Abstract
Background
Idiopathic nonspecific interstitial pneumonia (INSIP) presents with varying degrees of interstitial inflammation and fibrosis exhibiting a uniform appearance. Lack of knowledge on the underlying mechanisms of INSIP has contributed to few effective treatment strategies. Our study is designed to explore aberrantly expressed cytokines involvement in INSIP development.
Methods
Oligo GEArray was employed to detect the expression of cytokines in INSIP patients, and idiopathic pulmonary fibrosis (IPF) was setup as isotype control. Real-time PCR and immunohistochemistry analysis were used to further confirm the expression of abnormally expressed cytokines. The correlationship between cytokines expression and overall survival rate of patients with IPF and INSIP were analyzed.
Results
From microarray detection, transforming growth factor-beta-1 (TGF-β1), fibroblast growth factor 10 (FGF10), and platelet derived growth factor (PDGF) were predominantly up-regulated in patients with INSIP. Real-time PCR and immunohistochemistry also showed these cytokines was abnormally expressed in INSIP. In addition to, the clinical relevance analysis demonstrated relatively lower expression of PDGF patients had longer overall survival rate than those with higher expression of PDGF.
Conclusions
Our study suggests that TGF-β1, FGF10, and PDGF are required for the pathogenesis of INSIP, and may therefore be ideal targets in INSIP treatment. Moreover, INSIP patients with lower expression of PDGF had better survival rate.
Keywords: Idiopathic nonspecific interstitial pneumonia (INSIP), idiopathic pulmonary fibrosis (IPF), transforming growth factor-beta-1 (TGF-β1), fibroblast growth factor 10 (FGF10), platelet derived growth factor (PDGF)
Introduction
Idiopathic nonspecific interstitial pneumonia (INSIP) is generally characterized by hyperplastic type II pneumocytes with inflammatory cell infiltration, alveolar septum uniformly broadening with or without fibrosis, collagen deposition, occasional fibroblastic foci and honeycomb appearance of the lung (1,2) . INSIP is a major sub-type of idiopathic interstitial pneumonias (IIPs), which is a diverse group of lung disorders of unknown etiology characterized by various degrees of alveolar inflammation and remodeled alveolar structure that often result in pulmonary fibrosis (3,4). INSIP and idiopathic pulmonary fibrosis (IPF) share similar histomorphology, the latter also as the main sub-type of IIPs, and typically exhibits chronic fibrosis interstitial pneumonia, fibroblastic foci, and honeycomb changes (5), even though these two diseases were thought have etiological and inheritance heterogeneity. At present, many patients with IIPs respond to corticosteroid therapy to a certain degrees, but few achieve completely remission. Therefore, more effect needs to explore new potential strategy treatment for patients with IIPs, including INSIP and IPF.
Previous studies have suggested that many cytokines, including interleukins (ILs), transforming growth factor-beta (TGF-β), alpha-smooth muscle actin (α-SMA), and BMP-7 have pro-fibrogenic effects (6-9). Since patients with IPF frequently exhibit fibrotic lesion and have poor prognosis, many studies have focused on the role of these cytokines in IPF, and few investigations uncover the aberrantly expressed cytokines involved in the pathogenesis of patients with INSIP. In addition to, the clinical outcome of abnormally expressed cytokines in patients with INSIP is also still unclear.
In this study, we hypothesized that various cytokines were abnormally produced in the patients with INSIP. We determined the expression profile of cytokines in INSIP, including IPF by Oligo GEArray. Our initial results were validated by tissue-array with immunohistochemistry analysis and real-time PCR. Finally, the clinical outcome of related cytokines was further analyzed in 22 cases of INSIP and 25 cases of IPF. Our study aimed to identify the involvement of critical cytokines in the advancement of INSIP, and clarify whether these cytokines are related with the survival rate of patients with INSIP and IPF.
Methods
Human subjects
This study was approved by the ethics committee of Tongji Hospital [(Tong) No. 183 Ethics]. All samples were collected from patients with biopsy-confirmed INSIP and IPF from year 1999 to 2009, and all patients provided informed consent. Criteria for inclusion of subjects: (I) Biopsies for Oligo GEArray taken by video-assisted thoracoscope surgery or small incision lung biopsy. The control biopsy samples were collected from normal tissues of benign pulmonary tumors. Specimens used for tissue array and immunohistochemistry included 25 cases of IPF and 22 cases of INSIP (4 cases are cellular type, 18 cases are fibrosing type), which were collected at the time of diagnostic surgical lung biopsy through Tongji Hospital and Shanghai Pulmonary Hospital, affiliated with Tongji University School of Medicine; (II) the criteria of diagnosis referred to the American Thoracic Society (ATS)/European Respiratory Society (ERS) classification guidelines on IIP in 2002 (2), ATS/ERS views on INSIP Classification and Diagnostic Criteria in 2008 (10), Guidelines for Diagnosis and Management of IPF in 2011 (11) and Update of the International Multidisciplinary Classification of the Idiopathic Interstitial Pneumonias in 2013 (12); (III) the final diagnosis involved in multiple-disciplinary discussion is mutually made by pathologists, clinician and radiologists, and except other known causes of interstitial lung disease (ILD); (IV) each case had integrated clinical, radiologic, and pathologic data, including at least the follow-up data of more than 5 years. Besides, all patients received Glucocorticoids treatment, the Glucocorticoid use as following: (I) a large dosage: 100–200 mg/d methylprednisolone via intravenous injection, then 40 mg/d per os after 10 days, the dosage of methylprednisolone could be reduce until 4 weeks; (II) ordinary dosage: 0.5 mg/kg prednisone via per os, 0.25 mg/kg per os after 4 weeks, 0.125 mg/kg per os after 8 weeks or 0.25 mg/kg per os q.o.d (13). The clinical information of all patients included in the study is shown in Table 1.
Table 1. The clinical features of INSIP and IPF.
| Clinical features | Cases (%) | |
|---|---|---|
| INSIP (n=22) | IPF (n=25) | |
| Age (years) | 50.86±9.8 | 58.16±10.7 |
| Median age | 49 | 58.5 |
| Range | 36–74 | 41–75 |
| Gender | ||
| Male | 10 (45.5) | 17 (73.3) |
| Female | 12 (54.5) | 8 (26.7) |
| Smoking history | 9 (40.9) | 10 (40.0) |
| Dust exposure | 8 (36.4) | 6 (24.0) |
| Symptoms | ||
| Polypnea after activity | 17 (77.2) | 22 (88.0) |
| Velcro rale* | 15 (68.2) | 24 (96.0) |
| Acropachia | 3 (13.6) | 13 (52.0) |
| Average survival rate (months) | 80.95±31.8 | 45.2±19.2 |
*, Inspiratory crackles. INSIP, idiopathic nonspecific interstitial pneumonia; IPF, idiopathic pulmonary fibrosis.
Oligo GEArray
To monitor the expression profile of cytokines in patients with INSIP, including IPF, Oligo GEArray was employed. We extracted total RNA from human samples (3 cases of IPF and INSIP, and 1 case of normal control) that were grown on plastic plates, using the RNA Stat-60 reagent, and converted RNA into biotin-labeled cRNA target probes for microarray hybridization using the True Labeling-AMP linear RNA amplification kit. The cRNA targets (2 µg of cRNA) were next hybridized with oligonucleotide probes, representing different cytokines, printed on a nylon membrane. The resulting products on arrayed membranes were detected by a chemiluminescent detection kit, and analyzed by GEArray Analyzer data analysis software.
Tissue array design and immunohistochemistry (IHC)
Before constructing tissue array (TMA), typical lesions of 25 cases of IPF and 22 cases of INSIP were evaluated under light microscope. Then the TMA was performed using a core diameter of 2 mm by Shanghai Outdo Biotech laboratory. Each slide contained 77 lesion cores and 1 normal tissue core. IHC of paraffin-embedded sections was carried out using a standard streptavidin-biotinylated alkaline phosphatase (ABC-AP, DakoCytomation, Hamburg, Germany) method. The following antibodies were used: TGF-β1 (Santa Cruz, 1:100), fibroblast growth factor 10 (FGF10) (Santa Cruz, 1:100), platelet derived growth factor (PDGF) (Santa Cruz, 1:100). Under low-power magnification (100×), positive staining cells were screened and images of five representative fields were then captured under high-power magnification (400×) in Leica DMLA light microscope (Leica Microsystems, Wetzlar, Germany). The positive cell density of each core was counted by two independent investigators blind to clinical outcome and knowledge of the clinicopathological data. Data were expressed as mean value (± SE) of the triplicate cores taken from each patient.
Quantitative real-time reverse transcription polymerase chain reaction (QRT-PCR)
Total RNA was extracted from 50 mg biopsy using Trizol plus kit (TaKaRa, Japan). First-strand cDNA synthesis was done using Promega kit. Synthesized cDNA was used for QRT-PCR analysis using LightCycler (Roche, Switzerland) following the manufacturer’s instructions. TGF-β1, FGF10, and PDGF primers were specifically designed by Biosune Bio-Technology Co., Ltd (China). β-actin was used as the internal control. The Nucleotide sequences for primers: TGF-β1: 5'-CGACTACTACGCCAAGGAG-3', 5'-GAGAGCAACACGGGTTCAG-3'; FGF10: 5'-AGAGCGACCCTCACATCAAG-3', 5'-TCGTTTCAGTGCCACATACC-3'; PDGF: 5'-CCTGCCCATTCGGAGGAAGAG-3’, 5'-TTGGCCACCTTGACGCGCG-3'; β-actin: 5'-CCTGTACGCCAACACAGTG-3', 5'-ATACTCCTGCTTGCTGATCC-3'. Amplifications were carried out in the 20 µL reaction mixtures in the following conditions 95 °C for 2 min and followed by 40 cycles of 95 °C for 20 s, 55 °C for 30 s and 72 °C for 40 s, and then 72 °C for 5 min. The same experiment was repeated 3 times and similar results were obtained. The relative mRNA expression level was calculated and statistically analyzed using delta-delta-Ct method.
