Abstract
Sepsis is a systemic immune response responsible for considerable morbidity and mortality. Molecular modeling of host-pathogen interactions in the disease state represents a promising strategy to define molecular events of importance for the transition from superficial to invasive infectious diseases. Here we used the Gram-positive bacterium Streptococcus pyogenes as a model system to establish a mass spectrometry based workflow for the construction of a stoichiometric surface density model between the S. pyogenes surface, the surface virulence factor M-protein, and adhered human blood plasma proteins. The workflow relies on stable isotope labeled reference peptides and selected reaction monitoring mass spectrometry analysis of a wild-type strain and an M-protein deficient mutant strain, to generate absolutely quantified protein stoichiometry ratios between S. pyogenes and interacting plasma proteins. The stoichiometry ratios in combination with a novel targeted mass spectrometry method to measure cell numbers enabled the construction of a stoichiometric surface density model using protein structures available from the protein data bank. The model outlines the topology and density of the host-pathogen protein interaction network on the S. pyogenes bacterial surface, revealing a dense and highly organized protein interaction network. Removal of the M-protein from S. pyogenes introduces a drastic change in the network topology, validated by electron microscopy. We propose that the stoichiometric surface density model of S. pyogenes in human blood plasma represents a scalable framework that can continuously be refined with the emergence of new results. Future integration of new results will improve the understanding of protein-protein interactions and their importance for bacterial virulence. Furthermore, we anticipate that the general properties of the developed workflow will facilitate the production of stoichiometric surface density models for other types of host-pathogen interactions.
Bacterial infections present an important healthcare challenge with a wide spectrum of disease severity. One of the most severe bacterial diseases is sepsis, a systemic immune response mainly caused by bacteria entering the bloodstream (1). Sepsis is among the leading cause of deaths in hospitals (2, 3), with a mortality rate ranging from 20% to 50% (1, 4) and a worldwide incidence of 200 to 300 per 100,000 every year (4). Furthermore, patients surviving sepsis have a drastic reduction in life quality and life expectancy (5–7), and in the United States alone, the cost associated with sepsis is 17 billion dollars (3). A growing concern in sepsis treatment is that even though the mortality rate of sepsis cases has been decreasing over the last decades, the total number of deaths is increasing (1, 8). In addition, antibiotic resistant bacteria constitute an increasing problem for the healthcare system (9–11), making bacterial infections harder to treat. In the transition from superficial to severe disease, bacterial pathogens have devised a multitude of defense strategies. Some of these defense strategies include protein-protein interactions to prevent for example complement deposition, avoiding antibody binding and phagocytosis (12). The complex nature of the host-pathogen interaction network calls for new strategies to integrate and visualize these interactions at a molecular level preferably using systems wide analysis with as accurate measurements as possible. In particular, there is an unmet need to determine stoichiometric relationships between interacting proteins at the level of a biological system and visualize these interactions in an accurate model to support the development of new preventive, diagnostic and treatment strategies for severe infectious disease.
The accurate analysis of host-pathogen interactions to determine the stoichiometric relationship between pathogen, surface proteins and interacting host proteins is an important step toward understanding bacterial immune evasion strategies. In the work presented here, we used the Gram-positive bacterium Streptococcus pyogenes as a model pathogen and human blood plasma as a model for interacting host proteins. S. pyogenes is one of the most important human pathogens responsible for a wide spectrum of superficial and severe diseases such as pharyngitis and sepsis (13). The bacteria have a well-characterized human blood plasma interaction proteome (14) of critical importance for the transition to invasive diseases. S. pyogenes is divided into serogroups based on the sequence variation of the most abundant and important surface bound virulence factor, the M-protein (15). So far there are more than 200 different types of M-proteins identified (16, 17), where the most common is the M1 serogroup for both pharyngitis and invasive S. pyogenes disease (18, 19). The M1-protein is a coiled-coil dimer of a 453 amino acid long protein, anchored in the bacterial cell wall (20, 21). The M1-protein has several known human plasma protein interactions, such as serum albumin (22, 23), fibrinogen (21, 24, 25) and immunoglobulin gamma (IgG)1 (12, 23, 26). In combination with the cell wall and surface associated proteins of the bacterium, the interactions between pathogen and host give rise to a complex network of bacterial and host protein interactions.
Mass spectrometry (MS) is the method of choice when measuring the complex dynamics of pathogen surface proteins and their interactions with host proteins (14, 22, 27–30). In bottom-up MS based proteomics techniques, trypsin is usually used to digest proteins into peptides that are separated using liquid chromatography (LC) and coupled online for MS analysis (31, 32). In targeted MS techniques, such as selected reaction monitoring (SRM), transitions for selected proteotypic peptides are exclusively and reproducibly measured using triple quadruple mass spectrometers (33, 34). Together with stable isotope labeled peptides (29, 35, 36), SRM can provide absolute protein quantification for several proteins present in the same sample. The isotope-labeled peptides behave the same as the unlabeled peptides in LC-MS (35), and because the labeled peptide is of known concentration, the unlabeled peptide concentration can be calculated and hence absolutely quantified. The protein concentration is then extrapolated from the concentration of the proteotypic peptides measured for that protein (37).
In this article, we have performed a comprehensive study of the stoichiometric relationship between adhered host proteins and a bacterial surface. The stoichiometric relationship in combination with a new technical improvement for counting the number of bacteria using SRM enabled the construction of the first model of S. pyogenes surface interaction network in a host environment. This model includes the stoichiometric relations between host proteins, surface proteins, and the surface of the pathogen and it visualizes the density of this plethora of interactions in a stoichiometric surface density model.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The S. pyogenes strains that were used for the construction of the stoichiometric density model are: SF370, a clinical isolate of the M1 serotype; and an isogenic M1-mutant of SF370, ΔM1 (38). Strains used for cell counting of S. pyogenes are: JM50, JM57, JM59, JM62, JM67, JM72, JM81, and JM85 are all of the M1 serotype and isolated from healthcare clinics in Sweden during 2012 (39). Single colonies were grown at 37 °C and 5% CO2 to exponential and stationary phase (only the strains used for cell counting) in 30g/L Todd-Hewitt broth (BD, Sparks, MD) and 6g/L yeast extract (BD). The cells were then harvested by centrifugation and resuspended in 20 mm Tris-HCl, 150 mm NaCl, pH 7.6 (TBS tablet, Medicago, Uppsala, Sweden), to an approximate concentration of 2 × 109 CFU/ml.
Selection of Reference Method for Bacterial Cell Counting
Four commonly used methods for counting bacteria, optical density, colony forming units (CFU), microscopy and flow cytometry, were evaluated. The first two methods are simple well-established methods of estimating cell count in a dilution series, optical density at 620 nm and counting the number of colonies (CFU) on an agar plate after 24-hour incubation. The third method, light field microscopy with phase contrast was performed using an Olympus CKX41 (Olympus Corporation, Tokyo, Japan) equipped with an INFINITY1 camera (Lumenera Corporation, Ottawa, ON, Canada), together with a Petroff-Hausser counting chamber and tryphan blue staining (Sigma-Aldrich, Steinheim, Germany). Microscopy images were analyzed automatically using a custom Adobe Photoshop CS6 procedure, in which the area of the dark bacteria is calculated in a selected area of the counting chamber, after removing the counting chamber reference lines. The method has an estimated Pearson's product-moment correlation of 0.989 (p < 3e-11), compared with manual counting. The fourth method, flow cytometry was performed using a BD Accuri C6 Flow Cytometer with a 488 nm filter, gating in FL1-A (488 nm) versus FSC-A, with a flow rate of 35 μl/min and a core size of 16 μm. Samples were stained using a fluorescent dye (SYTO™ BC, Thermo Fisher). Samples were collected until 30 μl or 30000 bacterial events. The bacterial population in FSC-A/FL1-A was moved toward higher FL1-A in the more concentrated dilutions. This higher dye absorbance could indicate aggregated bacteria, but no compensation was made for this.
