00400 | SLC33A1 | Acetyl-coenzyme A transporter 1 | 1.58 | 0.031 | No |
| P48723 | HSPA13 | Heat shock 70 kDa protein 13 | 1.99 | 0.000 | Yes |
| Q07021 | C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial | 0.53 | 0.024 | Yes |
| P48643 | CCT5 | T-complex protein 1 subunit epsilon | 1.78 | 0.049 | Yes |
| Nuclear proteins | |||||
| O00567 | NOP56 | Nucleolar protein 56 | 2.01 | 0.034 | Yes |
| O76021 | RSL1D1 | Ribosomal L1 domain-containing protein 1 | 1.93 | 0.046 | Yes |
| P23246 | SFPQ | Splicing factor, proline- and glutamine-rich | 1.58 | 0.031 | Yes |
| Cell membrane | |||||
| Q9ULI3 | HEG1 | Protein HEG homolog 1 | 1.82 | 0.047 | Yes |
| Q96B21 | TMEM45B | Transmembrane protein 45B | 1.52 | 0.044 | Yes |
| Cytoskeleton | |||||
| P08779 | KRT16 | Keratin, type I cytoskeletal 16 | 0.15 | 0.046 | Yes |
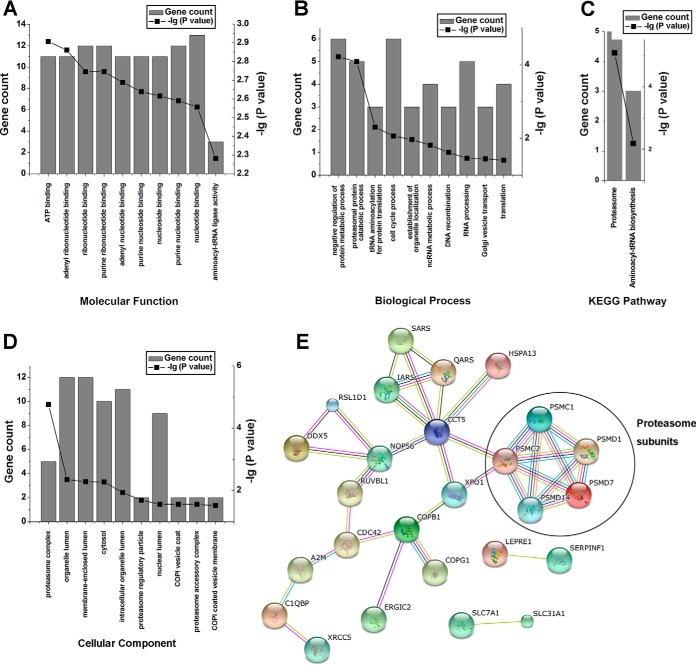
To gain insight into the potential role the exosomal proteins may play in modulation of HBV infection, GO enrichment, and KEGG Pathways analysis were performed on these 35 proteins that were differentially included into exosomes secreted from HepAD38 cells grown in the presence or in the absence of dox. The ATP- and adenyl ribonucleotide- binding were the top two Molecular Function categories that were predicted to be affected by HBV replication in HepAD38 cells (Fig. 3A). Likewise, proteins affecting the negative regulation of metabolic and proteasomal catabolic processes were predicted by the GO_BP analysis to be affected by HBV replication in HepAD38 cells (Fig. 3B). The KEGG pathway and GO_CC analyses identified significant enrichment for genes encoding proteins predicted to be involved in proteasome (Fig. 3C and 3D). A protein-protein interaction network generated by the online resource STRING, using the differentially included exosomal proteins as seeds, also showed that many proteasome subunits form tight connections with each other and with other differentially included exosomal proteins (Fig. 3E). These results indicated that HBV replication in HepAD38 significantly influenced the abundance of the proteasome complex proteins in secreted exosomes, which is in agreement with our results demonstrating a significant enhancement of the 26S proteasome complex proteins in HepAD38 (dox−)-exo.
Fig. 3.
Bioinformatic analyses of proteins differentially included in exosomes from HepAD38 (dox−/dox+) cells.
A, B, C, and D, GO enrichment and KEGG Pathways analysis of modified exosomal proteins. A, GO molecular function; B, GO biological processes; C, KEGG pathways, and D, GO cellular components. E, Protein-protein interaction networks of differential proteins. The graphs  show the total number of genes and
show the total number of genes and  show the -lg (p value) that fall into the designated Ingenuity classification.
show the -lg (p value) that fall into the designated Ingenuity classification.
Validation of Differential Proteins Present in HBV-derived Exosomes
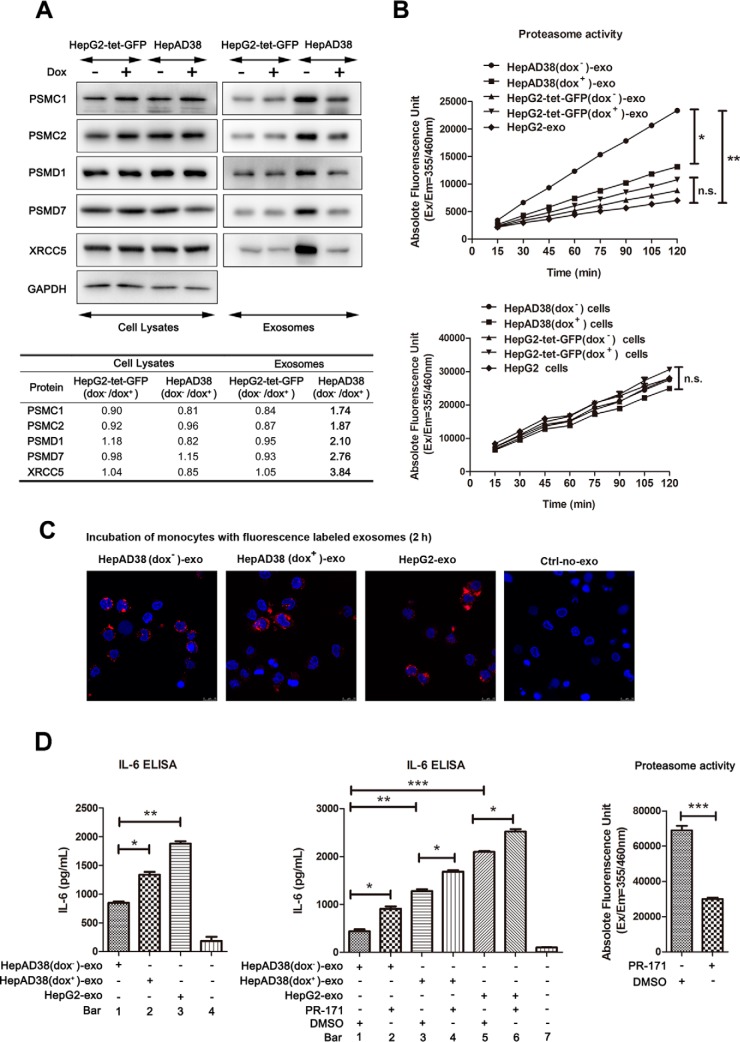
The observed changes in levels of proteins included into exosomes derived from HBV replicating HepAD38 cells may either reflect differences in the cellular expression of these proteins, or be a consequence of differential protein loading into the vesicles from cells as a result of HBV replication. Therefore, to get insights into the observed differences in exosomal protein content associated with HBV replication, we first verified the MS results using five representative proteins by Western immunoblotting. For this, the expression of these five proteins in whole cell lysates and exosome lysates from HepAD38 with or without HBV replication were compared. As a control, we also constructed a stably transfected, doxycycline-inducible GFP-expressing HepG2 cell line (HepG2-tet-GFP). In HepG2-tet-GFP cell line, the GFP expression is under the control of the promoter that can be modulated by doxycycline (dox), similarly to HBV replication in HepAD38 (supplemental Fig. S3). Consistent with the MS analyses, the levels of 26S protease regulatory subunit 4 (PSMC1), 26S protease regulatory subunit 7 (PSMC2), 26S proteasome non-ATPase regulatory subunit 1 (PSMD1), 26S proteasome non-ATPase regulatory subunit 7 (PSMD7), and X-ray repair cross-complementing protein 5 (XRCC5) were increased in exosomes secreted by HepAD38 supporting HBV replication (Fig. 4A). We further performed densitometry scanning of Western blots employing Image J software and the ratio between different samples was compared (Fig. 4A, lower table). The enhanced level of analyzed proteins was only observed in exosomes from HBV replicating dox− HepAD38 (>1.5-fold change), whereas their abundance in cell lysates and in HepG2-tet-GFP-exo were not changed by dox treatment (<1.2-fold change) as shown in Fig. 4A lower table, suggesting that these proteins were selectively packed into exosomes secreted from HBV replicating dox− cells. Among these proteins, the proteosome-associated subunits such as PSMC1, PSMC2, PSMD1, and PSMD7, were enhanced in exosomes from HBV-replicated cells.
