Abstract
Recent studies have shown that the LIM-homeodomain transcription factor Isl1 is required for the survival and differentiation of direct pathway striatonigral neurons during embryonic development. The downstream effectors of Isl1 in these processes are presently unknown. We show here that Foxo1, a transcription factor that has been implicated in cell survival, is expressed in striatal projection neurons (SPNs) that derive from the Isl1 lineage (i.e. direct pathway SPNs). Moreover, Isl1 conditional knockouts (cKOs) show a severe loss of Foxo1 expression at E15.5 with a modest recovery by E18.5. Although Foxo1 is enriched in the direct pathway SPNs at embryonic stages, it is expressed in both direct and indirect pathway SPNs at postnatal time points as evidenced by co-localization with EGFP in both Drd1-EGFP and Drd2-EGFP BAC transgenic mice. Foxo1 was not detected in striatal interneurons as marked by the transcription factor Nkx2.1. Conditional knockout of Foxo1 using Dlx5/6-CIE results in reduced expression of the SPN marker Darpp-32, as well as in the direct pathway SPN markers Ebf1 and Zfp521 within the embryonic striatum at E15.5. However, this phenotype improves in the conditional mutants by E18.5. Interestingly, the Foxo family members, Foxo3 and Foxo6, remain expressed at late embryonic stages in the Foxo1 cKOs unlike the Isl1 cKOs where Foxo1/3/6 as well as the Foxo1/3 target Bach2 are all reduced. Taken together, these findings suggest that Foxo-regulated pathways are downstream of Isl1 in the survival and/or differentiation of direct pathway SPNs.
Keywords: Basal ganglia, Cell survival, Neuronal subtype differentiation, Striatum
1. Introduction
The striatum (a.k.a. caudate and putamen) represents the principal component of the basal ganglia circuit and mediates its output through the striatal projection neurons (SPNs). These neurons utilize GABA as a neurotransmitter and comprise two separate pathways; the direct pathway that projects to the internal segment of the globus pallidus (GPi) and substantia nigra pars reticulata (SNr) and the indirect pathway which projects to the external segment of the globus pallidus (GPe) (Gerfen and Surmier, 2011). In addition to the distinct axonal targets that each of these output pathways target, they can be neurochemically defined by the expression of neuropeptides as well as dopamine receptors. In this respect the indirect pathway SPNs express enkephalin (Enk) and dopamine D2 receptors, while their direct pathway counterparts express substance P (SP) and dopamine D1 receptors (Gerfen and Surmeier, 2011). Balanced output between these pathways is known to be essential for normal movements and appropriate behaviors (Albin et al., 1989; Gerfen and Surmeier, 2011).
Despite the importance of the SPN output pathways, little is known about the mechanisms that regulate their development. A number of developmental control genes have been shown to play a role in the specification and/or differentiation of SPNs including the Gsx2, Ascl1 and Dlx transcription factors (Anderson et al., 1997; Toresson and Campbell, 2001; Yun et al., 2002, 2003; Waclaw et al., 2004; Long et al., 2009; Wang et al., 2009, 2011, 2013). The zinc finger transcription factors Ikaros and Helios as well as the early B-cell factor (Ebf1) have been shown to play crucial roles in the differentiation of the late born SPNs belonging to the matrix compartment (Garel et al., 1999; Martin-Ibanez et al., 2010, 2012). Studies in recent years have begun to shed light on the molecular genetic mechanisms that control the development of the direct and indirect SPN pathways. In this respect, Ikaros and Sp9 are required for the normal differentiation and survival of the indirect SPNs (Martin-Ibanez et al., 2010; Zhang et al., 2016). Conversely, Ebf1 (Garel et al., 1999; Lobo et al., 2006, 2008) and the LIM homeodomain protein, Isl1 (Ehrman et al., 2013; Lu et al., 2014) have been implicated in the development of the direct pathway (i.e. striatonigral) SPNs. In particular, Isl1 is required for the survival and ultimate differentiation of a subpopulation of direct pathway SPNs, already at embryonic stages (Ehrman et al., 2013; Lu et al., 2014). It is currently unclear how Isl1 regulates SPN survival, however, it is likely that it controls a genetic pathway which results in the expression of a survival factor in embryonic direct pathway neurons.
In this respect, the Foxo transcription factors have been implicated in numerous cellular processes including metabolism, differentiation, cell death and survival (Kousteni, 2012; Puthanveetil et al., 2013). Foxo genes (Foxo1, 3 and 6) have been shown to be expressed in either the developing or adult striatum, with Foxo1 being highest expressed at all stages examined (Hoekman et al, 2006). While Foxo1 has been shown to mediate neuronal cell death (Yuan et al., 2008; 2009; Zhou et al., 2015), it is required for survival of certain non-neuronal cells such as cardiomyocytes (Sengupta et al., 2011; Shao et al., 2014). We have investigated here whether Foxo1 (and its family members Foxo3/6) may function downstream of Isl1 to promote the survival and/or differentiation of direct pathway SPNs.
2. Material and Methods
2.1 Animals
Fate-mapping studies were carried out by mating ROSA-lox-stop-lox(lsl)-tdTomato (JAX stock#007914) mice with Isl1cre/+ mice (Srinivas et al., 2001), generously provided by T. Jessell (Columbia University). Isl1cre/+ mice were genotyped as previously described (Waclaw et al., 2010). Double transgenic embryos were collected on E15.5 and E18.5. To label the direct and indirect SPN pathways at postnatal stages, we used Drd1-EGFP and Drd2-EGFP BAC mice generated by the GENSAT consortium (Gong et al., 2003), as described in Ehrman et al. (2013). These mice were genotyped using EGFP PCR primers (Pei et al., 2011).
Isl1fl/+ mice (Mu et al., 2008) were used to generate the Isl1 conditional knockouts (cKOs) together with Dlx5/6-CIE mice (Stenman et al., 2003). These mice were genotyped as previously described (Ehrman et al., 2013). To generate Foxo1 cKOs, we crossed Foxo1fl/fl mice that were generously provided by R.A. DePinho (MD Anderson Cancer Center) with Dlx5/6-CIE mice. The Foxo1 mice were genotyped as described in Paik et al. (2007).
