ABSTRACT
CTCF-mediated chromatin interactions influence organization and function of mammalian genome in diverse ways. We analyzed the interactions among CTCF binding sites (CBS) at the murine TCRb locus to discern the role of CTCF-mediated interactions in the regulation of transcription and VDJ recombination. Chromosome conformation capture analysis revealed thymocyte-specific long-range intrachromosomal interactions among various CBS across the locus that were relevant for defining the limit of the enhancer Eb-regulated recombination center (RC) and for facilitating the spatial proximity of TCRb variable (V) gene segments to the RC. Ectopic CTCF binding in the RC region, effected via genetic manipulation, altered CBS-directed chromatin loops, interfered with RC establishment, and reduced the spatial proximity of the RC with Trbv segments. Changes in chromatin loop organization by ectopic CTCF binding were relatively modest but influenced transcription and VDJ recombination dramatically. Besides revealing the importance of CTCF-mediated chromatin organization for TCRb regulation, the observed chromatin loops were consistent with the emerging idea that CBS orientations influence chromatin loop organization and underscored the importance of CBS orientations for defining chromatin architecture that supports VDJ recombination. Further, our study suggests that in addition to mediating long-range chromatin interactions, CTCF influences intricate configuration of chromatin loops that govern functional interactions between elements.
KEYWORDS: antigen receptor loci, CTCF, VDJ recombination, chromatin loop, chromatin organization
INTRODUCTION
Spatial and temporal regulation of nuclear processes is intricately related to chromatin structure and organization since it can influence the interactions among regulatory elements. The mechanisms underlying these complex interactions are not completely understood. CTCF was identified to be the trans-acting factor that can organize an insulator and block enhancer-promoter interaction in a position dependent manner (1–3). Subsequently, CTCF has emerged as an important architectural protein that can influence interchromosomal and intrachromosomal interactions and impact nuclear functions largely in collaboration with cohesin (4, 5). Genome-wide investigations, including chromatin immunoprecipitation (ChIP), 4C, Hi-C, and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET), etc., have established an important role of CTCF in organization of topologically associated domains (TADs) (6, 7). In addition, CTCF binding sites (CBS) located within TADs can contribute to cell-type-specific chromatin loop organization by facilitating, as well as inhibiting, the interactions between regulatory elements. Locus-specific genetic analysis relying on deletion and inversion of CBS has revealed CBS orientation-dependent chromatin loop organization and its influence on transcriptional regulation (8–11). However, several aspects regarding the ability of CBS to organize chromatin and regulate function remain to be elucidated (12), and further exploration of the role of CTCF/cohesion complex in sub-TAD organization will be useful to unravel the varied role of CTCF in chromatin organization and function. We investigated the CBS orientation-dependent involvement of CTCF in chromatin organization and the regulation of the mouse TCRb locus that encodes the beta-chain of a/b T-cell receptors (TCRs).
TCRb locus and other antigen receptor (AgR) loci that encode TCRs and immunoglobulin chains provide a particularly useful context for exploring chromatin organization and function since higher-order chromatin organization is of critical importance for the regulation of AgR loci not only for transcription but also for VDJ recombination. These loci span large linear distances in the mouse genome ranging from 240 kb (Igl) to 2.75 Mb (Igh). ChIP-sequencing (ChIP-seq) analyses have revealed multiple CBS at AgR loci, more than a hundred each at large Igh, Igk, and TCRa/d loci, that also bind cohesin (13, 14). Transcription and VDJ recombination are precisely regulated by interaction of genetic elements, epigenetic mechanisms, and topological features. Hence, CTCF can potentially influence several aspects pertinent for such regulation.
Interactions of specific enhancers and promoters regulate the chromatin accessibility to RAG proteins that mediate nonhomologous end joining recombination between recombination signal sequences (RSS) associated with variable (V), diversity (D), and joining (J) segments (15). Despite the challenge posed by the large number of CBS at AgR loci and the possible redundancy in their action, deletion of specific CBS has revealed similar, as well as distinct, roles for CTCF in regulating enhancer-promoter interactions at AgR loci. CBS at IGCR1 at the Igh locus and Sis and Cer at the Igk locus serve as insulators to restrict the activity of enhancers (16, 17). At the TCRa/d locus, CTCF and cohesin organize a chromatin hub to suppress Ea-TCRd interactions but, interestingly, facilitate the interaction of Ea, TEA promoter, and promoters of Va segments in double-positive (DP) thymocytes (14, 18).
In addition, the recombination center (RC) at each locus, which encompasses D and J segments and preferentially binds RAG proteins (19), is located at large distances from the V segments. Hence, intrachromosomal interactions that facilitate the spatial proximity of various V segments with the RC are a necessary prerequisite for VDJ recombination at the AgR loci (20) and are manifested as “locus contraction,” toward which CTCF/cohesin complexes can contribute. At the Igh locus, a long-range interaction between IGCR1 and CBS located downstream of the 3′ regulatory region (3′RR) influences the Vh segments for VDJ recombination; the Vh usage gets altered upon the deletion of CBS from either of these elements (16, 21, 22). Further, CTCF/cohesin complex contributes to the organization of the Igh locus into rosettes whose spatial proximity to the RC provides equal opportunity for distal and proximal Vh segments for VDJ recombination (23–25). CTCF, Pax5, and YY1 have been proposed to contribute distinctly toward this process (26). In contrast to the Igh locus, the locus contraction at the TCRa/d locus was not significantly reduced upon conditional deletion of CTCF (14), indicating significant differences in the process of locus contraction at different loci in the context of their organization and/or presence of other tissue-specific trans-acting factors. In addition to facilitating Rag-mediated recombination at bona fide recombination signal sequences (RSS) of V, D, and J segments, CTCF-mediated chromatin loops have also been proposed to restrict the linear tracking of Rag complexes and thus minimize the off-target cleavage at cryptic RSS by Rag1 at Igh and TCRd loci (27, 28).
Multifunctional attributes of CTCF and varied functions of CTCF observed at different loci led us to investigate the CTCF-based chromatin interactions at the stringently regulated mouse TCRb locus. The relevance of CTCF-mediated chromatin organization is not well elucidated at TCRb, which spans ∼670 kb (Fig. 1A) and undergoes VDJ recombination in double-negative (DN) thymocytes to generate a repertoire of functional TCRb genes (29). Since it is much smaller than Igh and TCRa/d loci and is relatively simpler in organization of genic segments and regulatory elements, TCRb is likely to have similar as well as distinct requirements for functional chromatin organization. Further, the insertion of ectopic CTCF binding sites near the TCRb RC not only curtailed Eb activity, as predicted, but also severely impaired the usage of upstream V segments for VDJ recombination (30). Considering the facilitation of locus contraction by CTCF at other loci, the observed inhibition of VDJ recombination was rather intriguing. However, it was consistent with the importance of CTCF for locus organization relevant for VDJ recombination.
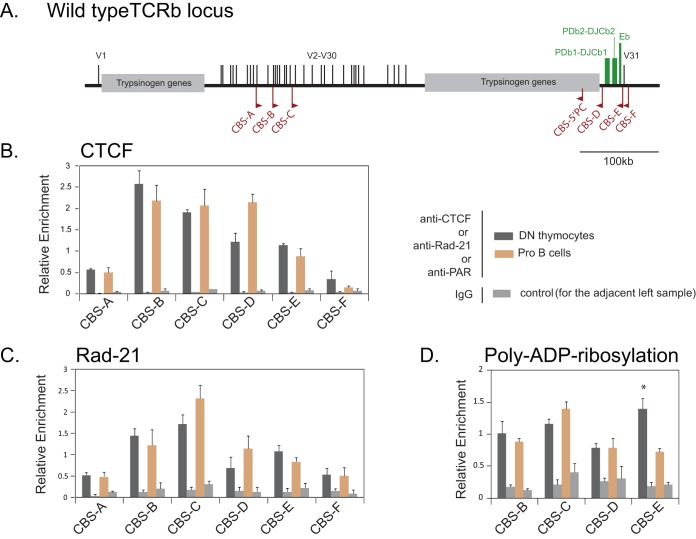
FIG 1.
Binding of CTCF, Rad-21, and poly(ADP-ribosyl)ated moieties at the CBS of the TCRb locus. (A) Schematic map of mouse TCRb locus showing the position of Trbv segments (V1 to V31) and the recombination center encompassing PDb1-DJCb1, PDb2-DJCb2, and Eb (green bars). The CBS selected for analysis are marked. The direction of the red flag indicates the orientation of the CBS. (B to D) ChIP-qPCR analysis to estimate the binding of CTCF, Rad-21, and poly(ADP-ribosyl)ated moieties at the CBS in DN thymocytes and ProB cells isolated from Rag1-deficient mice. In each case, the ChIP-qPCR signals were normalized to the values observed for CBS-3 of H19-ICR at the Igf2/H19 locus. Values represent the mean enrichments (plus the standard errors of the mean [SEM]) from three biological replicates. The significance of enrichment between DN and ProB cells was analyzed by using a Student t test (*, P < 0.05). Table S1 in the supplemental material provides the genomic coordinates of the analyzed CBS.
