Abstract
Purpose
Cancers may resist single-agent targeted therapies when the flux of cellular growth signals is shifted from one pathway to another. Blockade of multiple pathways may be necessary for effective inhibition of tumor growth. We document a case in which a patient with anaplastic thyroid carcinoma (ATC) failed to respond to either mTOR/PI3K or combined RAF/MEK inhibition, but experienced a dramatic response when both drug regimens were combined.
Experimental Design
Multi-region whole-exome sequencing of five diagnostic and four autopsy tumor biopsies was performed. Meta-analysis of DNA and RNA sequencing studies of ATC was performed.
Results
Sequencing revealed truncal BRAF and PIK3CA mutations, which are known to activate the MAPK and PI3K/AKT pathways respectively. Meta-analysis demonstrated 10.3% co-occurrence of MAPK and PI3K pathway alterations in ATC. These tumors display a separate transcriptional profile from other ATC, consistent with a novel subgroup of ATC.
Conclusions
BRAF and PIK3CA mutations define a distinct subset of ATC. Blockade of the MAPK and PI3K pathways appears necessary for tumor response in this subset of ATC. This identification of synergistic activity between targeted agents may inform clinical trial design in ATC.
Introduction
Anaplastic thyroid carcinoma (ATC) is among the most aggressive solid tumors, with an almost uniformly fatal prognosis. Most patients (70%) present with metastatic disease(1) and survive for an average of 4.2 months(2). Genomic interrogation of ATC patients has demonstrated a spectrum of mutated genes including TP53, NRAS, BRAF, and PIK3CA(3, 4). A subset of these patients carry BRAFV600E and activating PIK3CA mutations. Animal studies have shown that co-mutation of BRAF and PIK3CA is sufficient to robustly establish ATC in mice, whereas mutation of either gene alone is not(5). No effective systemic therapies exist for ATC(6), although recent reports of exceptional responders to single-agent targeted therapies(7, 8) provide support for the use of targeted therapies in this disease. Broadly, combinations of targeted therapies in patients with activating alterations in multiple oncogenic pathways have been incompletely characterized. Furthermore, tumor genomic heterogeneity, or lack thereof, in the context of exceptional responders has not yet been explored. Here we perform multi-regional genomic analysis of an exceptional responder to dual inhibition of the MAPK and PI3K/AKT pathways to determine genomic features driving this clinical phenotype.
Materials and Methods
Study Oversight
The tissue collection study and tumor profiling study was approved by the institutional review board of the Dana–Farber/Harvard Cancer Center (protocols #09-472 and #11-104). The patient provided written informed consent for treatment and genomic sequencing.
Pathology
All tumor samples underwent pathology review (VAP, JAB) prior to DNA extraction. Kinome assay was performed per manufacturer’s instruction by the Brigham and Women’s Hospital Clinical Pathology department. The Human Phospho-Kinase Array Kit was purchased from R&D systems (Minneapolis, MN) and used according to the manufacturer’s instructions.
DNA sequencing
Tumor genomic DNA was isolated from formalin-fixed paraffin embedded blocks of tissue using the QIAamp DNA FFPE Tissue Kit (Qiagen)(9). Normal DNA was isolated from whole blood. Targeted sequencing of 301 genes (442X average coverage) was performed on the initial tumor biopsy(10). Samples were sequenced on an Illumina HiSeq-2000 to an average of depth of 206X(9). Sequencing was performed with paired-end reads and an average read length of 76. The average rate of read alignment was 98.6%. FASTQ files were aligned to the reference genome and quality control metrics were computed using the Broad Institute Picard software suite version 3.
Somatic mutation calling
Somatic mutations were called with MuTect(11). OxoG artifacts were removed using the Broad Institute OxoG3 filter(12). Insertions and deletions (indels) were called with Strelka(13). All mutations were filtered against a panel of normal genomes sequenced at the same institute to remove sequencing artifacts.
Targeted Panel Sequencing of Anaplastic Thyroid Carcinoma
Specimens were collected under institutional protocol number 09-472. Samples were sequenced using a capture array of 300 known cancer genes(10). Sequencing and mutation calls were performed by the Center for Cancer Genome Discovery at DFCI.