For the staging of embryos, the morning of the vaginal plug was considered E0.5. Embryos were fixed in 4% paraformaldehyde overnight, washed in PBS, cryoprotected in 30% sucrose in PBS, and sectioned at 12 μm on a cryostat. Postnatal day 21 brains were fixed overnight in 4% paraformaldehyde, washed in PBS, cryoprotected in 20% sucrose in PBS, and sectioned at 35μm on a freezing sliding microtome. All of the mouse studies were approved by the Institutional Animal Care and Use Committee of the Cincinnati Children’s Research Foundation and were conducted in accordance with US National Institutes of Health guidelines.
2.2. Immunohistochemistry (IHC)
IHC (including tyramide amplification) was carried out as previously described (Waclaw et al., 2010). Primary antibodies were used at the following concentrations: rabbit anti-cleaved caspase 3 (1:250: Cell Signaling), goat anti-Darpp-32 (1:200; Santa Cruz), rabbit anti-dsRed (1:500: Clontech), rabbit anti-Ebf1 (1:500: Millipore), rabbit anti-EGFP (1:500; Thermo Fisher Scientific), goat anti-EGFP (1:5000, Abcam), rabbit anti-Foxo1 (1:500; Cell Signaling), rabbit anti-Foxo3 (1:333; Cell Signaling), rabbit anti-Foxo6 (1:1000; gift from A. Brunet, Stanford University) (Salih et al., 2012), rabbit anti-Foxp1 (1:4000; gift from E. Morrisey, University of Pennsylvania, Philadelphia, PA), goat anti-Isl1 (1:1000, R&D Systems), rabbit anti-Nkx2.1 (1:1000; Seven Hills Bioreagents). Diamino-benzidine (DAB) colorimetric reaction for bright-field immunostaining was developed as previously described (Waclaw et al., 2006). The following secondary antibodies were used for immunofluorescence: donkey anti-rabbit antibodies conjugated to Cy2 or Cy3 (Jackson ImmunoResearch); donkey anti-goat antibodies conjugated to Cy2 (Jackson ImmunoResearch); donkey anti-guinea pig antibodies conjugated to Cy3 (Jackson ImmunoResearch); donkey anti-mouse antibodies conjugated to Cy2 (Jackson ImmunoResearch). Tyramide Signal Amplification kits to Alexa Fluor 488 or Alexa Fluor 568 were purchased from ThermoFisher Scientific.
2.3. Quantification
Foxo1/tdTomato and Foxo1/Ebf1 double stains were quantified on confocal images taken at 400X power from E18.5 dorsolateral striatum on a Nikon C2 Confocal microscope. Images were taken from 4 serial sections of dorsolateral striatum in 3 Isl1 fate map embryos for Foxo1/tdTomato double stains or in 3 control and 3 Isl1 cKO embryos for the Foxo1/Ebf1 double stains.
In single IHC stained sections of Ebf1 (400X power), Foxo3 (400X power) and cleaved-caspase 3 (200X power) positive cells were counted individually in 3–4 striatal sections per embryo (at least 3 control, Isl1 cKO or Foxo1 cKO embryos analyzed) and are represented as cells/mm2.
Foxo1, Foxo6, and Darpp-32 single IHC stains were quantified by the area of striatal expression using ImageJ, as previously described (Waclaw et al., 2009; Ehrman et al., 2013). Striatal area was defined and calculated in ImageJ. Signal Intensity was measured after thresholding in at least 3 to 4 serial sections at mid-striatal levels per control/mutant group (at least 3 embryos for each genotype and embryonic stage). Average intensity per unit area was converted to % difference between mutant and control. The average intensity per unit area of control samples was set to 100%. Statistics were performed between control and Isl1 cKO or control and Foxo1 cKO using a Student’s unpaired t test.
2.4. In situ hybridization
The in situ hybridization procedure was performed as previously described (Toresson et al., 1999). Digoxigenin-labeled antisense probes against Bach2 (Clone ID: 4218490: Dharmacon) and Zfp521 (Clone ID: 5038671: Dharmacon) were used on 12 μm sections of E15.5 and E18.5 embryos.
3. Results
3.1. Foxo1 marks direct pathway SPNs at embryonic stages
Foxo1 is known to be expressed in the developing and mature striatum (Hoekman et al, 2006), however, it is unclear whether it is restricted to specific subtypes of striatal neurons such as direct versus indirect SPNs or interneurons. To address this, we made use of an Isl1 cre/loxP fate map strategy, which labels neuronal cell bodies and axons of the direct pathway as well as the cholinergic interneurons (Ehrman et al., 2013; Lu et al., 2014) (Fig. 1A and D). Immunostaining for Foxo1 in the fate mapped embryonic striatum, shows that a large portion of the Foxo1-positive neurons co-express tdTomato, indicating that they derive from the Isl1 lineage and at least, in part, represent direct pathway SPNs (Fig. 1B and E). In fact, quantification of the dorsolateral region of the striatum (the highest striatal Foxo1 expression area) reveals that 89% of the tdTomato+ neurons (i.e. Isl1 lineage) were Foxo1+ (753 out of 842 tdTomato+ cells analyzed from 3 embryos). Higher power views of dorsolateral striatum shows that very few of the Foxo1+ cells are single labeled (Fig. 1C and F). Indeed, quantification of this at E18.5 revealed that 87% of the Foxo1+ neurons co-localize tdTomato (753 out of 866 Foxo1+ cells analyzed from 3 embryos). While some of these single labeled cells may be due to incomplete recombination of the tdTomato reporter, not all Foxo1 single labeled striatal cells co-localize with the direct pathway marker/regulator Ebf1 (Fig. 1G–I) and therefore may represent other neuronal subpopulations such as interneurons or a subpopulation of indirect pathway SPNs. Interestingly, Foxo1 protein has been shown to shuttle between the nucleus and cytoplasm to affect specific cellular processes such as cell death versus cell survival (Yuan et al., 2008, 2009). Double staining with the pan SPN marker Foxp1 shows that in the embryonic striatum both nuclear and cytoplasmic expression is observed (sometimes within the same cell) (Fig. 1J–K).
Figure 1.
