Abstract
CYP1A1 functions in detoxifying xenobiotics but occasionally converts compounds into potent genotoxins. CYP1A1 activates polyaromatic hydrocarbons, such as benzo[a]pyrene 7,8 dihydrodiol (BaP-DHD), rendering them genotoxic. Particular alleles of CYP1A1, such as CYP1A1 I462V have been correlated with a higher incidence of breast and lung cancer, but it is unknown whether these variants express enzymes in vivo that are more potent in generating genotoxins. We individually expressed CYP1A1 (CYP1A1.1), CYP1A1 T461N (CYP1A1.4) and I462V (CYP1A1.2) alleles in wild-type and DNA repair deficient mutant strains of Saccharomyces cerevisiae (budding yeast) and asked which yeast strains exhibited the highest levels of carcinogen-associated genotoxicity after exposure to BaP-DHD, aflatoxin B1 (AFB1), and heterocyclic aromatic amines (HAAs). We measured carcinogen-associated recombination, Rad51 foci, and carcinogen-associated toxicity in a DNA repair mutant deficient in both nucleotide excision repair and recombinational repair. CYP1A1 activity was confirmed by measuring ethoxyresorufin-O-deethylation (EROD) activities. Our data indicate that CYP1A1 I462V allele confers the least carcinogen-associated genotoxicity, compared to CYP1A1; however, results vary depending on the chemical carcinogen and the genotoxic endpoint. We speculate that the cancer-associated risk of CYP1A1 I462V may be caused by exposure to other xenobiotics.
Keywords: CYP1A1 polymorphisms, food carcinogens, budding yeast, DNA damage
Introduction
CYP1A1 is a highly induced extrahepatic P450 enzyme whose expression varies greatly among individuals [1,2,3]. It metabolizes a broad range of xenobiotic and endogenous compounds including carcinogens, pharmaceuticals, and hormones, with a preference for aromatic substrates in a planar configuration [4]. Its importance in cancer etiology is underscored as the activator of the well-known polyaromatic hydrocarbon (PAH) benzo[a]pyrene, which is ultimately converted into BaP-dihydrodiol epoxide, the active metabolite that forms mutagenic N2-guanine DNA adducts [5]. CYP1A1 metabolically activates 7,12-dimethyl benzanthracene (DMBA, [4]), and particular food carcinogens, including aflatoxin B1 (AFB1, [6]) and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP, [4]). Alternatively, CYP1A1 converts anticancer drugs into potent therapeutics, such as aminoflavone [7] and dacarbazine [8]. Its metabolism of polyunsaturated fatty acids aids in controlling blood pressure [9]. Thus, CYP1A1’s metabolic functions have significance in cancer etiology, pharmacology, and cardiovascular physiology.
The importance of CYP1A1 in carcinogen metabolism has motivated investigators to identify CYP1A1 variants that are associated with disease. The CYP1A1 variant, CYP1A1*2C (CYP1A1 I462V [10]), has been correlated with the increased cancer incidence of adult leukemia [11], lung cancer [12], breast cancer [13,14,15,16], and renal cancer [17]. The CYP1A1 variant, CYP1A1*4 (CYP1A1 T461N, [18]), has been correlated with an increased risk for endometrial cancer [19]; see [20] for a list of CYP1A1 variants. CYP1A1 I462V is found in approximately 20% of the Japanese population and 9% of the Caucasian population while CYP1A1 T461N is found in 2–8% of the Caucasian population [21]. Considering that exposure to polyaromatic hydrocarbons (PAHs) is a risk factor for lung cancer (for review, see [22]), one hypothesis is that cancer-associated CYP1A1 alleles have higher catalytic activity in converting xenobiotics, such as PAHs, to potent genotoxins [23].
To test this hypothesis, investigators characterized enzymatic properties of CYP1A1 alleles expressed in vitro, and measured DNA adducts and quantified chromosomal abnormalities in human leukocytes from individuals bearing different CYP1A1 alleles. Enzymatic properties, such as Kcat and Km, support the notion that CYP1A1 enzymatic variants may have higher EROD activities but still lower activity towards BaP, compared to wild-type CYP1A1 ([24, 25]). However, other studies indicated that the m2 variant (CYP1A1*2C, 2454A>G, Ile462Val) is clearly associated with 6- to 12-fold higher enzymatic activity towards 17β-estradiol and estrone [26]. Studies using human lymphocytes indicate that particular CYP1A1 alleles, such as CYP1A1*2A, may confer higher levels of DNA PAH-adducts and chromosomal abnormalities, compared to individuals bearing the wild type allele [27]; however, there has been inconsistent reports on levels of PAH adducts in leukocytes from individuals bearing either CYP1A1*2C or *2B [28]. Differential expression of phase II enzymes that detoxify compounds, such as GSTM1, has been indicated to be an additional variable in evaluating CYP1A1 alleles [28] Thus, particular CYP1A1 alleles may confer higher levels of genotoxic activation depending on the exposure to the particular carcinogenic compound or substrate.
We used budding yeast to determine whether CYP1A1 alleles correlated to higher cancer incidence conferred higher levels of genotoxicity for particular carcinogens, compared to wild type. Yeast does not express Phase II enzymes that can rapidly detoxify the activated compounds nor does it express multiple P450s, as in the lung or liver, which can metabolically activate compounds. However, the expression of individual human P450s greatly enhances the genotoxicity of PAHs in yeast cells [29, 30, 31]. The full-length cDNA of the CYP1A1 was expressed and active enzyme was measured from yeast microsomal extracts [32]. We chose genotoxic endpoints that were particularly sensitive in detecting DNA damage, including growth in the rad4 rad51 mutant after carcinogen exposure, carcinogen-associated recombination, and carcinogen-associated Rad51 foci [33, 34]. We previously used this methodology to phenotype CYP1A2 polymorphisms [34]. Our studies indicate that phenotypic differences between CYP1A1 alleles depend on the xenobiotic.
2. Materials and Methods
2.1 Media
Standard media were used for the culture of yeast cells. YPD (yeast extract, peptone, dextrose), SC-TRP (synthetic complete lacking tryptophan), SC-URA (synthetic complete lacking uracil) and FOA (5-fluro-orotic acid) were described in Burke et al. [35].
2.2 Chemical Preparation
Stock solutions of 10 mM aflatoxin B1 (AFB1), 37.5 mM benzo[a]pyrene-7,8-dihydrodiol (BaP-DHD), 50 mM 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), and 100 mM 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) were dissolved into dimethyl sulfoxide (DMSO). Chemicals were purchased from Sigma (AFB1) and Santa Cruz Biotechnology (IQ, MeIQx, PhIP). BaP-DHD was obtained from the National Cancer Center Repository for Chemical Carcinogens (Kansas City, MO), as a powder. It was protected from light at −20oC, and stock solutions were made as required.