Plasma Adsorption
Human blood plasma (pooled healthy human plasma [Na-citrate], Innovative Research, Novi, MI) was mixed with bacteria, in a ratio 3:1, and the samples were incubated for 30 min at 37 °C, allowing plasma proteins to adsorb to the bacterial surface. This bacterial to plasma ratio is 10 times lower than the original plasma adsorption protocols (40, 41), but because the detection techniques have improved and to better replicate the disease state, the bacterial ratio was kept as low as possible. Bacteria were harvested after several washes in 20 mm Tris-HCl, 150 mm NaCl, and pH 7.6 through centrifugation (5000 × g). All samples were prepared in eight biological replicates.
Glycine Elution and Bacteria Digest
To elute the adsorbed proteins, the final cell pellets were prepared using two protocols. The first, glycine elution, has been described (12). Briefly, the bacterial pellet was resuspended in 0.1 m glycine (Sigma-Aldrich), pH 2.0, followed by incubation for 10 min, and the supernatants were neutralized to pH 7–8 with 1 m Tris (Ultrapure, Saveen Werner AB, Limhamn, Sweden). In the second protocol, bacteria digest, the pellet was resuspended in water (HPLC-graded, Sigma-Aldrich) and transferred to tube containing 0.1 mm Silica beads (Lysing Matrix tubes, Nordic Biolabs #6911100, Täby, Sweden). The bacteria were lysed with a cell disruptor (FastPrep96, MP Biomedicals, Santa Ana, CA). The in solution digestion (see below) was performed without transferring the liquid to new tube to minimize sample loss and enhance surface protein yield.
In-solution Digestion
Samples were dried completely through vacuum evaporation (miVac Duo, Genevac, Ipswich, UK) and resuspended in 8 m urea (Sigma-Aldrich) to denature the sample. The protein sample was reduced using tris (2-carboxyethyl)-phosphine (Sigma-Aldrich), at a final concentration of 10 mm and the samples were incubated at 37 °C for 60 min. The samples were incubated for 30 min in the dark at room temperature with 2-iodoacetamide (Sigma-Aldrich) at a concentration of 20 mm as the alkylating agent. Samples were diluted with 100 mm ammonium bicarbonate (Sigma-Aldrich) to a urea concentration of 0.73 m and digested with a final concentration of 3.6 ng/μl trypsin (Sequence grade modified trypsin Porcin, Promega, Madison, WI) over night (18 h). The reaction was stopped by adding formic acid (Sigma-Aldrich) to a final sample pH of 2–3.
C18 Peptide Clean Up
Vydac UltraMicroSpin® silica C18 300Å columns (#SUM SS18V, The Nest Group, Inc., Southborough, MA) was used for sample desalting, clean-up and concentrating peptides according to the manufacturer's instructions. To remove silica beads and cell debris, the samples prepared by bacteria digest were centrifuged at 15,000 × g for 10 min, and supernatant was used for C18 peptide clean-up.
Liquid Chromatography (LC) and Selected Reaction Monitoring (SRM)
Chromatographic separations of peptides were performed on an Easy-nLC II system (Thermo Fisher Scientific, San Jose, CA) with a nonlinear 30-min gradient of 5–10% acetonitrile (ACN, with 0.1% formic acid, UHPLC-graded, Fluka Analytical, Sigma-Aldrich) over 5 min, 10–20% ACN over 20 min, and 20–30% ACN over 5 min, using 15 cm 3 μm columns (Thermo Fisher Scientific). The SRM measurements were performed on a TSQ Quantiva triple quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with a nano electrospray ion source (EASY-spray, Thermo Fisher Scientific). TSQ global parameters were: polarity: positive; spray voltage: 2000V; and transfer tube temperature: 325 °C. TSQ scan parameters were: quadrupole resolution: 0.7 FWHM in both Q1 and Q3; cycle time: 1.7 s; CID gas (argon) 2mTorr; source fragmentation: 10V; Chrom filter: 3 s; and 788 transitions were measured simultaneously. The 788 transition consist of 76 peptides, using 4–6 assays per peptide, with a complementary labeled peptide making up a total of 768 transitions and with 20 additional transitions for retention time calibration peptides. The human proteins monitored by SRM are based on previous publications (14). A full list of proteins, peptides, transitions selected for monitoring, charge, and collision energy is listed in supplemental Table S1.
Isotope Labeled Peptides for Absolute Quantification
Peptides selected for targeted analysis was selected for the enriched binding, shown in a previous investigation (14), to the surface of S. pyogenes. Stable isotope labeled peptides (SpikeTides, JPT, Berlin, Germany; and AQUA QuantPro, Thermo Fisher Scientific) was used for absolute quantification of peptides. The linear range of the isotope labeled peptides were determined including the detection limit and the coefficient of determination (R2), listed in supplemental Table S1. The R2 was calculated between the log10 intensity and log10 peptide concentration. The log10 transform was used because of the large concentration range of the analyzed peptides, which would otherwise cause the correlation to be dominated by the high concentration peptides. The maximum measured value was 1 pmol/injection, which is within the linear range and higher than any value in the data.
Experimental Design and Statistical Rationale
The SRM data was analyzed in the software Skyline v3.1.0.7382 (University of Washington, Seattle, WA). Only peptides with stable performance of the isotope labeled standard across all samples were analyzed. The Skyline result value used for analysis was; ratio to standard value, where the standard is the isotope labeled peptide. Statistical analysis of the data is carried out by eight biological replicates for each sample; statistical significance was calculated using a Wilcoxon rank sum test with a probability less than 1% (p < 0.01). The raw data available at the server: http://www.peptideatlas.org/PASS/PASS00972. All Skyline data is publicly available in Panorama (University of Washington, WA) (42), at the server: https://panoramaweb.org/labkey/surface_density_model.url. The S. pyogenes cell-counting model was based on 8 M1 strains, harvested in both stationary and exponential phase, and diluted to five concentrations. The flow cytometry derived bacterial counts were used as reference value, whereas the intensity of each Baccus peptide was used as input data. First, both flow cytometry counts and MS intensity were log10 transformed. For each Baccus peptide, we estimate the number of bacterial cells (Apep) as, log10(Apep) = Wpep * log10(Spep), where Spep is the SRM peptide intensity and Wpep is a peptide-specific response factor. The Wpep values were calculated using R 3.0.1 and the function “rlm” in R-package MASS (version 7.3–29) using default parameters. The final bacterial estimation A, is computed as the median of the measured peptides; log10(Apep) = median(log10(Apep1), log10(Apep2), …, log10(ApepN)) where N corresponds to the number of Baccus peptides successfully measured during the analysis.