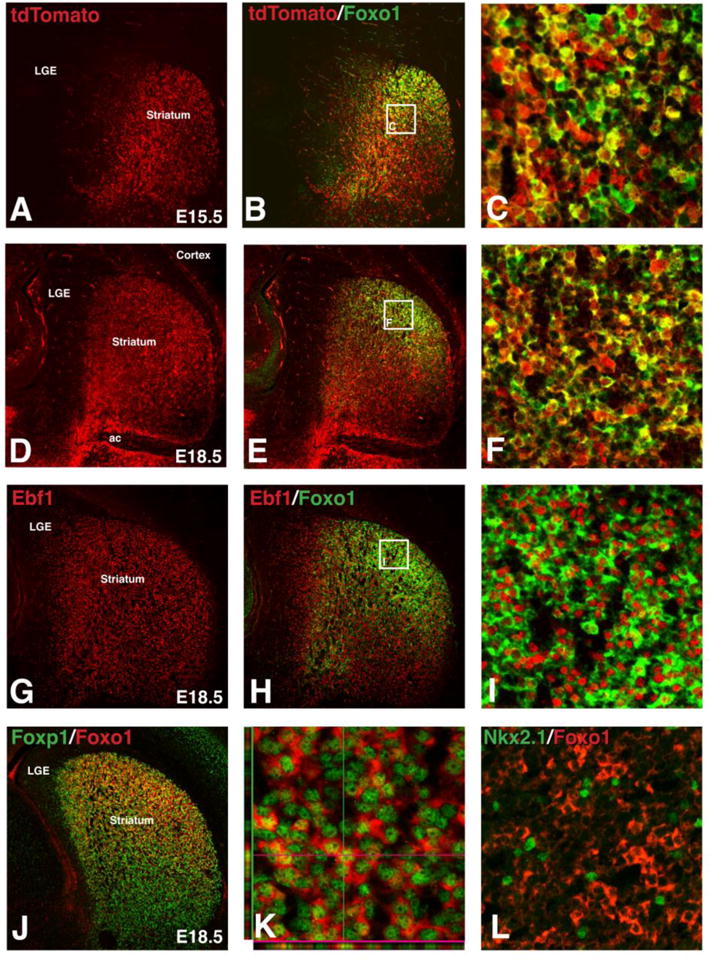
Foxo1 expression in embryonic direct pathway striatal projection neurons. Isl1 fate-map (Isl1cre/+;RosaCAG-tdTomato/+) shows tdTomato expression in direct pathway striatal projection neurons at E15.5 (A) and E18.5 (D). The majority of Foxo1+ cells are observed in Isl1 lineage striatal projection neurons at E15.5 (B–C) and E18.5 (E–F). Foxo1+ neurons are found in a subpopulation of Ebf1+ neurons (G–I) and Foxp1+ neurons at E18.5 (J–K). A selected confocal Z-plane shows Foxo1 protein in both the nucleus and cytoplasm of Foxp1+ developing striatal projections neurons (K). Foxo1 was not found in Nkx2.1+ striatal interneurons at E18.5 (L).
To determine whether striatal interneuron subtypes express Foxo1, we double stained for Nkx2.1, a marker of striatal interneuron subtypes from embryonic stages and into adulthood (Marin et al., 2000). Few if any Foxo1-positive striatal neurons co-expressed Nkx2.1 (Fig. 1L), indicating that Foxo1 is specific for SPNs and at least at embryonic time points, it is highly enriched in direct pathway SPNs.
3.2. Foxo1 is expressed in both direct and indirect pathway SPNs at postnatal stages
To examine whether the enrichment of Foxo1 in the direct pathway SPNs at embryonic stages is retained after birth, we examined the expression of Foxo1 in the striatum of Drd1-EGFP (direct pathway) and Drd2-EGFP (indirect pathway) BAC transgenic mice obtained from GENSAT (Gong et al., 2003). Double staining Foxo1 with the general SPN marker Darpp-32 (Anderson and Reiner, 1991) shows that Foxo1 is highly expressed in the cytoplasm including the neuronal cell bodies and neuropil within the striatum as well as in axons and terminals of the GPe, GPi and SNr at postnatal day (P)21 (Fig. 2A–B), suggesting that Foxo1 is expressed in both direct and indirect pathway SPNs within mature basal ganglia circuits. To confirm this, we colocalized Foxo1 with EGFP in cell bodies and fibers of Drd1-EGFP (Fig. 2C–E) and Drd2-EGFP (Fig. 2F–H) BAC transgenic mice. Foxo1 retains its largely restricted expression within the striatum of the postnatal brain but unlike the enriched expression in the direct pathway neurons during development it becomes a more general marker of the SPN output pathways in the adult, similar to Darpp-32.
Figure 2.