2.3 Plasmid constructions and site-specific mutagenesis
Standard molecular biology techniques [36] for DNA isolation and bacterial transformation were used to construct vector pRS424-CYP1A1 and pMF-human oxidoreductase (hOR). The human CYP1A1 was inserted into pRS424-CYP1A1 by subcloning the 2.3 kb CYP1A1 SmaI-SalI fragment from pSB229 into the SmaI and SalI sites of pRS424 [37]. pMF::hOR was constructed by inserting the BamH1 fragment containing the hOR gene in pSB229 into pMF [34] so that it is flanked by DNA sequences both centromere proximal and distal to TRP1. pRS424-CYP1A1 was introduced into yeast strains expressing hOR by selecting for Trp+ transformants.
Site-specific mutagenesis was performed using QuickChange kit (Stratagene) according to the manufacturer’s instructions. We constructed I462V using the forward primer TGCGTGAGACCGTTGCCCGCTGG, and the reverse primer CCAGCGGGCAACGGTCTCACCGA. We constructed T461N using forward primer TATCGGTGAGAACATTGCCCGCT and reverse primer AGCGGGCAATGTTCTCACCGATA. The specific mutations were then verified, and the entire gene was sequenced to verify that only the base substitutions specific for T461N and I462V were introduced.
2.4 Yeast strains
The genotypes of yeast strains used in this study are listed in Table 1. Strains used to measure AFB1-associated recombination or mutagenesis was isogenic to S288c; strains used for detecting Rad51 foci were derived from W303. For measuring AFB1-associated translocation frequencies, diploid strains contained trp1::hOR [34] and the recombination substrates [38], his3-Δ3’ and his3-Δ 5,’ as described [39, 40]. Plasmids containing CYP1A1 and alleles were introduced into yeast strains by selecting for Trp+ transformants.
Table 1.
Yeast Strains
| Strain (Synonym) | Genotype | Autonomous plasmid | Reference (Source) |
|---|---|---|---|
| YA101 (MCY726) | MATaura3-52 his3-Δ200 ade2-101 lys2-801 | M. Carlson | |
| BY4743 | MATa/MATα ura3Δ/- leu2Δ/- his3Δ1/- LYS2/lys2Δ0 met15Δ0/MET15 | wiki.yeastgenome.org/index.php/ Commonly_used_strains | |
| YB225 | MATa-inc ura3 rad4::KanMX | This laboratory [42] | |
| YB226 | MATa-inc trp1 ura3 his3 ade2 rad4::KanMX rad51::URA3 | This laboratory [42] | |
| YB318 | MATα ura3-52 his3-Δ200 ade2-n trp1-Δ1 gal3−leu2-3, 112 GAL1::his3-Δ5' trp1::his3-Δ3'::HOcs | This Laboratory [34] | |
| YB400 | MATa-inc trp1 ura3 his3 ade2 rad4::KanMX rad51 | Derived from YB226 [42] | |
| YB406 | MATaura3-52 trp1::hOR his3-Δ 200 ade2-101 lys2-801 | This Laboratory [34] | |
| LSY1957 | MATaYFP-rad51-I345T-URA3-RAD51 ADE2 leu2 trp1 ura3 his3 | L.Symington | |
| YB407 | MATα ura3-52, trp1::hOR his3-Δ200,ade2-101 lys2-801 | This Laboratory | |
| YB419 | MATaYFP-rad51-I345T-URA3-RAD51 ADE2 leu2 trp1::hOR ura3 his3 | Meiotic segregant of diploid cross of LSY1957 and YB407 | |
| YB420 | MATaYFP-rad51-I345T-URA3-RAD51 ADE2 leu2 trp1::hOR ura3 his3 | pRS424 | Trp+ transformant of YB419 |
| YB425 | MATα ura3-52 ura3-52 his3-Δ200 ade2-n trp1-Δ1 gal3−leu2-3, 112 GAL1::his3-Δ5' trp1::his3-Δ3'::HOcs rad4::kanMX | Derived from cross of YB318 and YB225 | |
| YB426 | MATa ura3-52, trp1::hOR his3-Δ200,ade2-101 lys2-801 rad4 ::KanMX | Derived from cross of YB407 and YB225 | |
| YB427 | MATa/MATα ura3-52/- his3-Δ200/- ade2-101/ade2-n trp1-Δ1/trp1::hOR leu2-3,112/leu2-801 GAL1::his3-Δ5' trp1::his3-Δ3'::HOcs rad4::kanMX/rad4 kanMX | Diploid cross of YB425 and YB426 | |
| YB428 | MATa/MATα ura3-52/- his3-Δ200/- ade2-101/ade2-n trp1-Δ1/trp1::hOR leu2-3,112/leu2-801 GAL1::his3-Δ5' trp1::his3-Δ3'::HOcs rad4::kanMX/rad4 kanMX | pRS424 CYP1A1 | Trp+ transformant of YB427 |
| YB429 | MATa/MATα ura3-52/- his3-Δ200/- ade2-101/ade2-n trp1-Δ1/trp1::hOR leu2-3,112/leu2-801 GAL1::his3-Δ5' trp1::his3-Δ3'::HOcs rad4::kanMX/rad4 kanMX | pRS424 CYP1A1 I462V | Trp+ transformant of YB427 |
| YB430 | MATa/MATα ura3-52/- his3-Δ200/- ade2-101/ade2-n trp1-Δ1/trp1::hOR leu2-3,112/leu2-801 GAL1::his3-Δ5' trp1::his3-Δ3'::HOcs rad4::kanMX/rad4 kanMX | pRS424 CYP1A1 T461N | Trp+ transformant of YB427 |
| YB431 | MATa-inc ura3 rad4::KanMX rad51 | PRS424 CYP1A1 | Trp+ transformant of YB400 |
| YB432 | MATa-inc ura3 rad4::KanMX rad51 | pRS424 CYP1A1 I462V | Trp+ transformant of YB400 |
| YB433 | MATa-inc ura3 rad4::KanMX rad51 | pRS424 CYP1A1 T461 | Trp+ transformant of YB400 |
| YB434 | MATaYFP-rad51-I345T-URA3-RAD51 ADE2 leu2 trp1::hOR ura3 his3 | pRS424 CYP1A1 | Trp+ transformant of YB400 |
| YB435 | MATaYFP-rad51-I345T-URA3-RAD51 ADE2 leu2 trp1::hOR ura3 his3 | pRS424 CYP1A1 I462V | Trp+ transformant of YB419 |
| YB436 | MATaYFP-rad51-I345T-URA3-RAD51 ADE2 leu2 trp1::hOR ura3 his3 | pRS424 CYP1A1 T461N | Trp+ transformant of YB419 |
Strains containing trpl::hOR were confirmed by PCR using the forward oligo AGGAGACAGACGTGGATCTCTCTG and the reverse oligo AAGCCAAACACACCCAGGAGACTA. Both MATa and MATα strains were made by genetic crosses to the S288c-related strains. Since it is easier to detect DNA damage-associated recombination in diploid strains [41], the indicated strains were made by diploid crosses, as indicated in Table 1.
The rad4 rad51 strains for measuring BaP-DHD and AFB1 sensitivity are derived from YB226, which contains his3 recombination substrates in tandem at TRP1 [38]. A Ura− derivative (YB400) of the rad4 rad51 strain (YB226, [42]) was selected on 5-fluoroorotic acid (FOA) medium.
