ABSTRACT
The Athabasca oil sand deposit is one of the largest single oil deposits in the world. Following surface mining, companies are required to restore soil-like profiles that can support the previous land capabilities. The objective of this study was to assess whether the soil prokaryotic alpha diversity (α-diversity) and β-diversity in oil sand soils reconstructed 20 to 30 years previously and planted to one of three vegetation types (coniferous or deciduous trees and grassland) were similar to those found in natural boreal forest soils subject to wildfire disturbance. Prokaryotic α-diversity and β-diversity were assessed using massively parallel sequencing of 16S rRNA genes. The β-diversity, but not the α-diversity, differed between reconstructed and natural soils. Bacteria associated with an oligotrophic lifestyle were more abundant in natural forest soils, whereas bacteria associated with a copiotrophic lifestyle were more abundant in reconstructed soils. Ammonia-oxidizing archaea were most abundant in reconstructed soils planted with grasses. Plant species were the main factor influencing α-diversity in natural and in reconstructed soils. Nitrogen deposition, pH, and plant species were the main factors influencing the β-diversity of the prokaryotic communities in natural and reconstructed soils. The results highlight the importance of nitrogen deposition and aboveground-belowground relationships in shaping soil microbial communities in natural and reconstructed soils.
IMPORTANCE Covering over 800 km2, land disturbed by the exploitation of the oil sands in Canada has to be restored. Here, we take advantage of the proximity between these reconstructed ecosystems and the boreal forest surrounding the oil sand mining area to study soil microbial community structure and processes in both natural and nonnatural environments. By identifying key characteristics shaping the structure of soil microbial communities, this study improved our understanding of how vegetation, soil characteristics and microbial communities interact and drive soil functions.
KEYWORDS: biodiversity, boreal forest soil, microbial community, nitrogen deposition, oil sands, plant community, soil restoration, soil microbiology
INTRODUCTION
The Athabasca oil sand deposit, located in the boreal forests of northern Alberta, is part of the largest single oil deposit in the world, with proven reserves of 166 billion barrels of bitumen and covering 142,200 km2 (1). Most of the bituminous sands (80%) can be extracted using in situ recovery methods (i.e., steam injection wells), but 20% of the resource, spreading over 4,800 km2 of land, is shallow and can only be recovered through open-pit mining (2, 3). To date, about 895 km2 of land has been disturbed by oil sand mining activity (1). Following surface mining, companies are required to restore soils that can achieve equivalent land capability (4, 5). After soil reconstruction, the area is revegetated. When reclamation in the area began in the 1980s, revegetation predominantly focused on erosion control and used both native and introduced grasses and shrubs. However, more recent revegetation practices use native tree species such as jack pine, white and black spruce, and aspen and understory shrubs such as blueberry and willow to reestablish a boreal forest plant community. Differences in soil organic matter composition, soil available N, and microbial communities between reconstructed oil sand soils and natural boreal forest soils of northern Alberta have been previously identified (6–9). As such, it is unlikely that these reconstructed forest soils will exactly mirror preexisting boreal forest soils, and novel soil ecosystems (10, 11) will most probably arise from the reconstruction efforts. Soil restoration efforts aiming at reestablishing soil functions, and chiefly nutrient cycling, rather than simply trying to replicate structural qualities of the previous soil ecosystem are seen as key to ensuring the long-term sustainability of reclaimed boreal forest landscapes (12).
From decomposition to nutrient cycling and greenhouse gas regulation, microbial communities are responsible for a plethora of functions in soils. Although some broad, or universal, biochemical processes such as glucose metabolism are highly conserved across living organisms, other processes such as nitrogen fixation or nitrification are carried out by a limited number of species spread over multiple phyla (13, 14). For these specialized processes, high microbial diversity can increase the probability that these functions will be retained in the event that a species is affected by disturbances (15).
The main phyla of bacteria found in boreal forest soils are Proteobacteria, followed by Bacteriodetes, Acidobacteria, Actinobacteria, Firmicutes, Planctomycetes, and Verrucomicrobia (16, 17). Environmental factors such as pH, moisture, base cation abundance, as well as the quality of available carbon, all influence soil microbial communities (18–21). For example, soils with near-neutral pH tend to have the highest microbial diversity and richness (18). Tree species affect the structure and abundance of soil microbial communities by influencing the chemical nature of litter and exudates, as well as mycorrhizal fungal symbionts (22–24). For example, in a common-garden experiment, Thoms et al. found that microbial communities varied with different tree diversity levels (25). However, the differentiation was related to the presence of specific tree species (Tilia and Acer), both of which produce base-rich litter; this appeared to exert the greatest influence on microbial communities.
The main natural disturbance in boreal forest ecosystems is wildfire (26, 27). Fire can cause shifts in soil microbial communities (28–30). Changes in soil microbial communities have been be linked to both direct and indirect effects of fire. Lysis of microbial cells and death of plant roots during fire causes changes in microbial communities; microorganisms especially sensitive to high temperature (i.e., Nitrobacter spp. and fungi) are more prone to lysis during fire (29). Postfire changes in soil characteristics (i.e., increased pH and temperature, reduced moisture, etc.) modify the structure of soil microbial communities. The increased soil pH 10 days after a prescribed fire in a Douglas fir forest in British Columbia (Canada) was thought to be the main factor responsible for the reduced abundance of Gram-positive bacteria, Gram-negative bacteria, actinomycetes, and arbuscular mycorrhizae, as well as the total bacterial biomass in soils after a fire (30). Ball et al. reported that forest fires increased charcoal content, gross nitrification rates, and the abundance of ammonia-oxidizing bacteria (28). Moreover, changes in the structure of bacterial community were still measurable more than 14 years after the original fire disturbance.
Previous studies contrasting microbial communities in reconstructed oil sand soils, ranging in age from 5 to 35 years, and natural boreal forest soil used denaturing gradient gel electrophoresis (DGGE) and phospholipid fatty acid (PLFA) profiling and showed differences in microbial community structure (9, 31). Although DGGE and PLFA do not allow for in-depth (species-level) characterization of microbial communities, it was possible to detect a general increase in Gram-negative bacteria in reconstructed soils compared to natural soils. Dimitriu and Grayston showed that abiotic variables, such as pH and soil moisture, rather than plant cover, were the main drivers of community structure in both reconstructed and natural soils (31). These previous studies in oil sands compared microbial communities in the top 15 cm of soils regardless of the presence or absence of a forest floor. It could be predicted that samples containing forest floor material, with high carbon and nutrient content, would have de facto greater microbial biomass. However, Dimitriu and Grayston found no differences in microbial community richness and structural diversity between samples containing exclusively mineral soil or exclusively forest floor material (31).
Given the pivotal role of microorganisms in soil functioning, the objective of this study was to assess whether prokaryotic diversity and structure (i.e., alpha diversity [α-diversity] and β-diversity) in oil sand soils reconstructed 20 to 30 years previously were similar to those found in natural boreal forest soils subjected to wildfire disturbance at approximately the same time. Specifically, we evaluated prokaryotic diversity and community structure in the top 12 cm of mineral soils of 20- to 30-year-old reconstructed and fire-disturbed boreal forest soils using massively parallel sequencing of 16S rRNA genes. In addition, we assessed the influence of three vegetation treatments (coniferous trees, deciduous trees, and grasses) used during reclamation, as well as the influence of understory plant species and soil chemical and physical characteristics on prokaryotic α- and β-diversity. We hypothesized (i) that α-diversity will be higher in natural soils, (ii) that β-diversity will differ between reconstructed and natural soils, and (iii) that pH and soil moisture will be the main drivers of prokaryotic α- and β-diversity.
RESULTS
Soil chemical, physical, and biological characteristics.
Microbial biomass carbon was significantly greater [F(3,16) = 4.613; P = 0.0165] in reconstructed soils planted with deciduous trees than in natural forest soils (Table 1). Soil nitrogen concentration [F(3,16) = 7.183; P = 0.0029], carbon concentration [F(3,16) = 8.649; P = 0.0012], C:N ratio [F(3,16) = 25.457; P = 2.48 × 10−6], and nitrogen deposition rates [F(3,16) = 52.184; P = 1.75 × 10−8] were all substantially higher in reconstructed soils. Soil pH [F(3,16) = 8.085; P = 0.0017] and soil moisture [F(3,16) = 2.608; P = 0.0874] were higher in reconstructed soils under grasses than in natural soils. The clay content of soil was similar at all sites [F(3,16) = 1.323; P = 0.3016] (Table 1).
TABLE 1.