Copy number determination
Relative copy number profiles were determined using ReCapSeg as described in the Broad’s GATK documentation (http://gatkforums.broadinstitute.org/categories/recapseg-documentation). Briefly, exome sequencing coverage data were normalized against exome coverage data from a panel of blood controls to generate probe level copy-level data that are subsequently segmented using circular binary segmentation(14). These segmented copy number profiles were paired with read counts at germline heterozygous sites to generate allele-specific relative copy number profiles using Allelic Capseg(15). Allele-specific copy number data was paired with exomic mutations as input to ABSOLUTE for final computation of discrete allele-specific copy number profiles(16).
Phylogenetic Tree Reconstruction
We used a force-calling strategy to recover evidence of any mutations that failed to reach the threshold of Mutect in a given biopsy at sites that were called confidently in other biopsies(15).
Phylogenetic trees were constructed from mutation data using an implementation of clonal ordering(17). Mutations were converted into a binary incidence-matrix depending on their absence/presence in each biopsy. These data were paired with a calculation of the power to detect each mutation in each biopsy given the purity, local ploidy and sequencing coverage. Mutations that were absent in one biopsy but present in others were marked as absent with a probability proportional to the power to detect the mutation (using independent sampling at each entry of the incidence matrix). A distance matrix was computed from the final incidence matrix using the following distance metric:
where corresponds to the binary vector of mutations in biopsy a and is the vector describing biopsy b. Hierarchical clustering of this distance matrix was performed using the complete linkage method in R. This procedure was performed 1,000 times to obtain a consensus tree. The reliability of specific splits was measured by their presence across permutations (Supplementary Figure 1). A given split was considered reliable across different trees if all of the members of the two branches are the same.
Chromosome arm level losses in each samples were recorded by manual review of the ABSOLUTE copy number plots (Supplementary Figure 2). Once one chromosome arm is lost, its alleles cannot be regained, thereby making these losses informative phylogenetic events. The phylogenetic tree in Figure 2 was generated by clonal ordering of these events, it is concordant with the tree built from the mutation data and allows for more accurate placement of several biopsies such as biopsies 4, 9 and 2.
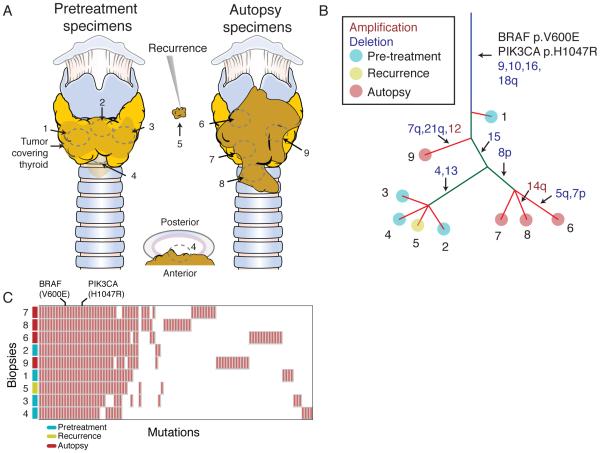
Figure 2.
Genomic features of an exceptional responder to combined MAPK and PI3K inhibition. A, Biopsy locations from the tumor following near-total thyroidectomy (left), at recurrence (middle) and at autopsy (right). B, Phylogenetic architecture of the tumor. Putative cancer driver mutations are annotated. Red font indicated copy number gain of the indicated chromosome, blue font indicates copy number loss. Branch lengths are not proportional to number of mutations. C, Mutations detected in each biopsy.
Results
A 62-year-old woman presented with a two-month history of dysphagia, throat irritation and a rapidly expanding neck mass. Fine needle aspiration of the dominant nodule demonstrated a high-grade malignancy suggestive of anaplastic thyroid carcinoma. The patient underwent near-total thyroidectomy to preserve the right recurrent laryngeal nerve and parathyroid. Only subtotal resection was possible due to tumor infiltration of the trachea. A right level VI lymph node dissection was performed as well. Pathologic analysis showed a 4.5 cm anaplastic thyroid carcinoma with extensive squamous differentiation and lymphovascular invasion. Tumor was present at the surgical resection margins. One week after surgery, the patient underwent a staging PET-CT with no evidence of FDG-avid metastatic disease. The patient received concurrent radiosensitizing chemoradiotherapy with carboplatin and paclitaxel. She was also started on a regimen of levothyroxine for TSH suppression. After completing chemoradiation, a re-staging PET-CT was performed and no residual disease was detected.