Strains used to detect Rad51 foci were derived from LSY1957, a gift of L. Symington [43]. This strain was crossed with YB407 and the meiotic segregant YB419 was obtained that contains both yfp-RAD51 and trpl::hOR.
2.5 Measuring carcinogen-associated recombination
To measure BaP-DHD and AFB1-associated recombination events, log phase yeast cells (A600 = 0.5–1) were centrifuged and concentrated five-fold in synthetic media (SC-URA or SC-TRP). Approximately 108 cells were exposed for four hours to indicated doses of BaP-DHD or AFB1, previously dissolved in DMSO. Cells were maintained in nutrient media (SC-URA or SC-TRP) during the carcinogen exposure and then washed twice in H2O. For measuring recombination, cells were directly plated on SC-HIS and an appropriate dilution was inoculated onto YPD to measure viability. Statistical significance was determined using the Student’s t-test.
To determine whether ionizing radiation stimulates the formation of Rad51 foci, cells were washed once in H2O and resuspended in 10 ml of H2O and placed in a 81 mm diameter Petri dish. Cells were irradiated at 6 krad using a Nordion 1.8kCi 137Cs irradiator (6 krad/hr).
After irradiation cells were concentrated in YPD medium and immobilized on glass slides.
2.6 Preparation of yeast microsomes and quantifying ethoxyresorufin O-deethylation (EROD) activities
Strains were grown to saturation in 75 mL SC-TRP media. The cells were pelleted, resuspended in 5 ml 50 mM Tris 1 mM EDTA (TE pH 7.4) buffer, incubated at room temperature for five minutes, and suspended in 0.6 M Sorbitol 50 mM Tris (pH 7.4). The cells were lysed by agitation in a bead beater at 4oC, and centrifuged for 10 minutes at 10000 × g. The supernatant from each sample was made 0.15 M in NaCl, and 20% in polyethylene glycol (MW 3350), and incubated on ice for at least one hour. Each tube was centrifuged at 10000 × g for 20 minutes. The supernatant was removed, and the resulting pellet was suspended in 50mM Tris (pH 7.4) and made 10% in glycerol; supernatants were stored in a -80oC freezer. Protein concentrations from yeast microsomes were determined by the Bradford assay, according to the manufacturer’s instructions. NADPH-dependent EROD activity was measured by resorufin fluorescence, as outlined by Mohammadi-Bardbori [44]. Final calculations were determined as picomoles resorufin/minute/mg protein.
2.7 Live cell epifluorescence and microscopy analysis
Cells for microscopic analysis were grown to early to mid-log phase overnight in synthetic medium. Strains harboring pRS424-CYP1A1 were cultured in SC-TRP. Cells were harvested by centrifugation and concentrated five-fold in the growth medium. Immobilization of cells was performed by mixing equal volumes of cell suspension and 1.4% low melt agarose plus growth medium solution. Cover slips were sealed with a wax mixture as described by Lisby et al. [45]. Slides were visualized using a Zeiss LSM 510 META confocal microscope.
2.8 Growth assays in 96 well plate to measure AFB1 and BaP-DHD sensitivity
In brief, individual saturated cultures were prepared for each yeast strain. Cell density was adjusted to ~0.8 × 107 cells/ml for all cultures. We maintained the cells in selective medium (synthetic dextrose lacking tryptophan). In each microtiter well, 95 μl of media and 5 μl of cells (8 × 104 cells) were aliquoted in triplicate for blank, control and experimental samples. For experimental samples, we added AFB1, dissolved in DMSO, for a final concentration of 100 nM and 10 μM. The microtiter dish was placed in a plate reader that is capable of both agitating and incubating the plate at 30o C, as previously described [33, 34]. We measured the A600 at 10 minute intervals, for a total period for 24 hrs, 145 readings. Data at 1hr intervals was then plotted. To avoid evaporation during the incubation, the microtiter dishes were sealed with clear optical tape.
2.9 Western blotting
Human CYP1A1 protein was detected in yeast microsomal extracts by Western blots, after semi-dry transfer from a SDS polyacrylamide gel onto nitrocellulose, similar to the protocol used to detect human CYP1A2 by Western blots. To detect CYP1A1, we used a mouse antibody raised against a Rat CYP1A2 protein (Abcam); this antisera contains significant cross-reactivity against human CYP1A1. We used a β-actin antibody (Abcam) that cross reacts with budding yeast actin (Act1), as a control.
3. Results
Previous studies indicated that human CYP1A1, when expressed on high-copy number expression vectors, can metabolically activate both AFB1 and BaP-DHD in budding yeast [29, 30]. We used genotoxic endpoints in budding yeast to detect differences in metabolic activation conferred by polymorphisms in the coding sequence of CYP1A1. By site-specific mutagenesis, CYP1A1 T461N and CYP1A1 I462V were individually introduced into the yeast vector pRS424-CYP1A1, and the entire CYP1A1 gene was sequenced to verify the individual base pair substitutions. To verify that equivalent levels of each CYP1A1 protein was expressed in each strain, we isolated yeast microsomes and performed Western blots, using a mouse antibody that recognized human CYP1A1; Act1 was used as an internal control and the ratio of signal (CYP1A1/ Act1) was measured and averaged for two Western blots. For strains expressing CYP1A1, CYP1A1 I462V, CYP1A1 T461N, we observed ratios of 1.3, 1.1, and 1.3, respectively. We conclude that equivalent levels of CYP1A1 protein are expressed in these strains expressing CYP1A1 alleles.
3.1 rad4 rad51 strains expressing CYP1A1 T461N and CYP1A1 I462V are less sensitive to AFB1 and BaP-DHD, compared to the strain expressing CYP1A1
Radiation sensitive cells defective in both recombinational repair and nucleotide excision repair are extremely sensitive to DNA damaging agents [33]. Sensitivity to chemical agents can be measured by growth curves, comparing growth in the presence and absence of the agent. We previously showed that rad4 rad51 cells expressing CYP1A2 are sensitive to 100 nM AFB1[33]. We exposed rad4 rad51 strains expressing CYP1A1, CYP1A1 T461N, and I462V, to AFB1, BaP-DHD, IQ and PhIP.