TCRb regulation recapitulates all key aspects underlying VDJ recombination at AgR loci. Enhancer (Eb) is the crucial cis-regulatory element that generates chromatin accessibility of an ∼25-kb region to establish the RC (31). Eb activates two promoters, PDb1 and PDb2, each linked to DJC clusters. Eb-mediated chromatin accessibility—marked by hyperacetylated histones, H3K4-trimethylated histone, and germ line transcription—leads to the recruitment of RAG proteins and D-to-J recombination that precedes V-to-DJ recombination. All the Trbv segments except V31 are located 300 to 400 kb upstream from the RC, are separated from it by intervening trypsinogen genes, and are brought into the vicinity of the RC via locus contraction. Mapping of long-range interactions in the context of regulatory elements has identified a tethering element, 5′PC, located ∼27 kb upstream of the RC and insulated from the activity of Eb by a chromatin barrier located upstream of PDb1. 5′PC binds CTCF to low levels and may facilitate the interaction of a subset of V segments with the RC by mechanisms that remain obscure (32).
Although CTCF is an important candidate that effects long range interactions, CBS that participate in and/or regulate long-range interactions at TCRb locus are not well defined. TCRb has 21 CBS identified by ChIP-seq (14). All of the CBS located in the domain of V segments are convergent to CBS in proximity of the RC and can potentially interact with them. This provides an interesting opportunity to examine the influence of CBS orientation, especially in the context of VDJ recombination. Intrachromosomal interactions at none of the AgR loci have been investigated in this context. We, therefore, analyzed the chromatin organization of the TCRb locus by chromosome conformation capture-quantitative PCR (3C-qPCR) assays and DNA-fluorescence in situ hybridization (FISH) in wild-type and mutant mice that carried ectopic CBS (30). Perturbation of CBS-based chromatin organization by ectopic CBS complements the CBS deletion- and inversion-based analyses of other loci (6). In addition to defining key aspects of the CTCF-mediated chromatin loop organization that is important for TCRb regulation, our analysis suggests that even relatively mild perturbation of CTCF-based chromatin organization can have severe functional consequences for gene regulation.
RESULTS
CTCF binding sites at the TCRb locus.
CTCF binding was evaluated by ChIP-seq in ex vivo thymocytes (DN2/3 stage) of Rag1-deficient mice (14). The 21 reported sites were scattered through the locus and exhibited various degrees of CTCF binding. Although each of the CBS present at the TCRb locus could be important from a regulatory perspective, we selected a subset of CBS (Fig. 1A) that were located in the proximity of the RC and the upstream Trbv segments and/or that exhibited high scores on the CBS prediction tool CTCFDB (33) compared to the position weight matrix (PWM) of the CTCF binding motif (REN-20) (34). CBS-A was in the vicinity of pseudogenes, while CBS-B and CBS-C flanked the V12.1 to V14 gene segments, most of which are functional and commonly used for VDJ recombination. CBS-D and CBS-E flanked the RC with CBS-E located in close proximity to Eb, which is the major regulatory element of the locus. CBS-F was downstream of V31.
CTCF is known to collaborate with cohesin to organize chromatin loops. CTCF and cohesin binding were verified at selected CBS by ChIP-qPCR. Five sites (Fig. 1A, CBS-A, -B, -C, -D, and -E) clearly bound CTCF, as well as cohesin, albeit to various degrees (Fig. 1B and C), but CBS-F was not enriched. In addition to CBS-B, CBS-C, and CBS-E, which were verified earlier (32), CBS-A and CBS-D also had a high scores based on the CBS prediction tool but were not verified earlier. In our ChIP-qPCR analysis, CBS-A and CBS-D exhibited significant CTCF and cohesin binding. CBS-D was particularly interesting as it is located upstream of the RC. The binding of neither CTCF nor cohesin was specific to the DN stage of development. Both double-positive (DP) thymocytes and ProB cells also showed CTCF and cohesin bound to these CBS (Fig. 1B and C; see also Fig. S3 in the supplemental material).
Poly(ADP-ribosyl)ation (PARylation) of CTCF was proposed to be critical to its insulator function in mammals (35) and its ability to act as a looping factor in Drosophila (36). ChIP-qPCR indicated PARylation at each of the CBS examined (Fig. 1D). Like CTCF and cohesin, the PARylation was also not specific to DN thymocytes at the CBS except at CBS-E. Although the PARylation was significantly greater in DN cells at CBS-E, it was also clearly evident in ProB cells. Overall, ChIP-qPCR analysis suggested that CBS-A, -B,- C, -D, and -E are all capable of participating in looping interactions.
Intrachromosomal interactions among CTCF binding sites.
The TCRb locus undergoes VDJ recombination specifically in DN thymocytes. We compared the contact frequencies among CBS by 3C-qPCR in Rag1-deficient DN thymocytes with ProB cells where TCRb does not undergo VDJ recombination. The absence of Rag1 ensured that the locus was in a state prior to recombination. We chose CBS-B and CBS-E as anchors since they bind CTCF and cohesin to high levels and are located centrally in the broad region encompassing V segments and downstream of the RC, respectively (Fig. 2A).
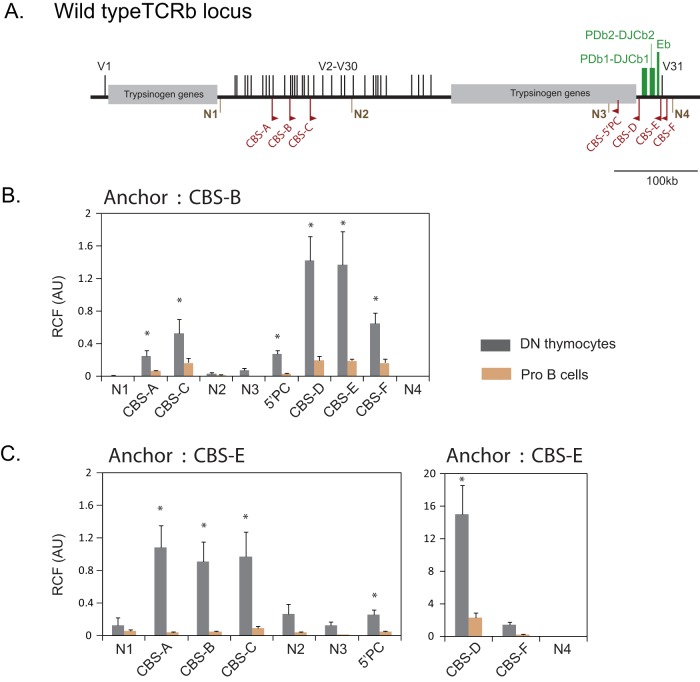
FIG 2.
Long-range interactions among CTCF binding sites detected by 3C-qPCR. (A) Schematic map showing the position of the analyzed CBS (CBS-A to CBS-F) relative to Trbv segments (V1 to V31) and the recombination center (RC). N1 to N4 are the regions included as negative controls for 3C analysis. (B) Spatial proximity of CBS-B to other CBS. (C) Spatial proximity of CBS-E to other CBS. Values represent the mean relative cross-linking frequency (plus the SEM) from three biological replicates. Significance was analyzed by using a Student t test (*, P < 0.05).
CBS-B interacted more strongly with CBS-D and CBS-E than with proximally located CBS-A and CBS-C (Fig. 2B) specifically in thymocytes, consistent with the idea that in most cases convergent CBS form a chromatin loop. CBS at 5′PC is also convergent to CBS-B and showed DN thymocyte-specific interaction with CBS-B, albeit to a lesser extent.
Taking CBS-E as an anchor, DN thymocytes exhibited a very high degree of interaction between CBS-D and CBS-E (Fig. 2C) despite their tandem orientation. The interaction was not merely due to linear proximity as it was significantly stronger than in ProB cells. The CBS-D and CBS-E interaction-based chromatin loop encompasses the PDb1-DJCb1-PDb2-DJCb2-Eb region that serves as the functional RC. The stability of the CBS-E/CBS-D interaction might be augmented by other DN thymocyte-specific protein factors that bind Eb, PDb1, and PDb2. In addition, two CBS that bind CTCF to low levels exist immediately upstream of PDb1 and Eb (14) and may augment the stability of the chromatin loop important for RC establishment. Importantly, CBS-D and CBS-E are located at the limits of the Eb-regulated domain, suggesting that their interaction defines the chromatin loop that acts as the RC.