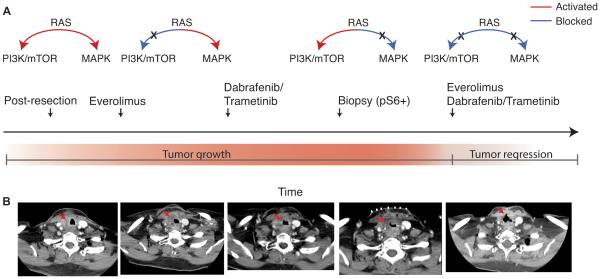
Nine months after completing chemoradiation, the patient’s tumor locally recurred. She received re-treatment with carboplatin and paclitaxel for palliation, but demonstrated radiographic evidence of progression. Next, the patient was placed on everolimus (an mTOR inhibitor) empirically. After four weeks, the patient showed disease progression by CT (Figure 1A-B) and everolimus was discontinued. Clinical targeted sequencing of a tumor biopsy from the original resection revealed BRAFV600E and PIK3CAH1047R mutations. On the basis of the BRAF mutation, therapy with combined dabrafenib and trametinib was initiated. After six weeks, the patient was evaluated by CT and her tumor had continued to grow. The patient developed stridor and a second biopsy was obtained. Kinome analysis was markedly positive for pS6, a marker of PI3K/MTOR pathway activation. Because the patient experienced minimal side effects with dabrafenib/trametinib, triple therapy with dabrafenib/trametinib/everolimus was initiated. Five weeks later, physical exam revealed reduced stridor, and the patient reported decreased shortness of breath. CT evaluation showed dramatic tumor shrinkage, with increased patency of the airway (Figure 1A-B).
Figure 1.
Clinical history of an exceptional responder to combined MAPK and PI3K inhibition. A, Graphical description of clinical events. B, Radiographic documentation of response. The patient’s tumor continued to grow through everolimus and dabrafenib monotherapies individually. When combined, the patient’s tumor showed a dramatic response.
Eight weeks after the initiation of dual pathway inhibition, the patient became acutely short of breath with worsening stridor and ultimately died. Clinically, fatal tracheomalacia secondary to rapid death of tumor cells was suspected. Autopsy revealed that approximately half of the tumor showed necrosis, fibrosis and calcification, consistent with a dramatic response to therapy. In addition, there was devitalization and erosion of adjacent cartilage.
Biopsies of the diagnostic resection and on-treatment autopsy tumor were obtained for genomic characterization and comparison with the diagnostic resection from 16 months prior. A total of 10 tumor biopsies that were subjected to genomic profiling, 9 of which were subjected to high-depth (mean 205X) whole-exome sequencing (WES). We identified a total of 99 coding mutations. A phylogenetic tree was constructed that revealed clonal and known activating BRAFV600E and PIK3CAH1047R mutations in all biopsies profiled (Figure 2A-C). The mutational structure of the phylogenetic tree demonstrated heterogeneity that was mostly restricted to mutations private to individual biopsies. Only 22 of 99 single nucleotide mutations were detected in all biopsies (Figure 2C). Of the remaining mutations, 54 were detected in only a single biopsy.
As with the mutational heterogeneity between biopsies, somatic copy number alterations differed. While all biopsies displayed clonal losses of chromosomes 9,10,16 and 18q, ten other chromosome-level copy number alterations were heterogeneous. Three of four pretreatment biopsies displayed clonal loss of chromosomes 4 and 13, whereas none of the biopsies obtained at autopsy displayed loss of these chromosome arms (Supplementary Figure 2). Only 3 of 5 biopsies from the post-resection on-treatment tumor clustered into a single branch of the phylogenetic tree, indicating that multiple subclones survived surgical resection and chemoradiation.
