Abstract
Background
Previous studies have suggested that IL4, IL13, and IL4R are associated with serum IgE levels and allergies, and common variants of these genes may alter cancer risk. To clarify these associations, we conducted a meta-analysis to investigate the associations of IL4, IL13, and IL4R polymorphisms with gastrointestinal cancer risk.
Methods
We used 27 eligible case–control studies describing the associations of six polymorphisms of IL4, IL13, and IL4R with gastrointestinal cancer risk to calculate summary odds ratios (ORs) and 95% confidence intervals (CIs) using five different genetic models. The Q-statistic and I2 statistic were calculated to examine heterogeneity.
Results
The IL4 rs2070874 T allele seems to be associated with an increased risk of gastrointestinal cancer (OR 1.11; 95% CI, 1.00–1.24 for T allele vs. C allele). This association was significant in studies conducted outside of Asia (OR 1.28; 95% CI, 1.03–1.58 for T allele vs. C allele) and in studies investigating the association with gastric cancer (OR 1.17; 95% CI, 1.03–1.34 for T allele vs. C allele). However, the IL4R rs1801275 heterozygote seems to be associated with a reduced risk of gastrointestinal cancer (OR 0.79; 95% CI, 0.65–0.96 for AG vs. AA). Other polymorphisms did not show any significant associations with gastrointestinal cancer risk in any of the genetic models and subgroup analyses.
Conclusions
Our results suggest that certain polymorphisms of IL4 and IL4R may affect susceptibility to gastrointestinal cancer. However, further studies are required to confirm these findings.
Keywords: Gastrointestinal cancer, Allergy, Polymorphism, Meta-analysis
Highlights
-
•
This meta-analysis included 27 eligible case–control studies.
-
•
IL4 rs2070874 T allele may increase the risk of gastrointestinal cancer.
-
•
IL4R rs1801275 variant may reduce the risk of gastrointestinal cancer.
-
•
Polymorphisms in IL4 and IL4R may affect susceptibility to gastrointestinal cancer.
Introduction
Previous epidemiologic studies have suggested that allergic disease may be associated with either a reduced or an increased risk of cancer.1, 2 Individuals with allergies have been found to have higher circulating levels of IgE, which is a product of T helper 2 (Th2) responses that are promoted in tissues by cytokines, such as IL4 and IL13. The IL4 and IL13 genes are located within the cytokine gene cluster on chromosome 5q31–31.1 They share a common IL4R chain on their receptors and are involved in the same signaling pathways.3 These cytokines may play a central role in allergies via stimulating IgE synthesis and reducing the production of pro-inflammatory cytokines by macrophages.2, 4 Therefore, polymorphisms of the IL4, IL13, and IL4R genes may disrupt the balance of the cytokine network and may be involved in allergies and various cancers.5, 6, 7
Gastrointestinal cancer is the leading cause of cancer-related death worldwide.8 Recently, several studies were conducted investigating the influence of these allergy-related polymorphisms on gastrointestinal cancer in different populations.9, 10 Because the effect of genetic polymorphisms on cancer risk is small, a single study may be underpowered to detect a true association. Meta-analysis takes advantage of reduced random error in the observed effect estimates and produces a single estimate with enhanced precision.11 Some previous meta-analyses were conducted to investigate the association of IL4 rs2243250 with either gastric cancer12 or colorectal cancer.9, 10 However, the findings of these meta-analyses were inconclusive, and other polymorphisms of the IL4/IL13 pathway may help to clarify the role of polymorphisms in these genes in gastrointestinal carcinogenesis. Therefore, to gain more comprehensive knowledge of the associations of polymorphisms of the IL4, IL13, and IL4R genes with gastrointestinal cancer risk, we conducted a meta-analysis using six polymorphisms of these genes.
Methods
Identification and eligibility of relevant studies
To identify all articles that explored the associations of six polymorphisms of IL4, IL13, and IL4R with gastrointestinal cancer risk, we conducted a literature search on PubMed and EMBASE from the year 2000 through February 3, 2016. Information on the selected polymorphisms is listed in Table 1. We used the following search terms: 1) “interleukin-4 (IL-4 or IL4), interleukin-13 (IL-13 or IL13), and interleukin-4R (IL-4R or IL4R)”; 2) “SNP, polymorphism, mutation, and variant”; 3) “cancer, tumor, carcinoma, and neoplasm”; and 4) “esophagus, gastric, colorectal, colon, rectal, hepatocellular, gallbladder, pancreatic, and gastrointestinal.” To identify additional studies, we also screened the references of retrieved publications. Searches were limited to human studies and publications written in English. We did not consider abstracts or unpublished reports.
Table 1.