Fig. 4.
Validation and functional analyses of differential proteins from HepAD38/HBV-derived exosomes. A, Western immunoblotting analysis of indicated proteins was performed using cellular extracts and exosomal preparations as described in the Experimental Procedures section. GAPDH was used as internal control for cell lysates, and Bradford assay was used to normalize the concentration of exosomal proteins. B, Proteasome activity of HepAD38 (dox−/dox+), HepG2-tet-GFP (dox−/dox+) and HepG2 derived exosomes and whole cell lysates. Proteasome activity assay of indicated exosomes (upper graph) and their parent cells (lower graph). X-axes represent the incubation time of protein lysates with the labeled substrate and y-axes represent absolute fluorescence unit. 2 μg exosome lysate or 10 μg cell lysate were incubated with the synthetic fluorogenic substrates Suc-Leu-Leu-Val-Tyr-AMC at 37 °C and the fluorescence was monitored by a plate reader with excitation and emission filters of 355 nm and 460 nm, respectively. The values were read per 15 min. The result shows that the fluorescence in HepAD38 (dox−)-exo was significantly higher than HepAD38 (dox+)-exo, HepG2-tet-GFP (dox−)-exo, HepG2-tet-GFP (dox+)-exo, and HepG2-exo (upper graph). Although no significant difference in the proteasome activity between the cell lysates from HepAD38 (dox−/dox+), HepG2-tet-GFP (dox−/dox+), and HepG2 were found (lower graph). C, HepAD38 (dox−/dox+) and HepG2 exosomes are actively internalized by monocytes. Confocal microscopy image of primary monocytes incubated for 2 h with purified PKH26-labeled exosomes (red). DAPI staining (blue) indicates the nucleus. D, The role of HepAD38 (dox−/dox+) and HepG2 exosomes in modulating the production of IL-6 by monocytes. 50 nm proteasome inhibitor PR-171 efficiently suppressed the proteasome activity of exosomes (right panel). 10 μg/ml of HepAD38 (dox−)-exo, HepAD38 (dox+)-exo, and HepG2-exo incubated with monocytes for 48 h. Culture supernatants were then analyzed by ELISA for IL-6 secretion (left graph). Human peripheral monocytes incubated with HepAD38 (dox−)-exo induced a significantly lower level of IL-6 secretion compared with IL-6 levels from HepAD38 (dox+)-exo and HepG2-exo. Then 10 μg/ml of indicated exosomes, pretreated with PR-171 or DMSO, were incubated with monocytes for 48 h, and culture supernatants were then analyzed by ELISA for IL-6 secretion. Irreversible inhibition of proteasomal activity within exosome restored the IL-6 induction in monocytes (middle panel). Note: (* p < 0.05, ** p < 0.01, *** p < 0.001). HepAD38 (dox−/dox+)-exo signifies exosomes from HepAD38 cells grown in the absence or presence of dox.
We therefore next examined whether there was a correlation between the increased abundance of proteasomal proteins in HepAD38 (dox−)-exo and the enhanced proteolytic activity in HepAD38 (dox−)-exo. To do this, we measured the proteolytic activity of exosome proteins by incubating them with fluorophore labeled substrate Leu-Leu-Val-Tyr-AMC and monitored changes in fluorescence (Fig. 4B). HepG2-tet-GFP (dox−/dox+) and HepG2 cell-derived exosomes were used as control (Fig. 4B, upper graph). Within ∼120 min of incubation, the fluorescence in HepAD38 (dox−)-exo remained at a steady and significant increase, whereas only a slow increase was seen in HepAD38 (dox+)-exo, HepG2-tet-GFP (dox−)-exo, HepG2-tet-GFP (dox+)-exo and HepG2-exo (Fig. 4B, upper graph). As a control we also incubated the cell lysates from either HepAD38 (dox−), HepAD38 (dox+), HepG2-tet-GFP (dox−), HepG2-tet-GFP (dox+), or HepG2 with the labeled substrate. We observed no significant difference in the proteasome activity within the host cells (Fig. 4B, lower graph), which was consistent with our previous results that HBV replication does not affect changes in the expression of the proteosomal proteins but rather induces a selective packing of these proteins into exosomes (Fig. 4A). Based on these results, it is reasonable to assume that proteasome subunit proteins packed into exosomes from HBV replicating cells may confer, upon their transfer, the proteolytic activity of the recipient cells.