Microarray data analysis
Hierarchical clustering of 127 cytokines were performed using the software CLUSTER 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and displayed by the software Java TreeView (http://www.yiiframework.com/forum/index.php?/topic/9180-the-tree-view/). For the network analysis, all genes were uploaded into the STRING 9.0 database (Search Tool for the Retrieval of Interacting Genes) to analyze the Protein-Protein interaction (PPI). Based on the neighborhood, gene fusions, co-occurrence, co-expression, experiments, and literature mining, STRING database provides information on both experimental and predicted interactions. In this study, we constructed the PPI network based on confidence score of 0.4, which implied that all possible interactions with low level of confidence were extracted from the database and as many as possible were considered, and we used Cytoscape v2.8.3 software for visual analysis of the constructed networks.
Statistical analysis
All measurement data were expressed as mean ± SD, the difference among groups compared using ANOVA, enumeration data were analyzed by chi-square test. Kaplan-Meier method was employed to evaluate survival curve, and the log-rank test was used to compare survival time among groups. The test results were reported as 2-tailed P values, where P<0.05 was considered to be statistically significance.
Results
Identification of cytokines involved in the pathogenesis of INSIP
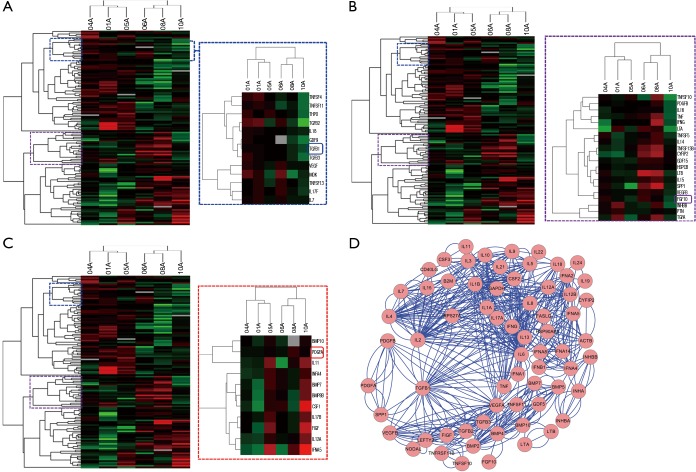
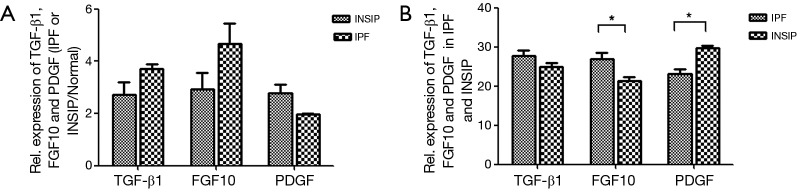
To identify cytokines involved in the pathogenesis of INSIP, with IPF as isotype control, Oligo GEArray that profiled 127 cytokines was employed. The analysis identified 109 cytokines as differentially expressed more than 2-fold with P value <0.05 between normal and lesion tissues. These included cytokines involved in regulation of cell cycle and proliferation, growth factor activity, and protein biosynthesis (Table S1). Specifically, TGF-β1, FGF10, and PDGF were dramatically up-regulated in patients with INSIP (Figure 1A,B,C), and were found closely related with pathogenesis of INSIP (Figure 1D). Similar phenotype was also found in patients with IPF. The quantitative analysis of these cytokines from GEArray is shown in Figure 2A. Interestingly, we found that the expression of TGF-β1 and FGF10 were higher in IPF than in INSIP, while PDGF was expressed at a higher level in patients with INSIP, though there have no statistical significance between INSIP and IPF groups.
Figure 1.
The hierarchical clustering and network analysis of cytokines microarray in IIPs patients. (A-C) The hierarchical clustering of 127 cytokines in patients were analyzed by cluster 3.0 and displayed by the software Java TreeView. Cytokines are color-coded according to their inheritance status (IPF: n=3, INSIP: n=3); (D) the network map of cytokines tightly involved in advancement of patients with IPF and INSIP. IIP, idiopathic interstitial pneumonia; IPF, idiopathic pulmonary fibrosis; INSIP, idiopathic nonspecific interstitial pneumonia.
Figure 2.
Relative expression of TGF-β1, FGF10, and PDGF in patients. (A) The quantification of microarray results, TGF-β1 and FGF10 were relatively higher in IPF, while PDGF was highly expressed in INSIP; (B) the expression of above genes in INSIP and IPF were confirmed by RT-PCR, and the experiment was independently repeated three times, *P<0.05. TGF-β1, transforming growth factor-beta-1; FGF10, fibroblast growth factor 10; PDGF, platelet derived growth factor; INSIP, idiopathic nonspecific interstitial pneumonia; IPF, idiopathic pulmonary fibrosis; RT-PCR, reverse transcription polymerase chain reaction.
TGF-β1, PDGF, and FGF10 expression were increased in INSIP patients
To confirm above results, we also performed RT-PCR to detect the expression of TGF-β1, PDGF, and FGF10 in patients with INSIP, including IPF. Consistence with results of Oligo GEArray, these genes was obviously increased in patient samples regardless of INSIP and IPF comparing the normal tissue (P<0.05). Their expressions were also individually analyzed in both INSIP and IPF patients. As demonstrated in Figure 2B, PDGF was relatively overexpressed in INSIP, whereas TGF-β1 and FGF10 were highly expressed in IPF (P<0.05), indicating the potential existence of a predominant expression axis of these cytokines in different subtypes of IIPs. Taken together, these results suggested that TGF-β1, FGF10, and PDGF were abnormally expressed in IIPs disease.
The expression and location of TGF-β1, PDGF, and FGF10 in INSIP by IHC detection
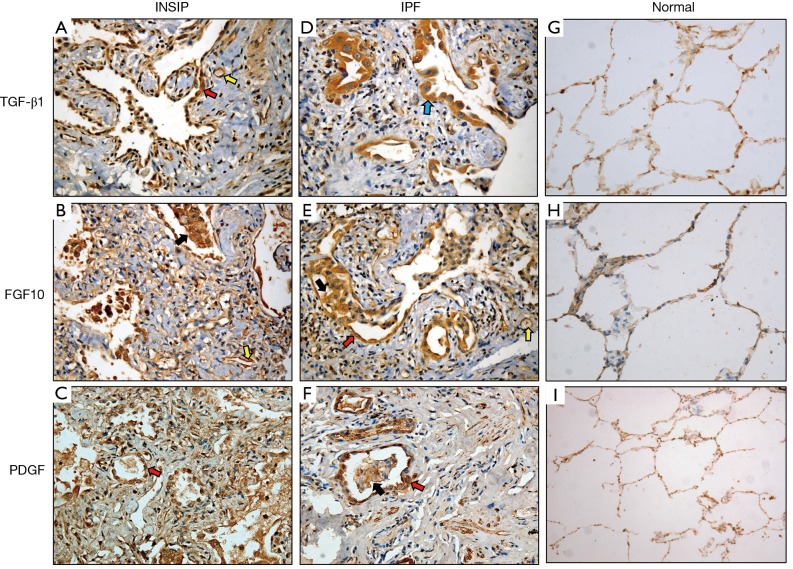
Since we observed dramatically increased expression of TGF-β1, PDGF, and FGF10 in both INSIP and IPF at the transcriptional level, we analyzed 22 cases of INSIP and 25 cases of IPF to determine the expression of these cytokines by immunohistochemical analysis. Comparing with normal lung tissues, TGF-β1, PDGF, and FGF10 were highly expressed in various type of cells in both INSIP and IPF (Figure 3A-I), including bronchial epithelial cells, alveolar epithelial cells, macrophage in alveolus and its mesenchyme, vascular endothelial cells, fibroblast (FB) and smooth muscle cells. Importantly, we observed that PDGF was more strongly expressed in patients with INSIP than patients with IPF (Figure 3C,F). Of note, the alveolar macrophages (AM) showed stronger expression of these cytokines than other type of cells. Consistently, through quantification of these cytokines, TGF-β1 and FGF10 were highly expressed in IPF than in INSIP, while PDGF was strongly expressed in INSIP (Table 2), which was consistent with the results of GEArray and RT-PCR. Based on these results, we hypothesized that there was likely a priority of the effect of TGF-β1, PDGF, and FGF10 in INSIP and IPF, and PDGF may be the predominant disease-promoting factor in INSIP, while TGF-β1 and FGF10 may be more important for progression of IPF, although they were all highly expressed in both IPF and INSIP.
Figure 3.
The expression of TGF-β1, FGF10, and PDGF in INSIP and IPF by immunohistochemistry detection. (A-C) TGF-β1, FGF10, and PDGF were strongly expressed in alveolar epithelial cells (red arrow), vascular endothelial cells (yellow arrow), alveolar macrophages (black arrow) of patients with INSIP (Envison ×200); (D-F) above cytokines also highly expressed in various type of cells in patients with IPF, including bronchial epithelial cells (blue arrow), alveolar epithelial cells (red arrow), vascular endothelial cells (yellow arrow), alveolar macrophages (black arrow) (Envison ×200); (G-I) the expression of TGF-β1, FGF10 and PDGF in normal lung tissues (Envison ×200). TGF-β1, transforming growth factor-beta-1; FGF10, fibroblast growth factor 10; PDGF, platelet derived growth factor; INSIP, idiopathic nonspecific interstitial pneumonia; IPF, idiopathic pulmonary fibrosis.