Selected physical, chemical, and biological characteristics of studied soils
| Treatment | Avg ± SDa |
|||||||
|---|---|---|---|---|---|---|---|---|
| pH | Soil moisture (%) | Total nitrogen (%) | Total carbon (%) | C:N ratio | Microbial biomass carbon (g C kg soil−1) | Clay content (%) | Nitrogen deposition (kg N ha−1 yr−1) | |
| Coniferous | 6.5 ± 0.2AB | 20.2 ± 12.1AB | 0.35 ± 0.22AB | 7.6 ± 4.7A | 21.9 ± 2.5A | 1.31 ± 0.37AB | 2.61 ± 0.52A | 12.5 ± 2.5A |
| Deciduous | 6.4 ± 0.3AB | 22.9 ± 6.3AB | 0.41 ± 0.14A | 7.6 ± 2.2A | 19.0 ± 2.9AB | 1.53 ± 0.84A | 1.87 ± 0.90A | 14.0 ± 1.4A |
| Grassland | 7.0 ± 0.3A | 25.3 ± 8.1A | 0.39 ± 0.13A | 7.5 ± 1.8A | 19.8 ± 2.1A | 0.93 ± 0.44AB | 1.59 ± 0.50A | 12.5 ± 0.0AB |
| Natural | 6.1 ± 0.3B | 12.1 ± 1.4B | 0.04 ± 0.008B | 0.39 ± 0.13B | 10.1 ± 1.8B | 0.53 ± 0.29B | 3.43 ± 2.69A | 3.0 ± 0.5B |
Uppercase characters indicate significant differences among treatments (α < 0.05).
Soil microbial community structure.
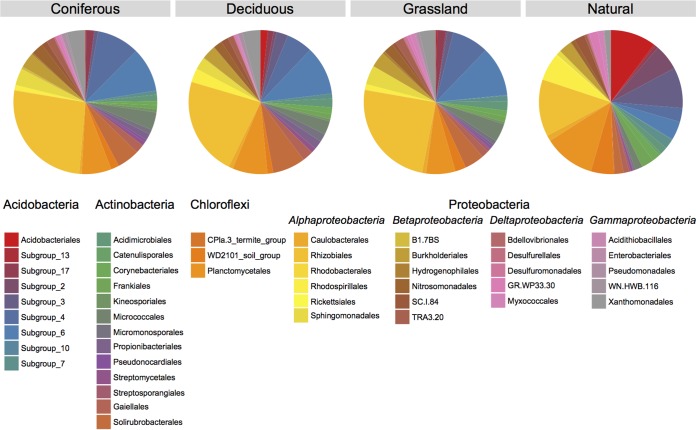
On average, 16,363 ± 6,035 reads per sample were kept after low-quality, ambiguous, chimeric, singletons, and chloroplastic and mitochondrial reads were removed from the database. After subsampling the database to the lowest number of reads found in one sample (3,234 reads), reads were grouped in 181 ± 50 operational taxonomic units (OTU) per sample on average (see Table S1 in the supplemental material). Rarefaction curves for each sample reached an asymptote at approximately 1,500 sequences (see Fig. S2 in the supplemental material). As such, the rarefaction curves suggest that the depth of sequencing was adequate. When the OTU were grouped at the order level, the most abundant orders in reconstructed soils were Rhizobiales (Alphaproteobacteria), followed by subgroup 6 (Acidobacteria), Planctomycetales (Planctomycetes), subgroup 4 (Acidobacteria), and Xanthomonadales (Gammaproteobacteria). The most abundant orders in natural soils were Rhizobiales, followed by Planctomycetales, Acidobacteriales (Acidobacteria), subgroup 3 (Acidobacteria), and Rhodospirillales (Alphaproteobacteria) (Fig. 1).
FIG 1.
Proportion of bacterial orders belonging to the most abundant phyla (>10% of the sequences) in reconstructed soils planted using coniferous, deciduous, and grass species and in natural forest soils.
α-Diversity.
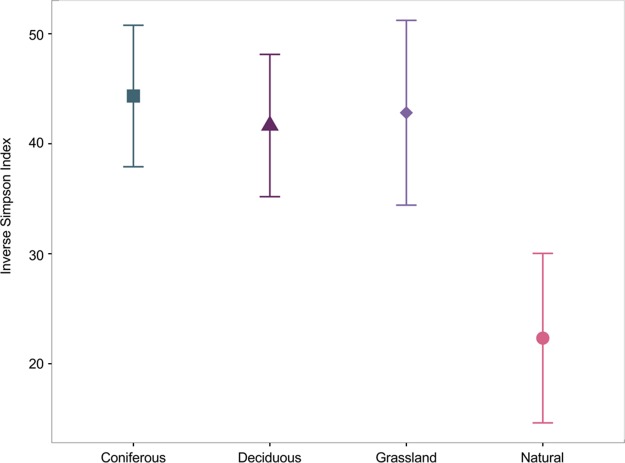
α-Diversity indices in reconstructed and natural soils were not significantly different [F(3,16) = 2.016; P = 0.1523], although reconstructed soils had a higher α-diversity (Fig. 2). The C:N ratio, the abundance of Pinus banksiana, Medicago sativa, Salix bebbiana, and Vaccinium myrtilloides significantly correlated with α-diversity. All variables were positively correlated with α-diversity except for P. banksiana abundance, which was negatively correlated with α-diversity (Table 2).
FIG 2.
Inverse Simpson index (average ± standard error) in reconstructed soils planted using coniferous, deciduous, and grass species and in natural forest soils.
TABLE 2.
Coefficients and significance of the multiple linear regression explaining α-diversity in the studied soilsa
| Variable | Coefficient (βi) | t statistic | Pb |
|---|---|---|---|
| C:N ratio | 1.25 | 1.80 | 0.0938† |
| Pinus banksiana | −0.48 | −3.17 | 0.0067** |
| Medicago sativa | 0.72 | 3.11 | 0.0076** |
| Salix bebbiana | 4.15 | 2.49 | 0.0266* |
| Vaccinium myrtilloides | 2.08 | 2.57 | 0.0224* |
| Intercept | 11.47 | 0.86 | 0.4064 |
The regression model used was as follows: α-diversity = β1 C:N ratio – β2 Pinus banksiana + β3 Medicago sativa + β4 Salix bebbina + β5 Vaccinium myrtilloides + β0 + εi. F-statistic = 9.83 on 5 and 14 degrees of freedom; adjusted R2 = 0.699; P = 0.0003393.
**, P < 0.01; *, P < 0.05; †, P < 0.1.
β-Diversity.
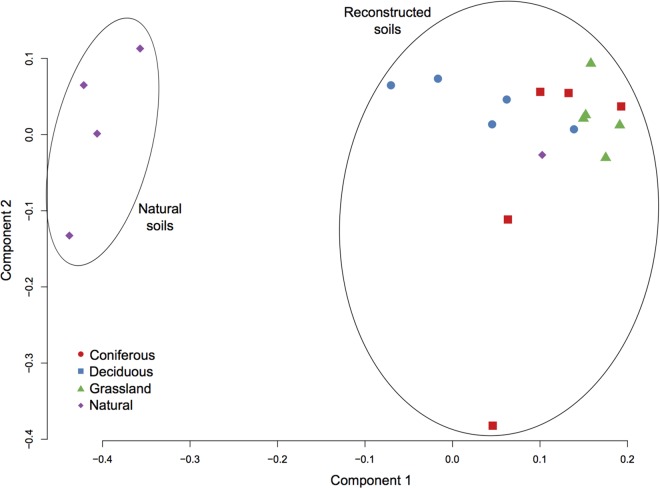
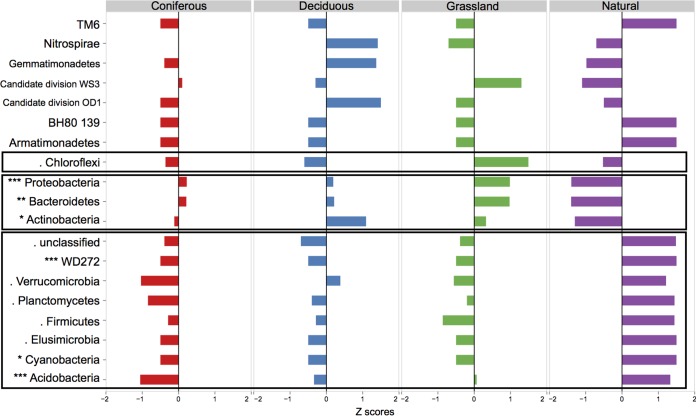
Microbial community structure differed between reconstructed and natural soils at the OTU level [F(3,16) = 7.358; P < 0.001] (Fig. 1 and 3). Within reconstructed soils, those planted with grasses harbored different communities than those planted with either deciduous [F(8,9) = 4.763; P = 0.005] or coniferous trees [F(8,9) = 1.915; P = 0.06] (Fig. 3). When the OTU were grouped at the phyla level, Proteobacteria, Bacteroidetes, and Actinobacteria were more abundant in reconstructed soils than in natural soils, whereas Acidobacteria, Cyanobacteria, Elusimicrobia, Firmicutes, Planctomycetes, Verrucomicrobia, and WD272 were less abundant in reconstructed soils. Finally, the phylum Chloroflexi was more abundant in reconstructed soils planted with grasses compared to reconstructed soils planted with trees and natural soils (Fig. 4 and Table 3; the F statistics and P values are presented in Table S2 in the supplemental material).
FIG 3.
Principal coordinate analysis (scaling 1) based on the OTU in reconstructed soils planted using coniferous, deciduous, and grass species and in natural forest soils.
FIG 4.
Z-scores of bacterial phyla in reconstructed soils planted with coniferous, deciduous, and grass species and in natural forest soils. The first rectangle from the bottom groups phyla that are more abundant in natural forest soils, the second rectangle groups phyla that are more abundant in reconstructed soils, and the third rectangle groups phyla that are more abundant in reconstructed soils planted with grasses (***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1).
TABLE 3.