The Role of Exosomal Proteasome Activity in Modulating the Production of Proinflammatory Mediators
Enrichment of proteasome subunit proteins in exosomes from HBV-replicating HepAD38 prompted us to assess the role of exosomes in delivering these proteins to the recipient cells. The proteasome proteases serve as pivotal regulators of LPS-induced inflammation in macrophages and function in modulation of cytokine production (40). Therefore, we examined if HepAD38 (dox−)-exo, rich in the proteasomal subunit proteins, influence the production of cytokines such as IL-6 in monocytes. To demonstrate this we first examined if exosomes from HepAD38 (dox+), HepAD38 (dox−) and HepG2 were internalized by monocytes. MACS purified monocytes were incubated with equal amount of fluorescence labeled exosomes for 2 h at 37 °C and examined by confocal microscopy. Results showed that exosomes from the three preparations were actively internalized by monocytes (Fig. 4C). Next, we examined if exosomes from HepAD38 (dox+), HepAD38 (dox−) and HepG2 could differentially modulate IL-6 production by monocytes. For this, monocytes were incubated with exosomes for 48 h and IL-6 production in culture supernatants was measured by ELISA. The results showed that exosomes from HepAD38 (dox+), HepAD38 (dox−), and HepG2 could all induce monocytes to produce IL-6 (Fig. 4D, left graph bar 1, 2, 3 vs. 4). However, the HepAD38 (dox−)-exo induced a significantly lower level of IL-6 secretion compared with HepAD38 (dox+)-exo and HepG2-exosomes (Fig. 4D left graph bar 1 vs. 2 vs. 3), suggesting that higher proteosome activity in exosomes may suppress IL-6 production by monocytes. To further corroborate on this, we used an irreversible proteasome inhibitor PR-171 to inhibit the proteolytic activity of exosomes. Exosomes from HepAD38 (dox+), HepAD38 (dox−) and HepG2 were pretreated with 50 nm of PR-171 for 1 h. As a control exosomes from each group were also pretreated with DMSO, a vehicle used for preparing stock solution of PR-171. After incubation with PR-171 or DMSO, exosomes were washed extensively and incubated with monocytes for 48 h. Pretreatment of exosomes with PR-171 could enhance IL-6 production in monocytes compared with pretreatment with DMSO (Fig. 4D middle graph bar 2, 4, 6 vs. 1, 3, 5, respectively). These results indicated that high level of proteasomes in exosomes could downregulate IL-6 production by monocytes, while inhibiting the proteasome activity in exosomes could release this downregulation. As a control, we also determined the biological activity of PR-171. 50 nm of PR-171 inhibited proteosomal activity of exosomes by more than 50% compared with the DMSO treated exosomes (Fig. 4D, right graph).
Taken together, these data suggest that exosomes may contribute to the delivery of proteasome subunit proteins to monocytes, and in turn influence IL-6 production by monocytes.
DISCUSSION
Exosomes are increasingly recognized as important mediators of cell-cell communication and accumulating evidence suggests that viruses can manipulate the infection process by secretion of specific viral and cellular components to exosomes (6). Although a few papers demonstrated that HBV utilizes some host-cell MVB (multivesicular bodies) functions (41), the influence of HBV replication on the content of exosomes secreted from HBV infected hepatocytes is not fully understood, and the potential role of exosomes in mediating the interactions between hepatocytes and their surrounding hepatic milieu remains largely unknown. In this study, we used proteomics approaches to evaluate the differences in protein content of exosomes secreted by the HepAD38 cell line with or without HBV replication. 1412 exosome protein groups were identified, including proteins involved in vesicle-mediated transport, establishment of protein location, immune effector related process as well as proteasome pathway. Exosomes are equivalent to cytoplasm enclosed in a lipid bilayer with the external domains of transmembrane proteins exposed to the extracellular environment (42). Exosomes contain membrane and cytosolic proteins. Recently, some studies have focused on analyzing proteins displayed on the outer membrane surface of exosomes (43). And it have been suggested that these exosomal surface proteins might have a great biomarker value because of their potential direct involvement in physiological function of exosomes. In this study, however, HepAD38 exosomes were isolated by ultracentrifugation and IP. Hence, the 1412 exosomal proteins that we identified include both the proteins inside the exosomes as well as those associated with the outside surfaces of exosomes. Bioinformatic analysis revealed that 32.98% exosomal proteins were annotated as plasma membrane-associated proteins. Some of these membrane-associated proteins may be anchored on the surface of exosomes and might be more directly involved in physiological function of hepatocyte-derived exosome. These results provide insights into the potential function of exosomes in HBV-host interaction.
Exosomes secreted by hepatocytes contribute to the maintenance of proper liver function by acting as coordinators of communication between different liver cell types. Mass spectrometry (MS)-based high-throughput bioanalytical methods have been used for qualitative and quantitative analysis of exosomes secreted by primary rat hepatocytes (23) and HCC cell lines (10) (24). Zhao et al., analyzed changes in exosome protein contents induced by hepatitis B virus in Huh 7 cells using SILAC labeling and LC-MS/MS (10) and reported identification and quantification of 399 proteins. He's group comprehensive characterization of the exosomal proteome contents derived from three HCC cell lines (HKCI-C3, HKCI-8, and MHCC97L) and from an immortalized hepatocyte line (MIHA), led to the identification of 278 proteins in HCC-derived exosomes. In contrast to these previous studies, the proteomics data sets obtained using the HepAD38 exosomes identified the highest number of proteins (1412 protein groups) including 68.6% (273/398) exosomal proteins detected in Huh7 cells and 46.9% (130/277) exosome proteins detected in HCC cell lines (HKCI-C3, HKCI-8 and MHCC97L). The Venn diagram produced a clear comparison of proteomics data set generated in this study versus in the proteomics data sets described in literature (Fig. S4). More importantly and in contrast to the previous studies, we have identified exosomal proteins distinctive to hepatocytes such as liver carboxylesterase 1 (CES1), liver form of glycogen phosphorylase (PYGL), liver form of fatty acid-binding protein (FABP1) and hepatocyte cell adhesion molecule (HEPACAM). In addition, we identified the presence of hepatocytes growth factor (HGF/MET) in exosomes secreted from HepAD38 cells, albeit it was also reported by others (24). These proteins are important for the proper function of the liver and their inclusion in exosomes secreted by liver cell can influence status of liver microenvironment. For example, CES1 is involved in the detoxification of xenobiotics and in the activation of ester and amide prodrugs. Human plasma carboxylesterase 1 was reported to be a novel serologic biomarker candidate for hepatocellular carcinoma (44). PYGL is an important allosteric enzyme in carbohydrate metabolism (45). FABP1 plays a role in lipoprotein-mediated cholesterol uptake in hepatocytes (46). HEPACAM is involved in regulating cell motility and cell-matrix interactions and may inhibit cell growth through suppression of cell proliferation (47). The hepatocyte growth factor (HGF) is a multifunctional cytokine with important roles in cell proliferation, survival, motility and morphogenesis. The HGF is involved in several biological processes such as liver regeneration and carcinogenesis (48). It has been demonstrated that HGF levels are increased during active HBV infection and enhance hepatocyte survival through activation of signaling pathways promoting interactions with extracellular matrix (49). Therefore, delivery of HGF and other liver-associated proteins, as identified in this study, via exosomes to cells in the liver may significantly modulate local changes within the liver microenvironment.