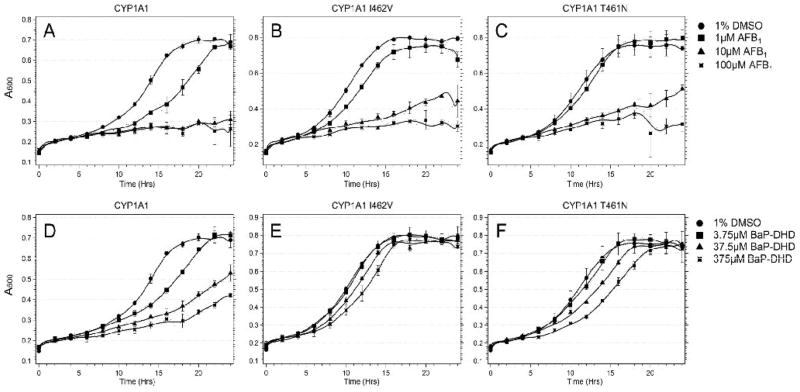
Results showing differences between strains expressing different CYPs were most notable for cells after chronic exposure to BaP-DHD and AFB1 (Figure 1). Cells expressing CYP1A1 I462V (YB432, see Table 1) were only sensitive to AFB1 and BaP-DHD at higher exposure levels, while cells expressing CYP1A1 (YB431) were sensitive at all exposure levels. After exposure to 10 μM AFB1, growth was equivalently reduced in cells expressing CYP1A1 I462V or CYP1A1 T461N, while growth was reduced the most (~65%) in cells expressing CYP1A1. After exposure to 37.5 uM BaP-DHD, growth was reduced the least in cells expressing CYP1A1 I462V, at an intermediate level for cells expressing CYP1A1 T461N (YB433), and at a maximum level for cells expressing CYP1A1. We conclude from these data that all three CYPs activate both BaP-DHD and AFB1, but growth of cells expressing CYP1A1 I462V were least affected by chronic BaP-DHD exposure.
Figure 1.
Growth curves of the DNA repair rad4 rad51 mutant expressing CYP1A1 polymorphisms after exposure to BaP-DHD and AFB1. Approximately 105 cells were inoculated in each well in a 96-well plate platform; yeast growth in each well was monitored in a Tecan Plate reader and over a period of twenty-four hours. Absorbance (A600) is plotted against time (hrs). The columns include the rad4 rad51 strain (YB431) expressing CYP1A1 (Left), the rad4 rad51 strain (YB432) expressing CYP1A1 I462V, and the rad4 rad51 strain (YB433) expressing CYP1A1 T461N. The first row (panels A, B, C) is growth curves of cells from each strain after exposure DMSO or indicated doses of AFB1. The second row (panels D, E, F) is growth curves of cells after exposure to DMSO or BaP-DHD. Error bars represent standard deviation (SD), where N = 2.
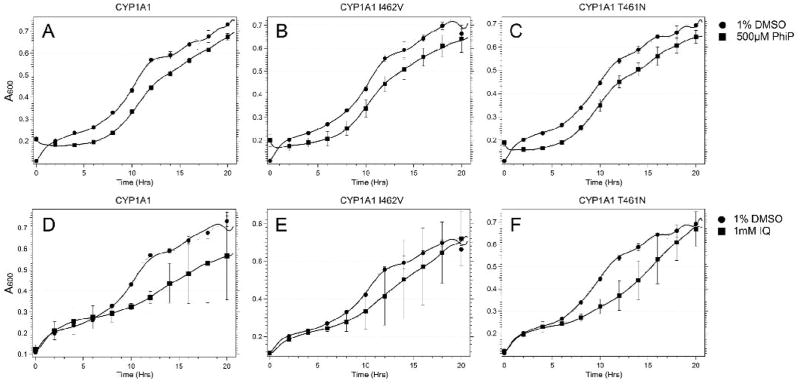
Expression of CYP1A1, CYP1A1 T461N and CYP1A1 I462V also reduced growth of the rad4 rad51 strain after chronic exposure to IQ and PhIP (Figure 2). Differences between strains were most notable after exposure to 10 mM IQ. Although a decrease in viability of the rad4 rad51 mutant expressing all three CYP1A1 alleles after PhIP exposure was detected, differences between these strains were not observed.
Figure 2.
Growth curves of the DNA repair rad4 rad51 mutant expressing CYP1A1 polymorphisms after exposure to IQ and PhIP. Using the protocol described in Panel A, yeast growth in each well was monitored in a Tecan Plate reader over a period of twenty hours. Absorbance (A600) is plotted against time (hrs). The columns include the rad4 rad51 strain (YB431) expressing CYP1A1 (Left), the rad4 rad51 strain (YB432) expressing CYP1A1 I462V, and the rad4 rad51 strain (YB433) expressing CYP1A1 T461N. The first row (panels A, B, C) curves of cells from each strain after exposure to DMSO or indicated doses of PhIP. The second row (panels D, E, F) is growth curves of cells after exposure to DMSO or IQ. Error bars represent standard deviation (SD), N = 2.
3.2 CYP1A1 I462V confers lower levels of carcinogen-associated recombination compared to CYP1A1 and CYP1A1 T461N
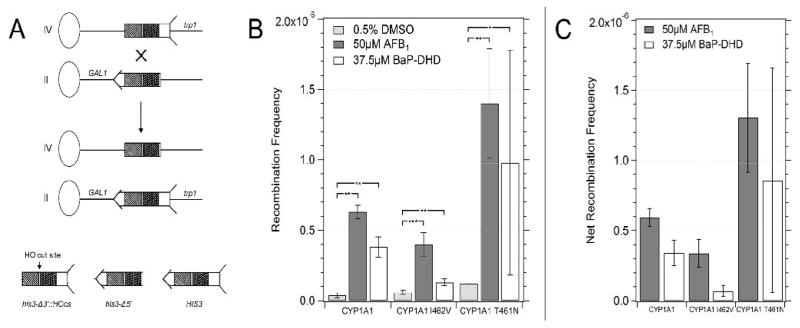
Expression of CYP1A1 and the human oxidoreductase (hOR) is sufficient to metabolically activate either AFB1 or BaP-DHD into potent yeast recombinagens [30]. We previously expressed CYP1A1 in a nucleotide excision repair (NER) mutant and observed enhanced recombination [29], resulting from a deficiency in repair of DNA bulky adducts. Here, we expressed in CYP1A1, CYP1A1 T461N and CYP1A1 I462V in a rad4 mutant and measured recombination between two his3 fragments, located on chromosomes II and IV (Figure 3). The frequencies of spontaneous recombination varied between 3.5 × 10−8 and 2 × 10−7 among strains, consistent with previously observed variations [38]. After exposure to 50 μM AFB1, we observed the highest level of recombination in cells expressing CYP1A1 T461N (YB430, fourteen-fold difference). However, cells expressing CYP1A1 (YB428) exhibited a higher-fold increase in recombination (seventeen-fold), due to lower levels of spontaneous recombination. Cells expressing CYP1A1 I462V exhibited the lowest frequency of AFB1-associated recombination. The survival in all the strains after acute AFB1 exposure was approximately 60–70%. Since there are essentially equivalent levels of CYP1A1 protein, compared to yeast actin, in all strains, we conclude that CYP1A1 I462V is the least efficient in activating AFB1 into a recombinagen.
Figure 3.
Frequencies of AFB1-associated recombination in diploid cells expressing CYP1A1 (YB428), CYP1A1 I462V (YB429), CYP1A1 T461N (YB430). Figure in panel A is a model of the recombination assay. The oval represents a centromere and the line represents the chromosome; the left arm of the chromosome is not shown for simplicity. The his3 fragment is shown with arrow and feathers. The shaded areas represent shared homology. An ”X” denotes where a cross-over event would occur. The product of the recombination event is shown below where CEN2 is linked to the long arm of chromosome IV and CEN4 is linked to the long arm of chromosome II. Figure in panel B shows the recombination frequencies after cells were exposed to DMSO (solid column), 50 μM of AFB1 (horizontally striped column) and 37.5 μM of BaP-DHD (diagonally-striped column). Figure in panel C shows the net recombination frequencies, which are obtained by subtracting the spontaneous recombination frequency from the carcinogen-associated frequency. The CYP1A1 allele is indicated on the X axis. Error bars represent standard deviation (SD), N = 3.