Phyla and other divisions that are more abundant in either reconstructed soils, natural soils, or grassland soils
| Phylum | Division(s) |
|---|---|
| Abundant in reconstructed soils | |
| Actinobacteria | Class Thermoleophilia |
| Bacteroidetes | Classes Cytophagia, Sphingobacteriia |
| Proteobacteria | Classes Alphaproteobacteria, Gammaproteobacteria |
| Class Verrucomicrobiae (phylum Verrucomicrobia) | |
| Uncultured bacterial clones JG30.KF.CM66, JG37.AG.4; class Ktedonobacteria (all in phylum Chloroflexi) | |
| Abundant in natural soils | |
| Acidobacteria | Class Acidobacteria |
| Cyanobacteriaa | Class Melainabacteriaa |
| Elusimicrobiaa | Class Elusimicrobiaa |
| Firmicutesc | Class Bacillic |
| Planctomycetes | Class Phycisphaerae |
| Verrucomicrobia | Classes Spartobacteri, Verrucomicrobiaeb |
| Class Deltaproteobacteria (phylum Proteobacteria) | |
| Abundant in grassland soils | |
| Chloroflexi | Classes Anaerolineae,d Caldilineae,e Chloroflexia; uncultured bacterial clone KD496 |
Absent from reconstructed soils.
Absent from natural soils.
Absent from grassland soils.
Absent from natural soils and reconstructed soils planted with coniferous species.
Absent from natural soils and reconstructed soils planted with deciduous species.
Within phyla that were relatively more abundant in reconstructed soils, the bacterial classes Thermoleophilia, Cytophagia, Sphingobacteria, Alphaproteobacteria, and Gammaproteobacteria were still more abundant in reconstructed soils, but the class Deltaproteobacteria was more abundant in natural soils (Table 3 and see Fig. S3 in the supplemental material; the F statistics and P values are presented in Table S3).
Within phyla that were more abundant in natural soils, the bacterial classes Acidobacteria, Melainabacteria, Elusimicrobia, Bacilli, Phycisphaerae, and Spartobacteria were more abundant in natural soils, but the class Verrucomicrobiae was more abundant in reconstructed soils (Table 3 and see Fig. S4 in the supplemental material; the F and P values are presented in Table S3). The classes Melainabacteria and Elusimicrobia were absent from reconstructed soils and class Bacilli was absent from grassland soils (Table 3 and see Fig. S4 in the supplemental material; the F statistics and P values are presented in Table S3).
Within the phylum Chloroflexia, which was more abundant in grassland soils, the classes Anaerolinea, Caldilineae, and Chloroflexia were more abundant in grassland soils, but the class Ktedonobacteria was more abundant in natural soils (Table 3 and see Fig. S5 in the supplemental material; the F statistics and P values are presented in Table S3).
There were relatively fewer archaea in reconstructed soils planted with trees than in reconstructed soils planted with grass and natural forest soils [F(3,16) = 13.810; P = 0.00001] (see Fig. S6 in the supplemental material). The archaeal phylum Euryarchaeota was found only at one natural site (N1; see Fig. S7 in the supplemental material). The phylum Thaumarchaeota was found in both reconstructed soils and in natural forest soils, but Thaumarchaeota were significantly more abundant in reconstructed soils planted with grass and in natural soils [F(3,16) = 8.90; P = 0.00105]. Within the phylum Thaumarchaeota, the class soil crenarchaeotic group (also known as the 1.1b group) was more abundant in reconstructed soils planted with grasses than in natural soils [F(3,16) = 3.870; P = 0.029]. In contrast, the archaeal class terrestrial group (also known as the 1.1c group) of the phylum Thaumarchaeota was only present in natural soils and was therefore significantly more abundant in these soils than in reconstructed soils [F(3,16) = 14.231; P = 8 × 10−5] (see Fig. S7 in the supplemental material).
Relationships between environmental variables and prokaryotic communities.
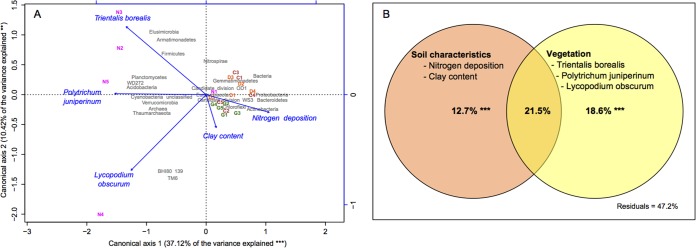
Canonical redundancy analysis indicated that distribution of phyla in the studied soils could be significantly explained by estimated nitrogen deposition, soil clay content, and the abundance of the mosses Lycopodium obscurum and Polytrichum juniperinum and the woodland perennial Trientalis borealis [F(5,14) = 4.495, P = 0.001; Fig. 5A]. Vegetation was positively correlated with the microbial communities that were abundant in the natural soils, whereas nitrogen deposition and soil clay content were positively correlated with the microbial communities that were more abundant in reconstructed soils (Fig. 5A). The variables included in the model explained 52.8% of the variation in microbial communities at the phylum level (Fig. 5B). Alone, the abundance of L. obscurum, P. juniperinum, and T. borealis explained 18.6% of the variation [F(3,14) = 5.551; P = 0.001]; nitrogen deposition and soil clay content explained 12.7% of the same variation [F(2,14) = 3.150; P = 0.001]. Jointly, these variables explained 21.5% of the variation [F(5,14) = 5.250; P = 0.001] (Fig. 5B).
FIG 5.
Canonical redundancy analysis (scaling 3) (A) and partition of variation (B) showing relations between site and soil characteristics and bacterial phyla in the studied soils (***, P < 0.001; **, P < 0.01).
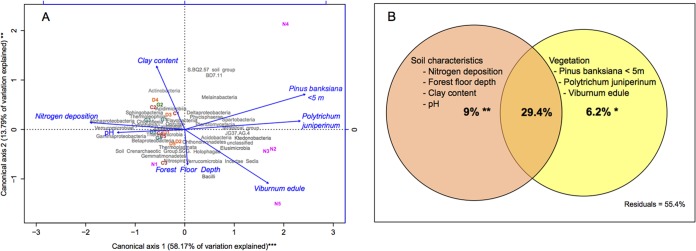
Slightly different variables explained the distribution of classes of microbial communities in the studied soils. Forest floor depth, pH, and abundance of trees and shrubs, P. banksiana (<5 m), and Viburnum edule further added to the already contributing nitrogen deposition, clay content and abundance of P. juniperinum variables [F(7,12) = 2.790; P = 0.001] (Fig. 6A). Vegetation was positively correlated with microbial classes abundant in natural soils, whereas chemical variables were positively correlated with the microbial classes abundant in reconstructed soils (Fig. 6A). These variables explained 44.6% of the variation in microbial communities at the class level (Fig. 6B). Alone, the abundance of P. banksiana (<5 m; only present in natural sites), P. juniperinum (only present in two of the 15 reconstructed sites), and V. edule explained 6.2% of the variation [F(3,12) = 1.563; P = 0.03]; nitrogen deposition, soil clay content, soil pH, and forest floor depth explained 9% of the same variation [F(4,12) = 1.650; P = 0.007]. Jointly, these variables explained 29.4% of the variation in prokaryotic microbial communities at the class level [F(7,12) = 3.186; P = 0.001] (Fig. 6B).
FIG 6.
Canonical redundancy analysis (scaling 3) (A) and partition of variation (B) showing relations between site and soil characteristics and bacterial classes in the studied soils (***, P < 0.001; **, P < 0.01).
DISCUSSION
Prokaryotic α-diversity.
In contrast to our hypothesis, the α-diversity did not significantly differ between reconstructed and natural soils, although the average inverse Simpson index was consistently higher in reconstructed soils. Comparing only data from mineral soils, Dimitriu and Grayston also found similar α-diversity indices in both natural, undisturbed boreal forest and reconstructed soils across a broader range of ages (3 to 29 years) (31). One reason that might explain this lack of difference may be the variety of materials used for soil reconstruction in the AOSR (peat and stockpiled mineral horizons) that, once mixed together, create a heterogeneous medium that offer a variety of niches to the microbial communities.
Bacterial β-diversity.
As predicted, bacterial community structures differed between reconstructed and natural soils. The order Rhizobiales was the most abundant order in all the studied soils, although it was more abundant in reconstructed soils. The family Hyphomicrobiaceae dominated the identified sequences of Rhizobiales. This family can utilize N2, NO3−, and NH3 and is thus an important player in the N cycle, especially for denitrification processes (32).