Currently, only a limited number of models are available to investigate the changes induced in the host cell by HBV replication. Infection of HepaRG cell or primary cultures of human hepatocytes with HBV, or transfection of human hepatoma cell line with HBV replicon plasmids (10, 50), results in a low infection and replication rate. Stable transfection of HepG2 cell line with HBV genome (HepG 2.2.15) prevents controlled studies of HBV effect on exosomal protein content. Therefore, for this study we chose the HBV-inducible cell line HepAD38, which allows a direct comparison of changes in cellular characteristics associated with HBV replication (11). We used a modified strategy for isolation of HepAD38-secreted exosomes, which led to a highly purified exosomal preparations with minimal contamination by other organelles as determined by Western immunoblotting, immunoprecipitation and electron microscopy. Using immunoprecipitation, we effectively removed potentially contaminating HBsAg particles and virus Dane particles from exosome preparations. The HepAD38-derived exosomal proteome content was then comprehensively characterized using ion-intensity based label-free quantitative proteomic strategy. The MS analysis of exosome preparations from dox− and dox+ HepAD38 cells confirmed the presence of exosomal protein markers. In addition, a few HBV proteins, including core, S, P protein were also identified (supplemental Table S1). We assume that HBV encoded proteins may be specifically packed into exosomes secreted by HBV infected hepatocytes and further study is required to define their role in the HBV infection process.
In order to elucidate the impact of HBV replication on the protein content of exosomes, ion intensity-based label-free strategies were applied to analyze and quantify exosomal proteins secreted by HepAD38 hepatoma-derived cell line. Among the total of 1412 exosome protein groups identified, 35 proteins were different between exosomes from cells grown in the presence or absence of dox. These differentially present proteins may therefore serve as potential biomarkers for HBV-induced hepatic alterations, and as potential modulators of the liver microenvironment. Among proteins differentially present in exosomes secreted from HBV-replicating and -nonreplicating cells there were proteins related to the HBV-induced pathogenetic process and proteins belonging to the 26S proteasome complex.
Although several proteins abundant in exosomes have been linked to pathogenetic conditions induced by HBV, none has been reported to be differentially included in exosomes in HBV replication dependent manner. For example, although the pigment epithelium-derived factor (PEDF) (36), cell division control protein 42 homolog (CDC42) (38), X-ray repair cross-complementing protein 5 (XRCC5/Ku86) (37), and versican core protein (VCAN) (39) were associated with progression of HCC, their levels in hepatocytes derived exosomes was not directly linked to HBV replication. In this study, we found, albeit at a low level, the presence of PEDF and CDC42 in HepAD38 (dox−)-exo. PEDF is a multifunctional secreted protein encoded by the SERPINF1 gene that has a potent antiangiogenic, antitumorigenic, and neurotrophic functions (9). Reduction of serum PEDF concentration is associated with the development of chronic liver diseases and may contribute to the progression of HCC (51) whereas the expression of CDC42 was reported to be higher in cancer tissues with HBV infection than in para-cancerous liver tissues and cancer tissues without HBV infection (38). Ku86, encoded by the XRCC5 gene, was increased in exosomes secreted by HepAD38 with HBV replication. Anti-Ku86 and -Ku86 antibodies are promising tumor markers for early detection and prognosis of HBV-related HCC (37). The significantly upregulated (by 3.46-fold) VCAN protein in HepAD38 exosomes secreted by HBV replicating cells was reported to be a potential biomarker for early-HCC diagnosis recently (39). Another protein identified in exosomes and used as biomarker is alpha-2-macroglobulin (A2M). It is reported that the pathological staging and extent of liver fibrosis in viral liver diseases can be estimated using blood markers including A2M (52). In our study, the increase of XRCC5 in HepAD38 (dox−)-exo was confirmed by Western immunoblotting. On the whole, the identification of differentially present exosomal proteins related to HBV-induced pathogenesis, clearly demonstrates that HBV replication in HepAD38 cells induces detectable changes in exosome protein contents.
This study has also identified several proteins upregulated in HepAD38 (dox−)-exo, which belong to the 26S proteasome complex including PSMD7, PSMD14, PSMC1, PSMD1, and PSMC2. The 26S proteasome belongs to the ubiquitin-proteasome system (UPS). In eukaryotic cells, the UPS is the major intracellular pathway for degradation and functional modification of cellular proteins (53). The alteration of proteasome subunits has been found in exosomes secreted by tumor associated macrophages (TAMs) (54), and in extracellular vesicles secreted by rat hepatocytes in response to lipopolysaccharide stimulation (55). It has been reported that viruses including HBV, can subvert or manipulate the UPS cellular machinery to favor viral propagation and to evade host immune response (53, 56). The proteasome subunit alpha type-7 (PSMA7), has been shown to specifically interact with HBX protein encoded by HBV genome (57). The HBV-encoded HBx protein was shown to be both a substrate and a potential inhibitor of the proteasome complex (57, 58). In this study, the levels of several subunits of the proteasome complex, including PSMC1, PSMC2, PSMD1, and PSMD7 were confirmed to be increased in HepAD38 (dox−)-exo compared with HepAD38 (dox+)-exo (Fig. 4A) and these proteins conferred enhanced proteasome activity in HepAD38 (dox−)-exo compared with HepAD38 (dox+)-exo (Fig. 4B). At the same time, the proteolytic activity of cell extracts showed no significant difference. These results suggest that the proteasome subunits were selectively packed into the vesicles, which upon secretion from HBV replicating cells may arm the recipient cells with enhanced proteasome activities.