Generating the Stoichiometric Surface Density Model
The surface model was generated using the program Maya (Autodesk, San Rafael, CA) with the plugin mMaya. The proteins are visualized as meshes, with the mesh quality set to high and the scale in all dimensions set to 0.1. The Protein Data Bank (PDB) identifiers of the proteins used in the model are listed in supplemental Table S3. As there is no full tertiary structure for any M-protein, we generated a model for the full-length protein. We assumed a uniform coiled-coil conformation over the length of the M1 homodimer; this allowed us to use the M1 fragment experimental crystal structure from PDB.id; 2XNX, (21) as a generic template for the length of the molecule. To constrain the modeling optimally, we repeatedly aligned the M and N chains of 2XNX (template) with the full-length M1-protein, UniProt id: Q99XV0, so that it partially overlapped with the proceeding alignment and minimized the sequence divergence between the sequence of the template and the query protein. Finally, we used the Rosetta multi-template modeling protocol (43) produce a total of 350 homology models. We selected models based on minimal Rosetta energy and an extended confirmation in agreement with Phillips et al. (44).
Electron Microscopy
Pelleted bacterial cells were resuspended in fixative (1.5% glutaraldehyde and 1.5% paraformaldehyde in 0.1 m Sörensen's phosphate buffer pH 7.2) and left at room temperature for 1h. After fixation, bacterial cells were rinsed in 0.1 m Sörensen's phosphate buffer pH 7.2, postfixed in 1% osmium tetraoxide (OsO4) for 1h, dehydrated using increasing concentration of acetone and embedded in Polarbed 812 (Polaron). Ultrathin sections (50–60 nm) were cut using a Leica EM UC7, and placed on thin-bar copper grids (Maxtaform H5) and stained with 4% uranyl acetate at 40 °C and 0.5% lead citrate at room temperature. The samples were examined with a Technai G2 Spirit electron microscope (FEI, Eindhoven, The Netherlands), operating at an excitation voltage of 100kV, and equipped with a Veleta (Olympus) 2kx2k CCD-camera. Size measuring of the bacteria was made using the Veleta software.
RESULTS
Determining Bacterial Concentration Using MS
The construction a stoichiometric surface density model of a host-pathogen interaction network in an ex vivo host environment, relies on reproducible protein quantification methods associated with high recovery. Here, we propose a targeted MS based workflow to capture and absolutely quantify the host proteins adhered to the bacterial surface along with the bacterial surface proteins and intracellular proteins. Absolute quantification provides the stoichiometric relationships between the surface proteins and the adhered host proteins, critical for constructing the model. The intracellular proteins are used to determine the bacterial cell numbers injected into the MS based on measured peptide intensities to calculate the protein copy numbers per cell (see Fig. 1A for a schematic overview of the experimental strategy).
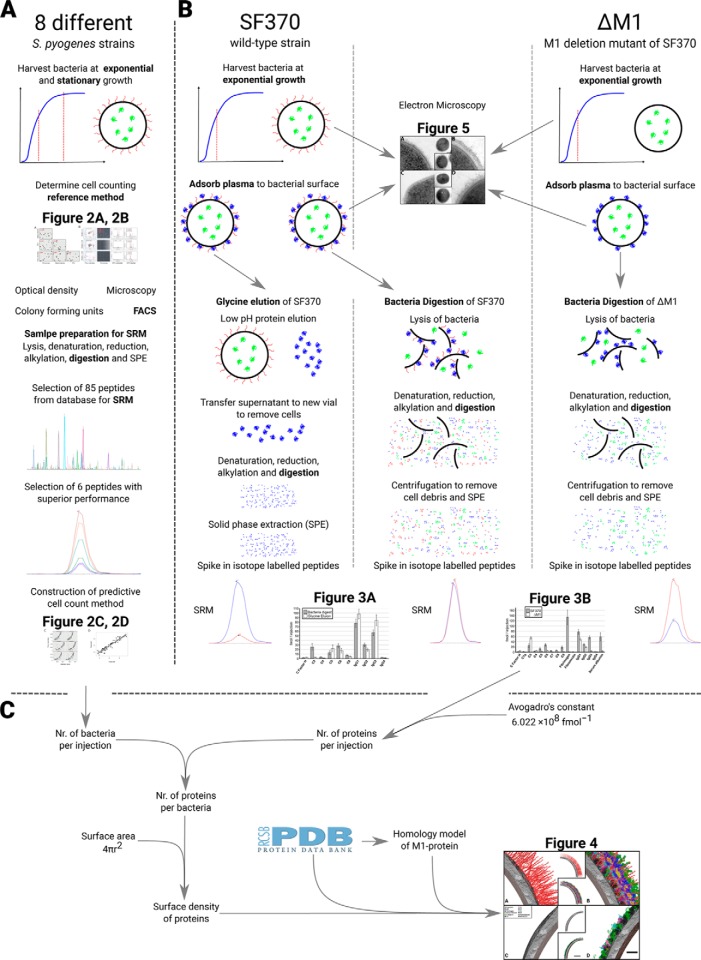
Fig. 1.
Schematic overview of the workflow. A detailed overview of the proposed workflow, from sample preparation to data handling and data analysis is shown. A, The selection of the Baccus peptides used in the bacterial cell counting method. B, Sample preparation methods and data analysis for the absolute quantification of proteins. The colors of proteins in the sample preparation: blue, plasma proteins; green, intracellular bacterial proteins; red, M1-protein, the main S. pyogenes surface proteins. C, Outline of the steps used for determining of the stoichiometric density model from the absolutely quantified data (B) and final construction of the surface density model.
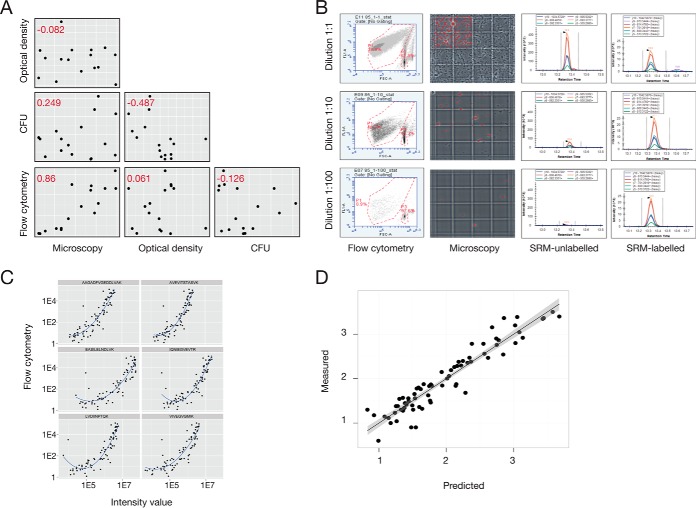
To determine bacterial cell numbers using measured peptide intensities requires a set of calibration samples with known bacterial cell numbers. We constructed a calibration set that was strain unspecific by serial dilution of eight different M1 S. pyogenes strain samples and growth phase unspecific by harvesting them in both exponential and stationary phase. Four widely used bacterial counting techniques were used to accurately determine the number of bacteria in these samples; optical density at 620 nm, colony forming units (CFU), light field microscopy with phase contrast using tryphan blue staining and flow cytometry with a bacterial DNA staining. We compared the bacterial concentrations in the diluted bacterial samples using the four techniques and found that optical density and CFU counts correlated poorly to the other methods (Pearson correlation coefficient from 0.249 to −0.487) (Fig. 2A). In contrast, the results from microscopy and flow cytometry techniques correlated reproducibly with each other (Pearson correlation coefficient of 0.86). Of the two reference methods, flow cytometry is easier to automate for a large number of samples and omits the manual interpretation associated with the microscopy technique. Hence, flow cytometry was selected as the reference bacterial counting technique.
Fig. 2.