Primary information for the six polymorphisms included in the meta-analysis.
| Gene | Function and pathway | Ch. location | rs number | Location | Trivial name | Base change | MAF in controls, meana |
|
|---|---|---|---|---|---|---|---|---|
| Asian | Non-Asian | |||||||
| IL4 | Th2 differentiation and IgE induction | 5q31.1 | rs2243250 | Promoter | C-590T | C > T | 0.78 | 0.17 |
| rs2070874 | 5′UTR | C-33T | C > T | 0.80 | 0.13 | |||
| IL13 | Th2 effector functions | 5q31 | rs1800925 | Promoter | C-1112T, C-1055T | C > T | 0.17 | 0.17 |
| rs20541 | Exon4 | G2044A, Arg130Gln | C > T | 0.19 | 0.40 | |||
| IL4R | α-chain of the IL4 and IL13 receptors | 16p12.1-p11.2 | rs1805010 | Exon5 | Ile50Val, I75V | A > G | 0.57 | 0.45 |
| rs1801275 | Exon12 | Gln551Arg, Q675R | A > G | 0.17 | 0.19 | |||
Ch, chromosome; IL4, interleukin-4; IL13, interleukin-13; IL4R, interleukin-4 receptor; MAF, minor allele frequency.
Frequency of variant allele in controls in studies included in this meta-analysis.
Studies included in this meta-analysis were required to meet the following criteria: 1) investigated the associations of IL4, IL13, and IL4R polymorphisms with gastrointestinal cancer risk; 2) used a case–control design; 3) provided genotype frequencies for cases and controls, so that odds ratios (ORs) with 95% confidence intervals (CIs) and a Hardy–Weinberg Equilibrium (HWE) could be calculated; and 4) featured a control population genotype distribution that did not deviate from HWE. Studies containing duplicated data were excluded. One study by El-Omar et al.13 presented genotype frequencies separately according to cancer type and was thus considered two studies in this meta-analysis.
Data extraction
Two investigators extracted the data independently and reached a consensus on all items. The following information was extracted from each article: author name, year of publication, country of study, source of controls, cancer type, genotype frequencies for cases and controls, and investigated polymorphisms.
Statistical analysis
The strengths of the associations between the polymorphisms and gastrointestinal cancer risk were measured using ORs with corresponding 95% CIs. Forest plots were used to illustrate the results of the included studies. Five different ORs were calculated using the following models: 1) homozygote comparison, 2) heterozygote comparison, 3) dominant genetic model, 4) recessive genetic model, and 5) allele comparison. This meta-analysis reported unadjusted pooled results. Before analysis, the genotype frequencies of the polymorphisms were assessed for HWE using a Chi-squared test. A p-value <0.01 was considered a significant deviation from HWE. The Q-statistic and I2 statistic were calculated to examine heterogeneity. The summary OR estimated for each study was calculated using either a fixed- or a random-effects model based on the Q-statistic.
Potential sources of heterogeneity were sought via subgroup analyses. For each genetic comparison, stratified analyses were performed according to cancer type, geographic location (Asia/non-Asia), and source of controls (hospital/population). Publication bias was investigated using a funnel plot, and funnel plot asymmetry was assessed using Egger's linear regression test.
All statistical analyses were performed using STATA software (version 11; Stata Corporation, College Station, TX, USA). Two-sided p-values <0.05 were considered statistically significant.
Results
Literature search and study characteristics
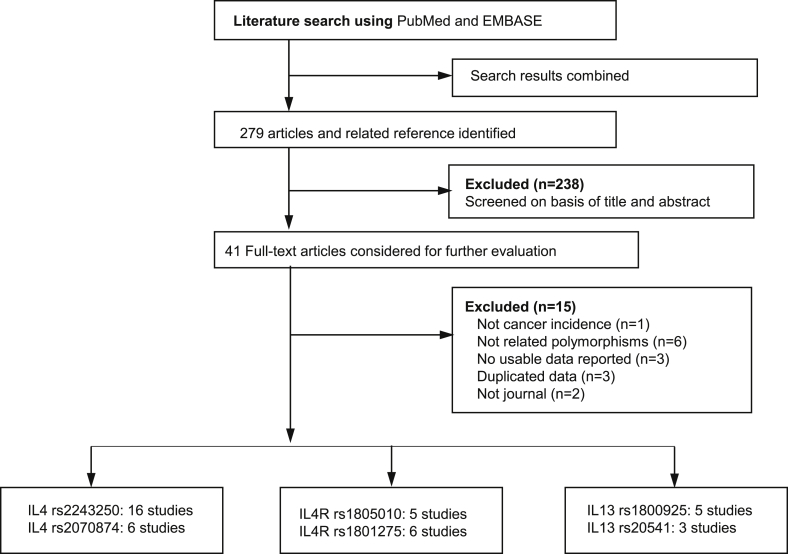
According to our search criteria, a total of 279 articles were retrieved. After screening these articles based on title and abstract, 238 articles were excluded. We then evaluated the full text of the remaining 41 articles, and 15 articles were excluded for several reasons. A study flow chart depicting the literature search and selection is presented in Fig. 1. Ultimately, we found 26 articles describing 27 studies on the associations of the IL4, IL13, and IL4R polymorphisms with gastrointestinal cancer that matched our inclusion criteria: IL4 rs2243250 (16 studies; 3783 cases/4895 controls) and rs2070874 (6 studies; 2202 cases/3388 controls), IL13 rs1800925 (5 studies; 1054 cases/1384 controls) and rs20541 (3 studies; 2162 cases/2568 controls), and IL4R rs1805010 (5 studies; 1215 cases/2004 controls) and rs1801275 (6 studies; 1013 cases/1065 controls).