Proteasome have been recognized as a potential therapeutic target in the treatment of inflammation and caner (40, 59). The proteasome proteases serve as pivotal regulators of LPS-induced inflammation in macrophages and function in modulation of cytokine production (40). Treatment of human intestinal epithelial cell line Caco-2 with proteasome inhibitors could stimulate IL-6 production (60). We reported that monocytes treated with HBsAg secreted 10-fold higher level of IL-6 compared with controls (61). We therefore examined the level of IL-6 produced by monocytes incubated with exosomes secreted from either HBV-replicating or -nonreplicating HepAD38 cells or from HepG2 controls. A lower IL-6 production was detected in cells incubated with exosomes secreted from HepAD38 (dox−) cells, which allowed HBV-replication, compared with exosomes from HepAD38 (dox+) or HepG2 cells. Pretreatment of exosomes from these three types of cells with proteasome inhibitor PR-171 followed by incubation with monocytes restored IL-6 production by monocytes. These results suggest that transmission of proteasome subunit proteins by HepAD38 (dox−)-exo might modulate the cytokine production in the recipient monocytes. It is becoming increasingly evident that the virally infected cells can manipulate the cellular environment via exosomes and as such contributing to viral persistence and pathogenesis. In this study, several proteins, such as HSP60, cell adhesion molecule 1, clusterin, tyrosine-protein kinase Lyn, and receptor-type tyrosine-protein phosphatase C were detected by mass spectrometry in HepAD38 derived exosomes (Table S4). These proteins were reported to participate in the regulation of cytokine production or cytokine-mediated signaling pathway (29–35) and therefore may contribute to the modulation of immune response during HBV infection. Our results indicate that exosomes of HBV-replicated HepAD38 cells might be a shuttle to deliver molecules such as immune effector related proteins and proteasome complex proteins to recipient monocytes and influence the cytokine production. A proposed model for biological relevance of the immune effector-related proteins and proteasome complex proteins carried by exosomes released by HBV infected cells is depicted in Fig. 5. In addition, besides proteins, miRNAs, can also be selectively packaged into exosomes and induce immune activation (14, 62). For example, exosomes derived from ethanol-treated hepatic Huh7.5 cells contain high level of miRNA-122, which can be delivered via exosomes to the recipient's monocytes resulting in increased levels of pro-inflammatory cytokines production following LPS stimulation (62). In addition to proteins, the role of other biomolecules carried by hepatocyte produced exosomes in modulation immune response needs to be more systematically and thoroughly investigated.
Fig. 5.
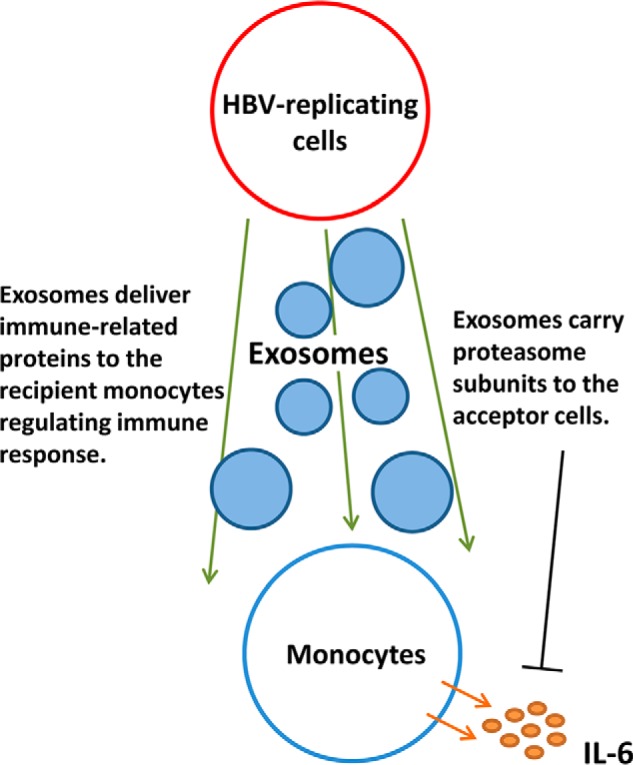
Proposed model for the role of exosomes secreted by HBV replicating cells in modulating the immune response through delivery of immune effector related proteins and proteaosme complex proteins to the acceptor cells.
Exosomes were discovered decades ago, and many reports described their role and functions in various viral infection processes. Accumulating evidence demonstrates that host exosome pathways are exploited by viruses and that virally modified exosomes contribute to viral spread, host immunity, and manipulation of the microenvironment. Exosomes from HCV-infected cells contain viral RNA that can facilitate infection of new cells, suggesting potential functions of exosomes in the spread of HCV (63). Exosomes secreted from EBV-infected cancer cells contain viral oncogene protein LMP1, which plays a role in altering the immune response (51). HIV-infected cells release exosomes loaded with viral proteins, including Gag and Nef that can trigger Nef-mediated apoptosis in bystander CD4+ T cells (39, 64). Exosomes were also implicated in NK cell dysfunction and regulation of innate immune responses during HBV infection (8, 9). In this study, several viral proteins including large S, core and P proteins were identified in exosomes secreted from HepAD38 cells sustaining HBV replication. Although these viral proteins carried by exosomes might play a role in HBV infection process, further studies are needed to address this issue. Our study also found that the proteasome subunit proteins were distinctively loaded into exosomes from HBV replicated HepAD38 cells and might modulate the production of pro-inflammatory molecules in the recipient monocytes. Hence, these HBV modified exosomes may play a critical role in trans-cellular communication and immune regulation. Our finding ultimately provides additional insights into the persistence of HBV infection and modulation of liver microenvironment.