Carcinogen-associated recombination was also observed in rad4 cells after exposure to BaP-DHD. We detected a ten-fold higher level of carcinogen-associated recombination in cells expressing CYP1A1, while cells expressing CYP1A1 I462V (YB429) only exhibited a two-fold higher level of recombination. Cells expressing CYP1A1 T461N also exhibited BaP-DHD associated recombination; however, there is no significant difference in recombination frequencies compared to cells expressing the other CYP1A1 alleles. Thus, the carcinogen-associated recombination is lower in cells expressing CYP1A1 I462V, compared to cells expressing CYP1A1. These data are consistent with the lower levels of BaP-DHD -associated toxicity observed in the rad4 rad51 strain (YB429) expressing CYP1A1 I462V.
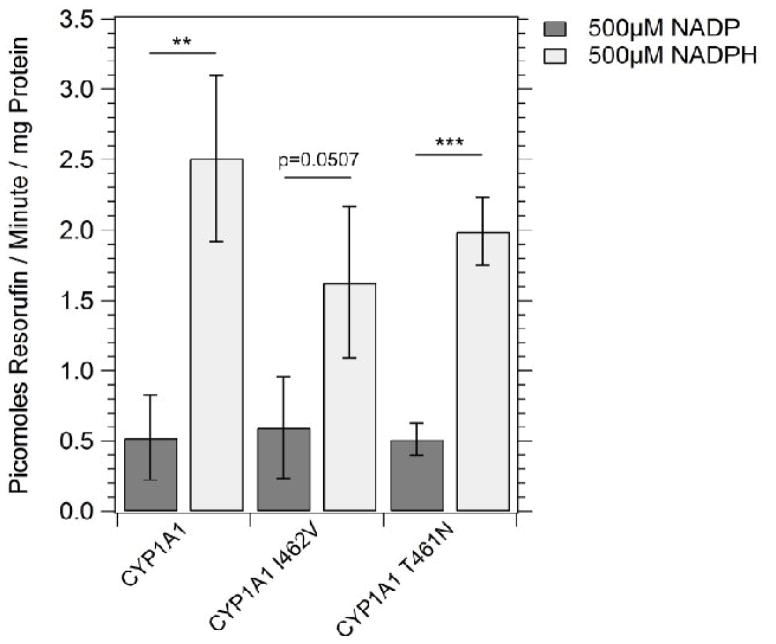
3.3 EROD activity in strains expressing CYP1A1, CYP1A1 T461N and CYP1A1 I462V
To verify that CYP1A1 variants were enzymatically active in yeast, we performed EROD assays ([44], Figure 4). We compared the EROD activity of cells expressing CYP1A1 I462V (YB429) with those of cells expressing CYP1A1 (YB428) and CYP1A1 T461N (YB430). NADPH-dependent activity was detected in microsome extracts of cells expressing all CYP1A1 variants (see supplemental Figure 1 for an example), and ranged from 0.5 pmol resorufin/min/mg to 2.5 pmol resorufin/min/mg (Figure 4). No EROD activity was detected in cells containing an empty vector (pRS424). We conclude that while EROD activity can be detected in cells expressing each CYP1A1 allele, cells expressing CYP1A1 I462V have comparable if not higher EROD activity compared to that detected in cells expressing other CYP1A1 alleles.
Figure 4.
Ethoxyresorufin-O-deethylase (EROD) activity measured in strains expressing CYP1A1 (YB428), CYP1A1 I462V (YB429), and CYP1A1 T461N (YB430). The measurements (pmol resorufin/min/mg protein) were made using yeast microsomes. Error bars represent standard deviation (SD), N =3.
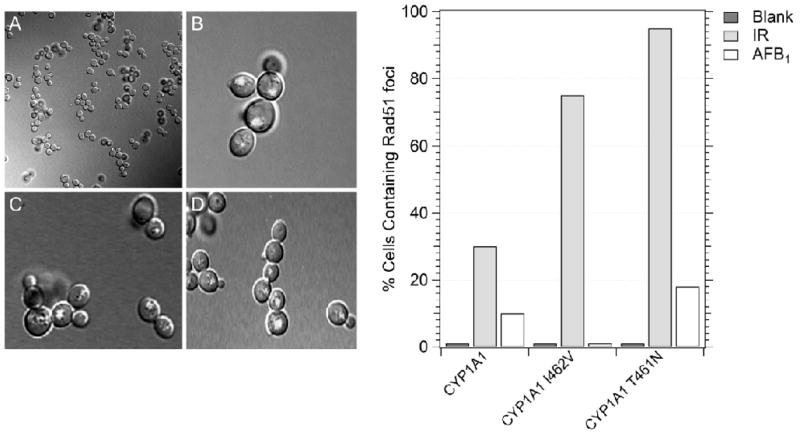
3.4 Rad51 foci appear after AFB1 exposure in cells expressing CYP1A1, CYP1A1 T461N and CYP1A1 I462V
We previously observed DNA damage-associated Rad51 foci in CYP1A2-expressing cells after exposure to AFB1 or γ rays (34). While radiation-associated Rad51 foci appeared in cells arrested in G2, AFB1-associated Rad51 foci appeared in both small-budded cells entering S phase and large dumb-bell shaped cells, indicative of G2- arrested cells. To observe Rad51 foci in cells expressing CYP1A1 alleles, we constructed a yfp-Rad51 strain in which hOR was integrated in the genome at the TRP1 locus. Less than one percent of cells (0/153) expressing either CYP1A1 (YB434), CYP1A1 T461N (YB436) or CYP1A1 I462V (YB435) were observed to exhibit Rad51 foci in the absence of DNA damage exposure, consistent with previous studies ([34], Figure 5). After exposure to ionization radiation, 30–90% of the irradiated cells from all strains exhibited Rad51 foci, and these cells were G2 arrested. In the strain that did not express P450, < 1% of the cells exhibited Rad51 foci after AFB1-exposure. The percentage of AFB1-exposed cells expressing either CYP1A1 or CYP1A1 T461N that exhibited Rad51 foci was essentially the same (12–13%), but significantly different from the number of Rad51 foci from cells that were not exposed (P < 0.05, Chi square). However, in the strain that expressed CYP1A1 I462V, <5% of the AFB1-exposed cells exhibited Rad51 foci. These data indicate that CYP1A1-mediated activation that confers the highest level of AFB1-associated recombination also correlates with the presence of Rad51 foci.
Figure 5.
Qualitative analysis of yfp-Rad51 foci in cells expressing CYP1A1 (YB434), CYP1A1 I462V (YB435), and CYP1A1 T461N (YB436). Rad51 foci were identified by the appearance of yellow fluorescent foci in the confocal microscope. Left panel shows A) non-irradiated cells containing pRS424-CYP1A1, B) Rad51 foci in AFB1-treated cells containing CYP1A1, C) Rad51 foci irradiated cells containing CYP1A1, D) Rad51 foci in AFB1-treated cells containing CYP1A1 T461N. The graph shows the percentage of CYP1A1-expressing cells containing Rad51 foci after either no exposure, exposure to AFB1 or 10 Gy irradiation. The percentage of cells containing foci is indicated on the y axis and the CYP1A1-expressing strain is identified on the X axis.