Given the genotype-phenotype relationship in this case, we sought to determine the rate at which BRAF and PIK3CA are co-mutated in ATC more generally. We combined data obtained from targeted sequencing at our institution with two previously published next-generation sequencing studies of ATC(3, 4). BRAF was co-mutated with PIK3CA in 10.3% of patients (9/87, Figure 3A). We identified an additional two tumors that had co-mutation of other MAPK and PI3K pathway genes, one with ARAFp.S214F-PTENp.Q171H co-mutation and the other with MAPK1p.P319L-PTENp.R335X co-mutation. Unsupervised clustering of expression data from ATC samples, demonstrated that the tumors with BRAF/PIK3CA co-mutation clustered together (p=0.006, Figure 3A). These data suggest that BRAF/PIK3CA co-mutated tumors define a distinct biological subset of ATC.
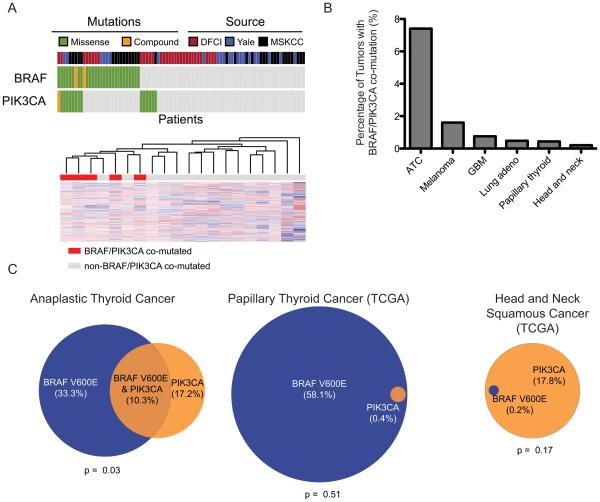
Figure 3.
Prevalence of BRAF/PIK3CA co-mutation. A, (top) Co-mutation of BRAF and PIK3CA in sequencing studies of anaplastic thyroid cancers. Each column of blocks represents an individual patient’s tumor. The source of the sequencing data, the presence of BRAF and PIK3CA mutations are represented from top to bottom. (bottom) Unsupervised clustering of expression data from ATC samples in MSKCC cohort. BRAF/PIK3CA co-mutated samples are indicated in red and cluster together (p = 0.006). B, Prevalence of BRAFV600E co-mutation with non-synonymous PIK3CA mutations across tumor types. Only tumor types with non-zero co-mutation rates are depicted (17 other tumor types had no cases of co-mutation). C, Overlap of BRAFV600E and PIK3CA mutations in head and neck cancers for which sequencing data is available. Circle area is proportional to the fraction of patients harboring mutations in the indicated gene.
We then analyzed 5,255 cancer exomes in the Cancer Genome Atlas (TCGA) to determine whether similar cancer genomes occurred outside of anaplastic thyroid carcinoma(17). We identified 11 tumors with BRAFV600E and non-synonymous PIK3CA mutations. Seven individuals were identified with BRAFV600E and PTEN co-mutation. Affected tumor types included melanomas (n=11), glioblastoma multiforme (n=2), lung adenocarcinoma (n=2), head and neck squamous cancers (n=1), and papillary thyroid cancers (n=2) (Figure 3B). While BRAF and PIK3CA were significantly co-mutated in ATC (p=0.03), this finding was not replicated across other head and neck cancers (Figure 3C).
Discussion
Preclinical ATC cell line and mouse models suggest that neither BRAFV600E nor PIK3CAH1047R mutations alone are sufficient to induce ATC-like tumors(18). However, mice bearing activating mutations of BRAF and PIK3CA develop tumors that resemble ATC. Similarly, mouse tumors and human cell lines bearing the same genetic architecture as our patient respond only to co-treatment with MAPK and mTOR inhibitors and not to either treatment alone(19, 20). These pre-clinical data anticipated the response observed in this patient. In this case, we were able to show that MAPK inhibition alone did not inhibit tumor growth while the PIK3CA pathway was still active providing a rationale for dual blockade, which resulted in tumor response. Taken together, these observations suggest that MAPK and PI3K/MTOR pathway blockade represents an effective dual therapy aimed at the founding oncogenic lesions in a subset of ATC. To our knowledge, this is the first instance of clinical response to dual pathway inhibition in ATC, and the prevalence of such genetic co-occurrence in this disease may warrant clinical trial development favoring combination therapies.