Table 2. The expression and location of cytokines in cells of INSIP and IPF patients ().
| Groups | Cases | TGF-β1 | PDGF | FGF10 |
|---|---|---|---|---|
| Bronchial epithelial cell | ||||
| INSIP group | 22 | 3.91±1.48*Δ1 | 5.41±1.65*Δ3 | 3.08±1.15*Δ5 |
| IPF group | 25 | 5.24±1.27Δ2 | 4.52±1.09Δ4 | 4.04±1.02Δ6 |
| Alveolar epithelial cell | ||||
| INSIP group | 22 | 3.86±1.39* | 5.32±1.67* | 3.16±1.14* |
| IPF group | 25 | 5.12±1.01 | 4.44±1.04 | 4.00±1.04 |
| Alveolar macrophage | ||||
| INSIP group | 22 | 4.32±1.13* | 6.68±1.99* | 4.52±1.12* |
| IPF group | 25 | 5.16±1.49 | 5.20±1.73 | 5.22±1.00 |
| Vascular endothelial cell | ||||
| INSIP group | 22 | 3.14±0.89* | 4.73±1.12* | 2.20±1.44* |
| IPF group | 25 | 4.44±1.23 | 4.04±0.68 | 3.00±1.21 |
| Fibroblast | ||||
| INSIP group | 22 | 2.68±0.89* | 4.77±1.07* | 2.12±1.83* |
| IPF group | 25 | 3.32±1.03 | 3.96±0.54 | 3.30±1.02 |
| Smooth muscle cell | ||||
| INSIP group | 22 | 1.41±1.05 | 4.41±1.05* | 1.04±0.61* |
| IPF group | 25 | 1.88±1.33 | 3.80±0.41 | 2.30±0.82 |
*, P<0.05, compared with IPF; Δ1, P=0.000, r=0.782; Δ2, P=0.000, r=0.885; Δ3, P=0.000, r=0.967; Δ4, P=0.000, r=0.930; Δ5, P=0.000, r=0.939; Δ6, P=0.000, r=0.980, compared with alveolar epithelial cell in original group. INSIP, idiopathic nonspecific interstitial pneumonia; IPF, idiopathic pulmonary fibrosis.
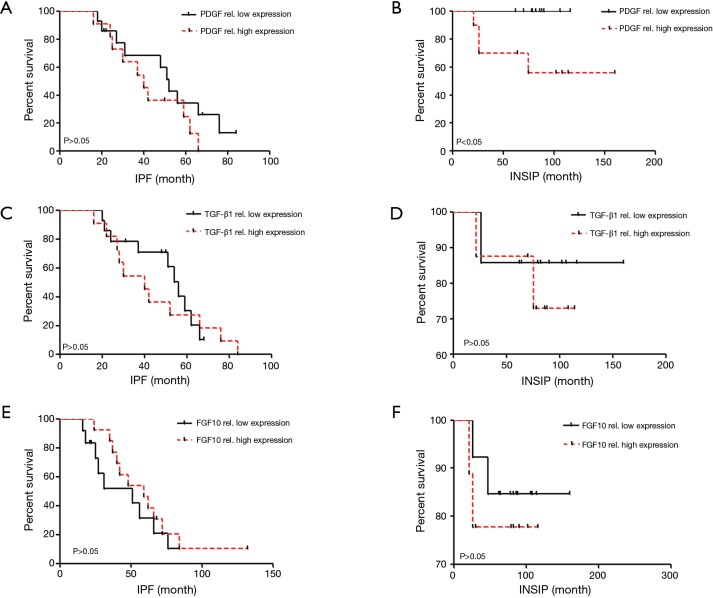
The relationship between PDGF and overall survival rate of patients with INSIP
Due to the relatively high expression of PDGF in patients with INSIP, we wonder whether its expression may be associated with the survival rate of patients. Additionally, the relationship between TGF-β1 and FGF10 expression with the patients’ survival rate were also studied. As mentioned in previously, each case had integrated clinical follow-up data of more than five years. Interestingly, we found that patients with above average PDGF expression survived better than those with PDGF expression below the average level. However its expression levels had no significant correlation with the survival rate of IPF patient (Figure 4A,B). Additionally, PDGF is not an independent prognostic factor of INSIP patients. Besides, our data also showed that TGF-β1 expression also had no significant correlation with patients’ survival rate in INSIP or IPF as well as FGF10 correlation analysis (Figure 4C,D,E,F). Taken together, our data suggests there is significant correlation ship between PDGF expression and survival rate of patients with INSIP.
Figure 4.
The correlation of TGF-β1, FGF10, and PDGF expression and the patients’ survival rate. (A,B) Lower expression of PDGF was closely associated with better survival rate of INSIP rather than IPF patients (*P<0.05); (C-F) the relationship between differential expression level of TGF-β1, FGF10 and the survival time of IPF and INSIP patients, there no statistical significance among these groups. TGF-β1, transforming growth factor-beta-1; FGF10, fibroblast growth factor 10; PDGF, platelet derived growth factor; IPF, idiopathic pulmonary fibrosis; INSIP, idiopathic nonspecific interstitial pneumonia.
Discussion
Our results demonstrated that IIPs, including IPF and INSIP, are associated with many abnormally-expressed cytokines. Oligo GEArray identified several cytokines which appeared to be important in the pathogenesis and advancement of INSIP and IPF. In further analysis, we found that TGF-β1, FGF10, and PDGF were dominantly up-regulated in patients with INSIP, as well as IPF. These results were also confirmed by RT-PCR and IHC. Interestingly, TGF-β1 and FGF10 were preferentially increased in IPF than that in INSIP, while PDGF was increasingly expressed in INSIP, indicating there was likely a priority-effect of these cytokines in the progression of IPF and INSIP. Importantly, we found that the negative correlationship between PDGF expression and overall survival rate of patients with INSIP.
Recently, high throughput technique was employed to screen potential therapeutic targets and biomarkers for IIPs. Kaminski et al. described global changes of gene expression in IPF by using the reductionist “cherry picking” and quantitative “systems” approach (6). Yang et al. evaluated transcriptional signatures between sporadic IIPs and familial IIPs, and CXCL12 was identified as a key regulator in the pathogenesis of the disease (8). Similarly, we screened the expression of 127 various cytokines in INSIP by Oligo GEArray, with IPF as isotype control. Expectedly, many cytokines were disorderly expressed in INSIP, including ILs, tumor necrosis factor, osteogenesis protein families and so forth. It is noteworthy that TGF-β1, FGF10, and PDGF were dominantly over-expressed in disease, suggesting they might be disease-drivers of INSIP and IPF. Several studies uncovered TGF-β1 as a well-known pro-fibrogenic factor (7,14-17), and pirfenidone or nintedanib could be used in IPF-involved dysfunction of TGF-β1 (18-21). But little is known about FGF10 and PDGF in the pathogenesis of pulmonary fibrosis and their therapeutic potential in INSIP and IPF, especially as studies have lacked human clinical relevance evidence that may identify the role of these cytokines in IIPs.
To clarify the clinical outcome of TGF-β1, FGF10, and PDGF in INSIP, we used tissue array and IHC to detect their expression in 22 cases of INSIP and 25 cases of IPF subjects. Similar to the report by Gu et al., which identified TGF-β1 and FGF as highly expressed in these two diseases, and localized in alveolar epithelial cells, AM and bronchial epithelium (17), we found that TGF-β1 and FGF10 were strongly expressed in these cells in our subjects. Interestingly, all these cytokines were predominantly expressed in AM, indicating AM are a major resource for the production of these cytokines. Lemaire et al. also found that AM isolated from lung fibrosis in rats induced by asbestos, releases a FGF which persisted over time (22). Importantly, we discovered that PDGF, which was more strongly expressed in INSIP. Intriguingly, some studies indicate that PDGF can promote the proliferation of fibroblasts (23). In bleomycin-induced mice, PDGF was significantly increased in murine pulmonary tissues (24), and target its expression could availably prevent the progress of fibrosis. In human studies, similar results show that imatinib, which specifically inhibits PDGF tyrosine kinase (25), could obviously improve the pulmonary function in IPF patients (26). Accordingly, our study also suggests that PDGF may a potential target for pulmonary fibrosis, especially for INSIP patients since its expression was negatively associated with the survival rate of patients with INSIP in our study. However, further analysis indicates that PDGF is not an independent prognostic factor for INSIP patients. This may due to the relatively small sample size. Thus the value of PDGF in the prognosis of INSIP need verified in more large samples in future studies.
In summary, our study investigated cytokine expression in INSIP and IPF subjects. To the best of our knowledge, most previous studies that have explored the potential therapeutic target or mechanisms of interstitial pneumonia have used cells or animal models, but few studied at the human pathologic level, especially for Asian population. Through analyzing gene expression profiling, we found that the expression of TGF-β1, FGF10, and PDGF were all increased in patients. Importantly, our results suggests that a potential priority effect exists among these cytokines in INSIP and IPF, whereby PDGF may be more important in the pathogenesis of INSIP, whereas TGF-β1 and FGF10 may be more critical for the advancement of IPF. Importantly, our findings suggest that lower expression of PDGF is associated with better overall survival rate of patients with INSIP.
Acknowledgements
Funding: This work was supported by the Science and Technology Commission Foundation of Key Medical Research Foundation of Shanghai, China (034119868; 09411951600), Key Medical Research Foundation of the Health Bureau of Shanghai, China (20134034) and National Nature Science Foundation of China (81570053).
We sincerely thank Yudong Zhang, Jun Gu for their skillful technique support.
