Abstract
Context:
The underlying molecular alterations causing sporadic parathyroid adenomas that drive primary hyperparathyroidism have not been thoroughly defined.
Objective:
The aim of the study was to investigate the occurrence of somatic mutations driving tumor formation and progression in sporadic parathyroid adenoma using whole-exome sequencing.
Design:
Eight matched tumor-constitutional DNA pairs from patients with sporadic parathyroid adenomas underwent whole-exome capture and high-throughput sequencing. Selected genes were analyzed for mutations in an additional 185 parathyroid adenomas.
Results:
Four of eight tumors displayed a frame shift deletion or nonsense mutation in MEN1, which was accompanied by loss of heterozygosity of the remaining wild-type allele. No other mutated genes were shared among the eight tumors. One tumor harbored a Y641N mutation of the histone methyltransferase EZH2 gene, previously linked to myeloid and lymphoid malignancy formation. Targeted sequencing in the additional 185 parathyroid adenomas revealed a high rate of MEN1 mutations (35%). Furthermore, this targeted sequencing identified an additional parathyroid adenoma that contained the identical, somatic EZH2 mutation that was found by exome sequencing.
Conclusion:
This study confirms the frequent role of the loss of heterozygosity of chromosome 11 and MEN1 gene alterations in sporadic parathyroid adenomas and implicates a previously unassociated methyltransferase gene, EZH2, in endocrine tumorigenesis.
Parathyroid tumors causing primary hyperparathyroidism (pHPT) are common, occurring in 2.1% of postmenopausal women (1). Sporadic (nonfamilial) pHPT occurs predominantly due to a single hyperfunctioning parathyroid adenoma (85%) but can be due to multiglandular (including four gland hyperplasia) disease in 10–15% of cases (2); rarely it is due to carcinoma (<1%) (3).
Allelic loss [loss of heterozygosity (LOH)] of chromosomal loci may identify tumor suppressor genes in neoplasia. LOH at the multiple endocrine neoplasia type 1 (MEN1) locus on chromosome band 11q13 has been demonstrated in approximately 25–40% of sporadic parathyroid adenomas. Somatic mutations of the MEN1 gene are found in 12–21% of adenomas, or about 50% of those tumors with LOH at 11q13 (4). These findings indicate that mutational aberrations in MEN1 contribute to parathyroid tumorigenesis but also raise the possibility that 11q13 may harbor an additional parathyroid tumor suppressor gene. Apart from the loss of the MEN1 locus, comprehensive LOH and comparative genomic hybridization studies of parathyroid adenomas have identified locations for several other candidate tumor suppressor genes such as 1p, 1q, 6q, 9p, 11p, and 15q (5). Although inactivating somatic mutations of HRPT2/CDC73 have been identified in a subset of mainly malignant parathyroid carcinomas, they are not commonly altered in benign parathyroid adenomas (6). Likewise, germline mutations in CASR have been associated with familial forms of pHPT, but the gene has not been found to be somatically mutated in sporadic forms of the disease (7). Furthermore, loss of the well-characterized tumor suppressor genes p53 and RB do not appear to contribute to the development of parathyroid adenomas (8, 9). To date, no gene other than MEN1 has been proven by somatic mutation to be frequently altered in parathyroid adenomas, leaving the majority of sporadic instances of the disease unexplained.
Apart from alterations in MEN1, genetic abnormalities in other genes appear to occur very rarely. For this reason, the contributions of epigenetic changes to tumorigenesis have been examined previously. Prior studies have shown that epigenetic modifications, like methylation of the MEG3 locus, often occur in conjunction with LOH across the locus to result in loss of expression (10). Similarly, DNA methylation of a number of other genes has been demonstrated in parathyroid adenomas (11, 12). Although the approach of this study will be unable to detect such epigenetic drivers of tumorigenesis, many microRNA are included in the exome capture array and are analyzed very carefully for mutations. This is important since mutations and expression differences in microRNA have been speculated to play a role in cancers. In fact, differential microRNA expression profiles have been reported to distinguish parathyroid adenomas from parathyroid carcinomas (13). Despite sometimes high frequencies of occurrence, the direct effects of epigenetic modifications on disease are often very difficult to determine without extensive functional studies.
To identify genetic events that may be contributing to parathyroid tumorigenesis, we applied a similar approach to that used previously to identify somatic mutations common in sporadic adrenal aldosterone-producing adenomas via exome sequencing (14). Our study provides, to our knowledge, the first comprehensive evaluation of genetic alterations in parathyroid tumors via whole-exome capture and high-throughput sequencing. The study supports the role of somatic mutations in parathyroid tumorigenesis of the MEN1 and EZH2 genes, both critical for proper histone methyltransferase activity.
Materials and Methods
Cases and samples
Eight cases with sporadic pHPT were included in the study using whole-exome sequencing. Inclusion criteria were inappropriate elevation of PTH in relation to serum calcium, normal serum creatinine levels, and no history of familial hyperparathyroidism or exposure to calcimimetic therapy (Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org). All parathyroid tumors were carefully evaluated and dissected by an experienced endocrine pathologist before use in the study. Preliminary Sanger sequencing determined that all eight cases were free of germline variants in MEN1, thereby confirming the sporadic nature of the cases. Tissues from an additional 185 parathyroid adenomas were obtained via the Yale Pathology Tissue Services (Supplemental Table 2). Tissues were snap frozen using liquid nitrogen and stored at −80 C until processing. Written informed consent from patients and approval by the local institutional review board were obtained.