Fig. 1.
Flowchart depicting the literature search and selection.
The allele frequency of each polymorphism in the controls is listed in Table 1. The frequency of the T allele in IL4 rs2243250 and rs2070874 was higher in the controls from Asian countries than in the controls from non-Asian countries (0.78 vs. 0.17 for rs2243250; 0.80 vs. 0.13 for rs2070874). Table 2 presents the characteristics of the studies included in the meta-analysis. The studies were conducted using either population or hospital controls in various countries and investigated gastrointestinal cancers, including esophageal,13, 14 gastric,13, 15, 16, 17, 18, 19, 20, 21, 22, 23 colorectal,24, 25, 26, 27, 28, 29, 30, 31, 32 hepatocellular,33, 34 and pancreatic cancer.35, 36, 37
Table 2.
Characteristics of the studies included in the meta-analysis.
| First author (year) | Country | Cancer type | Number of cases/controls | Source of control | SNPs |
|---|---|---|---|---|---|
| El-Omar (2003)13 | USA | Gastric | 112/209 | P | rs2243250 |
| El-Omar (2003)13 | USA | Esophageal | 90/209 | P | rs2243250 |
| Wu (2003)15 | Taiwan | Gastric | 220/230 | H | rs2243250, rs1805010, rs1801275 |
| Lai (2005)16 | China | Gastric | 123/162 | P | rs2243250 |
| Cozar (2007)24 | Spain | Colorectal | 96/174 | H | rs2243250 |
| Garcia-Gonzalez (2007)17 | Spain | Gastric | 404/404 | P | rs2243250 |
| Landi (2007)25 | Spain | Colorectal | 281/269 | H | rs2243250, rs2070874, rs1805010, rs1801275 |
| Olson (2007)36 | USA | Pancreatic | 405/212 | H | rs2243250, rs1801275 |
| Yannopoulos (2007)26 | Greece | Colorectal | 95/108 | P | rs2243250 |
| Crusius (2008)18 | 10 European countries |
Gastric | 244/1160 | P | rs2243250, rs2070874, rs1805010 |
| Suchy (2008)27 | Poland | Colorectal | 350/350 | H | rs2243250 |
| Wilkening (2008)28 | Sweden | Colorectal | 304/582 | P | rs2243250 |
| Zambon (2008)19 | Italy | Gastric | 142/171 | H | rs2243250, rs1805010, rs1801275 |
| Ando (2009)20 | Japan | Gastric | 330/190 | H | rs2243250, rs1805010 |
| Ko (2009)21 | Korea | Gastric | 81/324 | P | rs2243250, rs2070874 |
| Scola (2009)35 | Italy | Pancreatic | 48/131 | H | rs1801275, rs1800925 |
| Wu (2009)44 | China | Gastric | 1042/1099 | P | rs2070874 |
| Lee (2010)29 | Korea | Colorectal | 170/130 | P | rs1801275 |
| Sainz (2012)30 | Sweden | Colorectal | 1789/1771 | P | rs20541 |
| Walczak (2012)31 | Poland | Colorectal | 191/205 | P | rs1800925 |
| Sun (2013)14 | China | Esophageal | 365/369 | H | rs1800925 |
| Lu (2014)33 | China | Hepatocellular | 154/170 | P | rs2243250, rs2070874 |
| Pan (2014)22 | China | Gastric | 308/308 | P | rs2243250 |
| Yu (2014)32 | China | Colorectal | 299/296 | P | rs2070874 |
| Cotterchio (2015)37 | Canada | Pancreatic | 172/566 | P | rs20541 |
| Deng (2015)34 | China | Hepatocellular | 192/192 | P | rs1800925, rs20541 |
| Yin (2015)23 | China | Gastric | 234/465 | H | rs1800925 |
H, hospital; P, population.