Bacterial cell counting using SRM. Construction of a stoichiometry density models relies on measurement of the number of cells injected into the MS to determine copy numbers per cell. A calibration set of samples was constructed based on serial dilutions of eight different strains and the cells counted using four established counting methods followed by SRM analysis and refinement using isotope labeled reference peptides. A, Comparison of cell counts from four standard bacterial counting methods, where the red numbers are the respective Pearson's correlation coefficient. B, Visualization of the flow cytometry, microscopy and SRM data. C, Correlation of the intensity values between flow cytometry and intensity values obtained via SRM for each of the 6 Baccus peptides. D, Performance of predicted versus measured amount of bacterial cells, based on the median intensity of the Baccus peptides measured.
In the next step, we analyzed lysates from all samples in the calibration set using SRM. The construction of the bacterial counting method required the selection of tryptic peptides with high signal to noise ratios. As a starting point, we selected peptides based on their detectability in previously established large-scale MS repositories of S. pyogenes proteome data (27) and then ranked according to a previously published SRM-peptide assay score (27). Based on that score, 85 peptides with 6 transitions each were selected yielding a total of 510 transitions. Based on the peptide detectability, preservation and stability the 85 peptides was further refined to a final selection of 6 peptides, termed Baccus peptides (supplemental Table S1). All the diluted bacterial samples in the calibration set were spiked with corresponding isotope labeled reference peptides for the Baccus peptides, serving as internal standards, and re-analyzed using SRM. We observed a linear trend between the peptide intensities and the cell numbers previously determined with flow cytometry from the same samples (Fig. 2B and 2C). This linearity decreased at low amounts of bacteria, roughly 102 bacteria/μl, representing the limit of detection of the techniques. The mean intensity values of the measured peptides were fitted across a linear regression of known intensities and correspond to bacterial concentrations (Fig. 2D). The correlation between Baccus peptide intensities and the number of cells makes it possible to estimate bacterial cells injected into the MS from S. pyogenes derived samples.
Optimization of Samples Preparation Protocol for Absolute Protein Quantification
The standard method for recovering surface bound proteins from bacteria, referred to as glycine elution, relies on low pH for protein elution followed by removal of the intact bacteria. The glycine elution protocol has however, the disadvantage of only recovering surface bound host proteins, whereas covalently attached host proteins, bacterial surface proteins and the intracellular bacterial proteins are lost. We modified the elution protocol to achieve the highest level of recovery for all surface attached host proteins together with all the bacterial proteins. In the modified protocol, referred to as bacterial digest, cells with adhered human plasma proteins are directly lysed followed by denaturation, reduction, alkylation and digestion (Fig. 1B). The modified protocol retains both surface and intracellular bacterial proteins as well as all attached host proteins, including any potentially covalently bound host proteins. To determine the recovery of human proteins, we adhered plasma proteins to the S. pyogenes surface and prepared samples in eight biological replicates in three different sets; glycine elution (traditional protocol) of the wild-type SF370 strain, the bacterial digest protocol of SF370, and the bacterial digest protocol of a M1-protein deficient isogenic mutant strain (ΔM1) (Fig. 1B). All sample sets were analyzed with SRM using a single transition list (supplemental Table S1). The host proteins included in the transition list were selected based on previously published MS analysis of adhered plasma proteins to the surface of S. pyogenes (14). A particular focus was placed on proteins that were enriched on the bacterial surface compared with their normal levels in healthy blood plasma as these proteins are likely to represent specific protein interactions to the S. pyogenes surface (14). In addition, several known surface associated virulence factors and intracellular bacterial proteins used for counting the number of bacterial cells were included in the transition list. The final transition list contained 1–3 proteotypic peptides per protein and 4–6 fragment ions per peptide, and comprised of 788 transitions associated to 76 peptides and 56 proteins (supplemental Table S1). All samples were spiked with an absolutely quantified stable isotope labeled peptide standard of all targeted 76 selected peptides. Only peptides with consistent intensity between samples were considered for further analysis, because the spiked amount of a specific labeled peptide was the same for all samples.
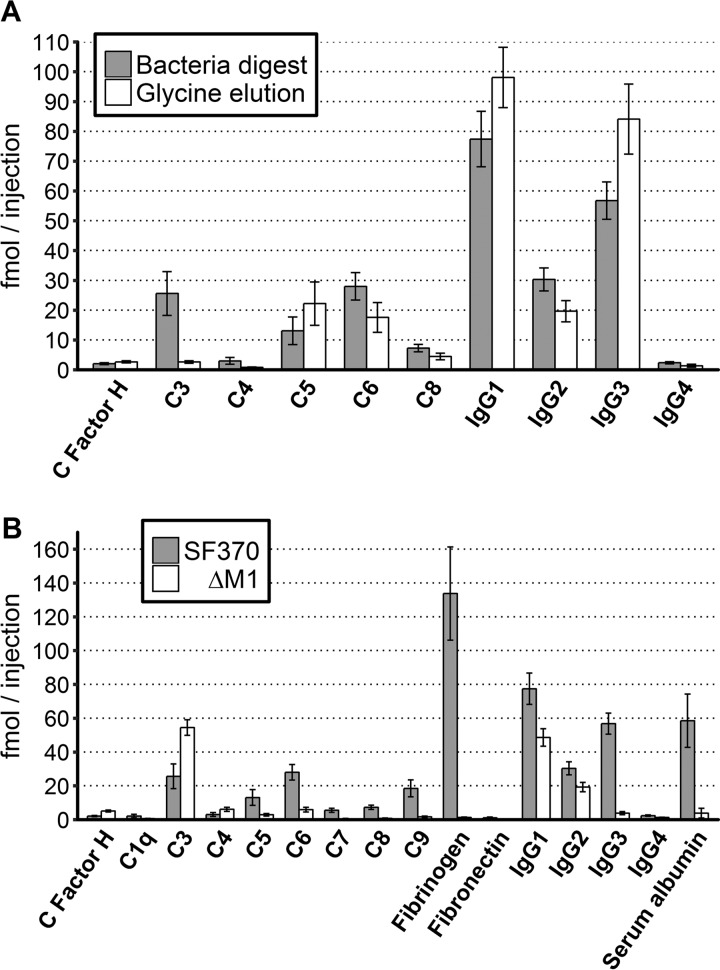
Overall, the bacterial digest protocol performs similar to the glycine elution protocol for the majority of proteins even though the bacterial digest protocol results in a considerably more complex peptide mixture (supplemental Table S2). However, the bacterial digest protocol also improves significantly (p < 0.01) the recovery of six adhered human proteins, such as the potentially covalently attached complement component 3 (C3) (45–47) and C4, with a 9.89- and 3.69-fold increase respectively (Fig. 3A). In contrast, there were four statistically significant proteins with a minor decrease, such as IgG3 and C5, with a 1.48- and 1.70-fold increase in the glycine elution protocol compared with the bacterial digest protocol. We conclude that there are relatively small differences in the recovery between the two protocols. Furthermore, the bacterial digest protocol can improve the recovery of covalently attached host proteins and importantly recovers the intracellular and surface attached bacterial proteins as shown in supplemental Table S2.
Fig. 3.
Significant differences in protein abundance levels in the SRM data. The absolute quantification of the plasma proteins binding to the surface of S. pyogenes, measured using SRM. A, Two methods were used to elute surface bound human plasma proteins from the SF370 strain; bacteria digest (gray bars) and glycine elution (white bars). B, Wild type strain SF370 (gray bars) and an M1-deletion mutant ΔM1 (white bars). Only human proteins with significant difference (p > 0.01) between the two elution methods are shown in (A) or the two strains shown in (B) are included in the figure. The y-axis displays the absolute quantified value of each protein in femtomole per injection into the MS. The error bars represent the standard deviation of the eight replicates.
