Abstract
Context:
Primary ovarian insufficiency (POI), or premature ovarian failure, results from ovarian follicle depletion with a consequent elevation of FSH levels before age 40 years. We identified a family in which 9 women in 3 consecutive generations developed menopause at approximately age 30 years. We hypothesized a genetic cause with a dominant mode of inheritance.
Design:
This was a family-based genetic study and a replicate group of women with POI.
Setting:
The study was conducted at an academic medical center.
Patients:
Seven affected women and an obligate carrier and 7 unaffected family members were genotyped. The genes of interest were also sequenced in 38 unrelated women with POI.
Intervention:
The DNA from 7 family members was subjected to whole-exome sequencing. The genotypes of interest were confirmed and genotypes of additional family members and unrelated women with POI were determined using Sanger sequencing.
Main Outcome Measure:
A high-impact, deleterious variant that segregated appropriately with POI in the family was required.
Results:
A heterozygous stop codon (Ser429X) was identified in the eukaryotic translation initiation factor 4E nuclear import factor 1 (eIF4ENIF1) in the proband and all affected women but not in the unaffected family members. The chance that such a high-impact, deleterious variant would segregate appropriately among the affected and unaffected relatives by chance is very low (P < .05). There were no additional mutations identified in eIF4ENIF1 or eIF4E in 38 unrelated women with POI.
Conclusion:
Data demonstrate a new gene associated with dominantly inherited POI. These results highlight the importance of translation initiation factors and their regulators in ovarian function.
Primary ovarian insufficiency (POI), previously termed premature ovarian failure, is characterized by early ovarian follicle depletion, resulting in low estradiol levels and an elevated FSH level. It affects 1% of women before the age of 40 years (1). The list of genetic causes of idiopathic POI is expanding, yet the etiology in a majority of the clinical cases remains unknown (1, 2).
We studied a large family with a history of POI, in which multiple affected women in 3 generations developed menopause at approximately 30 years of age. Therefore, a dominant mode of inheritance was hypothesized. Whole-exome sequencing demonstrated a heterozygous nonsense mutation in the eukaryotic translation initiation factor 4E nuclear import factor 1 (eIF4ENIF1) (3). The transport protein plays an important role in repressing translation by eukaryotic translation initiation factor 4E (eIF4E), a rate-limiting factor in translation initiation (4). It also appears to play an important role in ovarian germ cell development (5).
Materials and Methods
Case reports
The proband presented at age 29 years with 1 year of secondary amenorrhea after stopping oral contraceptives to become pregnant. Her physical examination was normal, with a height of 157 cm, normal skeletal development, and a normal neurologic examination. Her FSH level was elevated at 70 IU/L. Karyotype and FMR1 repeat length were normal and adrenal cortical antibodies were not detected.
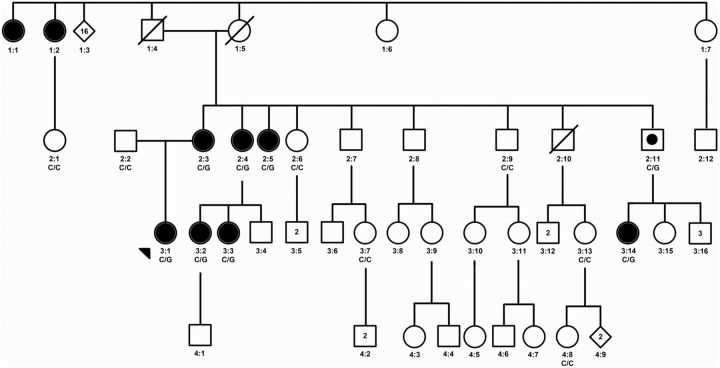
The proband belongs to a large family with an extensive history of POI, in which the affected arm of the family is of French Canadian ethnicity (Figure 1). Affected women experienced POI, resulting in early menopause (family member, age at menopause: 1:1, 33 y; 1:2, 35 y; 2:3, 29 y; 2:4, 30 y; 2:5, 35 y; 3:2, 30 y; 3:3, 30 y; 3:14, 35 y). An autosomal dominant inheritance pattern was hypothesized.