CONCLUSIONS
Overall, in this study by employing highly sensitive LC-MS/MS analyses in conjunction with HepAD38 cell line that supports HBV replication in a tightly controlled manner, we demonstrated that exosomes secreted from hepatocytes are packed with a large number of proteins involved in various biological processes. Exosomes shed by hepatocytes is a valuable source of information about intracellular communication in the liver microenvironment. More importantly, our study demonstrates that the level of some cellular proteins present in HepAD38-secreted exosomes could be modulated by HBV replication. In particular, we discovered that the proteasome subunit proteins were distinctively loaded into exosomes form HBV replicated HepAD38 cells and could modulate the production of pro-inflammatory molecules in the recipient monocytes. Based on our findings, it is reasonable to assume that HBV modified exosomes could convey signals to immune cells and may mediate trans-cellular communication and immune regulations. This may be a significant mechanism through which HBV maintains a persistent infection within the host liver.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD004724.
Supplementary Material
Acknowledgments
We thank the members of the Yuan's Lab for helpful discussions related to this work. We thank Kristin Rosowski, Birgit Korte, and Stefanie Tautges for their technical assistance during the proteomics experiments conducted in Sitek's Lab.
Footnotes
Author contributions: Zhenghong Yuan, Jieliang Chen and Xiaonan Zhang designed research; Xiaofang Jia preformed research; Dominik A. Megger and Barbara Sitek contribute to the MS analysis and label free data analysis; Zhong Fang and Jin Li contribute to monocytes separation; Min Wu and Yaming Li contribute to the construction of HepG2-tet-GFP cell line; Qiaofang Chu contribute to culturing HepAD38 cell line; Lijun Zhang contribute to the bioinformatic analysis; Xiaofang Jia and Maya Kozlowski wrote the paper, Jieliang Chen and Xiaonan Zhang contribute to proof reading the manuscript and providing critical comments.
* This work was supported by The National Natural Science Foundation of China (81461130019) to Zhenghong Yuan; the German Research Foundation (SFB/Transregio TRR60 to Zhenghong Yuan (A1) and to Barbara Sitek (Z3)); the National Natural Science Foundation of China (91542207) to Zhenghong Yuan; the Natural Science Foundation of Shanghai (13ZR1460100) to Xiaofang Jia; The National High-tech R&D Program (863 Program) (2014AA021403) and the National Natural Science Foundation of China (81271834) to Lijun Zhang.
 This article contains supplemental material.
This article contains supplemental material.
Authors Email: Xiaofang Jia: rain9205@163.com; Jieliang Chen: cjlid@163.com; Dominik A. Megger: dominik.megger@rub.de; Xiaonan Zhang: xnzhang80@hotmail.com; Maya Kozlowski: mayakozlow@163.com; Lijun Zhang: zhanglijun1221@163.com; Zhong Fang: clock-22@163.com; Jin Li: ldj127945@163.com; Qiaofang Chu:12111010070@fudan.edu.cn; Min Wu: wummi1981@163.com; Yaming Li: yamingli2012@gmail.com; Barbara Sitek: barbara.sitek@rub.de; Zhenghong Yuan: 13916306983@163.com.
1 The abbreviations used are:
- HBV
- hepatitis B virus
- HCC
- hepatocellular carcinoma
- LNPCs
- liver nonparenchymal cells
- LSECs
- liver sinusoidal endothelial cells
- FA
- formic acid
- CD63
- CD63 antigen
- LAMP2
- lysosome-associated membrane protein 2
- TSG101
- tumor susceptibility gene 101
- Cox IV
- cytochrome c oxidase IV
- EEA1
- early endosome antigen 1
- PSMD7
- 26S proteasome non-ATPase regulatory subunit 7
- XRCC5
- X-ray repair cross-complementing protein 5
- PEDF
- pigment epithelium-derived factor
- CDC42
- cell division control protein 42 homolog
- VCAN
- Versican core protein
- A2M
- Alpha-2-macroglobulin
- TEM
- transmission electron microscopy
- TAMs
- tumor associated macrophages
- MVB
- multivesicular bodies
- PTA
- phosphotungstic acid
- PBMCs
- peripheral blood mononuclear cells
- CHB
- chronic hepatitis B.
REFERENCES
- 1. Trepo C., Chan H. L., and Lok A. (2014) Hepatitis B virus infection. Lancet 384, 2053–2063 [DOI] [PubMed] [Google Scholar]
- 2. Schweitzer A., Horn J., Mikolajczyk R. T., Krause G., and Ott J. J. (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386, 1546–1555 [DOI] [PubMed] [Google Scholar]
- 3. Zhou T., Guo H., Guo J.-T., Cuconati A., Mehta A., and Block T. M. (2006) Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res. 72, 116–124 [DOI] [PubMed] [Google Scholar]
- 4. Jia S., Zocco D., Samuels M. L., Chou M. F., Chammas R., Skog J., Zarovni N., Momen-Heravi F., and Kuo W. P. (2014) Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Rev. Mol. Diagn. 14, 307–321 [DOI] [PubMed] [Google Scholar]
- 5. Imani Fooladi A. A., and Mahmoodzadeh H. H. (2014) Biological functions of exosomes in the liver in health and disease. Hepat. Mon. 14, e13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meckes D. G., Jr. (2015) Exosomal communication goes viral. J. Virol. 89, 5200–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M., Zhou X., and Yuan Z. (2013) Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 14, 793–803 [DOI] [PubMed] [Google Scholar]
- 8. Yang Y., Han Q., Hou Z., Zhang C., Tian Z., and Zhang J. (2016) Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol. Immunol. May 30. doi: 10.1038/cmi.2016.24 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kouwaki T., Fukushima Y., Daito T., Sanada T., Yamamoto N., Mifsud E. J., Leong C. R., Tsukiyama-Kohara K., Kohara M., Matsumoto M., Seya T., and Oshiumi H. (2016) Extracellular vesicles including exosomes regulate innate immune responses to Hepatitis B virus infection. Front. Immunol. 7, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao X., Wu Y., Duan J., Ma Y., Shen Z., Wei L., Cui X., Zhang J., Xie Y., and Liu J. (2014) Quantitative proteomic analysis of exosome protein content changes induced by hepatitis B virus in Huh-7 cells using SILAC labeling and LC-MS/MS. J. Proteome Res. 13, 5391–5402 [DOI] [PubMed] [Google Scholar]