M1-protein Dependent Interactions with Human Plasma Proteins
The M-protein is the most important virulence factor attached to the surface of S. pyogenes, responsible for the binding of several well-characterized plasma proteins (12, 21–26). To determine M1-protein dependent protein interactions at the surface of S. pyogenes, we used SRM to analyze the eight biological replicates of the bacterial digest protocol for the wild-type strain, SF370 and the M1-protein deficient isogenic mutant ΔM1 (38). The SRM results confirm that the most abundant adhered plasma proteins on the wild-type strain are fibrinogens (134 fmol/injection), IgG subclasses 1–3 (77–30 fmol/injection), and serum albumin (58 fmol/injection), followed by complement protein C3 and members of the complement system membrane attack complex (C5–9) (supplemental Table S2). The protein binding to the surface of the ΔM1 strain was drastically reduced compared with the wild-type strain, as shown in Fig. 3B, where human proteins with statistically significant (p < 0.01) differences are shown. There was as expected a significant decrease of the known M1-protein binding human plasma proteins such as fibrinogen, fibronectin, all subclasses of IgG and serum albumin (Fig. 3B). For fibrinogen and albumin, the differences were drastic with a more than 100- and 15-fold reduced binding to the ΔM1 strain compared with the wild-type SF370, whereas the differences in IgG subclasses 1, 2, and 4 binding were more moderate with an increased binding to the wild-type strain of 1.59, 1.58, and 1.85-fold, respectively. This can be expected as antibodies from these IgG subclasses can be specifically bound to other surface proteins than the M1-protein. In contrast, IgG3 binds 14.8-fold less to the surface of the wild-type strain, indicating a stronger correlation between the M1-protein and IgG3 subclass antibodies. In addition, we observe several additional proteins that bind in decreased amounts to the surface of the ΔM1 strain, such as all proteins of the complement membrane attack complex (C5–9). Only three proteins are found in higher amounts on the mutant strain, ΔM1; complement factor H and complement protein C3 and C4, all with a 2.48- to 2.07-fold increase. Complement factor H and C4 are found at low amounts at the surface of the wild-type strain as well as the mutant strain (<6.1fmol/injection), whereas complement protein C3 is found at relatively high amounts on the surface of both strains (>25fmol/injection).
Construction of a Stoichiometric Surface Density Model of S. pyogenes
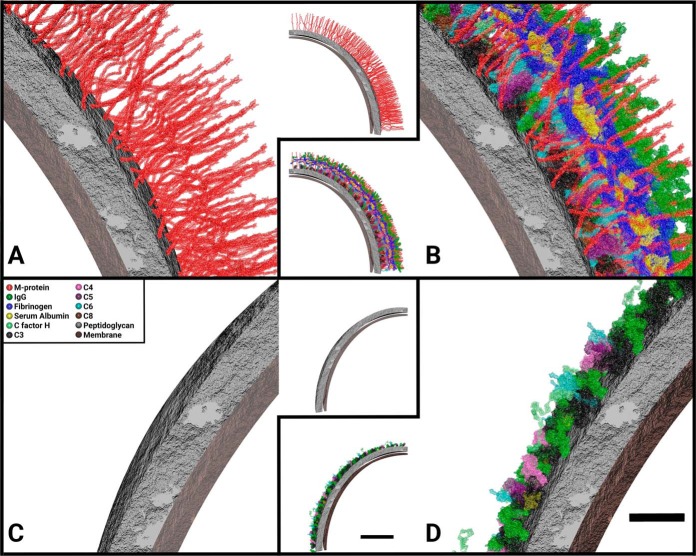
In the final step of the workflow (Fig. 1C), we used the absolute quantified S. pyogenes surface proteins, their stoichiometric relationship to interacting host plasma proteins and the proteins tertiary structure obtained from the Protein Data Bank (PDB). In addition, electron micrographs for validation purposes were used to construct a host-pathogen interaction model, referred to as a stoichiometric surface density model, as shown in Fig. 4. The model was created in the 3D visualization program Autodesk Maya. Known and proposed host-pathogen interactions for the measured proteins were used as binding sites of host proteins to bacterial surface (supplemental Table S3). In this model, we estimated the thickness of S. pyogenes peptidoglycan layer to 20 nm as previously shown (48). Furthermore, we assumed even distribution of the M1-protein in the peptidoglycan layer, based on the EM micrographs in Fig. 5, and from previously published work (12, 44, 49). In our surface model, we only include proteins with an available atomic model deposited in the PDB. The M1-protein is the only surface bound streptococcal protein for which part of the structure has been determined. As the deposited atomic model for the M1-protein only encompasses one third of the full-length protein, we use a computational model of the full-length protein based on homology modeling to generate an accurate surface model. Consequently, the surface of S. pyogenes is simplified in that it only has one surface protein, the M1-protein and hence the ΔM1 strain without any plasma bound to surface has a smooth appearance (Fig. 4C). The stoichiometry of protein and bacteria was calculated using the absolute quantified protein values, femtomole per injection, multiplied with Avogadro's constant to gain the number of proteins per injection, which was transformed to copy numbers per cell based on the cell numbers obtained via measuring the Baccus peptides as outlined above (Fig. 1A). To measure the protein density on the bacterial surface we measured the average diameter of the bacterium to 560 nm in the electron micrographs, and assumed that streptococci are spherical, thus the surface area of a bacterium is 9.85e5 nm2. For representation purposes, 1/32 of the streptococcal surface is shown in Fig. 4.
Fig. 4.
Molecular anatomy of bacterial surfaces using SRM data. Appearance of the stoichiometric surface density models of S. pyogenes with or without plasma proteins adsorbed to the surface: A, The wild-type strain SF370; B, SF370 with plasma adsorbed to surface; C, The SF370 M1-deletion mutant ΔM1; D, ΔM1 with plasma adsorbed to surface. A 1/32 piece of the surface, 30800 nm2, 3% of total bacterial area, is represented in the figures (inserts). In order to visualize the 3D image a close-up figure for each of the inserts is provided, depicting a smaller part of the image. A legend with the colors of the visualized proteins can be found in C and is consistent throughout the other panels. Scale bars, 20 nm (close-ups) and 100 nm (inserts).
Fig. 5.
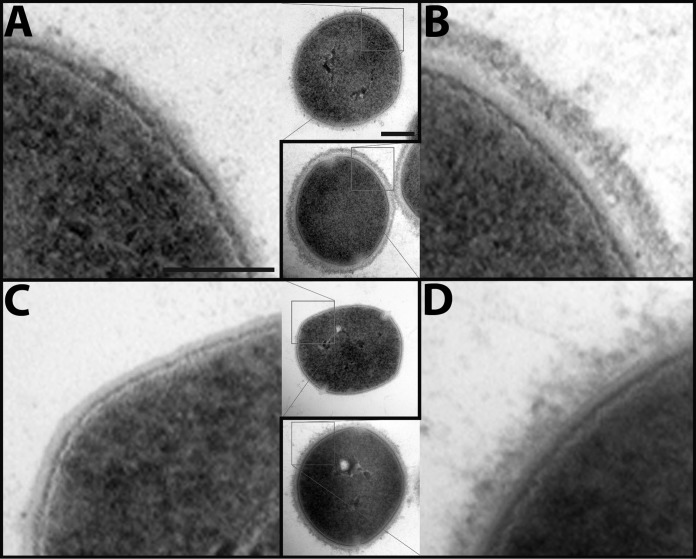
Electron micrographs of the S. pyogenes surface with and without adsorbed human plasma proteins. S. pyogenes cells from the same samples used for SRM were fixed and protein interaction networks is shown using electron microscopy: A, The wild-type strain SF370; B, SF370 with plasma adsorbed to surface; C, The SF370 M1 deletion mutant ΔM1; D, ΔM1 with plasma adsorbed to surface. The electron micrographs show a close-up of the bacterial surface, whereas the inserts depict one complete bacterium. Scale bars, 100 nm (close-ups) and 200 nm (inserts).
