Abstract
Burkholderia pseudomallei (Bpm) is a saprophytic rod-shaped gram-negative bacterium and the causative agent of melioidosis. This disease has previously been described as endemic in areas such as northern Australia and Southeast Asia, but, more recently, a better understanding of the epidemiology of melioidosis indicated that the disease is distributed worldwide, including regions of the Americas and Africa. A 16S-23S rDNA internal transcribed spacer (ITS) typing system has been developed for Bpm and has revealed that ITS types C, E, and hybrid CE are mainly associated with Australia and Southeast Asia while type G strains are more associated with cases of melioidosis in the Western Hemisphere. The purpose of the current study was to determine the in vitro and in vivo virulence profiles of the understudied Bpm type G strains Ca2009, Ca2013a, Mx2013, and 724644 and compared such phenotypes to the commonly studied Bpm type C strain K96243. We evaluated virulence by measuring invasion/uptake and survival of these Bpm strains in murine respiratory epithelial LA-4 cells and alveolar macrophage MH-S cells using different multiplicity of infections (MOIs of 1 and 10). We also calculated the lethal dose 50 values (LD50) in BALB/c mice that were inoculated intranasally with either Ca2009, Ca2013a, or Mx2013. Overall, the virulence and lethality phenotypes of Bpm type G strains were similar to the Bpm type C strain K96243. Additional comparative analyses between the Bpm ITS types may lead to a better understanding of the contribution of the ITS type to the epidemiology and ecology of Bpm strains.
Introduction
Burkholderia pseudomallei (Bpm) is a gram-negative, rod-shaped, motile, saprophytic, normally soil-dwelling bacterium and the causative agent of melioidosis worldwide. The common routes of infection are inhalation, cutaneous inoculation, and ingestion [1]. The inhalation exposure can lead to the most severe form of clinical disease. Localized clinical manifestations can result at the site of infection. For example, a pneumonia and/or flu-like illness may develop if a person inhales Bpm from the environment. In some melioidosis cases, localized infections have translocated to other sites of the body or advance to more severe systemic infections [2]. Acute, chronic, and recurrent melioidosis cases have been reported [3]. Currently, there is not vaccine commercially available for this disease and therapeutic treatments are only partially effective [4,5].
This bacterium can normally be found in tropical and sub-tropical regions of the world, with Thailand and northern Australia having some of the highest numbers of reported infections [6]. A recent study has estimated that there is an average of 165,000 human melioidosis cases per year worldwide, from which 89,000 people die [7]. Bpm is categorized by the Centers for Disease Control and Prevention (CDC) as a Tier 1 Select Agent, because of its high morbidity and mortality rates and potential to be used as a bioweapon [8] (https://www.selectagents.gov/selectagentsandtoxinslist.html). Recent studies have examined the prevalence of melioidosis cases within the Americas [9]. Melioidosis has been declared endemic in Puerto Rico [10,11] and Brazil [12,13]. Sporadic cases have been reported in North America, Central America, and South America [9].
Bacterial phylogenetic analysis, including multilocus sequence typing (MLST) [14] and 16S-23S rDNA internal transcribed spacer (ITS) typing [15], have been performed on Bpm and its near-neighbors. Interestingly, when examining the worldwide distribution of Bpm ITS types, Liguori et al. (2011) found a correlation between ITS types and their geographic origins. The ITS types C, E and hybrid CE were more associated with melioidosis endemic regions, such as northern Australia and Southeast Asia, while ITS type G was more associated with sporadic melioidosis cases in regions of Africa, North, Central, and South America [15,16]. Currently, a correlation between ITS type and disease attack rate or clinical presentation remains unknown.
Our aim in the current study was to determine if there is a difference in virulence or lethality of diverse and not so well characterized Bpm ITS type G strains while compared to a prototypic Bpm strain such as K96243, a ITS type C isolated within a region endemic for melioidosis. Our previous study using a standardized model of melioidosis infection provided a detailed, direct comparison of infection with different B. pseudomallei strains which let us understand their virulence potential [17]. Therefore, we compared the invasion, uptake and survival of four different ITS type G strains to the prototypical K96243 strain in murine lung epithelial (LA-4) and alveolar macrophage (MH-S) cell lines. We also determined the lethal doses 50 (LD50 values) of three ITS type G strains using a BALB/c mouse intranasal infection model. We report that the virulence and lethality of Bpm ITS type G strains when compared to the prototypical strain appeared to be similar in their ability to invade, survive, and infect mice. This information will improve our understanding of the biology and epidemiology of melioidosis using from the Western Hemisphere which have not been studied as extensively as strains from Australia and Southeast Asia.