When OTU were grouped on a phylum level, the phyla Actinobacteria, Bacteroidetes, and Proteobacteria were more abundant in reconstructed than in natural soils. The bacteria belonging to these phyla that were the most abundant in the studied soils have been categorized as copiotrophic, i.e., bacteria that thrive in nutrient-rich environments and are able to rapidly use a resource when available but are unable to survive in a nutritionally deprived environment. They contrast with the slow-growing oligotrophic bacteria, better adapted to nutrient-poor environments (33–35). The phyla Acidobacteria, Cyanobacteria, Elusimicrobia, Firmicutes, Planctomycetes, Verrucomicrobia, and WD272 were relatively less abundant in reconstructed soils and are predominantly recognized as oligotrophic bacteria. Even if this “copiotrophic-oligotrophic” spectrum is a simplification of reality, members of each category still tend to respond in similar ways to nutrient availability. For example, copiotrophic bacteria and especially Actinobacteria, Bacteroidetes, and Proteobacteria (particularly alpha-, beta-, and gammaproteobacteria) are more abundant when N (36–38) or labile C (34, 35, 39) are added to soil. In contrast, the abundance of Acidobacteria has been shown to decrease with N fertilization or N deposition (36, 39), as well as with increased soil C content (40) or C mineralization rates (34). It may be that the higher C and N content of reconstructed soils selects for copiotrophic bacteria. However, only estimated N deposition was positively correlated to Actinobacteria, Acidobacteria, Bacteroidetes, and Proteobacteria abundances in the studied soils. This suggests that the most important factor shaping microbial communities in these soils is not the total N or C content of the soils, but the availability of a new and readily available source of N through atmospheric deposition.
Influence of nitrogen deposition on β-diversity.
Estimated N deposition was positively correlated to the abundance Actinobacteria, Bacteroidetes, and Proteobacteria abundances in the studied soils. Historically low, throughfall N-deposition in the boreal forest of the Athabasca Oil Sands Region (AOSR) are higher near mining sites and decrease to background level with distance (41, 42). Ammonium deposition measured in the close vicinity of the mining sites (<3 km) varied between of 14.7 and 19.6 kg NH4+ ha−1 year−1, whereas the nitrate depositions were lower, ranging between 2.1 and 6.7 kg NO3− ha−1 year−1 (7, 41). These depositions were reduced to 0.81 kg NH4+ ha−1 year−1 and 0.27 kg NO3− ha−1 year−1 120 km away from the mining sites (41). Therefore, in the AOSR, nitrogen deposition represents a constant source of readily available N that seems to favor the growth of copiotrophic bacterial (Proteobacteria, Actinobacteria, and Bacteroidetes). According to the literature, these microorganisms are limited in their capabilities to degrade complex organic matter, which could limit decomposition processes in the reconstructed soils and favor C accumulation (38, 39, 43). In a study of gross N transformation rates at these same sites, ammonification rates from the labile organic N were also lower in reconstructed soils than in natural forest soils, although reconstructed soils had greater microbial biomass (44). This further suggests that microbes colonizing reconstructed soils could have a low competency to degrade organic matter.
Archaeal β-diversity.
Within the archaea identified, 3% were from the phylum Euryarchaeota (class Thermoplasmata), which were only identified in the natural site, N1. Not much is known about the class Thermoplasmata. Cultured species of this class are aerobic, heterotrophic, and thermoacidophilic and demonstrate optimum growth at pH 0.7 to 3 and a temperature of about 50°C (45). Site N1 was one of the more recently burned sites (in 1995) and therefore these archaeal Thermoplasmata may be remnants of extreme postfire conditions.
The remainder of the archaea identified (97%) belonged to the phylum Thaumarchaeota. Reconstructed soils planted with grass harbored more organisms from the Thaumarchaeota class soil Crenarchaeotic group (also known as group 1.1b) than reconstructed soils planted with trees. However, the soil Crenarchaeotic group only comprised 9% of the Thaumarchaeota sequences in the natural soils. All known members of the soil Crenarchaeotic group are autotrophic ammonia-oxidizing archaea (AOA) (46, 47). Nicol et al. demonstrated that the abundance of the bacterial amoA gene (one of the genes responsible for ammonia oxidation) declined under acidic conditions, while the archaeal amoA gene abundance increased in acidic conditions, demonstrating the potential importance of archaea (and especially of Thermoarchaeota) for N cycling in acidic soils (48).
The terrestrial group of Thaumarchaeota, restricted, in this study, to natural soils, seems to prefer even lower pH than organisms from the soil crenarchaeotic group (49–51). Organisms from the terrestrial group composed 29% of all 16S rRNA gene sequences in acidic soils of a mixed deciduous forest in Germany and were not detected in soils with a pH above 4.5 (52, 53). The canonical ordination of prokaryotic classes illustrates the influence of soil pH on the abundance of Thaumarchaeota classes in the present study: the abundance of the soil crenarchaeotic group was positively correlated with pH, while the abundance of the terrestrial group was negatively correlated with pH. Weber et al. found that, compared to the other members of the phylum Thaumarchaeota, organisms from the terrestrial group were not able to grow autotrophically (gaining energy from the oxidation of ammonia to nitrite) and therefore showed no evidence of ammonia oxidation during growth (47).
These results indicate that planting reconstructed soils with grass promotes ammonia-oxidizing archaea of the class soil crenarchaeotic group, which could stimulate the conversion of ammonium (NH4+) to nitrate (NO3−)—i.e., nitrification—in these soils. Nitrate is more mobile than NH4+ in soils and is more likely to be lost by leaching or gas emission (54). The greater abundance of AOA in reconstructed soils planted with grass may therefore have a significant effect on the nitrogen cycle in these soils. Indeed, higher total gross nitrification and net nitrification rates were measured, using the 15N pool dilution method, in these same grassland soils in the laboratory (44). However, autotrophic nitrification was not higher in the reconstructed soils planted with grass, which lends further support to the growing evidence that these soil crenarchaeotic group of archaea are not obligate autotrophic ammonia oxidizers (55) and might be growing mixotrophically (56) or heterotrophically in the studied soils.
Influence of plant species on α-diversity.
The plant species, more than the soil characteristics, was related to the bacterial α- and β-diversity in all the studied soils. The α-diversity was correlated with the abundance of M. sativa, S. bebbiana, V. myrtilloides, and P. banksiana. A diverse plant community can create a high level of heterogeneity in root exudate patterns capable of supporting a diverse microbial community (57, 58). The rhizosphere of M. sativa has been shown to harbor more bacteria than other grass species (59). Through its root exudates, M. sativa could, therefore promote α-diversity in reconstructed grassland soils. The fact that M. sativa was uniquely present in the reconstructed soils revegetated using grasses and especially in site G4, which had the highest α-diversity among all the studied soils (data not shown), supports this hypothesis. M. sativa also commonly host N-fixing bacteria (60), which could increase soil NH4+ content, N-cycling rates and the presence of ammonia oxidizers in grassland soils. Diverse root-exudate production in the rhizosphere could also result in the increased α-diversity of bacteria in the soils revegetated with S. bebbiana and V. myrtilloides. Both plants can develop symbiotic relationships with fungi: ectomycorrhizae with S. bebbiana and ericoid mycorrhizae with V. myrtilloides (61–63). Willows associate with specific ectomycorrhizal fungi, which, once established, increase bacterial diversity in the soil (62). Willows also produce a small, but highly labile pool of organic carbon that can support abundant copiotrophic proteobacteria (64). Similarly, after the formation of ericoid mycorrhizae on V. myrtilloides, a diverse set of bacteria, including biofilm bacteria, mycorrhizal helper bacteria, and plant growth-promoting rhizobacteria, can colonize and proliferate around the roots, increasing soil bacterial diversity (65). These results indicate that specific vegetation, through effects on root exudates and/or symbiotic mycorrhizal relationships, can create microenvironments which favor diverse microbial communities, even in reconstructed soils. The only type of vegetation that was associated with a reduction in bacterial α-diversity was P. banksiana. This may be related to its status as an early successional species in the boreal forest; P. banksiana was dominant in the most recently burned natural stands (site N1 [burned in 1995] and site N4 [burned in 1981]). P. banksiana also tends to dominate on the driest and most nutrient-poor sites, which may contribute to the low bacterial α-diversity in soils in these natural stands.
Influence of plant species on β-diversity.
Specific plant species were also correlated with the structure of microbial communities within the natural soils. The phyla Cyanobacteria, Planctomycetes, and Verrucomicrobia were positively correlated with the presence of P. juniperinum. Free-living cyanobacteria have been associated with bryophytes in boreal forests of Sweden (66–69) and can fix atmospheric N, which is then leaked to the moss (69, 70). The rates of N fixation in Swedish boreal forests increased with time since fire as moss reestablished, and with it, an increased presence of Cyanobacteria (66). The positive correlation between Cyanobacteria and the abundance of P. juniperinum in this study indicate that similar processes occur in the boreal forests of Canada. Verrucomicrobia and Planctomycetes were also positively correlated with the abundance of P. juniperinum. Perhaps these bacteria also colonize mosses. Planctomycetes are capable of anaerobic ammonium oxidation to dinitrogen with nitrate as an electron acceptor (annamox) (71, 72). The high N concentrations in the moss may stimulate colonization by annamox-capable bacterial species, such that the moss-bacterium association contributes to the N cycle beyond N fixation.
Influence of soil characteristics on β-diversity.
While plant species were positively correlated with microbial community structure (β-diversity) in natural soils, pH was positively correlated with the most abundant microbial communities in reconstructed soils (Actinobacteria, Bacteroidetes, and Proteobacteria). On a continental scale, soil pH has been shown to be an important driver of bacterial community structure, with Acidobacteria preferring acidic environments and Actinobacteria and Bacteroidetes preferring alkaline environments, while Proteobacteria have an optimum pH of 6 (73). Reconstructed soils were generally slightly more alkaline than their natural counterparts, which might have favored the growth of Actinobacteria, Bacteroidetes, and Proteobacteria.