Figure 1.
Family pedigree of women with POI. This figure shows the pedigree of the proband (arrow) and her family. Solid circles indicate the affected female family members with menopause between the ages of 29 and 35 years; open symbols represent the unaffected family members and a square with a solid circle indicates a male carrier. Available genotypes at coding position 1286 in eIF4ENIF1 are indicated below the symbols, with C/C wild-type variants and C/G wild-type and mutant variant coding for a stop codon in the protein.
The women were all of normal height (family member, height: 2:3, 152 cm; 2:4, 158 cm; 2:5, 155 cm; 3:2, 165 cm; 3:3, 152 cm; 3:14, 150 cm). Additional medical history in the affected women was notable for an aunt with hypothyroidism at age 70 years (2:5) and a cousin with 3 miscarriages before a successful pregnancy and delivery (3:2). There was no other history of autoimmune disease or medical problems.
Genotyping
DNA was extracted from whole blood or saliva from 14 members of the maternal family of the proband and her father (Figure 1). Whole-exome sequencing was performed on the proband, her father, and 5 affected female relatives to identify a causative mutation.
In addition, DNA was isolated from whole blood of 38 unrelated women with nonsyndromic POI. All of these women had amenorrhea for 6 months, an elevated FSH level before the age of 40 years and no karyotype abnormalities, FMR1 premutations, or positive adrenal cortical antibodies.
The study was approved by the Institutional Review Board of the Massachusetts General Hospital. All subjects provided written informed consent.
Genomic DNA was sheared to 200–300 bp using a Covaris acoustic adaptor, as previously described (6). Exons were captured using the Agilent 38Mb SureSelect version 2 oligonucleotide libraries (Agilent Technologies) and subjected to DNA sequencing on an IlluminaHiSeq2000 (Illumina), and genotype calls were made at targeted bases (6).
Data were processed with Picard (picard.sourceforge.ne) and local realignment to the human reference genome GRCh37/hg19 using Burrows-Wheeler aligner (7, 8). Variant calling was performed using GATK (7, 9) through the Analytical and Translational Genetics Unit Exome Browser (xBrowse; atgu.mgh.harvard.edu/xbrowse). We searched for heterozygous dominant variants with high impact (nonsense, essential splice site, and frame shift) shared by the proband and her affected family members but absent in the father and 6503 European or African American subjects in the Exome Variant Server (10), 1092 subjects of differing ethnicity in the 1000 Genomes Project (11), and 870 European Analytical and Translational Genetics Unit controls (6).
Results of whole-exome sequencing were confirmed by Sanger sequencing in all subjects. In addition, 6 women (1 affected and 5 unaffected) and 2 men (one obligate carrier and one with 2 unaffected daughters) were genotyped.
mRNA analysis
A fresh blood sample was obtained from 3 affected women and 3 unrelated control subjects for immediate mRNA isolation to evaluate the transcript expression levels of the wild-type C and mutant G alleles (QIAGEN RNeasy mini for blood). cDNA was reverse transcribed after deoxyribonuclease treatment (Invitrogen). PCR was performed to amplify a DNA segment bridging exons 9–11 of eIF4ENIF1. The product encompasses the mutation but results in a sequence that is different from that of the genomic DNA. Specifically, it contains a portion of exons 9, 10, and 11 but does not amplify genomic DNA (gDNA) because of the large introns, ensuring that only cDNA was represented in the resulting sequence. A ratio of the peak height of the G to C allele from cDNA in affected women was compared with that in controls and was compared with the peak height of the G to C allele from gDNA in affected women using a t test (12).
Genotyping in unrelated women with POI
The 5′ untranslated region and all coding exons in eIF4ENIF1 and eIF4E were sequenced in the 38 unrelated women with POI (Polymorphic DNA Technologies, Inc). Minor allele frequencies of single-nucleotide polymorphisms were compared with those in the National Heart, Lung, and Blood Institute Exome Sequencing Project using χ2 (10).