Outcome from eligible studies
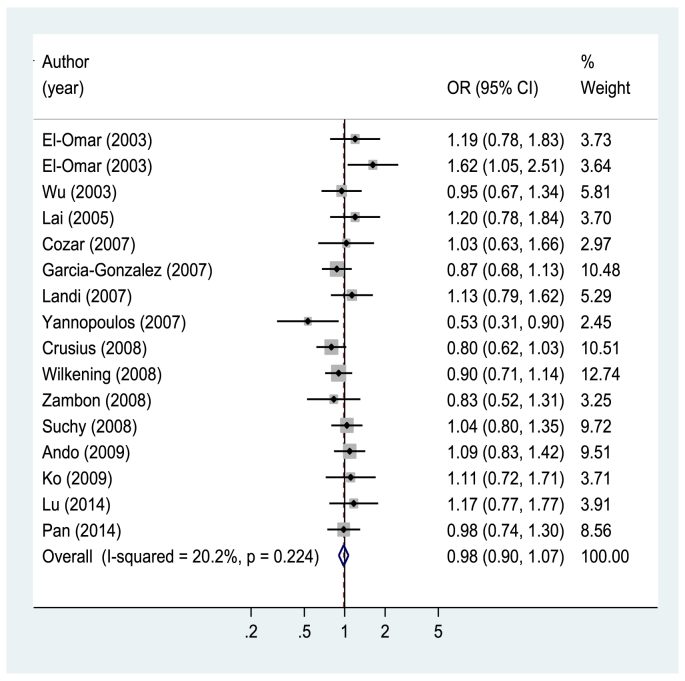
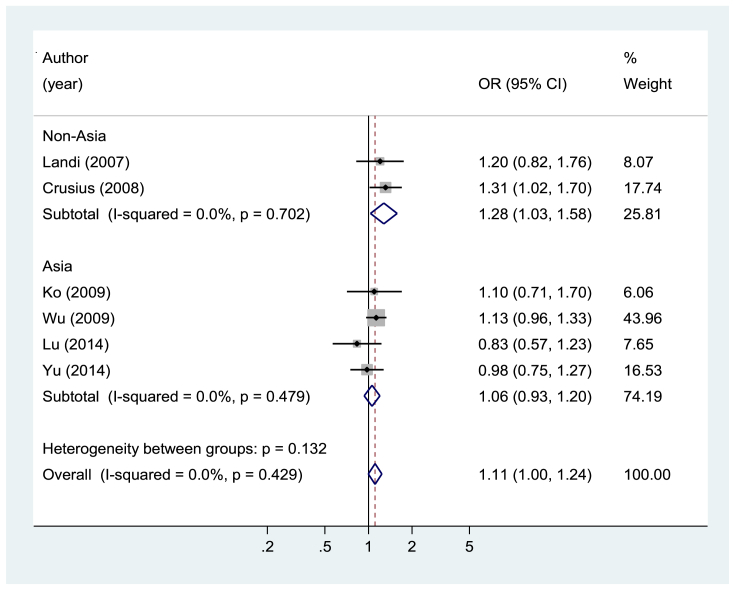
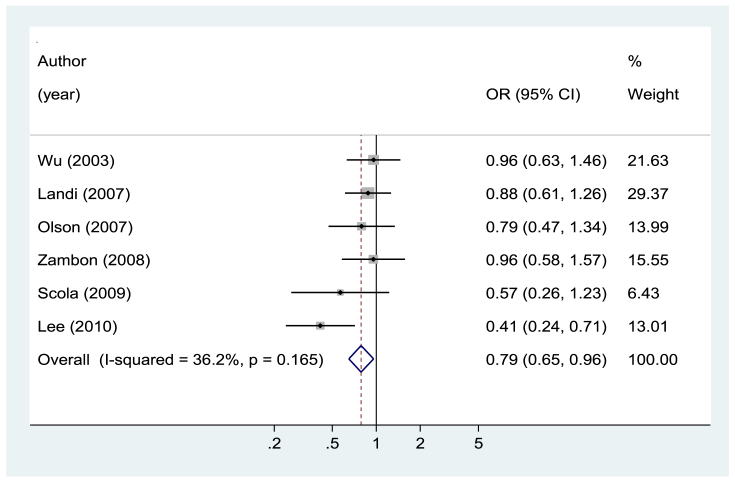
IL4 rs2243250 was not associated with gastrointestinal cancer risk for any of the genetic models and subgroup analyses (Fig. 2 and eTable 1). For IL4 rs2070874, an increased risk was observed for T allele carriers (OR 1.11; 95% CI, 1.00–1.24 for T allele vs. C allele), and this association was significant in studies conducted in studies outside of Asia (OR 1.28; 95% CI, 1.03–1.58 for T allele vs. C allele) and was significant in studies investigating the association with gastric cancer (OR 1.17; 95% CI, 1.03–1.34 for T allele vs. C allele) (Fig. 3 and eTable 1). Both IL13 rs20541 and rs1800925 were not associated with gastrointestinal cancer risk for any of the genetic models and subgroup analyses (eTable 2). For IL4R rs1801275, heterozygote seems to be associated with a reduced risk of gastrointestinal cancer (OR 0.79; 95% CI, 0.65–0.96 for AG vs. AA: OR 0.82; 95% CI, 0.68–0.99 for GG/AG vs. AA) (Fig. 4 and eTable 3). However, IL4R rs1805010 was not associated with gastrointestinal cancer risk for any of the genetic models and subgroup analyses (eTable 3).