Microbial community structure in reconstructed soils planted with grass.
Within reconstructed soils, sites planted with grasses had more bacteria from classes Anaerolineae, Caldilineae, and Chloroflexia of the phylum Chloroflexi than sites planted with either deciduous or coniferous trees. Members of the phylum Chloroflexi are slow-growing, heterotrophic bacteria, ubiquitous in natural ecosystems (74). Both Anaerolineae and Caldilineae are anaerobic (or facultative aerobic for Caldilineae) classes of Chloroflexi that have been found at depths of 140 m in both marine and terrestrial environments (74, 75). The phylum Chloroflexi and grassland soils were positively correlated with clay content in the redundancy analyses. Soil bulk density was not significantly higher in grassland than other soils (data not shown), although some soil compaction was noted during field sampling at grassland sites. Laidlaw showed that these grassland soils had the highest average proportion of microaggregates of any of the sites (76). These results suggest that the interaction between roots, clay content, compaction, and aggregation might increase the number of anaerobic microenvironments in grassland soils compared to the other reconstructed soils, which would favor anoxic classes of Chloroflexi (Anaerolineae and Caldilineae) (77).
Effect of disturbances, other than forest fire, on β-diversity in natural soils.
Natural site N1 had a different prokaryotic microbial community than the other natural soils, and the community was more similar to reconstructed soils. Site N1 burned in 1995 and was visited in 2012, prior to soil sampling. However, between summers of 2012 and 2013, the young stand was removed to establish a small access road. Consequently, in 2013, soil for this site was collected from an apparently undisturbed forest stand approximately 10 meters away from the newly constructed road. The finding that site N1 did not tightly group with the other reconstructed sites in the multivariate analyses suggests that there may have been altered environmental conditions resulting from the establishment of the access road at site N1.
In conclusion, using next-generation sequencing, we were able to distinguish the prokaryotic communities in reconstructed and natural forest soils in the AOSR. In contrast to our hypothesis, the α-diversity did not differ in reconstructed and natural soils and was actually higher in reconstructed soils. Vegetation cover, especially P. banksiana, M. sativa, S. bebbiana, and V. myrtilloides, was the main factor influencing α-diversity. As predicted, β-diversity differed between reconstructed and natural forest soils. Within reconstructed soils, those planted with grasses harbored a distinct microbial community than those planted with trees. Copiotrophic bacteria (Actinobacteria, Bacteroidetes, and Proteobacteria) were more abundant in reconstructed soils, whereas oligotrophic bacteria (Acidobacteria, Cyanobacteria, Elusimicrobia, Firmicutes, Planctomycetes, and Verrucomicrobia) were more abundant in natural forest soils. Ammonia-oxidizing archaea and anaerobic bacteria (from the classes Anaerolineae, Caldilineae, and Chloroflexia) were more abundant in reconstructed soils planted with grasses. Nitrogen deposition, pH, clay content, and plant species were the main variables influencing the structure of the communities. Taken together, these results indicated that soil and site characteristics, such as atmospheric N deposition, soil pH, and plant community engendered distinct prokaryotic communities in the reconstructed and natural forest soils and highlight the importance of aboveground-belowground relationships in shaping microbial communities in both natural and reconstructed soils.
MATERIALS AND METHODS
Study area.
The study area was situated in the Athabasca Oil Sands Region (AOSR) in northern Alberta, Canada (56°39′N, 111°13′W; altitude, 369 m). Short warm summers and long cold winters characterize the climate. The mean annual temperature is 1°C; mean monthly temperatures range from −17.4°C in January to 17.1°C in July. Mean annual precipitation is 418.6 mm, of which 316.3 mm occurs as rainfall during the growing season (78). Medium- to fine-textured Gray Luvisols (haplocryalfs according to the U.S. soil taxonomy) and Dystric Brunisols (Dystrocryepts) underlie landscapes shaped by the impact of Pleistocene ice activity, deglaciation, and postglacial modifications in upland areas. Organic soils (Cryaquepts) are found under wetland areas (79). The AOSR falls within the central mixed-wood region of the Canadian boreal forest. Dominant tree canopy species in upland landscapes are trembling aspen (Populus tremuloides Michx), white spruce (Picea glauca [Moench] Voss), and jack pine (Pinus banksiana Lamb) (79).
Oil sand mining activities involve forest harvest and the removal of surface soil materials, followed by the removal of 40 m of overburden material (approximate regional average) to expose the oil sand ore body. Salvaged soil materials are preferably used for reclamation of areas within the mine footprint that are ready for reclamation or are stockpiled for later use. The overburden is used for berm, dyke wall, or road construction or is deposited in a dedicated disposal area to create large-scale overburden landform units. The oil sand ore is transported to the extraction and upgrading facility. Oil sand soil reconstruction involves a number of cover designs, depending on the landform substrate being reclaimed. There are two main cover designs: one that uses only cover soil and the other consisting of a combination of cover soil and subsoil. Cover soil and subsoil materials are salvaged from surface soils within the mine development footprint. Only sites at which cover soil had been placed on top of overburden material were used in this study. The cover soil material used consisted of surface peat mixed with mineral soil material having a loam or coarser texture and is here referred to as “peat-mineral mix.” In the studied soils, the depth of the peat-mineral mix ranged from 20 cm to >100 cm. Early revegetation objectives in the AOSR were to establish native or introduced grass and shrub species to control erosion; however, oil sand operators are now required to revegetate using native trees and understory plant species, with the intention of promoting the reestablishment of a boreal forest community. During the period when the sites used in this study were reclaimed (20 to 30 years ago), 250 to 350 kg ha−1 of various proportions of N-P-K fertilizer was typically applied in the first year of revegetation. Some oil sand operators also applied fertilizer annually for four additional years.
Study sites.
Fifteen reconstructed sites and five natural forest sites were studied in the AOSR (n = 20; four treatments with five replicates each). The 15 reconstructed sites were previously studied by Sorenson et al. (26). Among them, five sites (C1 to C5) were planted with coniferous species (mostly P. glauca), five (D1 to D5) with deciduous species (mostly P. tremuloides), and five (G1 to G5) with grasses (Festuca sp., Elymus trachycaulus, and M. sativa). Soil from these sites was reconstructed 26 years (±6 years) prior to sampling (historical data provided by oil sand operators). For reference sites, we selected five natural sites (N1 to N5). These sites experienced a natural stand-replacing wildfire 26 years (±10 years) prior to sampling (historical data provided by the government of Alberta) (80). Natural boreal forests in this region experience such fires on average every 250 years (81, 82). Since the selected natural sites would be at approximately the same stage in the postdisturbance trajectory as the reconstructed sites, these sites were considered a more realistic reference for the reconstructed soils than would older natural forests. The five natural boreal forest sites were located 40 to 150 km south of Fort McMurray (Alberta, Canada), which in the context of the Canadian boreal forest is consider to be close to the mining sites (see Fig. S1 in the supplemental material). Soils at the natural sites were classified as Gleyed Eluviated Eutric Brunisol (soil N1), Eluviated Eutric Brunisol (soil N5), or Brunisolic Gray Luvisol (soils N2, N3, and N4).
At each site, one 10-m2 plot was subdivided into ten 1-m2 subplots, from which seven subplots were randomly selected. A 30-cm-deep soil pit was dug in each subplot and carefully described. The litter and forest floor layers were removed and the top 0 to 15 cm of the mineral part of the (bulk) soil was sampled (∼ 1 kg) at each subplot using a plastic trowel, and the seven subplot samples were pooled to produce one sample per plot (nsample = 20). A subsample (∼5 g) was immediately put on ice for microbial analyses and stored at −18°C until laboratory analyses were performed. The remaining samples were kept at 4°C for soil chemical and physical analyses. The removal of any surface organic material prior to sampling enabled us to directly compare the mineral substrates on which the organic layers develop and so determine how similar the reconstructed soils are to natural soils. If we assume that the same soil development factors are in play in reconstructed and naturally disturbed soils, the peat-mineral mix is the analogue of the mineral part of the natural soils on top of which a forest floor develops. Characteristics of the forest floor layers in reconstructed and natural soils in the AOSR have been described in previous research (83).
Concomitant to soil sampling, a vegetation survey was conducted (84) (detailed results are available in the supplemental material). Briefly, the vegetation within each 10-m2 plot was described, including cover by species and cover by vegetation type (nonvascular, graminoid, herbaceous, shrub, and tree). Tree density was not significantly different among reconstructed soils planted with deciduous species, reconstructed soils planted with coniferous species, and naturally disturbed sites (data not shown). Grassland sites were devoid of trees.
Laboratory analyses.
The soil moisture and bulk density at each site were determined using soil cores of known volume (78.5 cm3) from one of the subplots (85). Soil pH in water was determined in a 1:10 (soil:water) solution using a UB-10 pH meter (Denver Instruments, Bohemia, NY) (86). The soil total carbon and nitrogen concentrations were measured with an Elementar Vario El Cube elemental analyzer (Elementar, Hanau, Germany) using 15 mg of soil sieved (<0.5 mm) to ensure homogenization (87, 88). The C:N ratio was calculated by dividing total C content by total N content. Soil particle size of sand (60 μm to 2 mm) and clay (<2 μm) were determined in the Environmental Geoscience Research Centre at Trent University using laser diffraction with a Partica LA-950 instrument (Horiba, Ltd., Kyoto, Japan). Analyses were completed in triplicate after homogenizing the sample (89). Microbial biomass carbon was measured on the day of the sampling using the chloroform fumigation-extraction technique (modified from Tate et al. [90]). Unless otherwise mentioned, 25% of the samples were analyzed in duplicate and controls (AgroMAT AG-2 soil standard; SCP Science, Baie d'Urfé, Canada) were added. Given its suspected key role in shaping microbial communities, nitrogen deposition data at each site was obtained from the terrestrial monitoring database of the Wood Buffalo Environmental Association (91).