Table S1. The expressed profile of cytokines involved in the pathogenesis of patients with IPF and INSIP.
| Symbol | Description | GO term | 1A: standard value/control standard value | 4A: standard value/control standard value | 6A: standard value/control standard value | 5A: standard value/control standard value | 8A: standard value/control standard value | 10A: standard value/control standard value |
|---|---|---|---|---|---|---|---|---|
| RPS27A | Ribosomal protein S27a | Intracellular; protein biosynthesis; structural constituent of ribosome; ribosome | 0.6984812 | 0.88565386 | 1.15274426 | 0.71415456 | 1.21272104 | 1.169643001 |
| AREG | Amphiregulin (schwannoma-derived growth factor) | Integral to membrane; cell proliferation; extracellular space; growth factor activity; cell-cell signaling; cytokine activity | 0.36046713 | 0.54934792 | 1.94266824 | 0.13030534 | 0.60418898 | 1.82350768 |
| ARS | ARS component B | Extracellular; cytokine activity; biological process unknown | 1.08435248 | 1.51541266 | 0.88175234 | 0 | 0.62953952 | 0.915287115 |
| ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | ATP binding; hydrolase activity; transporter activity; hydrogen-transporting ATP synthase activity, rotational mechanism; hydrogen-transporting ATPase activity, rotational mechanism; ATP synthesis coupled proton transport; proton-transporting two-sector ATPase complex; proton transport | 0.57892632 | 0.81518827 | 1.20603347 | 0.21065414 | 1.18184632 | 0.870057203 |
| BMP1 | Bone morphogene tic protein 1 | Calcium ion binding; extracellular; proteolysis and peptidolysis; zinc ion binding; development; growth factor activity; cytokine activity; metallopeptidase activity; cartilage condensation; astacin activity; procollagen C-endopeptidase activity | 0.44907116 | 1.53291465 | 2.36971787 | 1.49313203 | 1.77526707 | 1.941768055 |
| BMP10 | Bone morphogene tic protein 10 | Extracellular; cell growth and/or maintenance; growth factor activity; cell-cell signaling; regulation of cell proliferation; cytokine activity; embryonic development; growth; cardioblast differentiation; embryonic heart tube development | 0.88372109 | 1.25930558 | 0.8686157 | 1.55617516 | 0 | 1.551193053 |
| BMP15 | Bone morphogene tic protein 15 | Extracellular; cell growth and/or maintenance; growth factor activity; cytokine activity; female gamete generation | 1.63601749 | 1.70915606 | 1.30182138 | 1.87381476 | 0.65265595 | 2.545671034 |
| BMP2 | Bone morphogene tic protein 2 | Extracellular; cell growth and/or maintenance; growth factor activity; skeletal development; cell-cell signaling; cytokine activity; growth | 0.41148814 | 0.53024484 | 1.28156685 | 0.67825423 | 0.89965975 | 1.225958449 |
| BMP3 | Bone morphogene tic protein 3 (osteogenic) | Growth factor activity; skeletal development; cell-cell signaling; cytokine activity | 0.4218214 | 0.80764214 | 0.67738821 | 0.58399011 | 0.8947683 | 0.926961445 |
| BMP4 | Bone morphogene tic protein 4 | Mesoderm development; signal transducer activity; growth factor activity; skeletal development; cytokine activity; growth | 0.68117675 | 0.94967571 | 1.35208101 | 0.40737043 | 0.37587385 | 1.032840747 |
| BMP5 | Bone morphogene tic protein 5 | Cellular component unknown; growth factor activity; skeletal development; cytokine activity; growth | 0.80459129 | 0.88061403 | 0.84268902 | 0.63724704 | 3.87536045 | 1.74188967 |
| BMP6 | Bone morphogene tic protein 6 | Growth factor activity; skeletal development; cytokine activity; growth | 0.4338411 | 0.45341569 | 1.28857593 | 0.4882698 | 1.85010961 | 1.305614283 |
| BMP7 | Bone morphogene tic protein 7 (osteogenic protein 1) | Growth factor activity; skeletal development; cytokine activity; growth | 0.60980249 | 1.16194954 | 1.70558442 | 2.43253239 | 1.17500204 | 3.415602872 |
| BMP8B | Bone morphogene tic protein 8b (osteogenic protein 2) | Extracellular; cell growth and/or maintenance; growth factor activity; skeletal development; cytokine activity; growth | 0.44606252 | 1.00370687 | 1.26870185 | 1.57379029 | 0.63197258 | 2.800244206 |
| CSF1 | Colony stimulating factor 1 (macrophage) | Integral to membrane; cell proliferation; positive regulation of cell proliferation; cell differentiation; hematopoiesis; macrophage differentiation; macrophage colony stimulating factor receptor binding | 0.39762743 | 0.85268102 | 1.69268968 | 1.62542045 | 0.62966387 | 7.966210029 |
| CSF2 | Colony stimulating factor 2 (granulocyte-macrophage) | Cell surface receptor linked signal transduction; development; extracellular space; cytokine activity; cellular defense response; granulocyte macrophage colony-stimulating factor receptor binding | 0.86439825 | 2.1039088 | 1.56077784 | 2.30494703 | 3.91822998 | 4.675454723 |
| CSF3 | Colony stimulating factor 3 (granulocyte) | Extracellular; immune response; positive regulation of cell proliferation; cell surface receptor linked signal transduction; development; extracellular space; cell-cell signaling; cytokine activity; interleukin-6 receptor binding; cellular defense response; granulocyte colony-stimulating factor receptor binding | 0.1869539 | 0.40499611 | 0.40866589 | 0.43818644 | 0.56068614 | 0.823405602 |
| CYFIP2 | Cytoplasmic FMR1 interacting protein 2 | Extracellular; immune response; chemokine activity | 0.21420331 | 0.25539393 | 1.30377059 | 0.44353184 | 1.90888968 | 0.527755359 |
| EBAF | Endometrial bleeding associated factor (left-right determination, factor A; transforming growth factor beta superfamily) | Growth factor activity; cell-cell signaling; cytokine activity; transforming growth factor beta receptor signaling pathway; transforming growth factor beta receptor binding; cell growth; oocyte axis determination | 1.21764497 | 0 | 1.15646718 | 2.06761498 | 0.81054292 | 0.924354838 |
| FAM3B | Family with sequence similarity 3, member B | Extracellular; cytokine activity; insulin secretion | 0.78531381 | 0.65905041 | 2.229818 | 1.28087945 | 4.95090772 | 4.701887783 |
| FAM3C | Family with sequence similarity 3, member C | Extracellular; cytokine activity; biological process unknown | 0.49964231 | 0.89692954 | 3.18000379 | 1.77520073 | 6.25169364 | 3.125564115 |
| FGF10 | Fibroblast growth factor 10 | Signal transduction; nucleus; regulation of cell cycle; extracellular space; heparin binding; organogenesis; growth factor activity; cell-cell signaling; response to stress; regulation oftranscription; fibroblast growth factor receptor signaling pathway; cell surface; protein-nucleus import, translocation; fibro blast growth factor receptor binding; embryonic limb morphogenesis; positive regulation of receptor mediated endocytosis; positive regulation of urothelial cell proliferation; urothelial cellproliferation | 3.19600254 | 4.89922147 | 5.88637612 | 2.51792617 | 4.16436653 | 2.088285455 |
| FIGF | C-fos induced growth factor (vascular endothelial growth factor D) | Cell proliferation; regulation of cell cycle; positive regulation of cell proliferation; membrane; extracellular space; growth factor activity; platelet-derived growth factor receptor binding; angiogenesis | 0.34008861 | 0.73039865 | 0.6914384 | 1.90344681 | 0.57572558 | 3.60552752 |
| FLT3LG | Fms-related tyrosine kinase 3 ligand | Integral to membrane; signal transduction; positive regulation of cell proliferation; soluble fraction; cytokine activity | 0.14434829 | 0.93506638 | 1.40548726 | 1.0582324 | 1.66671187 | 1.121110141 |
| GDF1 | Growth differentiation factor 1 | Extracellular; cell growth and/or maintenance; growth factor activity; cell differentiation; cytokine activity | 0.71304012 | 1.20313207 | 0.13961967 | 1.73922252 | 0.58195599 | 0.568924283 |
| GDF10 | Growth differentiation factor 10 | Growth factor activity; skeletal development; cytokine activity; transforming growth factor beta receptor signaling pathway | 0.19112295 | 0.4487812 | 0.51587711 | 0.59738119 | 1.10692823 | 1.029270193 |
| GDF11 | Growth differentiation factor 11 | Cellular component unknown; mesoderm development; growth factor activity; skeletal development; neurogenesis; cytokine activity; growth | 0.49771627 | 0.92771365 | 1.70170279 | 1.02288945 | 0.75853251 | 0.946061695 |
| GDF15 | Growth differentiation factor 15 | Signal transduction; extracellular; growth factor activity; cell-cell signaling; cytokine activity; transforming growth factor beta receptor signaling pathway | 1.16324212 | 1.25200137 | 2.17175906 | 1.03014713 | 2.08572453 | 1.630074383 |
| GDF2 | Growth differentiation factor 2 | Extracellular; cell growth and/or maintenance; growth factor activity; cytokine activity; growth | 0.67075334 | 1.78379253 | 1.15515494 | 1.24631474 | 0.9995993 | 1.320697065 |
| GDF3 | Growth differentiation factor 3 | Extracellular; cell growth and/or maintenance; growth factor activity; cytokine activity | 0.42614976 | 1.46617164 | 5.84659285 | 1.18540565 | 2.58311126 | 12.87213207 |