Materials and methods
All work with B. pseudomallei were conducted in CDC/USDA-approved and registered biosafety level 3 (BSL3) facilities at the University of Texas Medical Branch (UTMB), and experiments with select agents were performed in accordance with BSL3 standard operating practices. The animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol (IACUC # 0503014B) was approved by the Institutional Animal Care and Use Committee of the UTMB.
Bacterial strains
A thorough description of 16S-23S ribosomal DNA internal transcribed spacer (ITS) typing Burkholderia species is provided elsewhere [15]. The Bpm ITS type C K96243 strain was isolated in Thailand [18]. B. pseudomallei strains Ca2009, Ca2013a, Mx2013, and 724644 were all isolated from patients with exposure in the Western Hemisphere and represent different ITS type G strains [16]. These strains and their sources are listed in Table 1. Bpm strains were maintained on Luria-Bertani with 4% glycerol (LBG) agar at 37°C. Strains were incubated in LBG broth at 37°C and 200 RPM for 12 or 16 h before use in in vitro studies or in vivo experiments, respectively.
Table 1. B. pseudomallei ITS type G strains analyzed in this study [16].
| Strain Identification | Exposure Source | Date Isolated | MLST Type | ITS Type |
|---|---|---|---|---|
| Ca2009 | Resident of Mexico | 2009 | 95 | G |
| Mx2013 | Travel to Mexico | 2013 | 297 | G |
| Ca2013a | Iguana with melioidosis | 2013 | 518 | G |
| 724644 | Travel to Aruba | 2010 | 698 | G |
Animal studies
Female, 6-8-week-old BALB/c mice obtained from Harlan Laboratories (Indianapolis, IN, USA) were housed in microisolator cages under pathogen-free conditions. Animals were provided with rodent feed and water ad libitum and maintained on a 12 h light cycle. Before experiments, mice were afforded an adaption period of at least 1 week. Humane endpoints were strictly observed through daily monitoring throughout the study. Clinical symptoms (immobility, dyspnea, and paralysis) were monitored and animals were euthanized based on humane endpoints (moribund animals were over-anesthetized with isofluorane via the aerosol route followed by cervical dislocation). Some animals did die because of infection even though animals were monitored three times daily during the acute stages of infection and twice daily during the latter stages of infection. If an animal was found dead, this was reported to the attending veterinarian and documented.
The determination of the lethal dose 50 (LD50) of the B. pseudomallei strains was performed in BALB/c mice challenged via intranasal inoculation using a method similar to what was described previously [19]. To determine the LD50, four groups of mice (n = 8 mice per group) were inoculated with bacterial colony forming units (CFU) of 5, 50, 5 × 102 and 5 × 103 CFU/ml of Bpm strain Ca2009 or 1 × 102, 1 × 103, 1 × 104 or 1 × 105 CFU/ml for Bpm strains Ca2013 or MX2013. Due to technical difficulties, Bpm strain 724644 was not evaluated in the murine model. Mice were monitored and deaths recorded over a period of 24 days. The organs (lung, liver, and spleen) were collected, homogenized in 1 ml of PBS by using a tissue grinder (Covidien), and then the bacteria were enumerated by standard plate counts on LBG agar. For each strain, the LD50 was calculated using methods described by Reed and Muench [20].
Cell invasion/uptake and survival assays
Cellular uptake/invasion assays were similar to those previously performed [21,22,23]. Each assay was performed in quadruplicate. Murine respiratory epithelial LA-4 cells (ATCC® CCL-196™) or murine alveolar macrophage MH-S cells (ATCC® CRL-2019™) were incubated at 37°C with 5% CO2 in 24-well plates (Corning) at a concentration of 5 X 105 cells per well. The LA-4 cells and MH-S cells were grown in F-12K complete medium and RPMI-1640 complete medium, respectively, per manufacturer’s recommendations. B. pseudomallei suspensions were added to cells at a MOI of 1 or 10, followed by centrifugation at 250 xg for 5 min and incubation in 37°C with 5% CO2 for 1 h to determine uptake/invasion. One hour post-infection (hpi), the monolayers were washed twice with sterile Dulbecco's phosphate-buffered saline (DPBS) (Cellgro) and lysed with 0.1% Triton X-100 in PBS, and serial dilutions were plated and incubated at 37°C for 48 h. The percentage of uptake/invasion was calculated as: (invasion/ uptake CFU/ total inoculum CFU) X 100.