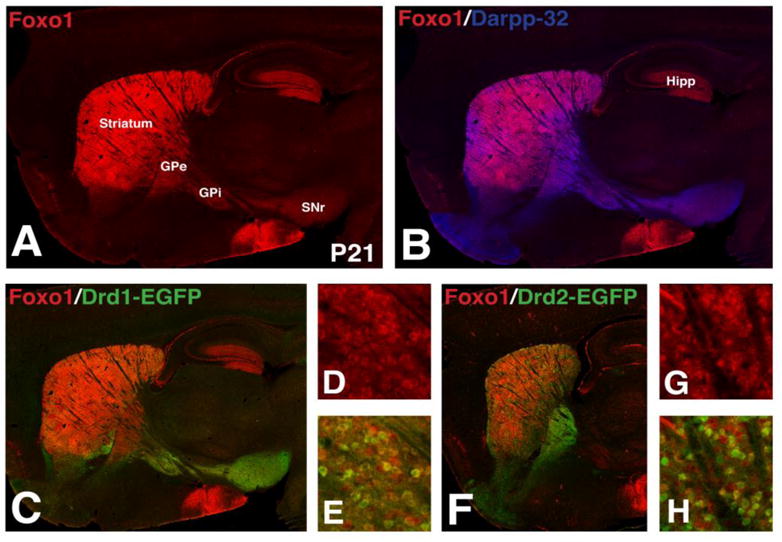
Foxo1 expression in direct and indirect pathway striatal projection neurons in the postnatal brain. Foxo1+ cells are found in the striatum and hippocampus (Hipp) at P21 (A). Foxo1+ striatal neurons express the striatal projection neuron marker Darpp-32 (B). Double immunofluorescence for Foxo1 and GFP show that Foxo1+ striatal neurons are found in both the Drd1-EGFP+ direct (C–E) and Drd2-EGFP+ indirect (F–H) striatal pathways at P21. GPe, external segment of the globus pallidus; GPi, internal segment of the globus pallidus; SNr, substantia nigra pars reticulata
3.3. Foxo1 is severely reduced in the Isl1 cKO striatum
Recent studies have shown that the LIM homeodomain protein Isl1 is required for the survival and differentiation of a subpopulation of direct pathway SPNs already at embryonic stages (Ehrman et al., 2013; Lu et al., 2014). Given that Foxo1 promotes survival in cardiomyocytes (Sengupta et al., 2011; Shao et al., 2014), it represents a good candidate to do the same in direct pathway SPNs downstream of Isl1. Indeed, at E15.5, the Isl1 cKO striatum is nearly devoid of Foxo1 expression (Compare Fig. 3B to 3A), with an 85% reduction in Foxo1 staining area compared to controls (Control 100% ± 22.6% and Isl1 cKO 15% ± 6.5%, p<.005, n=3). By E18.5, a moderate recovery of the Foxo1 staining is observed in the Isl1 cKO striatum (Fig. 3D), as compared to control (Fig. 3C), with a 55% reduction in staining (Control 100% ± 14.9% and Isl1 cKO 45% ± 3.1%, p<.05, n=3). As mentioned above, it appears that at perinatal time points Foxo1 is also expressed in the indirect pathway SPNs and, therefore, the partial recovery in the E18.5 Isl1 cKO striatum may reflect expression in this SPN subtype. To address this, we double stained for the direct pathway marker Ebf1, which remains, albeit in reduced numbers, in the Isl1 cKO striatum (Ehrman et al., 2013 and Fig. 3F) as compared to control (Fig. 3E). We observed a 33% reduction in the proportion of Ebf1+ cells that co-express Foxo1 in Isl1 cKOs compared to controls (Control 84% ± 0.6% and Isl1 cKO 57% ± 8.1%, p<.05, n=3). Despite the significant reduction in Foxo1 within the Isl1 mutant striatum, most of the remaining positive cells did co-express Ebf1 at E18.5 (Fig. 3F). Quantification in the dorsolateral striatum revealed that 92% of Foxo1+ cells co-expressed Ebf1 in the Isl1 cKO (948 out of 1028 Foxo1+ cells analyzed from 3 Isl1 cKOs), which was similar to controls at 95% (1652 out of 1731 Foxo1+ cells analyzed from 3 controls) suggesting that the partial recovery observed for Foxo1 expression (Fig. 3D) is largely confined to the direct pathway. These findings indicate Isl1 is required for normal expression of Foxo1 in direct pathway SPNs at embryonic stages and that this factor represents an attractive candidate to mediate the survival of at least a subpopulation of these neurons.
Figure 3.
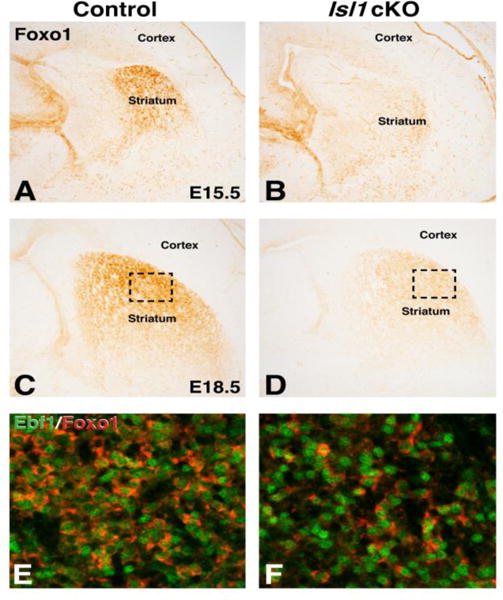
Foxo1 striatal expression is reduced in embryonic Isl1 conditional mutants. At E15.5, Isl1 conditional mutants show severe reduction of Foxo1 expression in the striatum (B) as compared to controls (A). Note the unaffected Foxo1 staining in MGE progenitors and blood vessels. Foxo1 expression remains reduced in E18.5 Isl1 conditional mutants (D) compared to controls (C). Boxed regions in in C and D represent the approximate striatal area where high power confocal images were taken in E and F that show Ebf1 (green) Foxo1 (red) double immunofluorescence in the control striatum (E) and the remaining Foxo1 + neurons that express Ebf1 in the Isl1 conditional mutant (F).
3.4. Foxo1 is required for the survival and differentiation of direct pathway SPNs at early stages of striatal neurogenesis
To examine whether direct pathway SPNs require Foxo1 for their normal survival and differentiation we generated Foxo1 cKOs by crossing Foxo1fl/fl mice (Paik et al., 2007) with Dlx5/6-CIE mice (Stenman et al., 2003) to recombine Foxo1 in ventral telencephalic derivatives, including the SPNs. The Dlx5/6-CIE mice drove efficient recombination of the Foxo1 locus in the ventral telencephalon, as evidenced by the complete loss of protein expression within the striatum of E15.5 Foxo1 mutants (compare Fig. 4B to A). Foxo1 expression was retained in the dorsomedial telencephalon and within blood vessels of the cKO, which is expected since the cre driven by the Dlx/5-CIE transgene has not been shown to be expressed in these cell types (Stenman et al., 2003).
Figure 4.