| GDF5 | Growth differentiation factor 5 (cartilage-derived morphogene tic protein-1) | Protein binding; growth factor activity; cell-cell signaling; cytokine activity; transforming growth factor beta receptor signaling pathway; growth | 0.09844814 | 0.76816607 | 1.50746376 | 1.1620905 | 3.08452196 | 3.312303835 |
| GDF8 | Growth differentiation factor 8 | Growth factor activity; muscle development; cytokine activity; transforming growth factor beta receptor signaling pathway; growth | 0 | 0 | 0 | 0 | 0 | 0 |
| GDF9 | Growth differentiation factor 9 | Extracellular; cell growth and/or maintenance; growth factor activity; cytokine activity; transforming growth factor beta receptor signaling pathway; female gamete generation | 1.5551875 | 1.88818857 | 0 | 1.51274806 | 0.93594492 | 0.554337305 |
| IFNA1 | Interferon, alpha 1 | – | 1.54500454 | 1.16844903 | 0.59446116 | 1.71518551 | 1.27340387 | 0.919061148 |
| IFNA13 | Interferon, alpha 13 | Extracellular; defense response; interferon-alpha/beta receptor binding | 1.82956475 | 1.08339289 | 1.27299482 | 1.85809945 | 0.72289898 | 1.125587879 |
| IFNA14 | Interferon, alpha 14 | Extracellular; defense response; hematopoietin/interferon-class (D200-domain) cytokine receptor binding | 1.18464548 | 0.91799836 | 1.20976248 | 1.22168442 | 1.50826849 | 1.573881337 |
| IFNA2 | Interferon, alpha 2 | Extracellular; induct ion of apoptosis; inflammatory response; cell surface receptor linked signal transduction; cell-cell signaling; defense response; interferon-alpha/beta receptor binding | 1.88261574 | 1.7431851 | 1.38292725 | 1.92205808 | 0.96346873 | 1.423555669 |
| IFNA4 | Interferon, alpha 4 | Extracellular; response to virus; cell-cell signaling; defense response; interferon-alpha/beta receptor binding | 1.17361593 | 1.06252523 | 1.92194785 | 2.76220278 | 1.33646694 | 2.699104604 |
| IFNA5 | Interferon, alpha 5 | Extracellular; defense response; hematopoietin/interferon-class (D200-domain) cytokine receptor binding | 1.23818065 | 1.65148932 | 1.27750868 | 14.6657583 | 6.37817411 | 33.94323995 |
| IFNA8 | Interferon, alpha 8 | Extracellular; defense response; hematopoietin/interferon-class (D200-domain) cytokine receptor binding | 0.114951 | 0.58061597 | 0.74191419 | 0.50904098 | 0.76927753 | 0.482888981 |
| IFNB1 | Interferon, beta 1, fibroblast | Extracellular; negative regulation of cell proliferation; cell surface receptor linked signal transduction; response to virus; Caspase activation; B-cell proliferation; defense response; natural killer cell activation; positive regulation of innate immune response; interferon-alpha/beta receptor binding; anti-inflammatory response; negative regulation of virion penetration; regulation of MHC class I biosynthesis | 1.77661581 | 0.70740493 | 1.18248114 | 1.72889506 | 1.12276541 | 0.984607959 |
| IFNG | Interferon, gamma | Regulation of cell growth; extracellular; immune response; cell surface receptor linked signal transduction; cell motility; cell-cell signaling; cytokine activity; interferon-gamma receptor binding | 1.7167365 | 0.67814498 | 1.88857383 | 1.93853225 | 7.61475888 | 0.973028806 |
| IFNK | Interferon, kappa | Extracellular; negative regulation of cell proliferation; response to virus; defense response; regulation of transcription; cytokine and chemokine mediated signaling pathway; natural killer cell activation; cellular defense response (sensu vertebrata); positive regulation of innate immune response; Interferon-alpha/beta receptor binding | 1.31719373 | 1.01248042 | 0.53252278 | 0.92103371 | 0.58158402 | 1.143468571 |
| IFNW1 | Interferon, omega 1 | Extracellular; response to virus; cell cycle arrest; defense response; interferon-alpha/beta receptor binding | 1.42434696 | 1.21945239 | 1.18832803 | 1.71327051 | 0.58954076 | 1.049929566 |
| IK | IK cytokine, down-regulator of HLA II | Immune response; extracellular space; soluble fraction; cell-cell signaling; cytokine activity | 1.64646668 | 1.8741568 | 1.06467638 | 1.10080838 | 0.38592034 | 0.599044565 |
| IL10 | Interleukin 10 | Extracellular; Immune response; anti-apoptosis; cell-cell signaling; B-cell proliferation; cytokine activity; B-cell differentiation; T-helper 2 type immune response; regulation of isotype switching; interleukin-10 receptor binding; cytoplasmic sequestering of NF-kappaB; hematopoiesis; immune cell chemotaxis; negative regulation of MHC class ii biosynthesis; negative regulation of t-cell proliferation; negative regulation of interferon-alpha biosynthesis; negative regulation of interferon-gamma biosynthesis; negative regulation of nitric oxide biosynthesis | 0.63884315 | 1.6675018 | 0.53983092 | 0.63422976 | 0.95138832 | 0.453962045 |
| IL11 | Interleukin 11 | Extracellular; positive regulation of cell proliferation; cell-cell signaling; plateletactivation; cytokine activity; B-cell differentiation; interleukin-11 receptor binding; adipocyte differentiation; megakaryocyte differentiation | 1.96323506 | 1.85343374 | 0.73993446 | 6.74102002 | 3.14383543 | 7.594803581 |
| IL12A | Interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) | – | 1.24653313 | 1.34306261 | 1.84622945 | 2.99207674 | 2.21844084 | 5.456309813 |
| IL12B | Interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) | Signal transducer activity; membrane; extracellular space; antimicrobial humoral response (sensu vertebrata); positive regulation of interferon-gamma biosynthesis; cytokine activity; hematopoietin/interferon-class (D200-domain) cytokine receptor activity; interleukin-12 receptor binding; T-helper cell differentiation; interferon-gamma biosynthesis; natural killer cell activation; positive regulation of activated T-cell proliferation; regulation of cytokine biosynthesis | 1.70500651 | 1.06834274 | 0.95909508 | 3.1452199 | 0.82850509 | 0.876244301 |
| IL13 | Interleukin 13 | Cell proliferation; signal transduction; extracellular; immune response; inflammatory response; signal transducer activity; soluble fraction; cell motility; antimicrobial humoral response (sensu vertebrata); cell-cell signaling; chemokine activity; interleukin-13 receptor binding | 1.21004911 | 1.62433632 | 0.72171694 | 0.75596692 | 0.66550881 | 0.642189933 |
| IL14 | Taxilin | Cell proliferation; extracellular; high molecular weight B-cell growth factor receptor binding | 0.52526398 | 0.88536095 | 1.06673542 | 0.71710052 | 1.42119473 | 0.658870839 |
| IL15 | Interleukin 15 | Golgi apparatus; cell proliferation; signal transduction; immune response; positive regulation of cell proliferation; membrane fraction; signal transducer activity; integral to plasmamembrane; extracellular space; endosome; cell-cell signaling; hematopoietin/interferon-class (D200-domain) cytokine receptor binding | 0.70213539 | 0.82335532 | 1.10137384 | 0.6390786 | 1.47449891 | 0.625214574 |
| IL16 | Interleukin 16 (lymphocyte chemoattractant factor) | Immune response; protein binding; extracellular space; chemotaxis; cytokine activity | 1.58139564 | 1.24719248 | 1.82767665 | 1.73220471 | 1.88149791 | 0.803103305 |
| IL17 | Interleukin 17 (cytotoxic T-lymphocyte-associated serine esterase 8) | Immune response; apoptosis; extracellular space; cell death; cell-cell signaling; cytokine activity; protein amino acid glycosylation | 9.80067608 | 7.4405083 | 1.39902053 | 5.41101957 | 1.64419825 | 3.46121765 |
| IL17B | Interleukin 17B | Immune response; signal transducer activity; cell-cell signaling; cytokine activity | 1.15724256 | 1.33395919 | 1.42822417 | 2.55111625 | 1.08124331 | 3.214249272 |
| IL17C | Interleukin 17C | Inflammatory response; cell surface receptor linked signal transduction; extracellular space; soluble fraction; cell-cell signaling; cytokine activity | 1.73479304 | 3.92625774 | 1.46620866 | 1.39194436 | 0.58171142 | 1.654175954 |
| IL17E | Interleukin 17E | Membrane; cytokine activity; interleukin-17E receptor binding; biological process unknown | 0.82060846 | 0.76920313 | 1.22449616 | 1.68156722 | 0.20627228 | 0 |
| IL17F | Interleukin 17F | Extracellular; cytokine activity; negative regulation of angiogenesis; cytokine biosynthesis; proteoglycan metabolism | 1.76371006 | 1.32606734 | 1.93859107 | 1.24426497 | 1.16497143 | 0.810622138 |
| IL18 | Interleukin 18 (interferon-gamma-inducing factor) | Extracellular; immune response; induction of apoptosis via death domain receptors; signal transducer activity; interleukin-1 receptor binding; cell-cell signaling; angiogenesis; interferon-gamma biosynthesis; positive regulation of activated T-cell proliferation; T-helper 2 type immune response; chemokine biosynthesis; granulocyte macrophage colony-stimulating factor biosynthesis; Interleukin-13 biosynthesis; Interleukin-2 biosynthesis; regulation of cell adhesion; sleep | 1.04899764 | 1.11062992 | 0.95779984 | 0.8448451 | 0.77969744 | 0.662700197 |
| IL19 | Interleukin 19 | Signal transduction; extracellular; immune response; cytokine activity; actin binding | 3.01819304 | 1.93573526 | 1.63277351 | 1.63144336 | 0.87518143 | 1.82184604 |