Validation of the Models Using Electron Microscopy
The distinct differences in the protein density between the wild-type strain SF370 and the mutant ΔM1 were validated using electron micrographs of the surface of both strains with and without plasma. The micrographs from the wild-type strains reveal the protruded M1-protein (44) extending from the cell wall (Fig. 5A), which is absent on the surface of ΔM1 (Fig. 5C). In addition, there is a clear change in the appearance of the bacterial surface when plasma proteins are adsorbed to SF370 (Fig. 5B) and ΔM1 (Fig. 5D). There is a dense outer layer of human plasma proteins mostly consisting of proteins such as fibrinogen, IgG and serum albumin surrounding the wild-type strain SF370 confirming the model (Fig. 4B). The outer layer of the ΔM1 strain has a distinctly different, less compact appearance, because of removal of the M1-protein and the almost complete removal of the M1-binding proteins. These electron micrographs support the overall network topology shown in the model. In addition, previously published work (12) can validate that the distribution of the M1-protein on the bacterial surface demonstrating the accuracy of this SRM-based bacterial cell counting method. Collectively, our MS measurements provide absolute protein quantities in combination with thin-section electron micrographs show that known S. pyogenes virulence factors and attached human plasma proteins form a dense structure around the bacterium and that this structure is dependent on the presence of the M1-protein. This classical view with electron micrographs of the bacterial surface providing relatively limited information of protein localization is further enhanced by the introduction of computational modeling using the precise data produced by SRM and quantified isotope labeled peptides.
In conclusion, the results presented in this work demonstrate a first stoichiometric surface density model of host-pathogen interactions using data from absolute protein quantification. The model captures the large differences in network topology between the surface structure of the wild-type and ΔM1 strain revealing a dense and a highly organized protein interaction network between S. pyogenes and human plasma proteins. Further improvement of this model, with new emerging data, will further facilitate the understanding the interactions between the host and the pathogen, which is essential for understanding the progression of S. pyogenes from superficial to severe disease.
DISCUSSION
In this article, we present a novel strategy for constructing a stoichiometric surface density model of a pathogen in a relevant host location by using stable isotope labeled peptides and SRM in combination with electron microscopy and advanced modeling. The model is based on S. pyogenes as a model system for the pathogen and human blood plasma as a model system for the host, to simulate a host-like ex vivo environment of sepsis. The samples were measured using SRM because of the high dynamic range, specificity and sensitivity associated with this technique. To further increase the analytical sensitivity, we used quantified and isotope labeled peptides for absolute quantification of proteins. We show that the bacteria digest protocol performs similarly to the standard protocol, glycine elution, for eluting the surface bound host proteins. This modified digestion method is specifically developed for bottom-up MS, as both interacting human proteins along with surface bound bacterial proteins are cleaved off into peptides using trypsin, at the same time as intracellular bacterial proteins is cleaved into peptides. In addition to the host proteins, the bacteria digest protocol provides all the bacterial surface and intracellular proteins required for constructing a stoichiometric surface density model of the host-pathogen complex (listed in supplemental Table S2).
Our study demonstrates that the method for capturing the intracellular and surface bound bacterial proteins along with the interacting host proteins enables measuring of the host interaction profile in the tested model system. As presented in Fig. 3A, one of the major improvements with bacteria digestion in contrast to the standard protocol, glycine elution, is the 9.89-fold increase of recovered complement component 3 (C3). It is likely that C3 binds to the streptococcal surface so tightly that the protein does not elute when using low pH and harsher methods are required to capture it. C3 is known to upon activation by C3 convertase bind covalently to saccharides on the cell wall of microorganisms (45–47), which would explain why C3 is not eluted by using low pH. C3 is also one of three proteins found to bind to higher extent on ΔM1 (the M1-deletion mutant) compared with SF370 (the wild-type strain), along with complement factor H and component 4. The increase in factor H on the surface of ΔM1 is contradictory to the studies stating that M1-protein and complement factor H binds to each other (50, 51). This increase of complement factor H could be explained by another observation made in literature, which states that complement factor H can bind to C3 (52, 53). This is also the case for C4 (54). Hence, it is possible that the increase of C3 results in increased binding of factor H and C4. The increase in C3 can potentially be further explained by the increase of surface accessibility because of lack of M1-protein and M-protein associated proteins such as fibrinogen (55), where these proteins may sterically hinder binding, as indicated in Fig. 5A and 5C. There are two advantages with the implementation of the bacterial digestion method. First, the method facilitates detection of the change in binding for C3; such change would be difficult or even impossible to detect using the standard method with low pH, because of the covalent binding of C3. Second, the method retains bacterial intracellular and surface associated proteins that are important for the construction of the model.
Even if C3 is found at higher concentration on the M1-protein deficient strain ΔM1, there are several important plasma proteins that are found at higher concentrations on the surface of the wild-type strain SF370. M1-protein is the most abundant surface bound virulence-factor of S. pyogenes (56). The amount of M1-protein on the surface of the wild-type strain versus the mutant is displayed in Fig. 4A and 4C, respectively, and in supplemental Table S3. Using absolute quantification and bacterial counting, we estimate that the M1-protein, a dimer, has over 8700 dimeric copies per bacterium on average, which is within the same range as have been detected before (57). Previously defined M1-protein interactions such as fibrinogen (21, 24, 25) and serum albumin (22, 23) are found to bind SF370 to a higher extent, this with higher quantitative accuracy than previously used. The results show that there is a ratio of 0.66 and 0.29, respectively, between fibrinogen and serum albumin, and the M1-protein on the wild-type strain, indicating that at least two thirds of the M1-proteins are associated with fibrinogen and/or serum albumin. A slightly higher ratio of 0.83 is observed for all subclasses of IgG. In contrast to fibrinogen and serum albumin, which are almost completely absent on the surface ΔM1, the reduction of IgG is considerably less dramatic for the mutant strain. The higher binding of all subclasses of IgG to the wild-type strain is expected, as most individuals have anti-M1 specific antibodies present in the plasma (12). In addition, the M1-protein harbors an IgG Fc binding domain (58). Consequently, the stoichiometric differences observed in IgG levels between the wild-type and the ΔM1 strain can be explained by antibodies associated with the M1-protein via a combination of Fab and Fc binding. The larger differences observed of IgG3 binding; 14.8 times more to SF370 than ΔM1, whereas the other subclasses bind only 1.58 to 1.86 times more, is interesting. These results imply that IgG3 does seemingly have a higher likelihood of binding epitopes on M1-protein than the other subclasses, as no differences in Fc binding affinity has been reported for the different subclasses (12). This phenomenon was previously observed (12), but the reason is not yet fully understood and it requires additional experimental work.