Fig. 2.
Forest plot of gastrointestinal cancer risk associated with IL4 rs2243250 (T allele vs. C allele). CI, confidence interval; OR, odds ratio.
Fig. 3.
Forest plot of gastrointestinal cancer risk associated with IL4 rs2070874 (T allele vs. C allele), stratified by geographic location. CI, confidence interval; OR, odds ratio.
Fig. 4.
Forest plot of gastrointestinal cancer risk associated with IL4R 1801275 (AG vs. AA). CI, confidence interval; OR, odds ratio.
Publication bias
Begg's funnel plot and Egger's test were performed to evaluate publication bias. The funnel plot of the selected studies showed significant symmetry, and the results of Egger's test indicated no significant publication bias (data not shown).
Discussion
To derive more precise conclusions about the associations between allergy-related polymorphisms and gastrointestinal cancer risk, we performed a comprehensive meta-analysis of 27 case–control studies that included six polymorphisms of IL4, IL13, and IL4R genes. In this meta-analysis, an increased risk of gastrointestinal cancer was observed in those carrying the IL4 rs2070874 T variant, whereas a reduced risk of gastrointestinal cancer was observed in those who were heterozygous for IL4R rs1801275.
The role of IL4 rs2243250 in carcinogenesis has been investigated in many studies, but the findings are still inconclusive. In previous meta-analyses, Sun et al.12 reported a possible positive association between gastric cancer and the IL4 rs2243250 T allele in Caucasians, but Li et al.10 and Wu et al.9 reported no association between IL4 rs2243250 and colorectal cancer risk. We also found no association of the IL4 rs2243250 T allele with gastrointestinal cancer. However, a positive association between IL4 rs2070874 T allele carriers and gastrointestinal cancer risk was observed. Crusius et al.18 studied a Caucasian population from 10 European countries and found a significant positive association for IL4 rs2070874 T allele carriers. Our findings may support the role of this polymorphism in gastrointestinal carcinogenesis. However, we also found a reduced risk of gastrointestinal cancer in IL4R rs1801275 heterozygote carriers, which is in contrast to the findings for IL4 rs2070874. This may be a chance finding, but the underlying mechanisms should be investigated further.
Polymorphisms of IL4, IL13, and IL4R are suggested to affect the level of IgE because these polymorphisms are associated with greater expression of these genes and cytokines.5, 38, 39 A meta-analysis by Wang et al.40 indicated that variants of IL4 (rs2243250 and rs2070874) and IL13 (rs1800925 and rs20541) were associated with an increased risk of asthma. In in vitro experiments, higher total serum IgE levels were observed in T allele carriers of IL4 rs2243250 and rs2070874 and IL13 rs1800925 and rs20541.41 Rosenwasser et al.39 reported that four single nucleotide polymorphisms tested in the IL4/IL13 pathway are suspected of altering the function of specific genes. Several mechanisms have been proposed to explain the role of IgE in carcinogenesis. It is possible that the capacity for Th2 responses to simultaneously promote and suppress natural surveillance may lead to inconsistent findings.42 High IgE reflects immune hyper-responsiveness, leading to the detection and eradication of dysregulated cells.2 However, Th2 cytokines, such as IL4 and IL13, suppress interferon-γ associated inflammatory Th1 and cytolytic responses. Thus, allergies may be positively associated with cancer.42
Several other factors may affect the association between the investigated polymorphisms and gastrointestinal cancer risk. In this meta-analysis, the positive association between IL4 rs2070874 T allele carriers and gastrointestinal cancer risk was stronger in studies conducted outside of Asia. The T allele frequency of IL4 rs2243250 and rs2070874 was very different between Asians and non-Asians; differences in the IL4 genetic background among ethnicities may explain the different roles of this gene in the same disease.33 In addition to this, the positive association with IL4 rs2070874 T allele carriers was only significant in studies investigating the association with gastric cancer, suggesting that the role of this polymorphism in carcinogenesis could differ by cancer type. Of the six included studies that evaluated this polymorphism, only