To determine the intracellular survival after initial uptake/invasion, after 1 h the monolayers were washed twice with sterile Dulbecco's phosphate-buffered saline (DPBS) (Cellgro) and replenished with complete medium containing 250 μg/ml kanamycin. At 3 h post inoculation the monolayers washed twice with sterile DPBS then lysed with 0.1% Triton X-100 in PBS, and serial dilutions were plated and incubated at 37°C for 48 h. The percentage of survival was calculated as: (survival CFU/ invasion/ uptake CFU.) X 100.
Statistics
Statistical analyses were performed using Graph Pad Prism 6 (La Jolla, CA, USA). One-way ANOVA with Dunnett’s test was used to compare type G strains to K96243. We also used Tukey’s multiple comparison test to compare each strain to all other strains. A p value of <0.05 was considered statistically significant.
Results
LA-4 lung epithelial cell invasion and survival
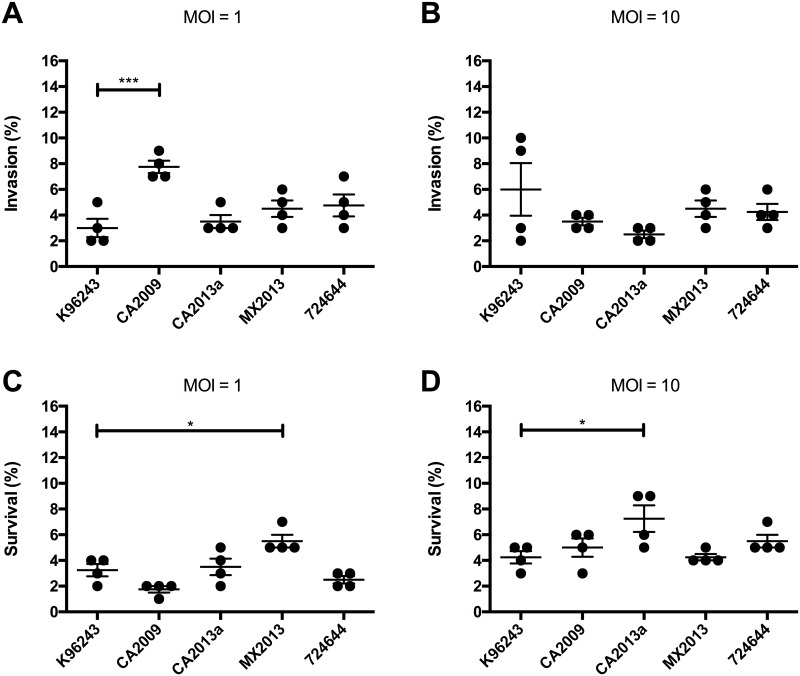
Cell invasion and intracellular survival were determined for the B. pseudomallei type G strains as well as K96243 in LA-4 mouse lung epithelial cells. At an MOI of 1, the Ca2009 strain had 4.75% higher cell invasion compared with K96243 (p<0.0005, Dunnett’s test) at 1 hpi (Fig 1A). When the MOI was increased to 10, there were no significant differences in invasion seen between the type G strains and K96243 (Fig 1B). Intracellular survival was determined 3 hpi based on the average number of bacteria that invaded the cell at 1 hpi. At an MOI of 1, strain Mx2013 had 2.25% higher survival (p<0.05) compared to K96243 (Fig 1C). When the MOI was increased to 10, strain Ca2013a had 3% higher survival (p<0.05) (Fig 1D). The invasion effect of an MOI of 1 was observed, when the type G strains where compared to each other (Tukey’s test)., The strain CA2009 invasion was significantly higher than CA2013a (p<0.005), MX2013 (p<0.05), and 724644 (p<0.05) (Fig 1A). However, no invasion differences was observed between this strains when a MOI 10 was used (Fig 1B). The effects of the choose MOI on survival are observed as well, sinceMX2013 had significantly increased survival compared to CA2009 (p<0.0005), CA2013a (p<0.05), and 724644 (p<0.005), with an MOI of 1 (Fig 1C). Also, when an MOI of 10 was used, the strain CA2013a showed a significantly increased survival compared to strain MX2013 (p<0.05) (Fig 1D)
Fig 1. Burkholderia pseudomallei invasion and intracellular survival in LA-4 murine lung epithelial cells.