Consistent with the possible role of other factors in stabilizing CBS-E to CBS-D interaction, the contact frequency of another CBS pair in tandem orientation (CBS-E and CBS-5′PC) was much lower. CBS-E also engaged in long-range interactions with CBS-A, CBS-B, and CBS-C to almost equivalent levels, specifically in DN thymocytes.
Thus, analysis of the CBS revealed their contribution to the higher-order organization of TCRb locus. Interactions among the selected CBS seemed to be important for the organization of the active RC, as well as for bringing the V segments to its proximity for VDJ recombination.
Influence of ectopic CTCF binding on intrachromosomal interactions.
To further investigate the relevance of the CTCF-mediated chromatin loop organization for the regulation of transcription and VDJ recombination, it was important to perturb it. We adopted a gain-of-function approach and analyzed the CBS interactions in a mutant allele, TCR-ins (Fig. 3A), wherein four ectopic CBS (CBS-ecto) were introduced by the insertion of H19-ICR derived from the imprinted Igf2/H19 locus via genetic manipulation (30). In the heterologous context of TCRb, H19-ICR binds CTCF upon maternal inheritance (37) as at its endogenous location (3, 38). Wild-type TCRb and TCR-mut were used as control alleles since TCR-mut had an H19-ICR-mut insertion wherein each of the CBS of H19-ICR are mutated. These mutations abrogate CTCF binding in vitro and in vivo (39). TCR-mut behaved like the wild-type allele during previous analyses of chromatin organization, transcription, and VDJ recombination (30, 37).
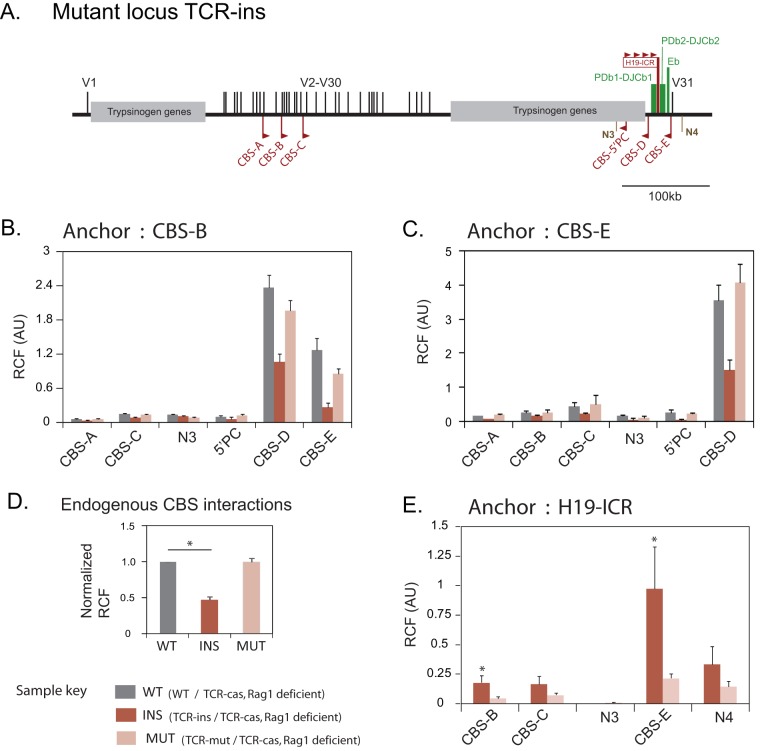
FIG 3.
Influence of ectopic CTCF binding sites on CBS interactions estimated by 3C-qPCR. (A) Schematic map of TCR-ins showing the positions of the CBS endogenous to TCRb (CBS-A to CBS-F) and the four ectopic CBS on H19-ICR inserted within the RC. N3 and N4 are the negative-control regions for 3C analysis. TCR-mut is similar to TCR-ins but incapable of binding CTCF at the ectopic CBS. (B and C) Long-range interactions among endogenous CBS in the presence or absence of ectopic CTCF binding using CBS-B and CBS-E as anchors. Values represent the mean (plus the SEM) of the relative cross-linking frequency (RCF) observed in three biological replicates. The differences between the interaction profiles of the three alleles were significant based on two-way ANOVA, followed by the F-test (P = 0.0026). (D) Extent of reduction in endogenous CBS interactions by TCR-ins. The mean RCF for each interaction in an allele (left and middle) was normalized to the wild-type TCRb allele (see Table S3 in the supplemental material). The graph shows the average (plus the SEM) of the normalized RCF at all test points for a given allele (excluding CBS-B-N3 and CBS-E-N3). Significance was tested by using a Student t test (*, P < 0.05). (E) Spatial proximity of ectopic CBS (H19-ICR) to endogenous CBS of TCRb. The values represent the mean relative cross-linking frequency (plus the SEM) from three biological replicates. Significance was analyzed by using a Student t test (*, P < 0.05).
Since TCR-ins could bind CTCF at ectopic CBS specifically upon maternal inheritance, we used allele-specific 3C-qPCR to ascertain the intrachromosomal interactions at the maternally inherited TCR-ins, TCR-mut, and wild-type TCRb alleles. In each case, the paternal allele was TCR-cas (37), which had the wild-type TCRb locus of Mus castaneus castaneus origin and hence had several single nucleotide differences compared to the maternal alleles (wild type, TCR-ins, or TCR-mut) of Mus musculus domesticus origin and were useful in designing various allele-specific PCR-based assays. The activity of the ectopic insulator in TCR-ins might render the HindIII site proximal to CBS-D inaccessible to HindIII digestion and interfere in 3C-qPCR-based interpretation. We verified that TCR-ins is digested as efficiently as the wild-type TCRb alleles (see Fig. S2 in the supplemental material). Further, allele-specific primers were designed and verified to detect 3C-qPCR-based interaction frequencies specifically on maternally inherited (domesticus) test alleles (see Table S2 in the supplemental material).
We examined the influence of the ectopic CTCF binding on the contact frequencies among endogenous CTCF binding sites using CBS-B and CBS-E as anchors (Fig. 3B and C). The CBS interactions appeared to be reduced in TCR-ins compared to wild-type TCRb and TCR-mut alleles. The contact frequencies were expected to vary due to two factors, i.e., allele type (wild type, TCR-ins, and TCR-mut) and CBS position. After excluding negative regions, two-way analysis of variance (ANOVA) established that besides the position of the CBS, allele type leads to significant variation (P = 0.0026), i.e., wild-type, TCR-ins, and TCR-mut alleles exhibit differences. Further analysis established that TCR-ins was different from the other alleles (wild type and TCR-mut).
To analyze the extent of TCR-ins dependent alteration in the contact frequencies, mean contact frequencies were normalized to the wild-type allele. The allelic profiles were confirmed to be significantly different (P < 0.0001) (see Table S3 in the supplemental material). ANOVA indicated that the extent of reduction observed in TCR-ins does not depend on the location of CBS. On average, a nearly 47% reduction was seen in TCR-ins compared to the wild-type allele (Fig. 3D; see Table S3 in the supplemental material). The interactions on TCR-mut were comparable to those observed with the wild-type TCRb allele. Reduced interaction of CBS-B with CBS-E and CBS-D did not lead to any enhancement in its ability to interact with CBS-A and CBS-C that are more proximal but not convergent.
Thus, ectopic CTCF binding influenced the interactions among endogenous CBS. It was plausible that CBS-ecto engaged with the endogenous CBS of TCRb, leading to reduced interactions among them. To explore this possibility, we examined the interaction of CTCF-bound H19-ICR with the CBS of the TCRb locus.
Since H19-ICR is located specifically on the mutated domesticus allele, the 3C-qPCR analysis using H19-ICR as an anchor was allele specific. In TCR-ins, H19-ICR was observed to interact with the endogenous CTCF binding sites (Fig. 3E). Notably, the proximally located and convergent CBS-E exhibited CTCF-dependent interaction with H19-ICR. CBS-D and H19-ICR interaction in TCR-ins could not be discerned by 3C-qPCR since they are separated by a single HindIII fragment. Hence, it is not clear whether CBS-E, CBS-D, and H19-ICR form a tripartite complex or whether any two of these elements are in contact at a given time. However, H19-ICR interaction with CBS-E and a concomitant reduction in CBS-D/CBS-E interaction suggests competition between H19-ICR and CBS-D for engaging with CBS-E. An interaction of H19-ICR with CBS-B or CBS-C in TCR-ins was unlikely due to the divergent orientation of CBS. H19-ICR exhibited a low level of contact with them, and this may explain the reduction in the interaction of CBS-B with CBS-A and CBS-C.