Sample preparation
High-molecular-weight genomic DNA was isolated from matched tumor and constitutional (blood) samples using standard protocols included in the commercially available QIAGEN DNeasy blood and tissue kit (QIAGEN, Venlo, The Netherlands). All specimens were quality control checked for purity using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA).
Whole-exome capture
Constitutional and tumor DNA was captured on a NimbleGen Sequence Capture 2.1M human exome array (NimbleGen, Madison, WI) following the manufacturer's protocols with modifications at the W. M. Keck Facility at Yale University. As previously described, (15), DNA was sheared and adaptors were ligated onto the resulting fragments. Fragments were amplified by ligation-mediated PCR, purified, and hybridized to the array. After washing, fragments were eluted and purified. Resulting fragments were then subjected to high-throughput sequencing.
High-throughput sequencing
Captured libraries were sequenced on the Illumina GA II sequencing system (Illumina, Inc., San Diego, CA) as 74-bp single-end reads and 74- or 99-bp paired-end reads following the manufacturer's protocols. Image analysis and base calling was performed by Illumina pipeline versions 1.3 and 1.4 with default parameters. The summary sequencing statistics for all eight discovery cohort tumor-constitutional pairs are included in Supplemental Table 3.
Analysis
Analysis of raw data from the Illumina sequencing was performed as previously (15). Sequence reads were mapped to the reference genome (hg18) using the Maq program (16). Reads outside the targeted sequences were discarded. Statistics on coverage were collected from the remaining reads using in-house perl scripts. For insertion/deletion detection, the Burrows-Wheeler Aligner was used to allow gapped alignment to the reference genome (17). SAMtools was used to call targeted bases with any base-call deviating from the reference sequence regarded as a potential variant (18). Variants given a SAMtools single-nucleotide polymorphism quality score of less than 100 were assumed to be false positives and thus excluded from further analysis. Identified variants were annotated based on novelty, impact on the encoded protein, and conservation using an automated pipeline. Somatic mutations were defined as those that were identified in tumor DNA but absent from constitutional DNA.
Mutation validation
Primer3 (http://frodo.wi.mit.edu/primer3/) was used to generate primers for PCR amplification of variants identified via exome sequencing or exons covered in additional screening. Amplification products of appropriate size were identified using agarose gel electrophoresis. Amplicons from constitutional and tumor DNA were sequenced using forward and reverse primers. Variants were confirmed by at least two independent sequences from different primers (Supplemental Tables 4 and 5).
Quantification of contamination using quantitative PCR
Surgically resected tumor tissue often contains constitutional cells surrounding the tumor in vivo. Although unavoidable, it is possible to estimate the level of contamination, which can facilitate correction for such during data analysis. Commercially available probes for the TaqMan custom single-nucleotide polymorphism genotyping assay (Applied Biosystems, Inc., Foster City, CA) were designed using the Applied Biosystems web site. Quantitative PCR using probes overlapping a nonsense MEN1 mutation identified in one of the tumors differentiated between mutant and wild-type alleles. Fluorophores FAM and VIC corresponded to amplification of the somatically-mutated tumor allele and the wild-type allele, respectively. This estimated the presence of tumor DNA in the constitutional sample to be negligible but constitutional DNA in the tumor sample to be 41.3 ± 1.0%. Sanger sequence traces for all heterozygous variants displayed a more pronounced wild-type peak compared with that of the mutant allele, which is consistent with the estimated level of contamination.
Results
Discovery cohort screening
Eight patients with sporadic parathyroid adenomas, in which no germline MEN1 mutations were present, were included in the discovery cohort for high-throughput analysis. Whole-exome capture and subsequent sequencing was performed on tumor and constitutional DNA samples from each patient as previously described (15). Once raw sequencing data were mapped to the reference human genome (hg18) and variants identified, tumor and constitutional sequences from each patient were cross-referenced to identify somatic mutations unique to each tumor genome.
In the eight tumors, whole-exome sequencing revealed a total of 440 high-quality, somatic, nonsynonymous variants in exons or splice sites (Supplemental Table 6). Variants were cross-referenced with more than 600 control exome sequences. This allowed the list to be narrowed down to 286 variants and then further to 251 variants once amino acid conservation of the mutated residue across 46 orthologs was accounted for. The remaining 251 variants were then examined using computational metrics and manual analysis of read alignments to reveal those mutations with a high likelihood of being false positives. PCR amplification and subsequent Sanger sequencing was performed on the remaining 94 variants, which confirmed 29 somatic mutations. This high false-positive rate can be attributed to our prioritization of sensitivity over specificity, which was done to maximize recovery of somatic, exonic variants.