| IL1A | Interleukin 1, alpha | Cell proliferation; immune response; cytoplasm; apoptosis; anti-apoptosis; negative regulation of cell proliferation; regulation of cell cycle; inflammatory response; signal transducer activity; extracellular space; interleukin-1 receptor binding; cell-cell signaling; chemotaxis | 0.92241657 | 2.3689492 | 1.7958075 | 1.79342661 | 1.68340863 | 1.482172667 |
| IL1B | Interleukin 1, beta | Cell proliferation; signal transduction; immune response; apoptosis; negative regulation of cell proliferation; regulation of cell cycle; inflammatory response; signal transducer activity; extracellular space; interleukin-1 receptor binding; antimicrobial humoral response (sensu vertebrata); cell-cell signaling | 1.69998232 | 1.59534836 | 2.28855983 | 1.25859418 | 11.2163116 | 1.721077691 |
| IL1F10 | Interleukin 1 family, member 10 (theta) | Extracellular; immune response; interleukin-1 receptor antagonist activity | 1.64501992 | 6.15108437 | 1.53726768 | 4.18960317 | 1.67940221 | 1.643218455 |
| IL1F5 | Interleukin 1 family, member 5 (delta) | Extracellular; immune response; interleukin-1 receptor antagonist activity | 1.49668861 | 7.90464734 | 1.45959404 | 3.85410687 | 1.31685159 | 4.478736387 |
| IL1F6 | Interleukin 1 family, member 6 | Extracellular; immune response; inflammatory response; interleukin-1 receptor binding | 1.35784015 | 1.39803102 | 0.70685224 | 1.81789403 | 0.22921418 | 0.926796096 |
| IL1F7 | Interleukin 1 family, member 7 (zeta) | Extracellular; immune response; interleukin-1 receptor antagonist activity | 1.52926815 | 1.70695734 | 1.27934853 | 1.20834604 | 0.31098844 | 1.094279548 |
| IL1F8 | Interleukin 1 family, member 8 (eta) | Extracellular; immune response; interleukin-1 receptor binding | 7.63819507 | 3.66816028 | 1.75098682 | 2.81217976 | 0.68113273 | 4.543417181 |
| IL1F9 | Interleukin 1 family, member 9 | Extracellular; immune response; cell-cell signaling; interleukin-1 receptor antagonist activity; response to pest, pathogen or parasite | 10.6074733 | 0.91372598 | 2.00002979 | 4.22983771 | 1.52435628 | 1.226817076 |
| IL2 | Interleukin 2 | – | 7.68503718 | 0.77761967 | 1.10690276 | 1.77467021 | 0.83628448 | 1.282571949 |
| IL20 | Interleukin 20 | Extracellular; immune response; interleukin-20 receptor binding; positive regulation of epidermal cell differentiation; positive regulation of keratinocyte differentiation; positive regulation of tyrosine phosphorylation of Stat3 protein; regulation of inflammatory response | 8.13303416 | 0.60438263 | 0.66200638 | 2.12157085 | 1.04528241 | 1.454492023 |
| IL21 | Interleukin 21 | Signal transduction; Nucleus; cytokine activity; lymph gland development | 3.93130568 | 1.93819257 | 0.50450821 | 1.88498697 | 0.28569141 | 1.256471497 |
| IL22 | Interleukin 22 | Extracellular; inflammatory response; acute-phase response; cell-cell signaling; interleukin-22 receptor binding | 2.24882983 | 1.00427292 | 0.82387008 | 2.06062241 | 0.70705756 | 1.627601657 |
| IL24 | Interleukin 24 | Extracellular; immune response; apoptosis; cytokine activity | 0.91127363 | 0.8669872 | 0.94603635 | 1.89331206 | 0.60682735 | 1.33606596 |
| IL26 | Interleukin 26 | Immune response; extracellular space; soluble fraction; antimicrobial humoral response (sensu vertebrata); cell-cell signaling; cytokine activity | 1.59430708 | 1.88389603 | 0.75213864 | 1.71489213 | 0.50874937 | 0.402468096 |
| IL3 | Interleukin 3 (colony-stimulating factor, multiple) | Cell proliferation; extracellular; immune response; positive regulation of cell proliferation; cell-cell signaling; neurogenesis; cytokine activity; interleukin-3 receptor binding | 1.31838154 | 1.96864535 | 1.67101865 | 1.69908821 | 0.59010334 | 2.797392551 |
| IL4 | Interleukin 4 | Cell proliferation; extracellular space; chemotaxis; interleukin-4 receptor binding; B-cell differentiation; T-helper 2 type immune response; cellular defense response; cholesterol metabolism; connective tissue growth factor biosynthesis; regulation of isotype switching | 9.63679381 | 1.16890173 | 0.81613764 | 3.7220775 | 2.20043825 | 1.565717883 |
| IL5 | Interleukin 5 (colony-stimulating factor, eosinophil) | Extracellular; immune response; inflammatory response; positive regulation of cell proliferation; cytokine activity; interleukin-5 receptor binding; hypersensitive response | 7.93629877 | 1.49643167 | 1.30074841 | 3.63138859 | 1.43450147 | 1.559777562 |
| IL6 | Interleukin 6 (interferon, beta 2) | Humoral immune response; negative regulation of cell proliferation; positive regulation of cell proliferation; cell surface receptor linked signal transduction; extracellular space; acute-phase response; cell-cell signaling; cytokine activity; interleukin-6 receptor binding | 3.4689024 | 1.01811895 | 3.96001262 | 1.05246174 | 2.00258984 | 4.185069361 |
| IL7 | Interleukin 7 | Extracellular; humoral immune response; positive regulation of cell proliferation; organogenesis; cell-cell signaling; interleukin-7 receptor binding | 2.27361139 | 1.96237312 | 2.19677239 | 1.5142724 | 1.44988391 | 1.418583441 |
| IL8 | Interleukin 8 | Immune response; intracellular signaling cascade; negative regulation of cell proliferation; G-protein coupled receptor protein signaling pathway; cell cycle arrest; extracellular space; soluble fraction; cell motility; cell-cell signaling; chemokine activity; chemotaxis; angiogenesis; regulation of cell adhesion; neutrophil chemotaxis; calcium-mediated signaling; interleukin-8 receptor binding; induction of positive chemotaxis; neutrophil activation; regulation of retroviral genome replication | 0.1335222 | 0.35942406 | 0.13590094 | 0.23689785 | 1.83093334 | 1.03244623 |
| IL9 | Interleukin 9 | Extracellular; immune response; inflammatory response; positive regulation of cell proliferation; cell-cell signaling; interleukin-9 receptor binding | 1.19731974 | 1.23996092 | 0.43462676 | 1.55514456 | 0.40694034 | 0.946526229 |
| INHA | Inhibin, alpha | Extracellular; protein binding; cell growth and/or maintenance; negative regulation of cell cycle; induction of apoptosis; cell surface receptor linked signal transduction; cell cycle arrest; growth factor activity; hormone activity; skeletal development; cell-cell signaling; cell differentiation; neurogenesis; cytokine activity; negative regulation of interferon-gamma biosynthesis; activin inhibitor activity; erythrocyte differentiation; hemoglobin biosynthesis; negative regulation of B-cell differentiation; negative regulation of follicle-stimulating hormone secretion; negative regulation of macrophage differentiation; negative regulation of phosphorylation; ovarian follicle development; positive | 1.32746285 | 1.2769295 | 0.88358932 | 1.04287598 | 0.67358137 | 0.730762637 |
| INHBA | Inhibin, beta A (activin A, activin AB alpha polypeptide) | Extracellular; protein binding; cell growth and/or maintenance; negative regulation of cell cycle; induction of apoptosis; mesoderm development; cell surface receptor linked signal transduction; cell cycle arrest; growth factor activity; hormone activity; skeletal development; cell-cell signaling; cell differentiation; Neurogenesis; defense response; cytokine activity; transforming growth factor beta receptor binding; negative regulation of interferon-gamma biosynthesis; growth; activin inhibitor activity; erythrocyte differentiation; hemoglobin biosynthesis; negative regulation of b-cell differentiation; negative regulation of follicle-stimulating hormone secretion; negative | 1.08073029 | 1.87227915 | 1.17236683 | 1.69650078 | 2.13238195 | 3.614676832 |
| INHBB | Inhibin, beta B (activin AB beta polypeptide) | Extracellular; protein binding; cell growth and/or maintenance; growth factor activity; hormone activity; cell-cell signaling; cell differentiation; defense response; cytokine activity; protein homodimerization activity; growth; negative regulation of follicle-stimulating hormone secretion; ovarian follicle development; positive regulation of follicle-stimulating hormone secretion; response to external stimulus; host cell surface receptor binding; negative regulation of hepatocyte growth factor biosynthesis | 3.59633698 | 1.68064664 | 1.09103593 | 1.25698631 | 2.60211969 | 0.677419361 |
| LTA | Lymphotoxin alpha (TNF superfamily, member 1) | Signal transduction; immune response; induction of apoptosis; membrane; cell-cell signaling; tumor necrosis factor receptor binding | 6.03872643 | 0.92303735 | 6.50682307 | 2.52759587 | 5.2510886 | 1.149333892 |
| LTB | Lymphotoxin beta (TNF superfamily, member 3) | Integral to membrane; signal transduction; immune response; cell-cell signaling; tumor necrosis factor receptor binding | 1.0086348 | 1.792797 | 6.96505111 | 1.09358159 | 5.41109041 | 0.882881616 |