Cells were infected with Bpm at a multiplicity of infection (MOI) of 1 (A) or 10 (B). After 1 hpi, cells were lysed and bacterial invasion was determined by dilution plating. Intracellular survival was determined using an MOI of 1 (C) or 10 (D) at 3 hpi. Intracellular survival percentages were calculated by dividing the number of bacteria recovered at 3 hpi after extracellular killing divided by the average number of cells that invaded in (A) or (B). Individual data points are shown as well as mean ± S.E.M. Levels of significance: *, p < 0.05; ***, p < 0.0005.
MH-S macrophage uptake and survival
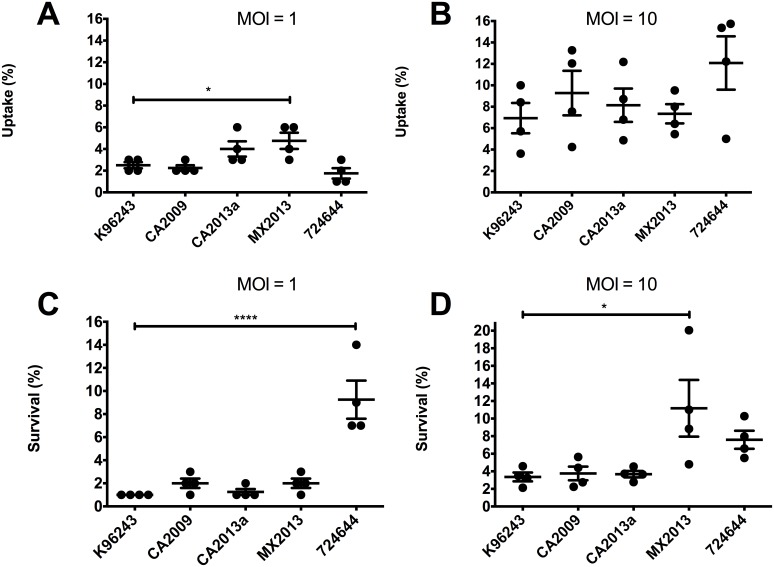
Cell uptake and intracellular survival was also measured in MH-S murine alveolar macrophages. At an MOI of 1, strain Mx2013 had 2.25% higher uptake compared to K96243 (p<0.05) (Fig 2A). At a MOI of 10, all strains had similar levels of uptake by MH-S cells. (Fig 2B). Intracellular survival at a MOI of 1 was 8.25% higher for strain 724644 compared to K96243 (p<0.0001) (Fig 2C). At a MOI of 10, strain 724644 trended towards increased intracellular survival (p = 0.221) but was not significant and strain Mx2013 had 7.813% higher survival (p<0.05). Between type G strains, MOI dependent differences in uptake and survival also could be observed. The strain MX2013 had significantly higher uptake in MH-S macrophages compared to strain CA2009 (p<0.05) and strain 724644(p<0.01) when a MOI of 1 was used (Fig 2A). Strain 724644 had significantly increased intracellular survival inside macrophages compared to all other strains tested (p<0.0001) (Fig 2C), and strain MX2013 had significantly increased survival inside of macrophages compared to strain CA2009 (p<0.05) and CA2013a (p<0.05) (Fig 2D).
Fig 2. Burkholderia pseudomallei uptake and intracellular survival in MH-S murine alveolar macrophages.
Cells were infected with Bpm at an MOI of 1 (A) or 10 (B). After 1 hpi, cells were lysed and bacterial uptake was determined by dilution plating. Intracellular survival was carried out using MOIs of 1 (C) or 10 (D). Intracellular survival percentages were calculated by dividing the number of bacteria recovered 3 hpi after extracellular killing divided by the average number of cells that were taken up in (A) or (B). Individual data points are shown as well as mean ± S.E.M. Levels of significance: *, p < 0.05; ***, p < 0.0005.