Four of the 29 mutations were found in MEN1 in four separate tumors (one nonsense mutation and three frame shift deletions; Table 1 and Supplemental Table 7). All of these mutations encode a truncated menin protein. Additionally, LOH spanning all of chromosome 11 was observed in all tumors with MEN1 mutations (locus 11q13). Regions were determined to be in LOH if they displayed differential minor allele frequencies between tumor and constitutional DNA (Supplemental Fig. 1). LOH was not limited to chromosome 11, with single instances of chromosomal loss spanning all of chromosomes 9, 13, 21, 22, and a portion of the short arm of chromosome 1 (Table 2 and Supplemental Fig. 1). Based on read coverage, all instances of LOH were determined to be copy neutral.
Table 1.
Somatic mutations identified by exome sequencing included in validation cohort screening
| Case | Chr | Position | Base change | LOH? | Gene | Effect on protein | AA position/length |
|---|---|---|---|---|---|---|---|
| PTH106 | 11 | 64331968 | G>A | Y | MEN1 | Q214X | 214/615 |
| 12 | 67328775 | C>T | N | RAP1B | R2C | 2/184 | |
| PTH107 | 11 | 64334027 | CG | Y | MEN1 | c.129_130delCG | 44/615 |
| 11 | 67937858 | C>T | Y | LRP5 | R877W | 877/1615 | |
| PTH108 | 11 | 64332147 | C | Y | MEN1 | c.460delG | 154/615 |
| PTH113 | 11 | 64333905 | AGAC | Y | MEN1 | c.249_252delGTCT | 85/615 |
| PTH122 | 7 | 148139661 | A>T | N | EZH2 | Y641N | 641/707 |
Chr, Chromosome; Y, yes; N, no; AA, amino acid.
Table 2.
Patterns of LOH in discovery cohort
| Case | Region of LOH | Somatic MEN1 mutation |
|---|---|---|
| PTH105 | chr1a; chr13 | — |
| PTH106 | chr11; chr21; chr22 | Q214X |
| PTH107 | chr11 | c.129_130delCG |
| PTH108 | chr9; chr11 | c.460delG |
| PTH113 | chr11 | c.249_252delGTCT |
| PTH120 | — | — |
| PTH122 | — | — |
| PTH125 | — | — |
Common regions of LOH are denoted in bold. Dashes indicate absence of either LOH or MEN1 mutation. Chr, Chromosome.
LOH occurred only across 0–103 Mb on the short arm of chromsome 1.
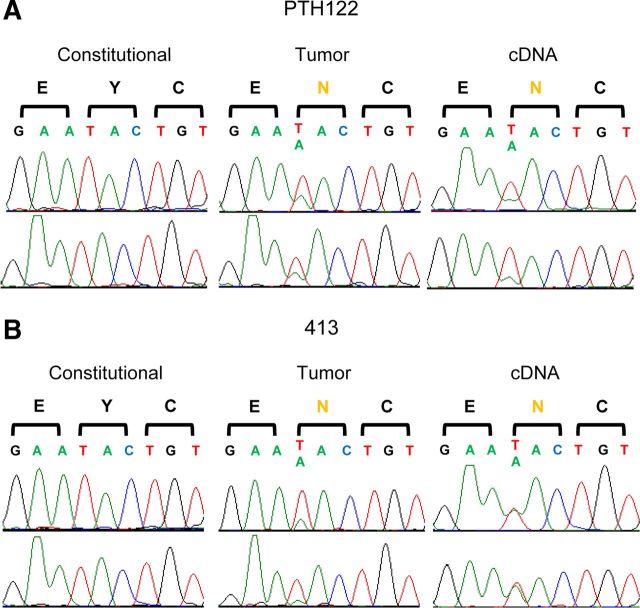
Although multiple tumors harbored MEN1 mutations, no recurrent alterations were identified in the discovery cohort. Despite being present in only single instances, several of the confirmed mutations had been previously associated with other types of tumors in the literature. For instance, one of the exome sequenced tumors harbored a heterozygous missense mutation in enhancer of zeste homolog 2 (EZH2) gene at a highly conserved residue (Y641N; Fig. 1A). This exact heterozygous mutation in EZH2 had been identified as common to B cell lymphomas (19).
Fig. 1.
Somatic mutations in EZH2. A, Sequences of constitutional genomic DNA, tumor genomic DNA, and tumor cDNA of EZH2 codons 640–642 in PTH122 of the discovery cohort. B, Sequences of constitutional genomic DNA, tumor genomic DNA, and tumor cDNA of EZH2 codons 640–642 in sample 413 from the large-scale EZH2 SET domain screen. Sequences from the forward direction are shown on the top row and the reverse direction are shown below.
A prior study found expression of an aberrantly spliced, internally deleted low-density lipoprotein receptor-related protein 5 (LRP5) mRNA and protein in the majority of sporadic parathyroid adenomas (20). LRP5 forms a complex with Wnt that negatively regulates β-catenin (CTNNB1). Although stabilizing mutations in CTNNB1 have been found in a subset of pHPT patients that possibly account for its reported overexpression, a separate study only identified one such mutation out of 180 tumors from this study's validation cohort (21). Although rare, these stabilizing mutations in CTNNB1 and loss-of-function mutations in LRP5, a negative regulator of CTNNB1, may result in similar effects, which could account for the apparent mutual exclusivity of these defects. These prior reports drew attention to a constitutional mutation in LRP5 (R877W) found in this study that resided at a highly conserved residue within the aforementioned, internally deleted region. This variant was heterozygous in the germline, but the wild-type allele was lost in the tumor due to LOH across chromosome 11. This tumor also displayed a frame shift deletion in MEN1, which may alone be sufficient for tumorigenesis. Due to its co-occurrence with a MEN1 mutation and the fact that LOH across chromosome 11 is frequently observed in these tumors, it is difficult to evaluate the possible role of this LRP5 variant in tumorigenesis without analyzing additional tumors for mutations in the gene.