Discussion
Allelic variants of CYP1A1, such as CYP1A1 I462V and CYP1A1 T461N, are alleged risk factors for specific cancers, including endometrial, lung, and breast cancer. The two alleles result from amino acid substitutions that map near the active site of the enzyme [46] , and CYP1A1 I462V has been reported to be overly represented in cancer patients [3, 47]. Considering that environmental agents are risk factors for lung and breast cancers, one hypothesis is that these CYP1A1 alleles confer a higher risk for cancer because they encode enzymes that have higher metabolic activities. We expressed CYP1A1 alleles in budding yeast, exposed cells to P450-activated carcinogens and measured different genotoxic endpoints that indicate DNA damage. This is the first time genotoxic endpoints have been directly measured in yeast to phenotype CYP1A1 alleles.
Two main conclusions can be derived from our results: 1) CYP1A1 I462V confers less carcinogen-associated genotoxicity compared to that conferred by the expression of the other alleles in yeast, and 2) phenotypic differences in CYP1A1-mediated activation are carcinogen dependent. These conclusions may appear to contradict the notion that CYP1A1 I462V is a risk factor for cancers associated with xenobiotic exposures. However, we offer two different explanations: 1) higher levels of genotoxicity may be detected in cells expressing CYP1A1 I462V after exposure to chemicals not tested in this study, and 2) in particular human tissues, other CYPs, not coupled to phase II metabolism, may activate genotoxic compounds when CYP1A1 activity is low. Thus, we speculate that altered CYP1A1 metabolism may be a risk factor for environmentally-related cancer for multiple reasons.
CYP1A1 I462V, compared to CYP1A1 and CYP1A1 T461N, conferred lower levels of carcinogen-associated genotoxicity, as indicated by multiple endpoints. The most salient data were growth curves in the presence of BaP-DHD, AFB1-associated recombination, and carcinogen-associated Rad51 foci. Although growth curves of cells expressing CYP1A1 I462V and CYP1A1 T461N in the presence of AFB1 did not appear different, CYP1A1 also metabolizes AFB1 to generate a hydroxylated form (AFM1, [48]) and it is unclear whether the AFB1-associated metabolites that initiate recombination and those that reduce growth rates are the same. In vitro experiments are necessary to elucidate the enzymatic properties of CYP1A1 variants when AFB1 is a substrate.
Differences in CYP1A1-mediated activation depend on the carcinogen or substrate. While there was no observable difference in AFB1-associated genotoxic effects in cells expressing CYP1A1 T461N and CYP1A1 I462V, there was a significant difference between BaP-DHD-associated genotoxic effects in cells expressing CYP1A1 T461N and CYP1A1 I462V. In addition, we observed equivalent levels of EROD activity from extracts derived from CYP1A1 I462V cells, compared to those expressing other CYP1A1 alleles. Higher levels of CYP1A1 I462V-associated EROD activity but lower levels of BaP-associated and BaP-DHD metabolism have been observed in protein extracts derived from ectopic expression of these CYP1A1 variants in bacteria [24, 49]. Considering that amino acid substitutions in exon 7 of CYP1A1 map near the active site, we speculate that the enzyme encoded by CYP1A1 I462V may differentially accommodate different substrates.
Previous studies have suggested that CYP1A1 may have a protective effect against some carcinogens [50]. Nebert et al. [50, 51] reported that CYP1A1−/− mice exhibit higher toxicity to BaP; and suggested that CYP1B1 is responsible for metabolic activation of BaP into a potent carcinogen [51]. One explanation is that CYP1A1 and phase II enzymatic activities, such as GSTM, are closely coupled; thus, BaP is more rapidly hydroxylated and genotoxic intermediates detoxified and excreted, reducing the cancer risk for particular xenobiotic exposures [53, 54].
We suggest two reasons for why CYP1A1 I462V may be a risk factor for some cancers. First, due to its lower activity, xenobiotics are metabolized by other CYPs whose activities are not tightly coupled with phase II enzymes. Second, compared to wild type, CYP1A1 I462V more efficiently activates other compounds associated with cancer risk besides BaP. Supporting the first idea are observations that CYP1A1 I462V obtained from ectopic expression in bacteria is less active using BaP and BaP-DHD as substrates in vitro, compared to CYP1A1[50, 51]. Our data supports these observations by showing that, compared to yeast expressing CYP1A1, CYP1A1 I462V confers less BaP-DHD-associated genotoxicity. While the in vitro characterization of the CYP1A2 I462V enzyme may confound the interpretation of observations that leukocytes from individuals bearing CYP1A2 I462V have higher levels of DNA adducts [49], it is possible that exposure to other xenobiotics, besides BaP, may be responsible for the increased risk in carrier patients. Thus, additional carcinogens should be tested to identify which compounds are most genotoxic in yeast cells expressing CYP1A1 I462V.
Our studies are an extension of previous work illustrating that AFB1 is activated by CYP1A2 polymorphisms [34]. Here we have shown that both PAHs and heterocyclic aromatic amines (HAAs) can be activated by CYP1A1 alleles expressed in yeast, extending the phenotypic characterization of these alleles. The availability of multiple genotoxic endpoints in yeast allows for further investigation of CYP1A1-activated substrates and metabolites, with the goal of elucidating xenobiotics that trigger the highest levels of genotoxicity. These studies can be extended to include other CYP1A1 polymorphisms and phase II enzymes..
Supplementary Material
Highlights for Review.
Phenotypic characterization of carcinogen activation conferred by CYP1A1 alleles
CYP1A1 I462V expression confers lower levels of carcinogen-associated genotoxicity compared to CYP1A1 T461N and CYP1A1.
Expression of CYP1A1 1462V can be measured by EROD assays and is robust.