Intranasal LD50 determination
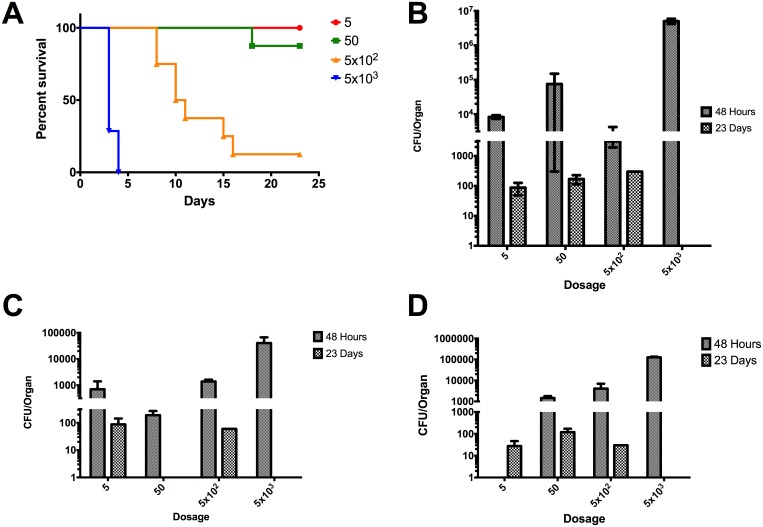
The LD50 studies were performed in BALB/c mice to evaluate virulence of Bpm strains Ca2009, Ca2013a, and Mx2013 in vivo using an intranasal model of infection. Bacterial burden in the lungs, liver, and spleen were measured at 2 days’ post-infection (dpi) and in surviving mice 23 dpi. It has previously been reported that the intranasal LD50 for K96243 is 312 CFU [24]. Strain Ca2009 had the lowest LD50 of the type G strains measured at 101 CFU and was the only strain to have a lower LD50 as compared to K96243 (Fig 3A). At 2 dpi, there were >103 CFU found in the lungs across all dose ranges given indicating productive replication within the lungs (Fig 3B). At 23 dpi among surviving animals, ~102 bacteria were present (Fig 3B). In both the liver and spleen of mice infected with Ca2009, bacteria were present indicating that strain Ca2009 can quickly disseminate within the host. The strain Ca2009 was also found at lower levels in both the liver and spleen of surviving mice at 23 dpi (Fig 3 panels C and D).
Fig 3. LD50 determination of B. pseudomallei CA2009 and colonization of lung, liver, and spleen in BALB/c mice.
(A) Percent survival of BALB/c mice (n = 8) infected via the intranasal route with the listed number of CFU of Bpm CA2009. LD50 = 101 CFU. Colonization of the lungs (B), livers (C), and spleens (D) were determined at 48 hpi and at 23 dpi in surviving mice. Values depicted in (B-D) are the mean ± S.E.M.
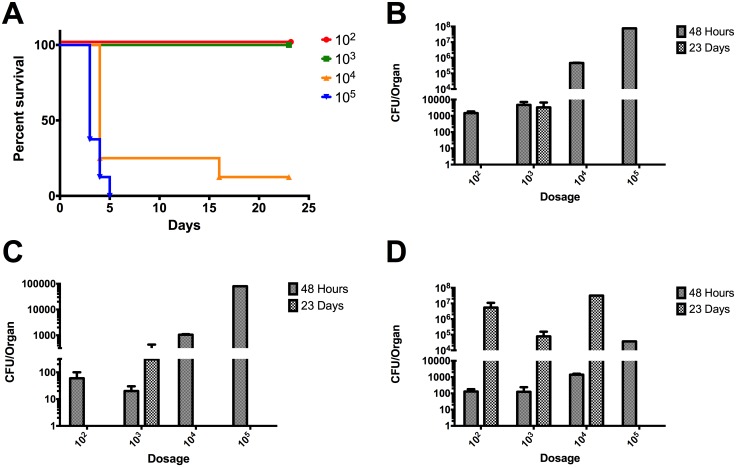
The LD50 for strain Ca2013a was 4,260 CFU which was the highest among strains tested (Fig 4). At 48 hpi, strain Ca2013a was present in the lung, liver and spleen (Fig 4 panels B, C, and D). At 23 dpi, Ca2013a was only present in the lungs and liver at the 103 CFU dose which resulted in 100% survival (Fig 4). Interestingly, in the spleen at 23 dpi, Ca2013a was found at high levels in surviving mice unlike the low levels seen for strain Ca2009 (Fig 4).
Fig 4. LD50 determination of B. pseudomallei CA20013a and colonization of lung, liver, and spleen in BALB/c mice.
(A) Percent survival of BALB/c mice (n = 8) infected via the intranasal route with the listed number of CFU of Bpm CA2013a. LD50 = 4,260 CFU. Colonization of the lungs (B), livers (C), and spleens (D) were determined at 48 hpi and at 23 dpi in surviving mice. Values depicted in (B-D) are the mean ± S.E.M.