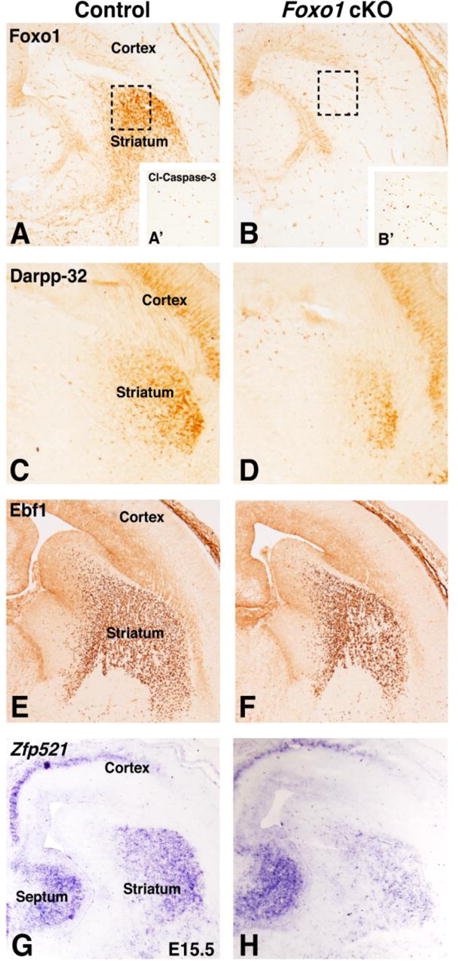
Ventral forebrain specific deletion of Foxo1 using Dlx5/6-CIE (Foxo1loxp/loxp;Dlx5/6-CIE called Foxo1 cKO). Foxo1 cKOs show a loss of Foxo1 expression at E15.5 (B) compared to controls (A). Note the Dlx5/6-CIE does not eliminate the ventricular zone Foxo1 staining in the dorsomedial cortex and blood vessels. Dashed boxes in A and B show the striatal region represented by cleaved caspase 3 staining in A′ and B′. Foxo1 cKOs show increased cleaved caspase3+ cells (B′) compared to controls (A′) and decreased striatal projection neuron markers Darpp-32 (compare D to C) as well as reductions in the direct pathway markers Ebf1 (compare F to E) and Zfp521 (compare H to G). Note the cortical Darpp-32 expression is unaffected in Foxo1 cKOs (compare cortex in D to E). In addition, note the cortical and septal expression of Zfp521 is unaffected in Foxo1 cKOs (compare F to E).
To determine if the loss of Foxo1 results in increased cell death within the mutant LGE/striatum at embryonic stages (i.e. E15.5) we stained for cleaved caspase-3. Dashed boxes in Fig. 4A and B represent the LGE/striatal area shown in Fig. 4A′ and B′ stained for cleaved caspase 3. We found a 2.8 fold increase in cleaved caspase 3 positive (i.e. dying) cells in the Foxo1 mutants as compared to controls (Foxo1 cKO = 257.9 cells/mm2 ± 13.9, Fig. 4B′ and Control = 92.2 cells/mm2 ± 10.3, n=3; Fig. 4A′, p<.001, n=3). This correlated well with a 48% reduction in Darpp-32 staining pattern in Foxo1 cKOs, (Fig. 4D) as compared to controls (Fig. 4C) (Control 100% ± 14.8% and Foxo1 cKO 52% ± 11.2%, p<.05, n=4). Thus, the observed increase in cell death in the Foxo1 cKO striatum may contribute to the loss of Darpp-32-expressing SPNs, however, it is also possible that Foxo1 is required for direct pathway SPN differentiation. To address this, we examined the expression of Ebf1 as well as Zfp521 gene expression, as markers of differentiating direct pathway SPNs (Garel et al., 1999; Lobo et al. 2006). We observed a 22% decrease in the striatal area as marked by Ebf1 staining in the Foxo1 cKO striatum (Fig. 4F) as compared to the controls (Fig. 4E) (Control 100% ± 14.6% and Foxo1 cKO 78% ± 2.3%, p<.05, n=3). Moreover, the density of Ebf1+ cells in the conditional mutant striatum was also reduced by 22% from the control striatum (compare Fig. 4F with E) (Control avg. 8398 ± 451 cells/mm2 and Foxo1 cKO avg. 6579 ± 319 cells/mm2, p<.05, n=3). Additionally, the expression of Zfp521, which also marks the maturing direct pathway SPNs in the control striatum (Fig. 4G), was notably reduced in the Foxo1 cKO striatum, but no change was seen in the septal or cortical expression domains (Fig. 4H). These findings suggest that Foxo1 is required for the survival/differentiation of direct pathway SPNs.
By E18.5, the expression of Darpp-32 recovered significantly in the Foxo1 cKO striatum (Fig. 5B) and appeared similar to the controls (Fig. 5A) (Control 100% ± 17.4% and Foxo1 cKO 87% ± 11.9%, p=.28, n=3). Foxo1 is one of four Foxo family members and Foxo3 and Foxo6 gene and/or protein expression has been previously observed in the developing striatum (Hoekman et al., 2006) as well as in the cortex and hippocampus (Hoekman et al, 2006; Paik et al., 2009; Salih et al., 2012; Webb et al., 2013). We detected both Foxo3 (Fig. 5C–D) and Foxo6 (Fig. 5E–F) in the Foxo1 cKO and control striatum at similar levels. These data suggest that while Foxo1 is required for the survival and differentiation of direct pathway SPNs at early stages of striatal development, the family members Foxo3/6 may play compensatory roles at later stages in the absence of Foxo1.
Figure 5.
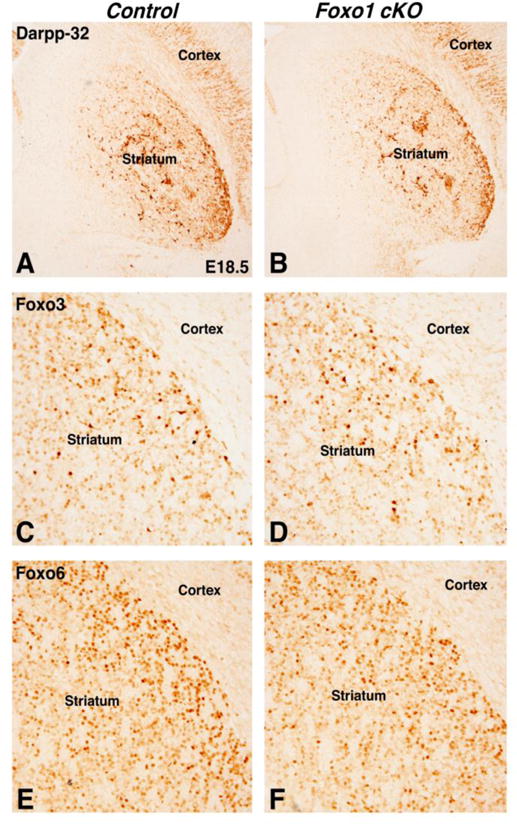
Foxo1 cKOs show improved striatal differentiation at E18.5. Darpp-32 expression is similar in control and Foxo1 cKOs at E18.5 (compare B to A). The Foxo family members, Foxo3 (compared D to C) and Foxo6 (compare F to E) are both expressed in the Foxo1 cKO striatum.