Results
Genotypes in POI family
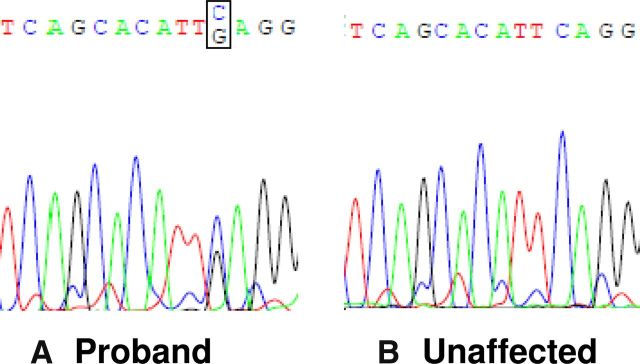
Analysis of the whole-exome sequencing data demonstrated a heterozygous mutation in eIF4ENIF1 (NM_019843) in the 6 affected women (3:1, 2:3, 2:4, 2:5, 3:2, 3:14) but not in the father (2:2) or the control group (6, 10, 11). The mutation in eIF4ENIF1 is located on chromosome 22, position 31850356, exon 10, coding base pair c. 1286C>G. The mutation results in a change from a serine to a stop (p. Ser429X). The mutation was confirmed by Sanger sequencing in all subjects (Figures 1 and 2). Of note, there are fewer than 10 rare nonsense, essential splice site, and frame shift variants per individual control in the data set (6, 10, 11). Therefore, the chance of finding a high-impact, deleterious variant in the proband that segregates appropriately among the affected and unaffected relatives is very low (P < .05).
Figure 2.
Genotypes of 2 women in a family with POI. This figure shows electropherograms and the gDNA sequence of the identified mutation in the proband (3:1) (A) and an unaffected family member (2:1) (B).
Sanger sequencing eIF4ENIF1 exon 10 in 5 additional unaffected female family members (2:1, 2:6, 3:7, 3:13, and 4:8) and one male with two unaffected daughters (2:9) demonstrated the normal sequence. An additional affected woman (3:3) and a male obligate carrier (2:11) were confirmed to carry the coding base pair change (c. 1286C>G) that results in a stop codon (Figure 1). There were no additional mutations in genes previously identified to cause POI that segregated with POI in the family.
cDNA sequences
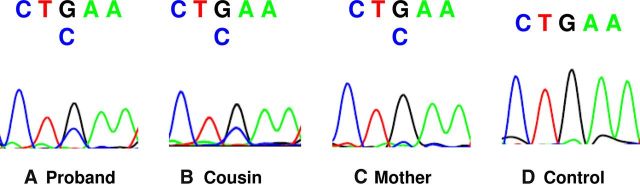
mRNA from white blood cells of the proband (3:1), mother (2:3), and cousin (3:3) was reverse transcribed into cDNA and subjected to PCR amplification. The mutant G allele was present at a variable proportion in cDNA from the 3 women (23.4, 40.9, and 45.9%). The average proportion of the G allele to the wild-type C allele in cDNA was reduced when compared with the proportion from gDNA in the carriers (36.7 ± 6.8 vs 61.8 ± 3.2%; P < .005), suggesting that the mRNA had undergone some nonsense-mediated decay. There was no G allele present in the mRNA from 3 unrelated controls, demonstrating that there was no sequence artifact (Figure 3).
Figure 3.
The figure shows electropherograms and the cDNA sequence of the proband (3:1) (A), the proband's cousin (3:3) (B), the proband's mother (2:3) (C), and a representative, unrelated control (D). The reverse DNA strand is shown, such that G is the wild-type allele and C is the nonsense allele. Variable amounts of the C allele are present in the affected women but not in the controls.