DNA extraction and amplification.
Soil DNA was extracted in triplicate using a PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA) with 0.25 g of soil. Triplicates were pooled and sent to Génome Québec Innovation Centre (Montréal, Quebec, Canada) for sequencing. The V4 region of the 16S rRNA gene was amplified with the 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSGGGTATCTAAT-3′) primers with expected length of 250 bp using the following program: 94°C for 3 min, followed by 35 cycles of 95°C for 45 s, 50°C for 60 s, and 72°C for 90 s, followed in turn by a final 10 min at 72°C. This primer pair is used for phylogenetic studies to target specifically bacterial and archaeal species (92, 93). PCR products (n = 20) were barcoded, pooled, and sequenced using an Illumina MiSeq sequencer with 250 bp on the forward and the reverse reads (Illumina, San Diego, CA).
Bioinformatics analyses.
A total of 736,710 paired-end Illumina sequences were obtained from Génome Québec from the 20 samples (36,836 ± 8,981 sequences per sample). They were analyzed using the mothur MiSeq SOP pipeline (94). Briefly, forward and reverse reads were merged. Nonambiguous sequences shorter than 500 bp were kept and aligned using SILVA, a manually curated rRNA sequence database (95). Sequences were further screened for chimeras using the chimera.uchime script (96). Singletons, chloroplasts, and mitochondrial sequences were removed from the data set using mothur's remove.lineage function. At the end of the screening process, 115,629 sequences were kept and clustered in 778 operational taxonomic units (OTU) with a 97% similarity using the dist.seqs and cluster (with the average neighbor algorithm) functions in mothur. We assigned the taxonomy of each OTU using the SILVA database (95) (scripts can be found in the supplemental material). The original reads obtained from Genome Quebec were deposited on MG-RAST's servers (http://metagenomics.anl.gov/linkin.cgi?project=mgp21608).
Calculations and statistics.
Good's coverage is an indicator of the depth of sequencing. It estimates the proportion of total species that are represented in a sample (94).
The microbial diversity at each site (α-diversity) was calculated using the inverse Simpson diversity index (97). Means and standard errors of the inverse Simpson index for each treatment are presented. Differences among treatment types (reconstructed using deciduous, coniferous, and grass species and natural forest soils) were assessed using one-way analysis of variance (ANOVA) with a permutation test (98). A multiple-linear-regression model was used to identify variables that significantly contribute to α-diversity. Theory and forward selection was used to choose significant variables (99, 100). Several models were tested and the model with the lowest AIC (Akaike Information Criterion) was kept (data not shown).
The β-diversity is the differentiation of microbial composition among habitats. It was assessed using different methods. A principal-coordinate analysis (PCoA) using a Bray-Curtis dissimilarity matrix on Hellinger-transformed community data was performed to visually assess whether sites with dissimilar vegetation harbored different microbial communities (101). Hellinger transformation of the community data provides an appropriate matrix of species abundances that eliminates the bias that species abundance matrices generate by emphasizing the weight of rare species in ordinations (102). PCoA was followed by an analysis of molecular variance, a nonparametric analysis of variance, to test whether the centers of the clouds of points representing treatments are more separated than the variation among samples of the same treatment (94).
We also evaluated β-diversity using one-way ANOVA with a permutation test on all phyla and classes (98) and by representing the data using Z-scores [Z-score = (x − x̄)/s, where x is the abundance of the phylum or class in the sample], x̄ is the mean abundance of the phylum, or class, using all samples, and s is the standard deviation of the abundance of the phylum, or class, using all samples. Represented are the Z-score means of each phylum or class for each treatment.
When significant differences were observed, post hoc Kruskal-Wallis tests were performed using the R package pgirmess (103). A canonical redundancy analysis (RDA) using Hellinger-transformed microbial data was performed to identify variables that significantly explained the distribution of microbial communities in the studied soils (99). Significant variables were selected using a forward selection algorithm: Packfor's forward.sel function (100). The RDA model, axis, and explanatory variables were tested using a permutation test (101). RDAs are presented using scaling 3. A partition of the variation was also performed to assess how much of the variation was explained by the soil and the vegetation characteristics (101). All statistical analyses were performed using R software (v3.3.1) (104).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeff Anderson and Meghan Laidlaw for assistance in the field and fruitful discussions about the project; Hannah Mei, Eli Rechtschaffen, Emily Mason, and Zack Wentz for indispensable help in the field and in the laboratory; and Sylvie Quideau, David Levy-Booth, and Jesse Shapiro for fruitful discussions. We are also grateful to Alice Chang (University of British Columbia [UBC]) for total C and N analyses and to the Génome Québec Innovation Centre for sequencing analyses. We also thank Marty Yarmuch (Syncrude), Christine Daly (Suncor), and Carmela Arevalo (Suncor) for granting access to the mining sites and for logistical help during the sampling campaign. We also thank three anonymous reviewers who helped to improve the manuscript.
This study was supported by a Natural Science and Engineering Research Council of Canada (NSERC) Collaborative Research and Development grant (Principal Investigator Sylvie Quideau) and an NSERC PGS-D scholarship, an FQRNT scholarship, UBC FYF, a UBC Forestry Strategic Recruitment Fellowship, an Edward W. Bassett Memorial Scholarship, a Mary and David Macaree Fellowship, and an Agricultural Institute of Canada Foundation fellowship to J.M. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03319-16.
REFERENCES
- 1.Government of Alberta. 2016. Alberta energy: facts and statistics. Government of Alberta, Edmonton, Alberta, Canada. [Google Scholar]
- 2.Government of Alberta. 2012. Oil sands: the resource 1–2. Government of Alberta, Edmonton, Alberta, Canada. [Google Scholar]
- 3.Government of Alberta. 2013. Oil sands: reclamation 1–2. Government of Alberta, Edmonton, Alberta, Canada. [Google Scholar]
- 4.Government of Alberta. 1993. Conservation and reclamation regulation (AR 115-1993). Government of Alberta, Edmonton, Alberta, Canada. [Google Scholar]
- 5.Powter C, Chymko N, Dinwoodie G, Howat D, Janz A, Puhlmann R, Richens T, Watson D, Sinton H, Ball K, Patterson B, Brocke L, Dyer R. 2012. Regulatory history of Alberta' s industrial land conservation and reclamation program. Can J Soil Sci 92:39–51. doi: 10.4141/cjss2010-033. [DOI] [Google Scholar]
- 6.Turcotte I, Quideau SA, Oh S-W. 2009. Organic matter quality in reclaimed boreal forest soils following oil sands mining. Org Geochem 40:510–519. doi: 10.1016/j.orggeochem.2009.01.003. [DOI] [Google Scholar]
- 7.Hemsley TL. 2012. Ecological response of atmospheric nitrogen deposition on reconstructed soils in the Athabasca Oil Sands Region. University of Alberta, Edmonton, Alberta, Canada. [Google Scholar]
- 8.Dimitriu PA, Prescott CE, Quideau SA, Grayston SJ. 2010. Impact of reclamation of surface-mined boreal forest soils on microbial community composition and function. Soil Biol Biochem 42:2289–2297. doi: 10.1016/j.soilbio.2010.09.001. [DOI] [Google Scholar]
- 9.Hahn AS, Quideau SA. 2013. Long-term effects of organic amendments on the recovery of plant and soil microbial communities following disturbance in the Canadian boreal forest. Plant Soil 363:331–344. doi: 10.1007/s11104-012-1306-4. [DOI] [Google Scholar]
- 10.Chazdon RL. 2008. Beyond deforestation: restoring degraded lands. Communities 1458:1458–1460. [DOI] [PubMed] [Google Scholar]
- 11.Hobbs RJ, Arico S, Aronson J, Baron JS, Cramer VA, Epstein PR, Ewel JJ, Klink CA, Lugo AE, Norton D, Ojima D, Richardson DM. 2006. Novel ecosystems: theoretical and management aspects of the new ecological world order. Glob Ecol Biogeogr 15:1–7. doi: 10.1111/j.1466-822X.2006.00212.x. [DOI] [Google Scholar]
- 12.Quideau SA, Swallow MJB, Prescott CE, Grayston SJ, Oh SW. 2013. Comparing soil biogeochemical processes in novel and natural boreal forest ecosystems. Biogeosciences 10:5651–5661. doi: 10.5194/bg-10-5651-2013. [DOI] [Google Scholar]