Another gene associated with neoplasia that was somatically mutated in one tumor was RAP1B. A member of the Ras oncogene family, RAP1B antagonizes Ras mitogenic signals by forming nonproductive complexes with effectors of Ras. A mutation in RAP1B had previously been associated with myelodysplastic syndromes (22). The identified variant in our cohort was a heterozygous missense mutation (R2C) in a tumor that also harbored a nonsense MEN1 mutation.
Validation cohort screening
In a cohort of only eight patients, the true frequency of rare mutations is difficult to determine. Therefore, tumor and constitutional DNA from 185 additional sporadic parathyroid adenomas was extracted and screened for the most promising mutations identified via exome sequencing. The best candidates for further sequencing were those that were likely damaging to protein function, at highly conserved residues in highly conserved genes, mutations absent from control exomes, and those previously associated in the literature with tumorigenesis or hormone secretion.
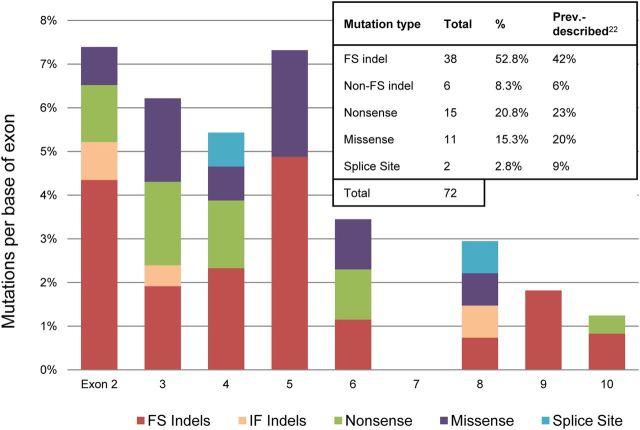
Because MEN1 mutations are frequently present in parathyroid adenomas and may fully account for tumorigenesis, the first screen covered all coding exons (2–10) of MEN1 in all 185 tumors. This screen brought the final tally to 72 somatic mutations identified in 68 of the total 193 tumors screened (35.2%) (Supplemental Table 8). This total includes four tumors that each displayed two separate mutations in the MEN1 gene. Most of these variants were undoubtedly damaging to protein function, with frame shift insertions/deletions and nonsense mutations accounting for 74% of MEN1 variants. The mutational breakdown is depicted in Fig. 2 (23). The mutation frequency in this cohort of 35.2% was much higher than the previously reported 12–21% (24, 25). It has been suggested that additional tumor suppressors may lie on 11q13 due to the observation that LOH at this locus has been observed more frequently than MEN1 mutations (25–40% compared with 12–21%, respectively). However, the higher MEN1 mutation rate observed in this study (and co-occurrence of MEN1 mutations with every instance of 11q13 LOH in the discovery cohort) suggests that MEN1 mutations and LOH may reliably co-occur, which indicates against the possibility of an additional tumor suppressor at this locus.
Fig. 2.
Results of the MEN1 mutation screen. Mutation types per base of exon are displayed across exons 2–10 for all 193 tumors. The table displays the total mutational breakdown across all exons compared with the findings from previous studies. FS indels and IF indels represent frame shift and in-frame insertions or deletions, respectively.
We next evaluated the SET [Su(var)3–9, Enhancer of Zeste, Trithorax] domain of EZH2 that was frequently mutated in myeloid and lymphoid malignancies (19, 26, 27). Screening of this domain covered four exons (16–19) across all 185 tumors. No mutations were identified in any of the tumors harboring MEN1 defects. However, one of the tumors with wild-type MEN1 displayed the exact same heterozygous mutation (Y641N) as previously identified in the discovery cohort (Fig. 1B). Somatic mutation at the same residue in even two separate tumors is very unlikely to occur by chance, which makes our identification of the somatic Y641N mutation in EZH2 highly significant (P = 4.13 × 10−10) (Supplemental Fig. 2). Furthermore, because identical somatic mutations strongly associated with B cell lymphoma were identified in separate tumors, they are likely driving tumorigenesis in a portion (two of 193) of parathyroid adenomas that cannot be attributed to defective MEN1.
We additionally sequenced all coding exons (2–7) of RAP1B and exons 9–12 of LRP5, which encompassed the entire region containing the previously discussed internal deletion. The initial RAP1B and LRP5 mutations observed in the discovery cohort were in tumors with MEN1 mutations. Therefore, only the MEN1-mutated cohort of 68 tumors was screened for variants in these two genes. However, no additional somatic variants in either gene were uncovered, indicating that the original variants are likely to be passenger mutations or very rare drivers of tumorigenesis.