The LD50 for strain Mx2013 was 1,330 CFU. At 48 hpi there were high levels of bacteria in the lungs similarly to both Ca2009 and Ca2013a strains (Fig 5B). Bacterial loads in the liver and spleen at 48 hpi were like those seen in both Ca2009 and Ca2013a (Fig 5 panels C and D). At 23 dpi, surviving mice had heavy colonization of the liver and spleen with 105 and 106 CFU, respectively (Fig 5 panels C and D).
Fig 5. LD50 determination of B. pseudomallei MX20013 and colonization of lung, liver, and spleen in BALB/c mice.
(A) Percent survival of BALB/c mice (n = 8) infected via the intranasal route with the listed number of CFU of Bpm MX2013. LD50 = 1,330 CFU. Colonization of the lungs (B), livers (C), and spleens (D) were determined at 48 hpi and at 23 dpi in surviving mice. Values depicted in (B-D) are the mean ± S.E.M.
Based on the in vivo data, strain Ca2009 caused the most acute infection. Surviving mice had less extensive chronic colonization of the liver and spleen compared to strains Ca2013a and Mx2013. The considerably higher LD50 values for Ca2013a and Mx2013 as well as high levels of colonization at 23 dpi suggest that these two strains result in a primarily chronic infection rather than acute.
Discussion
In this study, our goal was to assess whether the relatively understudied Bpm ITS type G strains were comparable in virulence and lethality to a more commonly studied Bpm ITS strain, such as the Bpm strain K96243 [18]. We accomplished this by measuring the invasion/uptake and survival abilities of Bpm ITS type G strains Ca2009, Ca2013a, Mx2013, and 724644 [16] and comparing them to the abilities of Bpm ITS type C strain K96243 in murine respiratory epithelial LA-4 cells and alveolar macrophage MH-S cells. We observed subtle differences between some of the strains studied when using a MOI of 1. We did not see any differences when we used an MOI of 10 in our studies. Studies by Welkos et al. [25] have previously performed a characterization of Bpm ITS types C and E strains using an in vitro J774.A1 murine-derived macrophage-like cell assay. This study suggested the existence of a potential inverse association between a strain’s virulence in mice, using an intraperitoneal route of infection, and their virulence in macrophages for at least a subset of B. pseudomallei strains. In the current study, we could not establish an inverse correlation and instead, the in vitro virulence phenotype of the Bpm ITS type G strains was similar, even though there were differences in the lethality using the in vivo intranasal model of infection. Because of the wide variability of the Burkholderia virulence properties [26], we strongly recommend that the selection of the tissue cultured cells used in the in vitro studies should directly correlate with those cells found in the organ where the in vivo dose is going to be delivered. As such, our study tried to standardize the type of cells used in the in vitro as well as the in vivo studies to provide more meaningful comparisons.
We also determined the LD50 values of Ca2009, Ca2013a, and Mx2013 in BALB/c mice inoculated by the intranasal route and compared those values to the LD50 of K96243, which we have determined in a previous study [24]. The LD50 values were calculated for Ca2009, Ca2013a, and Mx2013 ranged from 101 to 4,260. The previously determined LD50 value for K96243 was 312 [24]. The most virulent strain tested, CA2009, had low levels of colonization in surviving mice at 23 dpi (Fig 3). Interestingly, strains that exhibited increased intracellular survival such as CA2013a and MX2013 strains were less virulent in the in vivo studies. These strains were also able to colonize the lungs, livers, and spleens of the infected mice; which are often target organs during Bpm infection. Interestingly, mice infected with CA2013a and MX2013 had high levels of colonization at 23 dpi in the spleen. These two strains could be more skewed towards causing a chronic rather than acute infection but more studies are needed to determine whether these strains display specific tissue tropism and the infectivity is associated with an acute or chronic phase of the disease. The data collected in the current study clearly demonstrated that the Bpm ITS type G strains we examined are virulent. As indicated above, the differences observed in the in vitro studies did not seem to correlate to the mouse model of infection. The Bpm ITS type G strains studied have similar virulence and lethality when compared to Bpm ITS type C strain K96243 but have some subtle differences. Ours as well as other studies [19,25,27] provided the basis to develop a standardized murine model to directly compared different Bpm strains and; therefore, to perform future studies to test different medical countermeasures against Burkholderia species. Following our protocol, we have now tried to standardize in vitro and in vivo methods using murine cells derived from similar tissue origins to identify differences in the virulence phenotypes of pathogenic Bpm strains. It is our opinion that both the mouse model and murine alveolar cell lines in this study should be used in future studies to gain a better understanding of host-pathogen interactions during Burkholderia infections. Though the results of in vitro and in vivo experiments do not always agree, both types of tools are valuable for modeling the biology of infectious diseases.