Our analyses indicated that CTCF-bound H19-ICR interacts with the endogenous CBS of the TCRb locus. Whether these interactions are as frequent and/or as stable as those between the endogenous CBS could not be discerned. However, these interactions do impair the interactions among endogenous CBS of the TCRb locus.
Interactions among regulatory elements.
The observed CTCF-based interactions could potentially influence the establishment of the RC and locus organization that brings V gene segments to the proximity of RC: the two regulatory steps crucial for V-to-DJ recombination. Further, ectopic CTCF binding sites altered the CBS interaction profile. The influence of the altered CTCF-based interactions on the interactions of regulatory elements relevant for VDJ recombination was examined next.
In TCR-ins, the H19-ICR, with ectopically bound CTCF, was observed to interact with CBS-E with a concomitant reduction in interaction of CBS-E with CBS-D (Fig. 3B and C) that flank the RC. Based on the position dependence of enhancer blocking by CBS of H19-ICR (40), this configuration is predicted to generate a chromatin loop encompassing PDb2-DJCb2-Eb that can support Eb interaction with PDb2 and render PDb2-DJCb2 accessible for transcription and recombination. The excluded PDb1-DJCb1 region is likely to escape Eb-mediated chromatin changes. Consistent with this, our 3C-qPCR analysis indicated reduced Eb-PDb1 interaction, reduced PDb1-DJCb1 chromatin accessibility (37), and a marked reduction in DJCb1 recombination (30). As predicted, Eb-based activation of PDb2 and the linked DJCb2 cluster was unaltered. Effectively, a smaller but a functional RC is organized in the TCR-ins allele due to an interaction of CBS-ecto and CBS-E.
How does this “smaller” but functional RC participate in the long-range interactions that can potentially bring V segments to the proximity of the RC? To address this question, we examined the proximity of several V gene segments to the RC in wild-type and mutant alleles.
First, we analyzed the interaction of Eb with the HindIII fragments carrying the promoters of a few Trbv gene segments. We selected V4, V12.1, V19, and V26, which are highly used for V-to-DJ recombination (41), and V31, which is located downstream of the RC and was seen to be preferentially used in TCR-ins for recombination (30). The selected V regions were broadly scattered through the locus and flanked by CBS located at various distances (Fig. 4A) ranging from 500 bp to 11 kb. Moreover, V4 and V12.1 were a part of the distal V domain, and V19 and V26 are located in the proximal V domain, as defined earlier (31).
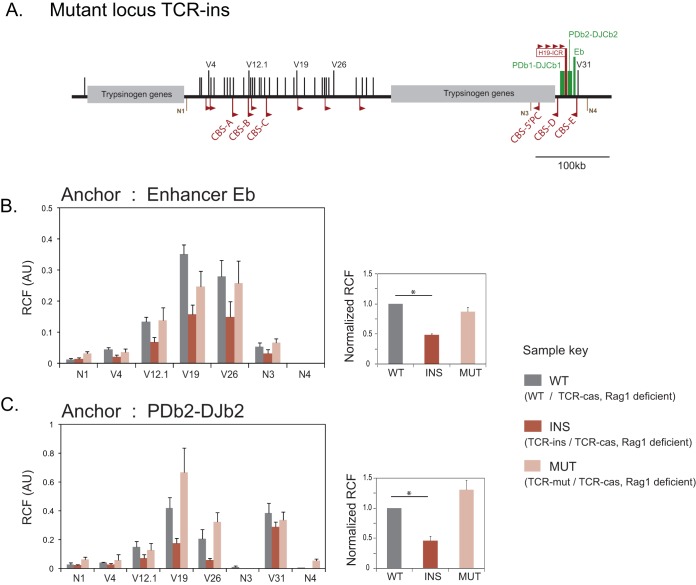
FIG 4.
Influence of ectopic CTCF binding sites on long-range interactions of Eb and PDb2-DJb2 estimated by 3C-qPCR. (A) Schematic map of TCR-ins showing the positions of Trbv segments (V1 to V31) and the RC. The Trbv segments chosen for analysis are marked. The closest CBS (upstream and downstream) to the analyzed V segments, as reported by ChIP-seq analysis (14), as well as ectopic CBS in TCR-ins, are shown as red triangles. N1 to N4 are the negative-control regions used for 3C analysis. TCR-mut is similar to TCR-ins but incapable of binding CTCF at the ectopic CBS. (B and C) (Left) Spatial proximity of Eb and PDb2-DJb2 to V segments. Values represent mean (+ the SEM) RCF from three biological replicates. The differences between the interaction profiles of the three alleles were significant based on two-way ANOVA, followed by the F-test (P < 0.001 for each anchor). (Right) Average (plus the SEM) RCF normalized to the wild-type TCRb allele (see Table S3 in the supplemental material) for Eb or PDb2-DJb2 interactions to the Trbv segments (V4, V12.1, V19, and V26) as shown in the left panels. Significance was analyzed by using a Student t test (*, P < 0.05).
Allele-specific 3C-qPCR analysis revealed contact frequencies of Eb with each of the V regions tested (Fig. 4B). Interaction with V4 and V12.1 was rather low compared to Eb-V19 and Eb-V26. No correlation was observed between the distance of the V regions from CBS and the V-Eb contact frequency. Further, interaction of Eb with each of the upstream V regions (V4, V12.1, V19, and V26) was reduced in the TCR-ins allele compared to the wild-type TCRb allele. Interactions of the V regions were inferred to be significantly different (P < 0.001) in the three alleles, and a reduction in interactions was specific to TCR-ins allele, thereby demonstrating CTCF dependence. Since Eb and V31 are located on adjacent fragments, their interaction could not be estimated by 3C-qPCR.
Although Eb is a part of the active RC, V-to-DJ recombination unambiguously requires the proximity of V segments to DJCb regions. In TCR-ins, the PDb2-DJCb2 region is functional and is preferentially used for V-DJ recombination (30). Hence, we performed 3C analysis, taking PDb2-DJb2 as an anchor (a HindIII fragment encompassing PDb2-Db2-Jb2.1-Jb2.7) in the wild-type, TCR-ins, and TCR-mut alleles.
Interactions of PDb2-DJb2 region with V regions closely resembled that of Eb for V4, V12.1, V19, and V26. V4 and V12.1 were observed to interact to a much lower degree than V19 and V26 (Fig. 4C). Further, the contact frequencies were significantly influenced by the allele (P < 0.001), and TCR-ins exhibited lowered interactions than those observed in control alleles, i.e., wild type and TCR-mut (Fig. 4C).
Using PDb2-DJb2 as an anchor, the proximity of V31 could also be discerned reliably as V31 is separated from PDb2 by ∼16 kb (encompassing four HindIII fragments). Interestingly, although upstream V segments were adversely affected for their interaction with the RC in TCR-ins allele, V31 interaction with PDb2-DJb2 was neither decreased nor increased in this scenario.
Finally, we analyzed the normalized means of interactions of upstream V regions (V4, V12.1, V19, and V26) with the RC discerned using Eb and PDb2-DJb2 as anchors during 3C analysis. In each case, two-way ANOVA demonstrated that irrespective of the V region chosen, its interaction with Eb was reduced to a similar extent (interaction reduced to 49%) in TCR-ins (Fig. 4B and C; see also Table S3 in the supplemental material). Even though the range of reduction observed due to TCR-ins was slightly wider for V to PDb2-DJb2 interactions (64% to 28% of the wild-type TCRb allele [average, 46%]), the variation was insignificant as determined by ANOVA (Fig. 4B and C; see also Table S3 in the supplemental material). Strikingly, the average reductions due to TCR-ins in Eb-V and PDb2-DJb2-V interactions were closely matched by the reductions observed for the interactions among CTCF binding sites (47%) (Fig. 3D). These data suggest that perturbations in CTCF-dependent interactions are closely reflected in the interactions of the RC with the V segments, even though the proximity of a V segment to CBS per se does not influence its interaction with the RC.
Influence of ectopic CTCF on TCRb locus compaction.