Discussion
MEN1 mutations are observed at a very high frequency in both familial and sporadic parathyroid adenomas as well as other endocrine tumors (28). This study found a higher mutation frequency of MEN1 than previously reported, suggesting an even more important role in parathyroid tumors than originally believed. The majority of the identified mutations were very severe, being either frame shift indels or nonsense mutations. There appeared to be an enrichment of these damaging mutations in the earlier exons of MEN1, which is consistent with its complete loss of function in endocrine neoplasias.
Despite its alteration in many cancers, the specific role of the ubiquitously expressed, nuclear-localizing (29) menin in the cell is largely unclear. Although this protein is highly conserved from mammals (30) to Drosophila (31), it shares no sequence similarity to any known gene (32). Menin has been demonstrated to play a role as a transcriptional regulator in the nucleus via interactions with histone deacetylases (33), histone methyltransferases (34), and transcription factors (35–37). It has also been shown to be involved in cell proliferation (38), apoptosis regulation (39), and DNA damage repair (40). The knockout mouse model has been used to mimic the pattern of MEN1 loss observed in MEN1 syndrome patients. Homozygous MEN1 knockout mice are unequivocally lethal during gestation, but heterozygous knockouts are viable and fertile. These MEN1+/− mice typically develop endocrine tumors between 9 and 12 months of age (41). When investigated, these tumors display inactivation of the remaining functional allele of MEN1 via LOH. However, a conditional, pancreas-specific, homozygous MEN1 knockout model shows a lag period of about 6 months before the earliest tumors are detectable (42). Such results suggest that additional somatic events may be required for clonal expansion to occur.
Neither this study nor others were able to find additional mutations common to tumors with both MEN1 alleles inactivated. There are several possible explanations for this observation. First, it could be that the inactivation of both copies of MEN1 is alone sufficient to drive tumorigenesis in the parathyroid gland. If so, additional aberrations present in these tumors could be passenger mutations randomly altered in the MEN1-defective cell that clonally expanded that do not contribute to tumor formation or progression. It is also possible that these rare mutations contribute a slight selective advantage to a cell with inactivated MEN1, which contribute to, but are not solely sufficient for, tumorigenesis. Another explanation for the absence of common mutations among MEN1-mutated tumors is that other factors are necessary for tumorigenesis once MEN1 is inactivated, but exome sequencing and previous studies failed to detect them. These could include copy number variations, epigenetic modifications, or alterations lying outside the exome, which would be undetectable using the current approach.
In the four discovery cohort tumors with functional copies of MEN1, no mutated genes were shared across tumors. However, additional screening in the validation cohort of 185 tumors was able to identify another tumor with an identical EZH2 mutation. The probability of identifying two tumors with somatic mutations in the same gene merely by chance is exceedingly low. Far less likely is the random occurrence of identifying somatic changes at the exact base in two separate tumors, which makes this finding highly significant. Although this single EZH2 mutation is likely to be a major driver of tumorigenesis in the tumors in which it is mutated, the fact that it is mutated in such a small percentage of parathyroid adenomas suggests that it plays a minor role in the total pathogenesis of the disease.
The tyrosine residue altered in our tumors is a key component of the EZH2 active site commonly mutated in B cell lymphomas. The active site resides in the SET domain of EZH2, which is the catalytic component of the PRC2 complex responsible for trimethylating H3K27, a mark of gene repression. Both increased and decreased activity of enzymes regulating H3K27 methylation have been linked to cancer, suggesting a fragile balance for maintenance of normal cell growth (43–45). Consistent with the specific EZH2 mutation identified in this study as being gain of function, in vitro functional studies demonstrated an increased ability of EZH2Y641N to trimethylate H3K27 (46). Given this result, it is interesting to speculate how gain-of-function mutations in EZH2 could phenocopy loss-of-function mutations in MEN1. MEN1 is a member of the MLL-containing histone H3 methyltransferase complex known to methylate the activation mark H3K4 (47), which is associated with activated gene transcription. Based on these opposite functions, it is possible that loss-of-function MEN1 mutations and gain-of-function EZH2 mutations could drive tumorigenesis via a common mechanism. For example, the loss of MEN1 is expected to result in a reduction in H3K4 methylation, which would result in an abnormal loss of expression of a set of genes. Alternatively, these same genes could be silenced through increased methylation of H3K27 by EZH2Y641N.
In summary, this study has identified novel mutations possibly contributing to tumorigenesis as well as provided the most comprehensive mutational profile of sporadic parathyroid tumors to date. However, further investigation into this topic is still needed. In some tumor samples, no candidate drivers of tumorigenesis shared among tumors were identified. Although genomic rearrangements have been previously implicated in many tumor types, including parathyroid adenomas (PTH-cyclin D1) (48), the approach used in this study would have difficulty detecting such cellular events. Moreover, driver mutations altered at a low frequency in these tumors may have been missed in our discovery cohort of eight tumors, necessitating further exome sequencing using larger cohorts. Alternatively to exome sequencing, tumor samples lacking MEN1 and EZH2 defects make excellent candidates for whole-genome sequencing because they may share variants outside the exome, which our approach was not able to identify. Even so, this study demonstrates the power of high-throughput sequencing to identify rare tumorigenic mutations that would not have been identified using traditional approaches. This confirmed the presence of mutations previously found in these tumors and also revealed novel candidates for endocrine tumorigenesis, which suggests mechanisms of sporadic tumorigenesis in the parathyroid gland. Ultimately these results also suggest an increasingly important role for chromatin-modifying methyltransferase genes in tumor formation and progression.