| MDK | Midkine (neurite growth-promoting factor 2) | Cell proliferation; signal transduction; regulation of cell cycle; extracellular space; heparin binding; growth factor activity; cell-cell signaling; cell differentiation; neurogenesis; cytokine activity | 2.94380443 | 4.66579429 | 5.27988906 | 1.2295348 | 0.82412643 | 1.679094814 |
| NODAL | Nodal homolog (mouse) | Development; growth factor activity; cytokine activity | 4.9657434 | 1.81908917 | 1.40132893 | 1.7936727 | 0.72246576 | 1.594742743 |
| PDGFA | Platelet-derived growth factor alpha polypeptide | Cell proliferation; regulation of cell cycle; cell surface receptor linked signal transduction; membrane; extracellular space; growth factor activity; cell-cell signaling; platelet-derived growth factor receptor binding | 1.9297869 | 2.02531946 | 1.99717728 | 2.48247569 | 3.45445054 | 2.414740302 |
| PDGFB | Platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | Cell proliferation; extracellular; cell growth and/or maintenance; regulation of cell cycle; membrane; response to wounding; growth factor activity; platelet-derived growth factor receptor binding | 0.83853903 | 0.97079553 | 0.95900676 | 0.85307547 | 1.31218767 | 0.441073007 |
| PTN | Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) | Cell proliferation; regulation of cell cycle; positive regulation of cell proliferation; extracellular space; heparin binding; growth factor activity; neurogenesis; cytokine activity; protein phosphatase inhibitor activity; transmembrane receptor protein tyrosine phosphatase signaling pathway | 1.5980138 | 1.24542208 | 0.87483804 | 0.91393046 | 1.16807059 | 0.985116858 |
| SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein, early T-lymphocyte activation 1) | Protein binding; anti-apoptosis; cell-matrix adhesion; extracellular matrix; growth factor activity; cell-cell signaling; cytokine activity; immune cell chemotaxis; integrin binding; positive regulation of T-cell proliferation; T-helper 1 type immune response; ossification; negative regulation of bone mineralization; regulation of myeloid blood cell differentiation; induction of positive chemotaxis; extracellular matrix (sensu Metazoa) | 1.87831749 | 3.00932904 | 5.74522182 | 0.62071966 | 12.4820223 | 1.80158236 |
| TGFA | Transforming growth factor, alpha | Cell proliferation; protein binding; protein-tyrosine kinase activity; regulation of cell cycle; signal transducer activity; integral to plasma membrane; extracellular space; soluble fraction; growth factor activity; cell-cell signaling; epidermal growth factor receptor activating ligand activity | 4.31496433 | 1.40547436 | 2.29988862 | 0.83922367 | 5.21695189 | 1.199012061 |
| TGFB1 | Transforming growth factor, beta 1 (Camurati-Engelmann disease) | Cell proliferation; anti-apoptosis; regulation of cell cycle; growth factor activity; cell-cell signaling; transforming growth factor beta receptor signaling pathway; transforming growth factor beta receptor binding; cell growth | 3.87456127 | 3.87868445 | 3.38612691 | 3.62205385 | 2.5987023 | 1.925692836 |
| TGFB2 | Transforming growth factor, beta 2 | 6.74713211 | 5.89042355 | 4.58762198 | 1.92222283 | 1.89256555 | 0.568599573 | |
| TGFB3 | Transforming growth factor, beta 3 | Cell proliferation; signal transduction; regulation of cell cycle; organogenesis; growth factor activity; cell-cell signaling; transforming growth factor beta receptor binding; cell growth | 5.09166362 | 6.78416947 | 4.71856209 | 4.15792631 | 3.33086612 | 1.843379256 |
| THPO | Thrombopoietin (myeloproliferative leukemia virus oncogene ligand, megakaryocyte growth and development factor) | Cell proliferation; extracellular; development; growth factor activity; hormone activity; cytokine activity; | 1.96988678 | 2.80676172 | 1.73069457 | 2.75051882 | 1.63698552 | 0.789714774 |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | Integral to membrane; signal transduction; immune response; regulation of transcription, DNA-dependent; apoptosis; anti-apoptosis; inflammatory response; response to virus; soluble fraction; cell-cell signaling; tumor necrosis factor receptor binding; leukocyte cell adhesion; necrosis | 1.40274268 | 1.33969398 | 2.29927615 | 2.06736798 | 4.45361783 | 0.795004677 |
| TNFRSF11 B | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | Signal transduction; extracellular; protein binding; apoptosis; receptor activity; skeletal development; cytokine activity | 2.43667195 | 1.07216513 | 0.51648813 | 1.81762159 | 1.09420177 | 0.76648775 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | Signal transduction; immune response; induction of apoptosis; apoptosis; positive regulation of I-kappaB kinase/NF-kappaB cascade; integral to plasma membrane; soluble fraction; cell-cell signaling; tumor necrosis factor receptor binding | 0.29859957 | 0.40336945 | 0.17101673 | 0.47425768 | 0.94754173 | 0.107686114 |
| TNFSF11 | Tumor necrosis factor (ligand) superfamily, member 11 | Extracellular; immune response; receptor activity; integral to plasma membrane; cell differentiation; tum or necrosis factor receptor binding; osteoclast differentiation | 1.84514703 | 1.58894407 | 0.76369915 | 1.55912449 | 1.15241823 | 0.537289446 |
| TNFSF12 | Tumor necrosis factor (ligand) superfamily, member 12 | Signal transduction; immune response; induction of apoptosis; apoptosis; integral to plasma membrane; angiogenesis; tumor necrosis factor receptor binding | 0.57298826 | 1.18267977 | 1.8772737 | 0.35582279 | 2.28919263 | 1.530630561 |
| TNFSF13 | Tumor necrosis factor (ligand) superfamily, member 13 | Signal transduction; immune response; positive regulation of cell proliferation; membrane; tumor necrosis factor receptor binding | 1.32075171 | 0.99920833 | 1.40025956 | 0.69674831 | 0.98571721 | 0.678497647 |
| TNFSF13B | Tumor necrosis factor (ligand) superfamily, member 13b | Cell proliferation; signal transduction; immune response; positive regulation of cell proliferation; integral to plasma membrane; soluble fraction; tumor necrosis factor receptor binding | 0.33414425 | 0.40715534 | 1.1565197 | 0.57007853 | 1.72446091 | 0.49062492 |
| TNFSF14 | Tumor necrosis factor (ligand) superfamily, member 14 | Integral to membrane; signal transduction; immune response; induction of apoptosis; tumor necrosis factor receptor binding; Caspase inhibitor activity; inhibition of Caspase activation; release of cytoplasmic sequestered NF-kappaB; T-cell proliferation; T-cell homeostasis | 1.56294653 | 1.72750559 | 0.74842096 | 2.28521965 | 1.01825948 | 0.973083683 |
| TNFSF15 | Tumor necrosis factor (ligand) superfamily, member 15 | Signal transduction; immune response; regulation of cell cycle; membrane; integral to plasma membrane; soluble fraction; tumor necrosis factor receptor binding | 1.66997676 | 1.44107317 | 0.90215988 | 1.84584106 | 0.93673721 | 1.080413403 |
| TNFSF18 | Tumor necrosis factor (ligand) superfamily, member 18 | Integral to membrane; signal transduction; immune response; anti-apoptosis; cell-cell signaling; tumor necrosis factor receptor binding | 1.82110465 | 1.26280989 | 0.56888294 | 1.59160549 | 0.76353241 | 0.610304864 |
| TNFSF4 | Tumor necrosis factor (ligand) superfamily, member 4 (tax-transcription ally activated glycoprotein 1, 34 kDa) | Signal transduction; immune response; positive regulation of cell proliferation; integral to plasma membrane; cell-cell signaling; tumor necrosis factor receptor binding | 0.85561258 | 0.69247961 | 0.36739997 | 0.43476557 | 0.49852032 | 0.189416647 |
| TNFSF5 | Tumor necrosis factor (ligand) superfamily, member 5 (hyper-IgM syndrome) | Signal transduction; immune response; anti-apoptosis; inflammatory response; integral to plasma membrane; soluble fraction; tumor necrosis factor receptor binding; CD40 receptor binding; B-cell proliferation; isotype switching; leukocyte cell adhesion; platelet activation | 0.56401054 | 1.26443741 | 1.05460831 | 0.86632133 | 1.70793484 | 0.636554156 |
| TNFSF6 | Tumor necrosis factor (ligand) superfamily, member 6 | Signal transduction; extracellular; immune response; induction of apoptosis; apoptosis; positive regulation of I-kappaB kinase/NF-kappaB cascade; integral to plasma membrane; cell-cell signaling; tumor necrosis factor receptor binding | 2.55725879 | 1.74148221 | 1.18322445 | 0.98551507 | 0.89388262 | 0.877481578 |
| TNFSF7 | Tumor necrosis factor (ligand) superfamily, member 7 | Cell proliferation; signal transduction; immune response; apoptosis; integral to plasma membrane; cell-cell signaling; tumor necrosis factor receptor binding | 1.79076183 | 1.86714014 | 1.11393837 | 0.96722807 | 0.66632963 | 1.07371487 |
| TNFSF8 | Tumor necrosis factor (ligand) superfamily, member 8 | Cell proliferation; signal transduction; immune response; induction of apoptosis; integral to plasma membrane; cell-cell signaling; tumor necrosis factor receptor binding | 16.6031255 | 16.1111832 | 4.28095125 | 11.2155931 | 4.62559223 | 5.704360903 |