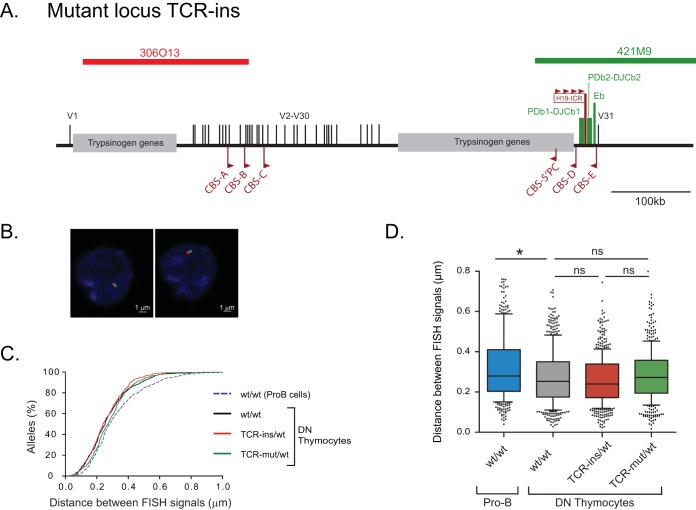
3C-qPCR revealed an approximately 50% reduction in the contact frequencies of upstream V segments with the RC. This could arise due to subtle changes in chromatin organization in all the nuclei or, alternatively, due to dramatic changes in locus organization in a subset of nuclei. DNA-FISH was carried out to distinguish between these possibilities since it affords visualization of the locus in individual nuclei.
Bacterial artificial chromosome (BAC) probes were used (Fig. 5A) to detect large-scale locus contractions that can impact the proximity of upstream V segments to the RC. A contracted state of the locus was evident in DN thymocytes (Fig. 5C and D) compared to the ProB cells, as reported earlier (32, 42). The degree of locus compaction visualized by FISH was similar between DN thymocytes derived from mice that bound CTCF at the CBS-ecto (TCR-ins/TCR-cas, Rag1 deficient) and control mice (+/TCR-cas, Rag1 deficient and TCR-mut/TCR-cas, Rag1 deficient). This was consistent with the 3C-qPCR analysis, which revealed an approximately 50% reduction in various interactions. Further, in the cumulative frequency plots (Fig. 5C), alleles from TCR-ins thymocytes closely followed the profile of thymocytes from control mice, suggesting that there was no dramatic loss in locus contraction in any subset of cells; the 50% reduction in spatial proximity of the RC with V segments, observed by 3C-qPCR, appeared to be distributed in the entire population of TCR-ins thymocytes.
FIG 5.
Influence of ectopic CBS on contraction of the TCRb locus. (A) Schematic diagram of the TCRb locus with the H19-ICR insertion showing V segments and recombination center and the BACs (306O13 and 421M9), labeled with Alexa Fluor 594 and Alexa Fluor 488, used as probes for 3D-FISH. (B) Confocal images from a representative nucleus (blue) showing the two TCRb alleles detected by the FISH-based signals. The distance between Alexa Fluor 594 (red) and Alexa Fluor 488 (green) FISH signals of an allele provides an estimate of its contraction. (C) Cumulative frequency distribution of the FISH signal distance in ProB cells and DN thymocytes of Rag1-deficient mice of specified genotypes. (D) Box plots showing median and 25th and 75th percentile values for the same data. Data have been collated from three independent experiments on cells with mice of the indicated genotypes. “n” represents the number of alleles analyzed. ProB cells (wt/wt, n = 436) and DN thymocytes (wt/wt, n = 562; TCR-ins/wt, n = 580; and TCR-mut/wt, n = 466) are represented. Significance was tested by one-way ANOVA, followed by a Tukey's post hoc test (*, P < 0.05; ns, not significant).
Taken together, the 3C-qPCR analysis and DNA-FISH analyses indicated that ectopic CTCF binding altered the TCRb locus organization, albeit not dramatically. Our data suggest that although CTCF may not be the only factor contributing to large-scale locus compaction, CTCF-mediated loops and the chromatin network they orchestrate can be critical in defining interactions between specific elements and determining the outcome of the recombination process.
Influence of altered chromatin organization on recombination efficiency.
Based on the 3C-qPCR, it was evident that interactions of regulatory elements critical for the activity of the RC were influenced by ectopic CTCF binding. We were curious to know whether this alters the recombination efficiency.
Reduced interaction of upstream V segments with the RC (Fig. 4) may predict their reduced usage for V-to-DJ recombination. This was observed during the previous functional analysis (30). The usage of V segments was analyzed in TCR-ins/TCRb-del mice, wherein the TCRb-del allele was unable to undergo V-to-DJ recombination. Hence, the altered usage of V segments was not informative about the altered efficiency of VDJ recombination. We argued that the relative efficiency of recombination between TCR-ins and wild-type TCRb alleles can be estimated if they are in direct competition, i.e., the paternal allele is competent to undergo VDJ recombination.
We analyzed thymocytes of heterozygous Rag1-sufficient mutant mice (TCR-ins/TCR-cas). Thus, in each cell, TCR-ins and wild type TCR-b alleles (TCR-cas) were expected to compete for VDJ recombination since they can support VDJ recombination (30). Thymocytes development in TCR-ins mutant mice was similar to control mice (Fig. 6B). Also, the transcriptionally activated state of V segments is a strong correlate of their usage for VDJ recombination (43). Germ line transcription of several upstream V segments (V4, V12.1, and V13.2) and V31 remained unaltered in TCR-ins (30; S. Shrimali et al., unpublished data). Thus, the transcriptional status of V segments did not appear to give any advantage or disadvantage to TCR-ins for VDJ recombination.
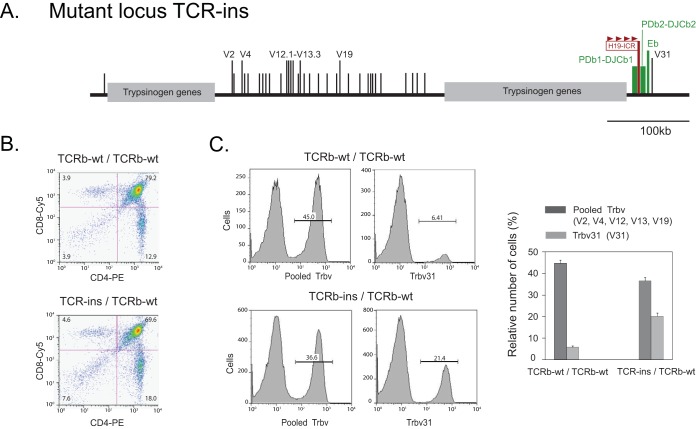
FIG 6.
Usage of Trbv segments for V-to-DJ recombination. (A) Schematic diagram of TCR-ins showing the insertion of ectopic CBS as H19-ICR within the RC. (B) Representative thymocyte developmental profiles, defined by cell surface markers CD4 and CD8, as analyzed by flow cytometry, in Rag1-sufficient mice which had maternally inherited wild-type TCRb or TCR-ins alleles. The paternal TCRb allele in each case was wild type. (C) Relative usage of Trbv segments for V-to-DJ recombination. Thymocytes were immunostained to detect the presence of CD4, CD8, and Trbv31 (V31) or CD4, CD8, and a pool of Trbv segments (V2, V4, V12, V13, and V19) on the cell surface. Histograms show representative fluorescence-activated cell sorting profiles, and numbers within them indicate the percentages of CD4-SP thymocytes positive for the V segment under investigation (Trbv31 or pooled Trbv). The bar graph shows the average (plus the SEM) for five such experiments.
Usage of various V segments in CD4-SP cells was discerned by detecting the expression of TCRb chains on cell surface (Fig. 6C and D). In control mice, V31 was utilized for V-to-DJ recombination in 6% of the CD4-SP cells and, together, the selected upstream V segments were used in 45% of the cells. In mutant mice, the usage of V31 was increased to 20% with a concomitant decrease in the usage of upstream V segments. Since TCR-ins exhibits enhanced usage of V31 for VDJ recombination (30), the skewed usage of V segments in heterozygous mice was informative about the choice of the allele (maternal TCR-ins or paternal wild type) that had undergone recombination.
Importantly, the usage of V31 in TCR-ins/TCR-cas mice increased ∼3.5-fold compared to +/TCR-cas control littermates (20% versus 6%). We had earlier observed an increase in usage of V31 in TCR-ins/TCRb-del mice by about 7- to 8-fold compared to +/TCRb-del control littermates (70% versus 9%) (30). Thus, in the presence of a competing wild-type allele (TCR-cas), the enhancement in V31 usage was half the enhancement in the absence of a competing allele (TCRb-del). This finding indicated that TCR-ins and TCR-cas alleles were equally efficient for VDJ recombination and competed effectively with each other. Clearly, the altered chromatin loopscape of TCR-ins (Fig. 3 and 4) did not lead to an enhanced or reduced VDJ recombination efficiency, even though distal V segments were hindered and V31 segments were preferred for V-to-DJ recombination on TCR-ins.