- 13.Bottomley PJ, Myrold DD. 2015. Biological N inputs, p 447–470. In Paul EA. (ed), Soil microbiology, ecology, and biochemistry, 4th ed Elsevier, Inc, New York, NY. [Google Scholar]
- 14.Robertson GP, Groffman PM. 2014. Nitrogen transformations, p 421–446. In Paul EA. (ed), Soil microbiology, ecology, and biochemistry, 4th ed Elsevier, Inc, New York, NY. [Google Scholar]
- 15.Schimel JP, Bennett J, Fierer N. 2005. Microbial community composition and soil nitrogen cycling: is there really a connection? In Bardgett RD, Usher MB, Hopkins DW (ed), Biological diversity and function in soils. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 16.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, Terhonen E, Koskinen K, Paulin L, Kasanen R, Asiegbu FO. 2014. Bacterial diversity and community structure along different peat soils in boreal forest. Appl Soil Ecol 74:37–45. doi: 10.1016/j.apsoil.2013.09.010. [DOI] [Google Scholar]
- 18.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansel CM, Fendorf S, Jardine PM, Francis CA. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl Environ Microbiol 74:1620–1633. doi: 10.1128/AEM.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockett BFT, Prescott CE, Grayston SJ. 2012. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol Biochem 44:9–20. doi: 10.1016/j.soilbio.2011.09.003. [DOI] [Google Scholar]
- 21.Ferrenberg S, O'Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR. 2013. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J 7:1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wardle DA. 2002. Communities and ecosystems. Linking the aboveground and belowground components. Princeton University Press, Princeton, NJ. [Google Scholar]
- 23.Grayston SJ, Prescott CE. 2005. Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol Biochem 37:1157–1167. doi: 10.1016/j.soilbio.2004.11.014. [DOI] [Google Scholar]
- 24.Prescott CE, Grayston SJ. 2013. Tree species influence on microbial communities in litter and soil: current knowledge and research needs. For Ecol Manage 309:19–27. doi: 10.1016/j.foreco.2013.02.034. [DOI] [Google Scholar]
- 25.Thoms C, Gattinger A, Jacob M, Thomas FM, Gleixner G. 2010. Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol Biochem 42:1558–1565. doi: 10.1016/j.soilbio.2010.05.030. [DOI] [Google Scholar]
- 26.Sorenson PT, Quideau SA, MacKenzie MD, Landhäusser SM, Oh SW. 2011. Forest floor development and biochemical properties in reconstructed boreal forest soils. Appl Soil Ecol 49:139–147. doi: 10.1016/j.apsoil.2011.06.006. [DOI] [Google Scholar]
- 27.Thomson MD. 1979. Ecological habitat mapping of the AOSERP study area (supplement): phase 1. Alberta Oil Sands Environmental Research Program, Edmonton, Alberta, Canada. [Google Scholar]
- 28.Ball PN, MacKenzie MD, DeLuca TH, Montana WEH. 2010. Wildfire and charcoal enhance nitrification and ammonium-oxidizing bacterial abundance in dry montane forest soils. J Environ Qual 39:1243. doi: 10.2134/jeq2009.0082. [DOI] [PubMed] [Google Scholar]
- 29.Hart SC, DeLuca TH, Newman GS, MacKenzie MD, Boyle SI. 2005. Postfire vegetative dynamics as drivers of microbial community structure and function in forest soils. For Ecol Manage 220:166–184. doi: 10.1016/j.foreco.2005.08.012. [DOI] [Google Scholar]
- 30.Switzer JM, Hope GD, Grayston SJ, Prescott CE. 2012. Changes in soil chemical and biological properties after thinning and prescribed fire for ecosystem restoration in a Rocky Mountain Douglas fir forest. For Ecol Manage 275:1–13. doi: 10.1016/j.foreco.2012.02.025. [DOI] [Google Scholar]
- 31.Dimitriu PA, Grayston SJ. 2010. Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb Ecol 59:563–573. doi: 10.1007/s00248-009-9590-0. [DOI] [PubMed] [Google Scholar]
- 32.Anderson CR, Condron LM, Clough TJ, Fiers M, Stewart A, Hill RA, Sherlock RR. 2011. Biochar induced soil microbial community change: implications for biogeochemical cycling of carbon, nitrogen, and phosphorus. Pedobiologia 54:309–320. doi: 10.1016/j.pedobi.2011.07.005. [DOI] [Google Scholar]
- 33.Koch AL. 2001. Oligotrophs versus copiotrophs. Bioessays 23:657–661. doi: 10.1002/bies.1091. [DOI] [PubMed] [Google Scholar]
- 34.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 35.Eilers KG, Lauber CL, Knight R, Fierer N. 2010. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem 42:896–903. doi: 10.1016/j.soilbio.2010.02.003. [DOI] [Google Scholar]
- 36.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C, Yan K, Tang L, Jia Z, Li Y. 2014. Change in deep soil microbial communities due to long-term fertilization. Soil Biol Biochem 75:264–272. doi: 10.1016/j.soilbio.2014.04.023. [DOI] [Google Scholar]
- 38.Freedman Z, Zak DR. 2014. Atmospheric N deposition increases bacterial laccase-like multicopper oxidases: implications for organic matter decay. Appl Environ Microbiol 80:4460–4468. doi: 10.1128/AEM.01224-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leff JW, Nemergut DR, Grandy AS, O'Neill SP, Wickings K, Townsend AR, Cleveland CC. 2012. The effects of soil bacterial community structure on decomposition in a tropical rain forest. Ecosystems 15:284–298. doi: 10.1007/s10021-011-9510-2. [DOI] [Google Scholar]
- 40.Nemergut DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR. 2010. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem 42:2153–2160. doi: 10.1016/j.soilbio.2010.08.011. [DOI] [Google Scholar]
- 41.Fenn ME, Bytnerowicz A, Schilling SL, Ross CS. 2015. Atmospheric deposition of nitrogen, sulfur and base cations in jack pine stands in the Athabasca Oil Sands Region, Alberta, Canada. Environ Pollut 196:497–510. doi: 10.1016/j.envpol.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Proemse BC, Mayer B, Fenn ME, Ross CS. 2013. A multi-isotope approach for estimating industrial contributions to atmospheric nitrogen deposition in the Athabasca Oil Sands Region in Alberta, Canada. Environ Pollut 182:80–91. doi: 10.1016/j.envpol.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Fontaine S, Bardoux G, Abbadie L, Mariotti A. 2004. Carbon input to soil may decrease soil carbon content. Ecol Lett 7:314–320. doi: 10.1111/j.1461-0248.2004.00579.x. [DOI] [Google Scholar]
- 44.Masse J, Prescott CE, Müller C, Grayston SJ. 2016. Gross nitrogen transformation rates differ in reconstructed oil-sand soils from natural boreal-forest soils as revealed using a 15N tracing method. Geoderma 282:37–48. doi: 10.1016/j.geoderma.2016.07.007. [DOI] [Google Scholar]
- 45.Angelov A, Liebl W. 2006. Insights into extreme thermoacidophily based on genome analysis of Picrophilus torridus and other thermoacidophilic archaea. J Biotechnol 126:3–10. doi: 10.1016/j.jbiotec.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Lin X, Handley KM, Gilbert JA, Kostka JE. 2015. Metabolic potential of fatty acid oxidation and anaerobic respiration by abundant members of Thaumarchaeota and Thermoplasmata in deep anoxic peat. ISME J 9:2740–2744. doi: 10.1038/ismej.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber EB, Lehtovirta-Morley LE, Prosser JI, Gubry-Rangin C. 2015. Ammonia oxidation is not required for growth of group 1.1c soil Thaumarchaeota. FEMS Microbiol Ecol 91:1–7. doi: 10.1093/femsec/fiu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance, and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 49.He JZ, Hu HW, Zhang LM. 2012. Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154. doi: 10.1016/j.soilbio.2012.06.006. [DOI] [Google Scholar]
- 50.Lehtovirta LE, Prosser JI, Nicol GW. 2009. Soil pH regulates the abundance and diversity of group 1.1c Crenarchaeota. FEMS Microbiol Ecol 70:367–376. doi: 10.1111/j.1574-6941.2009.00748.x. [DOI] [PubMed] [Google Scholar]
- 51.Oton EV, Quince C, Nicol GW, Prosser JI, Gubry-Rangin C. 2016. Phylogenetic congruence and ecological coherence in terrestrial Thaumarchaeota. ISME J 10:85–96. doi: 10.1038/ismej.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicol GW, Tscherko D, Embley TM, Prosser JI. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol 7:337–347. doi: 10.1111/j.1462-2920.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- 53.Kemnitz D, Kolb S, Conrad R. 2007. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol Ecol 60:442–448. doi: 10.1111/j.1574-6941.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 54.Robertson G, Groffman PM. 2007. Nitrogen transformations, p 341–364. In Paul EA. (ed), Soil microbiology, ecology, and biochemistry, 3rd ed Elsevier, Burlington, MA. [Google Scholar]
- 55.Mussmann M, Brito I, Pitcher A, Sinninghe Damste JS, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Muller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci U S A 108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:520–536. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PGL, van Veen JA. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Int J Gen Mol Microbiol 81:509–520. [DOI] [PubMed] [Google Scholar]