One of the conclusions that can be drawn from the data presented in this article is that a stoichiometric surface density model is a promising strategy to summarize and visualize all the data and that it represents in an intuitive way to display MS data (59), and for the first time with absolutely quantified data. It is interesting to note that the protein network is highly dense and highly organized around the M1-protein. Fig. 4 and the supplemental Figs. S1–13 illustrate how the M1-protein enables the subdivision of interacting proteins into layers based on the location of the protein binding interfaces. However, when designing the stoichiometric surface density model there were several parameters that we were required to simplify. Computational capacity has to be considered because this is correlated to the size models meshes; hence, 1/32 of the bacterium was used for model. Only proteins with known tertiary structure can be used in the model, this excludes proteins like the complement C9 from the model, even though it binds to the S. pyogenes surface. This is also the case for most streptococcal surface proteins; hence, we simplify the model of the streptococci so that it only includes the M1-protein on the surface. The binding sites of host proteins to the streptococcal surface is approximated from previous knowledge (21, 26), this could however be improved in the future and used to update the model. One example on how to find binding sites between host and pathogen proteins is cross-linking MS (60). Even though there are many different ways to improve the correctness this model, we are able to reproduce most of the features observed in the electron micrographs, for example, the tight layer on the surface of SF370 in plasma. Overall, we note that the modeling of the plasma interaction network for the wild-type strain resembles the structure shown in the electron micrographs. In contrast, the density without plasma (Fig. 5A) does not appears as the modeled surface (Fig. 4A), this is probably because of the thin section modeled or the resolution of the electron micrographs and the lack of S. pyogenes surface proteins other than M1-protein. However, the distribution of the M1-protein on the S. pyogenes surface is supported by previously published work (12) where higher magnification in the electron micrographs reveal a M1 distribution similar distribution in the model. In addition, visualizing a stoichiometric surface density model using Autodesk Maya provides the possibility to group the molecules, and to visualize the binding pattern of separate interactions (supplemental Figs. S1–13). The visualization also demonstrates that the absolute quantification in combination with the bacterial counting gives reliable values in proteins per bacterium, because steric hindrance is not an issue and that the proteins form a dense layer similar to the micrographs.
The current version of the stoichiometric surface density model of S. pyogenes and human blood plasma represents a first draft of a host-pathogen interaction network model. Future work is required to improve the accuracy of the model when. These improvements include for example more precise knowledge regarding binding sites of proteins to the bacterial surface and structural characterization of additional proteins, which to date have an unknown tertiary structure. The method presented here is not exclusive for the model system of S. pyogenes and human blood plasma. The method could possibly be extended to organelles and cytoplasm; virus and saliva or human cells and plasma, the list can be made long of biologically interesting units that interacts with its surrounding with an unknown surface interaction density model. However, if a new model of host and pathogen was selected other protein interactions occur and new peptides must be selected for SRM analysis. Furthermore, analysis of a new pathogen or organism (other than S. pyogenes) would demand a new method to estimate the cell count, preferably by MS. We believe that the analytical accuracy of the analyzed proteins along with the stoichiometric density model provided in this article outlines a novel attempt to visualize the complexity of host-pathogen interactions to enhance the understanding of these complex interactions involved in the development of severe and invasive infectious diseases.
DATA AVAILABILITY
The raw mass spectrometry data is available at the server: http://www.peptideatlas.org/PASS/PASS00972. All Skyline data is publicly available at Panorama (University of Washington, WA), at the server: https://panoramaweb.org/labkey/surface_density_model.url.
Supplementary Material
Footnotes
Author contribution: K. S., O. K. and J. M. conceived and designed the experiments. K. S., O. K. and L. H. performed the experiments. K. S. and O. K. performed the MS-analysis and bioinformatics analysis. K. S., O. K., L. H., J. T. and J. M. analysed the data. O. K. and J. T. created the SRM cell counting method. K. S. constructed the surface density model. L. M. did the homology model of the M1-protein. K. S., O. K., L. H., J. T., L. M and J. M. wrote the paper.
* This work was supported in part by the Swedish Research Council (project 621-2012-3559), the Swedish Foundation for Strategic Research (grant FFL4), the Crafoord Foundation (grant 20100892), Stiftelsen Olle Engkvist Byggmästare, the Wallenberg Academy Fellow KAW (2012.0178), European research council starting grant (ERC-2012-StG-309831) and the Medical Faculty, Lund University. Lund University Bioimaging Center (LBIC), Lund University is gratefully acknowledged for providing experimental resources. L.M. was funded by SNSF project grant 200021_16018. There is no conflict of interests within this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- IgG
- immunoglobulin gamma
- LC
- liquid chromatography
- SRM
- selected reaction monitoring
- CFU
- colony forming units
- MS
- Mass spectrometry.
REFERENCES
- 1. Martin G. S., Mannino D. M., Eaton S., and Moss M. (2003) The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 2. Alberti C., Brun-Buisson C., Goodman S. V., Guidici D., Granton J., Moreno R., Smithies M., Thomas O., Artigas A., Le Gall J. R., and European Sepsis Group (2003) Influence of systemic inflammatory response syndrome and sepsis on outcome of critically ill infected patients. Am. J. Respiratory Crit. Care Med. 168, 77–84 [DOI] [PubMed] [Google Scholar]
- 3. Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., and Pinsky M. R. (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 4. Jawad I., Luksic I., and Rafnsson S. B. (2012) Assessing available information on the burden of sepsis: global estimates of incidence, prevalence and mortality. J. Glob. Health 2, 010404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davis J. S., He V., Anstey N. M., and Condon J. R. (2014) Long term outcomes following hospital admission for sepsis using relative survival analysis: a prospective cohort study of 1,092 patients with 5 year follow up. PloS One 9, e112224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dick A., Liu H., Zwanziger J., Perencevich E., Furuya E. Y., Larson E., Pogorzelska-Maziarz M., and Stone P. W. (2012) Long-term survival and healthcare utilization outcomes attributable to sepsis and pneumonia. BMC Health Serv. Res. 12, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heyland D. K., Hopman W., Coo H., Tranmer J., and McColl M. A. (2000) Long-term health-related quality of life in survivors of sepsis. Short Form 36: a valid and reliable measure of health-related quality of life. Crit. Care Med. 28, 3599–3605 [DOI] [PubMed] [Google Scholar]
- 8. Seymour C. W., Rea T. D., Kahn J. M., Walkey A. J., Yealy D. M., and Angus D. C. (2012) Severe sepsis in pre-hospital emergency care: analysis of incidence, care, and outcome. Am. J. Respiratory Crit. Care Med. 186, 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies J., and Davies D. (2010) Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hawkey P. M., and Jones A. M. (2009) The changing epidemiology of resistance. The J. Antimicrob. Chemother. 64, Si3–Si10 [DOI] [PubMed] [Google Scholar]
- 11. de Kraker M. E., Jarlier V., Monen J. C., Heuer O. E., van de Sande N., and Grundmann H. (2013) The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infection 19, 860–868 [DOI] [PubMed] [Google Scholar]
- 12. Nordenfelt P., Waldemarson S., Linder A., Mörgelin M., Karlsson C., Malmström J., and Björck L. (2012) Antibody orientation at bacterial surfaces is related to invasive infection. J. Exp. Med. 209, 2367–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carapetis J. R., Steer A. C., Mulholland E. K., and Weber M. (2005) The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 14. Sjöholm K., Karlsson C., Linder A., and Malmström J. (2014) A comprehensive analysis of the Streptococcus pyogenes and human plasma protein interaction network. Mol. bioSystems 10, 1698–1708 [DOI] [PubMed] [Google Scholar]