Our data suggest that in the wild-type TCRb locus, upstream V segments and V31 engage competitively with the RC. In TCR-ins, the access of upstream V segments to the RC is curtailed due to some reduction in long-range interactions. Also, the reconfiguration of the RC might be more conducive for the recombination of DJCb2 with V31. Consequently, V31 gets a competitive advantage over the upstream V segments for VDJ recombination. Since the usage of a V segment is a net result of the collation of productive VDJ recombination events during the given developmental window, the competitive advantage of V31 plausibly enhances its usage, even though its contact frequency with the RC is not enhanced in TCR-ins (Fig. 4).
DISCUSSION
The critical role of CTCF in chromatin organization in the mammalian genome is undisputed (6, 12). Our analysis of the CTCF-based chromatin loops of the TCRb locus identifies possible roles for specific CTCF binding sites in the regulation of transcription and recombination. Importantly, even a relatively modest perturbation of the CTCF-based long-range chromatin loop organization drastically influenced the regulation of VDJ recombination.
All structural and regulatory components of TCRb locus are encompassed within a single topologically associated domain (TAD) (44). Hence, all of the CTCF-cohesin binding sites are relevant for intralocus organization and regulation. Each one of them also exhibited poly(ADP-ribosyl)ation, which is important for chromatin looping interactions (36). It is likely that some or all CBS of other AgR loci exhibit poly(ADP-ribosyl)ation, an aspect not yet reported.
We observed highest contact frequency between CBS that flank the RC. These CBS also interacted with the CBS located more than 400 kb upstream, in the vicinity of the V segments. In contrast, the contact frequencies among CBS located interspersed in the V segment domain were substantially lower, suggesting that they are not spatially clustered and/or that their interactions are very dynamic. Hence, it appears that CBS-D/CBS-E interaction contributes to the establishment and/or stabilization of the RC to which the upstream V segments are recruited by the interspersed CBS. The comparable contact frequency of CBS-E with CBS-A, CBS-B, and CBS-C suggests that upstream CBS interact with the RC in a dynamic and stochastic manner. The orientations of various CBS conform to this overall interaction pattern. However, CBS flanking the RC are oriented in tandem, and hence their interaction is likely to be more dynamic, as suggested for other loci (5, 45). The observed strong 3C-qPCR signal may be a result of this interaction achieved in a large majority of cells and/or stabilization by other thymocyte-specific and Eb-recruited proteins. In addition, CBS-5′PC is also in the reverse orientation. The interaction of CBS-5′PC with CBS-B or CBS-E was not very high, and it is likely that the tethering function of 5′PC (32) involves some additional molecular determinants beyond CTCF. A map of the CBS interaction profiles sets the stage for investigating in the future whether CBS-E, CBS-D, and CBS-5′PC cooperate or compete to engage with upstream CBS and how these interactions are regulated.
Ectopic CBS effectively competed with CBS-D for interaction with CBS-E. A reduced CBS-D/CBS-E interaction accompanied the functional exclusion of PDb1-DJCb1 region from the RC. This was evidenced by a severe reduction in Eb-PDb1 looping interaction, as well as by a lack of chromatin accessibility and an enormous decrease in the usage of the DJCb1 cluster in VDJ recombination (30, 37). The competitive advantage of CBS-ecto could be due to the convergent orientation of the CBS-ecto with respect to CBS-E augmented by the clustering of four CBS on H19-ICR. Nonetheless, a functional RC, as efficient for VDJ recombination as the RC in the wild-type allele, was organized by CBS-ecto and CBS-E.
Endogenous CBS-mediated interactions emerged to be important to facilitate V-to-RC interactions at TCRb as at other larger loci such as Igh (23). They were significantly reduced by ectopic CTCF binding. The extent of reduction in CBS interactions in TCR-ins (about 50%) was remarkably similar to the reduction in the V-to-RC interactions. The inability of the RC of TCR-ins to interact efficiently with upstream V segments indicates that CBS-ecto could not functionally replace CBS-D and its flanking region for this aspect. It is likely that CBS-D and/or CBS-E and their flanks bind additional, currently unidentified factors that stabilize the V-RC interactions.
The reduced proximity between upstream V segments and RC was consistent with, and partially explained, the most interesting outcome of the ectopic CTCF-based perturbation of the TCRb locus, i.e., the altered choice of V segments for VDJ recombination (30). Interestingly, even though the upstream V-to-RC spatial proximity was reduced only by about 50% in TCR-ins, the reduction in upstream V-to-DJ recombination was dramatic (30). Conversely, usage of downstream V31 was enormously enhanced without any change in its spatial proximity to the RC. This suggested that in addition to proximity, the precise relative configuration of the chromatin loop encompassing the RC and the V segments in TCR-ins was influenced by the altered CBS interactions and impacted RSS-mediated recombination. Based on functional analysis, we have earlier proposed that CTCF can both facilitate and inhibit RSS-mediated recombination (30, 37). Our 3C-qPCR-based analysis of the chromatin interactions in wild-type and mutant TCRb alleles supports this contention. In close parallel to our observations, insertion of a single ectopic CBS at the TCRa/d locus between Trdd2 and Trdv5 led to a reduced recombination between them concomitant with their segregation into separate chromatin loops (46). How segregation of RSS into different chromatin loops influences their synapsis remains elusive. However, an analysis of CBS contacts at the b-globin locus, in conjunction with thermodynamic considerations, suggested a significant impact of CTCF-based chromatin loops on the collision dynamics between LCR and target genes (47). By analogy, in the context of VDJ recombination, “collision dynamics” between RC and V segments are likely to influence synapsis between the RSS of recombining segments and influence recombination.
Notably, convergent CBS and tandem CBS fold the intervening chromatin differentially (48). Although tandem CBS (CBS-D/CBS-E) can create a coiled loop in the wild-type locus, convergent CBS (CBS-ecto/CBS-E) are likely to generate a stem-loop in TCR-ins, leading to differences in the relative configuration and/or dynamics of chromatin loops. Although it is not possible to visualize chromatin at this scale, mathematical simulations show that contact between two sites (e.g., CBS) on a polymer (e.g., chromatin) influences the interactions among elements located within and in the vicinity of the loop (49, 50). A stable loop sterically excludes interactions with the rest of the polymer. Hence, plausibly, a smaller and/or differently configured chromatin loop of TCR-ins RC is less flexible and conformationally restrictive. Its inability to align appropriately with the juxtaposed chromatin loop encompassing the upstream V segments may reduce the ability of V segments to gain spatial proximity to the RC in TCR-ins and/or prevent synapsis. This suggests that the CBS flanking the RC not only establish/stabilize the RC but are possibly involved in configuring the specific loops that facilitate the RSS-mediated synapsis and recombination. Since CBS flank the RC of all AgRs, CBS direction-dependent chromatin folding might define the dynamics of RC at all AgR loci.
The reduced availability of upstream V segments to recombine with DJCb2 region in TCR-ins, as well as an altered RC that is more conducive for V31 recombination with DJC2, is likely to give a competitive advantage to V31 for the usage in VDJ recombination, even in the absence of any enhancement in its contact frequency with the RC. The relative contributions of the two factors cannot be ascertained.
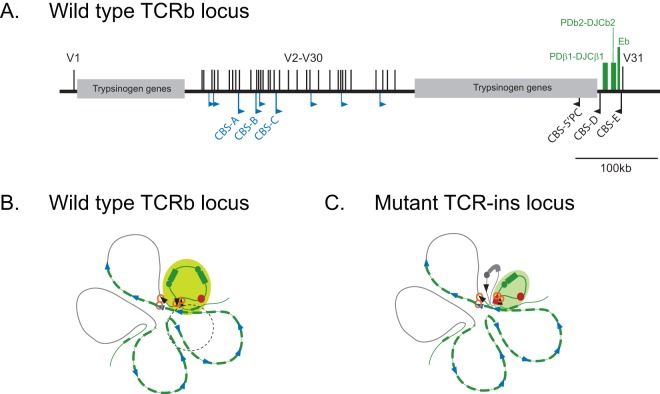
More recently, a CBS orientation-dependent chromatin loop organization was suggested to arise due to chromatin extrusion (11). It is interesting to examine the intrachromosomal interactions at TCRb locus in this context (Fig. 7). Extrusion of chromatin might start at various sites across the locus, and the presence of multiple CBS in the forward orientation, interspersed with V segments, and a few CBS in a reverse orientation near the RC, may generate an ensemble of chromatin loops in individual DN thymocytes. A chromatin loop domain, established by the extrusion of chromatin between CBS-E and CBS-D encompassing PDb1-DJCb1-PDb2-DJCb2 and Eb, would facilitate Eb-based activation of the RC. Depending on the occupancy of CBS by CTCF and the dynamics of the extrusion complexes, larger chromatin loops, encompassing a few V segments and the RC, might also be established. We suggest that the various CBS at TCRb might act as “punctuating marks” of various strengths that act stochastically during the dynamic process of extrusion. Consequently, a few V regions might be dynamically located in spatial proximity to the RC and/or within the same extruded chromatin domain and hence be used for VDJ recombination. Various CBS orientations and their interactions observed during this analysis are consistent with the possibility of spatial proximity of V segments to the RC brought about by chromatin extrusion.