| TNFSF9 | Tumor necrosis factor (ligand) superfamily, member 9 | Cell proliferation; signal transduction; immune response; apoptosis; integral to plasma membrane; cell-cell signaling; tumor necrosis factor receptor binding | 0.42683846 | 0.7782374 | 0.43051526 | 0.49961541 | 0.43147894 | 0.387778271 |
| VEGF | Vascular endothelial growth factor | Cell proliferation; signal transduction; extracellular; regulation of cell cycle; positive regulation of cell proliferation; homophilic cell adhesion; membrane; soluble fraction; heparin binding; growth factor activity; response to stress; vascular endothelial growth factor receptor binding; angiogenesis | 0.34493609 | 0.43924221 | 0.48077441 | 0.27217503 | 0.29507061 | 0.282455082 |
| PUC18 | PUC18 plasmid | – | N/A | N/A | N/A | N/A | N/A | N/A |
| VEGFB | Vascular endothelial growth factor B | Cell proliferation; regulation of cell growth; signal transduction; extracellular; regulation of cell cycle; positive regulation of cell proliferation; membrane; heparin binding; growth factor activity; vascular endothelial growth factor receptor binding | 0.59763504 | 0.75386562 | 1.02955742 | 0.6218807 | 0.85459142 | 0.466257659 |
| BLANK | – | – | N/A | N/A | N/A | N/A | N/A | N/A |
| AS1R2 | Artificial sequence 1 related 2 (80% identity) (48/60) | – | 1.5539686 | 1.22295593 | 1.32009457 | 0.98773529 | 0.52224864 | 0.591712439 |
| AS1R1 | Artificial sequence 1 related 1 (90% identity) (56/60) | – | 0.70736887 | 1.57696778 | 1.45720527 | 1.17608313 | 0.63025505 | 0.741073004 |
| AS1 | Artificial sequence 1 | – | 1.91282061 | 1.79068382 | 1.16574042 | 1.28525566 | 0.18929651 | 0.40325771 |
| GAPD | Glyceraldehyde-3-phosphate dehydrogenase | Cytoplasm; oxidoreductase activity; glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity; glucose metabolism; glycolysis | 0.56074995 | 0.98897125 | 1.07548536 | 0.9254465 | 1.22393987 | 0.759341513 |
| B2M | Beta-2-microglobulin | Extracellular; immune response | 0.74661134 | 0.89668368 | 0.95250045 | 0.94572891 | 0.98168609 | 0.84607305 |
| HSPCB | Heat shock 90 kDa protein 1, beta | ATP binding; protein binding; cytoplasm; heat shock protein activity; protein folding; TPR domain binding; nitric-oxide synthase regulator activity; positive regulation of nitric oxide biosynthesis; unfolded protein binding; response to unfolded protein; ATP binding; protein binding; cytoplasm; heat shock protein activity; protein folding; TPR domain binding; nitric-oxide synthase regulator activity; positive regulation of nitric oxide biosynthesis; unfolded protein binding; response to unfolded protein | 0.72168097 | 0.90224818 | 1.1021151 | 0.77865346 | 1.05306214 | 0.792455408 |
| ACTB | Actin, beta | – | 0.99999138 | 0.99998711 | 0.99999156 | 1 | 1 | 0.999990471 |
| BAS2C | Biotinylated artificial sequence 2 complementary sequence | – | 1.16477022 | 1.05712391 | 1.10977505 | 1.04786113 | 0.40201837 | 0.545022077 |
| BAS2C | Biotinylated artificial sequence 2 complementary sequence | – | 0.92847709 | 1.0361002 | 1.04182707 | 1.01571793 | 0.62977964 | 0.72145099 |
INSIP, idiopathic nonspecific interstitial pneumonia; IPF, idiopathic pulmonary fibrosis; N/A, not applicable.
Ethical Statement: This study was approved by the ethics committee of Tongji Hospital [(Tong) No. 183 Ethics]. And all patients provided informed consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Daniil ZD, Gilchrist FC, Nicholson AG, et al. A histologic pattern of nonspecific interstitial pneumonia is associated with a better prognosis than usual interstitial pneumonia in patients with cryptogenic fibrosing alveolitis. Am J Respir Crit Care Med 1999;160:899-905. 10.1164/ajrccm.160.3.9903021 [DOI] [PubMed] [Google Scholar]
- 2.American Thoracic Society .; European Respiratory Society. American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med 2002;165:277-304. [DOI] [PubMed] [Google Scholar]
- 3.Swigris JJ, Kuschner WG, Kelsey JL, et al. Idiopathic pulmonary fibrosis: challenges and opportunities for the clinician and investigator. Chest 2005;127:275-83. [DOI] [PubMed] [Google Scholar]
- 4.Green FH. Overview of pulmonary fibrosis. Chest 2002;122:334S-339S. 10.1378/chest.122.6_suppl.334S [DOI] [PubMed] [Google Scholar]
- 5.Fang X, Luo B, Yi X, et al. Usual interstitial pneumonia coexisted with nonspecific interstitial pneumonia, What's the diagnosis? Diagn Pathol 2012;7:167. 10.1186/1746-1596-7-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaminski N, Rosas IO. Gene expression profiling as a window into idiopathic pulmonary fibrosis pathogenesis: can we identify the right target genes? Proc Am Thorac Soc 2006;3:339-44. 10.1513/pats.200601-011TK [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray LA, Chen Q, Kramer MS, et al. TGF-beta driven lung fibrosis is macrophage dependent and blocked by Serum amyloid P. Int J Biochem Cell Biol 2011;43:154-62. 10.1016/j.biocel.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 8.Yang IV, Burch LH, Steele MP, et al. Gene expression profiling of familial and sporadic interstitial pneumonia. Am J Respir Crit Care Med 2007;175:45-54. 10.1164/rccm.200601-062OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu P, Luo B, Yi X, et al. The expressions and meanings of BMP-7 and TGF-β in idiopathic pulmonary fibrosis and idiopathic nonspecific interstitial pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2014;37:664-70. [PubMed] [Google Scholar]
- 10.Travis WD, Hunninghake G, King TE, Jr, et al. Idiopathic nonspecific interstitial pneumonia: report of an American Thoracic Society project. Am J Respir Crit Care Med 2008;177:1338-47. 10.1164/rccm.200611-1685OC [DOI] [PubMed] [Google Scholar]
- 11.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011;183:788-824. 10.1164/rccm.2009-040GL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Travis WD, Costabel U, Hansell DM, et al. An official American Thoracic Society/European Respiratory Society statement: Update of the international multidisciplinary classification of the idiopathic interstitial pneumonias. Am J Respir Crit Care Med 2013;188:733-48. 10.1164/rccm.201308-1483ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu XY, Yi XH, Li HP, et al. A study on the efficacy of glucocorticoid therapy for idiopathic nonspecific interstitial pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2010;33:593-6. [PubMed] [Google Scholar]
- 14.Wei X, Xia Y, Li F, et al. Kindlin-2 mediates activation of TGF-β/Smad signaling and renal fibrosis. J Am Soc Nephrol 2013;24:1387-98. 10.1681/ASN.2012101041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cell Mol Life Sci 2014;71:549-74. 10.1007/s00018-013-1349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin A, Jenkins G. Role of integrin-mediated TGFbeta activation in the pathogenesis of pulmonary fibrosis. Biochem Soc Trans 2009;37:849-54. 10.1042/BST0370849 [DOI] [PubMed] [Google Scholar]
- 17.Gu L, Xu WB, Liu HR, et al. Different cytokine profiles in usual interstitial pneumonia and nonspecific interstitial pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2003;26:350-3. [PubMed] [Google Scholar]
- 18.Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov 2012;11:790-811. 10.1038/nrd3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer SN, Gurujeyalakshmi G, Giri SN. Effects of pirfenidone on transforming growth factor-beta gene expression at the transcriptional level in bleomycin hamster model of lung fibrosis. J Pharmacol Exp Ther 1999;291:367-73. [PubMed] [Google Scholar]
- 20.Iyer SN, Gurujeyalakshmi G, Giri SN. Effects of pirfenidone on procollagen gene expression at the transcriptional level in bleomycin hamster model of lung fibrosis. J Pharmacol Exp Ther 1999;289:211-8. [PubMed] [Google Scholar]
- 21.Margaritopoulos G. Challenges in IPF diagnosis, current management and future perspectives: Patient case 2. Sarcoidosis Vasc Diffuse Lung Dis 2015;32 Suppl 1:38-9. [PubMed] [Google Scholar]
- 22.Lemaire I, Beaudoin H, Dubois C. Cytokine regulation of lung fibroblast proliferation. Pulmonary and systemic changes in asbestos-induced pulmonary fibrosis. Am Rev Respir Dis 1986;134:653-8. [DOI] [PubMed] [Google Scholar]
- 23.Walsh J, Absher M, Kelley J. Variable expression of platelet-derived growth factor family proteins in acute lung injury. Am J Respir Cell Mol Biol 1993;9:637-44. 10.1165/ajrcmb/9.6.637 [DOI] [PubMed] [Google Scholar]
- 24.Maeda A, Hiyama K, Yamakido H, et al. Increased expression of platelet-derived growth factor A and insulin-like growth factor-I in BAL cells during the development of bleomycin-induced pulmonary fibrosis in mice. Chest 1996;109:780-6. 10.1378/chest.109.3.780 [DOI] [PubMed] [Google Scholar]
- 25.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med 1996;2:561-6. 10.1038/nm0596-561 [DOI] [PubMed] [Google Scholar]
- 26.Distler JH, Manger B, Spriewald BM, et al. Treatment of pulmonary fibrosis for twenty weeks with imatinib mesylate in a patient with mixed connective tissue disease. Arthritis Rheum 2008;58:2538-42. 10.1002/art.23694 [DOI] [PubMed] [Google Scholar]