Acknowledgments
This research was supported by grants R21ES015954 and R15 ES023685-01 (MF) from the National Institutes of Health. We thank Autumn Smith for her technical support, Gary Schools for expertise with the confocal microscope, Patricia Zupan for quantifying Rad51 foci, and Nick St. John for assistance with the Western blots. We thank Xinxin Ding for carefully reading this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ding X, Kaminsky LS. Human Extrahepatic Cytochromes P450: Function in Xenobiotic Metabolism and Tissue-Selective Chemical Toxicity in the Respiratory and Gastrointestinal Tracts. Ann Rev of Pharmacology and Toxicology. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 2.Ioannides C, Lewis DF. Cytochromes P 450 in the bioactivation of chemicals. Curr Top Med Chem. 2004;4:1767–1788. doi: 10.2174/1568026043387188. [DOI] [PubMed] [Google Scholar]
- 3.Androutsopoulos VP, Tsatsakis AM, Spandidos DA. Cytochrome P450 CYP1A1: wider roles in cancer progression and prevention. BMC Cancer. 2009;9:187. doi: 10.1186/1471-2407-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badal S, Delgoda R. Role of the modulation of CYP1A1 expression and activity in chemoprevention. J Appl Toxicol. 2014;7:743–753. doi: 10.1002/jat.2968. [DOI] [PubMed] [Google Scholar]
- 5.Alexandrov K, Cascorbi I, Rojas M, Bouvier G, Kriek E, Bartsch H. CYP1A1 and GSTM1 genotypes affect benzo[a]pyrene DNA adducts in smokers' lung: comparison with aromatic/hydrophobic adduct formation. Carcinogenesis. 2002;23:1969–1977. doi: 10.1093/carcin/23.12.1969. [DOI] [PubMed] [Google Scholar]
- 6.Guengerich FP, Johnson WW, et al. Activation and detoxication of aflatoxin B1. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1998;402:121–128. doi: 10.1016/s0027-5107(97)00289-3. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Q, Sha X, Liu J, Heath E, Lorusso P, Li J. Association of Human Cytochrome P450 1A1 (CYP1A1) and Sulfotransferase 1A1 (SULT1A1) Polymorphisms with Differential Metabolism and Cytotoxicity of Aminoflavone. Molecular Cancer Therapeutics. 2010;9:2803–2813. doi: 10.1158/1535-7163.MCT-10-0597. [DOI] [PubMed] [Google Scholar]
- 8.Reid JM, Kuffel MJ, Miller JK, Rios R, Ames MM. Metabolic activation of dacarbazine by human cytochromes P450: the role of CYP1A1, CYP1A2, and CYP2E1. Clin Cancer Res. 1999;5:2192–2197. [PubMed] [Google Scholar]
- 9.Agbor LN, Wiest EF, Rothe M, Schunck WH, Walker MK. Role of CYP1A1 in modulating the vascular and blood pressure benefits of omega-3polyunsaturated fatty acids. J Pharmacol Exp Ther. 2014;351 doi: 10.1124/jpet.114.219535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi S-I, Watanabe J, Nakachi K, Kawajin K. PCR detection of an A/G polymorphism within exon 7 of the CYP1A1 gene. Nucl Acids Res. 1991;19:4797. doi: 10.1093/nar/19.17.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Zhao Q, Zhai Y-J, He H-R, Yang L-H, Gao F, … Ma X-C. Genetic polymorphisms of CYP1A1 and risk of leukemia: a meta-analysis. OncoTargets and Therapy. 2015;8:2883–2902. doi: 10.2147/OTT.S92259. http://doi.org/10.2147/OTT.S92259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan P, Wang Q, Qian Q, Wei SZ, Yu LK. CYP1A1 MspI and exon7 gene polymorphisms and lung cancer risk: an updated meta-analysis and review. J Exp Clin Cancer Res. 2011;30:99. doi: 10.1186/1756-9966-30-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin A, Kang D, Choi J, Lee K, Park S, Noh D, Yoo K. Cytochrome P450-1A1 (CYP1A1) polymorphisms and breast cancer risk in Korean women. Exp Mol Med Experimental & Molecular Medicine. 2007;39:361–366. doi: 10.1038/emm.2007.40. (n.d.) [DOI] [PubMed] [Google Scholar]
- 14.Diergaarde B, Potter J, Jupe E, Manjeshwar S, Shimasaki C, Pugh T, Defreese DC, Gramling BA, Evans I, White E. Polymorphisms in Genes Involved in Sex Hormone Metabolism, Estrogen Plus Progestin Hormone Therapy Use, and Risk of Postmenopausal Breast Cancer. Cancer Epidemiology Biomarkers & Prevention. 2008;17:1751–1759. doi: 10.1158/1055-9965.EPI-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sergentanis TN, Economopoulos KP. Four polymorphisms in cytochrome P450 1A1 (CYP1A1) gene and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122:459–69. doi: 10.1007/s10549-009-0694-. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Ramírez O, Pérez-Morales R, Castro C, Flores-Díaz A, Soto-Cruz K, Astorga-Ramos A, Rubio J. Polymorphisms of catechol estrogens metabolism pathway genes and breast cancer risk in Mexican women. The Breast. 2013;22:335–343. doi: 10.1016/j.breast.2012.08.004. (n.d.) [DOI] [PubMed] [Google Scholar]
- 17.Meng FD, Ma P, Sui CG, Tian X, Jiang YH. Association between cytochrome P450 1A1 (CYP1A1) gene polymorphisms and the risk of renal cell carcinoma: a meta-analysis. Sci Rep. 2015;5:8108–8114. doi: 10.1038/srep08108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cascorbi I, Brockmöller J, Roots I. A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res. 1996;56:4965–4969. [PubMed] [Google Scholar]
- 19.Esteller M, Garcia A, Martinez-Palones JM, Xercavins J, Reventos J. Germ line polymorphisms in cytochrome-P450 1A1 (C4887 CYP1A1) and methylenetetrahydrofolate reductase (MTHFR) genes and endometrial cancer susceptibility. Carcinogenesis. 1997;18:2307–2311. doi: 10.1093/carcin/18.12.2307. [DOI] [PubMed] [Google Scholar]
- 20.http://www.cypalleles.ki.se/cyp1a1.htm
- 21.Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, Baranova H, Bathum L, Benhamou S, Boffetta P, Bouchardy C, Breskvar K, Brockmoller J, Cascorbi I, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–1248. [PubMed] [Google Scholar]
- 22.Wild CP. Environmental exposure measurement in cancer epidemiology. Mutagenesis. 2009;24:117–125. doi: 10.1093/mutage/gen061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K. Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev. 2000;9:3–28. [PubMed] [Google Scholar]
- 24.Zhang ZY, Fasco MJ, Huang L, Guengerich FP, Kaminsky LS. Characterization of purified human recombinant cytochrome P4501A1-Ile462 and -Val462: assessment of a role for the rare allele in carcinogenesis. Cancer Res. 1996;56:3926–3933. [PubMed] [Google Scholar]
- 25.Schwarz D, Kisselev P, Honeck H, Cascorbi I, Schunck W-H, Roots I. Co-expression of human cytochrome P4501A1 (CYP1A1) variants and human NADPH-cytochrome P450 reductase in the baculovirus/ insect cell system. Xenobiotica: the fate of foreign compounds in biological systems. 2001;31(6):345–356. doi: 10.1080/00498250110055947. [DOI] [PubMed] [Google Scholar]
- 26.Kisselev P, Schunck W-H, Roots I, Schwarz D. Association of CYP1A1 Polymorphisms with Differential Metabolic Activation of 17β-Estradiol and Estrone. Cancer Res. 2005;65:2972–2978. doi: 10.1158/0008-5472.CAN-04-3543. [DOI] [PubMed] [Google Scholar]