a study by Crusius et al.18 found a significant positive association for IL4 rs2070874 T allele carriers, particularly among patients with non-cardia intestinal type gastric cancer infected with H. pylori in a Caucasian population. However, other studies showed no association. Even though differences in study design (e.g., sample size and ethnicity) may affect the inconsistent findings between studies, the intrinsic heterogeneity of gastrointestinal carcinogenesis, sub-classified by anatomic location and histologic changes, should also be considered when interpreting the findings.43 Finally, different environmental and lifestyle factors can interact with gene polymorphisms and strengthen or weaken the effect of the studied polymorphisms.40 Gene–gene interactions may also contribute to the complexity of genetic diseases.5 An analysis of genotype data from a large population of German children showed that when polymorphisms of IL4, IL13, IL4R, and STAT6 were combined, the risk of high serum IgE levels increased by 10.8 times, and the risk of the development of asthma increased by 16.8 times compared with the effect of any individual polymorphism.41
The present findings should be interpreted with caution because of some limitations of the meta-analysis. First, the quality of a meta-analysis depends on the quality of the original studies. In our case, all studies were retrospective case–control studies, some of which involved small sample sizes and used hospital-based controls. Second, we evaluated only two polymorphisms per gene, which may limit our ability to elucidate the role of related cytokines in gastrointestinal cancer risk. Third, some inevitable publication bias may exist because only published studies were retrieved, although the funnel plot and Egger's test did not reveal significant publication bias. Finally, the numbers of published studies collected in our analysis were not large enough for comprehensive analysis. Considering all of these factors, future studies should be designed to overcome these limitations.
In conclusion, we investigated the role of six potentially functional variants of the IL4, IL13, and IL4R genes in gastrointestinal cancer risk based on the hypothesis that total serum IgE levels may affect carcinogenesis. The results of this meta-analysis indicated that some of these cytokine polymorphisms may affect susceptibility to gastrointestinal cancer. Therefore, case–control studies with larger sample sizes and multi-ethnic groups are needed to further investigate these associations in detail. Moreover, detailed gene–gene and gene–environment interaction data are needed for a comprehensive understanding of the association between the studied polymorphisms and cancer risk.
Conflicts of interest
None declared.
Acknowledgments
This work was supported by the National Cancer Center, Korea (1410260), and the National Research Foundation of Korea (2015R1C1A2A01053728).
Footnotes
Peer review under responsibility of the Japan Epidemiological Association.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.je.2016.06.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
eTable 1. Meta-analysis of the polymorphisms of IL4 and gastrointestinal cancer risk.
eTable 2. Meta-analysis of the polymorphisms of IL13 and gastrointestinal cancer risk.
eTable 3. Meta-analysis of the polymorphisms of IL4R and gastrointestinal cancer risk.
References
- 1.Josephs D.H., Spicer J.F., Corrigan C.J., Gould H.J., Karagiannis S.N. Epidemiological associations of allergy, IgE and cancer. Clin Exp Allergy. 2009;43:1110–1123. doi: 10.1111/cea.12178. [DOI] [PubMed] [Google Scholar]
- 2.Jensen-Jarolim E., Achatz G., Turner M.C. AllergoOncology: the role of IgE-mediated allergy in cancer. Allergy. 2008;63:1255–1266. doi: 10.1111/j.1398-9995.2008.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X.X., Li F.X., Wu Y.S., Wu D., Tan J.Y., Li M. Association of TGF-beta1, IL-4 and IL-13 gene polymerphisms with asthma in a Chinese population. Asian Pac J Allergy Immunol. 2011;29:273–277. [PubMed] [Google Scholar]
- 4.de Vries J.E. The role of IL-13 and its receptor in allergy and inflammatory responses. J Allergy Clin Immunol. 1998;102:165–169. doi: 10.1016/s0091-6749(98)70080-6. [DOI] [PubMed] [Google Scholar]