Genotypes in unrelated women with POI
The results of Sanger sequencing the 5′ untranslated and coding exons of eIF4ENIF1and eIF4E, the protein it binds, in 38 women with nonsyndromic POI are presented in Table 1. There were no rare exonic sequence variants identified. Three single nucleotide variants (rs14990298, rs72905084, and rs62323192) in eIF4E and 4 in eIF4ENIF1 (rs5753627, rs9621258, rs2273251, and rs5997988) were found at frequencies similar to those in the control populations (all P > .05) (10). The only novel variant was found in an intron in eIF4E and is of unclear significance (Table 1).
Table 1.
Variants in eIF4E and eIF4ENIF1 Identified in Women With POI
| Chrom Number | Chrom Pos (in HG19) | Target Name | Ref. Base | Variant Found | Variant Name | Variant Function | Reference Codon | Variant Codon | Nucleotide Change | Ethnicitya | POI MAFb | Control MAF | P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 99 850 297 | EIF4E exon 1 | T | TC | rs14990298 | Intronic | EA | 0.014 | 0.0029 | .1 | |||
| 4 | 99 850 284 | EIF4E exon 1 | G | C | rs72905084 | Intronic | AA | NA | 0.053 | NDc | |||
| 4 | 99 808 254 | EIF4E exon 5 | C | T | rs62323192 | Coding, Synon. | GAC = Asp | GAT = Asp | 375C>T | EA | 0.263 | 0.287 | .6 |
| 4 | 99 808 225 | EIF4E exon 5 | G | Ad | Novel | Intronic | EA | 0.014 | NA | ND | |||
| 22 | 31 859 859 | EIF4ENIF1 exon 5 | T | C | rs5753627 | Coding, Synon. | GTT = Val | GTC = Val | 393T>C | EA | 0.041 | 0.078 | .2 |
| 22 | 31 844 270 | EIF4ENIF1 exon 13 | C | T | rs9621258 | Intronic | EA | 0.014 | 0.057 | .1 | |||
| 22 | 31 843 567 | EIF4ENIF1 exon 14 | C | G | rs2273251 | Intronic | AA/EA | 0.276 | 0.202 | .2 | |||
| 22 | 31 838 085 | EIF4ENIF1 exon 17 | C | T | rs5997988 | Coding, Synon. | AGC = Ser | AGT = Ser | 1704C>T | AA/EA | 0.434 | 0.447 | .8 |
Abbreviations: AA, African American woman; Chrom, Chromosome; EA, European Americans; MAF, minor allele frequency; NA, not applicable; ND, not determined; Pos, position; Ref, reference; Synon, synonymous. The frequencies of the variants identified in 38 women with POI are not different from those in the control population, with the exception of one novel intronic variant in eIF4E of unclear significance.
Ethnicity refers to the ethnicity of the subject(s) with POI that carries the variant allele.
The POI minor allele frequency (POI MAF) is calculated as the number of variant alleles in a total of 74 alleles for variants found in European American women, 76 alleles for those found in both African American and European American women, and NA for the allele found in the single African American woman in the group.
ND signifies that the P value was not applicable because the variant was found in the only African American subject or was a novel change.
The novel variant in eIF4E exon 5 is a splice site region variant with a marginally decreased donor site (www.mutationtaster.org).
Discussion
We identified an autosomal dominant nonsense mutation in the eIF4ENIF1 gene, which segregates with primary ovarian insufficiency (menopause age 29–35 y) in a large family. The mRNA in white blood cells from 3 affected women demonstrated nonsense mutant transcript at a decreased proportion compared with that in gDNA. Therefore, two mechanisms for ovarian insufficiency in eIF4ENIF1 nonsense mutations can be hypothesized. Either haploinsufficiency of eIF4ENIF1 or the production of a small amount of the truncated protein may account for ovarian insufficiency in these women. There were no mutations in eIF4ENIF1 or eIF4E in 38 unrelated women with nonsyndromic POI.