FIG 7.
Model depicting the chromatin loop configuration of the TCRb locus that might arise by chromatin extrusion in wild-type and mutant loci. (A) Linear map of the TCRb locus showing the relative positions of the RC, V segments, and a subset of endogenous CBS of TCRb locus. (B) 2D depiction of the chromatin loop configuration of the wild-type locus. From the ensemble of possible combinations that can organize the CTCF-dependent loops, one conformation is compared wherein two extrusion complexes have been considered. A CBS-D and CBS-E (black arrowheads)-based chromatin loop encompasses Eb (red circle) and promoters PDb1 and PDb2 (green circles), each linked to a DJC cluster (green rectangles). Eb-based activation of this domain creates the recombination center (yellow-green circle), leading to D-to-J recombination. The second extrusion complex involves CBS-5′PC and any one of the CBS (blue arrowheads) located in the domain (green lines) encompassing V segments. Consequently, depending on the CBS that halts the extrusion, some V segments are extruded away, while a few are brought to the vicinity of the RC. The flexibility of the chromatin loop of the RC (depicted as a black dashed circle), as well as of the chromatin loop encompassing the V segments, allow alignment of the RSS elements leading to V-to-DJ recombination. Extrusion of V segments proximal to the RC is likely to favor the utilization of distal V segments. Domains encompassing inert trypsinogen genes are indicated as gray lines. (C) 2D depiction of the chromatin loop configuration as altered in TCR-ins. Due to the interaction of CBS-ecto (red triangles) with CBS-E, the Eb-activated RC is smaller, as well as oriented differently (shaded green oval), and/or is less flexible. The PDb1-DJCb1 region (gray circles and rectangle) is excluded from the RC. The other extrusion process that brings V to the RC proximity may or may not be affected, depending on the dynamics of extrusion. However, due to the altered orientation and/or flexibility of the RC, the upstream V segments, despite reasonable proximity to the RC, are not able to align appropriately for V-to-DJ recombination. The spatial proximity of V31 (a green dash downstream of Eb) does not change and, being suitably aligned, it is used for V-to-DJ recombination in the wild-type TCR-b locus, as well as TCR-ins. In each case, the loops are not in proportion to the linear span of DNA and, for clarity, the RC is shown to be significantly larger. Also, additional proteins that may stabilize some of the looped configurations are not depicted since they have not yet been identified.
In this scenario, ectopic CBS with a forward orientation in TCR-ins might halt chromatin extrusion (Fig. 7C) and restrict Eb-based activation to PDb2-DJCb2, rendering PDb1-DJCb1 inactive as was observed (37). Also, consistent with earlier observations (43), the distance of CBS from the V segment is not likely to govern its usage in a correlative manner. However, interference in the CBS-based chromatin organization is expected to impact the usage of V segments for VDJ recombination. This was observed for TCR-ins (30). The usage of upstream V segments was reduced in TCR-ins, as would be predicted if the ectopic CBS halted the extrusion of upstream chromatin and thus interfered in enhancing the spatial proximity of V segments to the RC. Thus, our findings support the relevance of extrusion-based chromatin organization (11) in the context of VDJ recombination. However, several details regarding chromatin extrusion remain to be elucidated (12). Their elucidation, as well as the impact of altered orientations of CBS at AgR loci, will help to further define the way(s) by which CTCF can contribute to regulation of VDJ recombination at TCRb and other AgR loci.
In conclusion, our results support a role for CTCF-mediated chromatin interactions in the regulation of the TCRb locus. The influence of CBS orientations was evident in their interactions and suggests the importance of CBS orientations for the regulation of the chromatin loopscape in defining functional domains that support transcription, as well as VDJ recombination. However, analysis of the inversion of endogenous CBS orientations at TCRb locus and other AgR loci by genetic manipulations will be required to confirm this. Ectopic CTCF binding led to modest alterations in chromatin organization but dramatically influenced transcription and VDJ recombination. This suggests that, apart from regulating the proximity between distantly located genomic elements, CTCF binding also critically defines the finer configuration of the chromatin loops, whose size, orientation, and dynamics can potentially modulate transcription and VDJ recombination at AgR.
Our observations have significant implications beyond the regulation of AgR loci since they indicate that even mild changes in the CTCF-based long-range chromatin architecture, as well as localized chromatin folding, which may arise due to enhanced or lowered CTCF binding at specific CBS for a variety of reasons, can have profound functional consequences for gene expression. This is particularly relevant for developmental decisions, as well as for cellular functions, since CTCF-based chromatin architecture is important for the transcriptional regulation of several genes in metazoan genomes.
MATERIALS AND METHODS
Mice.
Mice were used as approved by the Institutional Animal Ethics Committee of National Institute of Immunology, New Delhi, India. C57BL6, Rag1-deficient, and OT2-Tg mice were procured from Jackson Laboratories, USA, and maintained as inbred strains. Mutant mice, carrying TCR-ins, TCR-mut, and TCR-cas alleles, were derived as described previously (30, 37) and bred with Rag1-deficient mice (B6.129S7-Rag1tm1Mom/J; Jackson Laboratories) to get the mutant alleles in a Rag1-deficient background.
Cells.
Ex vivo DN thymocytes were isolated from 4- to 6-week-old mice of different genotypes, as mentioned above. ProB cells were derived from the bone marrow of Rag1-deficient mice. ProB cells, stained with anti-B220-biotinylated antibody (clone RA3-6B2; BD Biosciences), were purified using streptavidin microbeads and MACS separation unit (catalog no. 130-048-102 and 130-042-201; Miltenyi Biotec) according to the manufacturer's protocol.
ChIP-qPCR assay.
ChIP assays were performed with anti-CTCF (catalog no. 07-729; Millipore) and anti-Rad-21 and anti-PAR (catalog no. ab992 and ab14459; Abcam) antibodies. IgG (catalog no. Pp64B; Millipore) served as a negative control for each. Briefly, ex vivo thymocytes or pro-B cells were used for ChIP as described previously (51). For anti-PAR ChIP, a Diagenode kit (catalog no. C01010072) was used. ChIP samples were analyzed by PCR at control regions (see Fig. S1 in the supplemental material) before proceeding to ChIP-qPCR. ChIP-qPCR analysis was carried out on ABI Prism SDS7000 system (Applied Biosystems) using SYBR green, and relative enrichments were calculated as described previously (51). Primer sequences are available upon request.
3C-qPCR and allele-specific 3C-qPCR assays.
3C assays were performed and analyzed as described previously (52). Briefly cells used in study were cross-linked, lysed to release nuclei, and digested overnight with 800 to 1,000 U of HindIII. Digestion efficiency was calculated as described previously (52). To compare the digestion efficiency of maternally inherited TCR-ins to the TCRb wild-type allele, allele-specific primers were used (see Fig. S2 in the supplemental material). Chromatin digested with high efficiency was diluted, ligated, and used for DNA purification to generate the 3C library. 3C libraries obtained from DN and ProB cells were analyzed by a TaqMan probe-based 3C-qPCR assay using the standard curve method on an ABI Prism SDS7000 system (Applied Biosystems). A standard curve was generated using BAC clones encompassing the TCRb locus (RP23-306O13, RP23-354-C19, RP24-322P20 or RP23-342K14, and RP23-421M9), the Igf2/H19 locus (RP24-251H17), and the Ercc locus (MSMG01-426I12) and relative cross-linking frequencies (RCFs) were calculated using the enhancer-promoter interactions at Ercc as a control. Allele-specific 3C-qPCR analysis, as described earlier (37), relied on single nucleotide differences between Mus musculus domesticus and Mus castaneus castaneus. Primers were designed to specifically amplify the domesticus DNA. Allele-specific amplification was verified for each amplicon (see Table S2 in the supplemental material). Probe and primer sequences are available upon request.
3D DNA-FISH and analysis.