Acknowledgments
We thank the patients whose participation made this study possible and the staff of the Yale West Campus Genomics Center and the Endocrine Surgical Laboratory (Clinical Research Centre, University Hospital, Uppsala, Sweden) for helpful suggestions.
This work was supported by an Ohse Research Award. R.P.L. is a paid scientific adviser to Merck and is an investigator of the Howard Hughes Medical Institute. T.C. is a Doris Duke-Damon Runyon Clinical Investigator supported in part by the Damon Runyon Cancer Research Foundation and the Doris Duke Charitable Foundation. The funders of this work had no role in the study design, data collection and analysis, decision to publish, or preparation of this manuscript.
Disclosure Summary: The authors declare they have nothing to disclose.
Footnotes
- CTNNB1
- Complex with Wnt that negatively regulates β-catenin
- EZH2
- enhancer of zeste homolog 2
- LOH
- loss of heterozygosity
- LRP5
- lipoprotein receptor-related protein 5
- MEN1
- multiple endocrine neoplasia type 1
- pHPT
- primary hyperparathyroidism
- SET
- Su(var)3–9, Enhancer of Zeste, Trithorax.
References
- 1. Lundgren E , Rastad J , Thrufjell E , Akerströöm G , Ljunghall S. 1997. Population-based screening for primary hyperparathyroidism with serum calcium and parathyroid hormone values in menopausal women. Surgery 121:287–294 [DOI] [PubMed] [Google Scholar]
- 2. Verdonk CA , Edis AJ. 1981. Parathyroid “double adenomas”: fact or fiction? Surgery 90:523–526 [PubMed] [Google Scholar]
- 3. Lee PK , Jarosek SL , Virnig BA , Evasovich M , Tuttle TM. 2007. Trends in the incidence and treatment of parathyroid cancer in the United States. Cancer 109:1736–1741 [DOI] [PubMed] [Google Scholar]
- 4. Carling T. 2001. Molecular pathology of parathyroid tumors. Trends Endocrinol Metab 12:53–58 [DOI] [PubMed] [Google Scholar]
- 5. Palanisamy N , Imanishi Y , Rao PH , Tahara H , Chaganti RS , Arnold A. 1998. Novel chromosomal abnormalities identified by comparative genomic hybridization in parathyroid adenomas. J Clin Endocrinol Metab 83:1766–1770 [DOI] [PubMed] [Google Scholar]
- 6. Shattuck TM , Välimäki S , Obara T , Gaz RD , Clark OH , Shoback D , Wierman ME , Tojo K , Robbins CM , Carpten JD , Farnebo LO , Larsson C , Arnold A. 2003. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med 349:1722–1729 [DOI] [PubMed] [Google Scholar]
- 7. Arnold A , Shattuck TM , Mallya SM , Krebs LJ , Costa J , Gallagher J , Wild Y , Saucier K. 2002. Molecular pathogenesis of primary hyperparathyroidism. J Bone Miner Res 17(Suppl 2):N30–N36 [PubMed] [Google Scholar]
- 8. Cryns VL , Rubio MP , Thor AD , Louis DN , Arnold A. 1994. p53 abnormalities in human parathyroid carcinoma. J Clin Endocrinol Metab 78:1320–1324 [DOI] [PubMed] [Google Scholar]
- 9. Cryns VL , Thor A , Xu HJ , Hu SX , Wierman ME , Vickery AL , Benedict WF , Arnold A. 1994. Loss of the retinoblastoma tumor-suppressor gene in parathyroid carcinoma. N Engl J Med 330:757–761 [DOI] [PubMed] [Google Scholar]
- 10. Gejman R , Batista DL , Zhong Y , Zhou Y , Zhang X , Swearingen B , Stratakis CA , Hedley-Whyte ET , Klibanski A. 2008. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab 93:4119–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carling T , Du Y , Fang W , Correa P , Huang S. 2003. Intragenic allelic loss and promoter hypermethylation of the RIZ1 tumor suppressor gene in parathyroid tumors and pheochromocytomas. Surgery 134:932–939; discussion 939–940 [DOI] [PubMed] [Google Scholar]
- 12. Starker LF , Svedlund J , Udelsman R , Dralle H , Akerström G , Westin G , Lifton RP , Björklund P , Carling T. 2011. The DNA methylome of benign and malignant parathyroid tumors. Genes Chromosomes Cancer 50:735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Milne TA , Hughes CM , Lloyd R , Yang Z , Rozenblatt-Rosen O , Dou Y , Schnepp RW , Krankel C , Livolsi VA , Gibbs D , Hua X , Roeder RG , Meyerson M , Hess JL. 2004. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci USA 102:749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi M , Scholl UI , Yue P , Björklund P , Zhao B , Nelson-Williams C , Ji W , Cho Y , Patel A , Men CJ , Lolis E , Wisgerhof MV , Geller DS , Mane S , Hellman P , Westin G , Åkerström G , Wang W , Carling T , Lifton RP. 2011. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 331:768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi M , Scholl UI , Ji W , Liu T , Tikhonova IR , Zumbo P , Nayir A , Bakkaloğlu A , Ozen S , Sanjad S , Nelson-Williams C , Farhi A , Mane S , Lifton RP. 2009. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 106:19096–19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H , Ruan J , Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H , Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H , Handsaker B , Wysoker A , Fennell T , Ruan J , Homer N , Marth G , Abecasis G , Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morin RD , Johnson NA , Severson TM , Mungall AJ , An J , Goya R , Paul JE , Boyle M , Woolcock BW , Kuchenbauer F , Yap D , Humphries RK , Griffith OL , Shah S , Zhu H , Kimbara M , Shashkin P , Charlot JF , Tcherpakov M , Corbett R , Tam A , Varhol R , Smailus D , Moksa M , Zhao Y , Delaney A , Qian H , Birol I , Schein J , Moore R , Holt R , Horsman DE , Connors JM , Jones S , Aparicio S , Hirst M , Gascoyne RD , Marra MA. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42:181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Björklund P , Akerström G , Westin G. 2007. An LRP5 receptor with internal deletion in hyperparathyroid tumors with implications for deregulated WNT/β-catenin signaling. PLoS Med 4:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Starker LF , Fonseca A , Akerstrom G , Bjorklund P , Westin G , Carling T. 11 May 2012. Evidence of a stabilizing mutation β-catenin encoded by CTNNB1 exon 3 in a large series of sporadic parathyroid adenomas. Endocrine 10.1007/s12020-012-9690-3 [DOI] [PubMed] [Google Scholar]
- 22. Gyan E , Frew M , Bowen D , Beldjord C , Preudhomme C , Lacombe C , Mayeux P , Dreyfus F , Porteu F , Fontenay M. 2005. Mutation in RAP1 is a rare event in myelodysplastic syndromes. Leukemia 19:1678–1680 [DOI] [PubMed] [Google Scholar]
- 23. Lemos MC , Thakker RV. 2008. Multiple endocrine neoplasia type 1 (MEN1): analysis of 1336 mutations reported in the first decade following identification of the gene. Hum Mutat 29:22–32 [DOI] [PubMed] [Google Scholar]
- 24. Carling T , Correa P , Hessman O , Hedberg J , Skogseid B , Lindberg D , Rastad J , Westin G , Akerström G. 1998. Parathyroid MEN1 gene mutations in relation to clinical characteristics of nonfamilial primary hyperparathyroidism. J Clin Endocrinol Metab 83:2960–2963 [DOI] [PubMed] [Google Scholar]
- 25. Heppner C , Kester MB , Agarwal SK , Debelenko LV , Emmert-Buck MR , Guru SC , Manickam P , Olufemi SE , Skarulis MC , Doppman JL , Alexander RH , Kim YS , Saggar SK , Lubensky IA , Zhuang Z , Liotta LA , Chandrasekharappa SC , Collins FS , Spiegel AM , Burns AL , Marx SJ. 1997. Somatic mutation of the MEN1 gene in parathyroid tumours. Nat Genet 16:375–378 [DOI] [PubMed] [Google Scholar]
- 26. Nikoloski G , Langemeijer SM , Kuiper RP , Knops R , Massop M , Tönnissen ER , van der Heijden A , Scheele TN , Vandenberghe P , de Witte T , van der Reijden BA , Jansen JH. 2010. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 42:665–667 [DOI] [PubMed] [Google Scholar]
- 27. Ernst T , Chase AJ , Score J , Hidalgo-Curtis CE , Bryant C , Jones AV , Waghorn K , Zoi K , Ross FM , Reiter A , Hochhaus A , Drexler HG , Duncombe A , Cervantes F , Oscier D , Boultwood J , Grand FH , Cross NCP. 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42:722–726 [DOI] [PubMed] [Google Scholar]
- 28. Bassett JH , Forbes SA , Pannett AA , Lloyd SE , Christie PT , Wooding C , Harding B , Besser GM , Edwards CR , Monson JP , Sampson J , Wass JA , Wheeler MH , Thakker RV. 1998. Characterization of mutations in patients with multiple endocrine neoplasia type 1. Am J Hum Genet 62:232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guru SC , Goldsmith PK , Burns AL , Marx SJ , Spiegel AM , Collins FS , Chandrasekharappa SC. 1998. Menin, the product of the MEN1 gene, is a nuclear protein. Proc Natl Acad Sci USA 95:1630–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stewart C , Parente F , Piehl F , Farnebo F , Quincey D , Silins G , Bergman L , Carle GF , Lemmens I , Grimmond S , Xian CZ , Khodei S , Teh BT , Lagercrantz J , Siggers P , Calender A , Van de Vem V , Kas K , Weber G , Hayward N , Gaudray P , Larsson C. 1998. Characterization of the mouse Men1 gene and its expression during development. Oncogene 17:2485–2493 [DOI] [PubMed] [Google Scholar]
- 31. Guru SC , Prasad NB , Shin EJ , Hemavathy K , Lu J , Ip YT , Agarwal SK , Marx SJ , Spiegel AM , Collins FS , Oliver B , Chandrasekharappa SC. 2001. Characterization of a MEN1 ortholog from Drosophila melanogaster. Gene 263:31–38 [DOI] [PubMed] [Google Scholar]