- 58.Ding GC, Piceno YM, Heuer H, Weinert N, Dohrmann AB, Carrillo A, Andersen GL, Castellanos T, Tebbe CC, Smalla K. 2013. Changes of soil bacterial diversity as a consequence of agricultural land use in a semi-arid ecosystem. PLoS One 8:e59497. doi: 10.1371/journal.pone.0059497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miethling R, Wieland G, Backhaus H, Tebbe CC. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb Ecol 40:43–56. doi: 10.1007/s002480000021. [DOI] [PubMed] [Google Scholar]
- 60.Carlsson G, Huss-Danell K. 2003. Nitrogen fixation in perennial forage legumes in the field. Plant Soil 253:353–372. doi: 10.1023/A:1024847017371. [DOI] [Google Scholar]
- 61.Bell TH, El-Din Hassan S, Lauron-Moreau A, Al-Otaibi F, Hijri M, Yergeau E, St-Arnaud M. 2014. Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J 8:331–343. doi: 10.1038/ismej.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bell TH, Cloutier-Hurteau B, Al-Otaibi F, Turmel MC, Yergeau E, Courchesne F, St Arnaud M. 2015. Early rhizosphere microbiome composition is related to the growth and Zn uptake of willows introduced to a former landfill. Environ Microbiol 17:3025–3038. doi: 10.1111/1462-2920.12900. [DOI] [PubMed] [Google Scholar]
- 63.Kourtev PS, Ehrenfeld JG, Häggblom M. 2002. Exotic plant species alter the microbial community structure and function in the soil. Ecology 83:3152–3166. doi: 10.1890/0012-9658(2002)083[3152:EPSATM]2.0.CO;2. [DOI] [Google Scholar]
- 64.Männistö MK, Kurhela E, Tiirola M, Häggblom MM. 2013. Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol Ecol 84:47–59. doi: 10.1111/1574-6941.12035. [DOI] [PubMed] [Google Scholar]
- 65.Robertson SJ, McGill WB, Massicotte HB, Rutherford PM. 2007. Petroleum hydrocarbon contamination in boreal forest soils: a mycorrhizal ecosystems perspective. Biol Rev 82:213–240. doi: 10.1111/j.1469-185X.2007.00012.x. [DOI] [PubMed] [Google Scholar]
- 66.Zackrisson O, DeLuca TH, Nilsson MC, Sellstedt A, Berglund LM. 2004. Nitrogen fixation increases with successional age in boreal forests. Ecology 85:3327–3334. doi: 10.1890/04-0461. [DOI] [Google Scholar]
- 67.Gundale MJ, Bach LH, Nordin A. 2013. The impact of simulated chronic nitrogen deposition on the biomass and N2 fixation activity of two boreal feather moss-cyanobacteria associations. Biol Lett 9:3–7. doi: 10.1098/rsbl.2013.0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rousk K, Jones DL, Deluca TH. 2013. Moss-cyanobacteria associations as biogenic sources of nitrogen in boreal forest ecosystems. Front Microbiol 4:150. doi: 10.3389/fmicb.2013.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeLuca TH, Zackrisson O, Nilsson M-C, Seastedt A. 2002. Quantifying nitrogen-fixation in feather moss carpets of boreal forests. Nature 419:917–920. doi: 10.1038/nature01051. [DOI] [PubMed] [Google Scholar]
- 70.Rousk K, Jones DL, DeLuca TH. 2014. Moss-nitrogen input to boreal forest soils: tracking 15N in a field experiment. Soil Biol Biochem 72:100–104. doi: 10.1016/j.soilbio.2014.01.031. [DOI] [Google Scholar]
- 71.Mulder A, Graaf AA, Robertson LA, Kuenen JG. 1995. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol 16:177–184. doi: 10.1111/j.1574-6941.1995.tb00281.x. [DOI] [Google Scholar]
- 72.Hayatsu M, Tago K, Saito M. 2008. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr 54:33–45. doi: 10.1111/j.1747-0765.2007.00195.x. [DOI] [Google Scholar]
- 73.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamada T, Sekiguchi Y. 2009. Cultivation of uncultured chloroflexi subphyla: significance and ecophysiology of formerly uncultured chloroflexi “subphylum I” with natural and biotechnological relevance. Microbes Environ 24:205–216. [DOI] [PubMed] [Google Scholar]
- 75.Breuker A, Köweker G, Blazejak A, Schippers A. 2011. The deep biosphere in terrestrial sediments in the Chesapeake Bay area, Virginia, USA. Front Microbiol 2:1–13. doi: 10.3389/fmicb.2011.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laidlaw M. 2015. Soil carbon stabilization under three reclaimed vegetation types in the Alberta oil sands. University of British Columbia, Vancouver, British Columbia, Canada. [Google Scholar]
- 77.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. 2013. Microscale determinants of bacterial diversity in soil. FEMS Microbiol Rev 37:936–954. doi: 10.1111/1574-6976.12023. [DOI] [PubMed] [Google Scholar]
- 78.Environment Canada. 2015. Canadian climate normals, 1821–2000: Fort McMurray. Government of Canada, Edmonton, Alberta, Canada: http://climate.weather.gc.ca/climate_normals/results_1981_2010_e.html?stnID=2519&lang=f&StationName=Fort&SearchType=Contains&stnNameSubmit=go&dCode=1. [Google Scholar]
- 79.Natural Regions Committee. 2006. Natural regions and subregions of Alberta. Government of Alberta, Edmonton, Alberta, Canada. [Google Scholar]
- 80.Government of Alberta. 2015. Historical wildfire database. Government of Canada, Edmonton, Alberta, Canada: http://wildfire.alberta.ca/resources/historical-data/historical-wildfire-database.aspx. [Google Scholar]
- 81.Bergeron Y, Gauthier S, Kafka V, Lefort P, Lesieur D. 2001. Natural fire frequency for the eastern Canadian boreal forest: consequences for sustainable forestry. Can J For Res 31:384–391. doi: 10.1139/x00-178. [DOI] [Google Scholar]
- 82.Binkley D, Fisher RF. 2013. Ecology and management of forest soils, 4th ed Wiley-Blackwell, West Sussex, United Kingdom. [Google Scholar]
- 83.Rowland SM, Prescott CE, Grayston SJ, Quideau SA, Bradfield GE. 2009. Recreating a functioning forest soil in reclaimed oil sands in northern Alberta: an approach for measuring success in ecological restoration. J Environ Qual 38:1580–1590. doi: 10.2134/jeq2008.0317. [DOI] [PubMed] [Google Scholar]
- 84.Anderson J. 2014. Organic matter accumulation in reclaimed soils beneath different vegetation types in the Athabasca oil sands. University of British Columbia, Vancouver, British Columbia, Canada. [Google Scholar]
- 85.Blake GR, Hartge KH. 1986. Bulk density, p 363–375. In Klute A. (ed), Method of soil analysis. 1. Physical and mineralogical methods, 2nd ed ASA-SSSA, Madison, WI. [Google Scholar]
- 86.Thomas GW. 1996. Soil pH and soil acidity, p 475–490. In Sparks DL. (ed), Methods of soil analysis. 3. Chemical methods. ASA-SSSA, Madison, WI. [Google Scholar]
- 87.Rutherford PM, McGill WB, Arocena JM, Figueiredo CT. 2008. Total nitrogen, p 239–250. In Carter MR, Gregorich EG (ed), Soil sampling and methods of analysis, 2nd ed Canadian Society of Soil Science/CRC Press, Boca Raton, FL. [Google Scholar]
- 88.Skjemstad JO, Baldock JA. 2008. Total and organic carbon, p 225–237. In Carter MR, Gregorich EG (ed), Soil sampling and methods of analysis, 2nd ed Canadian Society of Soil Science/CRC Press, Boca Raton, FL. [Google Scholar]
- 89.Konert M, Vandenberghe J. 1997. Comparison of laser grain size analysis with pipette and sieve analysis: a solution for the underestimation of the clay fraction. Sedimentology 44:523–535. [Google Scholar]
- 90.Tate KR, Ross DJ, Feltham CW. 1988. A direct extraction method to estimate soil microbial C: effects of experimental variables and some different calibration procedures. Soil Biol Biochem 20:329–335. doi: 10.1016/0038-0717(88)90013-2. [DOI] [Google Scholar]
- 91.Davis M, Bajwa K, Person R. 2015. Predicted spatial variations of sulphur and nitrogen compound concentrations and deposition in the AOSR, p 51–63. In Clair TA, Percy KE (ed), Assessing forest health in the Alberta Oil Sands Region. Wood Buffalo Environmental Association, Fort McMurray, Alberta, Canada. [Google Scholar]
- 92.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caporaso G. 2011. Earth Microbiome Project: 16S rRNA amplification protocol. http://press.igsb.anl.gov/earthmicrobiome/protocols-and-standards/16s/.
- 94.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simpson EH. 1949. Measurement of diversity. Nature 163:688–688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 98.Legendre P. 2007. anova.1way. Université de Montreal, Montreal, Quebec, Canada. [Google Scholar]
- 99.Legendre P, Legendre L. 2012. Numerical ecology, 3rd ed Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 100.Dray S, Legendre P, Blanchet G. 2011. packfor: forward selection with permutation (Canoco p.46). R package version 0.0-8. https://rdrr.io/rforge/packfor/.
- 101.Borcard D, Gillet F, Legendre P. 2011. Numerical ecology with R. Springer, New York, NY. [Google Scholar]
- 102.Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 103.Giraudoux P. 2016. pgirmess: data analysis in ecology. R package version 1.6.5. https://rdrr.io/cran/pgirmess/.
- 104.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.