- 15. Caparon M. G., Stephens D. S., Olsen A., and Scott J. R. (1991) Role of M protein in adherence of group A streptococci. Infection Immunity 59, 1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cole J. N., Barnett T. C., Nizet V., and Walker M. J. (2011) Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiology 9, 724–736 [DOI] [PubMed] [Google Scholar]
- 17. Beall B., Facklam R., and Thompson T. (1996) Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34, 953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cunningham M. W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steer A. C., Law I., Matatolu L., Beall B. W., and Carapetis J. R. (2009) Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 9, 611–616 [DOI] [PubMed] [Google Scholar]
- 20. McNamara C., Zinkernagel A. S., Macheboeuf P., Cunningham M. W., Nizet V., and Ghosh P. (2008) Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science 319, 1405–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Macheboeuf P., Buffalo C., Fu C. Y., Zinkernagel A. S., Cole J. N., Johnson J. E., Nizet V., and Ghosh P. (2011) Streptococcal M1 protein constructs a pathological host fibrinogen network. Nature 472, 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malmström J., Karlsson C., Nordenfelt P., Ossola R., Weisser H., Quandt A., Hansson K., Aebersold R., Malmström L., and Björck L. (2012) Streptococcus pyogenes in human plasma: adaptive mechanisms analyzed by mass spectrometry-based proteomics. J. Biol. Chem. 287, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kronvall G., Simmons A., Myhre E. B., and Jonsson S. (1979) Specific absorption of human serum albumin, immunoglobulin A, and immunoglobulin G with selected strains of group A and G streptococci. Infection Immunity 25, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kantor F. S. (1965) Fibrinogen Precipitation by Streptococcal M Protein. I. Identity of the Reactants, and Stoichiometry of the Reaction. J. Exp. Med. 121, 849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanford B. A., Davison V. E., and Ramsay M. A. (1982) Fibrinogen-mediated adherence of group A Streptococcus to influenza A virus-infected cell cultures. Infection Immunity 38, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Åkesson P., Schmidt K. H., Cooney J., and Björck L. (1994) M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem. J. 300, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlsson C., Malmström L., Aebersold R., and Malmström J. (2012) Proteome-wide selected reaction monitoring assays for the human pathogen Streptococcus pyogenes. Nat. Commun. 3, 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., and White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Picotti P., Bodenmiller B., Mueller L. N., Domon B., and Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson L., and Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 31. Aebersold R., and Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 32. Yates J. R., Ruse C. I., and Nakorchevsky A. (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11, 49–79 [DOI] [PubMed] [Google Scholar]
- 33. Lange V., Picotti P., Domon B., and Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Picotti P., and Aebersold R. (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 35. Gerber S. A., Rush J., Stemman O., Kirschner M. W., and Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., and Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuster B., Schirle M., Mallick P., and Aebersold R. (2005) Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 6, 577–583 [DOI] [PubMed] [Google Scholar]
- 38. Abbot E. L., Smith W. D., Siou G. P., Chiriboga C., Smith R. J., Wilson J. A., Hirst B. H., and Kehoe M. A. (2007) Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 9, 1822–1833 [DOI] [PubMed] [Google Scholar]
- 39. Malmstrom L., Bakochi A., Svensson G., Kilsgard O., Lantz H., Petersson A. C., Hauri S., Karlsson C., and Malmstrom J. (2015) Quantitative proteogenomics of human pathogens using DIA-MS. J. Proteomics 129, 98–107 [DOI] [PubMed] [Google Scholar]
- 40. Janulczyk R., Iannelli F., Sjoholm A. G., Pozzi G., and Bjorck L. (2000) Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J. Biol. Chem. 275, 37257–37263 [DOI] [PubMed] [Google Scholar]
- 41. Pandiripally V., Gregory E., and Cue D. (2002) Acquisition of regulators of complement activation by Streptococcus pyogenes serotype M1. Infection Immunity 70, 6206–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sharma V., Eckels J., Taylor G. K., Shulman N. J., Stergachis A. B., Joyner S. A., Yan P., Whiteaker J. R., Halusa G. N., Schilling B., Gibson B. W., Colangelo C. M., Paulovich A. G., Carr S. A., Jaffe J. D., MacCoss M. J., and MacLean B. (2014) Panorama: a targeted proteomics knowledge base. J. Proteome Res. 13, 4205–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song Y., DiMaio F., Wang R. Y., Kim D., Miles C., Brunette T., Thompson J., and Baker D. (2013) High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phillips G. N. Jr., Flicker P. F., Cohen C., Manjula B. N., and Fischetti V. A. (1981) Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc. Natl. Acad. Sci. U.S.A. 78, 4689–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Law S. K., Lichtenberg N. A., and Levine R. P. (1979) Evidence for an ester linkage between the labile binding site of C3b and receptive surfaces. J. Immunol. 123, 1388–1394 [PubMed] [Google Scholar]
- 46. Sim R. B., Twose T. M., Paterson D. S., and Sim E. (1981) The covalent-binding reaction of complement component C3. Biochem. J. 193, 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gadjeva M., Dodds A. W., Taniguchi-Sidle A., Willis A. C., Isenman D. E., and Law S. K. (1998) The covalent binding reaction of complement component C3. J. Immunol. 161, 985–990 [PubMed] [Google Scholar]
- 48. Vollmer W., Blanot D., and de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 49. Fischetti V. A. (2016) M Protein and Other Surface Proteins on Streptococci. In: Ferretti J. J., Stevens D. L., and Fischetti V. A., eds. Streptococcus pyogenes: Basic Biology to Clinical Manifestations, The University of Oklahoma Health Sciences Center, Oklahoma City: [PubMed] [Google Scholar]
- 50. Horstmann R. D., Sievertsen H. J., Knobloch J., and Fischetti V. A. (1988) Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. U.S.A. 85, 1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blackmore T. K., Fischetti V. A., Sadlon T. A., Ward H. M., and Gordon D. L. (1998) M protein of the group A Streptococcus binds to the seventh short consensus repeat of human complement factor H. Infection Immunity 66, 1427–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jokiranta T. S., Westin J., Nilsson U. R., Nilsson B., Hellwage J., Lofas S., Gordon D. L., Ekdahl K. N., and Meri S. (2001) Complement C3b interactions studied with surface plasmon resonance technique. Int. Immunopharmacol. 1, 495–506 [DOI] [PubMed] [Google Scholar]
- 53. Soames C. J., and Sim R. B. (1997) Interactions between human complement components factor H, factor I and C3b. Biochem. J. 326, 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muller-Eberhard H. J., Polley M. J., and Calcott M. A. (1967) Formation and functional significance of a molecular complex derived from the second and the fourth component of human complement. J. Exp. Med. 125, 359–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carlsson F., Sandin C., and Lindahl G. (2005) Human fibrinogen bound to Streptococcus pyogenes M protein inhibits complement deposition via the classical pathway. Mol. Microbiol. 56, 28–39 [DOI] [PubMed] [Google Scholar]
- 56. Fischetti V. A. (1989) Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2, 285–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Whitnack E., and Beachey E. H. (1985) Biochemical and biological properties of the binding of human fibrinogen to M protein in group A streptococci. J. Bacteriol. 164, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Heath D. G., and Cleary P. P. (1989) Fc-receptor and M-protein genes of group A streptococci are products of gene duplication. Proc. Natl. Acad. Sci. U.S.A. 86, 4741–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takamori S., Holt M., Stenius K., Lemke E. A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P., Müller S. A., Rammner B., Gräter F., Hub J. S., De Groot B. L., Mieskes G., Moriyama Y., Klingauf J., Grubmüller H., Heuser J., Wieland F., and Jahn R. (2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 60. Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmström L., and Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry data is available at the server: http://www.peptideatlas.org/PASS/PASS00972. All Skyline data is publicly available at Panorama (University of Washington, WA), at the server: https://panoramaweb.org/labkey/surface_density_model.url.