As stated, our study was examining the interactions of the relatively understudied Bpm type G strains with murine cell lines and in a murine model of melioidosis. We wanted to evaluate if a possible difference in virulence and/or lethality between ITS type G strains and other ITS type strains could be linked to the unique epidemiology and ecology of ITS type strains that has been identified [15]. In our analysis, no such association can be made. Numerous studies have been performed to identify the mechanisms that other more well studied Bpm ITS type strains utilize to survive and replicate intracellularly within mammalian cells [28,29]. Future studies should include an assortment of ITS types to ensure that a bias is not created by just examining commonly used or prototypical Bpm type strains. In our case, we also considered it was important to evaluate the Bpm strains within murine alveolar cell lines instead of non-alveolar cell lines. We have used MH-S and LA-4 cell lines before when examining host-pathogen interactions between B. mallei and host cells [23]. Exploring host-pathogen interactions more often in cell lines which are more biologically relevant to the respiratory environment in conjunction with animal models of respiratory infection will also assist in avoiding unknown biases and strengthen the credibility of future studies.
Acknowledgments
We want to thank Jay Gee and Mindy Elrod of the CDC for sharing Burkholderia pseudomallei type G strains, providing information incorporated in Table 1, and for critical reading of the manuscript. We also want to thanks the Torres laboratory for assistance with experiments.
Data Availability
All relevant data are within the paper.
Funding Statement
The study was partially funded by pilot funds from University of Texas Medical Branch. There was no additional external funding received for this study which was supported by pilot funds from UTMB.
References
- 1.Currie BJ (2015) Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med 36: 111–125. 10.1055/s-0034-1398389 [DOI] [PubMed] [Google Scholar]
- 2.Currie BJ (2003) Melioidosis: an important cause of pneumonia in residents of and travellers returned from endemic regions. Eur Respir J 22: 542–550. [DOI] [PubMed] [Google Scholar]
- 3.Lewis ER, Torres AG (2016) The art of persistence-the secrets to Burkholderia chronic infections. Pathog Dis 74: pii: ftw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatcher CL, Muruato LA, Torres AG (2015) Recent Advances in Burkholderia mallei and B. pseudomallei Research. Curr Trop Med Rep 2: 62–69. 10.1007/s40475-015-0042-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estes DM, Dow SW, Schweizer HP, Torres AG (2010) Present and future therapeutic strategies for melioidosis and glanders. Expert Rev Anti Infect Ther 8: 325–338. 10.1586/eri.10.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limmathurotsakul D, Peacock SJ (2011) Melioidosis: a clinical overview. Br Med Bull 99: 125–139. 10.1093/bmb/ldr007 [DOI] [PubMed] [Google Scholar]
- 7.Limmathurotsakul D, Golding N, Dance DAB, Messina JP, Pigott DM, et al. (2016) Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nature Microbiology 1: 15008. [DOI] [PubMed] [Google Scholar]
- 8.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM (2002) Public health assessment of potential biological terrorism agents. Emerg Infect Dis 8: 225–230. 10.3201/eid0802.010164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benoit TJ, Blaney DD, Doker TJ, Gee JE, Elrod MG, et al. (2015) A Review of Melioidosis Cases in the Americas. Am J Trop Med Hyg 93: 1134–1139. 10.4269/ajtmh.15-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doker TJ, Sharp TM, Rivera-Garcia B, Perez-Padilla J, Benoit TJ, et al. (2015) Contact investigation of melioidosis cases reveals regional endemicity in Puerto Rico. Clin Infect Dis 60: 243–250. 10.1093/cid/ciu764 [DOI] [PubMed] [Google Scholar]
- 11.Christenson B, Fuxench Z, Morales JA, Suárez-Villamil RA, Souchet LM (2003) Severe community-acquired pneumonia and sepsis caused by Burkholderia pseudomallei associated with flooding in Puerto Rico. Bol Asoc Med P R 95: 17–20. [PubMed] [Google Scholar]