- 32. Chandrasekharappa SC , Guru SC , Manickam P , Olufemi SE , Collins FS , Emmert-Buck MR , Debelenko LV , Zhuang Z , Lubensky IA , Liotta LA , Crabtree JS , Wang Y , Roe BA , Weisemann J , Boguski MS , Agarwal SK , Kester MB , Kim YS , Heppner C , Dong Q , Spiegel AM , Burns AL , Marx SJ. 1997. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 276:404–407 [DOI] [PubMed] [Google Scholar]
- 33. Kim H , Lee JE , Cho EJ , Liu JO , Youn HD. 2003. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res 63:6135–6139 [PubMed] [Google Scholar]
- 34. Hughes CM , Rozenblatt-Rosen O , Milne TA , Copeland TD , Levine SS , Lee JC , Hayes DN , Shanmugam KS , Bhattacharjee A , Biondi CA , Kay GF , Hayward NK , Hess JL , Meyerson M. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13:587–597 [DOI] [PubMed] [Google Scholar]
- 35. Agarwal SK , Guru SC , Heppner C , Erdos MR , Collins RM , Park SY , Saggar S , Chandrasekharappa SC , Collins FS , Spiegel AM , Marx SJ , Burns AL. 1999. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell 96:143–152 [DOI] [PubMed] [Google Scholar]
- 36. Kaji H , Canaff L , Lebrun JJ , Goltzman D , Hendy GN. 2001. Inactivation of menin, a Smad3-interactng protein, blocks transforming growth factor type β signaling. Proc Natl Acad Sci USA 98:3837–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heppner C , Bilimoria KY , Agarwhal SK , Kester M , Whitty LJ , Guru SC , Chandrasekharappa SC , Collins FS , Spiegel AM , Marx SJ , Burns AL. 2001. The tumor suppressor protein menin interacts with NF-κB proteins and inhibits NF-κB-mediated transactivation. Oncogene 20:4917–4925 [DOI] [PubMed] [Google Scholar]
- 38. Ratineau C , Bernard C , Poncet G , Blanc M , Josso C , Fontanière S , Calender A , Chayvialle JA , Zhang CX , Roche C. 2004. Reduction of menin expression enhances cell proliferation and is tumorigenic in intestinal epithelial cells. J Biol Chem 279:24477–24484 [DOI] [PubMed] [Google Scholar]
- 39. Schnepp RW , Mao H , Sykes SM , Zong WX , Silva A , La P , Hua X. 2004. Menin induces apoptosis in murine embryonic fibroblasts. J Biol Chem 279:10685–10691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin S , Mao H , Schnepp RW , Sykes SM , Silva AC , D'Andrea AD , Hua X. 2003. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res 63:4204–4210 [PubMed] [Google Scholar]
- 41. Harding B , Lemos MC , Reed AA , Walls GV , Jeyabalan J , Bowl MR , Tateossian H , Sullivan N , Hough T , Fraser WD , Ansorge O , Cheeseman MT , Thakker RV. 2009. Multiple endocrine neoplasia type 1 knockout mice develop parathyroid, pancreatic, pituitary and adrenal tumours with hypercalcaemia, hypophosphataemia and hypercorticosteronaemia. Endocr Relat Cancer 16:1313–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crabtree JS , Scacheri PC , Ward JM , McNally SR , Swain GP , Montagna C , Hager JH , Hanahan D , Edlund H , Magnuson MA , Garrett-Beal L , Burns AL , Ried T , Chandrasekharappa SC , Marx SJ , Spiegel AM , Collins FS. 2003. Of mice and MEN1: insulinomas in a conditional mouse knockout. Mol Cell Biol 23:6075–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martinez-Garcia E , Licht JD. 2010. Deregulation of H3K27 methylation in cancer. Nat Genet 42:100–101 [DOI] [PubMed] [Google Scholar]
- 44. van Haaften G , Dalgliesh GL , Davies H , Chen L , Bignell G , Greenman C , Edkins S , Hardy C , O'Meara S , Teague J , Butler A , Hinton J , Latimer C , Andrews J , Barthorpe S , Beare D , Buck G , Campbell PJ , Cole J , Forbes S , Jia M , Jones D , Kok CY , Leroy C , Lin ML , et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41:521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Varambally S , Dhanasekaran SM , Zhou M , Barrette TR , Kumar-Sinha C , Sanda MG , Ghosh D , Pienta KJ , Sewalt RG , Otte AP , Rubin MA , Chinnaiyan AM. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624–629 [DOI] [PubMed] [Google Scholar]
- 46. Yap DB , Chu J , Berg T , Schapira M , Cheng SW , Moradian A , Morin RD , Mungall AJ , Meissner B , Boyle M , Marquez VE , Marra MA , Gascoyne RD , Humphries RK , Arrowsmith CH , Morin GB , Aparicio SA. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117:2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu X , Hua X. 2008. Menin, histone 3 methyltransferases, and regulation of cell proliferation: current knowledge and perspective. Curr Mol Med 8:805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arnold A , Kim HG , Gaz RD , Eddy RL , Fukushima Y , Byers MG , Shows TB , Kronenberg HM. 1989. Molecular cloning and chromosomal mapping of DNA rearranged with the parathyroid hormone gene in a parathyroid adenoma. J Clin Invest 83:2034–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]