Given that Isl1 is required for the survival and differentiation of direct pathway SPNs and that Isl1 is required for the expression of Foxo1 in direct pathway SPNs at E15.5 (Fig. 3), we examined the expression of Foxo3 and Foxo6 in the Isl1 cKO striatum. Besides Foxo1, Foxo6 is expressed in the E15.5 and E18.5 striatum (Fig. 6A and 6C) and is reduced in Isl1 cKO at both stages (Fig. 6B and 6D). Quantification revealed a 38% reduction in Foxo6 staining at E18.5 in Isl1 cKO compared to controls (Control 100% ± 4.5% and Isl1 cKO 62% ± 15.0%, p<.05, n=4). Foxo3 is expressed later in development (after E15.5), and is also reduced in the Isl1 cKO striatum at E18.5 (compare Fig. 6F to 6E) with a 62% reduction in striatal Foxo3 positive cells (Control avg. 180 cells/mm2 ± 32.1 and Isl1 cKO avg. 68.5 cells/mm2 ± 9.6%, p<.05, n=3). Our data suggest that Isl1 cKO striatum is Foxo deficient during development (Fig. 3 and 6).
Figure 6.
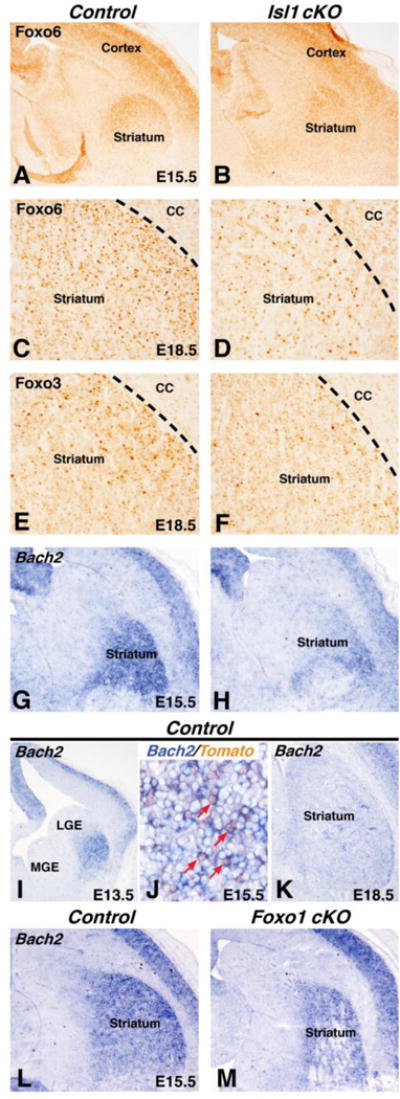
Isl1 conditional mutants are deficient in Foxo signaling. Isl1 conditional mutants show reduced expression of Foxo6 at E15 (compare B to A) and E18 (compare D to C). Foxo3 is also reduced in the Isl1 conditional mutant striatum at E18.5 (compare F to E). The Foxo1/Foxo3 target gene Bach2 is expressed in the developing striatum at E13 (I), E15 (G), and E18.5 (K). Double labeling with in situ for Bach2 and IHC for tdTomato show that Bach2 expression is observed in Isl1 fate-map (Isl1cre/+; RosaCAG−tdTomato/+) tdTomato+ neurons at E15.5 (red arrows in J). Isl1 conditional mutants show reduced Bach2 expression in the striatum at E15.5 (compare H to G). Bach2 is modestly reduced in the E15.5 Foxo1 cKO striatum (M) as compared to control (L). Note that the level of expression in the Foxo1 cKO striatum (M) appears higher than that observed in the Isl1 cKO striatum (H).
A recent report has identified core Foxo1 and Foxo3 target genes (Webb et al., 2013). Of these core genes, we found that the transcription factor Bach2 is robustly expressed in the developing striatum at E13 (Fig. 6I) and E15 (Fig. 6G). By E18.5, Bach2 expression is down regulated with only scattered cells within the striatum (Fig. 6K). Interestingly, combined in situ hybridization for Bach2 and immunostaining for tdTomato in the Isl1 fate map at E15.5, shows that Bach2 is expressed in the Isl1 (i.e. direct pathway) striatal lineage (red arrows in Fig. 6J). Moreover, Bach2 is reduced in the Isl1 cKO striatum at E15.5 (compare Fig. 6H to G) suggesting that the Foxo deficiency in these conditional mutants leads to a considerable reduction in the expression of this Foxo core target gene (Webb et al., 2013). However, we cannot rule out the possibility that Isl1 directly regulates Bach2 expression. We next examined the expression of Bach2 in the Foxo1 cKO striatum and found a more modest reduction (Fig. 6M), as compared to control (Fig. 6L), than that observed in the Isl1 cKO (Fig. 6H). Thus Bach2 represents a novel transcription factor gene expressed in developing direct pathway SPNs that may be a core direct target of Foxo factors and/or Isl1 itself.
4. Discussion
We have identified the forkhead transcription factor Foxo1 as a candidate downstream effector of Isl1 during development of the direct pathway SPNs. Our results indicate that Foxo1 expression is enriched in the Isl1 lineage of direct pathway SPNs at embryonic stages and is reduced in Isl1 cKOs during striatal development. Unlike at embryonic stages, Foxo1 becomes more broadly expressed in the adult striatum in both the direct and indirect pathway. Using a conditional knockout approach restricted to the ventral telencephalon (i.e. Dlx5/6-CIE mice), we identified that Foxo1 is required for the survival and/or differentiation of early-generated direct pathway SPNs. Interestingly, at late embryonic stages, Darpp-32 expression in the Foxo1 cKO striatum appears similar to controls and this is coincident with the expression of other Foxo family members, Foxo3 and Foxo6. In contrast, Isl1 cKOs exhibit reduced striatal Darpp-32 expression (Ehrman et al., 2013) and reductions in all striatal Foxo family members (Foxo1/3/6). Moreover, we identified that Bach2, which is a transcription factor and candidate Foxo core target gene, is expressed in the developing direct pathway SPNs and is also severely reduced in the Isl1 cKO striatum. Our results suggest that Foxo family members and their respectively regulated genes function downstream of Isl1 in the survival and/or differentiation of, at least a subpopulation of, direct pathway SPNs.