- 12.Rolim DB, Vilar DC, de Góes Cavalcanti LP, Freitas LB, Inglis TJ, et al. (2011) Burkholderia pseudomallei antibodies in individuals living in endemic regions in Northeastern Brazil. Am J Trop Med Hyg 84: 302–305. 10.4269/ajtmh.2011.10-0220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brilhante RS, Bandeira TJ, Cordeiro RA, Grangeiro TB, Lima RA, et al. (2012) Clinical-epidemiological features of 13 cases of melioidosis in Brazil. J Clin Microbiol 50: 3349–3352. 10.1128/JCM.01577-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, et al. (2003) Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 41: 2068–2079. 10.1128/JCM.41.5.2068-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liguori AP, Warrington SD, Ginther JL, Pearson T, Bowers J, et al. (2011) Diversity of 16S-23S rDNA internal transcribed spacer (ITS) reveals phylogenetic relationships in Burkholderia pseudomallei and its near-neighbors. PLoS One 6: e29323 10.1371/journal.pone.0029323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gee JE, Allender CJ, Tuanyok A, Elrod MG, Hoffmaster AR (2014) Burkholderia pseudomallei type G in Western Hemisphere. Emerg Infect Dis 20: 682–684. 10.3201/eid2004.130960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey S, Yeager LA, Blumentritt CA, Vijayakumar S, Sbrana E, et al. (2014) Comparative Burkholderia pseudomallei natural history virulence studies using an aerosol murine model of infection. Sci Rep 4: 4305 10.1038/srep04305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, et al. (2004) Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 101: 14240–14245. 10.1073/pnas.0403302101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mott TM, Vijayakumar S, Sbrana E, Endsley JJ, Torres AG (2015) Characterization of the Burkholderia mallei tonB Mutant and Its Potential as a Backbone Strain for Vaccine Development. PLoS Negl Trop Dis 9: e0003863 10.1371/journal.pntd.0003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed LJ, Muench H (1938) A simple method for estimating fifty percent end points. Am J Epidemiol 27: 493–497. [Google Scholar]
- 21.Brett PJ, Burtnick MN, Snyder DS, Shannon JG, Azadi P, et al. (2007) Burkholderia mallei expresses a unique lipopolysaccharide mixture that is a potent activator of human Toll-like receptor 4 complexes. Mol Microbiol 63: 379–390. 10.1111/j.1365-2958.2006.05519.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burtnick MN, Brett PJ, Nair V, Warawa JM, Woods DE, et al. (2008) Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect Immun 76: 2991–3000. 10.1128/IAI.00263-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitlock GC, Valbuena GA, Popov VL, Judy BM, Estes DM, et al. (2009) Burkholderia mallei cellular interactions in a respiratory cell model. J Med Microbiol 58: 554–562. 10.1099/jmm.0.007724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Judy BM, Taylor K, Deeraksa A, Johnston RK, Endsley JJ, et al. (2012) Prophylactic application of CpG oligonucleotides augments the early host response and confers protection in acute melioidosis. PLoS One 7: e34176 10.1371/journal.pone.0034176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welkos SL, Klimko CP, Kern SJ, Bearss JJ, Bozue JA, et al. (2015) Characterization of Burkholderia pseudomallei Strains Using a Murine Intraperitoneal Infection Model and in vitro Macrophage Assays. PLoS One 10: e0124667 10.1371/journal.pone.0124667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kespichayawattana W, Intachote P, Utaisincharoen P, Sirisinha S (2004) Virulent Burkholderia pseudomallei is more efficient than avirulent Burkholderia thailandensis in invasion of and adherence to cultured human epithelial cells. Microb Pathog 36: 287–292. 10.1016/j.micpath.2004.01.001 [DOI] [PubMed] [Google Scholar]
- 27.Wiersinga WJ, de Vos AF, de Beer R, Wieland CW, Roelofs JJ, et al. (2008) Inflammation patterns induced by different Burkholderia species in mice. Cell Microbiol 10: 81–87. 10.1111/j.1462-5822.2007.01016.x [DOI] [PubMed] [Google Scholar]
- 28.David J, Bell RE, Clark GC (2015) Mechanisms of Disease: Host-Pathogen Interactions between Burkholderia Species and Lung Epithelial Cells. Front Cell Infect Microbiol 5: 80 10.3389/fcimb.2015.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willcocks SJ, Denman CC, Atkins HS, Wren BW (2016) Intracellular replication of the well-armed pathogen Burkholderia pseudomallei. Curr Opin Microbiol 29: 94–103. 10.1016/j.mib.2015.11.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.