- 5.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 6.Rosenwasser L.J., Klemm D.J., Dresback J.K. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25(Suppl 2):74–78. doi: 10.1111/j.1365-2222.1995.tb00428.x. discussion 95–96. [DOI] [PubMed] [Google Scholar]
- 7.Heinzmann A., Mao X.Q., Akaiwa M. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R., Naishadham D., Jemal A. Cancer statistics. CA Cancer J Clin. 2013;2013(63):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 9.Wu H., Hu J., Liu B., Tao Y., Zhou X., Yuan X. Lack of association between interleukin-4 -524C>T polymorphism and colorectal cancer susceptibility. Tumour Biol. 2014;35:3657–3662. doi: 10.1007/s13277-013-1484-6. [DOI] [PubMed] [Google Scholar]
- 10.Li Q., Wang Q., Xu X., Ren S., Wang L. Association between IL-4 -589C>T polymorphism and colorectal cancer risk. Tumour Biol. 2014;35:2675–2679. doi: 10.1007/s13277-013-1352-4. [DOI] [PubMed] [Google Scholar]
- 11.Kavvoura F.K., Ioannidis J.P. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 12.Sun Z., Cui Y., Jin X., Pei J. Association between IL-4 -590C>T polymorphism and gastric cancer risk. Tumour Biol. 2014;35:1517–1521. doi: 10.1007/s13277-013-1209-x. [DOI] [PubMed] [Google Scholar]
- 13.El-Omar E.M., Rabkin C.S., Gammon M.D. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193–1201. doi: 10.1016/s0016-5085(03)00157-4. [DOI] [PubMed] [Google Scholar]
- 14.Sun J.M., Li Q., Gu H.Y., Chen Y.J., Wei J.S., Zhu Q. Interleukin 10 rs1800872 T>G polymorphism was associated with an increased risk of esophageal cancer in a Chinese population. Asian Pac J Cancer Prev. 2013;14:3443–3447. doi: 10.7314/apjcp.2013.14.6.3443. [DOI] [PubMed] [Google Scholar]
- 15.Wu M.S., Wu C.Y., Chen C.J., Lin M.T., Shun C.T., Lin J.T. Interleukin-10 genotypes associate with the risk of gastric carcinoma in Taiwanese Chinese. Int J Cancer. 2003;104:617–623. doi: 10.1002/ijc.10987. [DOI] [PubMed] [Google Scholar]
- 16.Lai K.C., Chen W.C., Jeng L.B., Li S.Y., Chou M.C., Tsai F.J. Association of genetic polymorphisms of MK, IL-4, p16, p21, p53 genes and human gastric cancer in Taiwan. Eur J Surg Oncol. 2005;31:1135–1140. doi: 10.1016/j.ejso.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Gonzalez M.A., Lanas A., Quintero E. Gastric cancer susceptibility is not linked to pro-and anti-inflammatory cytokine gene polymorphisms in whites: a Nationwide Multicenter Study in Spain. Am J Gastroenterol. 2007;102:1878–1892. doi: 10.1111/j.1572-0241.2007.01423.x. [DOI] [PubMed] [Google Scholar]
- 18.Crusius J.B., Canzian F., Capella G. Cytokine gene polymorphisms and the risk of adenocarcinoma of the stomach in the European prospective investigation into cancer and nutrition (EPIC-EURGAST) Ann Oncol. 2008;19:1894–1902. doi: 10.1093/annonc/mdn400. [DOI] [PubMed] [Google Scholar]
- 19.Zambon C.F., Basso D., Marchet A. IL-4 -588C>T polymorphism and IL-4 receptor alpha [Ex5+14A>G; Ex11+828A>G] haplotype concur in selecting H. pylori cagA subtype infections. Clin Chim Acta. 2008;389:139–145. doi: 10.1016/j.cca.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Ando T., Ishikawa T., Kato H. Synergistic effect of HLA class II loci and cytokine gene polymorphisms on the risk of gastric cancer in Japanese patients with Helicobacter pylori infection. Int J Cancer. 2009;125:2595–2602. doi: 10.1002/ijc.24666. [DOI] [PubMed] [Google Scholar]
- 21.Ko K.P., Park S.K., Cho L.Y. Soybean product intake modifies the association between interleukin-10 genetic polymorphisms and gastric cancer risk. J Nutr. 2009;139:1008–1012. doi: 10.3945/jn.108.101865. [DOI] [PubMed] [Google Scholar]
- 22.Pan X.F., Wen Y., Loh M. Interleukin-4 and -8 gene polymorphisms and risk of gastric cancer in a population in Southwestern China. Asian Pac J Cancer Prev. 2014;15:2951–2957. doi: 10.7314/apjcp.2014.15.7.2951. [DOI] [PubMed] [Google Scholar]
- 23.Yin J., Wang X., Wei J. Interleukin 12B rs3212227 T>G polymorphism was associated with an increased risk of gastric cardiac adenocarcinoma in a Chinese population. Dis Esophagus. 2015;28:291–298. doi: 10.1111/dote.12189. [DOI] [PubMed] [Google Scholar]
- 24.Cozar J.M., Romero J.M., Aptsiauri N. High incidence of CTLA-4 AA (CT60) polymorphism in renal cell cancer. Hum Immunol. 2007;68:698–704. doi: 10.1016/j.humimm.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Landi S., Bottari F., Gemignani F. Interleukin-4 and interleukin-4 receptor polymorphisms and colorectal cancer risk. Eur J Cancer. 2007;43:762–768. doi: 10.1016/j.ejca.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Yannopoulos A., Nikiteas N., Chatzitheofylaktou A., Tsigris C. The (-590 C/T) polymorphism in the interleukin-4 gene is associated with increased risk for early stages of corolectal adenocarcinoma. Vivo. 2007;21:1031–1036. [PubMed] [Google Scholar]