Despite a significant direct pathway SPN phenotype in the Isl1 cKO, a small portion of direct pathway SPNs remain in these mutants and are marked by Ebf1 expression (Ehrman et al., 2013; Lu et al., 2014). At early stages of striatogenesis (e.g. E15.5), Foxo1 expression is nearly undetectable in the Isl1 cKO striatum, however, by E18.5 a modest recovery of expression is observed. Interestingly, the improved Foxo1 expression is found within the remaining Ebf1-expressing direct pathway SPNs in the mutant. Therefore, while Foxo1 is downstream of Isl1 in early born direct pathway SPNs, other factors (including Ebf1) may be upstream of it in late stage direct pathway SPNs, suggesting that Foxo regulated pathways could play an essential role in the survival of embryonic direct pathway SPNs throughout striatal neurogenesis.
Even though Foxo1 is known to play multiple roles in cellular homeostasis within multiple organs (and cell types) in the body (Kousteni, 2012; Puthanveetil et al., 2013), its highly localized expression within the developing forebrain and, in particular, the forming striatum is rather remarkable (Hoekman et al, 2006; data shown here). At embryonic time points, a clear bias for Foxo1 expression in the direct pathway SPNs is observed. By postnatal stages, the striatal enrichment of Foxo1 expression remains, however, it is no longer specific to the direct pathway, being expressed also in the indirect pathway SPNs. Moreover, Foxo1 cKOs show a clear reduction in direct pathway SPN survival/differentiation, at least at early stages of striatogenesis. At late stages, a recovery of direct pathway SPN development is observed in these mutants and this correlates with the expression of the Foxo1 family member Foxo6 (and to a lesser extent Foxo3). These two factors are also expressed in the developing striatum from E15 onward, although at considerably lower levels than Foxo1 (Hoekman et al., 2006). Thus, Foxo1/3/6 may have redundant roles in the survival/differentiation of direct pathway SPNs. To examine this, it will be important to generate Foxo1/6 (double) and Foxo1/3/6 (triple) mutants to determine if the striatal phenotype resembles (or is more severe than) that in the Isl1 cKO mutants.
A recent study has identified a core set of gene targets for both Foxo1 and Foxo3 by comparing known Foxo1 target genes in B and T cells with newly identified Foxo3 targets in postnatal neural progenitor cells (see Table S3 in Webb et al., 2013). The core consists of 64 genes and includes the Bach2 gene, which encodes for a transcriptional regulator. We show here that Bach2 is highly enriched in the embryonic striatum and is significantly downregulated at perinatal stages. Bach2 is required for survival and differentiation of Foxp3+ T regulatory cells (Kim et al., 2014) and therefore may mediate the survival/differentiation of direct pathway SPNs downstream of Isl1 and Foxo1/3/6. Given the expression of Foxo1 in both the direct and indirect pathway SPNs at postnatal time points, we would assume multiple roles for Foxo regulated pathways in SPNs, however, the restricted expression of Bach2 in embryonic SPNs may implicate this factor as a regulator of direct pathway SPN survival/differentiation either directly downstream of Isl1 or Foxo1/3/6 (or both).
It is known that Foxo1 can regulate different cellular processes such as cell death versus survival depending on its subcellular localization (i.e. nucleus versus cytoplasm) (Yuan et al., 2008, 2009). Although Foxo1 is a nuclear (i.e. transcription) factor, phosphorylation by Akt leads to its shuttling from the nucleus to the cytoplasm (Accili and Arden, 2004; Van Der Heide et al., 2004) where it regulates cellular processes (e.g. autophagy) in a non-transcriptional manner (Medema and Jäättelä, 2010; Zhao et al., 2010). We observed Foxo1 expression in both the nuclear and cytoplasmic compartments of the embryonic direct pathway SPNs, however, at postnatal stages we observed predominant cytoplasmic (i.e. axons and terminals) Foxo1 expression in both direct and indirect pathway SPNs. It remains unclear in which subcellular compartment Foxo1 participates in the survival of direct pathway SPNs. Unlike Foxo1, Foxo6 is almost always nuclear (Jacobs et al., 2003). Considering that we see Foxo6 (and Foxo3) down-regulated in Isl1 cKO striatum and maintained at late embryonic stages in the in Foxo1 cKO striatum, this might suggest that the survival effect of Foxo factors downstream of Isl1 may be mediated via transcription. In this way, Isl1 positively regulates the expression of Foxo1/3/6 (either directly or indirectly) and within the nucleus these factors directly regulate the expression of Bach2, which may serve as a common survival/differentiation factor for the majority of direct pathway SPNs. Future experiments aimed at restricting the subcellular localization of Foxo factors (nuclear versus cytoplasmic) and the conditional striatal inactivation of Bach2 will help to test this hypothesis.
Highlights.
-
-
Foxo1 is enriched in embryonic direct pathway striatal projection neurons (dSPNs)
-
-
Isl1 is upstream of Foxo family members Foxo1/3/6 in dSPNs
-
-
loss of Foxo1 results in cell death and differentiation defects in embryonic dSPNs
-
-
we propose that Isl1 is upstream of Foxo regulated pathways in dSPN development
Acknowledgments
We thank R.A. DePinho for the Foxo1 floxed mice as well as X. Mu and W.H. Klein for providing the Isl1 floxed mice. We also thank A. Brunet (Foxo6) and E. Morrisey (Foxp1) for providing antibodies.