The first postulated mechanism for ovarian insufficiency in eIF4ENIF1 nonsense mutations is haploinsufficiency that may result in decreased mRNA degradation and increased mRNA stability. eIF4ENIF1 contains a binding site for eIF4E, a ubiquitously expressed eukaryotic translation initiation factor. eIF4E is expressed in small quantities in the cytoplasm and is therefore a rate-limiting factor for cap-dependent translation initiation (3, 13). eIF4ENIF1 has been demonstrated to repress translation by binding to eIF4E (4, 14). It also targets eIF4E to P bodies, cytoplasmic bodies containing factors that degrade eIF4E-bound mRNA (4, 14). Furthermore, depleting eIF4ENIF1 from cells results in increased mRNA stability (4). It is possible that increased mRNA results in toxicity to the ovarian follicle, as has been postulated in the mechanism of fragile X-associated primary ovarian insufficiency (15). Alternatively, there may be increased translation of specific mRNAs that result in oocyte apoptosis. Of note, eIF4E also plays a role in oncogenesis, and overexpression of eIF4E can lead to malignant transformation (13, 16). However, there are no reported cancers in these familial carriers of the nonsense mutation in eIF4ENIF1.
eIF4ENIF1 is expressed in the human ovary (www.ncbi.nih.gov/geo). Furthermore, previous studies of eIF4ENIF1 homologues, Drosophila Cup and mouse Clast4 proteins, provide functional evidence that haploinsufficiency of an eIF4E transport protein causes oocyte disruption. The Drosophila Cup protein contains a short eIF4E binding motif that is highly homologous to that in eIF4ENIF1 and acts as a nucleocytoplasmic shuttling protein (17). Cup is found only in oocytes and is required for the correct accumulation and localization of eIF4E to the posterior cytoplasm of developing oocytes (17). Cup mutations result in abnormal oocyte growth and dominant disruption of meiotic chromosome segregation (17, 18). Murine Clast4 protein is 89% homologous to eIF4ENIF1 (5). Like Cup, Clast4 is expressed in developing oocytes and interacts with eIF4E, consistent with a role in the translational control of oocyte development (5). Taken together, the data are consistent with a role for eIF4ENIF1 in the development of a normal oocyte complement.
In addition to its role in translation regulation, eIF4ENIF1 contains one nuclear localization signal and 2 nuclear export signals, thus acting as a nucleocytoplasmic shuttling protein responsible for the nuclear import of eIF4E (3). Approximately one quarter of eIF4E is found in the nucleus colocalized with splice factors, suggesting that eIF4E also plays a role in mRNA processing and maturation (19). Truncating eIF4ENIF1 before the nuclear export signals, at approximately the same location as the nonsense mutation in the current family, results in an inability of eIF4ENIF1 to leave the nucleus (3). Therefore, if the truncated protein were made in the ovary, it is possible that eIF4ENIF1 gets stuck in the nucleus and causes cell death. Although we cannot test this hypothesis without ovarian tissue to perform mRNA expression and protein analyses, we did demonstrate white blood cell eIF4ENIF1 mRNA expression of the nonsense allele. Nonsense-mediated mRNA decay occurs when a stop codon is inserted into a protein greater than 50 bases before the last exon-exon junction (20). Therefore, a proportion of the mRNA containing the stop codon would be eliminated from the cell before translation of the protein (20), as suggested by the reduced proportion of the G allele in cDNA compared with gDNA. Given that the nonsense transcript is present in white blood cells, albeit at a reduced proportion, it is possible that a reduced amount of the truncated protein is translated. Thus, nuclear sequestration may play a role in ovarian insufficiency in this family as an alternative or additional mechanism to that hypothesized to result from haploinsufficiency.
The autosomal dominant mutation in eIF4ENIF1 adds further evidence that factors involved in mRNA translation are important for ovarian germ cell development and/or function. Recessive mutations in the eukaryotic initiation factor 2B (eIF2B) genes cause POI associated with white matter disease (21). It is postulated that the loss of eIF2B results in increased apoptosis in the brains of patients who carry mutations and/or an abnormal cellular response to stress, and the same mechanisms may affect the ovary (21). Unlike patients with recessive mutations in eIF2B genes, there is no evidence for neurological abnormalities in the family reported here, even in the affected women older than the age of 80 years.