- 11. Ladner S. K., Otto M. J., Barker C. S., Zaifert K., Wang G. H., Guo J. T., Seeger C., and King R. W. (1997) Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41, 1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Megger D. A., Bracht T., Kohl M., Ahrens M., Naboulsi W., Weber F., Hoffmann A. C., Stephan C., Kuhlmann K., Eisenacher M., Schlaak J. F., Baba H. A., Meyer H. E., and Sitek B. (2013) Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol. Cell. Proteomics 12, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bracht T., Schweinsberg V., Trippler M., Kohl M., Ahrens M., Padden J., Naboulsi W., Barkovits K., Megger D. A., Eisenacher M., Borchers C. H., Schlaak J. F., Hoffmann A. C., Weber F., Baba H. A., Meyer H. E., and Sitek B. (2015) Analysis of disease-associated protein expression using quantitative proteomics-fibulin-5 is expressed in association with hepatic fibrosis. J. Proteome Res. 14, 2278–2286 [DOI] [PubMed] [Google Scholar]
- 14. Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A. J., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Wurdinger T., and Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mayer G., Stephan C., Meyer H. E., Kohl M., Marcus K., and Eisenacher M. (2015) ProCon - PROteomics CONversion tool. J. Proteomics 129, 56–62 [DOI] [PubMed] [Google Scholar]
- 17. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 18. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, SD447–SD452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu Z., Zhang Z., Doo E., Coux O., Goldberg A. L., and Liang T. J. (1999) Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73, 7231–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S., Chen Z., Hu C., Qian F., Cheng Y., Wu M., Shi B., Chen J., Hu Y., and Yuan Z. (2013) Hepatitis B virus surface antigen selectively inhibits TLR2 ligand-induced IL-12 production in monocytes/macrophages by interfering with JNK activation. J. Immunol. 190, 5142–5151 [DOI] [PubMed] [Google Scholar]
- 21. Zhou T., Guo H., Guo J. T., Cuconati A., Mehta A., and Block T. M. (2006) Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res. 72, 116–124 [DOI] [PubMed] [Google Scholar]
- 22. Hosseini H. M., Fooladi A. A., Nourani M. R., and Ghanezadeh F. (2013) The role of exosomes in infectious diseases. Inflamm. Allergy Drug Targets 12, 29–37 [DOI] [PubMed] [Google Scholar]
- 23. Conde-Vancells J., Rodriguez-Suarez E., Embade N., Gil D., Matthiesen R., Valle M., Elortza F., Lu S. C., Mato J. M., and Falcon-Perez J. M. (2008) Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 7, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He M., Qin H., Poon T. C., Sze S. C., Ding X., Co N. N., Ngai S. M., Chan T. F., and Wong N. (2015) Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 36, 1008–1018 [DOI] [PubMed] [Google Scholar]
- 25. Severi T., Ying C., Vermeesch J. R., Cassiman D., Cnops L., Verslype C., Fevery J., Arckens L., Neyts J., and van Pelt J. F. (2006) Hepatitis B virus replication causes oxidative stress in HepAD38 liver cells. Mol. Cell. Biochem. 290, 79–85 [DOI] [PubMed] [Google Scholar]
- 26. Mathivanan S., Fahner C. J., Reid G. E., and Simpson R. J. (2012) ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, SD1241–SD1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heymann F., and Tacke F. (2016) Immunology in the liver–from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 13, 88–110 [DOI] [PubMed] [Google Scholar]
- 28. Longatti A. (2015) The dual role of exosomes in Hepatitis A and C virus transmission and viral immune activation. Viruses 7, 6707–6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cohen-Sfady M., Nussbaum G., Pevsner-Fischer M., Mor F., Carmi P., Zanin-Zhorov A., Lider O., and Cohen I. R. (2005) Heat shock protein 60 activates B cells via the TLR4-MyD88 pathway. J. Immunol. 175, 3594–3602 [DOI] [PubMed] [Google Scholar]
- 30. Osterloh A., Kalinke U., Weiss S., and Fleischer Band Breloer M. (2007) Synergistic and differential modulation of immune responses by Hsp60 and lipopolysaccharide. J. Biol. Chem. 282, 4669–4680 [DOI] [PubMed] [Google Scholar]
- 31. Yokota S., Minota S., and Fujii N. (2006) Anti-HSP auto-antibodies enhance HSP-induced pro-inflammatory cytokine production in human monocytic cells via Toll-like receptors. Int. Immunol. 18, 573–580 [DOI] [PubMed] [Google Scholar]
- 32. Boles K. S., Barchet W., Diacovo T., Cella M., and Colonna M. (2005) The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood 106, 779–786 [DOI] [PubMed] [Google Scholar]
- 33. Xie Z., Harris-White M. E., Wals P. A., Frautschy S. A., Finch C. E., and Morgan T. E. (2005) Apolipoprotein J (clusterin) activates rodent microglia in vivo and in vitro. J. Neurochem. 93, 1038–1046 [DOI] [PubMed] [Google Scholar]
- 34. Infantino S., Jones S. A., Walker J. A., Maxwell M. J., Light A., O'Donnell K., Tsantikos E., Peperzak V., Phesse T., Ernst M., Mackay F., Hibbs M. L., Fairfax K. A., and Tarlinton D. M. (2014) The tyrosine kinase Lyn limits the cytokine responsiveness of plasma cells to restrict their accumulation in mice. Sci. Signal. 7, era77. [DOI] [PubMed] [Google Scholar]
- 35. Windheim M., Southcombe J. H., Kremmer E., Chaplin L., Urlaub D., Falk C. S., Claus M., Mihm J., Braithwaite M., Dennehy K., Renz H., Sester M., Watzl C., and Burgert H. G. (2013) A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. Proc. Natl. Acad. Sci. U.S.A. 110, SE4884–SE4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsumoto K., Ishikawa H., Nishimura D., Hamasaki K., Nakao K., and Eguchi K. (2004) Antiangiogenic property of pigment epithelium-derived factor in hepatocellular carcinoma. Hepatology 40, 252–259 [DOI] [PubMed] [Google Scholar]
- 37. Xu Y., Liu A. J., Gao Y. X., Hu M. G., Zhao G. D., Zhao Z. M., and Liu R. (2014) Expression of Ku86 and presence of Ku86 antibody as biomarkers of hepatitis B virus related hepatocellular carcinoma. Dig. Dis. Sci. 59, 614–622 [DOI] [PubMed] [Google Scholar]
- 38. Chang C., Huang S., Lin H., Wu C., and Wang C. (2007) Different expression of apoptotic proteins between HBV-infected and non-HBV-infected hepatocellular carcinoma. Hepatogastroenterology 54, 2061–2068 [PubMed] [Google Scholar]