- 27.Georgiadis P, Topinka J, Vlachodimitropoulos D, Stoikidou M, Gioka M, Stephanou G, Autrup H, Demopoulos NA, Katsouyanni K, Sram R, Kyrtopoulos SA. Molecular Epidemiology and Cancer Prevention: Interactions between CYP1A1 polymorphisms and exposure to environmental tobacco smoke in the modulation of lymphocyte bulky DNA adducts and chromosomal aberrations. Carcinogenesis. 2005;26:93–101. doi: 10.1093/carcin/bgh294. [DOI] [PubMed] [Google Scholar]
- 28.Lodovici M, Luceri C, Guglielmi F, Bacci C, Akpan V, Fonnesu ML, Boddi V, Dolara P. Benzo(a)pyrene diolepoxide (BPDE)-DNA adduct levels in leukocytes of smokers in relation to polymorphism of CYP1A1, GSTM1, GSTP1, GSTT1, and mEH. Cancer Epidemiol Biomarkers Prev. 2004;13:1342–1348. [PubMed] [Google Scholar]
- 29.Keller-Seitz M, Certa U, Sengstag C, Wurgler F, Sun M, Fasullo M. Transcriptional response of the yeast to the carcinogen Aflatoxin B1: Recombinational repair involving RAD51 and RAD1. Mol Biol Cell. 2004;15:4321–4336. doi: 10.1091/mbc.E04-05-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengstag C, Weibel B, Fasullo M. Genotoxicity of aflatoxin B1: evidence for a recombination-mediated mechanism in Saccharomyces cerevisiae. Cancer Res. 1996;56:5457–5465. [PMC free article] [PubMed] [Google Scholar]
- 31.Fasullo M, Sun M, Egner P. Stimulation of sister chromatid exchanges and mutation by aflatoxin B1-DNA adducts in Saccharomyces cerevisiae requires MEC1 (ATR), RAD53, and DUN1. Mol Carcinog. 2008:608–615. doi: 10.1002/mc.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengstag C, Würgler FE. DNA recombination induced by aflatoxin B1 activated by cytochrome P450 1A enzymes. Mol Carcinog. 1994;11:227–235. doi: 10.1002/mc.2940110408. [DOI] [PubMed] [Google Scholar]
- 33.Fasullo M, Chen Y, Bortcosh W, Sun M, Egner PA. Aflatoxin B(1)-Associated DNA Adducts Stall S Phase and Stimulate Rad51 foci in Saccharomyces cerevisiae. J Nucleic Acids. 2010;1:456–487. doi: 10.4061/2010/456487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fasullo M, Smith A, Egner P, Cera C. Activation of aflatoxin B1 by expression of human CYP1A2 polymorphisms in Saccharomyces cerevisiae. Mutat Res Genet Toxicol Environ Mutagen. 2014;761:18–26. doi: 10.1016/j.mrgentox.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burke D, Dawson D, Stearns T. Methods in yeast genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Press; New York, NY: 2000. [Google Scholar]
- 36.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1 Short Protocols in Molecular Biology. 4. Vol. 1. Wiley; New York: 1999. [Google Scholar]
- 37.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fasullo MT, Davis RW. Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc Natl Acad Sci U S A. 1987;84:6215–6219. doi: 10.1073/pnas.84.17.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fasullo M, Bennett T, AhChing P, Koudelik J. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol Cell Biol. 1998;18:1190–1200. doi: 10.1128/mcb.18.3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasullo MT, Davis RW. Direction of chromosome rearrangements in Saccharomyces cerevisiae by use of his3 recombinational substrates. Mol Cell Biol. 1988;8:4370–4380. doi: 10.1128/mcb.8.10.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fasullo M, Dave P, Rothstein R. DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae. Mutat Res. 1994;314:121–133. doi: 10.1016/0921-8777(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 42.Dong Z, Fasullo M. Multiple recombination pathways for sister chromatid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes. Nucleic Acids Res. 2003;31:2576–2585. doi: 10.1093/nar/gkg352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fung CW, Mozlin AM, Symington LS. Suppression of the double-strand-break- repair defect of the Saccharomyces cerevisiae rad57 mutant. Genetics. 2009;181:1195–1206. doi: 10.1534/genetics.109.100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohammadi-Bardbori A. Assay for quantitative determination of CYP1A1 enzyme activity using 7-ethoxyresorufin as standard substrate (EROD assay) Protocol Exchange Nature. 2014 doi: 10.1038/protex.2014.043. [DOI] [Google Scholar]
- 45.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 46.Lewis BC, Mackenzie PI, Miners JO. Comparative homology modeling of human cytochrome P4501A1 (CYP1A1) and confirmation of residues involved in 7-ethoxyresorufin O-deethylation by site-directed mutagenesis and enzyme kinetic analysis. Arch Biochem Biophys. 2007;468:58–69. doi: 10.1016/j.abb.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Androutsopoulos VP, Spyrou I, Ploumidis A, et al. Expression Profile of CYP1A1 and CYP1B1 Enzymes in Colon and Bladder Tumors. In: Afarinkia K, editor. PLoS ONE. Vol. 8. 2013. pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neal GE, Eaton DL, Judah DJ, Verma A. Metabolism and toxicity of aflatoxins M1 and B1 in human-derived in vitro systems. Toxicol Appl Pharmacol. 1998;151:152–158. doi: 10.1006/taap.1998.8440. [DOI] [PubMed] [Google Scholar]
- 49.Schwarz D, Kisselev P, Honeck H, Cascorbi I, Schunck W-H, Roots I. Differential metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-dihydrodiol by human CYP1A1 variants. Carcinogenesis. 2001;22:453–459. doi: 10.1093/carcin/22.3.453. doi:10:1093/carcin/22.3.453. [DOI] [PubMed] [Google Scholar]
- 50.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nature Reviews Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 51.Nebert DW, Shi Z, Gálvez-Peralta M, Uno S, Dragin N. Oral Benzo[a]pyrene: Understanding Pharmacokinetics, Detoxication, and Consequences—Cyp1 Knockout Mouse Lines as a Paradigm. Molecular Pharmacology. 2013;84:304–313. doi: 10.1124/mol.113.086637. http://doi.org/10.1124/mol.113.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moorthy B, Chu C, Carlin DJ. Polycyclic Aromatic Hydrocarbons: From Metabolism to Lung Cancer. Toxicol Sci. 2015;145:5–15. doi: 10.1093/toxsci/kfv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rojas M, Cascorbi I, Alexandrov K, Kriek E, Auburtin G, Mayer L, Kopp-Schneider A, Roots I, Bartsch H. Modulation of benzo[a]pyrene diolepoxide–DNA adduct levels in human white blood cells by CYP1A1, GSTM1 and GSTT1 polymorphism. Carcinogenesis. 2000;21(1):35–41. doi: 10.1093/carcin/21.1.35. [DOI] [PubMed] [Google Scholar]
- 54.Agudo A, Peluso M, Sala N, Capellá G, Munnia A, Piro S, Marín F, Ibáñez R, Amiano P, Tormo MJ, Ardanaz E, Barricarte A, Chirlaque MD, Dorronsoro M, Larrañaga N, Martínez C, Navarro C, Quirós JR, Sánchez MJ, González CA. Aromatic DNA adducts and polymorphisms in metabolic genes in healthy adults: findings from the EPIC-Spain cohort. Carcinogenesis. 2009;30:968–976. doi: 10.1093/carcin/bgp062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.