FISH probes (BACs RP23-306O13, and RP23-421M9) were labeled with Alexa Fluor 594 and Alexa Fluor 488, respectively, using FISHTAG DNA green and red kits (catalog no. F32947 and F32949; Invitrogen) by nick translation. Thymocytes and ProB cells were resuspended in phosphate-buffered saline and used for three-dimensional (3D) DNA-FISH analysis, as described previously (53). Posthybridization analysis was carried out by confocal microscopy on a Leica TCS-SP5 II system. The images were acquired with a 63×, 1.4-numerical-aperture oil immersion objective lens at a 256-by-256 resolution. Optical sections separated by 0.21 μm were collected, and only cells with FISH signals from both alleles were analyzed. 3D constructs and distance measurements were made using Imaris 7.5 software (Bitplane).
VDJ recombination assay.
Ex vivo thymocytes were stained with anti-CD4 (clone RM4-5), anti-CD8 (clone 53-6.7), and anti-Trbv31–FITC (clone 14-2) antibodies or a cocktail of six FITC-conjugated antibodies (anti-Trbv2 [clone KT4], anti-Trbv4 [clone B21.5], anti-Trbv12.1/12.2 [clone MR9-4], anti-Trbv13.1/13.2 [clone MR5-2], anti-Trbv13.3 [clone 1B3.3], and anti-Trbv19 [clone RR4-7]) from the mouse Vb TCR screening panel (catalog no. 557004; BD Bioscience). Flow cytometric analysis was performed on a BD FACSCalibur apparatus.
Supplementary Material
ACKNOWLEDGMENTS
BAC clone MSMG01-426I12 was provided by RIKEN BRC through the National Bio-Resource Project of MEXT, Japan. All other BAC clones were procured from BACPAC Resources, CHORI, USA. We thank Ramesh Yadav and Inderjeet Singh for technical assistance and Neelaram for help with microscopy.
This study was supported by intramural grants at NII and extramural grants to M.S. from Department of Biotechnology, Government of India. P.R. thanks CSIR, India, for the Senior Research Fellowship.
P.R. and A.K. performed ChIP analysis. P.R. and M.J. carried out 3C-qPCR analysis. A.S. carried out the FISH analysis. M.S. conceived of the study, designed experiments, and prepared the manuscript. All authors contributed to the design of the experiments and the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00557-16.
REFERENCES
- 1.Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 2.Bell AC, West AG, Felsenfeld G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387–396. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 3.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 4.Merkenschlager M, Odom DT. 2013. CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 5.Ong CT, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghirlando R, Felsenfeld G. 2016. CTCF: making the right connections. Genes Dev 30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holwerda SJ, de Laat W. 2013. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci 368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W. 2015. CTCF binding polarity determines chromatin looping. Mol Cell 60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A 112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merkenschlager M, Nora EP. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 13.Degner SC, Wong TP, Jankevicius G, Feeney AJ. 2009. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol 182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih HY, Verma-Gaur J, Torkamani A, Feeney AJ, Galjart N, Krangel MS. 2012. TCRa gene recombination is supported by a TCRa enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci U S A 109:E3493–E3502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. 2010. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med 207:2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature 477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeiro dA, Hendriks RW, Stadhouders R. 2015. Dynamic control of long-range genomic interactions at the immunoglobulin κ light-chain locus. Adv Immunol 128:183–271. doi: 10.1016/bs.ai.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schatz DG, Ji Y. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol 11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 20.Proudhon C, Hao B, Raviram R, Chaumeil J, Skok JA. 2015. Long-range regulation of V(D)J recombination. Adv Immunol 128:123–182. doi: 10.1016/bs.ai.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birshtein BK. 2012. The role of CTCF binding sites in the 3′ immunoglobulin heavy chain regulatory region. Front Genet 3:251. doi: 10.3389/fgene.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, Werling U, Hou H Jr, Will B, Steidl U, Scharff M, Edelman W, Feeney AJ, Birshtein BK. 2012. Germline deletion of Igh 3′ regulatory region elements hs5, 6, and 7 (hs5-7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol 188:2556–2566. doi: 10.4049/jimmunol.1102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossen C, Mansson R, Murre C. 2012. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol 30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 24.Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. 2011. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. 2011. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell 147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerasimova T, Guo C, Ghosh A, Qiu X, Montefiori L, Verma-Gaur J, Choi NM, Feeney AJ, Sen R. 2015. A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation. Genes Dev 29:1683–1695. doi: 10.1101/gad.263871.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW. 2015. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell 163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L, Frock RL, Du Z, Hu J, Chen L, Krangel MS, Alt FW. 2016. Orientation-specific RAG activity in chromosomal loop domains contributes to TCRd V(D)J recombination during T cell development. J Exp Med 213:1921–1936. doi: 10.1084/jem.20160670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikes ML, Oltz EM. 2012. Genetic and epigenetic regulation of TCRb gene assembly. Curr Top Microbiol Immunol 356:91–116. doi: 10.1007/82_2011_138. [DOI] [PubMed] [Google Scholar]
- 30.Shrimali S, Srivastava S, Varma G, Grinberg A, Pfeifer K, Srivastava M. 2012. An ectopic CTCF-dependent transcriptional insulator influences the choice of Vβ gene segments for VDJ recombination at TCRβ locus. Nucleic Acids Res 40:7753–7765. doi: 10.1093/nar/gks556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majumder K, Bassing CH, Oltz EM. 2015. Regulation of TCRb gene assembly by genetic, epigenetic, and topological mechanisms. Adv Immunol 128:273–306. doi: 10.1016/bs.ai.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Majumder K, Koues OI, Chan EA, Kyle KE, Horowitz JE, Yang-Iott K, Bassing CH, Taniuchi I, Krangel MS, Oltz EM. 2015. Lineage-specific compaction of TCRb requires a chromatin barrier to protect the function of a long-range tethering element. J Exp Med 212:107–120. doi: 10.1084/jem.20141479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao L, Zhou M, Cui Y. 2008. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res 36:D83–D87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg AP, Lobanenkov V, Klenova E, Ohlsson R. 2004. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet 36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 36.Ong CT, Van Bortle K, Ramos E, Corces VG. 2013. Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila. Cell 155:148–159. doi: 10.1016/j.cell.2013.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varma G, Rawat P, Jalan M, Vinayak M, Srivastava M. 2015. Influence of a CTCF-dependent insulator on multiple aspects of enhancer-mediated chromatin organization. Mol Cell Biol 35:3504–3516. doi: 10.1128/MCB.00514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10:853–856. doi: 10.1016/S0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 39.Schoenherr CJ, Levorse JM, Tilghman SM. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet 33:66–69. [DOI] [PubMed] [Google Scholar]
- 40.Gaszner M, Felsenfeld G. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 41.Wilson A, Marechal C, MacDonald HR. 2001. Biased V beta usage in immature thymocytes is independent of DJ beta proximity and pT alpha pairing. J Immunol 166:51–57. doi: 10.4049/jimmunol.166.1.51. [DOI] [PubMed] [Google Scholar]
- 42.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. 2007. Reversible contraction by looping of the TCRa and TCRb loci in rearranging thymocytes. Nat Immunol 8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 43.Gopalakrishnan S, Majumder K, Predeus A, Huang Y, Koues OI, Verma-Gaur J, Loguercio S, Su AI, Feeney AJ, Artyomov MN, Oltz EM. 2013. Unifying model for molecular determinants of the preselection Vβ repertoire. Proc Natl Acad Sci U S A 110:E3206–E3215. doi: 10.1073/pnas.1304048110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, Michalski P, Piecuch E, Wang P, Wang D, Tian SZ, Penrad-Mobayed M, Sachs LM, Ruan X, Wei CL, Liu ET, Wilczynski GM, Plewczynski D, Li G, Ruan Y. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Zhao L, Alt FW, Krangel MS. 2016. An ectopic CTCF binding element inhibits TCRD rearrangement by limiting contact between Vδ and Dδ gene segments. J Immunol 197:3188–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Junier I, Dale RK, Hou C, Kepes F, Dean A. 2012. CTCF-mediated transcriptional regulation through cell type-specific chromosome organization in the beta-globin locus. Nucleic Acids Res 40:7718–7727. doi: 10.1093/nar/gks536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. 2014. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 36:163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doyle B, Fudenberg G, Imakaev M, Mirny LA. 2014. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput Biol 10:e1003867. doi: 10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh V, Srivastava M. 2008. Enhancer blocking activity of the insulator at H19-ICR is independent of chromatin barrier establishment. Mol Cell Biol 28:3767–3775. doi: 10.1128/MCB.00091-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 53.Deligianni C, Spilianakis CG. 2012. Long-range genomic interactions epigenetically regulate the expression of a cytokine receptor. EMBO Rep 13:819–826. doi: 10.1038/embor.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.