- 27.Suchy J., Klujszo-Grabowska E., Kladny J. Inflammatory response gene polymorphisms and their relationship with colorectal cancer risk. BMC Cancer. 2008;8:112. doi: 10.1186/1471-2407-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkening S., Tavelin B., Canzian F. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis. 2008;29:1202–1206. doi: 10.1093/carcin/bgn101. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y.S., Choi H.B., Lee I.K., Kim T.G., Oh S.T. Association between interleukin-4R and TGF-beta1 gene polymorphisms and the risk of colorectal cancer in a Korean population. Colorectal Dis. 2010;12:1208–1212. doi: 10.1111/j.1463-1318.2009.02080.x. [DOI] [PubMed] [Google Scholar]
- 30.Sainz J., Rudolph A., Hoffmeister M. Effect of type 2 diabetes predisposing genetic variants on colorectal cancer risk. J Clin Endocrinol Metab. 2012;97:E845–E851. doi: 10.1210/jc.2011-2565. [DOI] [PubMed] [Google Scholar]
- 31.Walczak A., Przybylowska K., Dziki L. The lL-8 and IL-13 gene polymorphisms in inflammatory bowel disease and colorectal cancer. DNA Cell Biol. 2012;31:1431–1438. doi: 10.1089/dna.2012.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y., Zheng S., Zhang S. Polymorphisms of inflammation-related genes and colorectal cancer risk: a population-based case-control study in China. Int J Immunogenet. 2014;41:289–297. doi: 10.1111/iji.12119. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y., Wu Z., Peng Q. Role of IL-4gene polymorphisms in HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2014;9:e110061. doi: 10.1371/journal.pone.0110061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Y., Xie M., Xie L. Association between polymorphism of the interleukin-13 gene and susceptibility to hepatocellular carcinoma in the Chinese population. PLoS One. 2015;10:e0116682. doi: 10.1371/journal.pone.0116682. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Scola L., Giacalone A., Marasa L. Genetic determined downregulation of both type 1 and type 2 cytokine pathways might be protective against pancreatic cancer. Ann N Y Acad Sci. 2009;1155:284–288. doi: 10.1111/j.1749-6632.2008.03686.x. [DOI] [PubMed] [Google Scholar]
- 36.Olson S.H., Orlow I., Simon J. Allergies, variants in IL-4 and IL-4R alpha genes, and risk of pancreatic cancer. Cancer Detect Prev. 2007;31:345–351. doi: 10.1016/j.cdp.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Cotterchio M., Lowcock E., Bider-Canfield Z. Association between variants in atopy-related immunologic candidate genes and pancreatic cancer risk. PLoS One. 2015;10:e0125273. doi: 10.1371/journal.pone.0125273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsuyasu H., Izuhara K., Mao X.Q. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet. 1998;19:119–120. doi: 10.1038/472. [DOI] [PubMed] [Google Scholar]
- 39.Rosenwasser L.J., Borish L. Promoter polymorphisms predisposing to the development of asthma and atopy. Clin Exp Allergy. 1998;28:13–15. doi: 10.1046/j.1365-2222.1998.028s5013.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z.D., Lian D., Shen J.L. Association between the interleukin-4, interleukin-13 polymorphisms and asthma: a meta-analysis. Mol Biol Rep. 2013;40:1365–1376. doi: 10.1007/s11033-012-2180-0. [DOI] [PubMed] [Google Scholar]
- 41.Kabesch M., Schedel M., Carr D. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117:269–274. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 42.Van Hemelrijck M., Garmo H., Binda E. Immunoglobulin E and cancer: a meta-analysis and a large Swedish cohort study. Cancer Causes Control. 2010;21:1657–1667. doi: 10.1007/s10552-010-9594-6. [DOI] [PubMed] [Google Scholar]
- 43.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process–first American cancer society Award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 44.Wu J., Lu Y., Ding Y.B. Promoter polymorphisms of IL2, IL4, and risk of gastric cancer in a high-risk Chinese population. Mol Carcinog. 2009;48:626–632. doi: 10.1002/mc.20502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.