In Drosophila, eIF4E-3 is a testis-specific member of the eIF4E family responsible for translation initiation during germ line development (22). Males without eIF4E-3 are infertile and have defects in meiosis (22). However, there is no information about eIF4ENIF1 homologues regulating eIF4E-3. Furthermore, although eIF4ENIF1 is expressed in testes (biogps.org), there is no apparent fertility phenotype in the men of the current family.
A stop codon in eIF4ENIF1 is associated with autosomal dominant POI in a large family. The gene plays an important role in oocyte development in organisms from Drosophila to mice. Although additional mutations were not found in unrelated women with POI, larger numbers should be assessed. The work represents the first evidence for the role of eIF4E translational repression in ovarian function in women.
Acknowledgments
We thank Susan Slaugenhaupt, PhD, and Maire Leyne, MS, MBA, for advice. We also thank the patients for their overwhelming interest, support, and participation in this study.
This work was supported by the Harvard Clinical and Translational Science Center Grant 1 UL1 RR025758-01.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- eIF4ENIF1
- eukaryotic translation initiation factor 4E nuclear import factor 1
- eIF4E
- eukaryotic translation initiation factor 4E
- gDNA
- genomic DNA
- POI
- primary ovarian insufficiency.
References
- 1. Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med. 2009;360:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Welt CK. Primary ovarian insufficiency: a more accurate term for premature ovarian failure. Clin Endocrinol (Oxf). 2007;68:499–509. [DOI] [PubMed] [Google Scholar]
- 3. Dostie J , Ferraiuolo M , Pause A , Adam SA , Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferraiuolo MA , Basak S , Dostie J , Murray EL , Schoenberg DR , Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Villaescusa JC , Allard P , Carminati E, et al. Clast4, the murine homologue of human eIF4E-transporter, is highly expressed in developing oocytes and post-translationally modified at meiotic maturation. Gene. 2006;367:101–109. [DOI] [PubMed] [Google Scholar]
- 6. Neale BM , Kou Y , Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DePristo MA , Banks E , Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H , Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKenna A , Hanna M , Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. National Heart, Lung, and Blood Institute GO Exome Sequencing Project (ESP). 2012. http://evs.gs.washington.edu/EVS/ Seattle, WA: Accessed October 2012. [Google Scholar]
- 11. Abecasis GR , Auton A , Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Styrkarsdottir U , Thorleifsson G , Sulem P, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517–520. [DOI] [PubMed] [Google Scholar]
- 13. Mamane Y , Petroulakis E , Rong L , Yoshida K , Ler LW , Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–3179. [DOI] [PubMed] [Google Scholar]
- 14. Andrei MA , Ingelfinger D , Heintzmann R , Achsel T , Rivera-Pomar R , Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sullivan SD , Welt C , Sherman S. FMR1 and the continuum of primary ovarian insufficiency. Semin Reprod Med. 2011;29:299–307. [DOI] [PubMed] [Google Scholar]
- 16. Lazaris-Karatzas A , Montine KS , Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. [DOI] [PubMed] [Google Scholar]
- 17. Zappavigna V , Piccioni F , Villaescusa JC , Verrotti AC. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Natl Acad Sci USA. 2004;101:14800–14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keyes LN , Spradling AC. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development. 1997;124:1419–1431. [DOI] [PubMed] [Google Scholar]
- 19. Dostie J , Lejbkowicz F , Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J Cell Biol. 2000;148:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilusz CJ , Wang W , Peltz SW. Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev. 2001;15:2781–2785. [DOI] [PubMed] [Google Scholar]
- 21. Fogli A , Rodriguez D , Eymard-Pierre E, et al. Ovarian failure related to eukaryotic initiation factor 2B mutations. Am J Hum Genet. 2003;72:1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hernandez G , Han H , Gandin V, et al. Eukaryotic initiation factor 4E-3 is essential for meiotic chromosome segregation, cytokinesis and male fertility in Drosophila. Development. 2012;139:3211–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]