Funding
This work was supported by the NIH grant MH090740 to KC and start-up funds provided by the Cincinnati Children’s Hospital Medical Center to RW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. Trends Neurosci. 1989;12:366–375. doi: 10.1016/0166-2236(89)90074-x. [DOI] [PubMed] [Google Scholar]
- Anderson KD, Reiner A. Immunohistochemical localization of DARPP-32 in striatal projection neurons and striatal interneurons: implications for the localization of D1-like dopamine receptors on different types of striatal neurons. Brain Res. 1991;568:235–243. doi: 10.1016/0006-8993(91)91403-n. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Ehrman LA, Mu X, Waclaw RR, Yoshida Y, Vorhees CV, Klein WH, Campbell K. The LIM homeobox gene Isl1 is required for the correct development of the striatonigral pathway in the mouse. Proc Natl Acad Sci U S A. 2013;110:E4026–4035. doi: 10.1073/pnas.1308275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Marin F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6:134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- Kim EH, Gasper DJ, Lee SH, Plisch EH, Svaren J, Suresh M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J Immunol. 2014;192:985–995. doi: 10.4049/jimmunol.1302378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousteni S. FoxO1, the transcriptional chief of staff of energy metabolism. Bone. 2012;50:437–443. doi: 10.1016/j.bone.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Yeh C, Yang XW. Pivotal role of early B-cell factor 1 in development of striatonigral medium spiny neurons in the matrix compartment. J Neurosci Res. 2008;86:2134–2146. doi: 10.1002/jnr.21666. [DOI] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KM, Evans SM, Hirano S, Liu FC. Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc Natl Acad Sci U S A. 2014;111:E168–177. doi: 10.1073/pnas.1319138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Jäättelä M. Cytosolic FoxO1: alive and killing. Nat Cell Biol. 2010;12:642–643. doi: 10.1038/ncb0710-642. [DOI] [PubMed] [Google Scholar]
- Martin-Ibanez R, Crespo E, Urban N, Sergent-Tanguy S, Herranz C, Jaumot M, Valiente M, Long JE, Pineda JR, Andreu C, Rubenstein JL, Marin O, Georgopoulos K, Mengod G, Farinas I, Bachs O, Alberch J, Canals JM. Ikaros-1 couples cell cycle arrest of late striatal precursors with neurogenesis of enkephalinergic neurons. J Comp Neurol. 2010;518:329–351. doi: 10.1002/cne.22215. [DOI] [PubMed] [Google Scholar]
- Martin-Ibanez R, Crespo E, Esgleas M, Urban N, Wang B, Waclaw R, Georgopoulos K, Martinez S, Campbell K, Vicario-Abejon C, Alberch J, Chan S, Kastner P, Rubenstein JL, Canals JM. Helios transcription factor expression depends on Gsx2 and Dlx1&2 function in developing striatal matrix neurons. Stem Cells Dev. 2012;21:2239–2251. doi: 10.1089/scd.2011.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, Hiller D, Jiang S, Protopopov A, Wong WH, Chin L, Ligon KL, DePinho RA. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Wang B, Chen G, Nagao M, Nakafuku M, Campbell K. Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proc Natl Acad Sci U S A. 2011;108:1675–1680. doi: 10.1073/pnas.1008824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthanveetil P, Wan A, Rodrigues B. FoxO1 is crucial for sustaining cardiomyocyte metabolism and cell survival. Cardiovasc Res. 2013;97:393–403. doi: 10.1093/cvr/cvs426. [DOI] [PubMed] [Google Scholar]
- Salih DA, Rashid AJ, Colas D, de la Torre-Ubieta L, Zhu RP, Morgan AA, Santo EE, Ucar D, Devarajan K, Cole CJ, Madison DV, Shamloo M, Butte AJ, Bonni A, Josselyn SA, Brunet A. FoxO6 regulates memory consolidation and synaptic function. Genes Dev. 2012;26:2780–2801. doi: 10.1101/gad.208926.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Molkentin JD, Paik JH, DePinho RA, Yutzey KE. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011;286:7468–7478. doi: 10.1074/jbc.M110.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D, Zhai P, Del Re DP, Sciarretta S, Yabuta N, Nojima H, Lim DS, Pan D, Sadoshima J. A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nat Commun. 2014;5:3315. doi: 10.1038/ncomms4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Campbell K. A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development. 2001;128:4769–4780. doi: 10.1242/dev.128.23.4769. [DOI] [PubMed] [Google Scholar]
- Toresson H, Mata de Urquiza A, Fagerstrom C, Perlmann T, Campbell K. Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. Development. 1999;126:1317–1326. doi: 10.1242/dev.126.6.1317. [DOI] [PubMed] [Google Scholar]
- Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw RR, Allen ZJ, 2nd, Bell SM, Erdelyi F, Szabo G, Potter SS, Campbell K. The zinc finger transcription factor Sp8 regulates the generation and diversity of olfactory bulb interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Waclaw RR, Ehrman LA, Pierani A, Campbell K. Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J Neurosci. 2010;30:6944–6953. doi: 10.1523/JNEUROSCI.5772-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw RR, Wang B, Campbell K. The homeobox gene Gsh2 is required for retinoid production in the embryonic mouse telencephalon. Development. 2004;131:4013–4020. doi: 10.1242/dev.01272. [DOI] [PubMed] [Google Scholar]
- Wang B, Waclaw RR, Allen ZJ, 2nd, Guillemot F, Campbell K. Ascl1 is a required downstream effector of Gsx gene function in the embryonic mouse telencephalon. Neural Dev. 2009;4:5. doi: 10.1186/1749-8104-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Lufkin T, Rubenstein JL. Dlx6 regulates molecular properties of the striatum and central nucleus of the amygdala. J Comp Neurol. 2011;519:2320–2334. doi: 10.1002/cne.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Long JE, Flandin P, Pla R, Waclaw RR, Campbell K, Rubenstein JL. Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J Comp Neurol. 2013;521:1561–1584. doi: 10.1002/cne.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Pollina EA, Vierbuchen T, Urban N, Ucar D, Leeman DS, Martynoga B, Sewak M, Rando TA, Guillemot F, Wernig M, Brunet A. FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang Y, Wang C, Xu Z, Liang Q, An L, Li J, Liu Z, You Y, He M, Mao Y, Chen B, Xiong ZQ, Rubenstein JL, Yang Z. The zinc finger transcription factor Sp9 is required for the development of striatopallidal projection neurons. Cell Rep. 2016;16:1431–1444. doi: 10.1016/j.celrep.2016.06.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, Konishi Y, Schaefer EM, Bonni A. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319:1665–1668. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Lehtinen MK, Merlo P, Villen J, Gygi S, Bonni A. Regulation of neuronal cell death by MST1-FOXO1 signaling. J Biol Chem. 2009;284:11285–11292. doi: 10.1074/jbc.M900461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Yun K, Garel S, Fischman S, Rubenstein JL. Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol. 2003;461:151–165. doi: 10.1002/cne.10685. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li H, Li X, Zhang G, Niu Y, Yuan Z, Herrup K, Zhang YW, Bu G, Xu H, Zhang J. The roles of Cdk5-mediated subcellular localization of FOXO1 in neuronal death. J Neurosci. 2015;35:2624–2635. doi: 10.1523/JNEUROSCI.3051-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]