Furthermore, extensive WES of tumor tissue obtained at various points during the patient’s treatment showed a notable absence of potential new driver mutations, despite the emergence of multiple subclones typically associated with poor response to therapy(21). It is unclear whether homogeneity among drivers is a common characteristic of ATC. We speculate that the driver homogeneity in this tumor, as revealed through it's phylogenetic structure, may have resulted in a durable response, but unfortunately the patient experienced a catastrophic complication from the rapid initial treatment response.
If supported with larger cohorts, the observation that certain targeted therapies have little efficacy as single agents, but are clinically synergistic in combination suggests that monotherapy clinical trial strategies may fail to recognize drugs that could help genomically defined patient populations. Combination clinical trials may enable repurposing of therapies and targets that were ineffective as monotherapies or in unstratified clinical subgroups.
Questions remain as to whether dual pathway blockade will be effective in ATC with different genotypes, or in other tumor types. Within ATC, RAS mutations are frequent and function in part by activating both the MAPK and PI3K pathways, suggesting that dual pathway blockade could help these patients as well. Furthermore, while no previously profiled tumor type shares the same frequency of BRAF/PIK3CA co-mutation as ATC, there has been broad interest in combination therapy trials targeting the MAPK and PI3K pathways in other tumor types. An extensive body of data supports the idea that PI3K pathway activation is frequently responsible for acquired resistance to BRAF inhibition and combination therapy is necessary for durable responses across a variety of cancers, particularly in melanoma (23, 24). On the basis of these data, MAPK/PI3K combination therapy is being evaluated in several early stage clinical combination therapy trials in multiple tumor types including melanoma and colorectal cancer(22). Despite this widespread hope, our findings contrast with the modest preliminary clinical data observed in other tumors types(23). This difference may be accounted for by the lower rate of BRAF/PIK3CA co-mutation in other tumor types. Alternatively, ATC may represent a tumor-type with unusually strong dependence on a limited number of mitogenic signals from its activated oncogenes.
Finally, the use of both targeted therapeutics individually and in concert were all off-label in this case. Off-label therapy administration is increasingly common in the oncology community with the identification of actionable targets and available inhibitors(25). In our case, no viable treatment alternatives were available and the evidence from in vitro laboratory data and results from our kinome analysis could justify the combination of drugs that have not been tested together in a clinical trial. While we do not encourage the use of untested drug combinations, this case illustrates the critical need to capture clinical actions being made as a result of cancer genotyping data and off-label use across the broader clinical oncology community in order to identify these outlier events for systematic study and connection with clinical trial development.
Supplementary Material
Statement of Significance.
Activating mutations in BRAF and PIK3CA define a subset of anaplastic thyroid carcinoma and activate the MAPK and PI3K/mTOR pathways respectively. We identified a patient who had an exceptional response to simultaneous inhibition of both of these pathways, while inhibition of either alone was insufficient. Analysis of the clonal architecture of the tumor revealed the truncal role of these mutations. To our knowledge, this is the first report of synergistic activity between oncogene-targeted agents in anaplastic thyroid cancer, and may have relevance in other cancer types harboring truncal activating mutations in these pathways.
Acknowledgements
We thank the patient and her family for participating in this study and allowing autopsy. We thank Maegan Harden for project management contributions and Stacey Gabriel for sequencing studies. We thank Cancer Research UK for designing the thyroid illustration that we have modified via Creative Commons License. The project described was supported by award Number T32GM007753 from the National Institute of General Medical Sciences (W.J.G.), NIH F30 CA192725-02 (W.J.G), NIH K08 CA188615-02 (E.M.V.), and the American Cancer Society (E.M.V.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Disclosures: Dr. Van Allen is a consultant for Roche Ventana, Takeda, and Third Rock Ventures. Dr. Van Allen receives research funding support from Bristol Myers Squibb. Dr Jochen Lorch receives research support from Novartis and Millennium and has been a consultant for Eisai. Dr. Beroukhim receives research funding support from and is a consultant for Novartis.