- 39. Lenassi M., Cagney G., Liao M., Vaupotic T., Bartholomeeusen K., Cheng Y., Krogan N. J., Plemenitas A., and Peterlin B. M. (2010) HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qureshi N., Morrison D. C., and Reis J. (2012) Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta 1823, 2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watanabe T., Sorensen E. M., Naito A., Schott M., and Kim Sand Ahlquist P. (2007) Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. U.S.A. 104, 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schorey JSandBhatnagar S. (2008) Exosome function: from tumor immunology to pathogen biology. Traffic 9, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hildonen S., Skarpen E., Halvorsen T. G., and Reubsaet L. (2016) Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci. Rep. 6, 36331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Na K., Lee E. Y., Lee H. J., Kim K. Y., Lee H., Jeong S. K., Jeong A. S., Cho S. Y., Kim S. A., Song S. Y., Kim K. S., Cho S. W., Kim H., and Paik Y. K. (2009) Human plasma carboxylesterase 1, a novel serologic biomarker candidate for hepatocellular carcinoma. Proteomics 9, 3989–3999 [DOI] [PubMed] [Google Scholar]
- 45. Agius L. (2015) Role of glycogen phosphorylase in liver glycogen metabolism. Mol. Aspects Med. 46, 34–45 [DOI] [PubMed] [Google Scholar]
- 46. Huang H., McIntosh A. L., Landrock K. K., Landrock D., Storey S. M., Martin G. G., Gupta S., Atshaves B. P., Kier A. B., and Schroeder F. (2015) Human FABP1 T94A variant enhances cholesterol uptake. Biochim. Biophys. Acta 1851, 946–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shao H., Gu Y., Ding J., Lu P., Ruan T., and Lu W. (2016) HEPACAM inhibited the growth and migration of cancer cells in the progression of non-small cell lung cancer. Tumour Biol. 37, 2621–2627 [DOI] [PubMed] [Google Scholar]
- 48. Fajardo-Puerta A. B., Mato Prado M., Frampton A. E., and Jiao L. R. (2016) Gene of the month: HGF. J. Clin. Pathol. 69, 575–579 [DOI] [PubMed] [Google Scholar]
- 49. Barreiros A. P., Sprinzl M., Rosset S., Hohler T., Otto G., Theobald M., Galle P. R., Strand D., and Strand S. (2009) EGF and HGF levels are increased during active HBV infection and enhance survival signaling through extracellular matrix interactions in primary human hepatocytes. Int. J. Cancer 124, 120–129 [DOI] [PubMed] [Google Scholar]
- 50. Gripon P., Rumin S., Urban S., Le Seyec J., Glaise D., Cannie I., Guyomard C., Lucas J., Trepo C., and Guguen-Guillouzo C. (2002) Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U.S.A. 99, 15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dukers D. F., Meij P., Vervoort M. B., Vos W., Scheper R. J., Meijer C. J., Bloemena E., and Middeldorp J. M. (2000) Direct immunosuppressive effects of EBV-encoded latent membrane protein 1. J. Immunol. 165, 663–670 [DOI] [PubMed] [Google Scholar]
- 52. Meckes D. G. Jr., and Raab-Traub N. (2011) Microvesicles and viral infection. J. Virol. 85, 12844–12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khan M. L., and Stewart A. K. (2011) Carfilzomib: a novel second-generation proteasome inhibitor. Future Oncol. 7, 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Columba Cabezas S., and Federico M. (2013) Sequences within RNA coding for HIV-1 Gag p17 are efficiently targeted to exosomes. Cell Microbiol. 15, 412–429 [DOI] [PubMed] [Google Scholar]
- 55. Rodriguez-Suarez E., Gonzalez E., Hughes C., Conde-Vancells J., Rudella A., Royo F., Palomo L., Elortza F., Lu S. C., Mato J. M., Vissers J. P., and Falcon-Perez J. M. (2014) Quantitative proteomic analysis of hepatocyte-secreted extracellular vesicles reveals candidate markers for liver toxicity. J. Proteomics 103, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Demo S. D., Kirk C. J., Aujay M. A., Buchholz T. J., Dajee M., Ho M. N., Jiang J., Laidig G. J., Lewis E. R., Parlati F., Shenk K. D., Smyth M. S., Sun C. M., Vallone M. K., Woo T. M., Molineaux C. J., and Bennett M. K. (2007) Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 67, 6383–6391 [DOI] [PubMed] [Google Scholar]
- 57. Zhang Z., Torii N., Furusaka A., Malayaman N., Hu Z., and Liang T. J. (2000) Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275, 15157–15165 [DOI] [PubMed] [Google Scholar]
- 58. Lu M., Huang B., Hanash S. M., Onuchic J. N., and Ben-Jacob E. (2014) Modeling putative therapeutic implications of exosome exchange between tumor and immune cells. Proc. Natl. Acad. Sci. U.S.A. 111, SE4165–SE4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Frankland-Searby S., and Bhaumik S. R. (2012) The 26S proteasome complex: an attractive target for cancer therapy. Biochim. Biophys. Acta 1825, 64–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pritts T. A., Hungness E. S., Hershko D. D., Robb B. W., Sun X., Luo G. J., Fischer J. E., Wong H. R., and Hasselgren P. O. (2002) Proteasome inhibitors induce heat shock response and increase IL-6 expression in human intestinal epithelial cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, SR1016–SR1026 [DOI] [PubMed] [Google Scholar]
- 61. Fang Z., Li J., Yu X., Zhang D., Ren G., Shi B., Wang C., Kosinska A. D., Wang S., Zhou X., Kozlowski M., Hu Y., and Yuan Z. (2015) Polarization of monocytic myeloid-derived suppressor cells by Hepatitis B surface antigen is mediated via ERK/IL-6/STAT3 signaling feedback and restrains the activation of T cells in chronic Hepatitis B virus infection. J. Immunol. 195, 4873–4883 [DOI] [PubMed] [Google Scholar]
- 62. Momen-Heravi F., Bala S., Kodys K., and Szabo G. (2015) Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Scientific Reports 5, 9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramakrishnaiah V., Thumann C., Fofana I., Habersetzer F., Pan Q., de Ruiter P. E., Willemsen R., Demmers J. A. A., Stalin Raj V., Jenster G., Kwekkeboom J., Tilanus H. W., Haagmans B. L., Baumert T. F., and van der Laan L. J. W. (2013) Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. U.S.A. 110, 13109–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fang Y., Wu N., Gan X., Yan W., Morrell J. C., and Gould S. J. (2007) Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLos Biol. 5, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD004724.