References
- 1.Tan RK, Finley RK, 3rd, Driscoll D, Bakamjian V, Hicks WL, Jr., Shedd DP. Anaplastic carcinoma of the thyroid: a 24-year experience. Head & neck. 1995;17:41–7. doi: 10.1002/hed.2880170109. discussion 7-8. [DOI] [PubMed] [Google Scholar]
- 2.Ito K, Hanamura T, Murayama K, Okada T, Watanabe T, Harada M, et al. Multimodality therapeutic outcomes in anaplastic thyroid carcinoma: improved survival in subgroups of patients with localized primary tumors. Head & neck. 2012;34:230–7. doi: 10.1002/hed.21721. [DOI] [PubMed] [Google Scholar]
- 3.Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Human molecular genetics. 2015;24:2318–29. doi: 10.1093/hmg/ddu749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. The Journal of clinical investigation. 2016;126:1052–66. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charles RP, Silva J, Iezza G, Phillips WA, McMahon M. Activating BRAF and PIK3CA mutations cooperate to promote anaplastic thyroid carcinogenesis. Molecular cancer research : MCR. 2014;12:979–86. doi: 10.1158/1541-7786.MCR-14-0158-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denaro N, Nigro CL, Russi EG, Merlano MC. The role of chemotherapy and latest emerging target therapies in anaplastic thyroid cancer. OncoTargets and therapy. 2013;9:1231–41. doi: 10.2147/OTT.S46545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosove MH, Peddi PF, Glaspy JA. BRAF V600E inhibition in anaplastic thyroid cancer. The New England journal of medicine. 2013;368:684–5. doi: 10.1056/NEJMc1215697. [DOI] [PubMed] [Google Scholar]
- 8.Wagle N, Grabiner BC, Van Allen EM, Amin-Mansour A, Taylor-Weiner A, Rosenberg M, et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. The New England journal of medicine. 2014;371:1426–33. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nature medicine. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacConaill LE, Garcia E, Shivdasani P, Ducar M, Adusumilli R, Breneiser M, et al. Prospective enterprise-level molecular genotyping of a cohort of cancer patients. The Journal of molecular diagnostics : JMD. 2014;16:660–72. doi: 10.1016/j.jmoldx.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic acids research. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 14.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 15.Stachler M, Taylor-Weiner A, Peng S, McKenna A, Agoston A, Odze R, et al. Paired Exome Analysis of Barrett’s Esophagus and Adenocarcinoma. Nature Genetics. 2015 doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFadden DG, Vernon A, Santiago PM, Martinez-McFaline R, Bhutkar A, Crowley DM, et al. p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1600–9. doi: 10.1073/pnas.1404357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandil E, Tsumagari K, Ma J, Abd Elmageed ZY, Li X, Slakey D, et al. Synergistic inhibition of thyroid cancer by suppressing MAPK/PI3K/AKT pathways. The Journal of surgical research. 2013;184:898–906. doi: 10.1016/j.jss.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 20.Jin N, Jiang T, Rosen DM, Nelkin BD, Ball DW. Dual inhibition of mitogen-activated protein kinase kinase and mammalian target of rapamycin in differentiated and anaplastic thyroid cancer. The Journal of clinical endocrinology and metabolism. 2009;94:4107–12. doi: 10.1210/jc.2009-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Temraz S, Mukherji D, Shamseddine A. Dual Inhibition of MEK and PI3K Pathway in KRAS and BRAF Mutated Colorectal Cancers. International journal of molecular sciences. 2015;16:22976–88. doi: 10.3390/ijms160922976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer chemotherapy and pharmacology. 2013;71:1395–409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- 24.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nature reviews Clinical oncology. 2013;10:143–53. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 25.Conti RM, Bernstein AC, Villaflor VM, Schilsky RL, Rosenthal MB, Bach PB. Prevalence of off-label use and spending in 2010 among patent-protected chemotherapies in a population-based cohort of medical oncologists. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1134–9. doi: 10.1200/JCO.2012.42.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.